Abstract
Genomic DNA in the eukaryotic cell nucleus is present in the form of chromatin. Histones are the principal protein component of chromatin and their post-translational modifications play important roles in regulating the structure and function of chromatin and thereby in determining cell development and disease. An understanding of how histone modifications translate into downstream cellular events is important from both developmental and therapeutic perspectives. However, biochemical studies of histone modifications require access to quantities of homogenously modified histones that cannot be easily isolated from natural sources or generated by enzymatic methods. In the last decade, chemical synthesis has proven to be a powerful tool in translating the language of histone modifications by providing access to uniformly modified histones and by the development of stable analogs of thermodynamically labile modifications. This review highlights the various synthetic and semisynthetic strategies that have enabled biochemical and biophysical characterization of site-specifically modified histones.
Keywords: epigenetics, chromatin, histones, histone tails, post-translational modification, histone code, crosstalk, solid-phase peptide synthesis, expressed protein ligation
Introduction
Since their initial discovery by Albrecht Kossel in 1884 (1), the critical role of histone proteins in regulating cellular events such as DNA transcription (2, 3), replication (4, 5), and repair has been firmly established (6). Through numerous genetic and proteomic studies in a range of organisms, we now know that instead of serving as an inert protein scaffold for organizing genetic material in the nucleus, histones and their associated proteins play dynamic roles throughout the cell cycle (7). Key determinants of histone function are the amino acid side-chains near the histone N-terminal tails that are frequently and generally reversibly modified by post-translational modifications (PTMs) (8). Along with DNA methylation, chromatin remodeling, and RNA interference, histone PTMs have been suggested to be part of the epigenetic (heritable through cell division but not genetically encoded) regulation that dictates cellular fate (9). Therefore, a mechanistic understanding of the specific roles for histone PTMs and for histone-associated proteins is essential for understanding the mechanisms underlying the epigenetic control of cellular activity. This review will focus on the various synthetic approaches that have been employed to interrogate the roles for histone PTMs in recent years. Studies aimed at identifying PTM binding proteins, elucidating the effect of PTMs on mononucleosome and chromatin structure, and investigating the biochemical crosstalk between PTMs will be highlighted.
The language of histone modifications
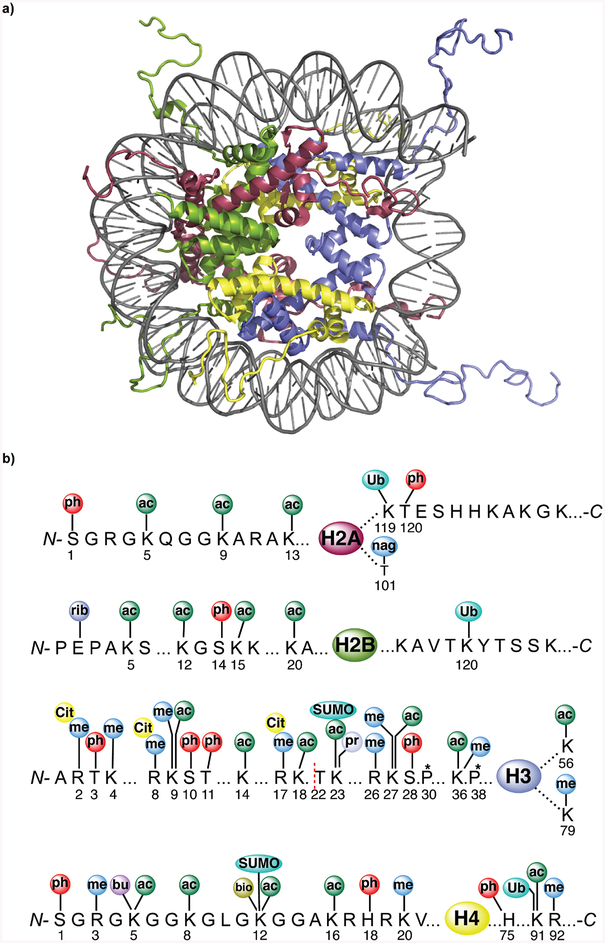
Genomic DNA in the nuclei of eukaryotic cells is packaged in the form of a nucleoprotein complex called chromatin (10). The fundamental repeating unit of chromatin is the nucleosome which consists of ~147 bp of double stranded left-handed superhelical DNA wrapped ~1.65 turns around an octameric complex composed of two copies each of the four core histones H2A, H2B, H3, and H4 (Figure 1a) (11). In the presence of divalent metal ions or additional packaging proteins, such as the linker histone H1, nucleosomes have been suggested to form 30-nm fibers with six nucleosomes per turn in a spiral (12) or a two-start helical arrangement (13). However, 30-nm fibers are rarely observed in vivo (14) and a dynamic molten polymer state has been proposed to be the predominant form of chromatin in most cells (15). The unstructured N-terminal tails of histones protrude from the nucleosome core and are therefore most accessible to histone-modifying enzymes. Not surprisingly, most PTMs of histones are observed in these tail regions, although some PTMs have also been observed in the globular core of the nucleosome. A large number of histone PTMs, also known as marks, have been detected at ~60 unique positions in the four core histones by modification-specific antibodies or mass spectrometric approaches (8). Known modifications of histone tails include methylation (16), phosphorylation (17), acetylation (18), citrullination (19), propionylation and butyrylation (20, 21), biotinylation (22), proline cis-trans isomerization (23), ubiquitylation (24), SUMOylation (25), glycosylation (26), ADP-ribosylation (27) and proteolysis (28) (Figure 1b).
Figure 1A. Structure of a mononucleosome and the diversity of histone modifications.
a, The structure of a mononucleosome at 1.9 Å resolution (PDB code 1KX5) showing individual histones and 147 bp of double stranded DNA. The histone N-termini (tails) protrude from the nucleosome and are sites for extensive post-translational modification. Red= H2A, green= H2B, blue= H3, yellow= H4. b, Schematic representation of modifications at the N- and C-termini and globular domains of the four core histones. Chemical groups are indicated as follows; ac, acetyl; cit, citrullyl; bio, biotinyl; but, butyryl; me, methyl; nag, β-N-acetylglucosaminyl; ph, phosphoryl; pr, propionyl; rib, ADP-ribosyl; SUMO, SUMOyl; and Ub, ubiquityl. Asterisks above Pro30 and Pro38 in the H3 tail indicate modification by cis-trans isomerization. The dashed line N-terminal to Thr 22 in H3 indicates the primary site of proteolysis by cathepsin-L. This figure was adapted from Ref 7.
On the basis of early observations that the chemical acetylation of histones greatly reduced their inhibitory effect on RNA synthesis by RNA polymerases from both calf thymus nuclei and Escherichia coli, Allfrey and co-workers suggested that chemical modifications of histones could offer a means of altering the transcriptional state at different loci along the chromosome (29). Subsequent genetic experiments in yeast and higher organisms established a correlation between the transcriptional state of genes and the modification of H3 and H4 N-terminal tails at specific lysine or serine residues by, acetylation (30, 31) and methylation or phosphorylation, respectively (32). This led Strahl and Allis to postulate a histone code hypothesis for the epigenetic regulation of cellular events (33). According to this hypothesis, several histone modifications on one or multiple histone tails may act in a combinatorial or sequential fashion to specify unique downstream functions such as transcriptional activation or silencing. Thus, the same primary histone sequence could dictate vastly different cellular outcomes by subtle changes in the spatio-temporal pattern of PTMs. In the decade since the histone code hypothesis was postulated, our understanding of gene regulation by histones has significantly increased by the discovery of many new PTMs and the identification of various proteins responsible for installing (writers), removing (erasers), and binding (readers) histone PTMs (34). At the same time, a bewildering array of combinatorial modifications have been discovered on all four core histones and their mechanistic roles appear to be highly context dependent, similar to words in a language (35–37). Chemical tools that will allow us to understand the grammatical rules of this language by enabling investigations of the precise mechanistic roles for histone PTMs are discussed below.
Synthetic peptide-based strategies
The utility of synthetic histone tail peptides to study the mechanism of histone modifying enzymes was first demonstrated in the 1970’s. However, certain limitations in peptide-based approaches were evident even at this early stage. For example, Allfrey and co-workers demonstrated that a synthetic peptide corresponding to residues 15–21 of histone H4 acetylated at Lys16 was not a substrate for histone deacetylases (HDACs) purified from calf thymus cells (38). However, a longer polypeptide containing residues 1–37 from H4 and uniformly acetylated at Lys12 and Lys16 was found to be a substrate for HDACs (39). This suggested that HDACs require a minimum portion of histone H4 for activity. Surprisingly, however, Parello and co-workers showed that an H4 tail peptide consisting of residues 14–21 and acetylated at Lys16 and Lys20 underwent deacetylation by calf thymus nuclear HDACs as long as the N- and C-termini were protected by acetyl and amide groups, respectively (40). Therefore, when assaying short N-terminal peptides with histone-modifying enzymes the choice of the peptide sequence may play a critical role in the outcome of biochemical experiments.
Grunstein and co-workers employed multiply acetylated yeast histone H4 peptides to demonstrate the role of acetylation in regulating interactions between H4 and the silencing information regulator protein Sir3 (41). In their study, H4 N-terminal peptides (residues 1–34) with or without acetylation at Lys5, Lys8, Lys12 and Lys16 were used to measure binding with a glutathione S-transferase (GST) tagged fragment of Sir3 (residues 503–970) by surface plasmon resonance (SPR). It was observed that the association rate constant (ka) decreased significantly with increasing levels of H4 acetylation. Moreover, a comparison of ka values revealed that the acetylation-induced disruption of H4-Sir3 binding was cumulative. The average degree of H4-Sir3 association decreased from ~65% to ~3% in going from the mono- to tetraacetylated H4 peptide. Although considerable redundancy was seen in the ability of acetylated lysines to prevent Sir3 binding in vitro- similar results were obtained irrespective of which lysine was acetylated and only the total extent of acetylation appeared critical- this was not the case in vivo where substitution of H4 Lys16 by the acetyllysine mimetic Gln had more pronounced effects on silencing than Lys-to-Gln mutations at positions 5, 8, or 12. This suggests that investigations of histone modifications in short peptides may not accurately reflect their effects in the context of chromatin.
Zhou and co-workers undertook an NMR titration experiment with a short H4 peptide (residues 1–12) acetylated at Lys8 and the bromodomain from the histone acetyltransferase (HAT) co-activator p300/CBP associated factor (P/CAF) (42). This revealed a Kd of ~350 μM and identified bromodomains as the first protein modules for binding acetylated lysines. Isothermal titration calorimetry (ITC) measurements with fully as well as partially acetylated H4 tail peptides (residues 1–36) have revealed the multivalent engagement of multiply acetylated lysines by two tandem bromodomains in the RNA polymerase II transcription factor D (TFIID) complex protein TAFII250/TAF1 (43). In this instance, the dissociation constants for both singly acetylated peptides (40 μM) and multiply acetylated peptides (1–5.8 μM) were significantly smaller than measured for P/CAF with a singly acetylated H4 peptide (42). Morinière et al. employed diacetylated H4 tail peptides in ITC measurements and X-ray crystallographic experiments to demonstrate the simultaneous binding of two acetyl marks at H4 Lys5 and Lys8 by a single bromodomain from the protein Brdt, a member of the bromodomain extraterminal (BET) family of proteins (44). Thus, experiments with short histone peptides have revealed many distinct mechanisms by which histone readers may recognize specific marks.
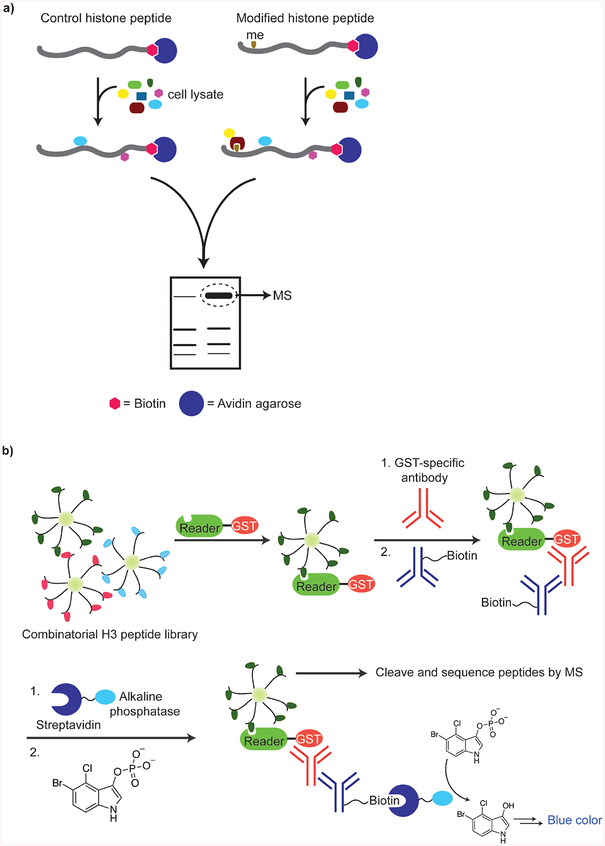
Two elegant examples of the potential for using site-specifically modified histone tail peptides in identifying methylation-state-specific histone binding proteins were presented by Allis and co-workers. The authors utilized biotinylated peptides (residues 1–20 of H3) to identify the proteins WD repeat-containing protein 5 (WDR5) and bromodomain and PHD finger transcription factor (BPTF) as readers partial to the H3 Lys4 dimethylated (45) and trimethylated states, respectively (46). In these studies, nuclear extracts from 293 cells were incubated with either unmodified, dimethylated, or trimethylated H3 Lys4 peptides to first identify protein binders. The specificity for each protein toward a distinct methylation state was subsequently tested in peptide pull-down assays with GST-tagged WDR5 or FLAG-tagged human BPTF (Figure 2a). Surprisingly, in subsequent experiments where binding affinities were measured by ITC or SPR, WDR5 did not demonstrate a pronounced preference for binding dimethylated H3 Lys4 over other methylation states (47, 48). In a high-throughput application of the biotinylated peptide approach, Gozani and co-workers synthesized a library of 63 biotinylated histone peptides of ~20 amino acids each that included all the known methyllysine modifications in histone H3 (49). The peptides were immobilized on a streptavidin-coated glass slide to generate a human epigenome peptide microarray platform (HEMP) that was employed to identify three novel methylated-H3 binding domains from a panel of ~70 domains from the Royal super-family of proteins. The H3 methylation-state specific interactions of these domains were further confirmed by pulldown assays with biotinylated peptides. Protein-peptide interactions that have dissociation constants greater than 300 μM cannot be detected by HEMP arrays, which may be a limitation in identifying weakly binding proteins. Similar biotinylated histone tail peptide libraries have also revealed the binding of a trimethylated H3 Lys4 peptide to plant homeodomain (PHD) fingers in the recombination activating gene 2 (RAG2) protein which is involved in V(D)J recombination (50), and in the tumor suppressor protein, inhibitor of growth 4 (ING4) (51).
Figure 2. Peptide-based strategies for studying histone modifications.
a, Synthetic biotinylated peptide-based unbiased strategy for identifying histone modification-specific readers. b, On-bead assay for histone tail library screening with GST-tagged histone-binding proteins.
Histone peptide libraries facilitate rapid high-throughput screening of a diverse set of modifications and are powerful tools to investigate the specificity determinants for writer, eraser and reader proteins in defined biochemical assays. In this regard, the direct synthesis of modified peptides in an array format is more efficient than immobilizing peptides post-synthesis. Therefore, Jeltsch and co-workers utilized SPOT synthesis, which involves the synthesis of peptides on cellulose filter-sheets by traditional fluorenylmethoxycarbonyl (Fmoc) chemistry (52), to generate an array of 420 mutant H3 tail peptides (residues 1–21) (53). The array was employed to test the substrate specificity of the H3 Lys9 methyltransferase Dim-5 from the fungi Neurospora crassa and revealed the key roles for Thr11 and Gly12 in conferring both specificity for Dim-5 activity and its ability to discriminate among several lysines in the H3 tail. SPOT synthesis was also employed to detect binding of the PWWP domain in the DNA methyltransferase Dnmt3a to the trimethylated H3 Lys36 mark (54). In another study, the Jeltsch group employed a commercially sourced 384-member CelluSPOT tail peptide array to identify the ADD domain in Dnmt3a as a reader for unmodified H3 Lys4 that leads to the preferential methylation of DNA bound to chromatin lacking modification at H3 Lys4 (55). CelluSPOT technology involves re-spotting peptides synthesized on a cellulose support onto glass slides and has the advantages of smaller spot sizes, a more robust glass surface, and identical peptide concentrations in duplicate spots (56).
Denu and co-workers have successfully applied a one-bead one-compound (OBOC) combinatorial peptide library generated by split-pool synthetic methodology toward identifying the specificity determinants for binding of the double tudor domain (DTD) of the human demethylase JMJD2A to the H4 tail (57). Assays with 512 unique 21-amino acid H4 tail peptides combinatorially modified by phosphorylation, acetylation, citrullination, and methylation, revealed that the DTD-H4 tail interaction was modulated to varying degrees by different PTMs. This was manifest in measurable binding affinities ranging from 1 μM to 1 mM. Importantly, this study highlighted the context dependence of specific histone modifications in recruiting downstream effector proteins such as demethylases. The effect of neighboring PTMs on recognition of the modification state at H3 Lys4 by several reader domains was revealed in recent studies by Garske et al (58). An OBOC library of 5000 combinatorially modified 10-amino acid long H3 tail peptides was tested with GST-tagged protein domains followed by GST-specific primary antibodies. The primary antibodies were detected with a biotinylated secondary antibody and finally with streptavidin-conjugated alkaline phosphatase. The compound 5-bromo-4-chloro-3-indolyl phosphate was employed as a substrate for the alkaline phosphatase, which resulted in formation of a blue precipitate on beads that retained GST-bound proteins (Figure 2b). The H3 recognition elements were then determined by mass spectral analysis of the peptides after cleavage from the resin. The binding of PHD fingers from the ING2, RAG2, BHC80 and AIRE proteins as well as the JMJD2 DTD to H3 Lys4 was affected by modifications at H3 Arg2, H3 Thr3 and H3 Thr6. Interestingly, phosphorylation at H3 Thr6 exhibited different effects on different reader domains. The occurrence in HeLa cells of this previously unreported mark was confirmed by mass spectrometric and antibody-based techniques. However, some combinations of histone modifications that are tested in such randomized combinatorial libraries may not exist in nature and the physiological relevance of novel hit sequences may require verification in vivo.
Bedford and co-workers employed the library approach in a reversed sense with the development of a chromatin-associated domain array (CADOR) chip bearing an array of 109 GST-tagged domains from histone-binding proteins that included bromo, chromo, tudor, PhD, SANT, SWIRM, MBT, CW and PWWP domains (59). Binding experiments with fluorophore-tagged H3 and H4 N-terminal peptides with varying degrees of methylation at H3 Lys4 or Lys9 and H4 Lys20 revealed six novel interactions between histone-binding domains and methylated histone tails.
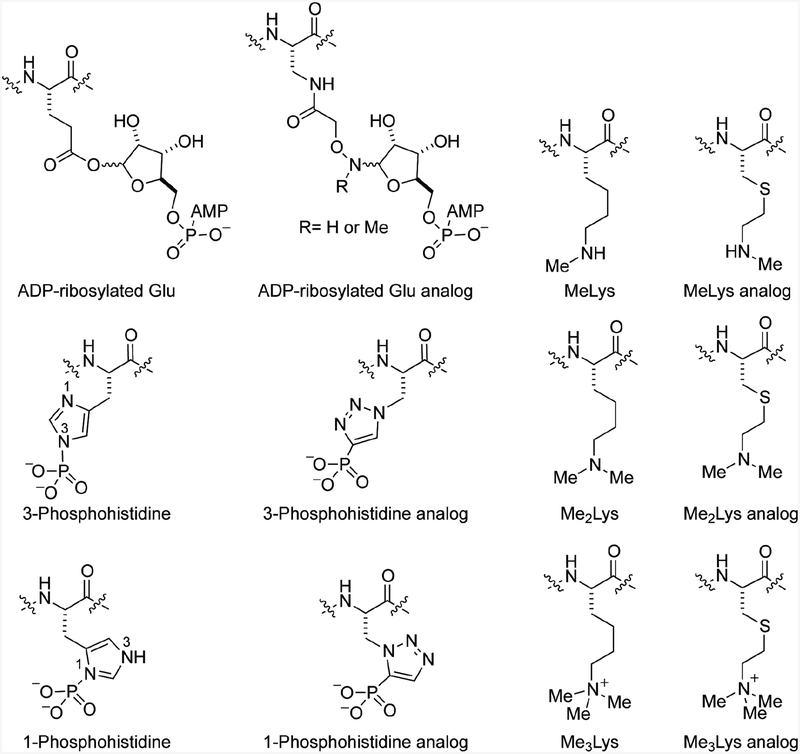
The design of complex peptide libraries with a greater diversity of modifications would benefit from novel synthetic methodologies to generate histone tails modified with chemical groups that are hard to access and characterize due to their inherent instability and/or rapid turnover in vivo. Muir and co-workers recently reported two impressive strategies to mimic histone peptides modified by ADP-ribosylation and histidine phosphorylation (Figure 3). To achieve the chemical ADP-ribosylation of an H2B peptide (residues 1–19) Moyle and Muir substituted the naturally occurring Glu2 with an amino-oxy side-chain, which formed an oxime linkage with the aldehyde group in ADP-ribose (60). This synthetic analog was demonstrated to effectively mimic mono-ADP-ribosylation. It was found to be a substrate for the enzyme poly(ADP-ribose) polymerase 1 (PARP1) and to bind the macro domain of the histone variant mH2A1.1 which is suggested to bind poly-ADP-ribosylated PARP1. The phosphorylation of histone H4 at His18 (pHis18) and His 75 (pHis75) has been correlated with increased DNA synthesis in vivo, and both 1-pHis and 3-pHis isomers have been observed (61, 62). However, the facile hydrolysis of pHis under acidic conditions and the isomerization of 1-pHis to the thermodynamically favored 3-pHis form are significant challenges toward mechanistic studies of the roles for histidine phosphorylation. With these hurdles in mind, Kee et al. employed Cu and Ru-catalyzed cycloaddition chemistry between t-butyloxycarbonyl azidoalanine and diethyl ethynylphosphonate to generate regioisomeric phosphoryltriazolylalanines (pTza) as stable non-isomerizable analogs of 3-pHis and 1-pHis, respectively (63). A histone H4 peptide (residues 14–23) bearing the 3-pTza isomer was successfully employed to generate an antibody specific for H4 pHis18. These reagents hold great promise toward understanding the biological roles for histidine phosphorylation in histones, and indeed in several prokaryotic and vertebrate proteins that are regulated by this transient modification.
Figure 3. Histone modifications and their synthetic analogs.
MeLys= monomethyl lysine, Me2Lys= dimethyllysine, Me3Lys= trimethyllysine.
Histone tail peptides have also been employed to measure the activity of histone-modifying enzymes by a variety of methods such as labeling of substrates with radioisotopes (64), enzyme-linked immunosorbent assays (ELISA) (65), and coupled-enzyme assays (66). Ghadiali et al. recently reported an alternative nanosensor-based approach for the detection of histone acetyltransferase (HAT) activity that employs a quantum-dot (QDot)-based Förster resonance energy transfer (FRET) donor (67). In this approach, a C-terminally hexahistidine-tagged H4 tail peptide was incubated with the HAT p300 in the presence of acetyl-CoA. The acetylated peptide was then bound to the surface of a CdSe/ZnS core/shell QDot by His6-mediated metal-affinity self-assembly. An Alexa Fluor 647 labeled acetyl-lysine specific monoclonal antibody was incubated with the acetylated peptide/QDot complex and the magnitude of QDot-Alexa Fluor 647 energy transfer was used to report on the degree of peptide acetylation. The nanosensor methodology permitted the detection of sub-nanomolar amounts of HAT activity and was also demonstrated to be useful for the assessment of the HAT inhibition activity of anacardic acid. A limitation of this methodology was that p300 failed to act on substrates that were pre-immobilized on the QDot, thus necessitating a two-step process of assaying in solution followed by subsequent immobilization of peptides on the QDot.
Although experiments conducted with histone peptides have afforded many insights regarding chromatin regulation, they cannot recapitulate the diverse roles for histone PTMs in the context of chromatin. In order to understand the mechanistic roles for histone marks in directing trans-tail histone modifications, modulating chromatin structure and facilitating chromatin remodeling, it is necessary to study these in the more physiologically relevant context of mononucleosomes and nucleosomal arrays. This requires the ability to synthesize full-length histones with the requisite site-specific modifications.
Native Chemical and Expressed protein Ligation
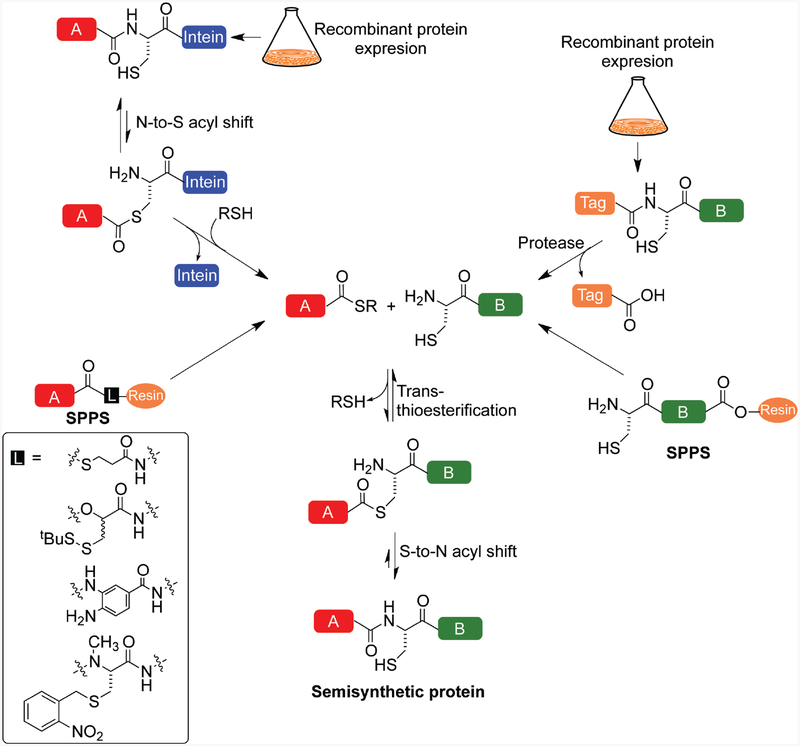
Kent and co-workers first reported the technique called Native Chemical Ligation (NCL) in 1994 (68). NCL is a chemoselective peptide ligation technique that allows the joining of two peptide fragments with unprotected side-chains by means of a native peptide bond (Figure 4). The only requirements for NCL are a C-terminal α-thioester in the N-terminal peptide and an N-terminal Cys residue in the C-terminal peptide. The peptide fragments may be obtained by solid-phase peptide synthesis employing either a t-butyloxycarbonyl (Boc) or N-(9-fluorenyl)methoxycarbonyl protecting group strategy (69). Several chemical linkers have also been reported that allow synthesis on the solid phase of C-terminally activated peptides that may be directly employed in chemical ligation reactions (Figure 4) (70–73). NCL is however limited by the ability to synthesize the N- and C-terminal peptide fragments in reasonable yields, which is challenging for proteins more than ~100 amino acids in length (74). Expressed protein ligation (EPL) is an extension of the NCL technique that significantly expands the range of proteins accessible for chemical modification (75). In EPL, the C-terminal α-thioester is obtained by thiolysis of a C-terminally fused intein (76, 77) rather than by synthesis, and hence any soluble protein can in principle be converted to its α-thioester form. The chemistry underlying ligation is identical in NCL and EPL and proceeds with an initial transthioesterification reaction between the peptide/protein thioester and a second peptide/protein bearing an N-terminal Cys. A thermodynamically favored S-to-N-acyl shift generates a native amide linkage between the two fragments, resulting in the full-length target protein (Figure 4). Based upon where the desired chemical modification is located in the final protein sequence, the synthetic and recombinant fragments employed in EPL may be reversed to enable ligation of a C-terminal expressed protein with an N-terminal synthetic peptide thioester. Since any number and type of chemical modifications can be introduced in the synthetic fragment, EPL essentially allows access to any physiologically relevant modified histone. The ability to reconstitute ~147 bp of DNA and core histones purified by high performance liquid chromatography (HPLC) into mononucleosomes (78, 79) has led to the application of EPL to numerous biochemical and biophysical studies of site-specifically modified nucleosomes as discussed below.
Figure 4. Native chemical and expressed protein ligation.
Strategies for combining fully synthetic, or, synthetic and recombinant peptides in order to generate full-length semisynthetic proteins. SPPS, solid-phase peptide synthesis. Inset are different chemical linkers that have been employed to obtain synthetic peptides with C-terminal thioesters.
Studies of histone phosphorylation
The first instance of application of EPL to histones was by Peterson and co-workers who used this technique to generate histone H3 phosphorylated at Ser10 (80). Their approach involved the synthesis of a fully protected N-terminal peptide fragment (residues 1–32) bearing the pSer10 residue on an acid-sensitive resin, release of the fully side-chain protected peptide from the resin with dilute acid and its subsequent coupling with benzyl mercaptan to generate the C-terminal thioester. The C-terminal Histone H3 fragment (residues 33–135) was expressed in E. coli and an N-terminal Cys exposed by Factor Xa protease cleavage. Ligation between the two halves yielded H3 with a Thr33Cys mutation as well as the desired pSer 10. The H3 pSer10 was then incorporated in nucleosomal arrays and efficiently remodeled by the yeast SWI/SNF remodeling complex, which indicated that H3 pSer10 does not affect nucleosome structure dramatically. Furthermore, kinetic analysis of the acetyltransferase activity of recombinant Gcn5 was undertaken. In contrast with results obtained with H3 pSer10 bearing peptide substrates, the HAT activity of the Gcn5-containing SAGA complex was not enhanced by H3 pSer10 in nucleosomal arrays. This difference between histone peptides and nucleosomal arrays as substrates for histone-modifying enzymes suggests that assays with chemically modified nucleosomes will enhance our understanding of the mechanistic effects of post-translational histone modifications in vivo.
Chiang et al. recently employed semisynthesis to generate a version of H2B that was phosphorylated at Ser10 and tetraacetylated at Lys5, Lys11, Lys12, and Lys15 (81). Several advances in their synthetic approach were the utilization of a thiol-protected 2-hydroxy-3-mercaptopropionic acid linker (72) and a diaminobenzoic acid linker that permitted the generation of peptide thioesters employing Fmoc chemistry (71). This avoided the possibility of epimerization at the C-terminus that occurs during the solution-phase activation of side-chain protected peptides (82). Furthermore, both Raney-nickel mediated (83) as well as a milder radical-mediated desulfurization strategy were employed to render the ligation product traceless by reverting the Cys employed for ligation to Ala which is present in the native sequence (84). Histones are ideal substrates for desulfurization as there is only one non-essential Cys (H3 Cys110) between the four core histones. Although tetraacetylated H2B was found to undergo phosphorylation at Ser14 by the mammalian sterile twenty-like 1 kinase (MST1), this could not be detected by an H2B pSer14-specific polyclonal antibody. This result highlights a possible limitation of antibody-based chromatin immunoprecipitation (ChIP) experiments (85) with biological samples as histones are highly modified in vivo and modifications at adjacent sites may interfere with antibody recognition at a specific site.
Studies of histone acetylation
In a landmark study, Peterson and co-workers employed EPL to generate histone H4 site-specifically acetylated at Lys16 (86). The acetylated histone was incorporated into nucleosomal arrays and its effects on MgCl2-induced compaction and the reversible self-association of arrays was investigated by sedimentation velocity analysis during ultracentrifugation and solubility measurements, respectively. Surprisingly, the single acetyl group in H4 was found to prevent both compaction and self-association of arrays to a similar extent as H4 in which residues 1–19 from the tail had been genetically removed. These results suggested a structural role for acetylation at Lys16 in preventing chromatin condensation and maintaining transcriptionally active euchromatin. In support of this hypothesis, the authors found that H4 Lys16 acetylation was enriched in the MgCl2-soluble fraction of micrococcal nuclease digested HeLa nuclei. Moreover, in budding yeast where most of the genome exists in a decondensed state, H4 Lys16 was found to largely exist in the acetylated state (87). This study was the first demonstration of the direct effect of a histone modification on chromatin structure.
EPL has enabled investigations of the effect of hyperacetylation in the H3 tail at Lys4, Lys9, Lys14, Lys18, and Lys23 on chromatin assembly and on the activity of histone-modifying enzymes (88). Interestingly, pentaacetylation of H3 did not hinder the ability of the remodeling and spacing factor (RSF) complex to mediate assembly of a chromatinized plasmid. Furthermore, pentaacetylated H3/H4 tetramers were also efficient substrates for the HDAC Sir1 and the lysine methyltransferase G9a. The divergent roles for acetylation on distinct histone tails toward nucleosome remodeling were demonstrated by Owen-Hughes and co-workers (89). Semisynthetic H3 tetraacetylated at Lys9, Lys14, Lys18 and Lys23 was found to preferentially bind the ATP-dependent nucleosome remodeling complex, remodel the structure of chromatin (RSC), and enhance the rate of chromatin remodeling by ~16-fold over unmodified chromatin. On the other hand, H4 tetraacetylated at Lys5, Lys8, Lys12 and Lys16 was found to inhibit nucleosome remodeling by the enzyme, Isw2. The effect of acetylation at lysines in the C-terminal region of histone H3 has also been investigated (90). Lys115 and Lys 122 in H3 are both involved in key histone-DNA contacts near the nucleosome dyad pseudo-symmetry axis. However, acetylation of H3 Lys115 was found to reduce DNA binding significantly more than acetylation at H3 Lys122 in nucleosome competitive reconstitution experiments. In contrast, acetylation at H3 Lys122 was found to increase the rate of thermal repositioning of nucleosomes by two-fold relative to the effect of acetylation at H3 Lys115. These results indicate that acetylation at different positions near the dyad interface may act through distinct mechanisms in vivo.
In order to investigate the effect of acetylation at Lys56 near the middle of the H3 primary sequence, Ottesen and co-workers recently reported the first total chemical synthesis of the 134 amino acid long histone (91). Key to their success was the utilization of an established sequential three-piece ligation strategy (92, 93) that permitted synthetic disconnections between residues 39–40 and 95–96. This reduced the length of the longest synthetic peptide fragment to 55 amino acids. Acetylation at H3 Lys56 increased the binding of LexA, a model DNA binding factor, to its DNA target site by ~3-fold presumably due to increased DNA unwrapping from the histone octamer. Interestingly, an H3 Lys56Gln mutation quantitatively mimicked the role of H3 Lys56 acetylation while similar Lys-to-Gln mutations did not recapitulate the effect of acetylation at H3 Lys115 or H3 Lys122 near the dyad axis. This revealed possible limitations in interpreting results from experiments employing Gln as a mimic of acetyllysine in vivo.
Studies of histone ubiquitylation
The modification of eukaryotic proteins by a single ubiquitin, termed monoubiquitylation, does not lead to their degradation by the 26S proteasome, but instead acts to initiate downstream signaling events. Mono-ubiquitylation of proteins was first discovered as a modification of histone H2A (94) and soon thereafter was also observed on histone H2B (24). The mono-ubiquitylation of histone H2B at Lys 120 (uH2B) is associated with DNA damage repair, transcription elongation, and methylation of H3 Lys4 and Lys79 by the human methyltransferases hSet1 and hDot1, respectively (95, 96). Biochemical interrogations of the mechanistic roles for uH2B have traditionally been limited by the low abundance (~1.5% of total H2B) and dynamic nature of this modification. In order to overcome these limitations, Muir and co-workers have pioneered several semisynthetic approaches for the site-specific chemical ubiquitylation of histones.
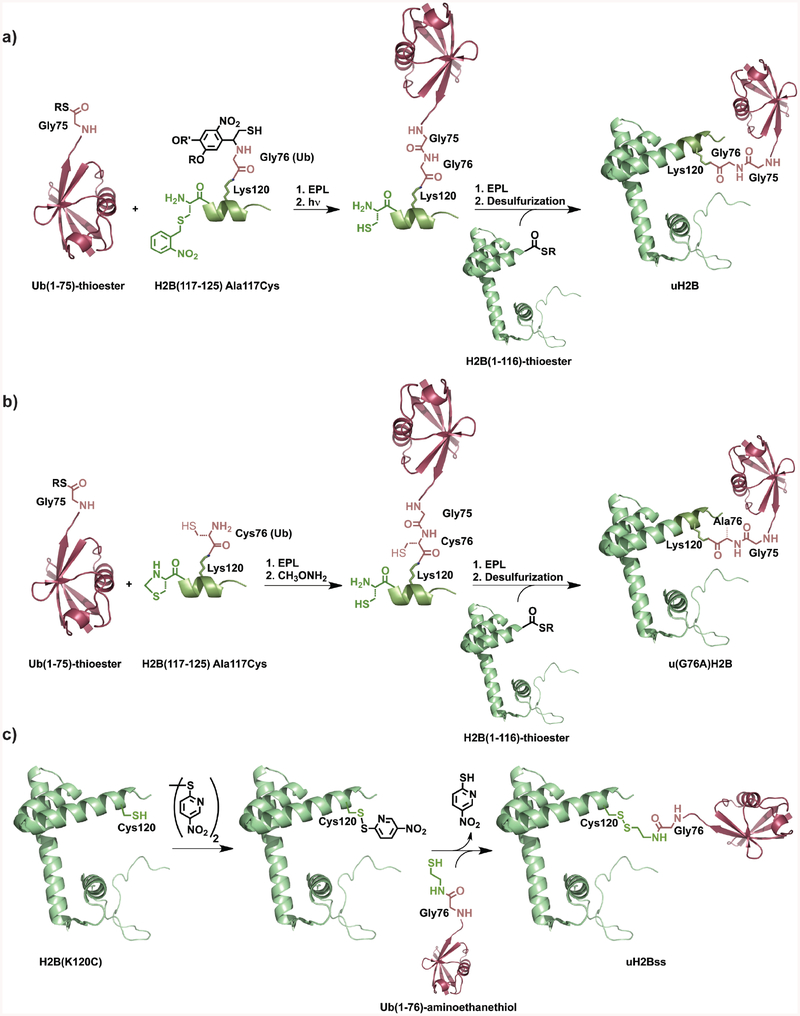
An initial EPL strategy employed a photolytically removable ligation-auxiliary (97) to undertake the traceless ubiquitylation of histone H2B (Figure 5a) (98). The synthetic strategy involved dividing H2B into a short synthetic C-terminal fragment and recombinant N-terminal α-thioester. In the first step, auxiliary-mediated ubiquitylation of the short peptide was accomplished at a Lys side-chain that corresponded to Lys120 in the full-length histone. Irradiation of this uH2B peptide product at 365 nm served to remove the ligation auxiliary as well as unmask a photoprotected N-terminal Cys. The ubiquitylated peptide was then ligated with the recombinant H2B thioester to generate full-length uH2B(A117C). Finally, Cys117 was desulfurized to generate the Ala117 present in native uH2B. Semisynthetic uH2B was successfully reconstituted in nucleosomes and tested in biochemical assays with hDot1L, a methyltransferase that is misregulated in several acute leukemias. This revealed that uH2B directly stimulates methylation of H3 Lys79 and was the first direct biochemical evidence for crosstalk between marks on two distinct histones. Despite the success of this strategy, a significant limitation toward its routine application for protein ubiquitylation was the requirement for a ligation auxiliary that was achieved in nine synthetic steps. Hence, an alternate methodology for histone ubiquitylation was developed that bypassed the need for ligation auxiliaries by incorporating a Gly76Ala mutation at the C-terminus of ubiquitin that permitted ubiquitylation at Cys (Figure 5b) (99). Surprisingly, the u(G76A)H2B mutant was detected by a uH2B-specific antibody and was also a substrate for the ubiquitin C-terminal hydrolase, UCHL3. u(G76A)H2B also stimulated hDot1L activity to a similar degree as wild-type uH2B. Structure-activity relationship studies with ubiquitylated nucleosomes obtained by this simplified method revealed an unusual mechanistic role for ubiquitin in that the canonical hydrophobic patch, which is important for binding ubiquitin-interacting motifs in proteins (100), was not critical for stimulating hDot1L activity.
Figure 5. Strategies for histone ubiquitylation.
Chemoselective strategies for synthesizing native and mutant forms of uH2B and a functionally equivalent analog. a, Photocleavable auxiliary-mediated histone ubiquitylation. b, Cysteine-mediated histone ubiquitylation. c, Disulfide-directed histone ubiquitylation.
In order to investigate the effect of mono-ubiquitylation at other Lys residues near the H2B C-terminus on hDot1L activity, a disulfide-directed ubiquitylation strategy was also developed (101). This synthetic strategy utilizes the intein-mediated introduction of a sulfhydryl group at the C-terminus of ubiquitin and its subsequent use to form an asymmetric disulfide with an H2B Lys-to-Cys mutant (Figure 5c). Disulfide-directed ubiquitylation of H2B at Lys120 (uH2Bss) or at Lys125 was found to stimulate methylation at H3 Lys79 by hDot1L. Furthermore, the protein Nedd8 that shares 55% sequence identity with ubiquitin was also found to stimulate methylation when disulfide-linked to Lys120. These experiments revealed an unprecedented plasticity in the mechanism of hDot1L activation and also suggested that multiple sites of modification on the nucleosome may have overlapping biochemical roles. Fierz et al. have employed the ready reversibility of the disulfide-linkage under reducing conditions to demonstrate that uH2Bss reversibly inhibits MgCl2-mediated compaction and higher order fiber formation in nucleosomal arrays (102).
Amber suppression and cysteine modification strategies
Two methodologies that are complementary to chemical ligation for generating modified histones are amber suppression mutagenesis (103) and cysteine-directed protein modification (104, 105). Schulz and co-workers have evolved a mutant Methanococcus jannaschii tyrosyl amber suppressor tRNACUA and tyrosyl-tRNA synthetase pair to site-specifically incorporate (Se)-phenylselenocysteine in histone H3 in response to the amber codon, TAG in E. coli (106). Oxidative elimination of phenylselenic acid yielded the Michael-acceptor dehydroalanine, which underwent nucleophilic addition with N-acetylated or N-methylated derivatives of 2-aminoethanethiol to yield thioether-containing analogs of acetylated and methylated lysine side-chains, respectively. An alternate method developed by Chin and co-workers employed an evolved pyrrolysl-tRNA synthetase/ tRNACUA pair from Methanosarcina barkeri to incorporate N-ε-acetyl-l-lysine (107) and N-ε-Boc-N-ε-methyl-L-lysine in histone H3 (108). In the latter instance, acidolytic deprotection of the Boc protecting group unmasked the desired N-methylated lysine side-chain. Alternate strategies to incorporate N-methyllysine have also been developed that employ a reversibly photocaged N-ε-methyl-l-lysine (109) or N-ε-allyloxycarbonyl-N-ε-methyl-l-lysine, which may be converted to N-methyllysine with a ruthenium catalyst in neutral aqueous solution (110). Recently, the incorporation of N-ε-Boc-l-lysine in response to an amber codon in histone H3 was used as an orthogonally protected form of lysine, which could be selectively deprotected after protecting the remaining Lys side-chains with an N-benzyloxycarbonyl group, and then reductively alkylated using formaldehyde and a dimethylamine borane complex to yield an N,N-dimethylated lysine side-chain (111).
The nucleophilic side-chain of Cys (pKa ~8.5) is an ideal site for selective modification of proteins at pH ~8–9. Shokat and co-workers have made use of the unique reactivity of Cys to generate N-methylated aminoethylcysteine analogs of N-methylated lysine side-chains that the authors term methyllysine analogs (MLAs) (Figure 3) (112). The relatively straightforward displacement chemistry involved in their generation underlies the popularity of the MLA approach and MLAs have been applied in a wide range of studies addressing the establishment of histone modifications (113), their interpretation by modification specific readers (114–116) as well as their propagation in chromatin (117).
Cysteine alkylation has also been employed to generate a thiocarbamate analog of acetyllysine, termed methylthiocarbonyl-thiaLys (MTCTK) (118). This analog was found to mimic acetylation at H4 Lys5 and Lys8 in binding the Brdt bromodomain, albeit with 2–4-fold less potency. Moreover, the MTCTK analog was not deacetylated by the class I histone deacetylase HDAC8 and the class III NAD-dependent deacetylase Sir2. This holds potential for the use of this analog in experiments with cell lysates where acetylated peptides would be turned over by HDACs. Finally, the ability to employ the MLA methodology to convert Cys residues into lysine analogs after chemical ligation also expands the scope for EPL by allowing peptide bond disconnections at highly abundant Lys residues in histones (119).
Conclusions and future directions
Histone PTMs and their associated writer, eraser, and reader proteins play critical and dynamic roles in regulating the structure and function of eukaryotic chromatin. Beginning with the synthesis of short modified histone tail peptides, chemists have developed increasingly sophisticated chemical tools to access homogenously modified full-length histones in order to understand their specific mechanistic roles. Several complementary methodologies, such as EPL, MLAs and amber codon suppression have rendered accessible synthetically challenging side-chain modifications such as ubiquitylation, and modifications in the histone globular core. These methods will permit studies of the mechanistic roles for other less understood histone modifications such as SUMOylation, which has been associated with transcription repression in both yeast and human cells (25, 120). The ability to reconstitute entirely synthetic or semisynthetic histones into mononucleosomes and nucleosomal arrays, dubbed designer chromatin (121), has enabled biochemical and biophysical investigations of histone modifications in a more physiologically relevant context. Most studies until now have employed homogenously modified nucleosomes where all copies of a specific histone bear the same modification. A more likely scenario in cells is that different degrees of modification may exist on two adjacent nucleosomes, or even on the two faces of the same nucleosome. Therefore, one of the challenges for the future will be to reconstitute asymmetric nucleosomal arrays with different degrees and combinations of modifications in order to study their roles in chromatin folding, remodeling, and replication. Initial progress toward this goal has been achieved by mixing modified histones in different ratios to generate asymmetric octamers (99, 122) and stitching together differently modified nucleosomes with a DNA ligase enzyme (98, 123). Thus, an amalgamation of chemical and enzymatic tools that permit the assembly and interrogation of increasingly complex nucleosomal arrays will no doubt be key to understanding the complex language of histone modifications.
Acknowledgements
C.C. and A.D. gratefully acknowledge financial support from the Department of Chemistry at the University of Washington, Seattle. We would also like to thank the reviewers for their insightful suggestions.
References
- 1.Kossel A, and Pringle H (1906) On protamines and histones, H-S. Z. Physiol. Chem 49, 301–321. [Google Scholar]
- 2.Shilatifard A (2006) Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression, Annu. Rev. Biochem 75, 243–269. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Carey M, and Workman JL (2007) The role of chromatin during transcription, Cell 128, 707–719. [DOI] [PubMed] [Google Scholar]
- 4.Unnikrishnan A, Gafken PR, and Tsukiyama T (2010) Dynamic changes in histone acetylation regulate origins of DNA replication, Nat. Struct. Mol. Biol 17, 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob Y, Stroud H, Leblanc C, Feng S, Zhuo L, Caro E, Hassel C, Gutierrez C, Michaels SD, and Jacobsen SE (2010) Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases, Nature 466, 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scully R (2010) A histone code for DNA repair, Nat. Rev. Mol. Cell. Biol 11, 164. [DOI] [PubMed] [Google Scholar]
- 7.Bhaumik SR, Smith E, and Shilatifard A (2007) Covalent modifications of histones during development and disease pathogenesis, Nat. Struct. Mol. Biol 14, 1008–1016. [DOI] [PubMed] [Google Scholar]
- 8.Kouzarides T (2007) Chromatin modifications and their function, Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- 9.Allis CD, Jenuwein T, Reinberg D, and Caparros M-L (2007) Epigenetics, First ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor. [Google Scholar]
- 10.Kornberg RD (1974) Chromatin structure: a repeating unit of histones and DNA, Science 184, 868–871. [DOI] [PubMed] [Google Scholar]
- 11.Davey CA, Sargent DF, Luger K, Maeder AW, and Richmond TJ (2002) Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution, J. Mol. Biol 319, 1097–1113. [DOI] [PubMed] [Google Scholar]
- 12.Robinson PJ, Fairall L, Huynh VA, and Rhodes D (2006) EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure, Proc. Natl. Acad. Sci. U.S.A 103, 6506–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, and Richmond TJ (2004) Nucleosome arrays reveal the two-start organization of the chromatin fiber, Science 306, 1571–1573. [DOI] [PubMed] [Google Scholar]
- 14.Woodcock C (1994) Chromatin fibers observed in situ in frozen hydrated sections. Native fiber diameter is not correlated with nucleosome repeat length, J. Cell Biol 125, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeshima K, Hihara S, and Eltsov M (2010) Chromatin structure: does the 30-nm fibre exist in vivo?, Curr Opin Cell Biol 22, 291–297. [DOI] [PubMed] [Google Scholar]
- 16.Murray K (1964) The Occurrence of Epsilon-N-Methyl Lysine in Histones, Biochemistry 3, 10–15. [DOI] [PubMed] [Google Scholar]
- 17.Stevely WS, and Stocken LA (1966) Phosphorylation of rat-thymus histone, Biochem. J 100, 20C–21C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips DM (1963) The presence of acetyl groups of histones, Biochem. J 87, 258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, and Coonrod SA (2004) Human PAD4 regulates histone arginine methylation levels via demethylimination, Science 306, 279–283. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, and Zhao Y (2007) Lysine propionylation and butyrylation are novel post-translational modifications in histones, Mol. Cell. Proteomics 6, 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K, Chen Y, Zhang Z, and Zhao Y (2009) Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software, J. Proteome Res 8, 900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, and Zempleni J (2005) K4, K9 and K18 in human histone H3 are targets for biotinylation by biotinidase, FEBS J. 272, 4249–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson CJ, Santos-Rosa H, and Kouzarides T (2006) Proline isomerization of histone H3 regulates lysine methylation and gene expression, Cell 126, 905–916. [DOI] [PubMed] [Google Scholar]
- 24.West MH, and Bonner WM (1980) Histone 2B can be modified by the attachment of ubiquitin, Nucleic Acids Res. 8, 4671–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shilo Y, and Eisenman R (2003) Histone sumoylation is associated with transcriptional repression, Proc. Natl. Acad. Sci. U.S.A 100, 13225–13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakabe K, Wang Z, and Hart GW (2010) Beta-N-acetylglucosamine (OGlcNAc) is part of the histone code, Proc. Natl. Acad. Sci. U.S.A 107, 19915–19920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogata N, Ueda K, and Hayaishi O (1980) ADP-ribosylation of histone H2B. Identification of glutamic acid residue 2 as the modification site, J. Biol. Chem 255, 7610–7615. [PubMed] [Google Scholar]
- 28.Duncan EM, Muratore-Schroeder TL, Cook RG, Garcia BA, Shabanowitz J, Hunt DF, and Allis CD (2008) Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation, Cell 135, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allfrey VG, Faulkner R, and Mirsky AE (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis, Proc. Natl. Acad. Sci. U.S.A 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, and Grunstein M (1995) Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast, Cell 80, 583–592. [DOI] [PubMed] [Google Scholar]
- 31.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, and Allis CD (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines, Nature 383, 269–272. [DOI] [PubMed] [Google Scholar]
- 32.Hagmann M (1999) How chromatin changes its shape, Science 285, 1200–1201, 1203. [DOI] [PubMed] [Google Scholar]
- 33.Strahl BD, and Allis CD (2000) The language of covalent histone modifications, Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 34.Ruthenburg AJ, Allis CD, and Wysocka J (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark, Mol. Cell 25, 15–30. [DOI] [PubMed] [Google Scholar]
- 35.Wu JI, Lessard J, and Crabtree GR (2009) Understanding the words of chromatin regulation, Cell 136, 200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J-S, Smith E, and Shilatifard A (2010) The language of histone crosstalk, Cell 142, 682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardner KE, Allis CD, and Strahl BD (2011) OPERating ON Chromatin, a Colorful Language where Context Matters, J. Mol. Biol 409, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krieger DE, Levine R, Merrifield RB, Vidali G, and Allfrey VG (1974) Chemical studies of histone acetylation. Substrate specificity of a histone deacetylase from calf thymus nuclei, J. Biol. Chem 249, 332–334. [PubMed] [Google Scholar]
- 39.Krieger D, Vidali G, Erickson B, Allfrey V, and Merrifield R (1979) Synthesis of diacetylated histone H4(1–37) for studies on the mechanism of histone deacetylation, Bioorg. Chem 8, 409–427. [Google Scholar]
- 40.Kervabon A, Mery J, and Parello J (1979) Enzymatic deacetylation of a synthetic peptide fragment of histone H4, FEBS Lett. 106, 93–96. [DOI] [PubMed] [Google Scholar]
- 41.Carmen AA, Milne L, and Grunstein M (2002) Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3, J. Biol. Chem 277, 4778–4781. [DOI] [PubMed] [Google Scholar]
- 42.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, and Zhou MM (1999) Structure and ligand of a histone acetyltransferase bromodomain, Nature 399, 491–496. [DOI] [PubMed] [Google Scholar]
- 43.Jacobson RH, Ladurner AG, King DS, and Tjian R (2000) Structure and function of a human TAFII250 double bromodomain module, Science 288, 1422–1425. [DOI] [PubMed] [Google Scholar]
- 44.Morinière J, Rousseaux S, Steuerwald U, Soler-López M, Curtet S, Vitte A-L, Govin J, Gaucher J, Sadoul K, Hart DJ, Krijgsveld J, Khochbin S, Müller CW, and Petosa C (2009) Cooperative binding of two acetylation marks on a histone tail by a single bromodomain, Nature 461, 664–668. [DOI] [PubMed] [Google Scholar]
- 45.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, and Allis CD (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development, Cell 121, 859–872. [DOI] [PubMed] [Google Scholar]
- 46.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, and Allis CD (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling, Nature 442, 86–90. [DOI] [PubMed] [Google Scholar]
- 47.Couture JF, Collazo E, and Trievel RC (2006) Molecular recognition of histone H3 by the WD40 protein WDR5, Nat. Struct. Mol. Biol 13, 698–703. [DOI] [PubMed] [Google Scholar]
- 48.Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, and Verdine GL (2006) Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex, Nat. Struct. Mol. Biol 13, 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, Casadio F, Bassi C, Amati B, Bedford MT, Guccione E, and Gozani O (2009) Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks, PLoS ONE 4, e6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matthews AGW, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, and Oettinger MA (2007) RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination, Nature 450, 1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, and Gozani O (2009) ING4 Mediates Crosstalk between Histone H3 K4 Trimethylation and H3 Acetylation to Attenuate Cellular Transformation, Mol. Cell 33, 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frank R (2002) The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports--principles and applications, J. Immunol. Methods 267, 13–26. [DOI] [PubMed] [Google Scholar]
- 53.Rathert P, Zhang X, Freund C, Cheng X, and Jeltsch A (2008) Analysis of the substrate specificity of the Dim-5 histone lysine methyltransferase using peptide arrays, Chem. Biol 15, 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, and Jeltsch A (2010) The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation, J. Biol. Chem 285, 26114–26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, and Jeltsch A (2010) Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail, Nucleic Acids Res. 38, 4246–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu C, and Li SS (2009) CelluSpots: a reproducible means of making peptide arrays for the determination of SH2 domain binding specificity, Methods Mol. Biol 570, 197–202. [DOI] [PubMed] [Google Scholar]
- 57.Garske AL, Craciun G, and Denu JM (2008) A combinatorial H4 tail library for exploring the histone code, Biochemistry 47, 8094–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garske AL, Oliver SS, Wagner EK, Musselman CA, LeRoy G, Garcia BA, Kutateladze TG, and Denu JM (2010) Combinatorial profiling of chromatin binding modules reveals multisite discrimination, Nat. Chem. Biol 6, 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, and Bedford MT (2006) Tudor, MBT and chromo domains gauge the degree of lysine methylation, EMBO Rep. 7, 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moyle PM, and Muir TW (2010) Method for the synthesis of mono-ADP-ribose conjugated peptides, J. Am. Chem. Soc 132, 15878–15880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smtih DL, Bruegger BB, Halpern RM, and Smith RA (1973) New histone kinases in nuclei of rat tissues, Nature 246, 103–104. [DOI] [PubMed] [Google Scholar]
- 62.Besant PG, and Attwood PV (2010) Histidine phosphorylation in histones and in other mammalian proteins, Methods Enzymol. 471, 403–426. [DOI] [PubMed] [Google Scholar]
- 63.Kee J-M, Villani B, Carpenter LR, and Muir TW (2010) Development of stable phosphohistidine analogues, J. Am. Chem. Soc 132, 14327–14329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poveda A, and Sendra R (2008) An easy assay for histone acetyltransferase activity using a PhosphorImager, Anal. Biochem 383, 296–300. [DOI] [PubMed] [Google Scholar]
- 65.Wynne Aherne G, Rowlands MG, Stimson L, and Workman P (2002) Assays for the identification and evaluation of histone acetyltransferase inhibitors, Methods 26, 245–253. [DOI] [PubMed] [Google Scholar]
- 66.Berndsen CE, and Denu JM (2005) Assays for mechanistic investigations of protein/histone acetyltransferases, Methods 36, 321–331. [DOI] [PubMed] [Google Scholar]
- 67.Ghadiali JE, Lowe SB, and Stevens MM (2011) Quantum-Dot-Based FRET Detection of Histone Acetyltransferase Activity, Angew. Chem. Int. Ed. Engl 50, 3417–3420. [DOI] [PubMed] [Google Scholar]
- 68.Dawson PE, Muir TW, Clark-Lewis I, and Kent SB (1994) Synthesis of proteins by native chemical ligation, Science 266, 776–779. [DOI] [PubMed] [Google Scholar]
- 69.Muralidharan V, and Muir TW (2006) Protein ligation: an enabling technology for the biophysical analysis of proteins, Nat. Methods 3, 429–438. [DOI] [PubMed] [Google Scholar]
- 70.Camarero JA, Adeva A, and Muir TW (2000) 3-thiopropionic acid as a highly versatile multidetachable thioester resin linker, Lett. Pept. Sci 7, 17–21. [Google Scholar]
- 71.Blanco-Canosa JB, and Dawson PE (2008) An efficient Fmoc-SPPS approach for the generation of thioester peptide precursors for use in native chemical ligation, Angew. Chem. Int. Ed. Engl 47, 6851–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.George EA, Novick RP, and Muir TW (2008) Cyclic peptide inhibitors of staphylococcal virulence prepared by Fmoc-based thiolactone peptide synthesis, J. Am. Chem. Soc 130, 4914–4924. [DOI] [PubMed] [Google Scholar]
- 73.Erlich LA, Kumar KSA, Haj-Yahya M, Dawson PE, and Brik A (2010) N-methylcysteine-mediated total chemical synthesis of ubiquitin thioester, Org. Biomol. Chem 8, 2392–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dawson PE, and Kent SB (2000) Synthesis of native proteins by chemical ligation, Annu. Rev. Biochem 69, 923–960. [DOI] [PubMed] [Google Scholar]
- 75.Muir TW, Sondhi D, and Cole PA (1998) Expressed protein ligation: a general method for protein engineering, Proc. Natl. Acad. Sci. U.S.A 95, 6705–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perler FB, Davis EO, Dean GE, Gimble FS, Jack WE, Neff N, Noren CJ, Thorner J, and Belfort M (1994) Protein splicing elements: inteins and exteins--a definition of terms and recommended nomenclature, Nucleic Acids Res. 22, 1125–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perler FB (1998) Protein splicing of inteins and hedgehog autoproteolysis: structure, function, and evolution, Cell 92, 1–4. [DOI] [PubMed] [Google Scholar]
- 78.Ausió J, and Moore SC (1998) Reconstitution of chromatin complexes from high-performance liquid chromatography-purified histones, Methods 15, 333–342. [DOI] [PubMed] [Google Scholar]
- 79.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, and Luger K (2004) Reconstitution of nucleosome core particles from recombinant histones and DNA, Methods Enzymol. 375, 23–44. [DOI] [PubMed] [Google Scholar]
- 80.Shogren-Knaak MA, Fry CJ, and Peterson CL (2003) A native peptide ligation strategy for deciphering nucleosomal histone modifications, J. Biol. Chem 278, 15744–15748. [DOI] [PubMed] [Google Scholar]
- 81.Chiang KP, Jensen MS, McGinty RK, and Muir TW (2009) A semisynthetic strategy to generate phosphorylated and acetylated histone H2B, ChemBioChem 10, 2182–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goodman M, and McGahren WJ (1967) Mechanistic studies of peptide oxazolone racemization, Tetrahedron 23, 2031–2050. [DOI] [PubMed] [Google Scholar]
- 83.Pentelute BL, and Kent SB (2007) Selective desulfurization of cysteine in the presence of Cys(Acm) in polypeptides obtained by native chemical ligation, Org. Lett 9, 687–690. [DOI] [PubMed] [Google Scholar]
- 84.Wan Q, and Danishefsky SJ (2007) Free-radical-based, specific desulfurization of cysteine: a powerful advance in the synthesis of polypeptides and glycopolypeptides, Angew. Chem. Int. Ed. Engl 46, 9248–9252. [DOI] [PubMed] [Google Scholar]
- 85.Kuo MH, and Allis CD (1999) In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment, Methods 19, 425–433. [DOI] [PubMed] [Google Scholar]
- 86.Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, and Peterson CL (2006) Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions, Science 311, 844–847. [DOI] [PubMed] [Google Scholar]
- 87.Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, and Smith DL (2003) Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4, Anal. Biochem 316, 23–33. [DOI] [PubMed] [Google Scholar]
- 88.He S, Bauman D, Davis JS, Loyola A, Nishioka K, Gronlund JL, Reinberg D, Meng F, Kelleher N, and McCafferty DG (2003) Facile synthesis of site-specifically acetylated and methylated histone proteins: reagents for evaluation of the histone code hypothesis, Proc. Natl. Acad. Sci. U.S.A 100, 12033–12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ferreira H, Flaus A, and Owen-Hughes T (2007) Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms, J. Mol. Biol 374, 563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, Edon A, Fishel R, Poirier MG, and Ottesen JJ (2009) Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding, J. Biol. Chem 284, 23312–23321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shimko JC, North JA, Bruns AN, Poirier MG, and Ottesen JJ (2011) Preparation of Fully Synthetic Histone H3 Reveals That Acetyl-Lysine 56 Facilitates Protein Binding Within Nucleosomes, J. Mol. Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bang D, and Kent SB (2004) A one-pot total synthesis of crambin, Angew. Chem. Int. Ed. Engl 43, 2534–2538. [DOI] [PubMed] [Google Scholar]
- 93.Ottesen JJ, Bar-Dagan M, Giovani B, and Muir TW (2008) An amalgamation of solid phase peptide synthesis and ribosomal peptide synthesis, Biopolymers 90, 406–414. [DOI] [PubMed] [Google Scholar]
- 94.Goldknopf IL, French MF, Musso R, and Busch H (1977) Presence of protein A24 in rat liver nucleosomes, Proc. Natl. Acad. Sci. U.S.A 74, 5492–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weake VM, and Workman JL (2008) Histone ubiquitination: triggering gene activity, Mol. Cell 29, 653–663. [DOI] [PubMed] [Google Scholar]
- 96.Kim J, Guermah M, McGinty RK, Lee J-S, Tang Z, Milne TA, Shilatifard A, Muir TW, and Roeder RG (2009) RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells, Cell 137, 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chatterjee C, McGinty RK, Pellois J-P, and Muir TW (2007) Auxiliary-mediated site-specific peptide ubiquitylation, Angew. Chem. Int. Ed. Engl 46, 2814–2818. [DOI] [PubMed] [Google Scholar]
- 98.McGinty RK, Kim J, Chatterjee C, Roeder RG, and Muir TW (2008) Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation, Nature 453, 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McGinty RK, Kohn M, Chatterjee C, Chiang KP, Pratt MR, and Muir TW (2009) Structure-activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B, ACS Chem. Biol 4, 958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hicke L, Schubert HL, and Hill CP (2005) Ubiquitin-binding domains, Nat. Rev. Mol. Cell Biol 6, 610–621. [DOI] [PubMed] [Google Scholar]
- 101.Chatterjee C, Mcginty RK, Fierz B, and Muir TW (2010) Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation, Nat. Chem. Biol 6, 267–269. [DOI] [PubMed] [Google Scholar]
- 102.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, and Muir TW (2011) Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction, Nat. Chem. Biol 7, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu CC, and Schultz PG (2010) Adding new chemistries to the genetic code, Annu. Rev. Biochem 79, 413–444. [DOI] [PubMed] [Google Scholar]
- 104.Chalker JM, Bernardes GJL, Lin YA, and Davis BG (2009) Chemical modification of proteins at cysteine: opportunities in chemistry and biology, Chem. Asian. J 4, 630–640. [DOI] [PubMed] [Google Scholar]
- 105.Messmore MJ, Fuchs DN, and Raines RT (1995) Ribonuclease A: Revealing structure-functiona relationships with semisynthesis, J. Am. Chem. Soc 117, 8057–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guo J, Wang J, Lee JS, and Schultz PG (2008) Site-specific incorporation of methyl- and acetyl-lysine analogues into recombinant proteins, Angew. Chem. Int. Ed. Engl 47, 6399–6401. [DOI] [PubMed] [Google Scholar]
- 107.Neumann H, Peak-Chew SY, and Chin JW (2008) Genetically encoding N(epsilon)-acetyllysine in recombinant proteins, Nat. Chem. Biol 4, 232–234. [DOI] [PubMed] [Google Scholar]
- 108.Nguyen DP, Alai MMG, Kapadnis PB, Neumann H, and Chin JW (2009) Genetically encoding N(epsilon)-methyl-L-lysine in recombinant histones, J. Am. Chem. Soc 131, 14194–14195. [DOI] [PubMed] [Google Scholar]
- 109.Wang YS, Wu B, Wang Z, Huang Y, Wan W, Russell WK, Pai PJ, Moe YN, Russell DH, and Liu WR (2010) A genetically encoded photocaged Nepsilon-methyl-L-lysine, Mol. Biosyst 6, 1557–1560. [DOI] [PubMed] [Google Scholar]
- 110.Ai HW, Lee JW, and Schultz PG (2010) A method to site-specifically introduce methyllysine into proteins in E. coli, Chem. Commun 46, 5506–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nguyen DP, Garcia Alai MM, Virdee S, and Chin JW (2010) Genetically directing varepsilon-N, N-dimethyl-L-lysine in recombinant histones, Chem. Biol 17, 1072–1076. [DOI] [PubMed] [Google Scholar]
- 112.Simon M, Chu F, Racki L, Delacruz C, Burlingame A, Panning B, Narlikar G, and Shokat K (2007) The Site-Specific Installation of Methyl-Lysine Analogs into Recombinant Histones, Cell 128, 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, Peng J, Cheng D, Sui G, Kaiser UB, Shi Y, and Shi YG (2010) Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation, Mol. Cell 39, 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, and Shilatifard A (2009) Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription, J. Biol. Chem 284, 7970–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kokura K, Sun L, Bedford MT, and Fang J (2010) Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion, EMBO J. 29, 3673–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, and Al-Sady B (2011) Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly, Mol. Cell 41, 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Druryiii WJ, Voigt P, Martin SR, Taylor WR, Marco VD, Pirrotta V, Reinberg D, and Gamblin SJ (2009) Role of the polycomb protein EED in the propagation of repressive histone marks, Nature 461, 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Huang R, Holbert MA, Tarrant MK, Curtet S, Colquhoun DR, Dancy BM, Dancy BC, Hwang Y, Tang Y, Meeth K, Marmorstein R, Cole RN, Khochbin S, and Cole PA (2010) Site-specific introduction of an acetyl-lysine mimic into peptides and proteins by cysteine alkylation, J. Am. Chem. Soc 132, 9986–9987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Allahverdi A, Yang R, Korolev N, Fan Y, Davey CA, Liu CF, and Nordenskiold L (2011) The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association, Nucleic Acids Res. 39, 1680–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, Johnson ES, and Berger SL (2006) Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications, Genes Dev 20, 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Allis CD, and Muir TW (2011) Spreading chromatin into chemical biology, ChemBioChem 12, 264–279. [DOI] [PubMed] [Google Scholar]
- 122.Li S, and Shogren-Knaak MA (2008) Cross-talk between histone H3 tails produces cooperative nucleosome acetylation, Proc. Natl. Acad. Sci. U.S.A 105, 18243–18248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Blacketer MJ, Feely SJ, and Shogren-Knaak MA (2010) Nucleosome interactions and stability in an ordered nucleosome array model system, J. Biol. Chem 285, 34597–34607. [DOI] [PMC free article] [PubMed] [Google Scholar]