Abstract
Many bacteria secrete cellulose, which forms the structural basis for bacterial multicellular aggregates, termed biofilms. The cellulose synthase complex of Salmonella typhimurium consists of the catalytic subunits BcsA and BcsB and several auxiliary subunits that are encoded by two divergently transcribed operons, bcsRQABZC and bcsEFG. Expression of the bcsEFG operon is required for full-scale cellulose production but the functions of its products are not fully understood. This work aimed to characterize the BcsG subunit of the cellulose synthase, which consists of an N-terminal transmembrane fragment and a C-terminal domain in the periplasm. Deletion of the bcsG gene substantially decreased the total amount of BcsA and cellulose production. BcsA levels were partially restored by the expression of the transmembrane segment, whereas restoration of cellulose production required the presence of the C-terminal periplasmic domain and its characteristic metal-binding residues. The high-resolution crystal structure of the periplasmic domain characterized BcsG as a member of the alkaline phosphatase/sulfatase superfamily of metalloenzymes, containing a conserved Zn2+-binding site. Sequence and structural comparisons showed that BcsG belongs to a specific family within alkaline phosphatase-like enzymes, which include bacterial Zn2+-dependent lipopolysaccharide phosphoethanolamine transferases such as MCR-1 (colistin resistance protein), EptA, and EptC and the Mn2+-dependent lipoteichoic acid synthase (phosphoglycerol transferase) LtaS. These enzymes use the phospholipids phosphatidylethanolamine and phosphatidylglycerol, respectively, as substrates. These data are consistent with the recently discovered phosphoethanolamine modification of cellulose by BcsG and show that its membrane-bound and periplasmic parts play distinct roles in the assembly of the functional cellulose synthase and cellulose production.
Keywords: alkaline phosphatase superfamily, biofilm formation, cellulose biosynthesis, extracellular matrix, virulence
Introduction
Bacterial cellulose, an exopolysaccharide with versatile biological roles, is produced by a variety of phylogenetically diverse bacteria [1]. In many of them, cellulose is required for biofilm formation that mediates environmental persistence, stress protection, and an anti-virulence phenotype [2–5]. Cellulose production is also important for microbial cell-cell interactions including bacterial-fungal interactions, adherence to surfaces, slowing-down of cell motility, interaction with amyloid fibers and protection against disinfectants [6–10]. Cellulose is a seemingly simple biopolymer that consists of glucose monomers bound into linear β-(1–4)-glucan chains and is resistant against hydrolysis by alkali and most strong acids. Despite this simple structure, biosynthesis of cellulose in different bacteria is carried out by at least three distinct operon classes, which are characterized by different auxiliary and accessory genes [1; 11–14] to produce macromolecules of amazingly different properties.
The core genes of all characterized cellulose biosynthesis operons code for the cellulose synthase catalytic subunit BcsA, an inner membrane protein with a cytosolic domain containing the active site, which together with BcsB, a periplasmic protein with a single BcsA-interacting C-terminal transmembrane domain, forms the enzymatically active cellulose synthase (Fig. S1; [15–17]). The active site of BcsA is blocked by a gating loop and requires second messenger cyclic diguanosine monophosphate (c-di-GMP) binding to the C-terminal PilZ domain to allow substrate access. In addition, bcsZ gene, encoding a periplasmic endoglucanase, is typically located either within the cellulose biosynthesis operon or in close vicinity [5]. Further on, bcsC, a gene predicted to encode an outer membrane pore, is part of class I and II bcs operons [1]. The function of various accessory genes, often specific to certain cellulose biosynthesis operons, is starting to become unraveled. For example, in class II operons that are found in many beta- and gamma-proteobacteria, the bcsEFG operon is adjacent to the bcsABZC operon. BcsE was recently shown to be a novel c-di-GMP receptor required for optimal cellulose biosynthesis in Salmonella enterica serovar Typhimurium (hereafter S. typhimurium) and Escherichia coli [18]. Bacterial two-hybrid assays have shown a strong interaction of the E. coli BcsG with the cellulose synthase subunit BcsA and the BcsF protein [13; 19]. Mutating the bcsG gene in E. coli and Salmonella resulted in severely disturbed cellulose synthesis, indicating a role of this protein in maintaining wild-type levels of cellulose [13; 18; 20]. More recently, BcsG was shown to participate in a chemical modification of the growing cellulose molecules in E. coli and S. typhimurium that results in production of cellulose with a phosphoethanolamine group added to every other glucosyl residue [19].
In this work, we further investigate the role(s) of the BcsG protein in cellulose biosynthesis and report the high-resolution crystal structure of its periplasmic domain. The crystal structure confirms that this domain is a member of the alkaline phosphatase/sulfatase enzyme superfamily, related to the membrane-anchored phosphoethanolamine and phosphoglycerol transferases. Mutational analyses demonstrated that the Ser278 residue, which is conserved in the BcsG family, is required for the catalytic activity of BcsG in vitro and optimal cellulose biosynthesis in vivo. However, the protein scaffold is required for production of wild-type levels of the cellulose synthase subunit BcsA. Thus, this work shows that BcsG is a multifunctional protein that, in addition to transferring a phosphoethanolamine headgroup from phospholipids to the nascent cellulose molecule, is involved in formation of the functional cellulose synthase complex.
Results
Functional characterization of BcsG
We reported recently that a polar bcsE mutant of Salmonella enterica serovar Typhimurium (hereafter S. typhimurium) lacking the biofilm extracellular matrix component curli fimbriae (ΔcsgBA mutant) displayed a smooth and nearly white (saw) colony morphotype when grown in the presence of the Congo red dye [18]. Such a phenotype is consistent with a lack of cellulose production [6; 18]. By contrast, a non-polar bcsE mutant in this ΔcsgBA background displayed a clearly diminished, but still prominent pdar (pink, dry and rough) morphotype suggesting a reduced but still noticeable production of cellulose [6; 18]. To dissect the contribution of genes downstream of bcsE in the bcsEFG operon to cellulose formation (Fig. 1A), we constructed bcsF and bcsG deletion mutants in S. typhimurium strain UMR1, a single-colony derivative of S. typhimurium ATCC14028, which displays highly regulated expression of curli and cellulose biofilm extracellular matrix components at 28 °C (Fig. 1B). In this wild-type strain background, the bcsG mutant grown on Congo red plates displayed a bdar, brown and rough, morphotype indicative of amyloid curli production. However, colonies of the bcsG mutant did not bind Calcofluor whitestain suggesting that the bcsG gene is required for cellulose biosynthesis (Fig. S2A). To avoid interference with curli fimbriae, we constructed bcsG mutants of the S. typhimurium strains MAE14 and MAE97, cellulose positive/curli negative UMR1 derivatives. The MAE14 bcsG derivative did not bind Calcofluor white and had white colonies on Congo red plates, which was similar to the previously characterized cellulose- and curli-negative control strain MAE50 [21], again suggesting that BcsG is required for cellulose biosynthesis (Fig. S2A; Fig. 1B). In the background of MAE97, an S. typhimurium strain that displays temperature-independent upregulated cellulose production due to a point mutation in the csgD promoter region which results in upregulation of the major biofilm regulator CsgD and the diguanylate cyclase AdrA [22], the bcsG mutant (Table S1) showed highly reduced, but still noticeable residual cellulose production. This suggested that while BcsG is required for optimal cellulose production, it is dispensable when cells are grown on salt-free LB agar plates (Fig. S2A; Fig. 1B). Investigation of cellulose production in MAE97 and its bcsG mutant by laser scanning microscopy on the single cell level confirmed the role of BcsG in cellulose production and cell-cell interactions leading to cell aggregation (Fig. 1C). In all strain backgrounds, the bcsG mutant could be complemented by expressing bcsG from plasmid pBAD30, although not entirely up to the wild-type level (Fig. 1B). Flagella-dependent swimming and swarming motility were not affected by bcsG deletion or overexpression (Fig. S2B and data not shown). As the phenotype of a bcsG mutant with respect to cellulose production is most pronounced in the MAE97 background, this strain was used as the basis for further studies.
Fig. 1. Cellulose biosynthesis operon structure and colony morphotypes of the bcsG deletion mutants in different backgrounds of Salmonella typhimurium ATCC14028–1s.
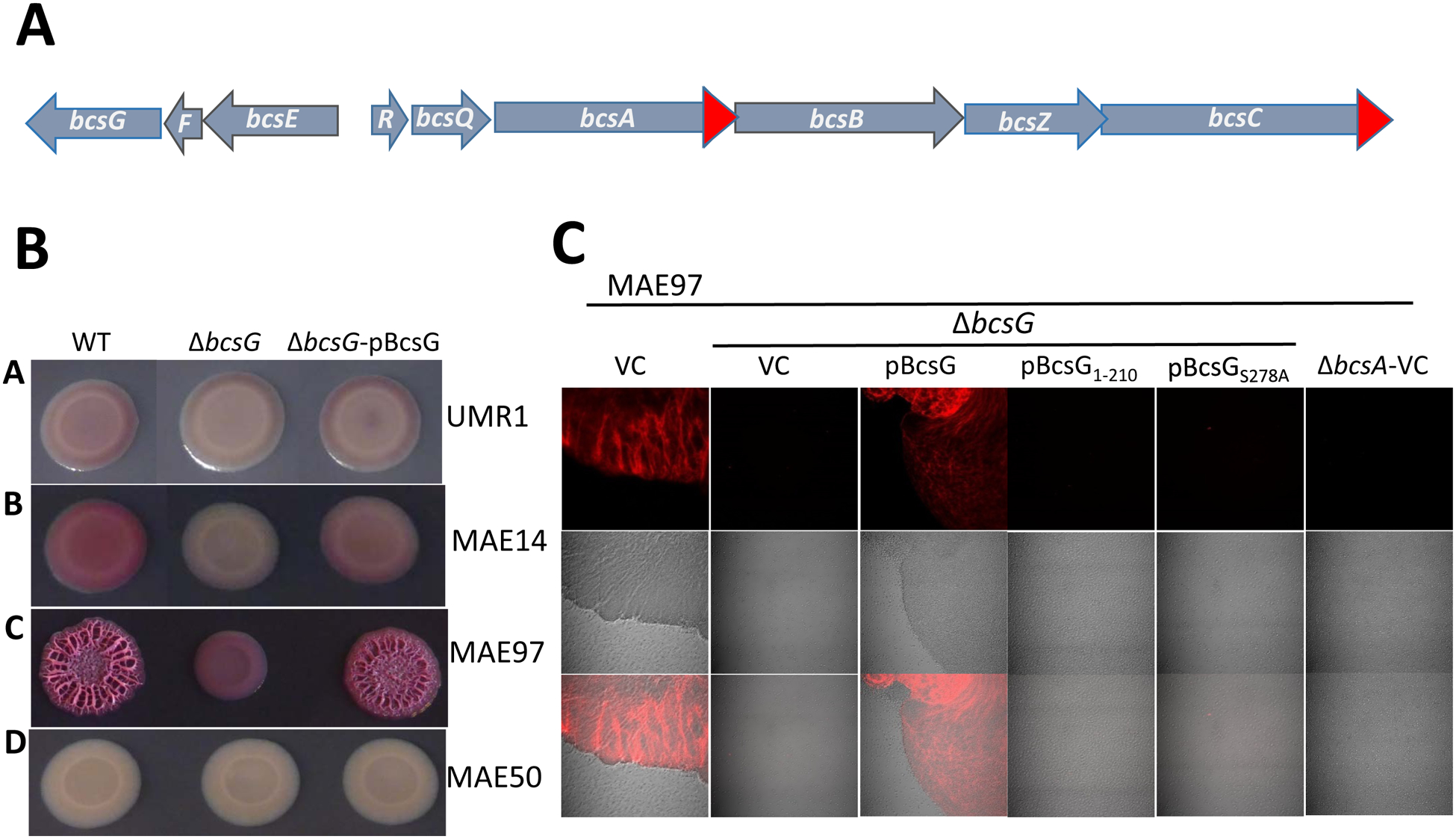
A. Organization of the bcsRQABZC and bcsEFG operons in both E. coli and S. typhimurium. Red arrows indicate addition of 3xFLAG-tags to the open reading frames of S. typhimurium used in this work.
B. Congo Red-stained colony morphotypes of S. typhimurium strains and their ΔbcsG derivatives. Strains used were UMR1 (wild type, A); MAE14 (cellulose positive/curli negative, B); MAE97 (semi-constitutive cellulose positive/curli negative, C), and MAE50 (UMR1 ΔcsgD, D), see Table S2 for the complete genotypes. WT indicates the wild type with respect to the bcsG gene, pBcsG indicates complementation with pBAD30 carrying the bcsG gene under the control of arabimose-dependent promoter. Cells were grown on salt-free LB agar plates for 24 h at 28 °C.
C. Cellulose production by agar-grown colonies of S. typhimurium strain MAE97 (bcsG+) and its ΔbcsG derivative as assessed by Calcofluor white staining and observed by confocal laser scanning microscopy. Cells grown on agar as in panel B were gently submerged in 0.001% Calcofluor white to preserve colony structure and aggregation patterns. The wild type MAE97 and the ΔbcsG mutant complemented with wild-type bcsG gene show major cell aggregation and cellulose production (displayed as false color image). The ΔbcsG mutant with pBAD30 alone (vector control, VC) or pBAD30 carrying either truncated (pBcsG1–210) or mutated (pBcsGS278A) bcsG gene dispersed into single cells with residual or no cellulose production. Top panel, fluorescence microscopy; middle panel, phase contrast; bottom panel, overlay of the two. 63x (oil) magnification. pBcsG, pBcsG1–210, and pBcsGS278A are wild-type bcsG and its variants cloned in pBAD30 with a C-terminal 8xHis tag. ΔbcsA strain was used as negative control.
The non-polar bcsF deletion mutant displayed only a minor reduction in cellulose biosynthesis (Fig. S3). The polar ΔbcsF mutant showed a more severe reduction in cellulose production, which could not be restored by the cloned bcsF gene. However, cellulose production in the polar ΔbcsF mutant could be restored by overexpressing the bcsG gene (Fig. S3). These data showed that the effect of the polar ΔbcsF mutation was primarily due to the decreased expression of bcsG, and that BcsG plays a much more significant role in cellulose biosynthesis than BcsF. Thus, we decided to focus on the bcsG gene product.
Next, we investigated the molecular basis of cellulose production stimulation by BcsG. Cellulose production on salt-free agar plates is stimulated by c-di-GMP that is produced by AdrA whose expression is triggered by the transcriptional activator CsgD [21; 22]. BcsG was not required for the production of CsgD (Fig. S4A). In order to assess whether BcsG is required for transcription, production, steady-state levels or degradation of the cellulose synthase core, we evaluated the effects of bcsG mutations on the expression of the catalytic subunit BcsA. Transcription of bcsA was not affected by the bcsG mutation as measured by a previously constructed MudJ transcriptional fusion in bcsA (Fig. S3B; [6; 23]). On the other hand, Western blot analysis showed that not only was the amount of BcsA-3xFLAG severely reduced in the bcsG mutant, in agreement with the downregulation of the pdar morphotype, but a number of proteolytic degradation products were observed (Fig. 2). However, the rate of BcsA degradation did not seem to be affected, as the half-life of BcsA-3xFLAG construct in the bcsG mutant was similar to that in the MAE97 wild-type background (Fig. 2B). Thus, as a working hypothesis, we conclude that BcsG aids the integration of BcsA into the cytoplasmic membrane.
Fig. 2. Effects of bcsG variants on the expression of the cellulose synthase subunit BcsA and cellulose production.
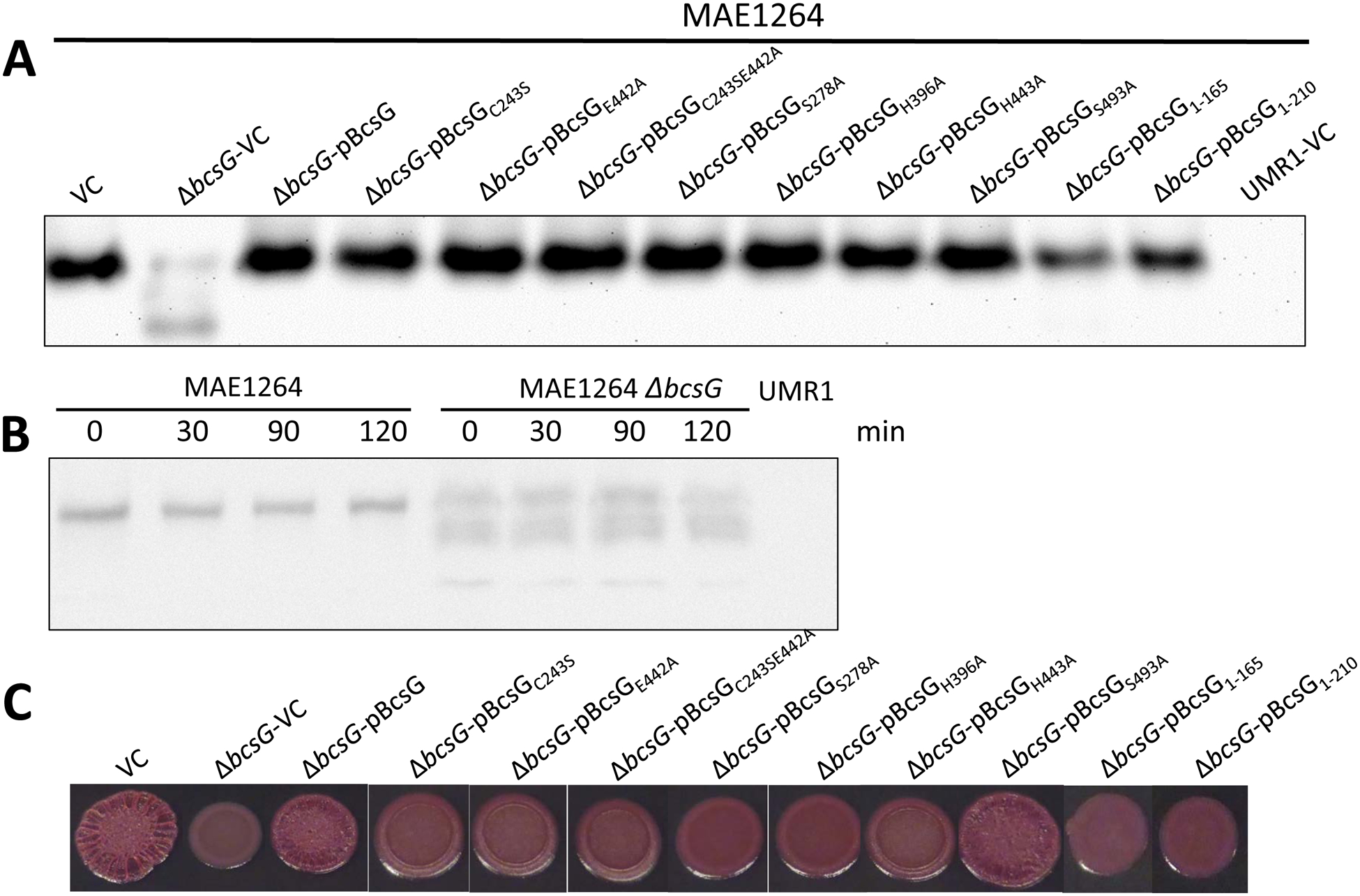
A. Expression of BcsA-3xFLAG in S. typhimurium strain MAE1264 (bcsG+ [5]) and its ΔbcsG mutant upon overexpression of bcsG and its variants. VC, pBcsG, pBcsG1–210, and pBcsGS278A are as in Fig. 1. pBcsGC243S, pBcsGE442A, pBcsGC243SE442A, pBcsGS278A, pBcsGH396A, pBcsGH443A, and pBcsGS493A are bcsG variants with mutations in predicted active site residues of the AlkP superfamily cloned in pBAD30 with a C-terminal 8x-His tag. Strain UMR1 with plasmid pBAD30 (UMR1-VC) without the 3xFLAG tag was used as a negative control. All samples contained equal amounts of cell extracts as judged by Coomassie staining.
B. Stability of chromosomally encoded BcsA in the presence (MAE1264) and the absence of bcsG. After translation was inhibited with chloramphenicol, the amount of the 3xFLAG-tagged BcsA subunit was quantified using anti-FLAG antibodies at indicated time points. Strain UMR1 without the 3xFLAG tag was the negative control. Owing to the lower expression of BcsA in the ΔbcsG mutant, 10-fold higher amounts of its cell extract than those for MAE1264 were applied on the gel.
C. Pdar colony morphotype (cellulose biosynthesis) of strains listed in panel A. Despite similar levels of the BcsA subunit (panel A), the wild-type (VC) level of cellulose production could only be observed upon overexpression of the wild-type BcsG or the BcsGS493A mutant. Cells were grown on CR salt-free LB agar plates for 24 h at 28 °C.
We then assessed whether additional gene products of the bcsABZC operon were affected when bcsG was deleted. Western blot analysis showed that expression of the cellulase BcsZ was inversely regulated compared to BcsA upon deletion of bcsG. BcsZ levels were upregulated upon deletion of bcsG and downregulated upon BcsG overexpression from the pBAD30 plasmid (Fig. 3A). The level of the putative outer membrane pore BcsC, on the other hand, was not affected by the expression of BcsG and/or its variants, neither in the wild type nor the ΔbcsG deletion mutant background (Fig. 3B). We conclude that BcsG specifically modulates the protein levels of BcsA and, potentially indirectly, BcsZ.
Fig. 3. Effects of bcsG variants on the expression of the cellulose synthase auxiliary subunits BcsZ and BcsC.
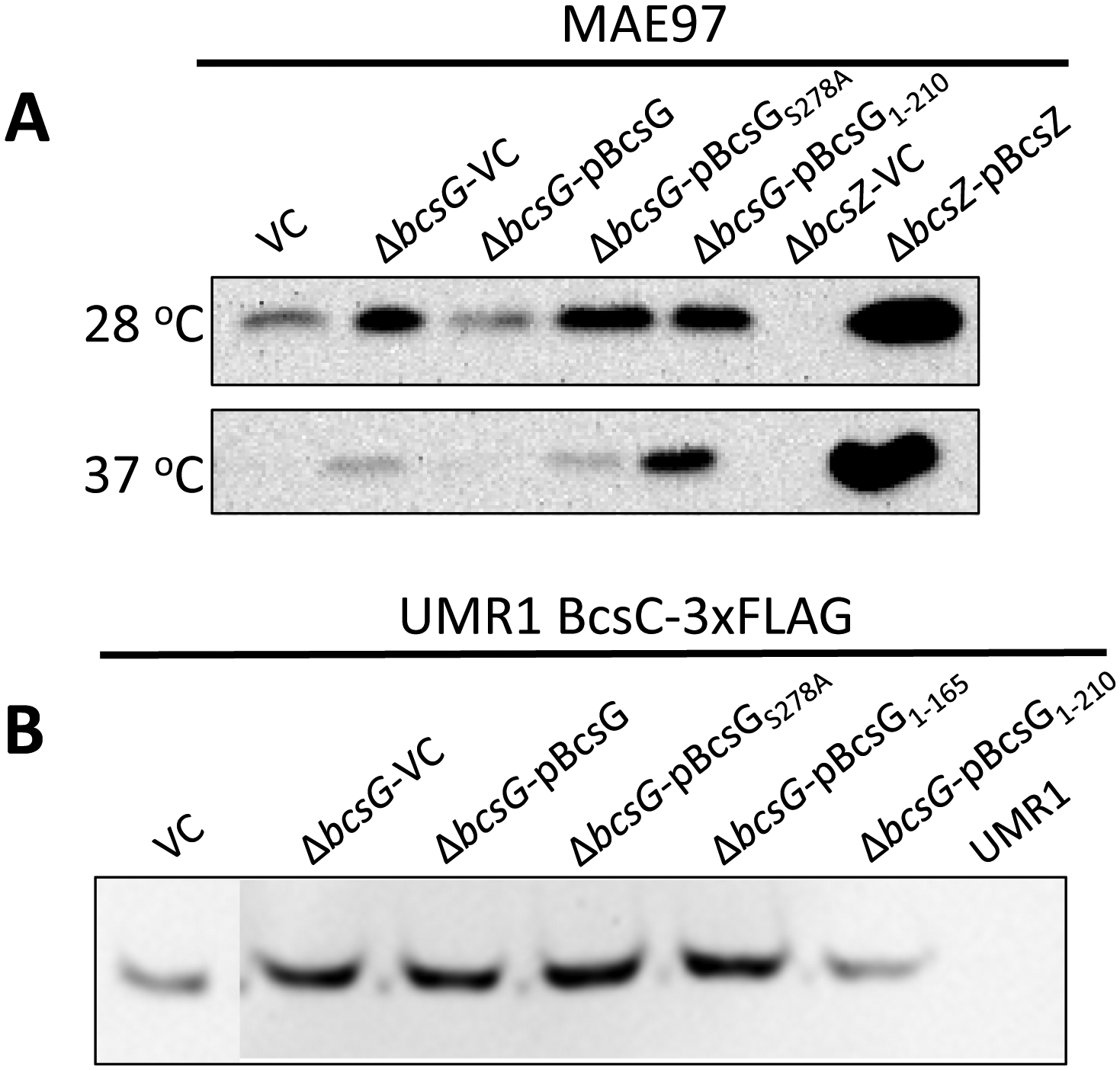
A. Production of the cellulase BcsZ in the ΔbcsG mutants of the cellulose synthase-positive strain MAE97. MAE97 bcsZ deletion strains with pBAD30 vector control (ΔbcsZ-VC) and with overexpressed BcsZ (ΔbcsZ-pBcsZ) were used as negative and positive controls, respectively. Detection of BcsZ production was by Western blot using a rabbit anti-BcsZ antibody.
B. Production of the putative outer membrane pore BcsC in S. typhimurium wild type and bcsG derivatives. Wild-type S. typhimurium UMR1 with BcsC-3xFLAG and its ΔbcsG mutant were complemented by the same plasmids as in panel A with the addition of the second truncated variant, pBcsG1–165. Detection of the 132.7 kDa BcsC-3xFLAG was by Western blot using a mouse anti-FLAG-tag antibody. UMR1 BcsC-3xFLAG with pBAD30 (vector control, left lane) and UMR1 without the FLAG tag (right lane) were used as positive and negative controls, respectively.
Cells were grown on salt-free LB agar plates for 24 h at either 28 °C (A, top lane) or 37 °C (A, bottom lane), or for 16 h at 28 °C (panel B).
The presence of BcsG is characteristic for class II cellulose biosynthesis operons [1]. The bcsG gene from S. typhimurium encodes a polypeptide chain of 559 amino acid residues. Sequence analysis suggests that the protein consists of two domains, an N-terminal part containing four or five transmembrane (TM) helices, which anchors the protein in the inner membrane (Fig. S5), and a periplasmic C-terminal domain, which belongs to the alkaline phosphatase/sulfatase (AlkP) superfamily [1] (Fig. 4A). The two domains are connected by a flexible linker region.
Fig. 4. Structure of BcsG.
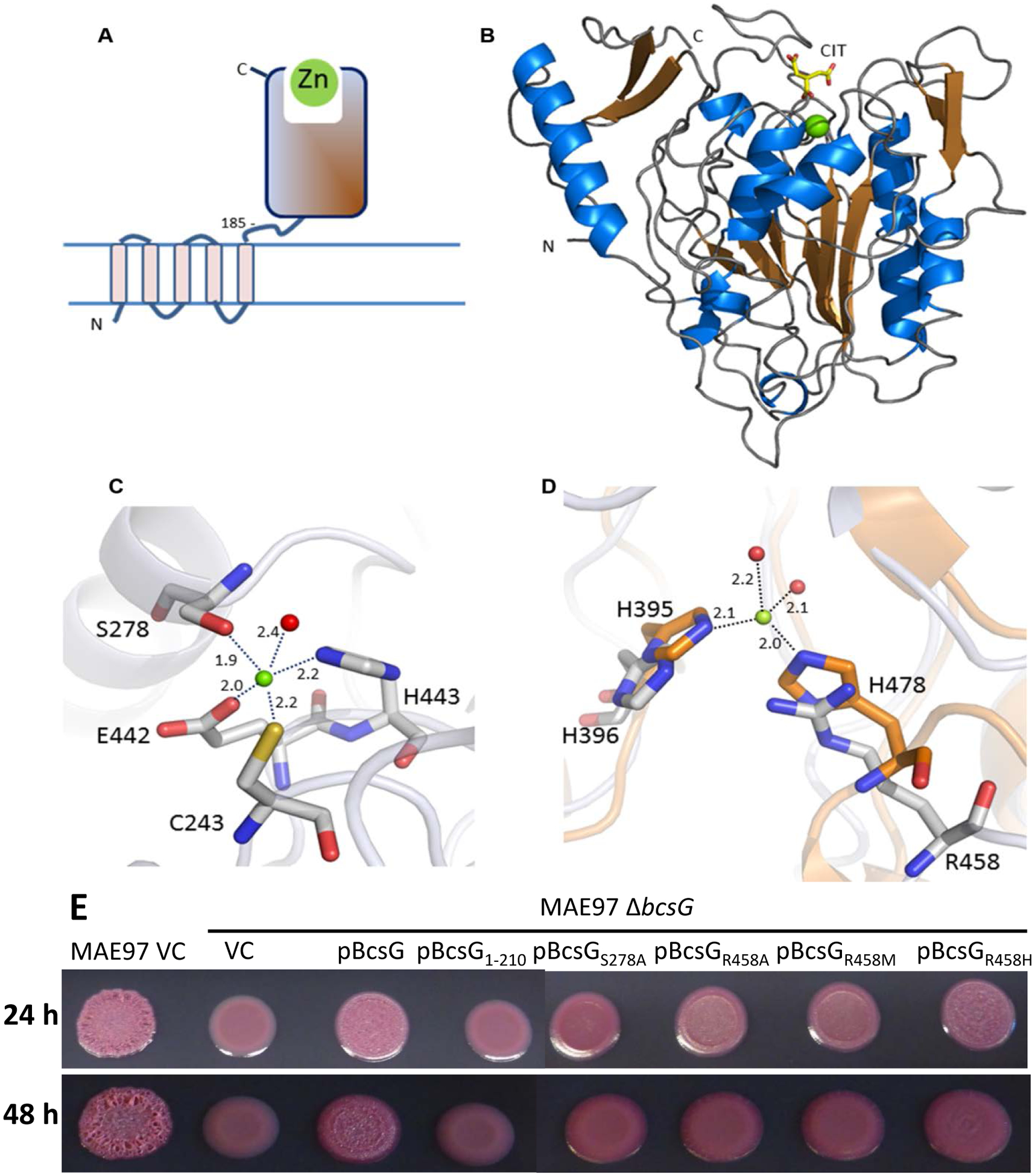
A. Domain structure of BcsG. The N-terminal transmembrane domain (residues 1–165) is linked to the C-terminal alkaline phosphatase-like domain (residues 185–559) via flexible inter-domain linker (residues 166–210). The fragment from aa 185 to 559 was used in the construct designed for crystallization. B. Schematic cartoon of the C-terminal alkaline phosphatase-type domain of BcsG. The zinc ion is shown as a green sphere and the citrate molecule bound close to the metal binding site as a stick-model. C. Metal binding site in BcsG. Distances between the Zn2+ and the coordinating atoms are indicated in Å. The water molecule is shown as a red sphere. D. View of the second Zn2+-binding site in the alkaline phosphatase family illustrated after superimposition of the phosphoethanolamine transferase MCR-2 (PDB code 5MX9) (orange carbon atoms) with BcsG (grey carbon atoms). While NmEptA contains a fully functional second Zn2+ site [35], one of the metal ligands, His478 in NmEptA, is replaced by Arg458 in BcsG, making binding of Zn2+ to this site less likely. E. Cellulose production in S. typhimurium strain MAE97 and its ΔbcsG mutants complemented with wild-type BcsG, the mutant lacking the alkaline phosphatase-type domain, and the mutants affecting the residues Ser278 and Arg458, shown in panels C and D. Other labels are as in Fig. 2. Cells were grown on salt-free LB agar plates for 24 h at 37 °C.
In order to investigate which part of BcsG is required for the formation of steady-state levels of membrane-bound BcsA and biosynthesis of cellulose, we created several different BcsG constructs. To this end, we engineered a BcsG version, BcsG1–165, with only the transmembrane domain; BcsG1–210, a version with the transmembrane domain and the flexible linker, and BcsGS278A, a full length BcsG construct with a mutation in Ser278, the predicted active-site nucleophile of the AlkP superfamily. Assessment of the BcsA protein levels upon expression of these constructs from plasmid pBAD30 revealed that the transmembrane domain was required for elevated BcsA protein levels. BcsA levels rose further with the addition of the linker in the BcsG1–210 construct and reached wild-type levels in the BcsGS278A variant (Fig. 2A). The truncated versions of BcsG, transmembrane-only (BcsG1–165) and transmembrane-only with a linker (BcsG1–210), were not detectable with anti-His-tag antibodies though (data not shown), despite an obvious effect on BcsA production. Pdar colony morphology, which is indicative of cellulose production, did not seem to correlate with BcsA protein levels in the truncated and point mutation versions. Upon overexpression of the BcsGS278A variant, wild-type levels of BcsA were produced. However, the morphotype presented as smooth and pink colonies, indicative of only minor residual cellulose production. In summary, these experiments showed that BcsG affects BcsA levels and cellulose production and that Ser278, the potential catalytic nucleophile of the extra-membrane domain of BcsG is required for full-scale cellulose production (Fig. 1 & 2).
Given the apparent requirement of the catalytic residue Ser278 of the BcsG periplasmic domain for effective cellulose production, despite essentially the same levels of the BcsA subunit, we constructed additional mutants with replacements in the active site residues of the AlkP superfamily (see below for details on the BcsG active site). Expression of these constructs in the bcsG deletion mutant at similar levels as the wild type protein (data not shown) showed that BcsG point mutants did not affect the level of the cellulose synthase BcsA, but showed only residual low-level cellulose production (Fig. 2A, C). This result showed that the periplasmic domain of BcsG is likely to have an enzymatic activity that is critical for full-scale cellulose production.
These data indicate that BcsG fulfills at least two different functions with respect to cellulose biosynthesis. First, the transmembrane domain of BcsG helps to maintain integrity of BcsA, presumably by aiding its integration into the cytoplasmic membrane. The presence of proteolytically degraded fragments of BcsA in the bcsG mutant (Fig. 2B) suggests that BcsG might also help in stabilizing BcsA in the membrane. Second, the catalytic activity of the periplasmic domain is required for optimal cellulose production. These observations prompted us to take a closer look at the periplasmic domain of BcsG. Given that AlkP superfamily enzymes are typically either phosphatases or phosphotransferases [24–27], we assumed that this domain could participate either in a modification of the BcsA subunit, anchoring BcsA in the membrane or to other cellulose synthase subunits, and/or in a modification of cellulose itself.
Structural characterization of BcsG
Topology of the BcsG N-terminal membrane domain
The UniProt entry Q7CPI7 describes BcsG from S. typhimurium (BCSG_SALTY) as having four TM segments. Use of several different software tools produced inconsistent results with 4 or 5 predicted TM helices and the location of the N-terminus either inside or outside the cytoplasm. Most of the tools, however, agreed on the coordinates of three C-terminal TM helices and the periplasmic location of the C-terminal domain (Fig. S5). An alignment of the N-terminal regions of BcsG proteins from diverse bacteria identified a likely cause of this discrepancy: the conserved lysine residue (Lys31 in BCSG_SALTY) whose presence in the hydrophobic core of the membrane seemed very unlikely. However, in BcsG, the positive charge of this lysine residue is likely to be balanced by the negative charge of membrane-embedded Asp82, which is also well conserved among BcsG homologs (Fig. S5A). Therefore, in accordance with the predictions of MEMSAT 3.0, OCTOPUS, TOPCONS and several other software tools, the N-terminal domain of BcsG was concluded to consist of five TM helices, accounting for the periplasmic location of its C-terminal domain. The predicted membrane topology and secondary structure of the N-terminal domain of BcsG was compatible with the recently solved structure of the membrane domain of Neisseria meningitidis lipid A phosphoethanolamine transferase NmEptA (previous designation LptA, UniProt: Q7DD94, Protein DataBank [PDB] entry 5fgn) [28]. Despite the lack of statistically significant sequence similarity (only ~12% of identical residues) between the respective membrane fragments, secondary structure prediction for BcsG included two short periplasmic helices between the third and fourth transmembrane segments and an ‘aromatic belt’ near the water-lipid interface, as seen in the NmEptA structure [28].
Overall structure of the BcsG C-terminal periplasmic domain
The crystal structure of the periplasmic domain of BcsG was determined using the expression construct that comprised residues Ala185–Gln559 of the UniProt entry Q7CPI7 (Fig. 4A) and included an N-terminal linker from the vector containing a factor Xa cleavage site and maltose binding protein. This construct expressed well and resulted in a soluble protein. After cleavage with factor Xa, the protein used for further studies consisted of 375 residues of BcsG, with eight additional amino acid residues from the N-terminal vector derived linker. Hereafter, amino acid residues of BcsG are listed under their numbers in the full-length protein (positions 185–559), whereas the linker residues are listed under their numbers in the cleaved construct. The integrity of the protein was verified by mass-spectrometry, giving a major peak corresponding to a mass of 42,081 Da, which compares well with the value calculated from the expected amino acid composition, 42,088 Da (Fig. S6). Analytical gel filtration resulted in a single peak with a mass of 54.7 kDa, indicating that the protein most likely is a monomer in solution (Fig. S7).
Initially the structure of BcsG was determined by molecular replacement (MR), using the predicted model obtained from Rosetta [29] as search template. The final model of BcsG, refined to a resolution of 1.55 Å with Rfree/R values of 0.166/0.198 (Table 1) comprises residues Ala185-Gly186-Asp187 and Gly192 – Gln559 of the BcsG sequence and Ile-Ser-Glu-Phe-Ser-Ser-Arg from the linker. Lack of electron density, probably due to disorder of the polypeptide chain prevented modelling of residues 188–191 of BcsG. BcsG proteins contain conserved cysteine residues in positions 243, 290, and 306. A mixed population with an oxidized and reduced disulfide bridge was observed between Cys290 and Cys306, with the oxidized species modeled at 0.4 occupancy. The side chain of Cys306 showed two different conformations, one engaged in the S-S-bridge formation and another with a weak hydrogen bond (3.3 Å) to the carbonyl oxygen of Asn303. Weakly defined electron density was also found at one loop region at the surface of the protein comprising residues 376–384. The model further contains one Zn2+ ion, two citrate and 316 water molecules (Fig. 4B).
Table 1.
Data collection and refinement statistics
| Data Collection | BcsG | BcsG Zn-SAD |
|---|---|---|
| PDB accession code | 5OJH | 5OLT |
| Beamline | P13, EMBL c/o DESY | P14, EMBL, c/o DESY |
| Space group | P3121 | P3121 |
| Cell dimensions | ||
| a, b, c (Å), α,β,γ (°) | 80.4, 80.4, 97.5 90.0, 90.0, 120.0 |
80.7, 80.7, 97.7 90.0, 90.0, 120.0 |
| Wavelength (Å) | 0.9763 | 1.278 |
| Resolution (Å)a | 69.7 – 1.55 (1.59–1.55) | 40.0 – 1.45 (1.53–1.45) |
| Rsym | 0.116 (0.790) | 0.068 (0.538) |
| Rmeas | 0.128 (0.830) | 0.072 (0.586) |
| Rpim | 0.056 (0.260) | 0.023 (0.206) |
| Total number of observations | 534385 (26343) | 1107780 (113316) |
| Total number unique | 53454 (2609) | 123431 (17487)b |
| I/σ(I) | 15.3 (3.3) | 21.3 (5.0) |
| Completeness (%) | 100 (99.9) | 97.3 (85.1) |
| Redundancy | 10.0 (10.0) | 9.0 (6.5) |
| Anomalous Correlation (%) | - | 39 (7) |
| SigAno | - | 1.135 (0.748) |
| Wilson B-factor (Å2) | 14.0 | 9.8 |
| Refinement | ||
| Resolution (Å) | 69.7– 1.55 (1.59–1.55) | 40.0–1.45 (1.49–1.45) |
| Number of reflections | 50696 | 60090 |
| Rwork /Rfree (%) | 16.6/19.8 (19.4/27.6) | 12.8/15.8 (18.1/21.2) |
| Number of atoms | ||
| Protein | 3095 | 3215 |
| Water molecules/Zn2+/citrate | 318/1/26 | 322/1/26 |
| B-factor (Å2) | ||
| Protein | 13.5 | 17.0 |
| Water molecules/Zn2+/citrate | 22.8/12.5/15.4 | 27.2/12.4/25.1 |
| r.m.s deviations | ||
| Bond lengths (Å) | 0.016 | 0.013 |
| Bond angles (°) | 1.747 | 1.565 |
| Ramachandran plot | ||
| Favored/allowed/outliers (%) | 97.7/1.8/0.5 | 97.4/2.3/0.3 |
Outer shell statistics are given in parentheses.
For the SAD data set, Friedel pairs are treated as different reflections.
During the structure determination of BcsG by MR it became clear that the domain contains a metal ion, which was subsequently identified by X-ray fluorescence and a low-resolution SAD data set as a zinc ion. A high-resolution data set from crystals obtained in the presence of additional ZnCl2 was collected at beam line P14, equipped with Compound Refractive Lenses, at the EMBL outstation at the absorption edge of Zn2+ (Table 1) to 1.45 Å resolution. These data allowed a straightforward structure determination using experimental phases. Refinement of the initial experimental model derived from SAD phasing (one Zn2+ ion per 382 residues) resulted in a structure with the same features as the model derived from phasing by MR. During the refinement of the model obtained from experimental phasing no use of the MR-derived structure was made to avoid bias. Superposition of the two models resulted in a r.m.s.d. of 0.1 Å, based on 368 equivalent Cα atoms.
The soluble periplasmic domain of BcsG forms a globular fold typical of the AlkP superfamily, with overall dimensions of 56×55×40 Å. The core of the domain contains a central seven-stranded mixed β-sheet sandwiched between several α-helices. A second, four-stranded anti-parallel β-sheet is formed by the C-terminal part of the chain and packs against one layer of α-helices and several of the loop regions connecting the secondary structural elements of the central core (Fig. 4B). The N-terminal linker region extends away from the globular core by 24 Å and forms important packing interactions with a neighboring molecule in the crystal lattice (see below). The metal binding site is located at the carboxyl end of the central β-sheet, facing an open, solvent accessible cavity, which presumably forms the substrate binding site. The N-terminus of the protein domain is located opposite to the metal binding site and is part of a predominantly hydrophobic surface of the protein. The proposed overall organization of the cellulose synthase complex interacting with BcsG is as shown in ref. [30].
Quaternary structure
The asymmetric unit of the trigonal crystals contains one BcsG molecule, giving a Matthew’s coefficient of 2.21 Å3/Da. An analysis of the crystal packing using PISA [31] does not reveal large interaction areas (>1000 Å2) between the molecules in the lattice and suggests a monomeric structure in the crystal. This agrees with the results from the gel filtration experiments that indicated monomeric structure in solution (Fig. S7).
An important packing interaction is made by the linker peptide derived from the expression plasmid. This part of the polypeptide chain extends about 24 Å from the core of the domain and forms tight interactions with another BcsG molecule in the crystal lattice (Fig. S8).
The atomic coordinates of the BcsG soluble domain were submitted to the DALI server [32] to search for homologous structures. The closest relatives were other members of the AlkP superfamily, with amino acid sequence identities of at best 14% and r.m.s.d. values in the range of 3.6 Å after superposition. The top homologs were an uncharacterized protein VP1736 from Vibrio parahaemolyticus (PDB accession code 3lxq, Z-score 24.3, sequence identity 11%), the periplasmic domain of the putative cardiolipin transporter PbgA from S. typhimurium (accession code 5i5f, Z-score 22.4, sequence identity 13%) [33], lipoteichoic acid synthase LtaS from Listeria monocytogenes (accession code 4uop, Z-score 22.1, sequence identity 13%) [34], the putative sulfatase SpAS2 from Silicibacter pomeroyi (accession code 4upl, Z-score 21.6, sequence identity 12%) and the lipooligosaccharide phosphoethanolamine transferase NmEptA (LptA) from Neisseria meningitidis (accession code 4kay, Z-score 20.3, sequence identity 11%) [28; 35]. Despite the low sequence identity, BcsG shares the overall α/β fold typical of this enzyme superfamily, with major structural differences found in loop regions.
The periplasmic domain of BcsG contains a single Zn2+ ion, already present in crystals obtained without the addition of Zn2+ salts during purification and crystallization. The identity of this metal ion was confirmed by an X-ray fluorescent scan and the structure analysis using data collected at the absorption edge of Zn2+(Fig. S9). The metal binding site is located at the C-terminal end of the central β-sheet at the same position as observed in other members of the AlkP superfamily (Fig. 4B). The metal ion is coordinated by the side chains of Cys243, Ser278, Glu442, His443 and a water molecule in a distorted trigonal bi-pyramidal ligand geometry (Fig. 4C). A search of the PDB did not reveal any metal site with this composition of ligands, neither in the AlkP superfamily (Table 2) nor in any other protein.
Table 2.
Properties of the BcsG-related members of the alkaline phosphatase superfamily
| Enzyme, source organism(s)a | PDB entry code | RMSD (Å), alignment lengthb | 5TM domain | Bound metal(s) | Active site residuec | Reference | |
|---|---|---|---|---|---|---|---|
| Me1 | Me2 | ||||||
| Phosphoethanolamine transferase | |||||||
| BcsG, S. typhimurium | 5OJH, 5OLT | None | Yes | Zn2+ | – | Ser | This work |
| LPS:phosphoethanolamine transferases MCR-1 and MCR-2 (mobile colistin resistance genes), E. coli | 5GOV, 5GRR, 5LRN, 5K4P, 5MX9 | 3.3–3.4, 257–260 | Yes | Zn2 | Zn2+ | Thr | [38; 39; 51; 52; 53] |
| LPS:phosphoethanolamine transferase EptA, N. meningitidis | 4KAY, 5FGN | 3.2–4.1, 258–269 | Yes | Zn2+ | (Zn2+)c | Thr | [28; 35] |
| LPS and FlgG protein phosphoethanolamine transferase EptC, C. jejuni | 4TN0 | 3.2, 254 | Yes | Zn2+ | – | Thr | [54] |
| Phosphoglycerol transferase | |||||||
| Lipoteichoic acid synthase LtaP, LtaS, B. subtilis, S. aureus, L. monocytogenes | 2W5Q, 2W8D, 4UOP, 4UOR | 3.7–3.9, 273–285 | Yes | Mn2+ | – | Thr | [34; 40; 55] |
| Sulfatases | |||||||
| Arylsulfatase, human, P. aeruginosa | 1FSU, 1HDH | 3.6–3.7, 269–279 | No | Ca2+ | – | fGlyd | [86; 87] |
| Iduronate-2-sulfatase, estrone sulfatase, human | 5FQL, 1P49 | 3.4–3.5, 269–279 | No | Ca2+ | – | fGlyd | [88; 89] |
| Glycosaminoglycan sulfatase; sulfoglucosamine hydrolase, human | 5G2T, 4MIV | 3.6–3.7, 249–279 | No | Zn2+ | – | fGlyd | [90; 91] |
| Alkaline phosphatase | |||||||
| Alkaline phosphatase, E. coli, H. salinarum, rat, bacterium Tab5 | 1ALK, 2X98, 4KJD, 2IUC | 3.2–3.3, 213–225 | No | Zn2 | Zn2 | Ser | [36] |
| Promiscuous phosphatases | |||||||
| Promiscuous phospho- and sulfoester hydrolase, B. caryophylli, R. leguminosarum | 2W8S, 2VQR | 3.2–3.4, 273–279 | No | Zn2+ Mg2+ | Fe2+ Ca2+ | fGlyd | [92; 93] |
| Phosphonoacetate hydrolase PhnA, S. meliloti | 3T02 | 3.6, 227 | No | Zn2+ | Zn2+ | Thr | [94] |
| Phosphoglyceromutase | |||||||
| Phosphoglycerate mutase, B. subtilis, S. aureus, B. stearothermophilus | 1O98, 2IFY, 4QAX | 3.3–3.4, 216–221 | No | Mn2+ | Mn2+ | Ser | [95; 96] |
| Phosphoglycerate mutase, L. mexicana, T. brucei | 3IGZ, 3NVL | 3.4–3.8, 232–239 | No | Co2+ | Co2+ | Ser | [97; 98] |
| Phosphopentomutase | |||||||
| Phosphopentomutase, B. cereus, S. mutans | 3M8Z, 4N7T | 3.1–3.2, 215–217 | No | Mn2+ | Mn2+ | Thr | [99] |
| No enzymatic activity | |||||||
| Cardiolipin transfer protein PbgA, S. typhimurium | 5I5D | 3.6, 277 | Yes | – | – | Thr | [33] |
The complete enzyme names and source organisms are available in the respective PDB entries.
Alignment lengths (amino acid residues) and RMSD (Å) from structural alignments of BcsG (PDB entry 5OJH) against the PDB obtained using DALI [32].
The second Zn2+ ion in NmEptA structure (PDB entry 4KAY) was seen only upon soaking the crystals in 2 mM ZnSO4 [35].
fGly indicates formylglycine residue, generated post-translationally from either Cys or Ser.
Members of the AlkP superfamily use from one to three different metal sites as part of their catalytic machinery, typically with two Zn2+ sites and in some cases a third Mg2+ binding site [36; 37]. In the lipid A phosphoethanolamine transferase NmEptA, the second Zn2+-binding site is formed by His383 and His465 and is only occupied upon soaking the crystals in ZnSO4 [28; 35], while in the closely related phosphoethanolamine transferases MCR-1 and MCR-2, which are responsible for colistin resistance in E. coli, the structurally corresponding residues are His395 and His478, respectively (in both MCR-1 and MCR-2) [38; 39]. In NmEptA, the ligand sphere is completed by two oxygen atoms from the phosphoryl group covalently bound to the active site nucleophile Thr280 [35], which corresponds to Ser278 in BcsG. In MCR-1 and MCR-2, one or two water molecules are bound in equivalent positions. One of these metal ligands, His383 is structurally equivalent to His396 in BcsG. However, in BcsG, the second conserved histidine metal ligand of site 2, His465 in NmEptA, is substituted by Arg458 (Fig. 4D). We did not find any signs of a second Zn2+ ion in the electron density maps. This could be due to the presence of the high amounts of citrate, preventing binding of Zn2+ to the second site. However, the His to Arg replacement of the site 2 metal ligand in BcsG suggests that this site may not be occupied by a metal ion. A similar loss of a metal binding to the Zn2+ site 1 has been observed in the crystal structure of the structurally related periplasmic domain of the putative cardiolipin transporter PbgA due to replacement of the metal ligand His443 by arginine [33]. Replacement of Arg458 by alanine or methionine resulted in mutants that were severely impaired in cellulose biosynthesis, while the Arg458His mutant showed some remaining cellulose formation (Fig. 4E). These findings suggest that Arg458 participates in substrate binding and/or catalysis, and that the positive charge of the guanidinium group is required, as replacement by the ionizable His residue retains some functionality.
Amino acid replacements of the Zn2+-binding residues Glu442 and His443 by alanine and of Cys243 by serine resulted in BcsG protein variants with perturbed cellulose biosynthesis as indicated by the colony morphology assay (Fig. 2C), suggesting that an intact Zn2+ site is important for functionality in vivo. Most pronounced effects were seen for the active-site mutant Ser278Ala and the second sphere ligand His396Ala variants, located at the protein surface. Replacements of these residues by alanine result in the expression of protein variants with a pdar morphotype similar to the truncated BcsG1–210. Although not a direct ligand of the site 1 metal ion, His396 is in close vicinity to this metal center and the putative substrate binding site and could therefore potentially play an important role in substrate binding and/or catalysis.
The periplasmic domain of BcsG is very similar to the lipoteichoic acid synthase LtaS, a phospholipid head group transferase, which transfers the phosphoglycerol group to synthesize lipoteichoic acid, a cell wall polymer of Gram-positive bacteria [34; 40; 41]. We therefore set up an enzymatic assay to assess whether BcsG can carry out phospholipid headgroup hydrolysis using fluorescent phospholipids as described previously [42]. To this end, we purified the periplasmic domain of BcsG as a MBP-pBcsG (periplasmic BcsG) fusion protein. Using fluorescent NBD-phosphatidylethanolamine (NBD-PE), a mimic of the most abundant phospholipid in bacterial membranes, and a thin layer chromatography plate assay, we could show that purified BcsG hydrolyzed this substrate, although with low activity, while the NBD derivative of the related phospholipid phosphatidylglycerol was not hydrolyzed (Fig. 5). Mn2+ also stimulated NBD-PE hydrolase activity, as well or even better than Zn2+ (Fig. 5A and data not shown).
Fig. 5. Enzymatic activity of BcsG.
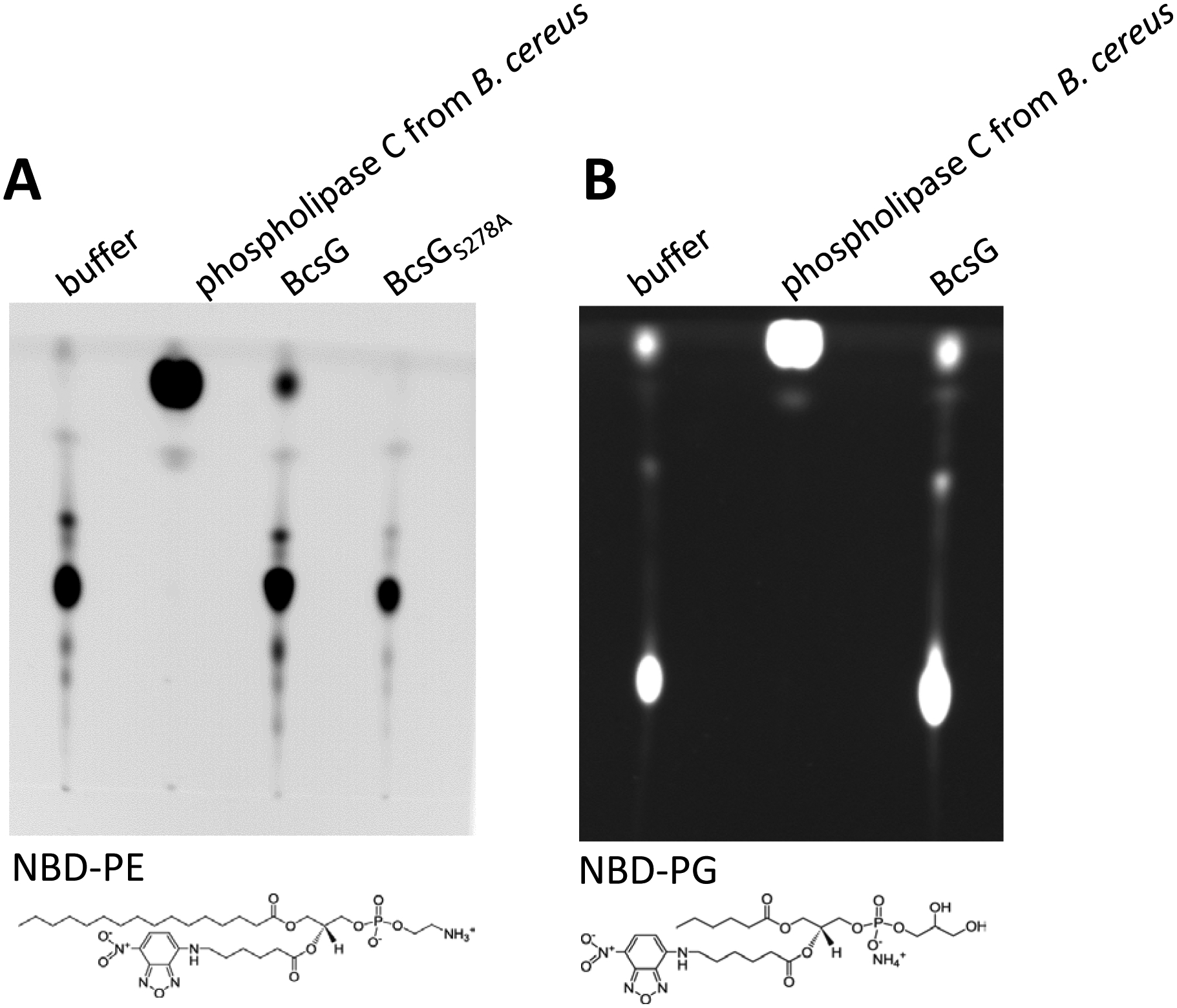
Hydrolysis of NBD-phosphatidylethanolamine (PE) by BcsG, but not the BcsGS278A variant. B. NBD-phosphatidylglycerol (PG) is not hydrolyzed by BcsG. The reaction was carried out in 300 μl of 10 mM sodium succinate pH 6.5/50 mM NaCl, 20 mM MnCl2 with 4.2 ng of purified NBD-PE or NBD-PG lipid and 400 μg MBP-BcsG fusion protein (or 2.5 units of phospholipase C from Bacillus cereus used as positive control) at 37 °C for 3 h. Substrates NBD-PE and NBD-PG are shown below the TLC plates. Each experiment was performed twice.
If BcsG is a phospho headgroup transferase with a similar mechanism as LtaS and NmEptA, the function of the conserved Ser278 residue might also be conserved. In both LtaS and NmEptA, a threonine in this position acts as a nucleophile in the transferase reaction and forms a covalently linked phosphorylated intermediate during catalysis. Therefore, we constructed a Ser278Ala mutant of the periplasmic domain of BcsG as an MBP-BcsGS278A fusion protein. This mutant did not show any activity with the fluorescent phospholipid substrate NBD-PE (Fig. 5).
Discussion
Production of bacterial cellulose in the fruit-degrading organism Komagataeibacter (formerly Acetobacter, Gluconacetobacter) xylinus has traditionally been investigated as an experimentally tractable model for the biosynthesis of cellulose in plants [43–45]. Today we know that cellulose is produced by numerous bacteria from different branches of the phylogenetic tree, including members of the family Enterobacteriaceae [6; 46]. All bacterial cellulose biosynthesis operons code for a functional cellulose synthase of two subunits, BcsA and BcsB, as core components of the cellulose biosynthesis pathway. The cellulose synthase complex BcsA/BcsB is localized at the cytoplasmic membrane and combines addition of new glucosyl residues to the nascent cellulose molecule with its export into the periplasmic (and eventually extracellular) space guided by BcsB [17]. The discrepancy between the in vitro and in vivo catalytic activity of the cellulose synthase led to the discovery of cyclic di-GMP as an activator of this enzyme [43]. Subsequently, cyclic di-GMP was identified as a nearly universal bacterial second messenger involved in a variety of bacterial signal transduction mechanisms including the lifestyle switch between motility and sessility and between chronic and acute infection [43; 47].
The functions of accessory genes of cellulose biosynthesis operons, are, however, still poorly understood [1]. For example, the bcsQ gene, which is found in class II and some class I bcs operons, encodes a putative MinD/ParA-like ATPase whose role in cellulose production is currently unknown but may involve proper positioning of the enzyme complex [48]. In Pseudomonas fluorescens, three or more auxiliary bcs genes (wssG, wssH, and wssI) are involved in cellulose acetylation [12]. In E. coli and Salmonella, the bcsEFG operon is required for cellulose production [7; 18; 49]. One of its products, BcsE, binds c-di-GMP with a higher affinity than the BcsA subunit [18] and interacts with BcsF and BcsG [13; 19]. It might participate in transmission of the signal to BcsA, although we did not notice any major effect of the bcsE and bcsF mutations on agar plate-produced cellulose in S. typhimurium. By contrast, inactivation of bcsG led to a significant decrease in cellulose production [18–20]..
In this work we show that the requirement of BcsG for full-scale production of cellulose also holds in S. typhimurium (Fig. 1). BcsG was apparently not required for the transcription of the bcsA gene and did not affect the stability of the membrane-associated BcsA subunit (Fig. 2). However, bcsG mutants had severely diminished levels of BcsA suggesting a role of BcsG in membrane insertion of BcsA (Fig. 2). These data correlate with the recent evidence of a direct interaction of BcsA and BcsG subunits, obtained by bacterial two-hybrid tests of protein-protein interactions [13; 19].
Further, diminished levels of BcsA were seen upon complementation of the ΔbcsG strain with plasmids carrying bcsG mutants lacking the soluble C-terminal domain of BcsG but not with constructs having point mutations in this C-terminal domain (Fig. 2). Thus, BcsA and BcsG likely interact through their membrane domains for membrane insertion, while a second interaction could occur via the periplasmic domain for cellulose modification. Besides its effects on BcsA, this study indicates that BcsG could also affect, directly or indirectly, the levels of the cellulase BcsZ IFig. 3A). Whether BcsG also affects the levels of the cellulose synthase subunit BcsB, which is required for the enzyme activity in vitro [15], remains to be tested in future experiments.
Sequence analysis of the N-terminal fragment of BcsG using a variety of software tools predicted the presence of either four or five TM helices. Sequence conservation suggested five TM helices, with the cytoplasmic location of the BcsG N-terminus and the periplasmic location of its C-terminal domain. This prediction is underpinned by the recent analysis of the translational fusions of BcsG [19]. Our data showed that the periplasmic domain of BcsG was dispensable for wild type steady state levels of the BcsA subunit, but its enzymatic activity was required for efficient cellulose biosynthesis on agar plates (Fig. 2C). As a member of the AlkP superfamily, this domain could possibly participate in modifying either the BcsA subunit, the synthesized cellulose itself, another periplasmic protein(s), peptidoglycan or osmoregulated periplasmic glucan [1]. While there is limited sequence similarity between BcsG and other members of the AlkP superfamily and no clustering with a specific activity or substrate preference, the experiments using fluorescently labelled phospholipids NBD-phosphatidylethanolamine and NBD-phosphatidylglycerol indicated BcsG to possess a phosphotransferase, rather than a phosphatase, activity. This is supported by the observation that BcsG does not hydrolyze p-nitrophenylphosphate, a non-specific substrate of acid and alkaline phosphatases (unpublished data). Recently, it was shown that BcsG acts as a phosphoethanolamine transferase, modifying cellulose in E. coli and S. typhimurium [19]. Our results are fully consistent with that conclusion, but also suggest an additional function of BcsG during the posttranscriptional assembly of the cellulose synthase complex, most likely by aiding the insertion of BcsA into the membrane (Fig. 2). Although mechanistically different, coupling of exopolysaccharide modification with polymerization has also been observed for alginate biosynthesis [50].
Structural analysis of the periplasmic domain of BcsG confirmed its membership in the AlkP superfamily. More specifically, BcsG proved to be a member of a family of enzymes that combine a periplasmic AlkP-like domain with an N-terminal membrane-bound domain that consists of 5 TM helices. Members of this family, besides BcsG, include previously characterized bacterial Zn2+-dependent phosphatidylethanolamine:lipopolysaccharide phosphoethanolamine transferases MCR-1, MCR-2, NmEptA, and EptC [28; 35; 38; 39; 51–54]. This family also includes Mn2+-dependent phosphatidylglycerol:lipoteichoic acid phosphoglycerol transferases LtaS/LtaP of Gram-positive bacteria [34; 40; 55]. Further, sequence comparisons also assigned to this family the E. coli OpgB (previously MdoB) and OpgE, which transfer phosphoglycerol and phosphoethanolamine residues to osmoregulated periplasmic glucans, previously named membrane-derived oligosaccharides [56–58], and Neisseria gonorrhoeae PptA protein, which modifies serine residues in type IV pilin PilE with phosphoethanolamine and, possibly, phosphocholine groups [59; 60]. Amino acid sequence and structural similarities among these proteins (Fig. S8) in combination with the identified substrates also indicate that the substrate range of these membrane-standing enzymes with periplasmic catalytic activity, including BcsG, may be broader than currently anticipated. Like BcsG, most of these enzymes contain a single Zn2+ ion in their active sites (Table 2) and utilize serine/threonine residues as the nucleophile during catalysis.
The genome of S. typhimurium, besides BcsG, encodes six other members of the phosphoethanolamine transferase family, all of which participate in modification of the outer membrane, osmoregulated periplasmic glucan and the exopolysaccharide cellulose (Table S3). At least three of them, EptA, EptB and CptA have been shown to function as phosphatidylethanolamine:lipopolysaccharide phosphoethanolamine transferases [61–63], with PmrC and EptB serving to mediate polymyxin resistance. OpgB functions as a phosphoglycerol transferase, while one more member of this family, PbgA (formerly YejM) participates in cardiolipin transfer from the inner to the outer membrane [33; 64]. PbgA has amino acid replacements in several metal-binding residues and its crystal structure does not contain any bound metal atoms. Accordingly, PbgA reportedly has lost the enzymatic activity, is unable to bind sn-glycerol-2-phosphate, and binds cardiolipin with its hydrophobic core and two Arg residues in the interdomain linker region [33; 64]. It should be noted that Lnt, another membrane-anchored periplasmic protein that catalyzes a somewhat similar reaction (also using phosphatidylethanolamine as a substrate but transferring its sn-1-acyl chain to the N-terminal cysteine residue of apolipoproteins), has a totally different structure and belongs to the CN hydrolase (nitrilase) family [65; 66].
In addition to chromosome-encoded phosphoethanolamine transferases, a rapidly increasing number of recent isolates of E. coli, Salmonella, Klebsiella, and other enterobacteria, including S. typhimurium, carry plasmid-borne mcr (mobile colistin resistance) genes [67; 68]. Their products MCR-1 and MCR-2, like BcsG, combine a 5TM N-terminal fragment with a periplasmic phosphoethanolamine transferase domain, whose structures (PDB entries 5lrn and 5grr, respectively) are very similar to that of BcsG (see Fig. S10). While phylogenetic analysis (Fig. S11) does not imply that BcsG is any closer to MCR-1 than to other phosphoethanolamine transferases, it highlights the diversity of such genes in Salmonella and E. coli (Table S3, Fig. S11) and illustrates ample possibilities for further divergence of the mcr genes [68–70].
In this work, we have shown that the two domains of the BcsG subunit of cellulose synthase perform at least two distinct functions in cellulose production in S. typhimurium. Its membrane domain is required for the positioning of the catalytic BcsA subunit (Fig. 2), which is consistent with the observations on the direct protein-protein interactions between BcsA and BcsG [13; 19]. The periplasmic domain of BcsG is an enzyme of the AlkP superfamily (Fig. 4), which uses phosphatidylethanolamine as substrate (Fig. 5) and, as recently demonstrated by Thongsomboon and colleagues, transfers the phosphoethanolamine group onto the growing cellulose molecule [19]. This enzymatic activity is required for the full-scale cellulose production and point mutations in Zn2+-binding residues decreases the amount of produced cellulose (Fig. 2). These data complement recent work on the functions of BcsG and the overall organization of the cellulose synthase complex of S. typhimurium and E. coli [13; 19]. Further work will be needed to evaluate the importance of the protein-protein interactions in the cellulose synthase holoenzyme and clarify the precise roles of other subunits, such as BcsE and BcsF.
Materials and Methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table S1. Escherichia coli and S. typhimurium was routinely grown on Luria-Bertani (LB) agar plates or in LB liquid culture supplemented with appropriate antibiotics at 37 °C overnight. The antibiotics used were ampicillin (100 μg ml−1), kanamycin (30 μg ml−1) and chloramphenicol (20 μg ml−1). For the expression of genes, 0.1 % arabinose was used.
Construction of mutants
The deletion mutant of bcsG was constructed by one-step gene inactivation as described [71]. Briefly, the primers with 40 bp of homology to the 5’ end and 3’ end of bcsG were used to amplify the chloramphenicol antibiotic resistance cassette from plasmid pKD3. The linear PCR fragment was purified and electroporated into UMR1 harboring plasmid pKD46. The mutants were selected on LB plates with chloramphenicol at 37°C. For the creation of 3xFLAG-tagged bcsG, 3xFLAG-Km was amplified from plasmid pSUB11 [72]. The bcsG mutant was transferred in a novel background with phage P22 HT105/1 int-201 transduction [73]. Site-directed mutagenesis was performed using QuikChange Site-Directed Mutagenesis Kit (Agilent).
Plasmid construction
Expression constructs used for complementation studies were based on the pBAD30 vector [74]. The coding sequence of bcsG was amplified using primers bcsG-comF and bcsG-comR (Table S2) from template S. typhimurium UMR1 (Table S1) genomic DNA adding a 8xHis tag. The PCR product was digested with HindIII and XbaI, and ligated into pBAD30. The ligation product was transformed into E. coli DH5α. Plasmids were verified by sequencing.
The construct used for recombinant protein expression was based on the predicted extra-membrane region of BcsG (UniProt code: Q7CPI7), comprising residues 185–559. This fragment encoding the soluble part of BcsG was amplified from UMR1 genomic DNA with primers bcsGpMAL2F and bcsGpMAL2R. The PCR product was digested with XbaI and HindIII, and ligated into vector pMAL-c2x (NEB) yielding pMAL::BcsG1. The resulting fusion construct includes an N-terminal maltose binding protein (MBP) tag and Factor-Xa cleavage site. Proteolytic removal of the MBP tag from recombinant protein leaves an N-terminal extension consisting of the amino acids ISEFGSSR from the linker region. All expression constructs were confirmed by DNA sequencing.
β-galactosidase assay
The β-galactosidase assay was performed as described previously [75] using bacterial cells grown on a salt-free LB plate with appropriate antibiotics and inducer. After 24 h incubation at 28 °C, cells were resuspended in cold working buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4, 0.05 M β-mercaptoethanol) and all samples were adjusted to OD600=0.1. β-galactosidase activity was calculated using the formula: units =1000 × {[OD420 - (1.75 × OD550)]/(t × V × OD660)} with t=reaction time in min; V=volume of cell suspension in assay in ml; OD420 and OD550 of reaction solution; OD660 of original cell suspension. Mean values are from at least two independent cultures in three technical replicates with error bars indicating the standard deviation.
Rdar morphotype and Calcofluor white binding assay
5 μl of an overnight culture suspended in PBS (OD600 of 5) were spotted onto salt-free LB agar plates supplemented with Congo red (40 μg ml−1) and Coomassie Brilliant Blue (20 μg ml−1) or Calcofluor white (fluorescence brightener 28) at 50 μg ml−1 and incubated at 28 °C for up to 48 h. While Congo red binds to amyloid structures such as curli fimbriae and the β-(1–4)-glucan cellulose, Calcofluor white only binds to cellulose. The development of the colony morphology and dye binding was analyzed over time. Upon visual inspection of Congo red binding, the cells were assigned to either of rdar (cellulose+/curli+), pdar (cellulose+/curli−), bdar (brown, dry and rough; cellulose−/curli+) or saw (cellulose−/curli−) morphotypes. Fluorescence of Calcofluor white bound to bacterial colonies was observed using a UV light source emitting at 366 nm. The colony morphotype and dye binding were only compared for the cells grown on the same plate.
Confocal microscopy
Cells grown on salt-free LB agar plates at 28 °C for 48 h were gently resuspended in water containing 0.001% Calcofluor white to maintain cell aggregation. Fluorescence imaging was carried out with an Olympus FV1000 confocal microscope. Digital image processing was executed using ImageJ software.
Western blot analysis
Cells (5 mg of wet weight) were harvested after 16 h-20 h growth on salt-free LB agar with 0.1 % arabinose and appropriate antibiotics at 28 °C. For detection of the membrane proteins BcsA and BcsC, 200 μl of urea buffer (8 M urea, 2% SDS, 11% glycerol, 62.5 mM Tris-HCl, pH 6.8) was added and sonicated four times (10 s, 3.5 μm amplitude) on ice as described previously [76; 77]. The sonicated extracts were examined with SDS-PAGE gels stained with Coomassie Blue to ensure loading of equal amounts of all sample proteins for the Western blots (with the exception of samples in Fig. 2B, where 10 times higher amounts of the cell extract from the bcsG mutant had to be loaded owing to the approx. 10-fold lower BcsA expression in the mutant cells). For Western blots, cell extracts were separated on SDS-PAGE gel and transferred onto a PVDF membrane (Millipore). BcsA-3xFLAG, BcsC-3xFLAG, and BcsG-3xFLAG were detected by using anti-FLAG-tag antibody (1:2000 for BcsA, 1:1000 for BcsC, and 1:2000 for BcsG; Sigma) and peroxidase-conjugated AffiniPure Goat Anti-Mouse IgG (1:3000, Jackson ImmuniResearch). Visualization of bands was performed using FUJI LAS1000-plus chemiluminescence imaging system (Fuji, Stamford, CT, USA).
Protein stability assay
MAE1264 (MAE97 bcsA-3xFLAG) and its ΔbcsG mutant without a Cm resistance cassette were grown in M9 medium at 28 °C for 10 h with 200 rpm shaking. Protein synthesis was stopped by adding 200 μg/ml chloramphenicol (time point 0) and aliquots were taken after 30, 90 and 120 min.
Protein production and purification
Recombinant MBP-BcsG was expressed in E. coli DH5α in RMG media consisting of tryptone (1 g/L), yeast extract (5 g/L), NaCl (5 g/L) and glucose (2 g/L, filter sterilized and added after autoclaving) supplemented with 100 μg/mL ampicillin. 1.5 L cultures were grown at 37°C until OD600 reached approx 0.6–0.8 and recombinant protein expression was induced by adding isopropyl-β-D-1-thiogalactopyranoside to 0.3 mM concentration. The incubation of these cultures proceeded at 27°C for 16 hours. The cells were pelleted by centrifugation (6000 g × 30 min), re-suspended in 20 mM Tris-HCl, 0.2 M NaCl pH 7.4 and treated with lysozyme (0.1 mg/mL), protease cocktail inhibitor (Roche, mini EDTA-free) and DNAseI (0.01 mg/mL) in the presence of 1 mM MgCl2. Sonication was then used to lyse the cells. Cell debris was sedimented by high-speed centrifugation (40905 g, 25 min) and the resulting crude lysate was filtered using a 0.2 μm syringe filter (Pall Corp.). The lysate was exposed to amylose-resin (New England Biolabs) pre-equilibrated for 20 minutes at 4 °C with 20 mM Tris-HCl, 0.2 M NaCl pH 7.4 in batch mode to allow efficient binding. The affinity matrix was collected in a plastic column and washed by three column volumes of buffer (20 mM Tris-HCl, 0.2 M NaCl pH 7.4). The MBP-BcsG protein was then eluted with buffer containing maltose (10 mM maltose, 20 mM Tris-HCl, 0.5 M NaCl pH 7.4). Maltose was removed by buffer exchange diafiltration (20 mM Tris-HCl, 0.2 M NaCl pH 7.4, Sartorius Vivaspin 20, 10 000 MWCO).
The MBP tag was removed prior to the crystallization by adding Factor-Xa protease in the presence of 5 mM CaCl2. Approximately 1 μg protease (Factor Xa, New England Biolabs) was added per 100 μg target protein and the solution was incubated for 16 hours at 25°C. The resulting cleavage mixture was exposed to the amylose resin in a gravity flow column (pre-equilibrated with 20 mM Tris-HCl, 0.2 M NaCl pH 7.4) to capture the unprocessed fusion protein and the MBP tag. The efficiency of the tag removal and the purity at this step were assessed by SDS-PAGE analysis. Factor Xa was removed using a HiTrap Benzamidine FF column (high sub, GE Healthcare #17-5144-01) pre-equilibrated and washed with 50 mM Tris-HCl, 0.5 M NaCl pH 7.4. A higher salt concentration was necessary during this stage to prevent binding of BcsG to the benzamidine column. The buffer was then changed into 20 mM Tris-HCl, 0.2 M NaCl pH 7.4 (Sartorius Vivaspin 20, 10 000 MWCO) and purified BcsG was concentrated to 23 mg/mL before flash freezing in liquid N2 and long-term storage at −80 °C.
Analytical gel filtration
200 μL of BcsG, diluted to 1 mg/mL, was loaded onto a S75 10/300 Superdex gel filtration column (GE Healthcare) equilibrated with 20 mM Tris-HCl, 0.2 M NaCl, pH 7.4. Calibration of the column was performed using ribonuclease-A (13.7 kDa), chymotrypsinogen-A (25 kDa), ovalbumin (43 kDa), albumin (67 kDa), catalase (232 kDa), ferritin (440 kDa) and Blue dextran (2 MDa).
Crystallization
Crystallization screening sitting drops were set up using Phoenix and Mosquito liquid handling robots in Corning 96-well 3550 trays. Generally, 0.1, 0.15 or 0.2 μL of protein was mixed with 0.2, 0.15 or 0.1 μL of the crystallization solution, respectively. Drops were equilibrated against 50 μL of crystallization solution in the reservoir, with incubation at 4 °C. BcsG (23 mg/mL), cleaved of the MBP tag, crystallized in Index HT condition B9 (Hampton Research, 1.8 M Tri-Ammonium citrate tribasic, pH 7.0). Phase separation was observed to occur within 7 days and rod-shaped crystals began to grow from the phase separated droplets within 12 days.
Crystallization in ZnCl2 supplemented conditions was achieved using hanging drops in Falcon 353047 24-well tissue culture plates. Generally, 1, 1.5, or 2 μL of the protein solution (23 mg/mL), supplemented with 6 mM ZnCl2, were mixed with 2, 1.5, or 1 μL of the crystallization solution (1.8 M Tri-Ammonium citrate tribasic, pH 7.0), respectively. Drops were equilibrated against 1 mL of crystallization solution in the reservoir at 4 °C.
Structure determination and refinement
An X-ray fluorescence scan data of crystals carried out on the EMBL beamline P14, on the PETRA III storage ring (DESY, Hamburg, Germany) showed the characteristic absorption edge feature expected for Zn2+ ions. X-ray diffraction data sets to 1.5 Å resolution were collected at EMBL beam lines P13 and P14 on PETRA III using crystals obtained with and without the addition of ZnCl2 (PDB codes 5OLT and 5OJH, respectively). The data set for the crystals obtained in the Zn-supplemented condition was collected at a wavelength of 1.2782 Å to optimize the anomalous contribution of Zn2+ to the diffraction data. X-ray images were processed using XDS [78] and scaled with AIMLESS from the CCP4 program suite [79]. Crystals of BcsG belong to space group P3121. Cell dimensions for the citrate condition were a=b= 80.4 Å and c=97.5 Å and for the zinc-supplemented condition a=b= 80.7 Å and c=97.6 Å. Statistics for the data processing are given in Table 1.
Initially, the structure of BcsG was determined by molecular replacement with the program PHASER [80] with a search model based on a model of the soluble C-terminal domain obtained from Robetta at the Rosetta server [29] spanning residues 207–559. The Robetta model was trimmed down to the core of the predicted α/β domain, residues 234–475 and all loops between secondary structure elements were removed. However, no structure solution was obtained. The search model was then further truncated to the central β-sheet, in total 69 residues, which gave a very weak possible solution, but the phasing power was insufficient to further extend this model. A second run of PHASER based on this solution and with an additional 18 residue helix as search model improved statistics and the space-group could be unambiguously assigned as P3121. The complete structure of the BcsG domain was bootstrapped from the Phaser model by alternating refinement with REFMAC5 [81], density improvement with Parrot [82] and model building in COOT [83]. The refined model comprises the whole construct including the linker region from the Factor Xa cleavage site except residues 188–191 that lack electron density. Towards the end of the refinement runs, water molecules and additional ligands were added to the model. Finally, anisotropic B-factor refinement resulted in Rfree/R values of 0.198/0.166 with expected model geometry (Table 1).
The crystal structure of BcsG was also independently determined using a Zn-SAD data set collected at beam line P14, equipped with Compound Refractive Lenses. SAD phasing at the Zn edge was carried out in SHELXC/D/E [84]. The correct substructure with one Zn2+ ion (CCall 27.6 and CCweak 26.2) was determined in under 100 trials by SHELXD and the electron density was further improved by density modification in SHELXE. An initial model was generated using the phased data by automatic model building in ARP/wARP [85], resulting in a model in which 96% of the sequence was docked. The ARP/wARP model was refined with REFMAC5, interspersed with model building using COOT. As the final steps, anisotropic B-factor refinement resulted in a model with Rfree/R values of 0.158/0.128 and good model parameters (Table 1).
In both crystal forms, obtained with and without addition of ZnCl2 during crystallization, the metal site was not completely occupied and occupancies were set at 0.75 during refinement. The absence of Zn2+ ions from this site might be due to the high concentration of the metal-chelating citrate under the crystallization condition. In fact, a citrate ion was found at the surface of the protein as a second sphere ligand of the Zn2+ ion. Another citrate molecule is located at the crystal packing interface between two BcsG molecules related by crystallographic symmetry. This citrate molecule is bound at the protein surface and interacts via hydrogen bonds with the side chains of His451 and Asn518. Citrate is located on a crystallographic two-fold axis and also interacts with corresponding residues from a neighboring BcsG molecule related by two-fold symmetry, thus contributing to crystal packing.
Enzyme activity assay
To assess the enzymatic activity of BcsG, phosphatidylethanolamine and phosphatidylglycerol derivatives labeled with a fluorophore (16:0–06:0 NBD-PE (1-palmitoyl-2-{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoethanolamine; Avanti 810153) and 16:0–06:0 NBD-PG (1-palmitoyl-2-{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-[phospho-rac-(1-glycerol)] (ammonium salt); Avanti 810163), respectively) were purified and used as potential substrates as previously reported [42]. Briefly, 100 μg NBD-PE or NBD-PG were spotted onto 250 μm-thick 60-Å silica TLC plates (Macherey-Nagel) and developed in a 60:20:2.7 CHCl3–MeOH–water solvent system for 15 min. The silica gel containing the major yellow fluorescent spot was scraped from TLC plates and added to 8 ml of a 1:1 methanol-chloroform mixture with vortexing to extract the lipids. The chloroform phases containing the purified NBD- lipids were dried for use.
For each reaction, 300 μl of 10 mM sodium succinate (pH 6.5, ionic strength 50 mM adjusted with NaCl) with approximately 4.2 ng purified NBD lipid was sonicated for solubilization. The reactions were initiated by the addition of 400 μg MBP fusion protein and incubated at 37 °C for 3 h. As a positive control, 2.5 units of phospholipase C (PLC) from Bacillus cereus (Sigma) was used. Visualization of fluorescent bands was performed using FUJI LAS1000-plus chemiluminescence imaging system (Fuji, Stamford, CT, USA).
Accession numbers
Crystallographic data for the two models were deposited with the Protein Data Bank under accession numbers 5OJH (MR derived structure) and 5OLT (Zn-SAD structure).
Supplementary Material
Acknowledgements
We gratefully acknowledge access to the Protein Science Facility (PSF), Karolinska Institutet, Stockholm, Sweden. We appreciate the assistance of Mark Gomelsky in initial protein purification and Sulman Shafeeq in microscopic analysis. Lei Sun and Fengyang Li received a scholarship from the Chinese Scholarship Council. Annika Cimdins was funded by the German Research Foundation (DFG; CI 239/1-1 and CI 239/2-1). This work was supported by the Röntgen-Ångström Cluster through the Swedish Research Council to G.S. (project number D0062401) and the Bundesministerium für Bildung und Forschung (BMBF) to T.R.S. (grant number 05K13YE1) U.R. was supported by Vetenskapsrådet (project number 621-2013-4809). M.Y.G. was supported by the NIH Intramural Research program at the U.S. National Library of Medicine.
References
- [1].Römling U, Galperin MY, Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 23 (2015) 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Matthysse AG, Role of bacterial cellulose fibrils in Agrobacterium tumefaciens infection. J. Bacteriol 154 (1983) 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Robledo M, Rivera L, Jimenez-Zurdo JI, Rivas R, Dazzo F, Velazquez E, Martinez-Molina E, Hirsch AM, Mateos PF, Role of Rhizobium endoglucanase CelC2 in cellulose biosynthesis and biofilm formation on plant roots and abiotic surfaces. Microb. Cell Fact 11 (2012) 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pontes MH, Lee EJ, Choi J, Groisman EA, Salmonella promotes virulence by repressing cellulose production. Proc. Natl. Acad. Sci. USA 112 (2015) 5183–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ahmad I, Rouf SF, Sun L, Cimdins A, Shafeeq S, Le Guyon S, Schottkowski M, Rhen M, Römling U, BcsZ inhibits biofilm phenotypes and promotes virulence by blocking cellulose production in Salmonella enterica serovar Typhimurium. Microb. Cell Fact 15 (2016) 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U, The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol 39 (2001) 1452–1463. [DOI] [PubMed] [Google Scholar]
- [7].Solano C, Garcia B, Valle J, Berasain C, Ghigo JM, Gamazo C, Lasa I, Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol 43 (2002) 793–808. [DOI] [PubMed] [Google Scholar]
- [8].Grantcharova N, Peters V, Monteiro C, Zakikhany K, Römling U, Bistable expression of CsgD in biofilm development of Salmonella enterica serovar typhimurium. J. Bacteriol 192 (2010) 456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Brandl MT, Carter MQ, Parker CT, Chapman MR, Huynh S, Zhou Y, Salmonella biofilm formation on Aspergillus niger involves cellulose--chitin interactions. PLoS One 6 (2011) e25553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zorraquino V, Garcia B, Latasa C, Echeverz M, Toledo-Arana A, Valle J, Lasa I, Solano C, Coordinated cyclic-di-GMP repression of Salmonella motility through YcgR and cellulose. J. Bacteriol 195 (2013) 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Saxena IM, Brown RM Jr., Identification of a second cellulose synthase gene (acsAII) in Acetobacter xylinum. J. Bacteriol 177 (1995) 5276–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Spiers AJ, Bohannon J, Gehrig SM, Rainey PB, Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol 50 (2003) 15–27. [DOI] [PubMed] [Google Scholar]
- [13].Krasteva PV, Bernal-Bayard J, Travier L, Martin FA, Kaminski PA, Karimova G, Fronzes R, Ghigo JM, Insights into the structure and assembly of a bacterial cellulose secretion system. Nat. Commun 8 (2017) 2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Maeda K, Tamura J, Okuda Y, Narikawa R, Midorikawa T, Ikeuchi M, Genetic identification of factors for extracellular cellulose accumulation in the thermophilic cyanobacterium Thermosynechococcus vulcanus: proposal of a novel tripartite secretion system. Mol. Microbiol (2018). [DOI] [PubMed] [Google Scholar]
- [15].Morgan JL, Strumillo J, Zimmer J, Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493 (2013) 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Morgan JL, McNamara JT, Zimmer J, Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat. Struct. Mol. Biol 21 (2014) 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].McNamara JT, Morgan JL, Zimmer J, A molecular description of cellulose biosynthesis. Annu. Rev. Biochem 84 (2015) 895–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fang X, Ahmad I, Blanka A, Schottkowski M, Cimdins A, Galperin MY, Römling U, Gomelsky M, GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol. Microbiol 93 (2014) 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Thongsomboon W, Serra DO, Possling A, Hadjineophytou C, Hengge R, Cegelski L, Phosphoethanolamine cellulose: A naturally produced chemically modified cellulose. Science 359 (2018) 334–338. [DOI] [PubMed] [Google Scholar]
- [20].Singletary LA, Karlinsey JE, Libby SJ, Mooney JP, Lokken KL, Tsolis RM, Byndloss MX, Hirao LA, Gaulke CA, Crawford RW, Dandekar S, Kingsley RA, Msefula CL, Heyderman RS, Fang FC, Loss of multicellular behavior in epidemic African nontyphoidal Salmonella enterica serovar Typhimurium ST313 Strain D23580. mBio 7 (2016) e02265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Römling U, Rohde M, Olsen A, Normark S, Reinkoster J, AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol 36 (2000) 10–23. [DOI] [PubMed] [Google Scholar]
- [22].Römling U, Sierralta WD, Eriksson K, Normark S, Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol 28 (1998) 249–264. [DOI] [PubMed] [Google Scholar]
- [23].Kader A, Simm R, Gerstel U, Morr M, Römling U, Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol 60 (2006) 602–616. [DOI] [PubMed] [Google Scholar]
- [24].Galperin MY, Bairoch A, Koonin EV, A superfamily of metalloenzymes unifies phosphopentomutase and cofactor-independent phosphoglycerate mutase with alkaline phosphatases and sulfatases. Protein Sci 7 (1998) 1829–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Galperin MY, Jedrzejas MJ, Conserved core structure and active site residues in alkaline phosphatase superfamily enzymes. Proteins 45 (2001) 318–324. [DOI] [PubMed] [Google Scholar]
- [26].Galperin MY, Koonin EV, Divergence and convergence in enzyme evolution. J. Biol. Chem 287 (2012) 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sunden F, AlSadhan I, Lyubimov A, Doukov T, Swan J, Herschlag D, Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution. J. Biol. Chem 292 (2017) 20960–20974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Anandan A, Evans GL, Condic-Jurkic K, O’Mara ML, John CM, Phillips NJ, Jarvis GA, Wills SS, Stubbs KA, Moraes I, Kahler CM, Vrielink A, Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding. Proc. Natl. Acad. Sci. USA 114 (2017) 2218–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim DE, Chivian D, Baker D, Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32 (2004) W526–W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Galperin MY, Shalaeva DN, A bacterial coat that is not pure cotton. Science 359 (2018) 276–277. [DOI] [PubMed] [Google Scholar]
- [31].Krissinel E, Henrick K, Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372 (2007) 774–797. [DOI] [PubMed] [Google Scholar]
- [32].Holm L, Laakso LM, Dali server update. Nucleic Acids Res. 44 (2016) W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dong H, Zhang Z, Tang X, Huang S, Li H, Peng B, Dong C, Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria. Sci. Rep 6 (2016) 30815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Campeotto I, Percy MG, MacDonald JT, Forster A, Freemont PS, Gründling A, Structural and mechanistic insight into the Listeria monocytogenes two-enzyme lipoteichoic acid synthesis system. J. Biol. Chem 289 (2014) 28054–28069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wanty C, Anandan A, Piek S, Walshe J, Ganguly J, Carlson RW, Stubbs KA, Kahler CM, Vrielink A, The structure of the neisserial lipooligosaccharide phosphoethanolamine transferase A (LptA) required for resistance to polymyxin. J. Mol. Biol 425 (2013) 3389–3402. [DOI] [PubMed] [Google Scholar]
- [36].Coleman JE, Structure and mechanism of alkaline phosphatase. Annu. Rev. Biophys. Biomol. Struct 21 (1992) 441–483. [DOI] [PubMed] [Google Scholar]
- [37].Coleman JE, Zinc enzymes. Curr Opin Chem Biol 2 (1998) 222–234. [DOI] [PubMed] [Google Scholar]
- [38].Ma G, Zhu Y, Yu Z, Ahmad A, Zhang H , High resolution crystal structure of the catalytic domain of MCR-1. Sci. Rep 6 (2016) 39540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Coates K, Walsh TR, Spencer J, Hinchliffe P, 1.12 Å resolution crystal structure of the catalytic domain of the plasmid-mediated colistin resistance determinant MCR-2. Acta Cryst. F73 (2017) 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lu D, Wörmann ME, Zhang X, Schneewind O, Gründling A, Freemont PS, Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. Proc. Natl. Acad. Sci. USA 106 (2009) 1584–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Percy MG, Gründling A, Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu. Rev. Microbiol 68 (2014) 81–100. [DOI] [PubMed] [Google Scholar]
- [42].Karatsa-Dodgson M, Wormann ME, Gründling A, In vitro analysis of the Staphylococcus aureus lipoteichoic acid synthase enzyme using fluorescently labeled lipids. J. Bacteriol 192 (2010) 5341–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ross P, Mayer R, Benziman M, Cellulose biosynthesis and function in bacteria. Microbiol. Rev 55 (1991) 35–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].McFarlane HE, Doring A, Persson S, The cell biology of cellulose synthesis. Annu. Rev. Plant Biol 65 (2014) 69–94. [DOI] [PubMed] [Google Scholar]
- [45].Kumar M, Turner S, Plant cellulose synthesis: CESA proteins crossing kingdoms. Phytochemistry 112 (2015) 91–99. [DOI] [PubMed] [Google Scholar]
- [46].Römling U, Molecular biology of cellulose production in bacteria. Res. Microbiol 153 (2002) 205–212. [DOI] [PubMed] [Google Scholar]
- [47].Römling U, Galperin MY, Gomelsky M, Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev 77 (2013) 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Le Quere B, Ghigo JM, BcsQ is an essential component of the Escherichia coli cellulose biosynthesis apparatus that localizes at the bacterial cell pole. Mol. Microbiol 72 (2009) 724–740. [DOI] [PubMed] [Google Scholar]
- [49].Serra DO, Richter AM, Hengge R, Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol 195 (2013) 5540–5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Moradali MF, Donati I, Sims IM, Ghods S, Rehm BH, Alginate polymerization and modification are linked in Pseudomonas aeruginosa. mBio 6 (2015) e00453–00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hu M, Guo J, Cheng Q, Yang Z, Chan EW, Chen S, Hao Q, Crystal structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for colistin resistance. Sci. Rep 6 (2016) 38793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hinchliffe P, Yang QE, Portal E, Young T, Li H, Tooke CL, Carvalho MJ, Paterson NG, Brem J, Niumsup PR, Tansawai U, Lei L, Li M, Shen Z, Wang Y, Schofield CJ, Mulholland AJ, Shen J, Fey N, Walsh TR, Spencer J, Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci. Rep 7 (2017) 39392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Stojanoski V, Sankaran B, Prasad BV, Poirel L, Nordmann P, Palzkill T, Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC Biol. 14 (2016) 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Fage CD, Brown DB, Boll JM, Keatinge-Clay AT, Trent MS, Crystallographic study of the phosphoethanolamine transferase EptC required for polymyxin resistance and motility in Campylobacter jejuni. Acta Cryst. D70 (2014) 2730–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schirner K, Marles-Wright J, Lewis RJ, Errington J, Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 28 (2009) 830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jackson BJ, Bohin JP, Kennedy EP, Biosynthesis of membrane-derived oligosaccharides: characterization of mdoB mutants defective in phosphoglycerol transferase I activity. J. Bacteriol 160 (1984) 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Fiedler W, Rotering H, Characterization of an Escherichia coli mdoB mutant strain unable to transfer sn-1-phosphoglycerol to membrane-derived oligosaccharides. J. Biol. Chem 260 (1985) 4799–4806. [PubMed] [Google Scholar]
- [58].Lequette Y, Lanfroy E, Cogez V, Bohin JP, Lacroix JM, Biosynthesis of osmoregulated periplasmic glucans in Escherichia coli: the membrane-bound and the soluble periplasmic phosphoglycerol transferases are encoded by the same gene. Microbiology 154 (2008) 476–483. [DOI] [PubMed] [Google Scholar]
- [59].Naessan CL, Egge-Jacobsen W, Heiniger RW, Wolfgang MC, Aas FE, Rohr A, Winther-Larsen HC, Koomey M, Genetic and functional analyses of PptA, a phospho-form transferase targeting type IV pili in Neisseria gonorrhoeae. J. Bacteriol 190 (2008) 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Anonsen JH, Egge-Jacobsen W, Aas FE, Borud B, Koomey M, Vik A, Novel protein substrates of the phospho-form modification system in Neisseria gonorrhoeae and their connection to O-linked protein glycosylation. Infect. Immun 80 (2012) 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lee H, Hsu FF, Turk J, Groisman EA, The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol 186 (2004) 4124–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Reynolds CM, Kalb SR, Cotter RJ, Raetz CR, A phosphoethanolamine transferase specific for the outer 3-deoxy-D-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J. Biol. Chem 280 (2005) 21202–21211. [DOI] [PubMed] [Google Scholar]
- [63].Tamayo R, Choudhury B, Septer A, Merighi M, Carlson R, Gunn JS, Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar typhimurium lipopolysaccharide core. J. Bacteriol 187 (2005) 3391–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Dalebroux ZD, Edrozo MB, Pfuetzner RA, Ressl S, Kulasekara BR, Blanc MP, Miller SI, Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17 (2015) 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Noland CL, Kattke MD, Diao J, Gloor SL, Pantua H, Reichelt M, Katakam AK, Yan D, Kang J, Zilberleyb I, Xu M, Kapadia SB, Murray JM, Structural insights into lipoprotein N-acylation by Escherichia coli apolipoprotein N-acyltransferase. Proc. Natl. Acad. Sci. USA 114 (2017) E6044–E6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lu G, Xu Y, Zhang K, Xiong Y, Li H, Cui L, Wang X, Lou J, Zhai Y, Sun F, Zhang XC, Crystal structure of E coli apolipoprotein N-acyl transferase. Nat. Commun 8 (2017) 15948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Muller-Pebody B, Ellington MJ, de Pinna E, Johnson AP, Hopkins KL, Woodford N, Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemother 71 (2016) 2300–2305. [DOI] [PubMed] [Google Scholar]
- [68].Lu X, Hu Y, Luo M, Zhou H, Wang X, Du Y, Li Z, Xu J, Zhu B, Xu X, Kan B, MCR-1.6, a new MCR variant carried by an IncP plasmid in a colistin-resistant Salmonella enterica serovar Typhimurium isolate from a healthy individual. Antimicrob. Agents Chemother 61 (2017) e02632–02616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kempf I, Jouy E, Chauvin C, Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 48 (2016) 598–606. [DOI] [PubMed] [Google Scholar]
- [70].Snesrud E, McGann P, Chandler M, The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. mBio 9 (2018) e02381–02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Datsenko KA, Wanner BL, One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97 (2000) 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L, Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 98 (2001) 15264–15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Schmieger H, Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet 119 (1972) 75–88. [DOI] [PubMed] [Google Scholar]
- [74].Guzman LM, Belin D, Carson MJ, Beckwith J, Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol 177 (1995) 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Miller JH. (1992). A short course in bacterial genetics : a laboratory manual and handbook for Escherichia coli and related bacteria, Cold Spring Harbor Laboratory Press, New York, N.Y. [Google Scholar]
- [76].Simm R, Morr M, Kader A, Nimtz M, Römling U, GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol 53 (2004) 1123–1134. [DOI] [PubMed] [Google Scholar]
- [77].Monteiro C, Papenfort K, Hentrich K, Ahmad I, Le Guyon S, Reimann R, Grantcharova N, Römling U, Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 9 (2012) 489–502. [DOI] [PubMed] [Google Scholar]
- [78].Kabsch W, XDS. Acta Cryst. D66 (2010) 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS, Overview of the CCP4 suite and current developments. Acta Cryst. D67 (2011) 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software. J. Appl. Crystallogr 40 (2007) 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA, REFMAC5 for the refinement of macromolecular crystal structures. Acta Cryst. D67 (2011) 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zhang KY, Cowtan K, Main P, Combining constraints for electron-density modification. Methods Enzymol. 277 (1997) 53–64. [DOI] [PubMed] [Google Scholar]
- [83].Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Cryst. D66 (2010) 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sheldrick GM, Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Cryst. D66 (2010) 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Langer GG, Hazledine S, Wiegels T, Carolan C, Lamzin VS, Visual automated macromolecular model building. Acta Cryst. D69 (2013) 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Boltes I, Czapinska H, Kahnert A, von Bulow R, Dierks T, Schmidt B, von Figura K, Kertesz MA, Uson I, 1.3 Å structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism of sulfate ester cleavage in the sulfatase family. Structure 9 (2001) 483–491. [DOI] [PubMed] [Google Scholar]
- [87].Bond CS, Clements PR, Ashby SJ, Collyer CA, Harrop SJ, Hopwood JJ, Guss JM, Structure of a human lysosomal sulfatase. Structure 5 (1997) 277–289. [DOI] [PubMed] [Google Scholar]
- [88].Demydchuk M, Hill CH, Zhou A, Bunkoczi G, Stein PE, Marchesan D, Deane JE, Read RJ, Insights into Hunter syndrome from the structure of iduronate-2-sulfatase. Nat. Commun 8 (2017) 15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Hernandez-Guzman FG, Higashiyama T, Pangborn W, Osawa Y, Ghosh D, Structure of human estrone sulfatase suggests functional roles of membrane association. J. Biol. Chem 278 (2003) 22989–22997. [DOI] [PubMed] [Google Scholar]
- [90].Cartmell A, Lowe EC, Basle A, Firbank SJ, Ndeh DA, Murray H, Terrapon N, Lombard V, Henrissat B, Turnbull JE, Czjzek M, Gilbert HJ, Bolam DN, How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans. Proc. Natl. Acad. Sci. USA 114 (2017) 7037–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Sidhu NS, Schreiber K, Propper K, Becker S, Uson I, Sheldrick GM, Gartner J, Kratzner R, Steinfeld R, Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA. Acta Cryst. D70 (2014) 1321–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Jonas S, van Loo B, Hyvonen M, Hollfelder F, A new member of the alkaline phosphatase superfamily with a formylglycine nucleophile: structural and kinetic characterisation of a phosphonate monoester hydrolase/phosphodiesterase from Rhizobium leguminosarum. J. Mol. Biol 384 (2008) 120–136. [DOI] [PubMed] [Google Scholar]
- [93].van Loo B, Jonas S, Babtie AC, Benjdia A, Berteau O, Hyvönen M, Hollfelder F, An efficient, multiply promiscuous hydrolase in the alkaline phosphatase superfamily. Proc. Natl. Acad. Sci. USA 107 (2010) 2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Agarwal V, Borisova SA, Metcalf WW, van der Donk WA, Nair SK, Structural and mechanistic insights into C-P bond hydrolysis by phosphonoacetate hydrolase. Chem. Biol 18 (2011) 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Nukui M, Mello LV, Littlejohn JE, Setlow B, Setlow P, Kim K, Leighton T, Jedrzejas MJ, Structure and molecular mechanism of Bacillus anthracis cofactor-independent phosphoglycerate mutase: a crucial enzyme for spores and growing cells of Bacillus species. Biophys. J 92 (2007) 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Roychowdhury A, Kundu A, Bose M, Gujar A, Mukherjee S, Das AK, Complete catalytic cycle of cofactor-independent phosphoglycerate mutase involves a spring-loaded mechanism. FEBS J. 282 (2015) 1097–1110. [DOI] [PubMed] [Google Scholar]
- [97].Nowicki MW, Kuaprasert B, McNae IW, Morgan HP, Harding MM, Michels PA, Fothergill-Gilmore LA, Walkinshaw MD, Crystal structures of Leishmania mexicana phosphoglycerate mutase suggest a one-metal mechanism and a new enzyme subclass. J. Mol. Biol 394 (2009) 535–543. [DOI] [PubMed] [Google Scholar]
- [98].Mercaldi GF, Pereira HM, Cordeiro AT, Michels PA, Thiemann OH, Structural role of the active-site metal in the conformation of Trypanosoma brucei phosphoglycerate mutase. FEBS J 279 (2012) 2012–2021. [DOI] [PubMed] [Google Scholar]
- [99].Panosian TD, Nannemann DP, Watkins G, Phelan VV, McDonald WH, Wadzinski BE, Bachmann BO, Iverson TM, Bacillus cereus phosphopentomutase is an alkaline phosphatase family member that exhibits an altered entry point into the catalytic cycle. J. Biol. Chem 286 (2011) 8043–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


