Abstract
Previously we have shown that the Japanese macaque gut microbiome differs not by obesity per se, but rather in association with high fat diet feeding. This held true for both pregnant dams, as well as their one-year old offspring, even when weaned onto a control diet. Here we aimed to examine the stability of the gut microbiome over time and in response to maternal and post-weaning high fat diet (HFD) feeding from 6 months of age, and at 1 and 3 years of age. In both cross-sectional and longitudinal specimens, we performed analysis of the V4 hypervariable region of the 16S rRNA gene on anus swabs collected from pregnant dams and their juveniles at age 6 months to 3 years (n=55). Extracted microbial DNA was subjected to 16S amplicon-based metagenomic sequencing on the Illumina MiSeq platform. We initially identified 272 unique bacterial genera, and multidimensional scaling (MDS) revealed samples to cluster by age and diet exposures. Dirichlet multinomial mixture modeling of microbiota abundances enabled identification of two predominant enterotypes to which samples sorted, characterized primarily by Treponema abundance, or lack thereof. Approximating the time of initial weaning (6 months), the Japanese macaque offspring microbiome underwent a significant state type transition which stabilized from 1 to 3 years of age. However, we also found the low abundance Treponema enterotype to be strongly associated with HFD exposure, be it during gestation/lactation or in the post-weaning interval. Examination of taxonomic co-occurrences revealed samples within the low Treponema cluster were relatively permissive (allowing for increased interactions between microbiota) whereas samples within the high Treponema cluster were relatively exclusionary (suggesting decreased interactions amongst microbiota). Taken together, these findings suggest that Treponemes are keystone species in the developing gut microbiome of the gut, and susceptible to HFD feeding in their relative abundance.
Keywords: microbiome, macaque, high fat diet, early development
Introduction
The developmental origins of health and disease (DOHaD) suggests that adverse conditions in early life contribute to later-in-life metabolic disease (Barker, 1990, 1995; Barker, 2004; Eriksson et al., 2001; Roseboom, de Rooij, & Painter, 2006; Schulz, 2010). We have previously demonstrated in both humans and non-human primates that epigenetic, metabolic, and microbial disruptions both co-occur with exposure to a maternal high fat diet, cannot be completely ameliorated with postnatal weaning onto a control diet, but do diminish when obese dams are reverted to a control diet prior to pregnancy (Aagaard-Tillery et al., 2008; Chu et al., 2016; Cox, Williams, Grove, Lane, & Aagaard-Tillery, 2009; Ma et al., 2014; Pace et al., 2018; Suter et al., 2011; Suter et al., 2014; Suter, Chen, et al., 2012; Suter, Sangi-Haghpeykar, et al., 2012; Suter, Takahashi, Grove, & Aagaard, 2013). Interestingly, dysbiosis of the gut microbiome has similarly been shown to occur in temporal association with metabolic disease in adults (Dao et al., 2016; Delzenne, Neyrinck, & Cani, 2011; Devaraj, Hemarajata, & Versalovic, 2013; Everard et al., 2013; Larsen et al., 2010; Ley, 2010; Qin et al., 2012; Tilg & Kaser, 2011; Turnbaugh et al., 2009; Turnbaugh, Bäckhed, Fulton, & Gordon, 2008; Turnbaugh et al., 2006; Turnbaugh & Gordon, 2009); however, it is unclear if the microbiome is cause or consequence of these associations. Therefore, understanding the developmental and ecological alterations that occur within the gut microbiome from early life into adolescence may have the potential for identifying and developing interventions aimed at curbing the rate and occurrence of adult onset obesity and related metabolic disease.
With this in mind, precisely when and how the offspring gut microbiome is seeded and maintained remains unclear at present. While it is evident that while the gut microbiome varies by days and weeks of post-natal age (Chu et al., 2017; Koenig et al., 2011; Wu et al., 2011; Yatsunenko et al., 2012), diet and timing of feeding has been demonstrated to significantly alter both the structure and function of the gut microbiome (David et al., 2013). Furthermore, examination of the gut microbiome in first year of life suggests a relatively dynamic environment with several key transitions, albeit the relative impact of dietary components versus cessation of breastfeeding is still unknown (Bäckhed et al., 2015). To this end, we have previously demonstrated that exposure to a maternal high fat diet through gestation and lactation persistently alters the offspring microbiome in our primate model, notably with a diminished abundance of Campylobacter spp. (Ma et al., 2014). The use of synbiotics failed to persistently ameliorate the dysbiosis resulting from maternal high fat diet exposure, and does not prevent the re-occurrence of dysbiosis when challenged up to 2.5 years after initiation of a post-weaning control diet (Pace et al., 2018).
Given the evidence to date suggesting that the primate gut microbiome community structure and its function is persistently influenced by a maternal HFD, understanding the developmental and ecological alterations in the offspring gut microbiome over time and under distinct dietary feeding is of high significance and importance. Identifying key taxa and their metabolic functions which are under the influence of the maternal diet would retain the promise of identifying impactful interventions with the potential to mitigate the footprint left by a maternal high fat diet. Thus, we aimed to examine the ecology of the offspring gut microbiome from early life through adolescence in a primate model of maternal high fat diet and obesity using 16S-amplicon based metagenomic sequencing and analytic modeling in a cohort of dams and their offspring sampled at 6 months, 1 year, and/or 3 years of age.
Methods
Study design
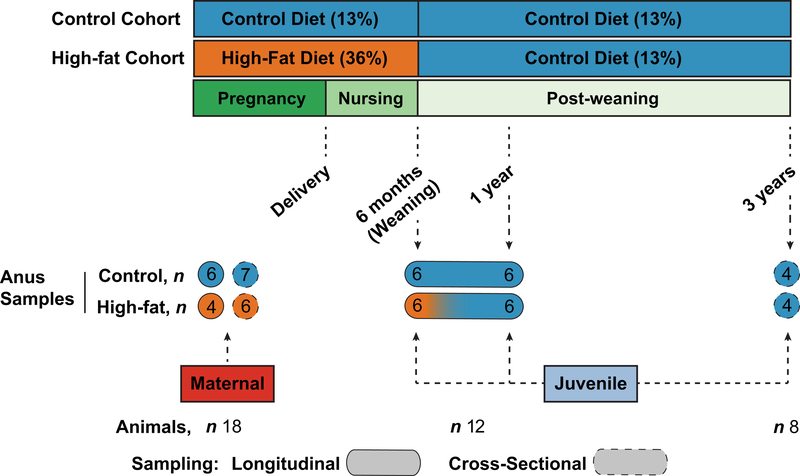
This is a longitudinal and cross-sectional analysis of non-human primate dams and their offspring (Figure 1). The use of Macaca fuscata by our group of investigators has been previously described ( Aagaard-Tillery et al., 2008; Cox et al., 2009; Ma et al., 2014; McCurdy et al., 2009; Pace et al., 2018; Suter et al., 2011; Suter, Chen, et al., 2012; Suter, Sangi-Haghpeykar, et al., 2012; Suter et al., 2013). Animals were socially housed within indoor/outdoor enclosures at the Oregon National Primate Research Center (ONPRC), meaning that for the duration in which they are co-housed animals their diets are shared. However, when their diets are switched (for example, post-weaning) then their social constructs will accordingly vary. Briefly, Japanese macaque dams were mated while consuming either a control (14% fat from soybean oil, Fiber-Balanced Monkey Diet 5052, Lab Diet, St. Louis, MO) or isocaloric high fat (36% diet from porcine and poultry fat, corn and fish oil, TAD Primate Diet – 5LOP, Test Diet, St. Louis, MO). Additionally, the high fat diet (HFD) group were supplemented with calorically dense treats (consisting of Glaxo powder/TAD pellets, peanut butter, honey, banana, and cornstarch). These diets continued through nursing resulting in offspring exposed to the maternal diet throughout gestation and lactation. After weaning (at approximately 6 to 7 months of age), offspring were either maintained on the maternal control diet (designated control/control) or weaned onto a control post-natal diet following maternal HFD feeding during gestation and lactation (high-fat/control). Administration of antibiotics preceding sample collection by 1 month did not occur in for any animal within the study. All methods were carried out in accordance with IACUC guidelines and regulations, and all experimental protocols were approved by the IACUC at ONPRC and Baylor College of Medicine. Notably, our use of both cross-sectional and longitudinal specimens reflects our overarching aims to minimize sedation to both dams and young offspring, and thus specimens were only collected when other a priori approved and scheduled clinical research procedures or necropsy were undertaken. All aspects of our research complied with the American Society of Primatologists Principles for the Ethical Treatment of Non-Human Primates.
Figure 1. Study design.
Japanese macaque dams consumed either a control diet (13% fat) or high-fat diet (36% fat) during gestation and lactation. Offspring were maintained on the maternal diet until weaning, after which time the offspring were either kept on the control diet (control/control) or switched from their maternal high-fat to the control diet (high-fat/control). Maternal samples were collected during the third trimester glucose tolerance test (GTT). Offspring infant or juvenile samples were collected at 6 months, 13 months, or 36 months during scheduled metabolic testing or necropsy. The study design was primarily cross-sectional with longitudinal sampling between the 6- and 13-month time points as indicated.
Gut microbiome sampling
When animals underwent metabolic testing (e.g. glucose-tolerance test or DEXA scan), Cesarean delivery (dams), or necropsy (36 month juveniles), anus samples comprised of stool from the high anus were collected using Catch-All Swabs (Epicentre, Madison, WI). This was primarily a cross-sectional study with a small subset of longitudinal sampling at between the 6- and 13-month timepoint. Swabs were vigorously swirled in MoBio PowerBead tubes (Qiagen, Germantown, MD), and samples were stored at −80°C prior to extraction. Microbial DNA was isolated using the MoBio PowerSoil protocol. Isolated DNA was subjected to 16S rDNA sequencing.
16S rRNA-amplicon based metagenomic sequencing and data processing
55 samples were sequenced on the Illumina MiSeq platform (2×250bp) of the 16S rRNA gene (V4 hypervariable region) at the Center for Microbiome and Metagenomic Research (CMMR) at BCM. The following primers were used: forward primer – 5’-GTGCCAGCMGCCGCGGTAA-3’; reverse primer – 5’-GGACTACHVGGGTWTCTAAT-3’. Raw reads were demultiplexed using idemp (https://github.com/yhwu/idemp). The demultiplexed data then had primers/adapters removed with cutadapt (Martin, 2013), and was further quality filtered and split into paired and unmatched reads with Trimmomatic (Bolger, Lohse, & Usadel, 2014). Quality filtered paired reads were then imported and processed with DADA2 (v1.6) (Callahan et al., 2016) in R (v3.4.3)(https://www.r-project.org/). Sequences were manually examined for drop off in sequencing quality and subsequently the forward and reverse reads were quality filtered and uniformly trimmed using the filterAndTrim() command. Error rates for both the forward and reverse reads were learned using the default settings. Sequence variants were inferred after sequence dereplication and paired reads merged to generate the amplicon sequence variants (ASVs). ASVs longer or shorter than the expected amplicon size were filtered out. Chimeric ASVs were identified using the command removeBimeraDenovo() using the consensus method. ASVs were assigned taxonomy with the assignTaxonomy() function using RDP’s naive Bayesian classifier against the provided Silva reference/training database (silva_nr_v128_train_set.fa.gz), with species level assignments also made using the addSpecies() function against the provided Silva species training data (silva_species_assignment_v128.fa.gz). The final ASV tables containing the abundance of each ASV in every sample were then imported into the phyloseq (v1.23.1) (McMurdie & Holmes, 2013) R package for downstream analysis. ASV tables were then filtered to remove non-Bacterial ASVs, including mitochondria and chloroplast, and unclassified Bacterial ASVs, as well as to remove samples with subsequent zero ASV counts.
Dirichlet multinomial mixture (DMM) modeling
The R package DirichletMultinomial(v1.20.0) (Holmes, Harris, & Quince, 2012) was used to describe the variability in the microbiome data and cluster samples into enterotypes based on the genus level ASV table. Model fit was determined based on the minimum Laplace goodness of fit.
Statistical analysis
Samples were rarefied to 19,944 reads for multidimensional scaling (MDS) and alpha diversity analyses using the phyloseq rarefy_even_depth function. Statistical analysis of beta diversity was performed with permutational multivariate analysis of variance (PERMANOVA) using the vegan (v2.5–4) adonis function and 999 permutations on the distance matrices, with either the variable of interest added to the model or with animal identity and nested diet entered as a factor into the models before adding other variables. To examine alpha diversity and taxa by age at the genus level, QIIME was utilized (Caporaso et al., 2010). Alpha diversity metrics examined included observed ASVs, Shannon’s diversity, Simpson’s diversity, Good’s Coverage, and Chao1. Statistical analysis of alpha diversity metrics was performed in Graphpad Prism (v7.0a, La Jolla, CA). Hierarchal clustering of taxa at the genus level was performed using Spearman’s correlation available from Morpheus (https://software.broadinstitute.org/morpheus) was utilized. Prior to analysis of hierarchal clustering, genus level taxonomy below 0.01% was removed along with taxa not identified to the genus level. Examination of taxonomic differences were performed using linear discriminant analysis (LDA) of effect size (LEfSe) (Segata et al., 2011). Alphas were set to 1 for Kruskal-Wallis and Wilcoxon analysis with an LDA score of 2.0 and utilization of all-against-all for multi-class analysis. Post-analysis, false discovery rate (FDR) correction was performed using R p.adjust. Significant taxa (FDR-corrected p<0.05) resulting from this analysis are displayed. LEfSe was performed on full genus level taxonomy tables generated from ASVs through QIIME (Caporaso et al., 2010).
Differential taxonomic features based on the DMM clusters were identified via DESeq2(v1.18.1) (Love, Huber, & Anders, 2014). PICRUSt2 (Langille et al., 2013) was used to predict functions for ASVs and ascribe taxonomic contributions to inferred functional pathways. Briefly, ASV sequences were placed into the PICRUSt2 reference phylogeny, followed by the mp method for hidden-state prediction. The final outputs (metagenome predictions and predicted pathway abundances and coverages – stratified and non-stratified) were then exported for further analysis using R. Except where noted, all statistical analyses were performed using R (version 3.4.3) and/or GraphPad Prism (GraphPad Software Inc., La Jolla, CA). The R packages factoextra (v1.0.5), pheatmap (v1.0.8), vegan (v2.5–2) (Oksanen, Blanchet, & Friendly, n.d.), phyloseq (v1.23.1) (McMurdie & Holmes, 2013), and ggplot2 (v3.0.0) (Wickham, 2009) were used to perform and visualize cluster analyses and ordinations.
Data availability
The 16S amplicon-based metagenomic sequence data generated from this analysis has been deposited in the Sequence Read Archive (SRA) under bioproject ID PRJNA508806.
Results
16S rRNA amplicon-based metagenomic sequencing
A total of 55 samples were collected over a period of four years, with each subject sampled at least once but not necessarily at all time points (due to timing of scheduled procedures and necropsy; Fig. 1). A total of 2,012,245 filtered reads were obtained for all samples (average of 36,586 reads per sample) with minimum and maximum read counts of 26,592 and 45,089, respectively (Supp. Table 1). There was no difference in the number of reads based on diet, or when samples were stratified by age groups (Table 1). Additionally, we utilized Good’s Coverage as a measure of sequence quality, which estimates the percentage of species represented in a given sample. For all samples, >99% of taxa were represented in the sample and thus indicating that high quality sequencing data had been generated among all subjects and their specimens (Supp. Table 1). Additionally, there were no significant differences in Good’s Coverage between diet or age groups (Table 1). From these data we identified 3,173 amplicon sequence variants (ASVs), corresponding to 272 unique genera which were comparable by animal age and diet allocation with respect to both the number of 16S sequence reads generated and their quality.
Table 1.
Average (± standard deviation) read counts and Good’s Coverage across maternal and offspring age strata and within diet cohorts, t test (Mann-Whitney).
| Read Count | Good’s Coverage | ||||||
|---|---|---|---|---|---|---|---|
| n | Control diet (n 29) | High-fat diet (n 26) | p | Control diet (n 29) | High-fat diet (n 26) | p | |
| Overall | 55 | 36,728±820 | 36,428±878 | 0.80 | 0.9996±0.0002 | 0.9996±0.0002 | 0.88 |
| Adults | 23 | 36,074±1,272 | 34,338±1,417 | 0.37 | 0.9996±0.0002 | 0.9998±0.00009 | 0.15 |
| 6 months | 12 | 36,893±1,836 | 36,061±1,074 | 0.70 | 0.9996±0.0002 | 0.9997±0.00004 | >0.99 |
| 13 months | 12 | 35,908±2,212 | 39,992±1,264 | 0.14 | 0.9996±0.0002 | 0.9994±0.0001 | 0.09 |
| 36 months | 8 | 39,833±834 | 36,966±3,150 | 0.41 | 0.9996±0.0001 | 0.9995±0.0002 | 0.49 |
Microbial community structure is associated with maternal diet and age
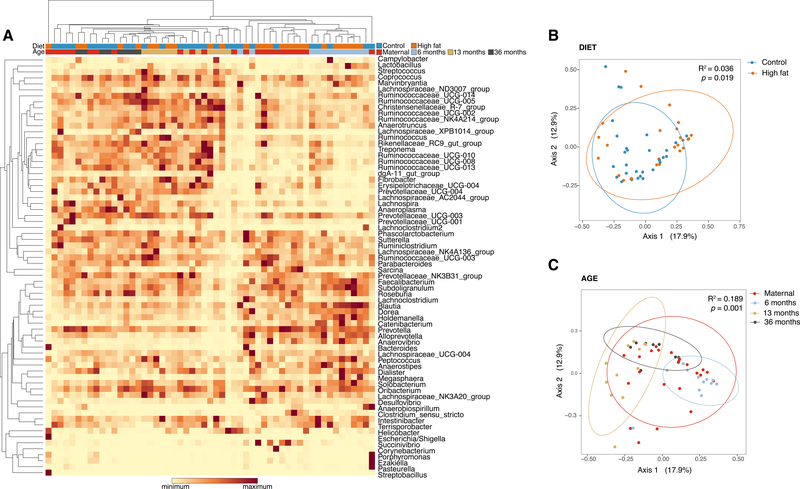
Upon initial examination of the microbial community and its composition, hierarchical clustering (Spearman’s correlation) demonstrated that samples were predominantly structured by age and diet (Fig. 2A). To confirm these findings in an independent analysis, we performed multidimensional scaling (MDS) and found that the offspring gut microbiome was structured by diet across all samples (Bray-Curtis, PERMANOVA F-model=1.96, R2=0.04, p=0.019) (Fig. 2B). Furthermore, the data demonstrated that post-natal age was also a driver of alterations in the gut microbiome (Bray-Curtis, PERMANOVA F-model=3.97, R2=0.19, p=0.001)(Fig. 2C) with separate clustering of maternal, 6 month juvenile, 13 month juvenile, and 36 month juvenile samples by both weighted (Fig. 2C) and unweighted metrics (Supp. Fig. 1). These differences were not attributable to maternal age, parity, pre-pregnancy weight, housing, nor body composition as there were no significant differences in these demographics between the offspring cohorts (Supp. Table 2).
Figure 2. The Japanese macaque gut microbiome community membership and structure varies by post-natal age and maternal diet.
Anal swabs were collected from at various time points (6-month infant (n=12), 13-month juvenile (n=12), 36-month juvenile (n=8), and adult dams (n=23) during the third trimester of pregnancy) in our primate model of maternal high fat diet and obesity. (A) Hierarchal clustering of all taxa to the genus level using Spearman’s correlation demonstrates that the gut microbiome are structured by age and diet. (B) Multidimensional scaling (MDS) demonstrates that beta diversity (Bray-Curtis) differs significantly when comparing high fat diet exposed animals to those only exposed to a control diet (p=0.019 by PERMANOVA, R2=0.036). (C) MDS examination of beta diversity (Bray-Curtis) by age demonstrates significant alterations (p=0.0001 by PERMANOVA, R2=0.189). When inputting the animal identification first into the PERMANOVA modeling, animal identification (with its nested diet) and age were retained after removing non-significant interaction terms (animal identification and nested diet: F model 1.29, R2 0.67, p=0.006; animal age: F-model 7.386, R2 0.104, p=0.001). Altogether, we find that the gut microbiome of the macaque is structured by age and maternal dietary exposures.
Since our cohort retained both cross-sectional and longitudinally sampled animals, we similarly analyzed our microbial community and its composition be inputting the animal identification first into the PERMANOVA modeling (see Methods). In doing so, animal identification (with its nested diet) and age were retained as a significant drivers of the alterations in the gut microbiome after removing non-significant interaction terms (animal identification and nested diet: F model 1.29, R2 0.67, p=0.006; animal age: F-model 7.386, R2 0.104, p=0.001). Similarly, the DMM cluster (see further below) was retained as significant (F-model of 4.07, R2 0.07, p=0.001).
Examination of alpha diversity revealed significant differences based on post-natal age (Kruskal-Wallis, Dunn’s corrected p<0.05 for Chao1 and observed ASVs, p>0.05 for Shannon and Simpson, Dunn’s corrected), with 6-month old juveniles observed to have a lower alpha diversity compared to the older juveniles (p<0.05) and adults when utilizing Dunn’s multiple comparisons test (Supp. Fig. 2). This difference in alpha diversity at 6 months of age was temporally associated with observed decreases in Bifidobacterium which occur post-weaning. Specifically, we found a significant decrease in Bifidobacterium after 6 months (Kruskal-Wallis, p=0.004; Dunn’s post-test p<0.05), which would be anticipated to be attributed more to the absence of maternal breastfeeding and milk consumption rather than the addition of non-milk dietary constituents. Additionally, we observed alterations in alpha diversity when comparing control and high fat diet exposures with lower alpha diversity associated with high fat diet (all samples, p<0.05 by Mann-Whitney for Chao1, Observed ASVs, and Shannon). With our results demonstrating that the gut microbiome could be stratified by age and diet, we next sought to employ Dirichlet multinomial mixture (DMM) modeling to examine if enterotypes and transition states could be identified within the macaque gut microbiome.
Dirichlet multinomial mixture modeling reveals two enterotypes that shift with dietary changes
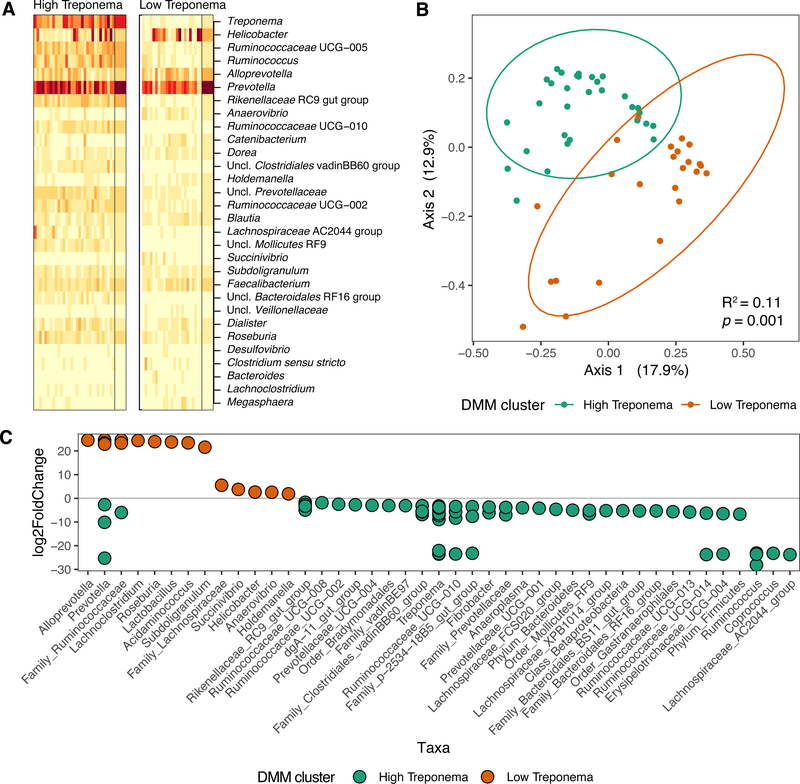
DMM modeling of microbiota abundances revealed that the offspring’s samples were partitioned into two dominant community states, or enterotypes (Fig. 3A). Both enterotypes retained high abundances of Prevotella (Wilcoxon rank sum test, U=282.0, p=0.13), with state type categorization being significantly associated with variations in the relative abundance of Treponema spp. (Wilcoxon rank sum test, U=741.0, p=3.79e-10, FDR adjusted p=1.03e-7). When we allowed the taxonomy to further define our clusters in an unbiased fashion, we found that state types were categorized into two enterotypes as a function of (cluster A) observed high abundance of Treponema and (cluster B) observed low abundance of Treponema. A majority of samples were classified within the high Treponema abundance enterotype (n=31, ~56%), with the remaining samples in the low Treponema abundance enterotype (n=24, ~44%). Although with a lower relative abundance, we did find Ruminococcus spp. significantly co-occurring in cluster A rather B (Fig. 3A, Wilcoxon rank sum test, U=491, p=2.97e-5). Intriguingly, we observed that the DMM modeled enterotypes explained less of the variation in the microbial taxonomic abundance than post-natal age, but more than that explained by variation in maternal diet alone (Bray-Curtis, PERMANOVA F-model=7.69, R2=0.11, p=0.001) (Fig. 3B) indicating shifts in enterotypes likely occur over time, but maternal diet has a persistent influence. Additionally, examination of alpha diversity by four independent measures (Observed, Shannon, Chao1, and Simpson) revealed the enterotypes to be significantly different (Mann-Whitney test, p< 0.0001), with the high Treponema abundance enterotype observed to have higher alpha diversity (Supp. Fig. 2). Given these observations in taxonomic structure and community variation by age and maternal diet, we next examined genus-level taxonomy with differential abundance using LEfSe.
Figure 3. Dirichlet multinomial mixture modeling reveals distinct enterotypes that differ over time and with maternal diet.
(A) Heatmap of the top 30 genera. The first DMM component (left) is characterized primarily by a high abundance of Treponema. In contrast, the second DMM component (right) is characterized mainly by a low abundance of Treponema. Narrow columns represent samples and the two broader columns represent component averages. Cell colors represent square-root counts, with lighter and darker colors corresponding to smaller and larger counts, respectively. (B) Multidimensional scaling (MDS) ordination of Bray-Curtis distance of samples demonstrates clustering based on DMM components (PERMANOVA, R2=0.11, p=0.001). (C) Differentially abundant taxa identified via DESeq2.
Enterotypes are differentially enriched for predicted metabolic pathways
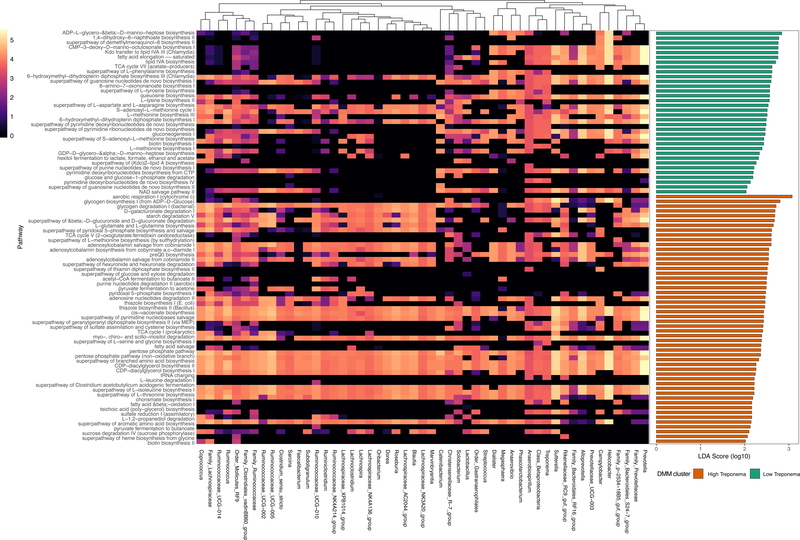
As enterotypes appeared to differ with respect to dietary exposures, we next utilized PICRUSt2 to infer the functional genetic potential of the macaque community state types. Altogether we identified 86 predicted pathways that were differentially enriched based on DMM cluster as determined via LEfSe (Fig. 4). The high Treponema abundance enterotype was enriched for 52 pathways, whereas the low Treponema abundance enterotype was enriched for 34 pathways (Fig. 4) when analyzed by LEfSe with an FDR correction. Within these enriched pathways, filtering taxa based on a contribution greater than the mean abundance revealed that, at least by 16S generated measures, as few as 51 taxa could be potentially driving these predicted functional distinctions (Fig. 4). Within the low Treponema abundance enterotype, we observed enrichment of fatty acid elongation. Within the high Treponema abundance enterotype, we instead observed an enrichment of pathways for the short chain fatty acid butanoate as well as an enrichment of preQ0 (prequeosine-0) biosynthesis.
Figure 4. Inferred bacterial functional pathways abundance and enrichment.
Differentially enriched pathways based on DMM cluster were determined with LEfSe (right). Heatmap demonstrates the pathway abundance (log10 transformed) within the 51 taxa that were found to have the greatest contribution to the enriched pathways.
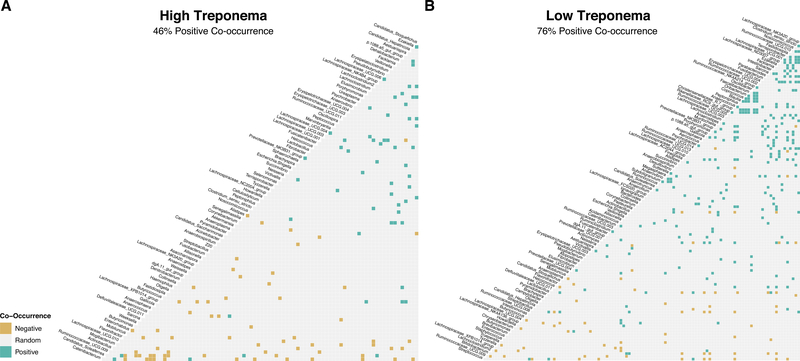
Species co-occurrence modeling
We next sought to determine if microbial ecological networks and their landmark species might vary when comparing the high and low Treponema clusters identified by DMM. We hypothesized that the relative abundance of Treponema defining these clusters might not just influence community composition and function, but also affect the permissiveness of microbial interactions. When separated by high Treponema or low Treponema cluster, we found that the low Treponema group appeared more permissive with a 76% positive co-occurrence between genera (Fig. 5). Conversely, we observed a 46% positive co-occurrence in the high Treponema group (Fig. 5), a finding which was accompanied by a lower number of co-occurrences (n = 124) when compared to the low Treponema (n = 335). These alterations in co-occurrence were significant (Fisher’s Exact Test, p=5.5−8, odds ratio 0.28). Altogether, this data is consistent with our prior observations and collectively suggests that high Treponema may be a more restrictive enterotype, which may promote a gut microbiome which is most closely taxonomically and functionally related to animals born to dams exclusively fed a control diet (hereafter referred to as “healthy”; Ma et al., 2014). We acknowledge that “healthy” and “dysbiotic” community states are not well defined in humans nor primates alike, and failure to revert to the control state may not, in fact, be dysbiotic but may represent an adaptive community. To begin to clarify this issue of adaptivity, we examined gut microbiome community structure as a true function of age and maternal diet.
Figure 5. Probabilistic model of species co-occurrence suggests the relative low abundance or absence of Treponema promotes and ecologic environment which is permissive to microbial interactions.
Co-occurrence modeling of (A) high Treponema and (B) low Treponema clusters show that high Treponema clusters have fewer positive co-occurrence interactions (46%) when compared to low Treponema clusters (76%). This difference is significant (p=5.5e−8) by Fisher’s Exact Test with an odds ratio of 0.28. Negative co-occurrences are highlighted in yellow while positive co-occurrences are highlighted in green.
Postnatal age structures the gut microbiome community
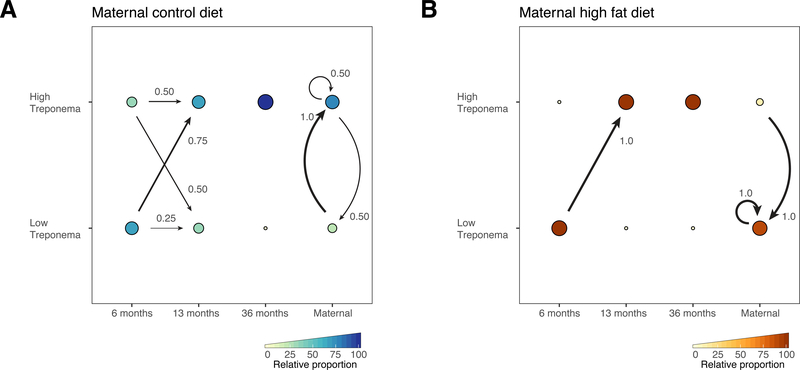
We next examined how enterotypes were distributed across time (age of offspring), with samples stratified by maternal dietary exposure. For juveniles with longitudinal sampling, we found all 6 month old juveniles on the maternal high fat diet transitioned to the high Treponema abundance enterotype at 13 months old (Fig. 6B). The 6-month-old juveniles on the maternal control diet had much more variable transitions, although a majority transitioned to the high Treponema abundance enterotype (Fig. 6A). All of the 3-year-olds, regardless of maternal diet, were observed to fall within the high Treponema abundance enterotype (Fig. 6). Interestingly, dams on the control diet were mainly observed to fall into both enterotypes, although animals with subsequent sampling (n=3) were observed to generally remain or transition to the high Treponema abundance enterotype (Fig. 6). In contrast, dams on the high fat diet with subsequent sampling (n=2) were primarily observed to occur or transition to the low Treponema abundance enterotype (Fig. 6). When we next examined if high fat diet exposure was associated with enterotype classification, we found the low Treponema abundance enterotype was relatively enriched in samples taken from animals exposed to a maternal high fat diet (Fisher’s exact test, p=0.06), which appeared driven mainly by maternal diet (Fisher’s exact test, p=0.003) as we found no significant difference in enterotype at 6 months, 1 or 3 years (Fisher’s exact test, p>0.05); this time interval would have allowed for co-housing variation. When examining taxonomic differences by LEfSe, we found that animals only exposed to a maternal control diet had few differentially abundant taxa when compared to those exposed to a maternal high fat diet, even when weaned and maintained on a control diet for as great as 2.5 years (Supplemental Fig. 3). While we did not see taxa overlapping between control or high fat diet groups within the low Treponema cluster, we did find that Ruminococcus spp., Fibrobacter, Anaeroplasma, and Lachnospiraceae spp. were abundant in the high Treponema cluster regardless of diet. Furthermore, when examining co-occurrence when stratified by DMM cluster and diet, we saw that Lachnospiraceae had many significant positive co-occurrences (p < 0.05) while Ruminococcus had significant negative co-occurrences (p < 0.05).
Figure 6. Community transitions in the gut microbiome of the Japanese macaque.
Proportions and transitions between and within DMM components over time stratified by maternal (A) control and (B) high fat dietary cohorts. Arrows indicate the transition probabilities in infant and juvenile offspring from 6 months to 13 months, and from subsequent samples taken from dams. Circle size and color, as detailed in the figure key, indicate the relative proportion of samples within each dietary cohort assigned to each DMM component. The very small circles indicate relative proportions of 0 (), except in the case of panel B where the small circle at 6 months indicates a relative proportion of 10%.
Conclusions
We found that the gut microbiome of Japanese macaque offspring from 6 months to 3 years of age was structured by both maternal diet and post-natal age. We have previously published that maternal diet persistently effects the offspring microbiome in this highly relevant primate model of maternal high fat diet and obesity (Ma et al., 2014; Pace et al., 2018). Here we provide further evidence to demonstrate that the macaque gut microbiome is a representative model for interrogating the human microbiome throughout early life. This is in agreement with prior human data (Yatsunenko et al., 2012). Furthermore, alterations in alpha diversity by age within the gut microbiome are also in agreement with prior human studies (Yatsunenko et al., 2012), and may be due to decreases in Bifidobacterium that occur post-weaning. Specifically, we found a significant decrease in Bifidobacterium after 6 months, most likely due to the absence of mother’s own breastmilk consumption.
In order to better model the gut communities beyond weaning, we employed DMM modeling and observed two main enterotypes characterized by high and low relative abundance of Treponema. In parallel with Treponema enrichment to high abundance, we observed co-enrichment of members of Clostridia, such as Ruminococcus and Lachnospiraceae ASVs. We find these findings to be intriguing since murine studies have demonstrated the effect of Clostridia on gut serotonin production and regulatory T lymphocyte induction (Atarashi et al., 2011; Gaboriau-Routhiau et al., 2009; Mathewson et al., 2016; Reigstad et al., 2014; Yano et al., 2015). Consistent with these observations were our findings (based on inferred metagenomics) that butanoate, a mediator of inflammation and modulator of gut serotonin production (Reigstad et al., 2014; Yano et al., 2015), temporally varied. To elucidate the mechanism of these interactions further, we examined the microbial interactions and ecology using co-occurrence modeling. We found that although Treponema per se did not have any significant detectable co-occurrences, positive co-occurrence events differed significantly between high and low Treponema groups (p = 5.5e−8 by Fisher’s Exact Test). These results suggest that Treponema may regulate the permissiveness of the overall commensal microbial community, most likely through an indirect mechanism to promote exclusion of detrimental microbes within a community. Altogether, the current study’s findings are consistent with our prior observations and collectively suggest that high Treponema (seen with a maternal control but not high fat diet) may be a more restrictive enterotype which, in turn, promotes a “healthy” gut microbiome (Ma et al., 2014; Pace et al, 2018). We acknowledge that “healthy” and “dysbiotic” community states are not well defined in humans nor primates alike, and failure to revert to the control state may not, in fact, be dysbiotic but may represent an adaptive community as evidenced by its community ecology.
With this in mind, we examined the ecologic mechanisms by which maternal dietary exposures influenced the high and low Treponema groups over time, even with switching the offspring to a control diet post-weaning for up to 2.5 years. We found that within offspring only exposed to a control diet (i.e., the dams were fed a control diet, and the offspring were weaned onto a control diet; control/control) we observed significantly greater variability in the transition between high and low Treponema enterotypes. This was particularly true in the 6 month to 13 month transition and between adults examined across multiple time points. This differed in high fat diet exposed animals, with all juveniles at 6 months transitioning to the high Treponema group by 13 months of age. Furthermore, adults consuming a high fat diet were more likely to remain in the low Treponema cluster. This is consistent with our prior data demonstrating that a high fat diet post-weaning diet was associated with a lower abundance of Treponema within the offspring gut microbiome (Ma et al., 2014). Finally, upon examining taxonomic differences stratified by DMM cluster and diet, we found that Clostridia (Ruminococcus and Lachnospiraceae) were differentially abundant in the high Treponema cluster regardless of diet. These findings reinforce the notion that in the macaque, Treponema may be regulating permissiveness of the commensal gut microbiome. Since the maternal diet significantly alters its relative abundance, the end result of the niche occupancy by Treponema is parlayed variation in the abundance of Clostridia taxa. Altogether, we found that the gut microbiome of the Japanese macaque is altered by age and maternal dietary exposure with a resultant alteration in Treponema and permissively associated Clostridia taxa (Ruminococcus and Lachnospiraceae).
Given our observations of maternal dietary influence, it is intriguing to postulate when this early life influence is occurring. We and others have previously demonstrated that we can employ metagenomics and 16S sequencing to detect and characterize a unique and low abundance, low biomass placental and neonatal microbiome (Aagaard et al., 2014; Amarasekara et al., 2015; Antony et al., 2015; Chu et al., 2016, Chu et al. 2017; Collado et al., 2016; Doyle et al., 2014; Jiménez et al., 2005; Leon et al., 2018; Prince et al., 2016; Satokari et al., 2009; Ardisonne et al., 2014; Bassols et al., 2016; Doyle et al., 2017; Gomez-Arango et al., 2017; Zheng et al., 2017; Rautava et al., 2012; Martinez et al., 2018; Borghi et al., 2018; Parnell et al., 2017). Furthermore, in an elegant set of experiments, Li et al (2019) recently demonstrated that memory CD4+ T cells are generated during intrauterine development in the human fetal intestine, which are phenotypicially similar to innate-like lymphocytes previously described in mice that are dependent on microbes for their maintenance (Prince et al., 2014). While there are a few investigators who have questioned the ability to reliably distinguish a placental microbiota from contaminant controls (de Goffau et al., 2018; Lauder et al., 2016; Leiby et al., 2018; Theis et al., 2019), it is not the aim nor intent of the current study to describe the potential origins or source of the primate offspring gut microbiome. Suffice it to say, ongoing and future studies in humans and primates are necessary to further clarify the source and seeding of the offspring microbiome.
There are inherent strengths and weaknesses to our study. First, our study design enables us to parse the influence of the maternal versus post-weaning diet, and further deconvolute the impact of post-natal age. Second, we have modeled the ecology of the developing gut microbiome in the primate as a function of both maternal diet and developmental age. These have collectively led to a series of novel and significant observations.
An evident potential weakness to our study is the small sample number. Fortunately, despite sample number limitations, by conducting a power analysis we determined we were able to robustly address our research questions. Specifically, the number of animals studied, number of samples sequenced, and depth of our sequencing enabled us to be adequately powered to reach statistical significance in our findings, as documented in our results and after controlling for multiple comparisons. Future studies related to the questions posed here may benefit from additional animals and further longitudinal sampling across a longer time interval. Additional limitations to our study include reliance on inferred metagenomics pathways, which is an acknowledged limitation with any 16S-based pathway analysis. Finally, since our animals were socially housed, and all animals in a single run shared the same diet, there is the potential for co-housing to confound our dietary analysis. However, given that there was a persistent impact on the offspring gut microbiome even when diet and co-housing was varied, the significant modifiers in our cohort were postnatal age and diet. Since juveniles exposed to both maternal control and high fat diets were co-housed when fed a post-weaning control diet, we conclude that co-housing alone was not the weighted effect modifier.
Nevertheless, based on the strength and significance of our findings reported herein, future examinations of the taxa and pathways involved using functional shotgun metagenomics to species and strain level resolution will enable determinations of how Treponema may be involved in regulating the commensal gut microbiome during crucial stages of development. Further, as we and the wider community of microbial ecologists and microbiome scientists continue to advance and refine culture-independent methodologies, we anticipate the inclusion of additional positive (e.g., mock communities) and negative controls in future studies will enable a more precise characterization of animal microbiomes. Knowing that in our animals our offspring are predisposed to metabolic disease as a result of maternal high-fat diet feeding, these and future studies are of great translational and public health importance.
Supplementary Material
Beta diversity metrics were rarefied to 19,964 (75% of minimum sample read count) and measured by Binary Jaccard (unweighted metric) using multidimensional scaling (MDS). (A) Maternal diet (p=0.002 by PERMANOVA) and (B) age (p=0.001 by PERMANOVA) significantly structured the gut microbiome. Orange: maternal high-fat diet, dark blue, maternal control diet, light blue: 6-month juvenile, gold: 13-month juvenile, charcoal: 36-month juvenile, and red: maternal.
Alpha diversity metrics were rarefied to 19,944 (75% of minimum sample read count) and measured by Chao1 (A), Observed ASVs (B), Shannon Diversity (C), and Simpson’s Diversity (D). Significant differences between age groups by Kruskal-Wallis with Dunn’s Correction were seen with Chao1 and Observed ASVs (p<0.005) but not with Shannon or Simpson’s Diversity. Additionally, significant differences were observed by diet (p<0.05, Mann-Whitney) with Chao1, Observed ASVs, and Shannon. All alpha diversity metrics demonstrated significance (p<0.0001, Mann-Whitney) when comparing enterotypes as identified by DMM modeling.
Exposure to a high fat maternal diet is associated with increased taxonomic differences between high and low Treponema clusters. LEfSe was utilized to examine taxonomic differences within clusters identified by DMM after stratification by dietary exposure. We found that primates only exposed to a control diet had fewer genera with differential abundance (A) when compared to primates exposed to a high fat diet (B).
Research Highlights
Maternal diet structures the postnatal gut microbiome in Japanese macaques.
The Japanese macaque gut microbiome communities are distinguished by the relative abundance of Treponema, largely as a consequence of permissivity versus exclusivity of other taxa.
Major transitions in the gut microbiome occur post-weaning likely due to cessation of nursing
Acknowledgements
The authors gratefully acknowledge the support of the NIH–NIDDK (R01 DK089201; K.M. Aagaard), the NIH-NIDDK (R24 DK090964), NIH-NICHD (R01 R01HD091731; K.M. Aagaard), NIH-IRACDA Award (NIGMS K12 GM084897; R.M. Pace), and NIH-NICHD NRSA Fellowship (NICHD F32 HD082969, A. L. Prince).
Footnotes
Competing interests
The authors declare no competing interests.
Conflicts of interest
The authors declare no conflicts of interest.
References
- Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, & Lane RH (2008). Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. Journal of Molecular Endocrinology, 41(2), 91–102. 10.1677/JME-08-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, & Lane RH (2008). Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. Journal of Molecular Endocrinology, 41(2), 91–102. 10.1677/jme-08-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, & Versalovic J. (2014). The placenta harbors a unique microbiome. Science Translational Medicine, 6(237), 237ra65 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasekara R, Jayasekara RW, Senanayake H, & Dissanayake VHW (2015). Microbiome of the placenta in pre-eclampsia supports the role of bacteria in the multifactorial cause of pre-eclampsia. Journal of Obstetrics and Gynaecology Research, 41(5), 662–669. 10.1111/jog.12619 [DOI] [PubMed] [Google Scholar]
- Antony KM, Ma J, Mitchell KB, Racusin D. a., Versalovic J, & Aagaard K. (2015). The preterm placental microbiome varies in association with excess maternal gestational weight gain. American Journal of Obstetrics and Gynecology, 212(5), 13–18. 10.1016/j.ajog.2014.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardissone AN, de la Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, … Neu J. (2014). Meconium Microbiome Analysis Identifies Bacteria Correlated with Premature Birth. PLoS ONE, 9(3), e90784 10.1371/journal.pone.0090784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, … Honda K. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science (New York, N.Y.), 331(6015), 337–41. 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, … Wang J. (2015). Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host & Microbe, 17(6), 852 10.1016/j.chom.2015.05.012 [DOI] [PubMed] [Google Scholar]
- Barker DJ (1990). The fetal and infant origins of adult disease. BMJ (Clinical Research Ed.), 301(6761), 1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ (1995). Fetal origins of coronary heart disease. BMJ (Clinical Research Ed.), 311(6998), 171–4. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7613432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJP (2004). The developmental origins of well-being. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 359(1449), 1359–66. 10.1098/rstb.2004.1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassols J, Serino M, Carreras-Badosa G, Burcelin R, Blasco-Baque V, Lopez-Bermejo A, & Fernandez-Real J-M (2016). Gestational diabetes is associated with changes in placental microbiota and microbiome. Pediatric Research, 80(6), 777–784. 10.1038/pr.2016.155 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, & Usadel B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30(15), 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi E, Massa V, Severgnini M, Fazio G, Avagliano L, Menegola E, … Borgo F. (2018). Antenatal Microbial Colonization of Mammalian Gut. Reproductive Sciences, 193371911880441 10.1177/1933719118804411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, & Holmes SP (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods, 13(7), 581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, … Knight R. (2010). QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7(5), 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, & Aagaard KM (2016). The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Medicine, 8(1), 77 10.1186/s13073-016-0330-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, & Aagaard KM (2017). Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nature Medicine, 23(3), 314–326. 10.1038/nm.4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Rautava S, Aakko J, Isolauri E, & Salminen S. (2016). Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Scientific Reports, 6(October 2015), 23129 10.1038/srep23129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Williams S, Grove K, Lane RH, & Aagaard-Tillery KM (2009). A maternal high-fat diet is accompanied by alterations in the fetal primate metabolome. American Journal of Obstetrics and Gynecology, 201(3), 281.e1–9. 10.1016/j.ajog.2009.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, … Lepage P. (2016). Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: Relationship with gut microbiome richness and ecology. Gut, 65(3), 426–436. 10.1136/gutjnl-2014-308778 [DOI] [PubMed] [Google Scholar]
- David L.a Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, … Turnbaugh PJ (2013). Diet rapidly and reproducibly alters the human gut microbiome. Nature, 505(7484), 559–563. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Goffau MC, Lager S, Salter SJ, Wagner J, Kronbichler A, Charnock-Jones DS, … Parkhill J. (2018). Recognizing the reagent microbiome. Nature Microbiology, 3(8), 851–853. 10.1038/s41564-018-0202-y [DOI] [PubMed] [Google Scholar]
- Delzenne NM, Neyrinck AM, & Cani PD (2011). Modulation of the gut microbiota by nutrients with prebiotic properties: consequences for host health in the context of obesity and metabolic syndrome. Microbial Cell Factories, 10(Suppl 1), S10 10.1186/1475-2859-10-S1-S10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S, Hemarajata P, & Versalovic J. (2013). The human gut microbiome and body metabolism: implications for obesity and diabetes. Clinical Chemistry, 59(4), 617–28. 10.1373/clinchem.2012.187617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle RM, Alber DG, Jones HE, Harris K, Fitzgerald F, Peebles D, & Klein N. (2014). Term and preterm labour are associated with distinct microbial community structures in placental membranes which are independent of mode of delivery. Placenta, 35(12), 1099–101. 10.1016/j.placenta.2014.10.007 [DOI] [PubMed] [Google Scholar]
- Doyle RM, Harris K, Kamiza S, Harjunmaa U, Ashorn U, Nkhoma M, … Klein N. (2017). Bacterial communities found in placental tissues are associated with severe chorioamnionitis and adverse birth outcomes. PLOS ONE, 12(7), e0180167 10.1371/journal.pone.0180167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson JG, Forsen T, Tuomilehto J, Osmond C, Barker DJP, Forsén T, … Barker DJP (2001). Early growth and coronary heart disease in later life: longitudinal study. BMJ (Clinical Research Ed.), 322(7292), 949 10.1136/bmj.322.7292.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, … Cani PD (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proceedings of the National Academy of Sciences of the United States of America, 1–6. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, … Cerf-Bensussan N. (2009). The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity, 31(4), 677–89. 10.1016/j.immuni.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, & Nitert MD (2017). Contributions of the maternal oral and gut microbiome to placental microbial colonization in overweight and obese pregnant women. Scientific Reports, 7(1), 2860 10.1038/s41598-017-03066-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes I, Harris K, & Quince C. (2012). Dirichlet multinomial mixtures: Generative models for microbial metagenomics. PLoS ONE, 7(2). 10.1371/journal.pone.0030126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E, Fernández L, Marín ML, Martín R, Odriozola JM, Nueno-Palop C, … Rodríguez JM (2005). Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Current Microbiology, 51(4), 270–4. 10.1007/s00284-005-0020-3 [DOI] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, … Ley RE (2011). Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America, 108 Suppl, 4578–85. 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes J.a, … Huttenhower C. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology, 31(9), 814–21. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N, Vogensen FK, van den Berg FWJ, Nielsen DS, Andreasen AS, Pedersen BK, … Jakobsen M. (2010). Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PloS One, 5(2), e9085 10.1371/journal.pone.0009085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, … Kruschke J. (2016). Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome, 4(1), 29 10.1186/s40168-016-0172-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiby JS, McCormick K, Sherrill-Mix S, Clarke EL, Kessler LR, Taylor LJ, … Bushman FD (2018). Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome, 6(1), 196 10.1186/s40168-018-0575-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon LJ, Doyle R, Diez-Benavente E, Clark TG, Klein N, Stanier P, & Moore GE (2018). Enrichment of Clinically Relevant Organisms in Spontaneous Preterm-Delivered Placentas and Reagent Contamination across All Clinical Groups in a Large Pregnancy Cohort in the United Kingdom. Applied and Environmental Microbiology, 84(14). 10.1128/AEM.00483-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE (2010). Obesity and the human microbiome. Current Opinion in Gastroenterology, 26(1), 5–11. 10.1097/MOG.0b013e328333d751 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12), 1–21. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, … Aagaard KM (2014). High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nature Communications, 5, 3889 10.1038/ncomms4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2013). Cutadapt removes adapter sequences from high-throughput sequencing reads kenkyuhi hojokin gan rinsho kenkyu jigyo. EMBnet.Journal, 17(1), 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martinez KA, Romano-Keeler J, Zackular JP, Moore DJ, Brucker RM, Hooper C, … Weitkamp J-H (2018). Bacterial DNA is present in the fetal intestine and overlaps with that in the placenta in mice. PLOS ONE, 13(5), e0197439 10.1371/journal.pone.0197439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T, … Reddy P. (2016). Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nature Immunology, 17(5), 505–13. 10.1038/ni.3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE, & Grove KL (2009). Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. The Journal of Clinical Investigation, 119(2), 323–35. 10.1172/JCI32661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, & Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One, 8(4), e61217 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet F, & Friendly M. (n.d.). Community Ecology Package. R Package Version 24–6. [Google Scholar]
- Pace RM, Prince AL, Ma J, Belfort BDW, Harvey AS, Hu M, … Aagaard KM (2018). Modulations in the offspring gut microbiome are refractory to postnatal synbiotic supplementation among juvenile primates. BMC Microbiology, 18(1), 28 10.1186/s12866-018-1169-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell LA, Briggs CM, Cao B, Delannoy-Bruno O, Schrieffer AE, & Mysorekar IU (2017). Microbial communities in placentas from term normal pregnancy exhibit spatially variable profiles. Scientific Reports, 7(1), 11200 10.1038/s41598-017-11514-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince AL, Ma J, Kannan PS, Alvarez M, Gisslen T, Harris RA, … Aagaard KM (2016). The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. American Journal of Obstetrics and Gynecology, 214(5), 627.e1–627.e16. 10.1016/j.ajog.2016.01.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince AL, Watkin LB, Yin CC, Selin LK, Kang J, Schwartzberg PL, & Berg LJ (2014). Innate PLZF + CD4 + αβ T Cells Develop and Expand in the Absence of Itk. The Journal of Immunology, 193(2), 673–687. 10.4049/jimmunol.1302058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, … Wang J. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature, 490(7418), 55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- Rautava S, Collado MC, Salminen S, & Isolauri E. (2012). Probiotics Modulate Host-Microbe Interaction in the Placenta and Fetal Gut: A Randomized, Double-Blind, Placebo-Controlled Trial. Neonatology, 102(3), 178–184. 10.1159/000339182 [DOI] [PubMed] [Google Scholar]
- Reigstad CS, Salmonson CE, Rainey JF, Szurszewski JH, Linden DR, Sonnenburg JL, … Kashyap PC (2014). Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology, 1(4), 1–9. 10.1096/fj.14-259598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseboom T, de Rooij S, & Painter R. (2006). The Dutch famine and its long-term consequences for adult health. Early Human Development, 82(8), 485–491. 10.1016/j.earlhumdev.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Satokari R, Grönroos T, Laitinen K, Salminen S, & Isolauri E. (2009). Bifidobacterium and Lactobacillus DNA in the human placenta. Letters in Applied Microbiology, 48(1), 8–12. 10.1111/j.1472-765X.2008.02475.x [DOI] [PubMed] [Google Scholar]
- Schulz LC (2010). The Dutch Hunger Winter and the developmental origins of health and disease. Proceedings of the National Academy of Sciences, 107(39), 16757–16758. 10.1073/pnas.1012911107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, & Huttenhower C. (2011). Metagenomic biomarker discovery and explanation. Genome Biology, 12(6), R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter MA, Ma J, Vuguin PM, Hartil K, Fiallo A, Harris RA, … Aagaard KM (2014). In utero exposure to a maternal high-fat diet alters the epigenetic histone code in a murine model. American Journal of Obstetrics and Gynecology, 210(5), 463.e1–463.e11. 10.1016/j.ajog.2014.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M.a, Chen A, Burdine MS, Choudhury M, Harris RA, Lane RH, … Aagaard KM (2012). A maternal high-fat diet modulates fetal SIRT1 histone and protein deacetylase activity in nonhuman primates. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology, 26(12), 5106–14. 10.1096/fj.12-212878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M.a, Sangi-Haghpeykar H, Showalter L, Shope C, Hu M, Brown K, … Aagaard KM (2012). Maternal high-fat diet modulates the fetal thyroid axis and thyroid gene expression in a nonhuman primate model. Molecular Endocrinology (Baltimore, Md.), 26(12), 2071–80. 10.1210/me.2012-1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M.a, Takahashi D, Grove KL, & Aagaard KM (2013). Postweaning exposure to a high-fat diet is associated with alterations to the hepatic histone code in Japanese macaques. Pediatric Research, 74(3), 252–8. 10.1038/pr.2013.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M, Bocock P, Showalter L, Hu M, Shope C, McKnight R, … Aagaard-Tillery K. (2011). Epigenomics: maternal high-fat diet exposure in utero disrupts peripheral circadian gene expression in nonhuman primates. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology, 25(2), 714–26. 10.1096/fj.10-172080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis KR, Romero R, Winters AD, Greenberg JM, Gomez-Lopez N, Alhousseini A, … Hassan SS (2019). Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomics. American Journal of Obstetrics and Gynecology, 220(3), 267.e1–267.e39. 10.1016/j.ajog.2018.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H, & Kaser A. (2011). Gut microbiome, obesity, and metabolic dysfunction. Journal of Clinical Investigation, 121(6), 2126–2132. 10.1172/JCI58109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Bäckhed F, Fulton L, & Gordon JI (2008). Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host & Microbe, 3(4), 213–23. 10.1016/j.chom.2008.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, & Gordon JI (2009). The core gut microbiome, energy balance and obesity. The Journal of Physiology, 587(Pt 17), 4153–8. 10.1113/jphysiol.2009.174136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, … Gordon JI (2009). A core gut microbiome in obese and lean twins. Nature, 457(7228), 480–4. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald M. Magrini V, Mardis ER, & Gordon JI (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 444(7122), 1027–31. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009). ggplot2: Elegant Graphics for Data Analysys. Springer. [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-YY-Y, Keilbaugh SA, … Lewis JD (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science (New York, N.Y.), 334(6052), 105–8. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, … Hsiao EY (2015). Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell, 161(2), 264–276. 10.1016/j.cell.2015.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, … Gordon JI (2012). Human gut microbiome viewed across age and geography. Nature, 486(7402), 222–7. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Xiao X, Zhang Q, Mao L, Yu M, Xu J, & Wang T. (2017). The Placental Microbiota Is Altered among Subjects with Gestational Diabetes Mellitus: A Pilot Study. Frontiers in Physiology, 8 10.3389/fphys.2017.00675 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Beta diversity metrics were rarefied to 19,964 (75% of minimum sample read count) and measured by Binary Jaccard (unweighted metric) using multidimensional scaling (MDS). (A) Maternal diet (p=0.002 by PERMANOVA) and (B) age (p=0.001 by PERMANOVA) significantly structured the gut microbiome. Orange: maternal high-fat diet, dark blue, maternal control diet, light blue: 6-month juvenile, gold: 13-month juvenile, charcoal: 36-month juvenile, and red: maternal.
Alpha diversity metrics were rarefied to 19,944 (75% of minimum sample read count) and measured by Chao1 (A), Observed ASVs (B), Shannon Diversity (C), and Simpson’s Diversity (D). Significant differences between age groups by Kruskal-Wallis with Dunn’s Correction were seen with Chao1 and Observed ASVs (p<0.005) but not with Shannon or Simpson’s Diversity. Additionally, significant differences were observed by diet (p<0.05, Mann-Whitney) with Chao1, Observed ASVs, and Shannon. All alpha diversity metrics demonstrated significance (p<0.0001, Mann-Whitney) when comparing enterotypes as identified by DMM modeling.
Exposure to a high fat maternal diet is associated with increased taxonomic differences between high and low Treponema clusters. LEfSe was utilized to examine taxonomic differences within clusters identified by DMM after stratification by dietary exposure. We found that primates only exposed to a control diet had fewer genera with differential abundance (A) when compared to primates exposed to a high fat diet (B).
Data Availability Statement
The 16S amplicon-based metagenomic sequence data generated from this analysis has been deposited in the Sequence Read Archive (SRA) under bioproject ID PRJNA508806.