 We have employed glass nanopore as a single molecule technique for direct sensing amyloidosis process of Aβ1–42 peptide, which of great significance in Alzheimer's disease.
We have employed glass nanopore as a single molecule technique for direct sensing amyloidosis process of Aβ1–42 peptide, which of great significance in Alzheimer's disease.
Abstract
We have developed a glass nanopore based single molecule tool to investigate the dynamic oligomerization and aggregation process of Aβ1–42 peptides. The intrinsic differences in the molecular size and surface charge of amyloid aggregated states could be distinguished through single molecule induced characteristic current fluctuation. More importantly, our results reveal that the neurotoxic Aβ1–42 oligomer tends to adsorb onto the solid surface of nanopores, which may explain its instability and highly neurotoxic features.
Alzheimer's disease (AD) is one of the most common types of neurodegenerative disease, affecting more than 50 million people all over the world.1 This disease causes specific pathological changes in the brain prior to cognitive impairment, usually with more than 10 years of a preclinical phase.2 One of the major pathological features of AD is the formation of neuritic plaques in the cerebral cortices, which mainly consist of amyloid β (Aβ) peptide of 39–43 amino acids in length.3,4 The Aβ peptides are generated from sequential proteolytic cleavage of amyloid precursor protein (APP) by using enzymes β-secretase and γ-secretase.5 Among them, Aβ1–42 and Aβ1–40 are the two major species of Aβ production, and the former species predominates in the neurotoxicity of neuritic plaques. It is known that soluble oligomers, not soluble monomers or insoluble fibrils, would disrupt long-term potentiation and directly induce cognitive impairment.6 Thus the oligomerization process of the Aβ1–42 peptide plays a significant role in AD development. Many techniques have been employed for the investigation of pathogenic oligomers, such as nuclear magnetic resonance (NMR) spectroscopy, transmission electron microscopy (TEM) and single-touch atomic force microscopy (AFM).7–9 It has been demonstrated that the large population of oligomers consists of Aβ1–42 pentamers/hexamers, with disc-shaped morphology of 10–15 nm width and 1.5–2.5 height, and a smaller proportion of pentamers/decamers with the same width and 3–5 nm height.10 However these methods could hardly monitor the dynamic aggregation of the Aβ1–42 peptide in aqueous solution at the single molecule level, much less the heterogeneity of small oligomers.
Nanopore technology has become a well-developed approach for single molecule sensing by providing detailed intrinsic molecular information regarding the geometric structure, surface charge and even the conformational properties.11 In general, biological nanopores are superior in the sensing of oligonucleotides, peptides, organic molecules and other small molecules, but not proteins and other large sized entities, owing to the 1–2 nm narrowest part of the pore lumen.12,13 Attributed to the size tunable features and easy fabrication process, glass nanopore sensors have broadened the application for single entity analysis.14–16 Traditional glass nanopore sensors are mainly based on the volume exclusion mechanism, which can be briefly described as the transient decrease of pore conductance resulting from the single entity traversing through the nanopore, exhibiting pulse like blockage signals on the ionic current.17 Under this mechanism, it is challenging to capture small molecules in a fairly large-scale pore due to the negligible volume exclusion effect.18 In recent years, researchers have identified that both electrophoresis and electroosmosis contribute greatly in charged nanopore sensing for proteins/peptides, which typically have complex tertiary structures and non-uniform surface charge.19,20 It has been suggested that the electroosmotic force may contradict the electrophoretic force and prevail in nanopore sensing by elaborately adjusting the surface charge of nanopores and analytes.21 Besides, the surface charge of the analyte could induce ion accumulation, which competes with the volume exclusion effect, resulting in the increase of ionic current. Thus many factors could be regulated in protein molecule sensing via glass nanopore sensors, indicating a broad and subtle approach for single protein analysis.
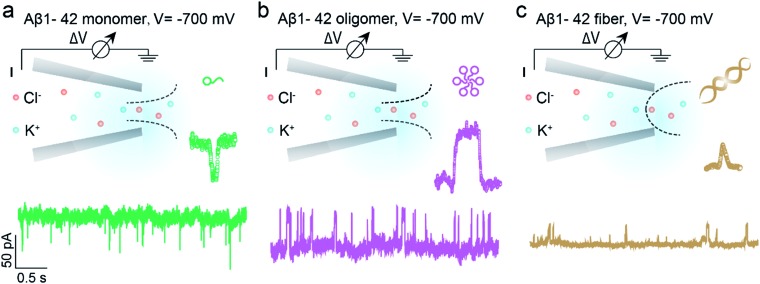
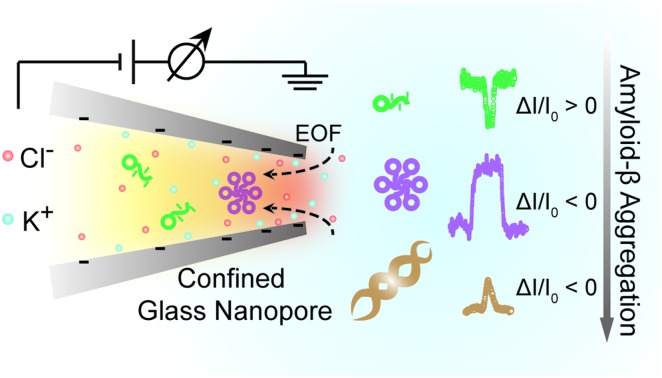
Benefitting from the advantages of single molecule characterization, nanopore-based analytics has been employed for the sensing of individual amyloid particles22 such as α synuclein,23,24 lysozyme,25,26 amyloid β,27–29 etc. For example, solid state nanopores were used to enable the translocation of amyloid β aggregates and further measure the size of individual oligomers in solution without chemical modification.29,30 The denatured lysozyme and its amyloid could also be discriminated with silicon nitride nanopores. In recent years, the morphology of protein aggregates could be discriminated by polymer coated conical nanopores,26,27 which further facilitated the investigation of amyloid growth, inhibition and enzymatic degradation.31 Moreover, under appropriate salt and pH conditions, bare quartz nanopores could be used for amyloid sensing based on resistive-pulse analysis.32 Herein, we propose a unique sensing method based on bare glass nanopores to achieve single amyloid molecule discrimination. In this process, both electrophoretic and electroosmotic forces contribute to the ionic current fluctuation. As shown in Fig. 1, three types of Aβ peptides including monomers (green), oligomers (purple) and fibres (orange) could be directly detected in aqueous solution under a biased voltage. And the differences in size and surface charge density of Aβ peptides would induce characteristic current fluctuations, exhibiting heterogeneity at the single molecule level. Furthermore, we proposed single molecule evidence for the toxic Aβ oligomer' hypothesis that the loosely packed structure of oligomers makes it easy to be adsorbed onto a solid surface.
Fig. 1. Schematic representation of single molecule monitoring of three types of Aβ1–42 peptides with glass nanopore sensors. Aβ1–42 monomers (a, green), oligomers (b, purple) and fibres (c, orange) could be distinguished in real time through the characteristic current fluctuations. The baseline currents in the nanopore sensing of Aβ1–42 monomers, oligomers and fibres are approximately –950 pA, –1670 pA and –1040 pA, which are obtained through three independent nanopores.
The prepared glass nanopore was first characterized by SEM imaging which shows a tip diameter of approximately 30 nm (Fig. S1†). A pair of Ag/AgCl electrodes was employed to apply a biased potential and the electrode fixed outside the nanopore was grounded. Aβ1–42 peptides in different aggregated states were added to the external buffer solution with three independent nanopores. The detailed preparation and characterization of Aβ1–42 peptides are described in the ESI† (Fig. S2 and S3†). Results in the bottom of Fig. 1 show the different pulse-like signals of Aβ1–42 peptides in the current traces under –700 mV; no obvious current fluctuation was observed under positive voltages. The current signals pointing downwards represented the current enhancement and those pointing upwards represented the current blockage under a negative biased potential. This indicated that the Aβ1–42 peptides were driven into the nanopore under negative potentials. Considering the negatively charged glass nanopore wall and the low ionic strength solutions (10 mM), the electroosmotic force would serve as a predominant force in our peptide sensing.33 Herein, using a low salt concentration solution would have the following advantages. On the one hand, alkaline solution with a low salt concentration could prevent further aggregation of Aβ1–42 peptides, stabilizing the peptide molecules during the nanopore sensing process. On the other hand, during the traversing of the negatively charged Aβ1–42 peptide through the nanopore orifice, both the volume exclusion and charge effect would induce current fluctuation. Therefore, apart from the general blockage of ionic current with Aβ1–42 oligomers (Fig. 1b, purple traces) and fibres (Fig. 1c, orange traces), the translocation of Aβ1–42 monomers through the nanopore could be captured from the transient current increase (Fig. 1a, green traces). We recognized such an observation to be caused by ion accumulation during the translocation of the monomer peptide. In detail, the surface charges carried by the traversing Aβ1–42 peptide would result in the redistribution of ions in the pore orifice, thereby increasing the ionic current. Meanwhile, the traversing peptide for volume occupation in glass nanopores would effectively reduce the transient conductance, resulting in ionic current blockages. In this case, Aβ1–42 monomers were highly negatively charged while having a small size, and notably, the ion accumulation effect serves as a dominating force during monomer sensing by the glass nanopore sensor. For the Aβ1–42 oligomer and fibre sensing, the volume exclusion effect was the primary effect and thus exhibited current blockage as shown in Fig. 1b and c.
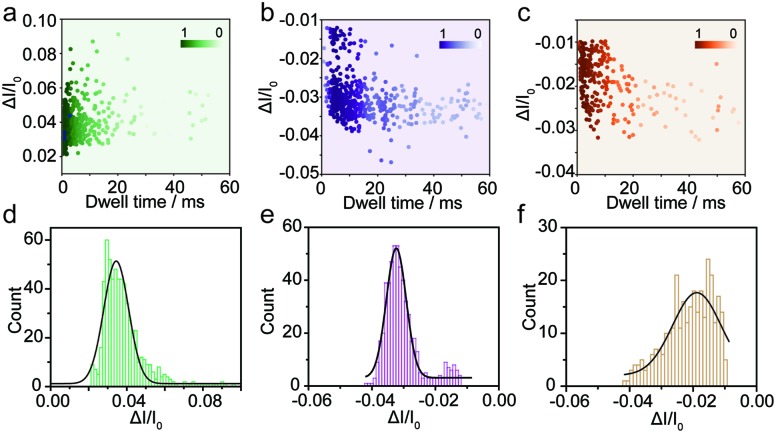
The spike-like signals of transient current were statistically analyzed under –700 mV. We use ΔI/I0 to denote the current amplitude, where ΔI and I0 represent the current fluctuation amplitude and the baseline current, respectively. Fig. 2a demonstrates the event scatter plot of monomer translocation which correlates the current amplitude versus peak dwell time. And the current amplitude (ΔI/I0) histogram was plotted and fitted with a single peak probability distribution of 0.034 (Fig. 2d). Given the short time scale of the event dwell time and the current increase distribution, the Aβ1–42 monomers were driven inward across the nanopore demonstrating single molecular behavior in aqueous solution. The translocation events of Aβ1–42 oligomers were statistically analyzed and are shown in Fig. 2b and e. The scatter plot of ΔI/I0versus dwell time and histograms indicate a fitted current amplitude of –0.032. Positive and negative values of ΔI/I0 represented ionic current enhancement and blockage respectively. Finite element method (FEM) simulation was employed to verify the ionic accumulation and blockage induced by Aβ1–42 monomers and oligomers respectively (Fig. S4†). Moreover, Fig. 2e shows the ΔI/I0 fitted well with a narrow Gaussian distribution, indicating that the Aβ1–42 oligomers were largely in a uniform geometry with comparable size, rather than with various sizes.34 Only a small portion of blockage under 0.02 may be due to the existence of small oligomers. Nevertheless, the Aβ1–42 fibre usually with μm or sub-μm long could hardly translocate through a 30 nm diameter nanopore. There are blockages in the ionic current baseline with a lower current amplitude (Fig. 1c). This would be generated by the collision events of Aβ1–42 fibres onto the nanopore orifice, which has been reported for large particle sensing with small pores.35 The statistical analysis is shown in Fig. 2c, with Fig. 2f representing a low current blockage of –0.019, which is typical in collision events. Although the fibre could hardly translocate across the nanopore due to the sub-μm morphology, it could move towards the orifice under the electroosmotic flow, blocking the ionic current in a short time scale. The collision events of Aβ1–42 fibres onto the nanopore orifice resulted in a dwell time of 6.32 ms to be longer than the direct translocation of monomers (Fig. S5†). The interactions between protein aggregates and the nanopore surface25 may prolong the dwell time as shown in Fig. S5.†
Fig. 2. Monitoring the three types of Aβ1–42 peptides using the glass nanopore sensors. Event scatter plots of ΔI/I0versus dwell time for the translocation of Aβ1–42 monomers (a), oligomers (b) and fibres (c) under –700 mV. The numbers of total events in each peptide sensing are 624, 554 and 314 respectively. Histograms showing the ΔI/I0 distributions of the measurement of Aβ1–42 monomers (d), oligomers (e) and fibres (f).
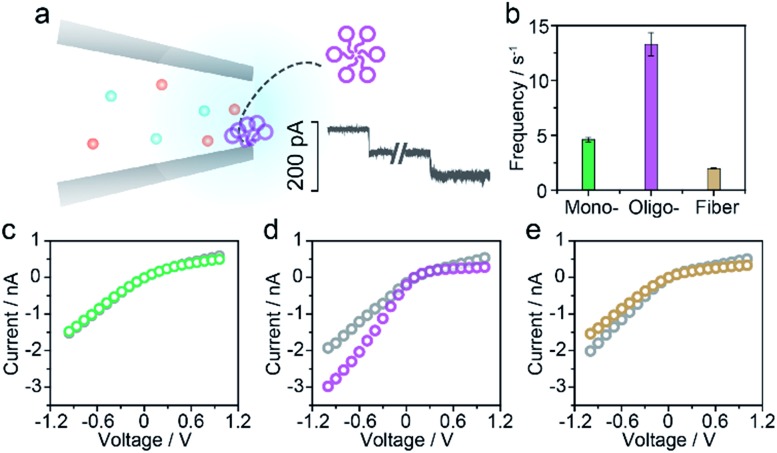
For the nanopore sensing of Aβ1–42 oligomers, a staircase-like current increase was observed (Fig. 3a) besides the spike-like blockages mentioned above. In addition, the frequency of spike-like signals of the Aβ1–42 oligomer was much higher than that of the Aβ1–42 monomer and fibres as demonstrated in Fig. 3b. Considering that the molar concentration of oligomers was not as high as that of monomers, the frequency of these three kinds of peptides was not mainly dominated by analyte concentration. I–V curves were then recorded and indicated the increase of negative charges on the nanopore sidewall, resulting in a stronger electroosmotic force in oligomer sensing. Glass nanopores with parallel conductance were employed for the sensing of three states of Aβ1–42 peptides and the experimental setup was the same. I–V curves were first recorded by filling and immersing the nanopore into a pure electrolyte (gray traces). The Aβ1–42 containing electrolyte was subsequently used to replace the external solution with continuous recording of the ionic current. After peptide sensing, the inside and outside solutions of glass nanopores were both substituted with the pure electrolyte. I–V curves were re-recorded and showed great difference between Aβ1–42 monomers (Fig. 3c), oligomers (Fig. 3d) and fibres (Fig. 3e). The adsorption of protein aggregates onto the nanopore has been investigated25 and great effort has been made to prevent nonspecific adsorption.26,29–31 Herein, only the Aβ1–42 oligomers showed an obvious adsorption, which could be confirmed by the increase in the ion current rectification ratio. The dynamic adsorption of oligomers onto the nanopore side would regulate the electroosmosis effect, which would enhance the frequency and provide detailed single molecular information. As previously studied, the ring-shaped Aβ1–42 oligomers could be stabilized at a low salt concentration, and their C-terminal region was loosely aggregated inside the ring.6 We hypothesized that hydrophobic C-terminals would adsorb onto the inner wall of the nanopore, exposing the hydrophilic negatively charged N-terminal outwards. Thus the inner wall of the nanopore features an increase in negative charges which contributes to the temporary staircase increase in the baseline ionic current. The accumulation of negative charges on the nanopore could enhance the electroosmotic force and increase the possibility of translocation of the Aβ1–42 oligomer greatly. It is widely known that the Aβ1–42 oligomer is a key factor in neurotoxicity in Alzheimer's disease, inhibiting synaptic plasticity and altering cell surface receptors. According to the presented findings, we would speculate that the adsorbent feature of oligomers may account for their instability and toxic mechanism of inhibiting synaptic plasticity and altering cell surface receptors.
Fig. 3. Aβ1–42 oligomer sensing with glass nanopores. (a) The typical step like current fluctuations induced by the adsorption of Aβ1–42 oligomers onto the nanopore sidewall. (b) Frequencies of the pulse-like signals for the sensing of three types of Aβ1–42 peptides. I–V curves of the glass nanopore measured in the pure electrolyte before (gray) and after Aβ1–42 monomer (c), oligomer (d) and fibre (e) sensing, respectively.
In summary, we employed glass nanopores as a single molecule technique for direct sensing of the amyloidosis process of the Aβ1–42 peptide, which is of great significance in Alzheimer's disease. The distinct current fluctuations in sensing Aβ1–42 monomers, oligomers and fibres were acquired to better understand molecular heterogeneity. Further analysis of the rectification of glass nanopores showed that the Aβ1–42 oligomers could be more easily adsorbed on the nanopore surface and we have proposed a potential adsorption mechanism. Our research shed light on the adsorption effect on the high neurotoxicity of Aβ1–42 oligomers, which would benefit the fundamental understanding of the pathogenesis of Alzheimer's disease.
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
This research was sponsored by the National Natural Science Foundation of China (61871183, 21922405, 21834001), the Shanghai Collaborative Innovation Center for Translational Medicine (TM201808), the Shanghai Sailing Program (19YF1411200) and the China Postdoctoral Science Foundation (2018M640349).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc03260f
References
- Yang X., Meisl G., Frohm B., Thulin E., Knowles T. P. J., Linse S. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E5849–E5858. doi: 10.1073/pnas.1803539115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R. A., Aisen P. S., Beckett L. A., Bennett D. A., Craft S., Fagan A. M., Iwatsubo T., Jack C. R., Kaye J., Montine T. J., Park D. C., Reiman E. M., Rowe C. C., Siemers E., Stern Y., Yaffe K., Carrillo M. C., Thies B., Morrison-Bogorad M., Wagster M. V., Phelps C. H. Alzheimer's Dement. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesné S., Koh M. T., Kotilinek L., Kayed R., Glabe C. G., Yang A., Gallagher M., Ashe K. H. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- Jack Jr C. R., Knopman D. S., Jagust W. J., Shaw L. M., Aisen P. S., Weiner M. W., Petersen R. C., Trojanowski J. Q. Lancet Neurol. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe D. J., Hardy J. EMBO Mol. Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyubin I., Walsh D. M., Lemere C. A., Cullen W. K., Shankar G. M., Betts V., Spooner E. T., Jiang L., Anwyl R., Selkoe D. J., Rowan M. J. Nat. Med. 2005;11:556–561. doi: 10.1038/nm1234. [DOI] [PubMed] [Google Scholar]
- Bitan G., Fradinger E. A., Spring S. M., Teplow D. B. Amyloid. 2005;12:88–95. doi: 10.1080/13506120500106958. [DOI] [PubMed] [Google Scholar]
- Lee S. J. C., Nam E., Lee H. J., Savelieff M. G., Lim M. H. Chem. Soc. Rev. 2017;46:310–323. doi: 10.1039/c6cs00731g. [DOI] [PubMed] [Google Scholar]
- Ma X., Liu L., Mao X., Niu L., Deng K., Wu W., Li Y., Yang Y., Wang C. J. Mol. Biol. 2009;388:894–901. doi: 10.1016/j.jmb.2009.03.054. [DOI] [PubMed] [Google Scholar]
- Ahmed M., Davis J., Aucoin D., Sato T., Ahuja S., Aimoto S., Elliott J. I., Van Nostrand W. E., Smith S. O. Nat. Struct. Mol. Biol. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y.-L., Gao R., Hu Y.-X., Long Y.-T. Small Methods. 2018:1700390. [Google Scholar]
- Cao C., Long Y. Acc. Chem. Res. 2018;51:331–341. doi: 10.1021/acs.accounts.7b00143. [DOI] [PubMed] [Google Scholar]
- Ying Y., Cao C., Hu Y., Long Y. Natl. Sci. Rev. 2018;5:450–452. [Google Scholar]
- Yu R.-J., Ying Y.-L., Gao R., Long Y.-T. Angew. Chem., Int. Ed. 2019;58:3706–3714. doi: 10.1002/anie.201803229. [DOI] [PubMed] [Google Scholar]
- Morris C. A., Friedman A. K., Baker L. A. Analyst. 2010;135:2190–2202. doi: 10.1039/c0an00156b. [DOI] [PubMed] [Google Scholar]
- Sha J., Si W., Xu W., Zou Y., Chen Y. Sci. China, Ser. E: Technol. Sci. 2015;58:803–812. [Google Scholar]
- Wang Y., Wang D., V Mirkin M. Proc. R. Soc. London, Ser. A. 2017;473:20160931. doi: 10.1098/rspa.2016.0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhanek M., Kemp J. T., Pourmand N., Davis R. W., Webb C. D. Nano Lett. 2005;5:403–407. doi: 10.1021/nl0480464. [DOI] [PubMed] [Google Scholar]
- Lan W. J., Kubeil C., Xiong J. W., Bund A., White H. S. J. Phys. Chem. C. 2014;118:2726–2734. [Google Scholar]
- Soskine M., Wloka C., Maglia G. Nat. Commun. 2017:1–13. doi: 10.1038/s41467-017-01006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firnkes M., Pedone D., Knezevic J., Döblinger M., Rant U. Nano Lett. 2010;10:2162–2167. doi: 10.1021/nl100861c. [DOI] [PubMed] [Google Scholar]
- Houghtaling J., List J., Mayer M. Small. 2018;14:1802412. doi: 10.1002/smll.201802412. [DOI] [PubMed] [Google Scholar]
- Wang H. Y., Gu Z., Cao C., Wang J., Long Y. T. Anal. Chem. 2013;85:8254–8261. doi: 10.1021/ac401496x. [DOI] [PubMed] [Google Scholar]
- Hu R., Diao J., Li J., Tang Z., Li X., Leitz J., Long J., Liu J., Yu D., Zhao Q. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep20776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balme S., Coulon P. E., Lepoitevin M., Charlot B., Yandrapalli N., Favard C., Muriaux D., Bechelany M., Janot J. M. Langmuir. 2016;32:8916–8925. doi: 10.1021/acs.langmuir.6b02048. [DOI] [PubMed] [Google Scholar]
- Giamblanco N., Coglitore D., Janot J. M., Coulon P. E., Charlot B., Balme S. Sens. Actuators, B. 2018;260:736–745. [Google Scholar]
- Wang H. Y., Ying Y. L., Li Y., Kraatz H. B., Long Y. T. Anal. Chem. 2011;83:1746–1752. doi: 10.1021/ac1029874. [DOI] [PubMed] [Google Scholar]
- Hu Y., Ying Y., Gu Z., Cao C., Yan B., Wang H.-F., Long Y.-T. Chem. Commun. 2016;52:5542–5545. doi: 10.1039/c6cc01292b. [DOI] [PubMed] [Google Scholar]
- Yusko E. C., Prangkio P., Sept D., Rollings R. C., Li J., Mayer M. ACS Nano. 2012;6:5909–5919. doi: 10.1021/nn300542q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusko E. C., Johnson J. M., Majd S., Prangkio P., Rollings R. C., Li J., Yang J., Mayer M. Nat. Nanotechnol. 2011;6:253–260. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamblanco N., Coglitore D., Gubbiotti A., Ma T., Balanzat E., Janot J. M., Chinappi M., Balme S. Anal. Chem. 2018;90:12900–12908. doi: 10.1021/acs.analchem.8b03523. [DOI] [PubMed] [Google Scholar]
- Martyushenko N., Bell N. A. W., Lamboll R. D., Keyser U. F. Analyst. 2015;140:4882–4886. doi: 10.1039/c5an00530b. [DOI] [PubMed] [Google Scholar]
- Kuo T. C., Sloan L. A., Sweedler J. V., Bohn P. W. Langmuir. 2001;17:6298–6303. [Google Scholar]
- Hu R., Rodrigues J. V., Waduge P., Yamazaki H., Cressiot B., Chishti Y., Makowski L., Yu D., Shakhnovich E., Zhao Q., Wanunu M. ACS Nano. 2018;12:4494–4502. doi: 10.1021/acsnano.8b00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., He X., Zhang K., Wang K., Yu P., Mao L. Chem. Sci. 2016:1–3. doi: 10.1039/c6sc02241c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.