Abstract
Emodin is a Chinese herb-derived compound that exhibits a variety of pharmacological benefits. Although emodin has been shown to inhibit growth of cancer cells, its antineoplastic function is incompletely understood. CD155 is a member of poliovirus receptor–related (PRR) family of adhesion molecules; it is constitutively expressed on many tumor cell lines and tissues and has diverse functions. CD155 has been reported to mediate activation of T cells via CD226 or inhibition of T cells via T-cell immunoreceptor with Ig and ITIM domains (TIGIT). In addition, CD155 may play a critical role through non-immunological mechanisms in cancer. In this study, we tested the ability of emodin to modulate CD155 expression in cancer cells. We found that emodin significantly decreased the expression of CD155 in tumor cells and inhibited tumor cell proliferation and migration, and induced cell-cycle arrest at G2/M phase. The tumor inhibitory effects of emodin were lost with CD155 knockdown. Furthermore, emodin was used to treat mice bearing B16 melanoma. It was shown that emodin attenuated tumor growth accompanied by suppressing CD155 expression. Therefore, we propose that emodin could inhibit tumor growth, and the antineoplastic properties of emodin are at least partially CD155 dependent. Our study provides new insights into the mechanisms by which emodin inhibits tumor growth.
Keywords: Emodin, adhesion molecule, CD155, tumor, herb-derived compound
INTRODUCTION
Adhesion molecules are transmembrane proteins responsible for a variety of biological signaling in cell regulation. Many studies have documented that alterations in expression of adhesion molecules correlate with the growth of primary or metastatic tumors. The adhesion molecule CD155, also called PVR or necl-5, is broadly distributed on epithelial and endothelial cells in many tissues [1]. Notably, CD155 is overexpressed on various tumors, including colorectal cancer [2], gastric cancer [3], ovarian cancers [4], neuroblastoma [5], myeloid leukemias [6], multiple myeloma [7] and melanoma [8]. Interactions between CD155 on tumor cells and CD226 on NK or T cells augment cell-mediated cytotoxicity and cytokine production [9–11]. However, TIGIT competes with CD226 for binding to CD155 and exerts potent inhibitory action in various subsets of immune cells [12–14]. In addition, CD155 is localized in the cell–matrix and cell–cell junctions, and it could inhibit cell adhesion and enhance cell migration [15]. Knockdown of CD155 resulted in markedly decreased invasion of glioblastoma [16]. Therefore, CD155 may be an attractive target for cancer therapy.
Emodin is a naturally occurring anthraquinone derivative that is found in several Chinese herbs, particularly in their roots and barks. These herbs have been wildly used as traditional medicines in many countries, especially in eastern Asia [17]. Emodin has been shown to possess a wide spectrum of pharmacological effects, such as anti-tumor, anti-inflammatory, antiviral, antibacterial, anti-allergic, anti-osteoporotic, anti-diabetic, immunosuppressive, neuroprotective and hepatoprotective activities [17]. Currently, a number of researchers are investigating its anti-tumor effects [18]. It has been shown to affect many different tumor cell lines and inhibit proliferation of leukemia, breast, colon, and lung carcinoma cells [19, 20].However, the molecular targets of emodin in tumor cells are elusive, and thus the mechanisms of its inhibitory effects on tumors are not completely understood.
Our laboratory has previously shown that emodin inhibited the growth and metastasis of breast tumors through inhibition of the macrophage-tumor cell feedforward tumor-promoting loop and macrophage M2-like polarization [21, 22]. In this study, we demonstrated that emodin may also inhibit tumor growth via downregulation of CD155 in cancer cells. We provide new insights into the mechanisms by which emodin inhibits tumor growth.
MATERIALS AND METHODS
Reagents
Emodin, a trihydroxy-anthraquinone, was purchased from Nanjing Langze Medicine and Technology Co. Ltd. and verified by NMR spectroscopy and mass spectrometry as we previously described [23].
Cell culture and siRNA transfection
B16-F10 melanoma and 4T1 cell lines were obtained from the American Type Culture Collection (ATCC, Mana ssas, VA). EO771 cells, developed from an ER+ spontaneous mammary adenocarcinoma [24, 25], were maintained in culture as previously described [22]. The cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM , Invitrogen , Gran d Island, NY) supplemented with 10% fetal bovine serum (FBS, Invitrogen ) and a combination of penicillin/streptomycin at 37°C in a humidified 5% CO2 atmosphere. Mouse CD155 siRNA and control siRNA (Qiagen, USA) were transfected into B16-F10, 4T1 and EO771 cells in 24-well plates using Lipofectamine 3000 (Invitrogen, CA, USA), according to the manufacturer’s protocol.
Flow cytometric analysis (FACS)
Cells were stained with anti-CD155 PE mAb, anti-CD3 APC mAb, and anti-NK1.1 APC mAb (all from eBioscience, San Diego, CA) in staining buffer (PBS containing 2% FBS) for 30 min on ice in the dark. Samples were washed twice with staining buffer, the mean fluorescence intensity (MFI) was analyzed by flow cytometry using a FACSAria flow cytometer and the FCS Express 4 software (De Novo Software, Glendale, CA, USA)
Cell isolation
Tumors were weighed, cut into small fragments (<3 mm) and digested in 5 ml of dissociation solution (RPMI 1640 medium supplemented with 10% FBS, Collagenase type I (200 U/ml) and DNase I (100 μg/ml)) for 60 min at 37°C. Erythrocytes were lysed with red blood cell lysing buffer (Sigma, St. Louis, MO). Cell suspensions were passed through 70-μm cell strainers, then washed and resuspended in staining buffer.
Quantitative real-time PCR (qPCR)
Total RNA was extracted using the TRIzol reagent (Invitrogen). RNA (1μg) was reverse-transcribed using iScript cDNA Synthesis Kit (Bio-Rad, Life Science). qPCR was conducted on a CFX96 system (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). All primers used for qPCR analysis were synthesized by Integrated DNA Technologies. All assays were conducted following the manufacturer’s instructions. The relative amount of target mRNA was determined using the comparative threshold (Ct) method by normalizing target mRNA Ct values to those of 18S RNA. PCR thermal cycling conditions were 3 min at 95°C, and 40 cycles of 15 s at 95°C and 58 s at 60°C. Samples were run in triplicate.
Wound healing assay
B16-F10, 4T1 cells and EO771 cells were seeded in 24-well plates and cultured until 70–80% confluent. A straight scratch was made using a pipette tip to form an artificial wound. The cells were treated with emodin and/or siRNA transfection, and the migration of cells across this artificial wound was assessed.
Proliferation and cell cycle analysis
B16-F10, 4T1 cells and EO771 cells (50%–60% confluent) were synchronized for overnight with the methods of serum starvation, and were treated with emodin (20 or 50 μM) for 24 h. Proliferation was evaluated by counting relative number of living cells after 24 h of 20 and 50μM emodin treatment, and trypan blue staining was used to count the alive cells. After washed twice with chilled PBS, cell pellet was resuspended in 150 μL cold PBS to which cold ethanol (450 μL) was added, then were incubated for 12 h at 4°C. After centrifuged at 1,000 rpm for 5 min, the pellet was washed twice with chilled PBS, and incubated with 100 μL RNase A (20 μg/mL) at 37°C for 30 min. The cells were then cooled down on ice for 10 min and incubated with 400 μL PI (50 μg/mL) for 30 min in the dark and were analyzed by flow cytometry. Data were further analyzed by using ModiFitLT software (Verity Software House) for cell cycle analysis.
Tumor models
C57BL/6 (8–12 weeks, female) were purchased from The Jackson Laboratory. They were housed at the University of South Carolina Animal Research Facility, and all procedures were approved by the Institutional Animal Care and Use Committee. To establish subcutaneous tumors in mice, 5×106 B16-F10 cells in 200 μl of PBS were implanted into the rear flanks of mice. Starting on Day 1, emodin (40 mg/kg) or vehicle (1% DMSO) was injected intraperitoneally in 1 ml PBS once daily. Tumor growth was monitored by measurement of tumor size with a caliper every other day. Tumor volume was determined by the formula: length×width2/2. At the experimental end point (15 days), mice were sacrificed, and tumors were removed, weighed and processed for FACS analysis.
Statistical analysis
Data were presented as mean ± SEM as indicated. Statistical significance was calculated using the Students’ t test (two-group comparison) or one-way ANOVA followed by post hoc Dunnett test (multi-group comparison) using the GraphPad Prism statistical program (GraphPad Prism; GraphPad Software, Inc.). P < 0.05 was considered significant.
RESULTS
Emodin inhibits CD155 expression in multiple cancer cell lines
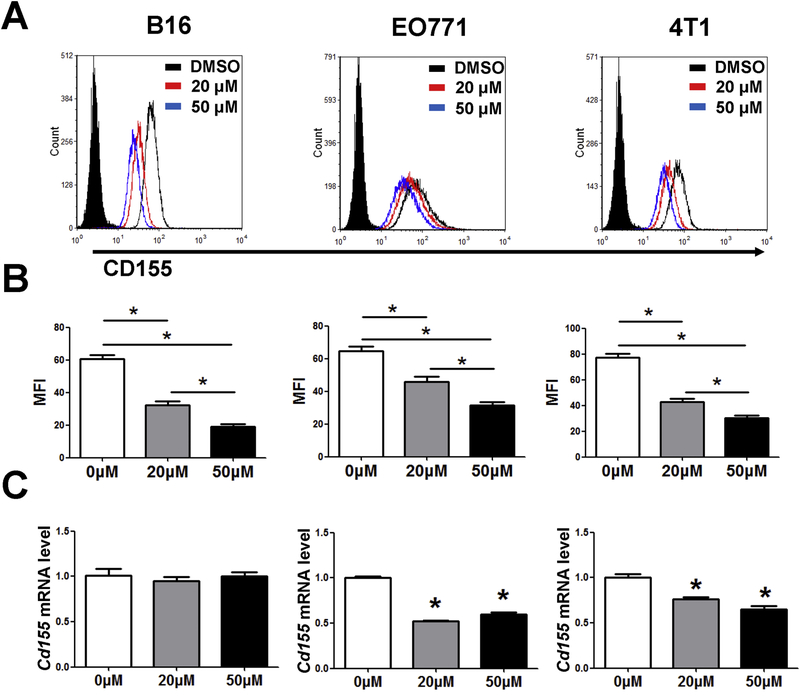
The alterations in expression of adhesion molecules have been shown to correlate with the growth of primary and metastatic tumors [26]. We hypothesized that emodin might regulate the expression of adhesion molecules in cancer cells, which may at least partially contribute to the tumor inhibitory effects of emodin observed in our previous studies [21, 22]. We examined the expression of CD155 in mouse B16-F10 melanoma, EO771 and 4T1 breast cancer cells. The results showed that CD155 expression in the cells treated with emodin was significantly lower than that in control group (Fig. 1A and B). The results also showed that both emodin treatment at 20 μM and 50 μM decreased CD155 mRNA in EO771 and 4T1 cells, but had no effect on the expression of CD155 mRNA in B16-F10 cells (Fig. 1C).
Figure 1.
Emodin decreases the expression of CD155 in cancer cells. (A) Emodin inhibited CD155 expression in B16-F10, 4T1 and EO771 cells. Cells were treated with or without 20 or 50 μM emodin for 24 h and then analyzed using flow cytometry. (B) Quantification of MFI is shown. *p<0.05. (C) Cells were treated with or without 20 or 50 μM emodin for 24h, and CD155 expression was analyzed by qPCR. Data were presented as means ± SEM of one of three independent experiments; n=3. *p<0.05 vs Control.
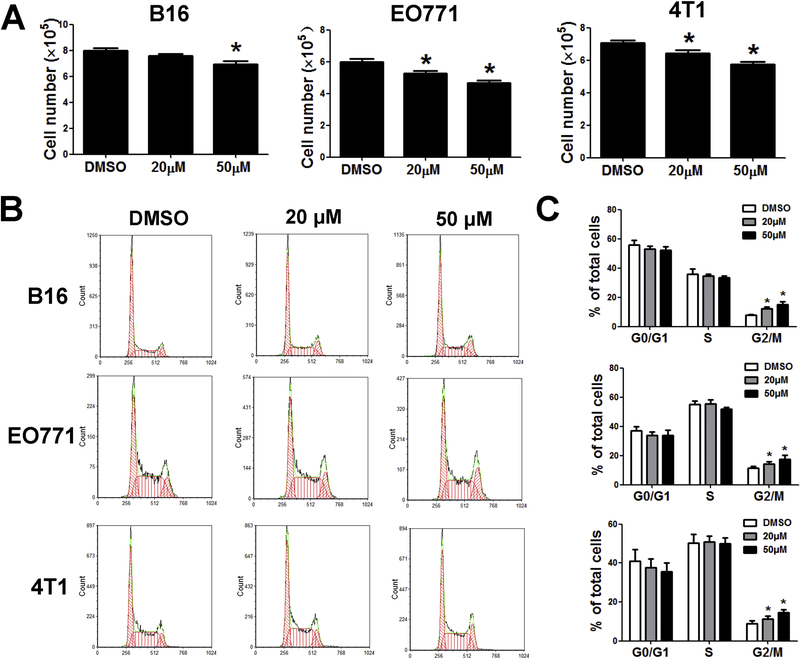
Emodin suppresses cell proliferation and induces G2/M-phase arrest in cancer cells
Emodin has been shown to have detrimental effects on tumor cells in culture; we thus evaluated the response of cancer cell lines to emodin. Figure 2A showed that emodin at 50 μM significantly reduced proliferation of B16, EO771 and 4T1 cells by 10–20%, while at 20 μM it only reduced proliferation of EO771 and 4T1. Cell cycle progression plays an important role in proliferation of cancer cells. Thus, we investigated cell phases of cancer cell lines in order to determine whether the inhibition of emodin on tumor cell proliferation was mediated by dysregulation of cell cycle. Figure 2B showed the changes in the cell cycle of cancer cells induced by emodin. The proportion of cancer cells in the G2/M-phase of the cell cycle was increased significantly by emodin in a dose-dependent manner as compared with the untreated cells (Fig. 2C). Our data demonstrated that emodin causes G2/M-phase arrest in the cell cycle of cancer cells.
Figure 2.
Emodin suppressed cell proliferation and induced G2/M-phase arrest. (A) Relative number of cells after 24 h of 20 and 50μM emodin treatment. Living cells were counted and compared with controls. Data were presented as means ± SEM of one of three independent experiments; n=3. *p<0.05 vs Control. (B) Emodin induced G2/M-phase arrest in tumor cells. The cell cycle distribution was monitored by flow cytometry. (C) Cell cycle results as means±SEM of one of three independent experiments; n=3. *p<0.05 vs Control.
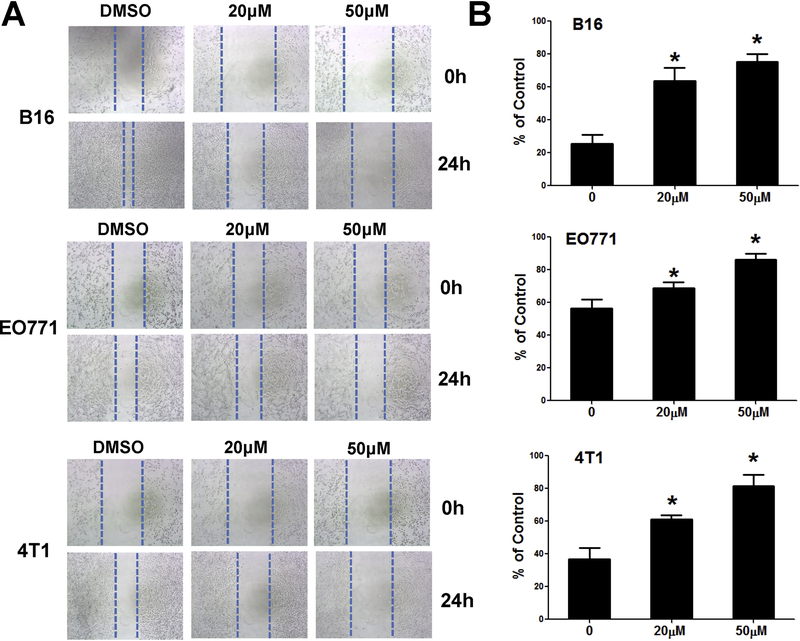
Emodin inhibits the migration of cancer cells
To examine the effects of emodin on tumor cell migration, cancer cells were treated with emodin. The motilities of B16-F10, EO771, and 4T1 cells were eamined using wound-healing assays. At 24 h after treatment, the wound in the emodin group had healed less compared to that in the control group (Fig. 3).
Figure 3.
Emodin inhibits migration of cancer cells. Effects of emodin on cancer cell migration were examined by a wound-healing assay. Representative images (A) and quantification of migration distance (B) are shown. Data were presented as means ± SEM of one of three independent experiments; n=3. *p<0.05 vs Control.
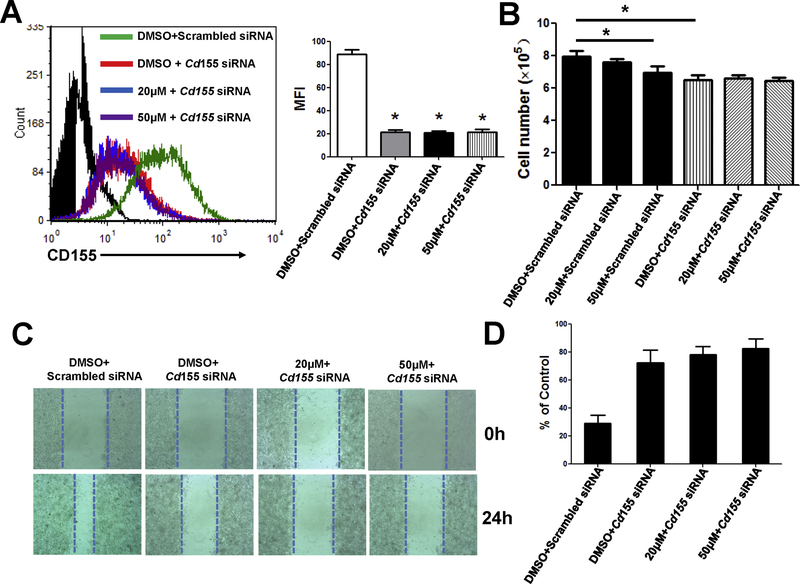
CD155 is involved in emodin-mediated tumor-suppressive effects
To examine the involvement of CD155 in anticancer activity of emodin, we transfected B16-F10 cells with CD155 small interfering RNAs (siRNA) for 48 h to knock down CD155 expression in the cells. We then treated the cells with 20 or 50 μM emodin for 24 h. Transfection of CD155 siRNAs significantly reduced CD155 expression in the cells, and emodin treatment did not further reduce CD155 expression (Fig. 4A). Silencing of CD155 in B16-F10 cells decreased the cell numbers, while emodin had no further effects (Fig. 4B). Furthermore, emodin could not further inhibit migration of the cancer cells with CD155 silencing (Figs. 4C–D). These results suggested that emodin’s inhibitory effects on cancer cell growth and migration are dependent on CD155 expression.
Figure 4.
Knockdown of CD155 abolished the effects of emodin on cancer cells. Cells were transfected with scrambled siRNA or siRNA targeting CD155 for 48 h, and treated with or without 20 or 50 μM emodin for 24 h. (A) Cell surface CD155 expression in B16-F10 cells was measured by flow cytometry. Representative histograms (left) and quantification of MFI (right) are shown. Data were presented as means ± SEM of one of three independent experiments; n=3. *p<0.05 vs Control. (B) Relative numbers of cells are shown. Living cells were counted and compared with controls. Data were presented as means ± SEM of one of three independent experiments; n=3. *p<0.05 vs Control. Cells were transfected with scrambled siRNA or siRNA targeting CD155 for 48 h, and then subjected to wound healing assays with or without 20 or 50 μM emodin for 24 h. Representative images (C) and quantification of migration distance (D) are shown. Data were presented as means ± SEM of one of three independent experiments; n=3. *p<0.05 vs all other groups.
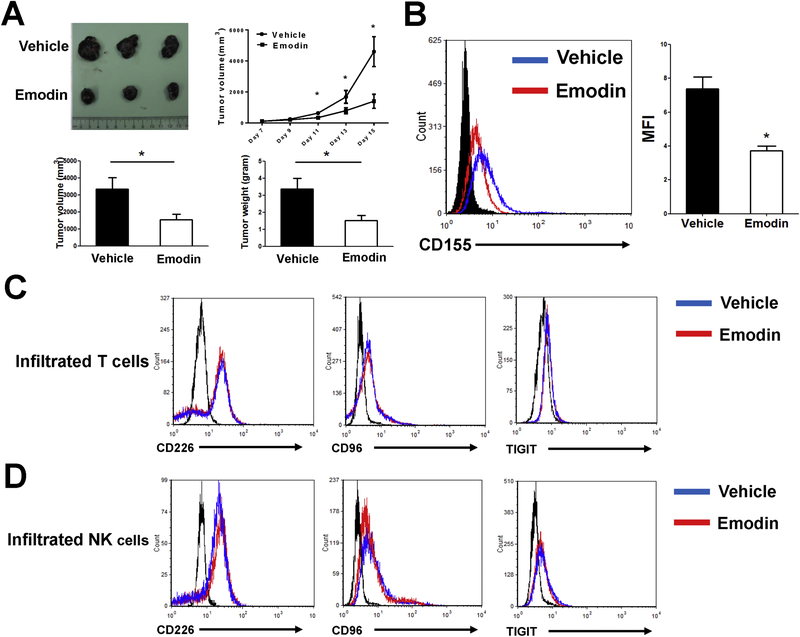
Emodin inhibited CD155 expression in vivo
To examine if emodin affects tumor development in vivo through acting on CD155, B16-F10 melanoma cells were subcutaneously inoculated into the rear flanks of WT mice. Emodin treatment (40 mg/kg i.p. once daily) began 1 day after tumor cell injection. Emodin caused a significant inhibition of primary tumor growth, and reduced tumor weight and tumor size at the endpoints when the mice were sacrificed on Day 15 post-inoculation (Fig. 5A). Moreover, emodin treatment resulted in decrease of CD155 in the tumor cells (Fig. 5B), which is similar to the results obtained from the in vitro study. Furthermore, we examined if emodin affects the expression of CD155 binding partners in NK cells and T cells in the tumors, including the costimulatory receptor CD226 and the coinhibitory receptors CD96 and TIGIT. We isolated T cells and NK cells from the tumors at the experimental endpoint and found that there was no difference in the expression of CD226, CD96 and TIGIT in the tumor infiltrated T cells (Fig. 5C) and NK cells (Fig. 5D) between the two groups.
Figure 5.
Emodin decreases the expression of CD155 in tumors in vivo. (A) Emodin inhibited growth of B16-F10 melanoma. Representative tumors (upper), tumor sizes (lower left) and weights (lower right) are shown. (B) Cell surface CD155 expression in B16-F10 cells. Cancer cells were acquired from B16-F10 tumors at the experimental endpoint and measured by flow cytometry. Data were presented as means ± SEM of one of two independent experiments; n=5. *p<0.05. (C and D) Cell surface CD226, CD96 and TIGIT expression in tumor-infiltrating T cells (C) and NK cells (D) from control and emodin treatment mice were analyzed by flow cytometry. Tumor-infiltrating T cells were gated from CD3+ population, and tumor-infiltrating NK cells were gated from NK1.1+ population.
DISCUSSION
Emodin has been reported to induce cancer cell apoptosis [27], inhibit metastasis [22], induce cell cycle arrest [28], and reverse multidrug resistance [29]. Our previous study showed that emodin could significantly inhibit the growth of early-stage breast cancer and effectively suppress metastatic tumor growth at the late stage by modulating the tumor microenvironment (TME) [21, 22, 30]. Although there were important reports that revealed multiple signaling pathways and molecules involved in the anti-cancer functions of emodin [18], and emodin in combination with chemotherapy could increase the therapeutic efficacy [31, 32], the detailed molecular mechanisms remained to be elucidated. In this study, our data showed emodin inhibited the tumor growth also through downregulation of CD155 in cancer cells. These results suggest that emodin inhibits tumor development by multiple mechanisms.
The majority of previous studies on emodin have been focused on emodin’s direct toxicity to tumor cells. In this study, we found that emodin could decrease some adhesion molecules which are normally associated with cell migrations and often up-regulated in tumors. Growing evidences indicate that alterations in the adhesion properties of tumor cells play a pivotal role in the development and progression of cancer [33]. CD155 is an immunoglobulin-like molecule which is barely detected in most normal tissues, but highly expressed in different type of tumors, including colon cancer, lung adenocarcinoma, melanoma, pancreatic cancer and glioblastoma[2, 8, 34, 35]. It has been reported to be involved in cell–cell adhesion via a heterophilic trans-interaction with nectin-3 and thus plays important roles in cell adhesion and migration [36–38]. CD155 overexpression contributes to tumor growth and metastasis [34, 39]. CD155 downregulation suppresses tumor cell proliferation and induces cell-cycle arrest at G2/M phase [35]. In this study, we examined if emodin inhibits CD155 expression in multiple cancer cell lines. We used 3 tumor cell lines, B16, 4T1 and EO771, in all of which CD155 is highly expressed. We found that emodin could significantly decrease the expression of CD155 in these cells and thereby inhibited their proliferation and migration. Therefore, we hypothesized that the anticancer activity of emodin is at least partially CD155 dependent. In support of this hypothesis, we observed that knockdown of CD155 by siRNA significantly abolished the effect of emodin on cancer cell proliferation in vitro. To further validate the involvement of CD155 in the inhibitory effects of emodin on tumor growth in vivo, we used a B16 melanoma model in mice. We found emodin could decrease CD155 expression in implanted B16 tumors and inhibit tumor growth. Taken together, these results indicated that CD155 is a new target of emodin in tumors.
CD155 can ligate with costimulatory molecule CD226, or coinhibitory molecules TIGIT and CD96, to exert a dual function in oncoimmunity [40]. Interaction of CD226 with CD155 triggers NK or T cell-mediated cytotoxicity [9, 41], while CD155/TIGIT or CD155/CD96 ligation could mediate inhibitory signaling in immune cells [12, 13, 42]. Emodin has been reported to exert a broad range of actions on the immune system. While our study in vivo showed that emodin did not have an effect on the expression of all above three CD155 receptors in T cells and NK cells, it is conceivable that emodin may affect T cell and NK cell function through reducing the binding of CD155 to these receptors, especially the coinhibitory molecules TIGIT and CD96. This aspect warrants further investigation.
The mechanism by which emodin suppresses the suppression of CD155 in tumor cells is currently unknown. Emodin has been reported to inhibit LPS-induced expression of proinflammatory cytokines in macrophages through suppressing Erk1/2 and p38 signaling [23]. It is also reported regulation of CD155 expression involves Raf-MEK-ERK-AP1 signaling [43]. Therefore, it is likely that emodin might use one of these signaling axes to regulate CD155 expression. However, we cannot exclude other mechanisms. For example, emodin treatment significantly suppressed the TWIST1-induced upregulation of CD44, which is associated with tumor initiation [44].
In conclusion, we propose a new mechanism by which emodin suppresses tumor growth. CD155 expression is upregulated in some cancers, and some chemotherapeutic agents can also upregulate CD155 on tumor cells [45]. Emodin can inhibit tumor cell proliferation and migration through decreasing their expression of CD155. Considering that emodin can act on both cancer cells and macrophages in the TME, our study provides a more comprehensive mechanistic insight into the anti-tumor activities of emodin.
Highlights.
Emodin decreases the expression of CD155 in cancer cells.
Emodin suppressed proliferation and migration of cancer cells.
The antineoplastic properties of emodin are at least partially CD155 dependent.
Acknowledgments
Financial support: The project supported by National Institutes of Health Grants HL116626, AT003961-8455 and CA216230 to D. Fan, Natural Science Basic Research Plan in Shaanxi Province of China (Program No. 2017JM8066 and 2018JM7043), and National Natural Science Foundation of China (81872315)
Abbreviations:
- TME
the tumor microenvironment
- TIGIT
T-cell immunoreceptor with Ig and ITIM domains
- PRR
poliovirus receptor-related family
Footnotes
Conflict of interest: Authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Iwasaki A, Welker R, Mueller S, Linehan M, Nomoto A, Wimmer E, Immunofluorescence analysis of poliovirus receptor expression in Peyer’s patches of humans, primates, and CD155 transgenic mice: implications for poliovirus infection, The Journal of infectious diseases 186(5) (2002) 585–92. [DOI] [PubMed] [Google Scholar]
- [2].Masson D, Jarry A, Baury B, Blanchardie P, Laboisse C, Lustenberger P, Denis MG, Overexpression of the CD155 gene in human colorectal carcinoma, Gut 49(2) (2001) 236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Iguchi-Manaka A, Okumura G, Kojima H, Cho Y, Hirochika R, Bando H, Sato T, Yoshikawa H, Hara H, Shibuya A, Shibuya K, Increased Soluble CD155 in the Serum of Cancer Patients, PloS one 11(4) (2016) e0152982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Carlsten M, Norell H, Bryceson YT, Poschke I, Schedvins K, Ljunggren HG, Kiessling R, Malmberg KJ, Primary human tumor cells expressing CD155 impair tumor targeting by down-regulating DNAM-1 on NK cells, J Immunol 183(8) (2009) 4921–30. [DOI] [PubMed] [Google Scholar]
- [5].Castriconi R, Dondero A, Corrias MV, Lanino E, Pende D, Moretta L, Bottino C, Moretta A, Natural killer cell-mediated killing of freshly isolated neuroblastoma cells: critical role of DNAX accessory molecule-1-poliovirus receptor interaction, Cancer research 64(24) (2004) 9180–4. [DOI] [PubMed] [Google Scholar]
- [6].Sanchez-Correa B, Morgado S, Gayoso I, Bergua JM, Casado JG, Arcos MJ, Bengochea ML, Duran E, Solana R, Tarazona R, Human NK cells in acute myeloid leukaemia patients: analysis of NK cell-activating receptors and their ligands, Cancer Immunol Immunother 60(8) (2011) 1195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fionda C, Abruzzese MP, Zingoni A, Soriani A, Ricci B, Molfetta R, Paolini R, Santoni A, Cippitelli M, Nitric oxide donors increase PVR/CD155 DNAM-1 ligand expression in multiple myeloma cells: role of DNA damage response activation, BMC cancer 15 (2015) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bevelacqua V, Bevelacqua Y, Candido S, Skarmoutsou E, Amoroso A, Guarneri C, Strazzanti A, Gangemi P, Mazzarino MC, D’Amico F, McCubrey JA, Libra M, Malaponte G, Nectin like-5 overexpression correlates with the malignant phenotype in cutaneous melanoma, Oncotarget 3(8) (2012) 882–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chan CJ, Andrews DM, McLaughlin NM, Yagita H, Gilfillan S, Colonna M, Smyth MJ, DNAM-1/CD155 interactions promote cytokine and NK cell-mediated suppression of poorly immunogenic melanoma metastases, J Immunol 184(2) (2010) 902–11. [DOI] [PubMed] [Google Scholar]
- [10].Gilfillan S, Chan CJ, Cella M, Haynes NM, Rapaport AS, Boles KS, Andrews DM, Smyth MJ, Colonna M, DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors, The Journal of experimental medicine 205(13) (2008) 2965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shibuya A, Campbell D, Hannum C, Yssel H, Franz-Bacon K, McClanahan T, Kitamura T, Nicholl J, Sutherland GR, Lanier LL, Phillips JH, DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes, Immunity 4(6) (1996) 573–81. [DOI] [PubMed] [Google Scholar]
- [12].Stanietsky N, Simic H, Arapovic J, Toporik A, Levy O, Novik A, Levine Z, Beiman M, Dassa L, Achdout H, Stern-Ginossar N, Tsukerman P, Jonjic S, Mandelboim O, The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity, Proceedings of the National Academy of Sciences of the United States of America 106(42) (2009) 17858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, Park S, Javinal V, Chiu H, Irving B, Eaton DL, Grogan JL, The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function, Cancer cell 26(6) (2014) 923–937. [DOI] [PubMed] [Google Scholar]
- [14].Inozume T, Yaguchi T, Furuta J, Harada K, Kawakami Y, Shimada S, Melanoma Cells Control Antimelanoma CTL Responses via Interaction between TIGIT and CD155 in the Effector Phase, The Journal of investigative dermatology 136(1) (2016) 255–63. [DOI] [PubMed] [Google Scholar]
- [15].Sloan KE, Stewart JK, Treloar AF, Matthews RT, Jay DG, CD155/PVR enhances glioma cell dispersal by regulating adhesion signaling and focal adhesion dynamics, Cancer research 65(23) (2005) 10930–7. [DOI] [PubMed] [Google Scholar]
- [16].Enloe BM, Jay DG, Inhibition of Necl-5 (CD155/PVR) reduces glioblastoma dispersal and decreases MMP-2 expression and activity, Journal of neuro-oncology 102(2) (2011) 225–35. [DOI] [PubMed] [Google Scholar]
- [17].Dong X, Fu J, Yin X, Cao S, Li X, Lin L, Ni J, Emodin: A Review of its Pharmacology, Toxicity and Pharmacokinetics, Phytother Res 30(8) (2016) 1207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wei WT, Lin SZ, Liu DL, Wang ZH, The distinct mechanisms of the antitumor activity of emodin in different types of cancer (Review), Oncology reports 30(6) (2013) 2555–62. [DOI] [PubMed] [Google Scholar]
- [19].Shrimali D, Shanmugam MK, Kumar AP, Zhang J, Tan BK, Ahn KS, Sethi G, Targeted abrogation of diverse signal transduction cascades by emodin for the treatment of inflammatory disorders and cancer, Cancer letters 341(2) (2013) 139–49. [DOI] [PubMed] [Google Scholar]
- [20].Dumit VI, Zerbes RM, Kaeser-Pebernard S, Rackiewicz M, Wall MT, Gretzmeier C, Kuttner V, van der Laan M, Braun RJ, Dengjel J, Respiratory status determines the effect of emodin on cell viability, Oncotarget 8(23) (2017) 37478–37490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Iwanowycz S, Wang J, Hodge J, Wang Y, Yu F, Fan D, Emodin Inhibits Breast Cancer Growth by Blocking the Tumor-Promoting Feedforward Loop between Cancer Cells and Macrophages, Molecular cancer therapeutics 15(8) (2016) 1931–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jia X, Yu F, Wang J, Iwanowycz S, Saaoud F, Wang Y, Hu J, Wang Q, Fan D, Emodin suppresses pulmonary metastasis of breast cancer accompanied with decreased macrophage recruitment and M2 polarization in the lungs, Breast cancer research and treatment 148(2) (2014) 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jia X, Iwanowycz S, Wang J, Saaoud F, Yu F, Wang Y, Hu J, Chatterjee S, Wang Q, Fan D, Emodin attenuates systemic and liver inflammation in hyperlipidemic mice administrated with lipopolysaccharides, Experimental biology and medicine (Maywood, N.J 239(8) (2014) 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Casey AE, Laster WR Jr., Ross GL, Sustained enhanced growth of carcinoma EO771 in C57 black mice, Proc Soc Exp Biol Med 77(2) (1951) 358–62. [DOI] [PubMed] [Google Scholar]
- [25].Nachat-Kappes R, Pinel A, Combe K, Lamas B, Farges MC, Rossary A, Goncalves-Mendes N, Caldefie-Chezet F, Vasson MP, Basu S, Effects of enriched environment on COX-2, leptin and eicosanoids in a mouse model of breast cancer, PLoS One 7(12) (2012) e51525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Makrilia N, Kollias A, Manolopoulos L, Syrigos K, Cell adhesion molecules: role and clinical significance in cancer, Cancer investigation 27(10) (2009) 1023–37. [DOI] [PubMed] [Google Scholar]
- [27].Li WY, Ng YF, Zhang H, Guo ZD, Guo DJ, Kwan YW, Leung GP, Lee SM, Yu PH, Chan SW, Emodin elicits cytotoxicity in human lung adenocarcinoma A549 cells through inducing apoptosis, Inflammopharmacology 22(2) (2014) 127–34. [DOI] [PubMed] [Google Scholar]
- [28].Zhang X, Chen Y, Zhang T, Zhang Y, Inhibitory effect of emodin on human hepatoma cell line SMMC-7721 and its mechanism, African health sciences 15(1) (2015) 97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen P, Liu Y, Sun Y, Chen C, Qi Y, Zhang Y, AZT and emodin exhibit synergistic growth-inhibitory effects on K562/ADM cells by inducing S phase cell cycle arrest and suppressing MDR1 mRNA/p-gp protein expression, Pharmaceutical biology 51(12) (2013) 1586–91. [DOI] [PubMed] [Google Scholar]
- [30].Iwanowycz S, Wang J, Altomare D, Hui Y, Fan D, Emodin Bidirectionally Modulates Macrophage Polarization and Epigenetically Regulates Macrophage Memory, The Journal of biological chemistry 291(22) (2016) 11491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Su YJ, Tsai MS, Kuo YH, Chiu YF, Cheng CM, Lin ST, Lin YW, Role of Rad51 down-regulation and extracellular signal-regulated kinases 1 and 2 inactivation in emodin and mitomycin C-induced synergistic cytotoxicity in human non-small-cell lung cancer cells, Molecular pharmacology 77(4) (2010) 633–43. [DOI] [PubMed] [Google Scholar]
- [32].Ko JC, Tsai MS, Kuo YH, Chiu YF, Weng SH, Su YC, Lin YW, Modulation of Rad51, ERCC1, and thymidine phosphorylase by emodin result in synergistic cytotoxic effect in combination with capecitabine, Biochemical pharmacology 81(5) (2011) 680–90. [DOI] [PubMed] [Google Scholar]
- [33].Okegawa T, Pong RC, Li Y, Hsieh JT, The role of cell adhesion molecule in cancer progression and its application in cancer therapy, Acta biochimica Polonica 51(2) (2004) 445–57. [PubMed] [Google Scholar]
- [34].Nakai R, Maniwa Y, Tanaka Y, Nishio W, Yoshimura M, Okita Y, Ohbayashi C, Satoh N, Ogita H, Takai Y, Hayashi Y, Overexpression of Necl-5 correlates with unfavorable prognosis in patients with lung adenocarcinoma, Cancer science 101(5) (2010) 1326–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nishiwada S, Sho M, Yasuda S, Shimada K, Yamato I, Akahori T, Kinoshita S, Nagai M, Konishi N, Nakajima Y, Clinical significance of CD155 expression in human pancreatic cancer, Anticancer research 35(4) (2015) 2287–97. [PubMed] [Google Scholar]
- [36].Ikeda W, Kakunaga S, Itoh S, Shingai T, Takekuni K, Satoh K, Inoue Y, Hamaguchi A, Morimoto K, Takeuchi M, Imai T, Takai Y, Tage4/Nectin-like molecule-5 heterophilically trans-interacts with cell adhesion molecule Nectin-3 and enhances cell migration, The Journal of biological chemistry 278(30) (2003) 28167–72. [DOI] [PubMed] [Google Scholar]
- [37].Ikeda W, Kakunaga S, Takekuni K, Shingai T, Satoh K, Morimoto K, Takeuchi M, Imai T, Takai Y, Nectin-like molecule-5/Tage4 enhances cell migration in an integrin-dependent, Nectin-3-independent manner, The Journal of biological chemistry 279(17) (2004) 18015–25. [DOI] [PubMed] [Google Scholar]
- [38].Sloan KE, Eustace BK, Stewart JK, Zehetmeier C, Torella C, Simeone M, Roy JE, Unger C, Louis DN, Ilag LL, Jay DG, CD155/PVR plays a key role in cell motility during tumor cell invasion and migration, BMC cancer 4 (2004) 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Morimoto K, Satoh-Yamaguchi K, Hamaguchi A, Inoue Y, Takeuchi M, Okada M, Ikeda W, Takai Y, Imai T , Interaction of cancer cells with platelets mediated by Necl-5/poliovirus receptor enhances cancer cell metastasis to the lungs, Oncogene 27(3) (2008) 264–73. [DOI] [PubMed] [Google Scholar]
- [40].Gao J, Zheng Q, Xin N, Wang W, Zhao C, CD155, an onco-immunologic molecule in human tumors, Cancer science (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tahara-Hanaoka S, Shibuya K, Onoda Y, Zhang H, Yamazaki S, Miyamoto A, Honda S, Lanier LL, Shibuya A, Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112), International immunology 16(4) (2004) 533–8. [DOI] [PubMed] [Google Scholar]
- [42].Chan CJ, Martinet L, Gilfillan S, Souza-Fonseca-Guimaraes F, Chow MT, Town L, Ritchie DS, Colonna M, Andrews DM, Smyth MJ, The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions, Nature immunology 15(5) (2014) 431–8. [DOI] [PubMed] [Google Scholar]
- [43].Hirota T, Irie K, Okamoto R, Ikeda W, Takai Y, Transcriptional activation of the mouse Necl-5/Tage4/PVR/CD155 gene by fibroblast growth factor or oncogenic Ras through the Raf-MEK-ERK-AP-1 pathway, Oncogene 24(13) (2005) 2229–35. [DOI] [PubMed] [Google Scholar]
- [44].Way TD, Huang JT, Chou CH, Huang CH, Yang MH, Ho CT, Emodin represses TWIST1-induced epithelial-mesenchymal transitions in head and neck squamous cell carcinoma cells by inhibiting the beta-catenin and Akt pathways, Eur J Cancer 50(2) (2014) 366–78. [DOI] [PubMed] [Google Scholar]
- [45].Soriani A, Zingoni A, Cerboni C, Iannitto ML, Ricciardi MR, Di Gialleonardo V, Cippitelli M, Fionda C, Petrucci MT, Guarini A, Foa R, Santoni A, ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype, Blood 113(15) (2009) 3503–11. [DOI] [PubMed] [Google Scholar]