Abstract
Due to recent medical and technological advances in neonatal care, infants born extremely premature have increased survival rates1,2. After birth, these infants are at high risk of hypoxic episodes due to lung immaturity, hypotension, and lack of cerebral flow regulation, and can develop a severe condition called encephalopathy of prematurity (EP)3. Over 80% of infants born before post conception week (PCW) 25 have moderate to severe long-term neurodevelopmental impairments4. The susceptible cell types in the cerebral cortex and the molecular mechanisms underlying associated gray matter defects in premature infants remain unknown. Here, we used human three-dimensional brain region-specific organoids to study the effect of oxygen deprivation on corticogenesis. We identified specific defects in intermediate progenitors, a cortical cell type associated with the expansion of the human cerebral cortex, and show that these are related to the unfolded protein response (UPR) and cell cycle changes. Moreover, we verified these findings in human primary cortical tissue and demonstrated that a small molecule modulator of the UPR pathway can prevent the reduction in intermediate progenitors following hypoxia. We anticipate that this human cellular platform will be valuable for studying environmental and genetic factors underlying brain injury in premature infants.
Extremely premature birth (before PCW28) coincides with critical biological events in the development of the central nervous system (CNS), including the formation of the expanded human cerebral cortex. EP is characterized by gray and white matter abnormalities and a reduction in cortical volume that correlates with neurodevelopmental outcomes, including cognitive and behavioral disorders1. A common pathogenic factor in EP appears to be perinatal hypoxia5 (often considered as decreases of PaO2 below 40 mmHg). However, the cellular substrates and the molecular mechanisms by which changes in oxygen tension lead to cortical gray matter defects in extremely premature infants are still not understood. This is primarily due to challenges in directly investigating the preterm human brain and difficulties in recapitulating the trajectory of human brain development and maturation in other species. Moreover, the unique cellular and molecular features underlying cortical development in humans6 underscore the need for personalized human models of brain development. Recent advances in cell reprogramming technologies as well three-dimensional (3D) cell differentiation methods make possible the non-invasive derivation of structures resembling regions of the developing human brain7.
Here, we leveraged a method that we have previously described8–10 to develop an in vitro human cellular model of hypoxic EP. To achieve this, we differentiated human induced pluripotent stem (hiPS) cells into brain-region specific organoids called human cortical spheroids (hCS)9–11. After ~10 weeks in vitro, hCS transcriptionally resemble the cerebral cortex at 19–24 PCW9,10, which corresponds to extreme prematurity. This developmental stage is characterized by extensive proliferation and neurogenesis in the cerebral cortex– a process that continues into the third trimester of pregnancy12–14. We exposed hCS to low oxygen concentrations in vitro and found a reduction in a specific population of cortical progenitors that are thought to contribute to the expansion of the primate cerebral cortex. Moreover, we found that pharmacologically modulating the unfolded protein response (UPR) can prevent this defect, and we used human primary tissue to validate these findings.
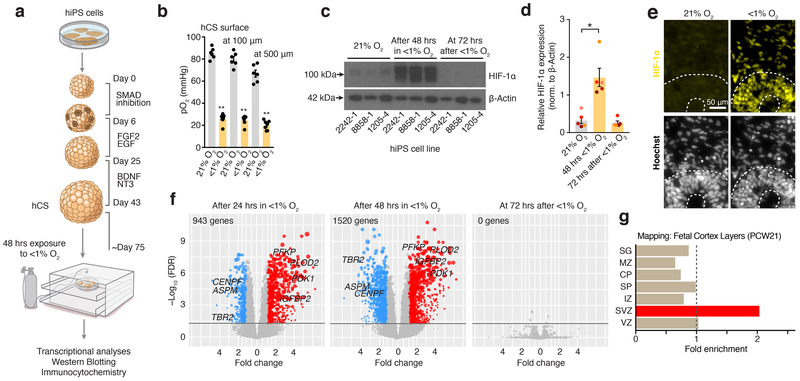
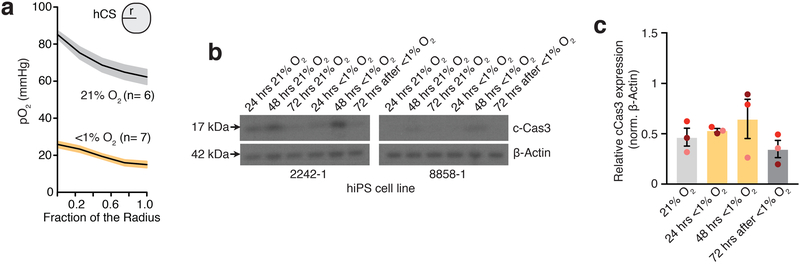
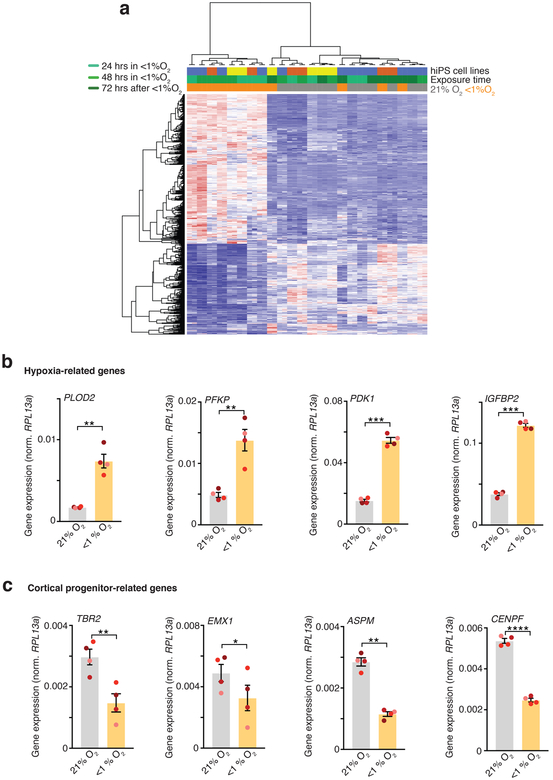
To develop a model of hypoxia during human corticogenesis, we differentiated hCS from 5 hiPS cell lines derived from 4 healthy subjects (Fig. 1a, Supplementary Table 1). We used a gas control chamber to expose hCS at approximately day 75 of differentiation to low oxygen tension (<1%) for 48 hours, followed by re-introduction to 21% O2. To monitor changes in oxygen partial pressure in hCS, we used a needle-type fiber optic oxygen microsensor. At the surface of hCS, the partial oxygen pressure (pO2) was ~85 mmHg, which is similar to pO2 in arterial blood, while in the center values were on average above 62 mmHg (Fig. 1b; Extended Data Fig. 1a). Exposure to <1% O2 for 48 hours resulted in a drop to ~25 mmHg at the hCS surface and ~20 mmHg in the hCS center, which is below the critical O2 tension in the brain (P< 0.0001)15. Whole-hCS Western blot analysis demonstrated that HIF-1α (hypoxia inducible factor-1 alpha), a key oxygen-labile protein in the hypoxia response, stabilized at 48 hours in low O2 (P= 0.02) and returned to previous levels following 72 hours of re-oxygenation (Fig. 1c, d; Supplementary Table 2). Similarly, immunocytochemistry in hCS cryosections indicated the expected nuclear localization of the HIF-1α protein (Fig. 1e). At the same time, the level of cell death as estimated by cleaved caspase 3 (c-CAS3) did not significantly increase during exposure to <1% O2 (P= 0.29; Extended Data Fig. 1b, c), suggesting that a hypoxia-like response was induced without massive cell death. We next investigated the transcriptional changes associated with exposure to <1% O2 by performing RNA sequencing at 24 and 48 hours (middle and end of <1% O2 exposure interval), as well as after 72 hours of re-oxygenation at 21% O2 (Fig. 1f). Hierarchical clustering of the gene expression profiles separates the <1% O2 exposed from unexposed samples at 24 hours and 48 hours, but not at 72 hours post re-oxygenation (time = 120 hours). Together, this suggests that exposure to low O2 levels resulted in a defined transcriptional profile and that after 72 hours at atmospheric O2, hCS reverted back to their expression state pre-low O2 exposure (Extended Data Fig. 2a). To identify genes associated with the response to low O2 we identified genes differentially expressed (DE) between the <1% O2 exposed hCS and 21% exposed hCS. We tested and controlled for potential confounding variables, such as genetic background, differentiation batch, and RNA-Seq quality metrics (Material and Methods). In total, we identified 943 DE genes at 24 hours, 1520 DE genes at 48 hours, and no DE genes after 72 hours of re-oxygenation (Fig. 1f; FDR ≤ 0.05, fold change ≥ 1.5; hCS from 3 hiPS cell lines in two differentiations experiments; Supplementary Table 3). Among the DE genes were transcripts associated with a hypoxic response, such as PLOD2 (P= 0.006), PFKP (P= 0.008), PDK1 (P= 0.0005), IGFBP2 (P= 0.0002) (Fig. 1f), which we validated by qPCR (Extended Data Fig. 2b; Supplementary Table 4). Interestingly, we also observed several genes associated with dorsal forebrain progenitors, such as EOMES (also known as TBR2) (P= 0.006) and EMX1 (P= 0.02), as well as the cell-cycle related genes ASPM (P= 0.002) and CENPF (P< 0.0001) (Fig. 1f; Extended Data Fig. 2c). To gain insights into potential cell-type-specific changes following low O2 exposure, we assessed whether the combined set of DE genes were enriched for genes identified to be specifically expressed in laser-microdissected regions of the developing human cortex16. Overlap of the hypoxia-induced DE in hCS (1754 unique genes) and subregions of the human cerebral cortex at PCW21 revealed two-fold enrichment for transcripts specifically expressed in the subventricular zone (SVZ) (P< 10–7, Fig. 1g), a proliferative region bordering the ventricular epithelium and that has undergone significant expansion in primates6 and where proliferation and neurogenesis continues until later stages of gestation12–14.
FIGURE 1. Human cellular model for studying changes in oxygen tension in hCS.
(a) Scheme illustrating the major stages in the generation of hCS from hiPS cells as described in Ref9. At day 74–78 of in vitro differentiation, hCS are exposed for 48 hours at <1% O2 in a gas-controlled culture chamber, and then maintained for another 72 hours at 21% O2. Control hCS are maintained at 21% O2 throughout. (b) O2 tension levels (pO2, mmHg) measured with optical microsensor at the surface and inside hCS (100 and 500 μm) before (n= 6 hCS) and at 48 hours of exposure to <1% O2 (n= 7); hCS from 3 hiPS cell lines; Kruskal-Wallis test, P< 0.0001, Dunn’s multiple comparison test, **P= 0.002, **P= 0.008, **P= 0.01. (c, d) Representative western blots and quantification of HIF-1α protein expression in hCS after 48 hours of exposure to <1% O2 and after 72 hours of reoxygenation versus unexposed (21% O2); normalized to β-actin (n= 5 differentiated hiPS cell lines with at least 2 hCSs per condition; Friedman’s test, P= 0.02, Dunn’s multiple comparison test versus 21% O2, *P= 0.02 for 48 hours and P> 0.99 for 72 hours). Data are mean ± s.e.m. Individual values are indicated by dots. Western blots were cropped to show the relevant bands; molecular weight (MW) markers are indicated on the right (in kD). See Source Data 1 for uncropped western blots and Supplementary Table 2 for quantifications. (e) Representative immunostaining of HIF-1α (yellow) in hCS exposed for 48 hours to <1% O2 versus 21% hCS. Experiment performed in 2 hiPS cell lines. Nuclei labeled by Hoechst staining. Scale bar, 50 μm. (f) Volcano plots showing the results of RNA-seq experiments after exposure to <1% O2 for 24 hours or 48 hours, as well as after 48 hours of <1% O2 followed by 72 hours of re-oxygenation (total time of 120 hours). Each dot represents a single gene, with genes significantly upregulated shown in red, genes significantly downregulated in blue, and non-significant genes in grey (determined based on FDR ≤ 0.05 and fold change ≥ 1.5). The size of the points corresponds to the difference in expression level between low-oxygen exposed hCS and unexposed hCS (difference of medians); n= 24 samples from hCS derived from 3 hiPS cell lines. (g) Overlap between hypoxia-related transcriptome changes in hCS and layer-specific transcriptome signatures in the developing human cortex at PCW21 as described in ref16. Strong enrichment is observed only in SVZ (p value corrected for multiple comparisons). SG, subpial granular zone; MZ, marginal zone; CP, cortical plate; SP, subplate; IZ, intermediate zone; VZ, ventricular zone.
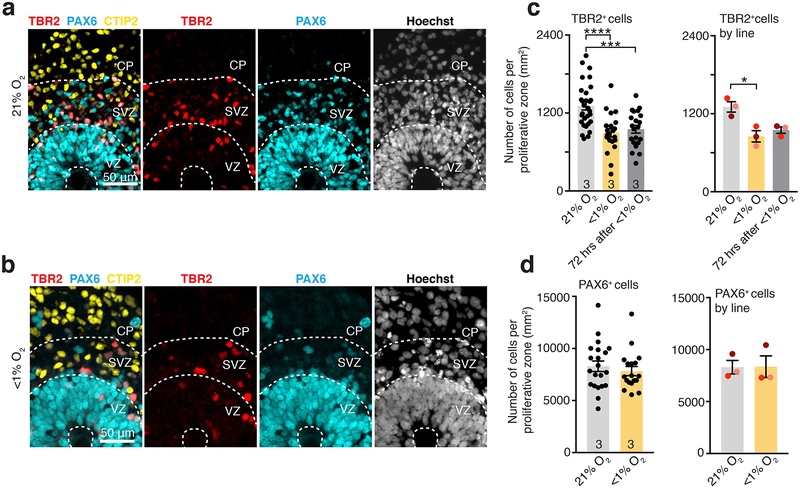
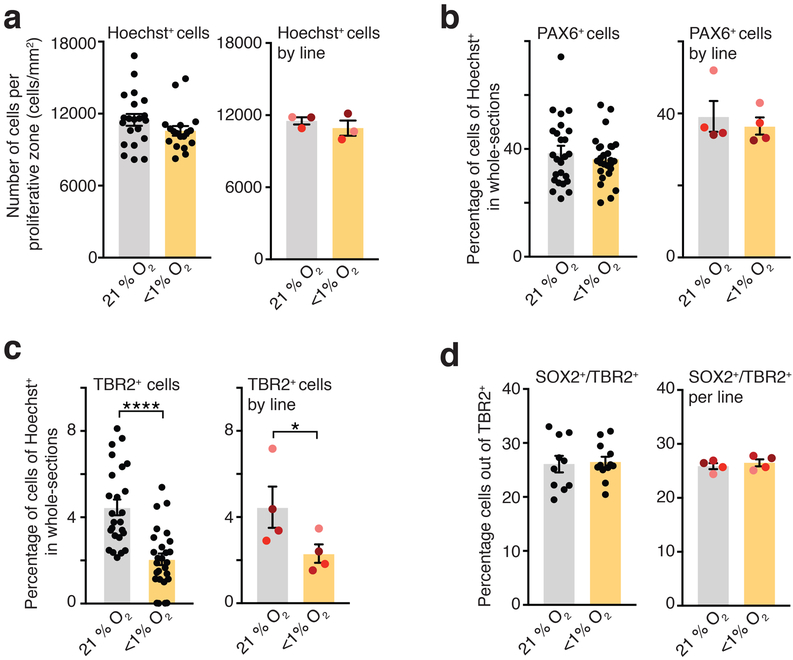
Based on this observation we next assessed whether low O2 exposure of hCS is associated with changes in the cortical progenitors present in the SVZ, such as intermediate progenitors that are characterized by the expression of the key transcription factor TBR2. At this in vitro stage, hCS display an internal cytoarchitecture consisting of PAX6+ radial glial cells organized in ventricular-like zones (VZ) around a lumen (Fig. 2a, b)9. These regions are surrounded by an intermediate region rich in TBR2+ cells resembling the SVZ, which is followed by a cortical plate (CP) region containing mostly deep-layer neurons expressing the marker CTIP2 (also known as BLC11B). We used immunocytochemistry in hCS cryosections for PAX6, TBR2 and CTIP2 to delineate the proliferative VZ-like and SVZ-like regions from the CP-like domain, as previously described in a cortical organoid system17. We quantified data from hCS derived from 3 hiPS lines in independent differentiations, and observed that the density of TBR2+ cells per proliferative region was reduced by ~35% in hCS exposed for 48 hours to <1% O2 (left: P< 0.0001; right: P= 0.03) and this persisted at 72 hours after re-oxygenation (left: P= 0.0003; right: P= 0.06) (Fig. 2c). In contrast, the density of PAX6+ cells in the same delineated regions was not affected after 48 hours of low O2 exposure (left: P= 0.44; right: P> 0.99; Fig. 2d), and the overall density of cells labeled with the nuclear dye Hoechst was unchanged (left: P= 0.09; right: P= 0.50, Extended Data Fig. 3a). This reduction in TBR2+ cells, but not in PAX6+ cells, was also observed when we counted the raw, total number of TBR2+ (left: P< 0.0001; right: P= 0.03) or PAX6+ cells (left: P= 0.44; right: P= 0.25) out of all Hoechst-labeled nuclei in multiple whole-cryosections of hCS (Extended Data Fig. 3b, c). Moreover, the affected population of TBR2+ cells was not the population of SVZ progenitors co-expressing SOX2 as investigated before in a mouse model18 (left: P= 0.82; right: P= 0.06, Extended Data Fig. 3d).
FIGURE 2. Proportion of TBR2+ cells in hCS exposed to low oxygen.
(a, b) Representative images of proliferative areas in hCS maintained in 21% O2 (upper) or exposed to <1% O2 for 48 hours (lower); scale bar 50 μm. The VZ–, SVZ– and CP–like areas are delineated by the pattern of expression of PAX6, TBR2, CTIP2 and the density and orientation of nuclei (labeled with Hoechst). Scale bar, 50 μm. (c) Quantification of the density of TBR2+ cells in hCS after 48 hours of exposure to <1% O2 and after 72 hours of reoxygenation versus unexposed (21% O2). (Left) Data shown as average across individual hCS proliferative zones from 3 hiPS cell lines per condition (n= 30 areas for 21% O2, n= 21 areas for <1% O2; n= 21 areas for re-oxygenation; one-way ANOVA, F2, 69= 13.64, P< 0.0001, Dunnett’s multiple comparison test versus 21% O2, ****P< 0.0001, ***P= 0.0003), or (Right) as average across different hiPS cell lines (n= 3 hiPS cell lines; one-way ANOVA, F2, 4= 8.38, P= 0.03, Dunnett’s multiple comparison test versus 21% O2, *P= 0.03, *P= 0.06; each line is shown in a different color). (d) Quantification of the density of PAX6 cells in hCS after exposure to 48 hours of <1% O2. (Left) Data shown as average across individual hCS proliferative zones from 3 hiPS cell lines per condition (n= 21 areas for 21% O2 versus n=18 areas for <1% O2; two-tailed Mann-Whitney U test, P= 0.44) or (Right) as average across different hiPS cell lines (n= 3 hiPS cell lines; Wilcoxon test, P> 0.99 for all comparisons; each line is shown in a different color).
Data are mean ± s.e.m., individual values are indicated by dots.
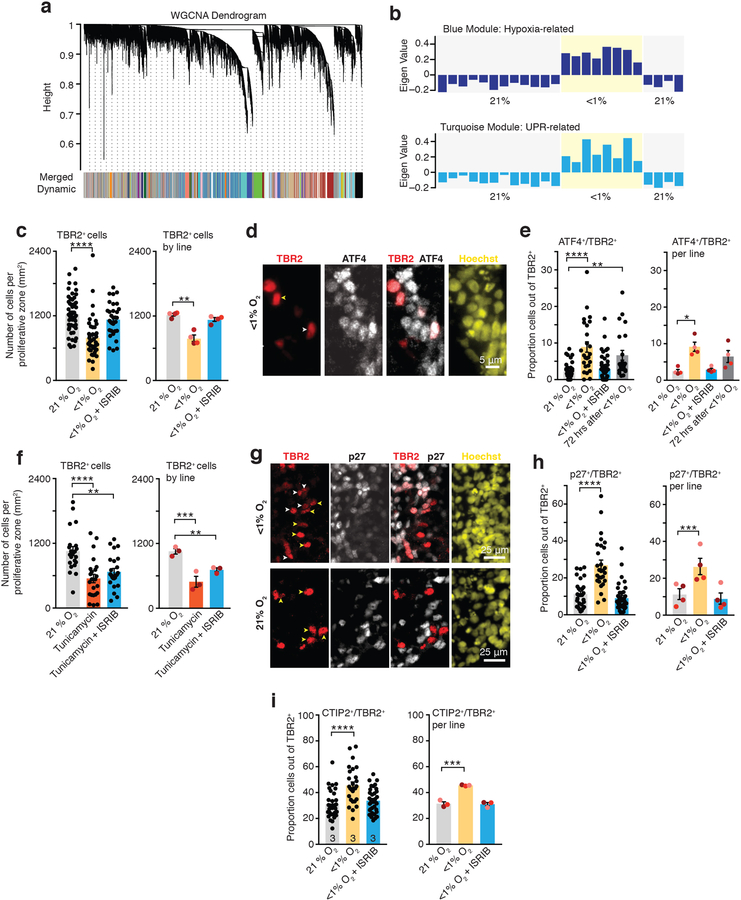
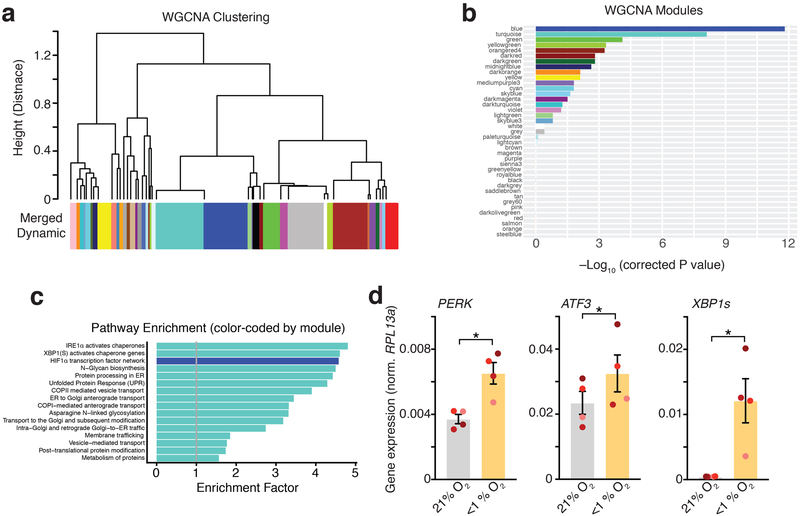
To gain insights into this potential cell-specific vulnerability of cortical progenitors following low O2 exposure, we turned back to the hypoxia-induced transcriptional changes in hCS. It has previously been demonstrated that the transcriptome has a reproducible co-expression structure that provides a framework for understanding disease biology19,20. Therefore, we applied Weighted Gene Co-expression Network Analysis (WGCNA)21 to identify modules of genes that changed with similar patterns following low O2 exposure in hCS (Fig. 3a). WGCNA identified 9 gene co-expression modules correlated with low O2 exposure (FDR ≤ 0.05). These are referred to by color label identifiers; each representing a cluster of genes with a common expression pattern across samples (Extended Data Fig. 4a, b). Blue and turquoise were the most strongly associated modules (Fig. 3b, Extended Data Fig. 4a, b). The blue module is enriched for genes associated with the hypoxic response pathway and genes regulated by HIF-1α (Extended Data Fig. 4c; FDR ≤ 0.0001). In contrast, the turquoise module was enriched for genes associated with the UPR, such as PERK (P= 0.03), ATF3 (P= 0.03), XBP1s (P= 0.04). Analysis by qPCR confirmed these changes (Extended Data Fig. 4d; Supplementary Table 4).
FIGURE 3. Unfolded protein response pathway in hCS exposed to low oxygen.
(a) Dendrogram for the Weighted gene co-expression network analysis (WGCNA), which identified genes with similar expression profiles and grouped them into modules (represented by colors). (b) Eigengene expression profiles (‘average’ expression profile of all module genes) for the top WGCNA modules associated with exposure of hCS to low O2 for 48 hours. The blue module (upper) contains genes strongly enriched for annotation in biological pathways related to the hypoxia response (HIF-1α transcription factor network), while the turquoise module (lower) is enriched for genes involved in the unfolded protein response (UPR) pathway. (c) Quantification of the density of TBR2+ cells in hCS after 48 h exposure to <1% O2 in the presence or absence of 10 nM ISRIB. Data are shown as averages across individual hCS proliferative zones from four hiPS cell lines per condition (left, n = 50 areas for 21% O2 versus n = 42 areas for <1% O2 versus n = 32 areas for <1% O2 with ISRIB; one-way ANOVA test, F2,21 = 15.85, P < 0.0001; Dunnett’s multiple-comparison test versus 21% O2, ****P < 0.0001 for <1% O2, P = 0.30 for <1% O2 + ISRIB) or as averages across different hiPS cell lines (right, n = 4 hiPS cell lines; one-way ANOVA test, F2,6 = 13.43, P = 0.006; Dunnett’s multiple-comparison test versus 21% O2, **P = 0.004 for <1% O2, P = 0.45 for <1% O2 + ISRIB; each line is shown in a different color). (d) Representative images of cells co-expressing ATF4 and TBR2 in hCS exposed to <1% O2 for 48 hours. Yellow arrow show example of TBR2+/ATF4–; white arrows show example of cells that are TBR2+/ATF4+. Scale bar, 5 μm. (e) Quantification of the proportion of cells co-expressing ATF4 and TBR2 in cryosections of hCS after exposure to 48 hours of <1% O2 in the presence or absence of 10 nM ISRIB, and after 72 hours of re-oxygenation. (Left) Data shown as average across individual hCS cryo-sections from 4 hiPS cell lines per condition (n= 36 sections for 21% O2, n= 25 sections for <1% O2, n= 48 sections for <1% O2 with ISRIB, n= 23 sections for re-oxygenation; Kruskal-Wallis test P< 0.0001, Dunn’s multiple comparison versus 21% O2, ****P< 0.0001, P> 0.99, **P= 0.001), or (Right) as average across different hiPS cell lines (n= 4 hiPS cell line; Friedman test P= 0.01, Dunnett’s multiple comparison test versus 21% O2, *P= 0.01, P> 0.99, P= 0.51; each line is shown in a different color). (f) Quantification of the density of TBR2+ cells in hCS after exposure for 48 hours to 1.2 μM of tunicamycin in the presence or absence of 10 nM ISRIB. (Left) Data shown as average across individual hCS proliferative zones from 3 hiPS cell lines per condition (n= 22 areas for 21% O2, n= 23 areas for tunicamycin, n= 24 areas for tunicamycin with ISRIB; one-way ANOVA, F2, 66= 12.69, P< 0.0001; Dunnett’s multiple comparison test versus 21% O2, ****P< 0.0001 for Tunicamycin; ***P=0.003 for Tunicamycin + ISRIB or (Right) as average across different hiPS cell lines (n= 3 hiPS cell line; one-way ANOVA F2, 4= 11.85, P= 0.02; with Dunnett’s multiple comparison test versus 21% O2, ***P= 0.01 for Tunicamycin ; *P= 0.03 for Tunicamycin + ISRIB; each line is shown in a different color). (g) Representative images of cells co-expressing p27 and TBR2 in cryosections of hCS after exposure to 48 hours of <1% O2 in the presence or absence of 10 nM ISRIB. Yellow arrow show example of TBR2+/p27–; white arrows show example of cells that are TBR2+/p27+. Scale bar, 25 μm. (h) Quantification of the proportion of cells co-expressing p27 and TBR2 in cryosections of hCS after exposure to 48 hours of <1% O2 in the presence or absence of 10 nM ISRIB. (Left) Data shown as average across individual hCS cryo-sections from 4 hiPS cell lines per condition (n= 27 sections for 21% O2 versus n= 28 sections for <1% O2 versus n= 36 sections for <1% O2 with ISRIB; Kruskal-Wallis test P< 0.0001, Dunn’s multiple comparison versus 21% O2, ****P<0.0001, P= 0.54), or (Right) as average across different hiPS cell lines (n= 4 hiPS cell line; one-way ANOVA F2, 6= 43.78, P= 0.0003, Dunnett’s multiple comparison test versus 21% O2, ***P= 0.0006, P= 0.43; each line is shown in a different color). (i) Quantification of the proportion of cells co-expressing CTIP2 (also known as BLC11B) and TBR2 in cryosections of hCS after exposure to 48 hours of <1% O2 in the presence or absence of 10 nM ISRIB. (Left) Data shown as average across individual hCS cryo-sections from 3 hiPS cell lines per condition (n= 30 sections for 21% O2, n= 27 sections for <1% O2, n= 34 sections for <1% O2 with ISRIB; one-way ANOVA, F2, 88= 12.31, P< 0.0001, Dunnett’s multiple comparison test versus 21% O2, ****P< 0.0001, P= 0.58), or (Right) as average across different hiPS cell lines (n= 3 hiPS cell line; one-way ANOVA F2, 6= 53.67, P= 0.0001, Dunnett’s multiple comparison test versus 21% O2, ***P= 0.0002, P= 0.96; each line is shown in a different color).
Data are mean ± s.e.m., individual values are indicated by dots.
The UPR pathway is a protective cellular response induced during periods of cellular stress that aims to restore protein homeostasis22. In certain cancer cells, hypoxia can activate components of the UPR pathway23. Interestingly, previous work has linked UPR to the generation of Tbr2+ intermediate progenitors in rodents24. To verify if the transcriptional changes in the UPR pathway are related to TBR2-related phenotype after low O2, we exposed hCS to a potent small molecule called ISRIB, which reverses with high specificity the effects of eIF2α phosphorylation and restores protein translation while maintaining some of the protective effects of the integrated cellular response and UPR25–27. We observed that adding 10 nM ISRIB during the 48 hours of <1% O2 exposure of hCS restored the density of TBR2+ cells (left: P= 0.3; right: P= 0.45 for <1% O2 + ISRIB versus 21% O2; Fig. 3c). Moreover, while the UPR-pathway related ATF4 transcription factor was co-expressed by a larger proportion of TBR2+ cells in hCS exposed to low O2 for 48 hours versus hCS maintained at 21% O2, this effect was restored with 10 nM ISRIB (left and right: P> 0.99) and was partially restored after re-oxygenation (left: P= 0.001; right: P= 0.51) (Fig. 3d, e). At the same time, radial glia expressing PAX6 did not show an increase in ATF4 co-expression (left: P= 0.70; right: P=0.48) and this remained unchanged by exposure to ISRIB (left: P= 0.54; right: P= 0.21) (Extended Data Fig. 5a). To further investigate the role of UPR, we exposed for 48 hours hCS to 1.2 μM of tunicamycin, which has been shown to induce UPR in the developing brain28. Similar to exposure to low O2, tunicamycin reduced the proportion of TBR2 cells (left: P< 0.0001; right: P= 0.01; Fig. 3f). Moreover, the proportion of PAX6+ cells was not affected by exposure to tunicamycin (left: P= 0.37; right: P= 0.2; Extended Data Fig. 5b). Together, these experiments indicate that a 48-hour exposure to low O2 leads to a reduction in TBR2+ intermediate progenitors but not in PAX6+ radial glia, and that this phenotype can be prevented by co-exposure to nanomolar concentration of a UPR-pathway modulator.
We did not observe significant cell death in hCS after 48 hours of hypoxia (Extended Data Fig. 1b, c; Supplementary Table 2, Source Data 2) and only ~0.1% of TBR2+ cells were c-CAS3+ in hCS cryosections (P> 0.99; Extended Data Fig. 5c). Moreover, the unique set of 815 DE genes downregulated across 24 and 48 hours are enriched for genes involved in cell cycle (143 total genes, fold-enrichment 2.47, Bonferroni FDR 6.5 × 10–21) (Fig. 1f). Therefore, we explored whether this reduction is related to changes in cell cycle. Co-staining with p27, a protein that regulates G1 and helps cells withdraw from cell cycle when they terminally differentiate, showed a 2.4-fold increase in the overlap with TBR2+ exposed to low O2 (left: P<0.0001; right: P= 0.0006), and this effect was prevented by co-exposure to ISRIB (Left: P= 0.54; right: P= 0.43) (Fig. 3g, h). At the same time, few PAX6+ radial glia in hCS co-expressed p27, and this proportion did not change following oxygen deprivation (left: P= 0.88; right: P= 0.67) or ISRIB exposure (left: P= 0.90; right: 0.94) (Extended Data Fig. 5d). To explore if this increase in p27 expression was associated with changes in proliferation, we co-stained TBR2+ cells with the cell cycle marker Ki67 (left: P= 0.91; right: P= 0.91) and the mitotic marker PH3 (left: P= 0.55; right: P= 0.56) but did not find differences after low O2 exposure (Extended Data Fig. 5e–g). Alternatively, these changes in p27 may be associated with cell cycle exit and early neural differentiation. Therefore, we quantified the proportion of TBR2+ cells that co-express the cortical marker CTIP2 and found an increase by ~50% in double-positive cells (left: P< 0.0001, right: P= 0.0002), which was prevented by 10 nM ISRIB exposure (left: P= 0.58; right: P= 0.96) (Fig. 3i). This suggests that TBR2+ cells may be particularly susceptible to premature differentiation after exposure to low O2 levels and that modulators of the UPR pathways could be used to restore hypoxia-related defects in cortical progenitor subtypes.
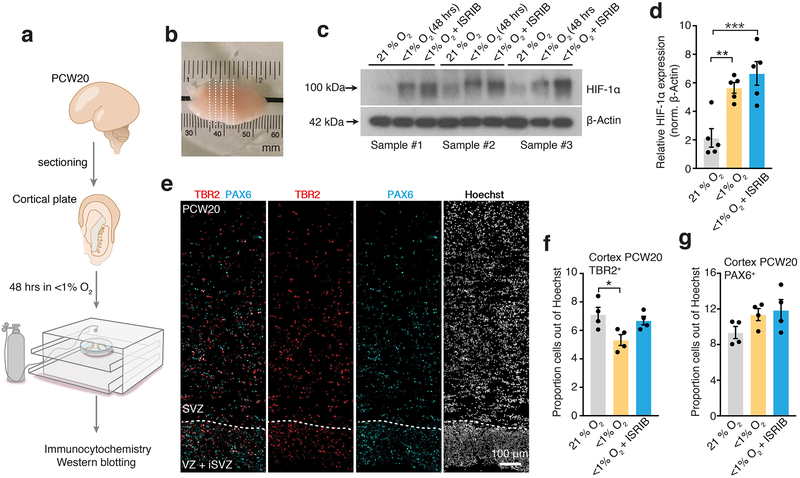
Lastly, to validate these findings in a more physiological system, we used primary human cortical tissue at PCW20, which we sectioned and then either exposed to <1% O2 or 21% O2 for 48 hours in a gas-controlled environmental chamber (Fig. 4a, b). Western blot analysis showed that HIF-1α protein stabilized at 48 hours in low O2 (P= 0.003) and this effect was present even after co-exposure to 10 nM ISRIB (P= 0.0007) (Fig. 4c, d; Supplementary Table 2, Source Data 3). We next performed immunostaining for PAX6 and TBR2 in cortical slices to delineate the proliferative VZ, SVZ+ oSVZ areas (Fig. 4e). We found a decrease in the proportion of TBR2+ cells following low O2 exposure (P= 0.02) and this was prevented by co-exposure to 10 nM ISRIB (P= 0.69) (Fig. 4f). At the same time, the proportion of PAX6+ cells was unchanged in the absence (P= 0.23) or presence of ISRIB (P= 0.12) (Fig. 4g).
FIGURE 4. Validation in primary human cortical tissue in vitro.
(a) Scheme showing sectioning and gas-chamber exposure to low oxygen of human cortical tissue (~PCW20). (b) Macroscopic image of cortical tissue and sectioning. Dashed lines indicate approximate regions of sectioning for slice culture. (c, d) Representative western blots and quantification of HIF-1α protein expression in human cortical sections (PCW20) after 48 hours of exposure to <1% O2 with or without 10 nM ISRIB normalized to β-actin (n= 5 slices; one-way ANOVA F2, 8= 19.39, P= 0. 0009, Dunnett’s multiple comparison test versus 21% O2, **P= 0.003, ***P= 0.0007). Western blots were cropped to show the relevant bands; molecular weight (MW) markers are indicated on the left (in kDa). See Source Data 3 for uncropped western blots and Supplementary Table 2 for quantifications. (e) Image of proliferative zones (VZ, SVZ) in cortical primary tissue delineated by expression of PAX6, TBR2 and Hoechst. Scale bar, 100 μm. (f) Quantification of density of TBR2+ cells in cryosections of primary human fetal tissue exposed after exposure for 48 hours to <1% O2 in the presence or absence of 10 nM ISRIB (n= 4 sections; one-way ANOVA F2, 9= 5.32, P= 0.02, Dunnett’s multiple comparison test versus 21% O2, *P= 0.02, P= 0.69). (g) Quantification of density of PAX6+ cells in cryosections of primary human fetal tissue exposed after exposure for 48 hours to <1% O2 in the presence or absence of 10 nM ISRIB (n= 4 sections; one-way ANOVA F2, 9= 2.29, P= 0.15, Dunnett’s multiple comparison test versus 21% O2, P= 0.23, P= 0.12).
Data are mean ± s.e.m. Individual values are indicated by dots.
In this study, we show how hiPS cell-derived 3D brain cultures can be used to model injury in the developing brain. There are several novel aspects of this work. First, we used a reproducible brain-region specific organoid model that recapitulates key features of the mid-gestation human cortex8–10 and we validate our findings in slices of human cortex. This platform could serve both as a model of hypoxic EP as well as a model of second trimester placental insufficiency. Recent data in human brain tissue demonstrates continued neurogenesis into the third trimester of pregnancy12–14. In fact, the risk for encephalopathy of prematurity is highest in extremely preterm infants and the neurodevelopmental consequences are most severe for this patient population4. Second, we find that TBR2+ progenitors, a population of amplifying cells that reside in the SVZ and that is thought to contribute to the expansion of the neocortex by increasing neuron number, are particularly affected by oxygen deprivation. In fact, TBR2+ progenitors are positioned close to blood vessels29 and increased oxygen tension in the rodent cortex results in expansion of the SVZ18. While other studies have investigated the role of oxygen on dissociated human radial glia30, the effect of hypoxia on human TBR2+ progenitors has not been explored. Future studies using this system should investigate the susceptibility of outer radial glia, which are more transcriptionally similar to radial glia, but reside in the expanded outer SVZ in humans31. Moreover, it remains to be established how changes in specific progenitors affect brain development as previous studies in premature brains have indicated an impact on glutamatergic neurogenesis and gray mater14. Third, we find that these cell-specific defects are related to the UPR, and in particular to PERK-eiF2α-ATF4 pathway. This ER stress-response pathway controls protein homeostasis and has been associated with hypoxia in cancer23, and more recently with cortical development32. Components of the UPR pathway correlate with milestones in corticogenesis, and changes in UPR in rodents affect Tbr2+ cells24. More specifically, loss of a component of the elongator complex leads to impaired translation speed, triggers UPR and reduces the number of Tbr2+ cells24. Therefore, hypoxia-related UPR changes in intermediate progenitors could result in cell cycle changes and premature neural differentiation. Fourth, we show that the UPR modulator ISRIB– a small molecule with good pharmacokinetic properties and that permeates the blood–brain barrier25, prevents the oxygen-related TBR2 defects in hCS and in human tissue. Future studies should validate this effect in other models to evaluate translational potential and investigate the effect of genotype on hypoxia susceptibility and recovery.
There are several limitations to our study. Our hCS system does not include immune cells and is not vascularized, and therefore the inflammation component of EP is not captured here33. Future assembloids models could incorporate microglia and other cells34 to study their contributions to EP. Although oxygen concentration in the cerebral circulation increases following premature birth35 and our goal was to model postnatal hypoxia in the premature brain, hCS derivation and maintenance are pursued at atmospheric oxygen levels. We have shown that hCS can be maintained in vitro for hundreds of days to model late stages of development to include astrocytes that resemble primary postnatal astrocytes11. Therefore, future studies could use this system to model brain injury at later in utero stages. Postmortem studies have indicated a significant loss of GABAergic interneurons in premature infants36,37. Our current model does not include interneurons, which are born in the ventral forebrain and have to migrate dorsally to integrate into cortical circuits38. Therefore, future studies could use forebrain assembloids that combine dorsal and ventral forebrain organoids to model migration39. Similarly, organoid models that include oligodendrocytes together with astrocytes and neurons could be combined with the hypoxia platform we are describing to model myelination defects40.
The use of 3D brain organoids or assembloids hold great in studying the interaction of environmental factors impacting brain development. Moreover, this personalized, scalable human system could dissect individual susceptibility and protective factors, and therefore, may ultimately identify therapeutics strategies to bridge the disconnect between medical advances and neurodevelopmental outcomes in neonatology.
MATERIALS & METHODS:
Culture of hiPS cells
The hiPS cell lines used in this study were validated using standardized methods as previously described9,39,41. Cultures were tested for and maintained mycoplasma free. A total of five control hiPS cell lines derived from fibroblasts collected from four subjects (three males and one females). Approval for this study was obtained from the Stanford IRB panel and informed consent was obtained from all subjects.
Generation of hCS from hiPSC
hPS cells were cultured on inactivated mouse embryonic fibroblast feeders (EmbryoMax PMEF; Millipore) in hPS cell medium containing DMEM/F12 (1:1, Life Technologies, 11330), knockout serum (20%, Life Technologies, 10828), non-essential amino acids (1 mM, Life Technologies, 11140), GlutaMax (1: 200, Life Technologies, 35050), β-mercaptoethanol (0.1 mM, Sigma-Aldrich, M3148), penicillin and streptomycin (1:100, Life Technologies, 15070), and supplemented with FGF2 (10 ng ml−1 diluted in 0.1% BSA; R&D Systems). The generation of hCS from hiPS cells was performed as previously described9. To initiate the generation of hCS, intact hiPS cells colonies were lifted from the plates using dispase (0.35 mg ml−1) and transferred into ultralow-attachment plastic dishes (Corning) in hPS cell medium supplemented with the two SMAD inhibitors dorsomorphin (5 μM, Sigma-Aldrich) and SB-431542 (10 μM, Tocris), and the ROCK inhibitor Y-27632 (10 μM, EMD Chemicals). For the first five days, the hPS cell medium was changed every day and supplemented with dorsomorphin and SB-431542. On the sixth day in suspension, neural spheroids were transferred to neural medium containing neurobasal-A (Life Technologies, 10888), B-27 supplement without vitamin A (Life Technologies, 12587), GlutaMax (1:100, Life Technologies), penicillin and streptomycin (1:100, Life Technologies) and supplemented with the growth factors EGF (20 ng ml−1; R&D Systems) and FGF2 (20 ng ml−1; R&D Systems) until day 24. From day 25 to 42, the neural medium was supplemented with the growth factors BDNF (20 ng ml−1, Peprotech) and NT3 (20 ng ml−1, Peprotech) with medium changes every other day. From day 43 onwards, hCS were maintained in unsupplemented neural medium with medium changes every four to six days. A step-by-step protocol describing hCS generation can also be found ref8.
Exposure of hCSs to low O2
hCSs derived from five hiPS cell lines (4 individuals) were used for low O2 exposure in a total of 9 independent experiments. At 74–78 days of in vitro differentiation, hCS maintained in 21% O2 and 5% CO2, were transferred to a C-chamber hypoxia sub-chamber (Biospherix) in neural media (without growth factors) for 48 hours. The media was previously equilibrated for ~16 hours at <1% O2, 5% CO2, 37°C. The level of O2 was controlled using a Proox 110 Compact Oxygen Controller (Biospherix) and a mixed CO2/N2 compressed gas source. After 48 hours, hCS were immediately collected for analyses or transferred to an incubator with 21% O2 and 5% CO2 for another 72 hours (re-oxygenation condition).
Exposure of hCSs to tunicamycin
At 74–78 days of in vitro differentiation, hCS were exposed to 1.2 μM tunicamycin (Sigma, T7765) for 48 hours at 21% O2 and 5% CO2. After 48 hours, hCS were collected for analyses as described for the hypoxia experiments.
Oxygen tension measurements
The oxygen optical microsensor OXB50 (50 μm, PyroScience, Aachen, Germany) was used to measure O2 tension in hCSs. This sensor measures changes in O2 tension by calculating the quenching of a probe (Red Flash Dye) situated at the tip of the sensor. The probe is excited with red light at a wavelength of 610–630 nm and shows an oxygen-dependent luminescence in the near infrared spectrum (760–790 nm). A two-point calibration was performed before measurements. Pure nitrogen gas and ambient air were set as 0 and 100% air saturation. Optical signals were sampled and digitized by the sensor’s compatible meter (FireStingO2, PyroScience) and the Oxygen Logger software (PyroScience). To determine the O2 levels at various depths, we mounted the sensor on a micromanipulator with a high-precision slide (<0.1 μm axial resolution) and applied 50 μm steps from the hCS surface towards the center.
Human tissue
Human tissue was obtained under a protocol approved by the Research Compliance Office at Stanford University. The tissue was immediately placed in RPMI media and processed within 16 hours of collection using a previously adapted protocol42. In brief, PCW20 frontal brain tissue was embedded in 4% low-melting point agarose in PBS and cut using a scalpel to obtain 2-mm thick sections. The sections were then placed in tissue-culture plates containing culture medium (66% BME, 25% Hanks, 5% FBS, 1% N-2, 1% penicillin, streptomycin and glutamine; all from Invitrogen) and 0.66% D-(+)-glucose (Sigma-Aldrich) and incubated in standard conditions (21% O2, 5% CO2, 37 °C) for 24 hours. Sections were subsequently exposed to <1% O2 and 5% CO2 in a chamber or maintained in 21% O2 and 5% CO2 for 48 hours. Samples were then collected and processed for analyses.
Cryopreservation
hCS were fixed in 4% paraformaldehyde (PFA) overnight. They were then washed in PBS, transferred to 30% sucrose for 48–72 hours. Subsequently, they were transferred into embedding medium (Tissue-Tek OCT Compound 4583, Sakura Finetek), snap-frozen on dry ice and stored at −80 °C. For immunohistochemistry, 10 μm thick sections were cut using a cryostat (Leica).
Slices of primary cortical tissue (2 mm thick) were fixed in 4% PFA for 48 hours. They were then washed in PBS and transferred to 30% sucrose for 5 days. Subsequently, they were moved to embedding medium (1:1 mixture of 30% sucrose and Tissue-Tek OCT Compound 4583, Sakura Finetek), snap-frozen on dry ice and stored at −80 °C. For immunohistochemistry, 40 μm thick cryosections were cut using a Leica cryostat.
Immunocytochemistry
Cryosections of hCS were washed with PBS to remove excess OCT and blocked in 10% normal donkey serum (NDS), 0.3% Triton X–100 diluted in PBS for 1 hour at room temperature. The sections were then incubated overnight at 4°C with primary antibodies diluted in PBS containing 2% NDS and 0.1% Triton X–100. PBS was used to wash off the primary antibodies and the cryosections were incubated with secondary antibodies in PBS with 2% NDS and 0.1% Triton X-100 for 1 hour. After secondary antibodies, 3 washes of 15 minutes were performed in PBS. The following primary antibodies were used for immunohistochemistry: anti-CTIP2 (rat, 1:300, Abcam, AB18465), p27 (rabbit, 1:100, Thermo Fisher Scientific, PA5–27188), ATF4 (rabbit,1:50, Cell Signaling, D4B8, 11815), HIF-1α (mouse,1:50, Santa Cruz sc-53546), PAX6 (rabbit, 1:300, Biolegend, PRB-278P), SOX2 (rabbit, 1:200, Cell Signaling 3579S), PH3 (rat, 1:500, Abcam AB10543), Ki67 (rabbit, 1:100, Novus Biological NB600–1252). For TBR2 staining (anti-TBR2, mouse, 1:100; R&D, MAB6166), antigen retrieval (using 10 mM Na+ citrate and heating to 95°C) was performed for 20 minutes and sections were blocked in 0.1% Triton X–100, 10% serum, and 0.2% gelatin for 1 hour. The sections were incubated with primary antibodies in blocking buffer overnight at 4°C, then washed with PBS and incubated with secondary antibodies diluted in blocking buffer for 1 hour at room temperature. After secondary antibodies, 3 washes of 15 minutes were performed in PBS.
Cryosections of primary cortical tissue were washed with PBS to remove excess OCT. Antigen retrieval (using 10 mM Na+ citrate and heating to 95°C) was performed for 20 minutes. Cryosections were blocked in 0.1% Triton X–100, 10% normal donkey serum (NDS) and 0.2% gelatin diluted in PBS for 4 hours at room temperature. The sections were then incubated for 24 hours at 4°C with primary antibodies diluted in blocking solution. Primary antibodies were washed off with 0.5% Triton X–100 diluted in PBS for 1 hour and subsequently incubated with secondary antibodies in blocking solution for 2 hours at room temperature. After secondary antibodies, 3 washes of 20 minutes were performed in PBS. The following primary antibodies were used for immunohistochemistry: PAX6 (rabbit, 1:300, Biolegend, PRB-278P), TBR2 (sheep, 1:100, Novus Biologicals, AF6166).
Secondary antibodies AlexaFluor 488, 568 and 647 (donkey anti– rabbit, rat, mouse or sheep) were used at 1: 500 dilution. Nuclei were visualized with Hoechst 33258 (LifeTechnologies, 1:10,000). Cryosections were mounted for microscopy on glass coverslips, using Aquamount (ThermoScientific) and imaged on a Zeiss M1 Axioscope (for hCS cryosections) or Keyence BZ-X700 microscope (for primary cortical tissue cryosections).
Immunocytochemistry quantifications
The density of TBR2+, PAX6+ and Hoechst+ cells was estimated in 10 μm thick hCS cryo-sections. The number of cells was quantified in circular areas around proliferative zones larger than 300μm2. The boundary between proliferative zones (VZ and SVZ) and CP was established based on the expression, cell orientation and density of nuclei (Hoechst), PAX6+ and CTIP2+ cells. Independently, the proportion of TBR2+ and PAX6+ cells were also quantified out of total number of cells (Hoechst+) in whole 10 μm thick cryosections of hCS from multiple hiPS cells lines.
Western Blot
hCS were rapidly lysed on ice with the RIPA lysis buffer system (Santa Cruz, sc-24948) after low O2 exposure. Whole cell lysates were then separated on gels (Invitrogen, NuPAGE 4–12% bis-tris) and transferred to a PVDF membrane (Millipore, Immobilon-P Membrane, 0.45 μm). The membranes were incubated at 4°C overnight with the following primary antibodies: HIF-1α (mouse, 1:1000, BD bioscience, 610958) β-actin (mouse, 1:20,000, Sigma, A5316) and c-CAS3 (rabbit, 1:500, Abcam, ab32042). Horseradish peroxidase(HRP) conjugated anti-mouse or anti-rabbit IgG were used as secondary antibodies. Signals were developed by chemiluminescence (Amersham, ECL Western blotting detection reagents, RPN2106). Bands were quantified using ImageJ software, normalized to background and to a β-actin control. Uncropped blots of representative Western blots are shown in Supplementary Data Figure and values from quantification for all blots used are listed in Supplementary Table 2.
Real-time quantitative PCR (qPCR)
mRNA was isolated using the RNeasy Mini kit and RNase-Free DNase set (Qiagen), and template cDNA was prepared by reverse transcription using the SuperScript III First-Strand Synthesis SuperMix for qRT–PCR (Life Technologies). qPCR was performed using Sybr Green (Roche) on a ViiA7 machine (Applied Biosystems, Life Technologies). Data was processed using the QuantStudio RT–PCR software (Applied Biosystems). Primers and sequences are listed in Supplementary Table 4.
RNA-Seq
mRNA was isolated using the miRNeasy micro Kit and RNase-Free DNase set (Qiagen). Library preparation was conducted by Macrogen (https://www.macrogenusa.com) using the TruSeq Stranded Total RNA LT Sample Prep Kit. Samples were then sequenced on the Illumina HiSeq 4000 System using paired-end, 101 base pair-long reads to a minimum of 40 million reads per sample. We used the STAR version 2.5.2b43 to align raw RNA-Seq reads to the GRCh37 human genome reference from the Illumina iGenome Project. We generated alignment quality control (QC) metrics with Picard (www.broadinstitute.github.io/picard)). Principal component analysis (PCA) of these metrics did not detect outliers. We then used HTSeq44 to generate gene level count data, and kept transcripts with at least one read in more than 50% of the samples. The CQN R package45 normalized the remaining 24,878 transcripts for gene length, GC content, and sample library size. We further removed genes whose expression value did not change across all samples or with average normalized expression value less than one (i.e. lowly expressed gene). We screened again with PCA of normalized gene expression but did not detect outliers.
We conducted hierarchical clustering and differential gene expression (DEX) analysis with normalized gene expression values. In order to identify confounding variables, we used a regression model selection approach. We built negative binomial regression models to model expressions of each gene separately. A forward stepwise algorithm selected the best model, defined as the model where the Bayesian information criterion (BIC) could not be improved by adding additional confounding variables. With this approach, we identified the three most impactful confounding variables: hiPS cell line, percentage of UTR bases, and percentage of intronic bases. We used edgeR46,47 to identify significant genes at each of the three time points (after 24 hours or 48 hours of <1% O2, and after 72 hours of re-oxygenation) and to remove the effects of the three confounding variables. We corrected p values for multiple comparisons using the Benjamini-Hochberg method and selected significant genes based on two criteria: false discovery rate (FDR) ≤ 0.05, and at least 50% increase or decrease in expression between control and hypoxia condition (i.e. 1.5-fold-change, up or down). As a final quality control step, we identified genes that are differentially expressed across time in the control samples (i.e. highly correlated with time) and removed them from the list of differentially expressed genes. As the fold-change threshold is somewhat arbitrary we list the entire set of genes with FDR ≤ 0.05 in the supplementary material, along with log2 fold-changes (Supplementary Table 3). We combined all significant genes identified at all three time points to generate a heatmap of all samples (heatmap3 R package). We used the Goseq48 R package as well as ToppGene49 to identify GO terms enriched in our significant gene list.
To examine the relationship between DEX and the developing human cortex, we used layer-specific data that was previously reported for PCW21 ref16. We used a hypergeometric test (two-sided) to assess enrichment, and corrected P values for multiple comparisons using the Bonferroni correction. We determined the background for these calculations as the number of genes overlapping between our RNA-seq data and the data reported in Miller et al.16. In order to ensure that our choice of a 1.5-fold-change threshold did not affect the results of these analyses, we also conducted exploratory analyses to assess whether the enrichment for SVZ transcripts is robust at different thresholds. We observe that this enrichment is stable across a wide range of fold-change thresholds (approximately 1.3 fold-change to 2.5 fold-change).
We conducted gene co-expression analysis following normalization for gene length and GC content (using CQN—see above). The WGCNA50 R package computed unsigned similarity matrix with a softpower of 12. We created gene modules based on hierarchical clustering results of the similarity matrix and summarized the “average” expression of all the genes in a given module with the first principal component of module expression (module “eigengene”). We then evaluate the relationship between these gene modules and experimental conditions (i.e., oxygen tension) by computing correlation coefficients to the first principal component. We identified a total of 9 significant gene co-expression modules based on two cutoffs: correlation coefficient ≥ 0.9 and FDR ≤ 0.05. We conducted GO enrichment analysis of the genes in each of these 9 significant modules with the R package Goseq48. Only two modules were significantly enriched for GO terms after FDR correction: the blue and turquoise modules.
Statistics
Data are presented as mean ± s.e.m., unless otherwise indicated. Distribution of the raw data was tested for normality of distribution; statistical analyses were performed accordingly using paired or unpaired t-test (two-sided), Mann–Whitney U-test, Wilcoxon rank-sum test, or the Friedman or ANOVA test with multiple comparison correction as indicated. Sample sizes were estimated empirically. GraphPad Prism v7 was used for statistical analyses.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Oxygen tension and cleaved caspase-3 in hCS following hypoxia.
(a) Oxygen tension (pO2, mmHg) measurements inside hCS from Fig. 1b, shown as a function of depth (data for each hCS was normalized as function its radius) (n= 6 hCS for 21% O2 and n= 7 hCS for <1% O2; from 3 hiPS cell lines). Shaded area indicates s.e.m. (b, c) Representative western blots and quantification of cleaved Cas3 (c-Cas3) in hCS after 24 hours and 48 hours of exposure to <1% O2 and at 72 hours after reoxygenation (one-way ANOVA, F3, 6= 1.56; P= 0.29); normalized to β-actin (n= 3 differentiated hiPS cell lines with two hCSs per condition; each line is shown in a different color). Western blots were cropped to show the relevant bands; molecular weight (MW) markers are indicated on the left (in kDa). Source Data 2 for uncropped western blots and Supplementary Table 2 for quantifications. Data are mean ± s.e.m. Individual values are indicated by dots.
Extended Data Fig. 2. RNA-seq clustering and qPCR validation.
(a) Hierarchical clustering of RNA-seq data showing clustering of samples based on exposure to oxygen concentration. Samples (n= 24) from hCS differentiated from 3 hiPS cell lines were collected at 24 hours or 48 hours after<1% O2, as well as 72 hours after re-oxygenation. We clustered based on all DE genes identified (1,754 unique genes). Clustering with all expressed genes results in a similar dendrogram (data not shown). (b, c) Validation by qPCR of hypoxia-related genes PLOD2 (two-tailed paired t-test, **P= 0.006), PFKP (two-tailed paired t-test, **P= 0.008), PDK1 (two-tailed paired t-test, ***P= 0.0005), IGFBP2 (two-tailed paired t-test, ***P= 0.0002), and cortical progenitor and cell cycle-related genes EOMES (also known as TBR2) (two-tailed paired t-test, **P= 0.006), EMX1 (two-tailed paired t-test, *P= 0.02), ASPM (two-tailed paired t-test, **P= 0.002), CENPF (two-tailed paired t-test, ****P< 0.0001), which were identified in the RNA-seq (n= 4 hiPS cell lines differentiated; each line is shown in a different color; expression normalized to the RPL13a housekeeping gene). Data are mean ± s.e.m. Individual values are indicated by dots.
Extended Data Fig. 3. Immunocytochemistry quantifications in hCS following hypoxia.
(a) Quantification of the density of Hoechst+ cells in proliferative areas in hCS after exposure to 48 hours of <1% O2. (Left) Data shown as average across individual hCS proliferative zones from 3 hiPS cell lines per condition (n= 21 areas for 21% O2 versus n= 18 areas for <1% O2; two-tailed Mann-Whitney test, P= 0.09), and (Right) as average across different hiPS cell lines (n= 3 hiPS cell line, two-tailed Wilcoxon test, P = 0.50). (b) Quantification of the proportion of PAX6+ cells in whole-cryosections of hCS after exposure to 48 hours of <1% O2. (Left) Data shown as average across whole-section of hCS from 4 hiPS cell lines per condition (n= 25 sections for 21% O2 versus n= 23 sections for <1% O2; two-tailed unpaired t-test, P= 0.44), or (Right) as average across different hiPS cell lines (n= 4 hiPS cell lines; right: two-tailed Wilcoxon-test, P= 0.25). (c) Quantification of the proportion of TBR2+ cells in whole-cryosections of hCS after exposure to 48 hours of <1% O2. (Left) Data shown as average across whole-section of hCS from 4 hiPS cell lines per condition (n= 25 sections for 21% O2 versus n= 27 sections for <1% O2; two-tailed Mann-Whitney U test, ****P< 0.0001), or (Right) as average across different hiPS cell lines (n= 4 hiPS cell lines; two-tailed paired t-test, *P= 0.03; each line is shown in a different color). (d) Quantification of the proportion of cells co-expressing SOX2 and TBR2 in cryosections of hCS after exposure to 48 hours of <1% O2. (Left) Data shown as average across individual hCS cryo-sections from 4 hiPS cell lines per condition (n= 10 sections for 21% O2, n= 12 sections for <1% O2; two-tailed unpaired t-test, P= 0.82; from 4 hiPS cell lines), or (Right) as average across different hiPS cell lines (n= 3 hiPS cell line; right: two-tailed paired t-test, P= 0.06; each line is shown in a different color). Data are mean ± s.e.m., individual values are indicated by dots.
Extended Data Fig. 4. WGCNA analysis and qPCR validation.
(a) Hierarchical clustering of WGCNA modules identified in the RNA-seq data. Clustering is based on the module eigengenes (‘average’ expression profile of all module genes. The turquoise and blue modules are very similar in overall eigengene expression pattern. (b) Statistical significance for correlation of each module with low O2 exposure (bars are labeled by the color of the modules). The blue and turquoise modules are highly associated with exposure (FDR ≤ 0.05). (c) Enrichment for pathways in the turquoise and blue modules (bars are labeled by the color of the modules in which they are enriched). Only pathways with Bonferroni-corrected FDR < 1 x 10-4 are shown. (d) Validation by qPCR of the UPR-related genes PERK (two-tailed paired t-test, *P= 0.03), ATF3 (two-tailed paired t-test, *P= 0.03), XBP1s (two-tailed paired t-test, *P= 0.04), which were identified in the RNA-seq (n= 4 hiPS cell lines; each line is shown in a different color; expression normalized to the RPL13a housekeeping gene). Data are mean ± s.e.m., individual values are indicated by dots.
Extended Data Fig. 5. Immunocytochemistry quantifications of UPR, cell cycle and proliferation in hCS.
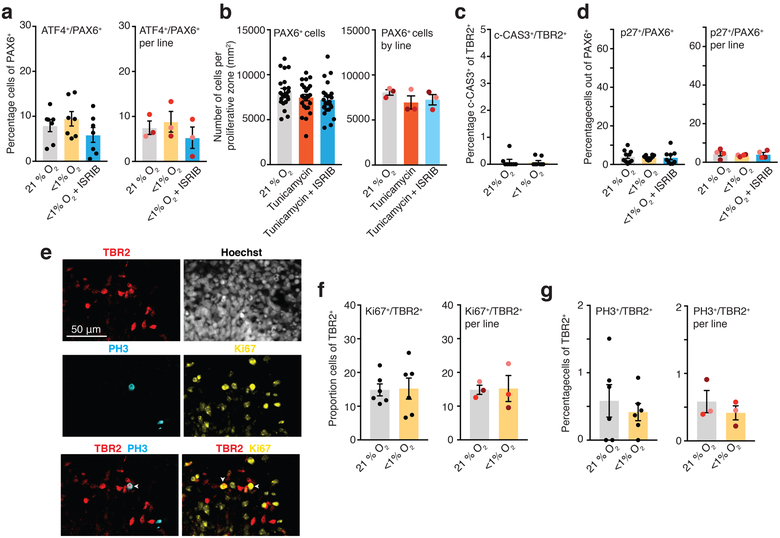
(a) Quantification of the proportion of cells co-expressing ATF4/PAX6 in cryo-sections of hCS after exposure to 48 hours of <1% O2 in the presence or absence of 10 nM ISRIB. (Left) Data shown as average across individual hCS cryo-sections from 3 hiPS cell lines per condition (n= 7 sections for 21% O2 versus n= 7 sections for <1% O2 versus n= 7 sections for <1% O2 with ISRIB; one-way ANOVA F2, 18= 1.37, P= 0.27; Dunnett’s multiple comparison test versus 21% O2, P= 0.70, P= 0.54), or (Right) as average across different hiPS cell lines (n= 3 hiPS cell lines; one-way ANOVA, F2, 4= 4.64, P= 0.09; Dunnett’s multiple comparison test versus 21% O2, P= 0.48, P= 0.21; each line is shown in a different color). (b) Quantification of the density of PAX6+ cells in hCS after exposure for 48 hours to 1.2 μM of tunicamycin in the presence or absence of 10 nM ISRIB. (Left) Data shown as average across individual hCS proliferative zones from 3 hiPS cell lines per condition (n= 22 areas for 21% O2, n= 23 areas for tunicamycin, n= 24 areas for tunicamycin with ISRIB; one-way ANOVA, F2,66 = 1.57, P= 0.21; Dunnett’s multiple comparison test versus 21% O2, P= 0.37, P= 0.50), or (Right) as average across different hiPS cell lines (n= 3 hiPS cell line; Friedman’s test, P = 0.19; Dunnett’s multiple comparison test versus 21% O2, P= 0.20, P= 0.08; each line is shown in a different color). (c) Proportion of TBR2+ cells that co-express c-CAS3 in whole cryo-sections of hCS maintained in 21% O2 or exposed to <1% O2 for 48 hours. (two-tailed Mann-Whitney U test, P> 0.99; n= 8 cryo-section from 2 hiPS cell lines). (d) Quantification of the proportion of cells co-expressing p27 and PAX6 in cryosections of hCS after exposure to 48 hours of <1% O2 in the presence or absence of 10 nM ISRIB. (Left) Data shown as average across individual hCS cryo-sections from 4 hiPS cell lines per condition (n= 10 sections for 21% O2 versus n= 10 sections for <1% O2 versus n= 9 sections for <1% O2 with ISRIB; one-way ANOVA, F2, 26= 0.10, P= 0.90 Dunnett’s multiple comparison test versus 21% O2, P= 0.88, P= 0.90), or (Right) as average across different hiPS cell lines (n= 4 hiPS cell line; one-way ANOVA F2, 6= 0.30, P= 0.74, Dunnett’s multiple comparison test versus 21% O2, P= 0.67, P= 0.94; each line is shown in a different color). (e) Representative images of cells co-expressing TBR2, Ki67 and PH3 in cryosections of hCS. White arrows show example of cells that are TBR2+/PH3+ or TBR2+/Ki67+. Scale bar, 50 μm. (f) Quantification of the proportion of cells co-expressing Ki67 and TBR2 in cryosections of hCS after exposure to 48 hours of <1% O2. (Left) Data shown as average across individual hCS cryo-sections from 3 hiPS cell lines per condition (n= 6 sections for 21% O2 versus n= 6 sections for <1% O2; two-tailed unpaired t-test, P= 0.91), or (Right) as average across different hiPS cell lines (n= 3 hiPS cell line; two-tailed paired t-test, P= 0.91; each line is shown in a different color). (g) Quantification of the proportion of cells co-expressing PH3 and TBR2 in cryosections of hCS after exposure to 48 hours of <1% O2. (Left) Data shown as average across individual hCS cryo-sections from 3 hiPS cell lines per condition (n= 6 sections for 21% O2 versus n= 6 sections for <1% O2; two-tailed unpaired t-test, P= 0.55), or (Right) as average across different hiPS cell lines (n= 3 hiPS cell line; two-tailed paired t-test, P= 0.56; each line is shown in a different color). Data are mean ± s.e.m., individual values are indicated by dots.
Supplementary Material
ACKNOWLEDGEMENTS
We thank W.E. Benitz, D.K. Stevenson, V. K. Bhutani and U. Francke for valuable scientific advice and discussions. This work was supported by the US National Institute of Health (NIH) BRAINS Award (MH107800), the MQ Fellow Award, the NYSCF Robertson Stem Cell Investigator Award, the Stanford Neurosciences Institute’s Human Brain Organogenesis Program and the Brain Rejuvenation Project, Stanford Bio-X, the Kwan Research Fund and the California Institute of Regenerative Medicine (CIRM) (to S.P.P.); the UCSF Weill Institute for Neurosciences (startup funds to A.J.W.); NIH R01MH108659 and R01MH108660 (to T.D.P.); and the NIH K12-HD000850 (Pediatric Scientist Development Program), the Association of Medical School Pediatric Department Chairs (AMSPDC) and Stanford Maternal and Child Health Research Institute Fellowship (to A.M.P.).
Data availability
Gene expression data are available in the Gene Expression Omnibus (GEO) under accession numbers GSE112137. The data that support the findings of this study are available on request from the corresponding author.
Footnotes
Competing interests
Stanford University has filed a provisional patent application that covers the generation of region-specific brain organoids from pluripotent stem cells (US Application Serial No. 15/158,408).
MAIN REFERENCES
- 1.Penn AA, Gressens P, Fleiss B, Back SA & Gallo V Controversies in preterm brain injury. Neurobiol Dis 92, 90–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volpe JJ Brain injury in premature infants: a complex amalgam of destructive and developmental disturbances. The Lancet. Neurology 8, 110–124 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volpe JJ The encephalopathy of prematurity--brain injury and impaired brain development inextricably intertwined. Semin Pediatr Neurol 16, 167–178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarjour IT Neurodevelopmental outcome after extreme prematurity: a review of the literature. Pediatr Neurol 52, 143–152 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Salmaso N, Jablonska B, Scafidi J, Vaccarino FM & Gallo V Neurobiology of premature brain injury. Nat Neurosci 17, 341–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sousa AMM, Meyer KA, Santpere G, Gulden FO & Sestan N Evolution of the Human Nervous System Function, Structure, and Development. Cell 170, 226–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasca SP The rise of three-dimensional human brain cultures. Nature 553, 437–445 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Sloan SA, Andersen J, Pasca AM, Birey F & Pasca SP Generation and assembly of human brain region-specific three-dimensional cultures. Nat Protoc (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasca AM, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nature methods 12, 671–678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoon SJ, et al. Reliability of human cortical organoid generation. Nature methods 16, 75–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sloan SA, et al. Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron 95, 779–790 e776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arshad A, et al. Extended Production of Cortical Interneurons into the Third Trimester of Human Gestation. Cereb Cortex 26, 2242–2256 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zecevic N, Chen Y & Filipovic R Contributions of cortical subventricular zone to the development of the human cerebral cortex. The Journal of comparative neurology 491, 109–122 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik S, et al. Neurogenesis continues in the third trimester of pregnancy and is suppressed by premature birth. J Neurosci 33, 411–423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carreau A, El Hafny-Rahbi B, Matejuk A, Grillon C & Kieda C Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J Cell Mol Med 15, 1239–1253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bershteyn M, et al. Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell 20, 435–449 e434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagenfuhr L, Meyer AK, Braunschweig L, Marrone L & Storch A Brain oxygen tension controls the expansion of outer subventricular zone-like basal progenitors in the developing mouse brain. Development 142, 2904–2915 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Stuart JM, Segal E, Koller D & Kim SK A gene-coexpression network for global discovery of conserved genetic modules. Science 302, 249–255 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B & Horvath S A general framework for weighted gene co-expression network analysis. Statistical applications in genetics and molecular biology 4, Article17 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Wang M & Kaufman RJ Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Fels DR & Koumenis C The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther 5, 723–728 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Laguesse S, et al. A Dynamic Unfolded Protein Response Contributes to the Control of Cortical Neurogenesis. Dev Cell 35, 553–567 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Sidrauski C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2, e00498 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sidrauski C, et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife 4, e07314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zyryanova AF, et al. Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B. Science 359, 1533–1536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, et al. Tunicamycin-induced unfolded protein response in the developing mouse brain. Toxicol Appl Pharmacol 283, 157–167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Javaherian A & Kriegstein A A stem cell niche for intermediate progenitor cells of the embryonic cortex. Cereb Cortex 19 Suppl 1, i70–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortega JA, Sirois CL, Memi F, Glidden N & Zecevic N Oxygen Levels Regulate the Development of Human Cortical Radial Glia Cells. Cereb Cortex 27, 3736–3751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollen AA, et al. Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Godin JD, Creppe C, Laguesse S & Nguyen L Emerging Roles for the Unfolded Protein Response in the Developing Nervous System. Trends in neurosciences 39, 394–404 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Jantzie LL & Robinson S Preclinical Models of Encephalopathy of Prematurity. Developmental neuroscience 37, 277–288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amin ND & Pasca SP Building Models of Brain Disorders with Three-Dimensional Organoids. Neuron 100, 389–405 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Soothill PW, Nicolaides KH, Rodeck CH & Gamsu H Blood gases and acid-base status of the human second-trimester fetus. Obstet Gynecol 68, 173–176 (1986). [PubMed] [Google Scholar]
- 36.Lacaille H, et al. Impaired Interneuron Development in a Novel Model of Neonatal Brain Injury. eNeuro 6(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson S, Li Q, Dechant A & Cohen ML Neonatal loss of gamma-aminobutyric acid pathway expression after human perinatal brain injury. J Neurosurg 104, 396–408 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paredes MF, et al. Extensive migration of young neurons into the infant human frontal lobe. Science 354(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birey F, et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marton RM, et al. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat Neurosci 22, 484–491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paşca SP, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med 17, 1657–1662 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lui JH, et al. Radial glia require PDGFD-PDGFRbeta signalling in human but not mouse neocortex. Nature 515, 264–268 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen KD, Irizarry RA & Wu Z Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 13, 204–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarthy DJ, Chen Y & Smyth GK Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young MD, Wakefield MJ, Smyth GK & Oshlack A Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology 11, R14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J, Bardes EE, Aronow BJ & Jegga AG ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langfelder P & Horvath S WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.