Abstract
Mycophenolic acid (MPA) is a potent natural product inhibitor of fungal and other eukaryotic inosine 5′‐monophosphate dehydrogenases (IMPDHs) originally isolated from spoiled corn silage. MPA is produced by the filamentous fungi Penicillium brevicompactum, which contains two IMPDHs, PbIMPDHA and PbIMPDHB, both of which are MPA‐resistant. The MPA binding sites of these enzymes are identical to MPA‐sensitive IMPDHs, so the structural determinants of resistance are unknown. Here we show that a single residue, Ser267, accounts for the MPA resistance of PbIMPDHA. Substitution of Ser267 with Ala, the residue most commonly found in this position in eukaryotic IMPDHs, makes PbIMPDHA sensitive to MPA. Conversely, Aspergillus nidulans IMPDH becomes MPA‐resistant when the analogous Ala residue is substituted with Ser. These substitutions have little effect on the catalytic cycles of either enzyme, suggesting the fitness costs are negligible despite the strong conservation of Ala at this position. Intriguingly, while only 1% of fungal IMPDHs contain Ser or Thr at position 267, these residues are found in the IMPDHs from several Aspergillus species that grow at the low temperatures also favored by Penicillium. Perhaps Ser/Thr267 is an evolutionary signature of MPA exposure.
Keywords: Aspergillus, guanine nucleotides, IMP dehydrogenase, IMPDH, Penicillium
1. INTRODUCTION
Mycophenolic acid (MPA) was the first purified antibiotic, originally isolated from spoiled corn silage by Gosio in 1893.1 MPA is an uncompetitive inhibitor of inosine 5′‐monophosphate dehydrogenase (IMPDH), thereby blocking guanine nucleotide biosynthesis.2 Despite its antibiotic origins, MPA is a poor inhibitor of prokaryotic IMPDHs (K ii typically ∼10 μM), but a potent inhibitor of fungal and other eukaryotic IMPDHs (K ii typically ∼10 nM). MPA and its morphilino ester prodrug (Cellcept) are currently used as immunosuppressant drugs.3
MPA is produced by certain Penicillium fungi, most notably Penicillium brevicompactum.4 The MPA biosynthetic cluster includes a gene encoding a MPA‐resistant IMPDH (PbIMPDHB) and expression of this gene is sufficient to provide self‐resistance.5 This enzyme is 103‐fold more resistant to MPA than IMPDHs from fungi such as Aspergillus nidulans (AnIMPDH) that do not produce the drug.6, 7 P. brevicompactum also contains a “normal” IMPDH gene encoding PbIMPDHA. This enzyme is also more resistant than typical fungal IMPDHs.6, 7 Curiously, the MPA binding sites of PbIMPDHA and PbIMPDHB are identical to those of MPA‐sensitive IMDPHs (Figure 1a). While some of the structural determinants of the PbIMPDHB resistance have been identified,7 the structural basis for the resistance of PbIMPDHA is unknown.
Figure 1.
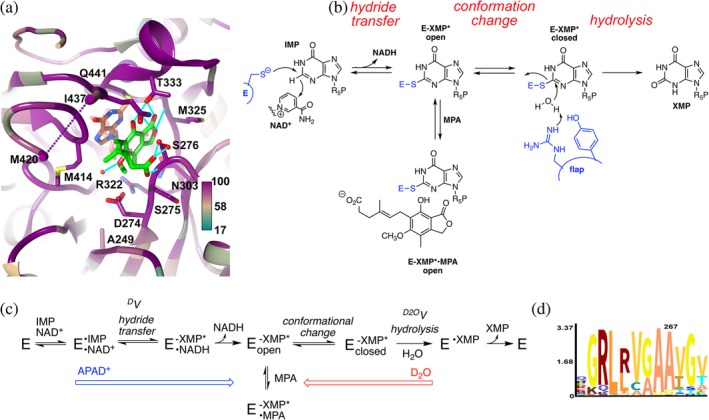
The inhibition of inosine 5′‐monophosphate dehydrogenase (IMPDH) by mycophenolic acid (MPA). (a) Structure of the MPA binding site in Cricetulus griseus (CgIMPDH2) colored by conservation (https://apps.spi-global.com/eProofing/VerifyTokenandAuthenticate.aspx?token=oIM5AEXhrAJLPSPI_PlusGnzGemOw&ChapterOrArticleOrBook=Article 27). The alignment included three MPA‐resistant IMPDHs (PbIMPDHA, PbIMPDHB, and ScIMPDH2) and seven MPA‐sensitive IMPDHs (AnIMPDH, CaIMPDH, PrIMPDHA, PrIMPDHB, ScIMPDH3, ScIMPDH4, and CgIMPDH2; alignment in Supporting Information). MPA, green; E‐XMP*, coral; K+, purple ball; residues within 4 Å of MPA and IMP are shown in stick. Ala249 is analogous to Ala267 in PbIMPDHA. (b). Mechanism of the IMPDH reaction. MPA traps E‐XMP* by binding in the vacant cofactor site after NADH dissociates, preventing the conformational change that brings the flap into the cofactor site for hydrolysis of E‐XMP*. (c). Kinetic mechanism of the IMPDH reaction. APAD+ has a lower affinity and higher redox potential than NAD+ (−0.258 V vs. −0.320),12 which will push the reaction toward E‐XMP*. D2O will make the hydrolysis step more rate limiting, which can increase the accumulation of E‐XMP*. (d). Logo graphic of conservation of sequence around position 267 in 20,000 fungal IMPDHs
IMPDH catalyzes the oxidation of inosine 5′‐monophosphate (IMP) to xanthosine 5′‐monophosphate (XMP) with the concomitant reduction of NAD+.2 This reaction is the rate‐limiting step in guanine nucleotide biosynthesis. The catalytic cycle of IMPDH involves two different chemical transformations (Figure 1b). In the redox step, the catalytic Cys attacks C2 of IMP and a hydride is transferred to NAD+, forming the covalent intermediate E‐XMP*. NADH is released and a mobile flap folds into the vacant cofactor site, bringing the conserved Arg‐Tyr dyad into position to hydrolyze E‐XMP*. MPA traps the intermediate E‐XMP* by binding in the cofactor site, competing with flap closure. Thus, in addition to the structure of the MPA binding site, the accumulation of E‐XMP* and the equilibrium between open and closed flap conformations modulate MPA susceptibility (Figure 1c).
Here we show that the MPA resistance of PbIMPDHA is determined by Ser267 (PbIMPDHA numbering). PbIMPDHA becomes more sensitive to MPA when Ser267 is substituted with Ala, the residue found in this position in most eukaryotic IMPDHs. Conversely, the substitution of Ala267 with Ser increases the MPA resistance of AnIMPDH. These substitutions have little effect on the catalytic cycles of either enzyme, suggesting the fitness costs are negligible despite the strong conservation of Ala at this position. Intriguingly, while only 1–2% of fungal IMPDHs contain Ser or Thr at position 267 (Figure 1d), these residues are found in the IMPDHs from several Aspergillus species that grow at the low temperatures also favored by Penicillium. Perhaps Ser/Thr267 is an evolutionary signature for MPA exposure.
2. RESULTS AND DISCUSSION
2.1. Residue 267 is a candidate determinant of MPA resistance
We identified Ser267 as a potential structural determinant of MPA resistance in PbIMPDHA. Substitutions at this position have previously been shown to account for MPA resistance in other fungi.8, 9 The IMPDH from a MPA‐resistant strain of Candida albicans contained an Ala to Thr substitution at the analogous position (residue 251; this enzyme will be referred to as CaIMPDH‐A251T).8 This mutation caused the value of K iapp for MPA to increase four‐fold, and appeared to shift the conformation of the flap toward the closed conformation.10 Similarly, the IMD2 gene from S. cerevisiae encodes a MPA‐resistant IMPDH (ScIMPDH2) which contains Ser at the analogous position (253); substitution of this Ser with Ala made ScIMPDH2 sensitive to MPA, and the reciprocal Ala to Ser mutation made the MPA‐sensitive ScIMPDH3 resistant.9 Position 267 is Ala in most fungal IMPDHs, and the surrounding sequence is highly conserved, implying that the structure of this region is constrained by function (Figure 1d). Thus, Ser267 might be the structural determinant of MPA‐resistance in PbIMPDHA. AnIMPDH‐A267S and PbIMPDHA‐S267A were constructed to test this hypothesis. The active sites of these two enzymes are conserved (Figure 1a, Alignment S1), and overall the sequences of the two enzymes are 80% identical.
2.2. Characterization of wild‐type IMPDHs
His‐tagged wild‐type and mutant enzymes were purified as described in the Methods. The wild‐type enzymes AnIMPDH and PbIMPDHA were more active than in our previous report, with values of k cat increased by factors of 2.6 and 3.9, respectively (Table 1, Figure 2 6, 7). We do not know why the original purification protocol resulted in enzyme with lower activity, though we suspect that metals leaching from the resin may be the culprit. The new protocol features a different Ni‐NTA resin and inclusion of EDTA in the dialysis buffer, as well as changes in the bacteria growth and induction protocol, any of which could account for the improved activity. The values of Km for both IMP and NAD+ and the value of K ii for NAD+ substrate inhibition were similar to previous reports. MPA is a potent inhibitor of AnIMPDH (Kii = 13 nM versus NAD+), in reasonable agreement with our previous report (Table 1). PbIMPDHA is MPA‐resistant (Ki = 47 nM), although more modestly than our previous report (the value of K ii is increased 3.5‐fold relative to that of AnIMPDH, versus 20‐fold in the previous report). In both AnIMPDH and PbIMPDHA, the MPA inhibition pattern best fit an uncompetitive mechanism with respect to both IMP and NAD+, as expected given MPA binds preferentially to E‐XMP* in other IMPDHs.11
Table 1.
Steady‐state kinetic parameters of fungal IMPDHs
| Enzyme | K m IMP (μM) | K m NAD+ (μM) | K ii NAD+ (mM) | k cat (s−1) | K ii MPA versus IMP (nM) | K ii MPA versus NAD+ (nM) | D V |
|
|
|---|---|---|---|---|---|---|---|---|---|
| AnIMPDH | 15 ± 1 | 120 ± 8 | 1.5 ± 0.1 | 1.9 ± 0.1 | 23 ± 1 | 13 ± 2 | 1.1 ± 0.1 | 2.3 ± 0.1 | |
| AnIMPDH‐A267S | 15 ± 1 | 190 ± 20 | 2.2 ± 0.2 | 1.9 ± 0.1 | 150 ± 30 | 77 ± 12 | 1.0 ± 0.1 | 1.8 ± 0.1 | |
| PbIMPDHA | 93 ± 5 | 400 ± 40 | 3.0 ± 0.4 | 2.7 ± 0.1 | 86 ± 18 | 47 ± 7 | 1.4 ± 0.1 | 1.5 ± 0.1 | |
| PbIMPDHA‐S267A | 79 ± 6 | 330 ± 40 | 2.6 ± 0.3 | 1.6 ± 0.1 | 35 ± 6 | 21 ± 3 | 1.4 ± 0.1 | 1.4 ± 0.3 |
Figure 2.
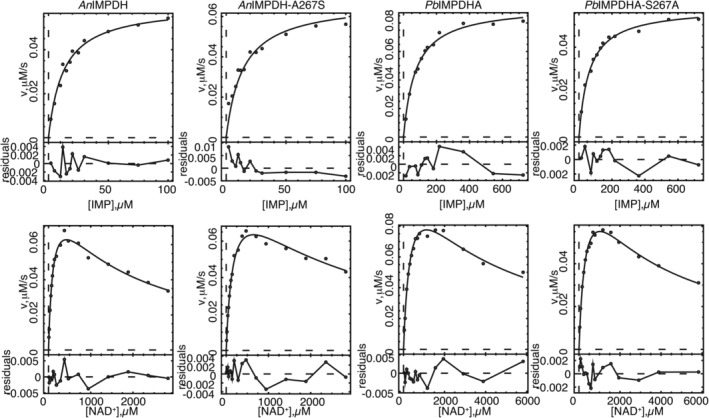
Steady state kinetics of fungal inosine 5′‐monophosphate dehydrogenases (IMPDHs). Conditions as described in Section 3
2.3. Ser267 is the determinant of MPA‐resistance of PbIMPDHA
The substitution of Ser267 with Ala made PbIMPDHA more sensitive to MPA by a factor of ∼2.2–2.5 (Table 1). This substitution decreased the value of k cat by less than a factor of 2, and had virtually no effect on the values of K m for IMP and NAD+. Conversely, AnIMPDH became ∼5.9–6.5‐fold more resistant to MPA when Ala267 was substituted with Ser. This substitution had no effect on the values of k cat and K m parameters. Thus residue 267 modulates MPA resistance in both protein contexts, with negligible cost to catalytic efficiency.
2.4. AnIMPDH and PbIMPDHA have different rate‐limiting steps
Residue 267 could determine MPA sensitivity by influencing the accumulation of E‐XMP*open, which in turn will depend on the release of NADH, the equilibrium between the open and closed conformations of the flap and the hydrolysis of E‐XMP* closed. We used isotope effects to interrogate the rate‐limiting step(s) for each enzyme to distinguish between these possibilities. No D V isotope effect was observed with AnIMPDH when 2‐[2H]‐IMP was the substrate, indicating that hydride transfer is not rate‐limiting (Figure 3, Table 1). AnIMPDH displayed a solvent isotope effect of 2.3, indicating that hydrolysis is partially rate‐limiting (Figure 4, Table 1; note = ∼4 when hydrolysis is fully rate limiting6). Perhaps product release or flap closure is also rate‐limiting for AnIMPDH.
Figure 3.
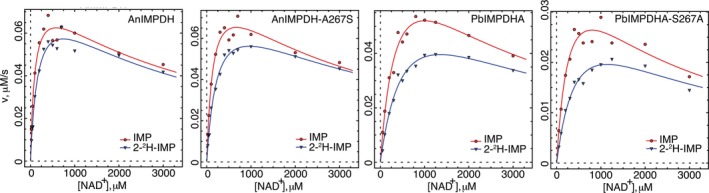
Primary isotope effects for the reaction of [2H]‐IMP with fungal IMPDHs. The data were fit to Equation (2). Conditions as described in Section 3. red, [1H]‐IMP; blue, [2H]‐IMP
Figure 4.
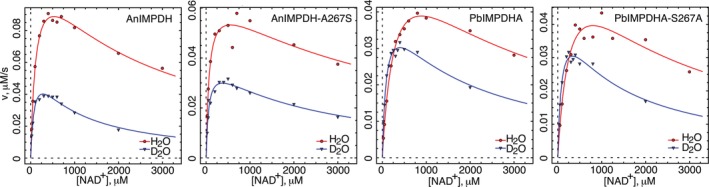
Solvent isotope effects for the fungal IMPDHs. The data were fit to Equation 2. Conditions as described in Section 3. Red, H2O; blue, D2O
In contrast, a small isotope effect (D V = 1.4) was observed with PbIMPDHA when 2‐[2H]‐IMP was the substrate, indicating that hydride transfer is partially rate‐limiting, as reported previously.6 A solvent isotope effect was also observed ( = 1.5), although the value was smaller than that of AnIMPDH. Thus both hydride transfer and hydrolysis of E‐XMP* are rate‐limiting in PbIMPDHA. Since different steps are rate‐limiting in the reactions of AnIMPDH and PbIMPDHA, the accumulation of E‐XMP* will also be different.
Substitutions at position 267 had little effect on the catalytic cycles of AnIMPDH and PbIMPDHA. No D V isotope effect was observed for AnIMPDH‐A267S, indicating that hydride transfer is not rate limiting, as observed in the wild‐type enzyme. A somewhat smaller isotope effect was observed for AnIMPDH‐A267S ( = 1.8), suggesting that hydrolysis might be less rate‐limiting. Faster hydrolysis should decrease the net accumulation of E‐XMP*, making the enzyme less susceptible to MPA. Faster hydrolysis can be a manifestation of a shift in the conformational equilibrium toward E‐XMP*closed, as occurs in CaIMPDH‐A251T.10
In contrast, the S267A substitution does not change the values of either D V or in PbIMPDHA, suggesting that the net accumulation of E‐XMP* is unchanged by the S267A mutation. Perhaps the mutation changes the equilibrium between the MPA‐sensitive conformation E‐XMP*open and MPA‐resistant conformation E‐XMP*closed.
2.5. Position 267 has little effect on hydrolysis of E‐XMP*
We further probed the effects of substitution of position 267 on the hydrolysis of E‐XMP* by determining the solvent isotope effects (Table 2). If hydrolysis is fully rate‐limiting ( = ∼46), then the presence of D2O will not increase the accumulation of E‐XMP*, and the value of K iapp will not change ( = 1; Figure 1c). However, if hydrolysis is partially rate‐limiting ( < 4), then D2O will increase the accumulation of E‐XMP*, resulting in stronger MPA inhibition ( > 1). For AnIMPDH and AnIMPDH‐A267S, the values of K iapp decreased in the presence of D2O, as expected given hydrolysis was partially rate‐limiting in both enzymes. However, while the solvent isotope effects suggested that hydrolysis might be less rate‐limiting for AnIMPDH‐A267S, a smaller effect ( = 1.5) was observed relative to AnIMPDH ( = 2.0). These observations suggest that residue 267 has little influence on the hydrolysis reaction in AnIMPDH.
Table 2.
Effect of increasing E‐XMP* on MPA inhibition. = K iapp(H2O)/K iapp(D2O). APAD K iapp = K iapp(NAD+)/K iapp(APAD+)
| Enzyme | K iapp(H2O)a (nM) | K iapp(D2O) (nM) |
|
K iapp(NAD+)a (nM) | K iapp(APAD+) (nM) | APAD K iapp | |
|---|---|---|---|---|---|---|---|
| AnIMPDH | 16 ± 2 | 8 ± 1 | 2.0 ± 0.2 | 24 ± 3 | 8.7 ± 0.8 | 2.8 ± 0.2 | |
| AnIMPDH‐A267S | 78 ± 6 | 51 ± 9 | 1.5 ± 0.2 | 73 ± 3 | 49 ± 2 | 1.5 ± 0.1 | |
| PbIMPDHA | 73 ± 4 | 27 ± 2 | 2.7 ± 0.1 | 110 ± 5 | 56 ± 2 | 1.6 ± 0.1 | |
| PbIMPDHA‐S267A | 28 ± 2 | 11 ± 6 | 2.5 ± 0.6 | 19 ± 2 | 12 ± 1 | 1.6 ± 0.1 |
Independent determinations for D2O and APAD+ experiments.
In the cases of PbIMPDHA and PbIMPDHA‐S267A, hydrolysis is less rate‐limiting compared to AnIMPDH, so greater values of were expected, and this was indeed observed (Table 2). Similar effects were observed in the wild‐type and mutant enzymes ( of ∼2.5 for both enzymes). Thus position 267 also has little influence on the hydrolysis step in PbIMPDHA.
2.6. Position 267 has little effect on hydride transfer and release of E‐XMP*
We evaluated the reactions of the fungal IMPDHs with the NAD+ analog APAD+ to further probe the effect of the mutations on the catalytic cycles. If NADH release is rate‐limiting, the hydride transfer step will be effectively at equilibrium [Figure 1(c)]. APAD+ has a higher redox potential than NAD+ (−0.258 V vs. −0.320), which will push the reaction toward E‐XMP*.12, 13 In addition, APADH is released more rapidly than NADH, which can further increase the accumulation of E‐XMP*.13 Therefore MPA will be a more potent inhibitor in the presence of APAD+ if NADH release is rate‐limiting.
MPA is indeed a more potent inhibitor of AnIMPDH in the presence of APAD+, by a factor of 2.8, indicating that NADH release is partially rate‐limiting for this enzyme. The APAD+ effect is reduced in AnIMPDH‐A267S, to only a factor of 1.5, suggesting that cofactor release is less rate‐limiting (Table 2). This finding was unexpected, because if cofactor release is less rate‐limiting, more E‐XMP* should accumulate, which would make AnIMPDH‐A267S more MPA sensitive than AnIMPDH. This conundrum can be explained if the mutation stabilized the closed conformation of the flap, so that E‐XMP* is not available to bind MPA. As noted above, the MPA resistance of CaIMPDH‐A251T was similarly ascribed to stabilization of the closed conformation.10
The APAD+ effects were smaller for PbIMPDHA than AnIPMPDH (APAD K iapp = 1.5 vs. 2.8, respectively). A similar value was observed for PbIMPDHA‐S267A (APAD K iapp = 1.3), indicating that the mutation had little effect on hydride transfer or NADH release. Thus the S267A mutation has little effect on the total accumulation of E‐XMP* in the PbIMPDHA scaffold, at least as can be detected with these methods. The effects of the S267A mutation can be explained by a shift in the conformational equilibrium to favor E‐XMP*open. Therefore, our experiments are consistent with a model wherein Ala267 favors the MPA‐sensitive conformation E‐XMP*open and Ser267 favors the MPA‐resistant conformation E‐XMP*closed.
2.6.1. The conservation of position 267
Position 267 is predominantly Ala in fungal and other eukaryotic IMPDHs, and the surrounding sequence is also highly conserved, suggesting that these residues have an important role in protein function (Figure 1d). We aligned the sequences of 20,000 fungal IMPDHs, and found that residue 267 is 91% Ala, 7.3% Gly, 1.2% Ser, 0.1% Thr and 0.1% Val. As noted above, in addition to PbIMPDHA and AnIMPDH, Ser267 is associated with MPA‐resistance in ScIMPDH2, and Thr267 causes MPA‐resistance in CaIMPDH.8, 9 These four proteins average 67% identity, ranging from 80 (AnIMPDH and PbIMPDHA) to 61% (AnIMPDH and ScIMPDH2). These observations suggest that Ser or Thr at position 267 will confer MPA resistance to most eukaryotic IMPDHs.
IMPDHs with Ser/Thr267 appear to be especially common in organisms that are likely to have encountered MPA. Penicillium fungi are often found in silage, and MPA has been detected in 10–42% of silage samples.14, 15, 16 Aspergillus fungi, which contain a single IMPDH gene, are also often found in silage, although these fungi typically prefer higher temperatures than Penicillium.14 Of the 27 Aspergillus IMPDHs included in the alignment (Alignment S2), seven are commonly found in spoiled silage: A. candidus, A. clavatus, A. fumigatus, A. glaucus, A. niger, A. oryzae, and A. terreus.14, 16, 17 Three of these fungi encode IMPDHs that contain Ser/Thr267 and are likely to be MPA‐resistant: A. clavatus, A. glaucus, and A. terreus. Two of these, A. clavatus and A. glaucus, prefer to grow at 25°C,17, 18, 19, 20 the temperature favored by most Penicillium,14, 16, 21 and therefore seem especially likely to have encountered MPA. Three additional Aspergillus fungi also contain IMPDHs with Ser/Thr267 that are likely MPA‐resistant: A. heteromorphus, A. steynii, and A. zonata (now known as Penicilliopsis zonata). These fungi also prefer to grow at low temperature, and therefore might well have encountered Pencillium producing MPA. Intriguingly, the IMPDHs of rice, barley, and corn also contain Ser267, and are likely to be MPA resistant. We suggest that Ser/Thr267 may be an evolutionary signature of MPA exposure.
3. MATERIALS AND METHODS
3.1. Materials
IMP disodium salt was purchased from MP Biochemical (Solon, OH). NAD+ free acid was purchased from Roche. DTT was purchased from GoldBio (St. Louis, MO). MPA, Tris, glycerol, EDTA, KCl, trimethylamine, and methanol were purchased from Fisher Scientific. D2O and DCl were purchased from Cambridge Isotope Laboratories, Inc. (Cambridge, MA). The pET28a plasmid p113 expressing PbIMPDHA and p115 expressing AnIMPDH were described previously.7 [2H]‐IMP was synthesized as previously described.12
3.2. Mutation construction
Site‐directed mutagenesis to construct plasmids expressing AnIMPDH‐A267S and PbIMPDHA‐S267A was performed using a two‐step method. A pair of forward and reverse primers (p115‐A267S: forward 5′‐CTG CTT CTA TTG GCA CCC GC‐3′, reverse 5’‐CAG CCG GAT CTA AGA GTA GGC‐3′; p113‐S267A: forward 5′‐CTG CTG CTA TCG GTA CCC GT‐3′, reverse 5′‐GTT AGC AGC CGG ATC TAC GC‐3′) was used on the wild‐type plasmid to introduce the mutation and produce a pair of 300‐bp megaprimers. Then megaprimers were used as the forward and reverse primers to complete the plasmid so that the mutation was ensured to be carried in most of the final plasmids. The entire IMPDH coding sequences were verified by sequencing (Genewiz, South Plainfield, NJ).
3.3. Protein expression and purification
Escherichia coli BL21 Δ guaB cells containing pET28a plasmids p115, p115‐A267S, p113, p113‐S267A expressing AnIMPDH, AnIMPDH‐A267S, PbIMPDHA, PbIMPDHA‐S267A, respectively, were grown in LB medium with kanamycin at 30°C. When the OD600 reached 0.8 to 1.0, the culture was induced with 0.1 mM IPTG and grown at 16°C overnight. The cell paste was collected by centrifugation at 4°C for 20 min at 5,000g in a Beckman JLA10.5 rotor, resuspended in phosphate buffer (50 mM K2HPO4, pH 8.0, 500 mM KCl, 5 mM imidazole, 0.1 mM TCEP, 10% glycerol) and sonicated on ice. After centrifugation at 9,000g for in a Beckman JA21 rotor, the supernatant was applied to Ni‐NTA Sepharose beads (GE). IMPDH was eluted with 250 mM imidazole and fractions were collected and dialyzed in Buffer A (50 mM Tris‐HCl, pH 8.0, 100 mM KCl, 3 mM EDTA, 1 mM DTT, 10% glycerol). IMPDH‐containing fractions were identified by SDS gel electrophoresis, concentrated to 100 μM, and flash‐frozen in a dry ice/acetone cooling bath. Enzymes were stored at −80°C. Enzyme concentrations were determined immediately after dialysis by measuring A280 with Buffer A dialysate as the blank.
3.4. Steady‐state kinetics
Steady‐state kinetics assays were performed in assay buffer (50 mM Tris‐HCl, pH 8, 100 mM KCl, and 1 mM DTT) with 20 nM enzyme at 25°C. The production of NADH was monitored by absorbance at 340 nm (ɛ340 = 6.22 mM−1 cm−1) on a Shimadzu UV‐2600 UV–Vis spectrophotometer. Steady‐state kinetic parameters were determined by collecting initial velocity data at varying concentrations of IMP (2 μM to 5 mM) or NAD+ (10 μM to 6 mM). All reactions were carried out in a total of 1 ml volume in 1 cm path length quartz cuvettes. Steady‐state initial rate data were combined into a single data set. The kinetic constants kcat, Km(IMP), Km(NAD), and Kii(NAD) were determined by regression analysis using Equations 1, 2 using Dynafit.22
| (1) |
| (2) |
3.5. MPA inhibition
The values of K i,app for MPA inhibition were determined by measuring the initial velocities at a varying concentrations of MPA (0–50 nM for both wild‐type AnIMPDH and the mutant AnIMPDH‐A267S, 0–200 nM for both wild‐type PbIMPDHA and the mutant PbIMPDHA‐S267A) at a fixed concentrations of IMP (500 μM for AnIMPDH and AnIMPDH‐A267S, 3 mM for PbIMPDHA and PbIMPDHA‐S267A) and NAD+ or APAD+ (500 μM for AnIMPDH and AnIMPDH‐A267S, 1 mM for PbIMPDHA and PbIMPDHA‐S267A). Enzyme (25 nM) was used. K iapp was determined using Equation (3), where [E] is total enzyme concentration, [I] is total inhibitor concentration, K iapp is the apparent inhibition constant, v i is the initial velocity in the presence of inhibitor at the concentration [I] and v 0 is the initial velocity in absence of inhibitor. SigmaPlot was used to fit the data.
| (3) |
3.6. Inhibition mechanism
MPDH activity was measured at varying concentrations of MPA (0–320 nM) at fixed concentrations of NAD+ (500 μM for AnIMPDH and AnIMPDH‐A267S, 1,250 μM for PbIMPDHA and PbIMPDHA‐S267A) with varying concentrations of IMP (3–100 μM for both AnIMPDH and AnIMPDH‐A267S, 30–600 μM for PbIMPDHA and PbIMPDHA‐S267A) and fixed IMP (1,000 μM for AnIMPDH and AnIMPDH‐A267S, 1,500 μM for PbIMPDHA and PbIMPDHA‐S267A) with varying NAD+ (30–2,500 μM for AnIMPDH and AnIMPDH‐A267S, 80–2,500 μM for PbIMPDHA and PbIMPDHA‐S267A). K iapp was determined by regression analysis using Equation (3) for each substrate concentration. The plots of K iapp against [S]−1 were linear with an intercept of K ii, demonstrating uncompetitive inhibition. K ii(MPA,IMP) and K ii(MPA,NAD+) were then determined by Equations 4, 5, respectively, using R (nlsLM package).
| (4) |
| (5) |
3.7. Primary deuterium isotope effects
Primary deuterium isotope effects were determined by collecting initial velocity data at fixed saturating concentrations of IMP (500 μM for AnIMPDH and AnIMPDH‐A267S, 3,000 μM for PbIMPDHA and PbIMPDHA‐S267A) or 2H‐IMP (250 μM for AnIMPDH and AnIMPDH‐A267S, 3,000 μM for PbIMPDHA and PbIMPDHA‐S267A) and varying concentrations of NAD+ (25–3,000 μM).
3.8. Solvent deuterium isotope effects
The assay buffers were prepared in H2O (pH = 8.0) or D2O (pD = 8.0). Solvent deuterium isotope effects were determined by collecting initial velocity data at fixed saturating concentrations of IMP (500 μM for AnIMPDH and AnIMPDH‐A267S, 3,000 μM for PbIMPDHA and PbIMPDHA‐S267A) and varying concentrations of NAD+ (25–3,000 μM).
3.9. Sequence alignment
Modern IMPDH sequences used in Hidden Markov Model (HMM) logo creation were identified using the Reference proteins (reseq_protein) database provided with the BLASTP algorithm using selected query sequences.23 Twenty thousand sequences that passed the BLASTP E‐value cutoff of 10−10 were downloaded along with unique Trichocomacaea family IMPDHs found with the non‐redundant database that passed the same E‐value cutoff. All sequences were aligned using the software package MAFFT,24 after which residues corresponding with PbIMPDHA residue 267 and the local residues around it were identified and separated using the alignment viewing software AliView,25 after which the HMM logo was created using Skylign.26
Supporting information
Data S1: Supporting Information: Alignments of (1) IMPDHs from MPA producers and nonproducers and (2) Alignment of Aspergillus IMPDHs.
ACKNOWLEDGMENTS
This work was funded by the National Institute of General Medical Sciences (GM054403 to LH).
Freedman R, Yu R, Sarkis AW, Hedstrom L. A structural determinant of mycophenolic acid resistance in eukaryotic inosine 5′‐monophosphate dehydrogenases. Protein Science. 2020;29:686–694. 10.1002/pro.3766
Rebecca Freedman and Runhan Yu contributed equally.
Funding information National Institute of General Medical Sciences, Grant/Award Number: GM054403
REFERENCES
- 1. Bentley R. Mycophenolic acid: A one hundred year odyssey from antibiotic to immunosuppressant. Chem Rev. 2000;100:3801–3826. [DOI] [PubMed] [Google Scholar]
- 2. Hedstrom L. IMP dehydrogenase: Mechanism of action and inhibition. Curr Med Chem. 1999;6:545–560. [PubMed] [Google Scholar]
- 3. van Gelder T, Hesselink DA. Mycophenolate revisited. Transpl Int. 2015;28:508–515. [DOI] [PubMed] [Google Scholar]
- 4. Regueira TB, Kildegaard KR, Hansen BG, Mortensen UH, Hertweck C, Nielsen J. Molecular basis for mycophenolic acid biosynthesis in Penicillium brevicompactum. Appl Environ Microbiol. 2011;77:3035–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hansen BG, Genee HJ, Kaas CS, et al. A new clade of IMP dehydrogenases with a role in self‐resistance in mycophenolic acid producing fungi. BMC Microbiol. 2011;11:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun XE, Hansen BG, Hedstrom L. Kinetically controlled drug resistance: How Penicillium brevicompactum survives mycophenolic acid. J Biol Chem. 2011;286:40595–40600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hansen BG, Sun XE, Genee HJ, et al. Adaptive evolution of drug targets in producer and non‐producer organisms. Biochem J. 2012;441:219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kohler GA, White TC, Agabian N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bact. 1997;179:2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jenks MH, Reines D. Dissection of the molecular basis of mycophenolate resistance in Saccharomyces cerevisiae . Yeast. 2005;22:1181–1190. [DOI] [PubMed] [Google Scholar]
- 10. Kohler GA, Gong X, Bentink S, et al. The functional basis of mycophenolic acid resistance in Candida albicans IMP dehydrogenase. J Biol Chem. 2005;280:11295–11302. [DOI] [PubMed] [Google Scholar]
- 11. Link JO, Straub K. Trapping of an IMP dehydrogenase‐substrate covalent interemediate by mycophenolic acid. J Am Chem Soc. 1996;118:2091–2092. [Google Scholar]
- 12. Wang W, Hedstrom L. The kinetic mechanism of human inosine 5′‐monophosphate type II: Random addition of substrates, ordered release of products. Biochemistry. 1997;36:8479–8483. [DOI] [PubMed] [Google Scholar]
- 13. Digits JA, Hedstrom L. Kinetic mechanism of Tritrichomonas foetus inosine‐5′‐monophosphate dehydrogenase. Biochemistry. 1999;38:2295–2306. [DOI] [PubMed] [Google Scholar]
- 14. Wambacq E, Vanhoutte I, Audenaert K, De Gelder L, Haesaert G. Occurrence, prevention and remediation of toxigenic fungi and mycotoxins in silage: A review. J Sci Food Agric. 2016;96:2284–2302. [DOI] [PubMed] [Google Scholar]
- 15. Zulkifli NA, Zakaria L. Morphological and molecular diversity of Aspergillus from corn grain used as livestock feed. HAYATI. J Biosci. 2017;24:26–34. [Google Scholar]
- 16. Ogunade IM, Martinez‐Tuppia C, Queiroz OCM, et al. Silage review: Mycotoxins in silage: Occurrence, effects, prevention and mitigation. J Dairy Sci. 2018;101:4034–4059. [DOI] [PubMed] [Google Scholar]
- 17. Panasenko VT. Ecology of microfungi. Bot Rev. 1967;33:189–215. [Google Scholar]
- 18. Ogundero VW. Toxigenic fungi and the deterioration of Nigerian poultry feeds. Mycopathologia. 1987;100:75–83. [DOI] [PubMed] [Google Scholar]
- 19. Meijer M, Houbraken JA, Dalhuijsen S, Samson RA, de Vries RP. Growth and hydrolase profiles can be used as characteristics to distinguish Aspergillus Niger and other black aspergilli. Stud Mycol. 2011;69:19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tarazona A, Gomez JV, Gavara R, et al. Risk management of ochratoxigenic fungi and ochratoxin a in maize grains by bioactive EVOH films containing individual components of some essential oils. Int J Food Microbiol. 2018;269:107–119. [DOI] [PubMed] [Google Scholar]
- 21. Li Y, Wadso L, Larsson L. Impact of temperature on growth and metabolic efficiency of Penicillium roqueforti‐correlations between produced heat. ergosterol content and biomass J Appl Microbiol. 2009;106:1494–1501. [DOI] [PubMed] [Google Scholar]
- 22. Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal Biochem. 1996;237:260–273. [DOI] [PubMed] [Google Scholar]
- 23. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. [DOI] [PubMed] [Google Scholar]
- 24. Yamada KD, Tomii K, Katoh K. Application of the MAFFT sequence alignment program to large data‐reexamination of the usefulness of chained guide trees. Bioinformatics. 2016;32:3246–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larsson A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wheeler TJ, Clements J, Finn RD. Skylign: A tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics. 2014;15:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sintchak MD, Fleming MA, Futer O, et al. Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid. Cell. 1996;85:921–930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: Supporting Information: Alignments of (1) IMPDHs from MPA producers and nonproducers and (2) Alignment of Aspergillus IMPDHs.


