Abstract
Objectives
To examine the in vitro selection of aztreonam/avibactam resistance among MBL-producing Klebsiella pneumoniae and to understand the mechanism of increased resistance.
Methods
The MICs of aztreonam were determined with and without avibactam (4 mg/L) using a broth microdilution method. Single-step and multi-step mutant selection was conducted on five MBL-producing K. pneumoniae strains, including two dual carbapenemase producers. Genomic sequencing and gene cloning were performed to investigate the mechanism of increased resistance.
Results
We examined the MICs for 68 MBL-producing K. pneumoniae isolates, including 13 dual carbapenemase producers. Compared with aztreonam alone, the addition of avibactam (4 mg/L) reduced the MICs for all isolates by >128-fold, with MIC50 and MIC90 values of 0.25 and 1 mg/L, respectively. One NDM-1-, OXA-48-, CTX-M-15- and CMY-16-positive ST101 K. pneumoniae strain was selected to be resistant to aztreonam/avibactam, with a >16-fold increase in MIC (>128 mg/L). WGS revealed that the resistant mutants lost the blaNDM-1 gene, but acquired amino acid substitutions in CMY-16 (Tyr150Ser and Asn346His). Construction of blaCMY-16 mutants confirmed that the substitutions (Tyr150Ser and Asn346His) were primarily responsible for the decreased susceptibility to aztreonam/avibactam. In addition, transfer of blaCMY-16 mutant (Tyr150Ser and Asn346His) plasmid constructs into certain clinical carbapenemase-producing isolates demonstrated >64-fold increased MICs of aztreonam/avibactam and aztreonam/avibactam/ceftazidime.
Conclusions
Aztreonam in combination with avibactam showed potent in vitro activity against MBL-producing K. pneumoniae. However, our study suggested the likelihood of aztreonam/avibactam resistance among MBL- and AmpC-co-producing strains and clinical practice should beware of the possibility of the emerging resistance.
Introduction
The global spread of carbapenem-resistant Enterobacteriaceae, especially carbapenem-resistant Klebsiella pneumoniae, has emerged as a major public health concern. Three major classes of carbapenemases are largely associated with the global spread of carbapenem-resistant Enterobacteriaceae: KPC (Ambler class A), MBL (Ambler class B, e.g. NDM, VIM and IMP) and OXA-48-like (Ambler class D) carbapenemases. Class A and D enzymes have serine-based hydrolytic activity, while class B enzymes require the presence of metal, i.e. zinc, for their activity.1
The newly FDA-approved diazabicyclooctane β-lactamase inhibitors avibactam and relebactam and an acyclic boronic acid β-lactamase inhibitor, vaborbactam, are potent mechanism-based inactivators of KPC,2,3 but do not inhibit MBLs, such as NDM.4 As such, the MBLs remain to be a clinical challenge for antimicrobial treatment. The combination of aztreonam and avibactam has shown inhibitory effects on MBLs (NDM, IMP or VIM), including strains co-harbouring KPC or OXA-48-like carbapenemases,5–9 and a clinical trial is ongoing using aztreonam/avibactam to treat serious infection due to MBL-producing Gram-negative bacteria (NCT03580044).
Currently, aztreonam/avibactam resistance is rarely observed among clinical isolates. Consequently, unravelling the molecular mechanism(s) underlying aztreonam/avibactam resistance, in order to guide appropriate clinical usage of this combination of antibiotics and to limit the emergence of resistance, is important. Here, we present in vitro susceptibility testing data for a collection of MBL-producing K. pneumoniae strains, including isolates co-producing KPC or OXA-48-like carbapenemases. Selected isolates were subjected to in vitro selection to study the molecular mechanism of aztreonam/avibactam resistance.
Materials and methods
Bacterial strains
Sixty-eight MBL-producing K. pneumoniae isolates were selected from the archived bacterial collection at the Center for Discovery and Innovation at Hackensack Meridian Health (HMH-CDI). The carbapenemase genotypes were previously characterized by PCR and Sanger sequencing. Among them, 55 strains were single carbapenemase producers, including 38 NDM-1, 5 VIM-1, 4 VIM-26, 3 NDM-7, 2 NDM-6, 1 VIM-4, 1 VIM-27 and 1 IMP-26. Thirteen were dual or triple carbapenemase producers, including two NDM-1- and OXA-48-, three VIM-1- and KPC-2-, two NDM-1- and OXA-181-, two NDM-5- and OXA-181-, two NDM-1- and KPC-2- and one NDM-5- and KPC-2-co-producing strains and one strain co-producing three carbapenemases (NDM-1, VIM-1 and OXA-244) (Table 1).
Table 1.
In vitro activities of aztreonam/avibactam against MBL-producing K. pneumoniae
| Strains (n) | CAZ |
IPM |
ATM |
CAZ/AVI |
ATM/AVI |
ATM/CAZ/AVI |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC range (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | MIC range (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | MIC range (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | MIC range (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | MIC range (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | MIC range (mg/L) | MIC50 (mg/L) | MIC90 (mg/L) | |
| All (68) | 128 to >128 | >128 | >128 | 4 to >32 | >32 | >32 | ≤0.5 to >128 | >128 | >128 | 16 to >64 | >64 | >64 | ≤0.25–8 | ≤0.25 | 1 | ≤0.125 | ≤0.125 | ≤0.125 |
| Single MBL (55) | 128 to >128 | >128 | >128 | 4 to >32 | 32 | >32 | ≤0.5 to >128 | >128 | >128 | 64 to >64 | >64 | >64 | ≤0.25–1 | ≤0.25 | 0.5 | ≤0.125 | ≤0.125 | ≤0.125 |
| Dual/triple carbapenemases (13) | >128 | >128 | >128 | 8 to >32 | >32 | >32 | ≤0.5 to >128 | >128 | >128 | 16 to >64 | >64 | >64 | ≤0.25–8 | 0.5 | 8 | ≤0.125 | ≤0.125 | ≤0.125 |
CAZ, ceftazidime; IPM, imipenem; ATM, aztreonam; AVI, avibactam.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using a standard broth microdilution method following CLSI guidelines.10 Avibactam was tested at a fixed concentration of 4 mg/L, in combination with 2-fold dilutions of aztreonam. When ceftazidime, aztreonam and avibactam were combined, aztreonam was fixed at 8 mg/L and avibactam was used at 4 mg/L, with 2-fold dilutions of ceftazidime.11 MICs were interpreted using 2018 CLSI breakpoints10 for all antimicrobial agents except for aztreonam/avibactam and ceftazidime/aztreonam/avibactam combinations. The testing was performed in duplicate on two different days. Quality control strains Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used in all testing.
In vitro selection
Single-step mutant selection was conducted as previously described.12 In brief, approximately 109 cfu from overnight broth cultures were plated on LB agar with aztreonam/avibactam at 2× to 16× MICs. Multi-step selection was performed by inoculation of ∼108 cfu in 2 mL of LB broth containing aztreonam/avibactam at 0.5× MIC and incubated for 24 h.13 This procedure was repeated daily, each time doubling the aztreonam concentration up to a maximum of 128 mg/L. Resistant variants were selected by plating a bacterial suspension on agar plates of corresponding aztreonam/avibactam levels. The MIC values for the mutants and parent strains were determined by the broth microdilution method. The same single-step and multi-step selections were used to select ceftazidime/aztreonam/avibactam resistance, with aztreonam and avibactam concentrations fixed at 8 and 4 mg/L, respectively.
Genome sequencing
Six aztreonam/avibactam-resistant mutant colonies and the parental strain Kp202 were selected for next-generation sequencing using Illumina HiSeq. Illumina raw reads were de novo assembled using SPAdes v3.11.1.14 MLST STs were determined using the K. pneumoniae MLST website (https://bigsdb.pasteur.fr/klebsiella), while the acquired resistance genes, including all β-lactamase-encoding genes, were determined by ResFinder 3.1.15 Core SNPs were identified using the method described previously.16 Outer membrane protein genes (ompK35 and ompK36) were examined using BLAST. In addition, the parental strain Kp202 was also subject to Nanopore sequencing using the MinION platform. A hybrid assembly was conducted by Unicycler using the combination of Illumina HiSeq and Nanopore sequencing reads.17
Construction of recombinant strains
The promoter and full length of the blaCMY-16 gene and its mutant variants (Tyr150Ser and Asn346His) were amplified from the parental and mutant strains using primers CMY-F-EcoRI (5'-CCGGAATTCCAAAACCGCAAGATATGTAATCA-3′) and CMY-R-XbaI (5'-CTAGTCTAGATTATTGCAGCTTTTCAAGAATGCGCC-3′). The PCR products were then cloned into plasmid vector pET28a, followed by electroporation into E. coli DH10B. In addition, the recombinant plasmids, namely pET28a-CMY16-wt, pET28a-Tyr150Ser and pET28a-Asn346His, were electroporated into five selected carbapenemase-producing K. pneumoniae strains. The recombinant strains were selected with 1 mg/L aztreonam and 4 mg/L avibactam and confirmed by PCR and Sanger sequencing.
Expression analysis
Bacterial strains were grown to mid-log phase and total RNA was prepared using a QIAGEN RNeasy Mini Kit. A total of 5 ng RNA was used in an RT–PCR assay using a QIAGEN QuantiTect SYBR Green RT–PCR Kit (Germantown, MD, USA). The oligonucleotides used to detect the expression of the CMY allele in K. pneumoniae were CMY-F (5′-GATGCAGGAGCAGGCTATTC-3′) and CMY-R (5′-CCGATCCTAGCTCAAACAGC-3′) and the control oligonucleotides to detect expression of the control gapA gene were gapA-F (5′-TTTCTGAGCAGCACGGAA-3′) and gapA-R (5′-ATAGTCATATGT TCCTCCA-3′).
Accession numbers
The complete genome sequences of Kp202 were submitted to GenBank under the accession numbers CP041082 to CP041089.
Results
Activity of aztreonam/avibactam against MBL-producing K. pneumoniae
Among 68 MBL-producing isolates, all were resistant to imipenem (MIC50 >32 mg/L) and 95.6% (n=65) of them were resistant to aztreonam (MIC50 >128 mg/L). Compared with the result of aztreonam alone, the addition of avibactam resulted in a >512-fold reduction of aztreonam MIC50 (≤0.25 mg/L). Two dual-carbapenemase-producing K. pneumoniae strains had the highest aztreonam/avibactam MIC of 8 mg/L. All isolates were resistant to ceftazidime/avibactam, with an MIC50 of >64 mg/L, while the addition of aztreonam significantly lowered the MIC (MIC50 ≤0.125 mg/L) for MBL-producing isolates, including the two strains with aztreonam/avibactam MIC of 8 mg/L.
In vitro selection
We selected five MBL-producing strains, including three single MBL producers (IMP-4-producing ST1307, VIM-1-producing ST147 and NDM-1-producing ST11 strains) and the two dual carbapenemase producers (NDM-1- and OXA-48-co-producing ST101 and NDM-1- and OXA-232-co-producing ST14 strains) with aztreonam/avibactam MIC of 8 mg/L, for the in vitro selection experiment. The single-step selection failed to yield any colonies at 2× MIC or higher concentrations of aztreonam/avibactam. For the in vitro multi-step selection, the dual-carbapenemase-producing ST101 strain (namely Kp202) was successfully selected and was able to grow at concentrations up to 16× MIC (128 mg/L). However, the selection of the other four isolates was not successful and bacterial growth was not observed at 2× MIC. In addition, selection using aztreonam/ceftazidime/avibactam was not successful either.
The in vitro-selected Kp202 mutant culture was plated on LB agar with concentrations of aztreonam/avibactam used in the selection experiment (e.g. 2× MIC growth was plated on 16 mg/L aztreonam/avibactam agar plates). Two single colonies recovered from plates with 32 (namely Kp202_32A and Kp202_32B), 64 (Kp202_64A and Kp202_64B) and 128 mg/L (Kp202_128A and Kp202_128B) aztreonam/avibactam were randomly selected and subjected to next-generation sequencing and susceptibility testing. Susceptibility testing showed that the six mutants all demonstrated aztreonam/avibactam MICs ≥128 mg/L, while the MICs of imipenem and meropenem were decreased by ∼4-fold (Table 2). Interestingly, the aztreonam/ceftazidime/avibactam MIC also increased >8-fold for all six mutant colonies (from <0.125 to ≥16 mg/L) (Table 2).
Table 2.
In vitro activity of different antibiotics against K. pneumoniae CMY-16 mutants
| Strain | β-Lactamase(s) | MIC (mg/L) |
||||||
|---|---|---|---|---|---|---|---|---|
| AMP | ATM | CAZ | IPM | MEM | ATM/AVI | CAZ/ATM/AVI | ||
| K. pneumoniae strain Kp202 and the selected mutants | ||||||||
| Kp202 | NDM-1, OXA-48, CTX-M-15, CMY-16, SHV-1, TEM-1, OXA-10, SCO-1 | >1024 | 1024 | >1024 | 16 | 16 | 8 | ≤0.125 |
| Kp202_32A | OXA-48, CTX-M-15, CMY-16 (Tyr150Ser), SHV-1, TEM-1, OXA-10, SCO-1 | >1024 | 1024 | 512 | 4 | 4 | >256 | 16 |
| Kp202_32B | OXA-48, CTX-M-15, CMY-16 (Tyr150Ser), SHV-1, TEM-1, OXA-10, SCO-1 | >1024 | 1024 | 1024 | 4 | 4 | 128 | 32 |
| Kp202_64A | OXA-48, CTX-M-15, CMY-16 (Asn346His), SHV-1, TEM-1, OXA-10 | >1024 | 1024 | 512 | 4 | 4 | >256 | 16 |
| Kp202_64B | OXA-48, CTX-M-15, CMY-16 (Tyr150Ser), SHV-1, TEM-1, OXA-10, SCO-1 | >1024 | 1024 | 1024 | 4 | 4 | 128 | 32 |
| Kp202_128A | OXA-48, CTX-M-15, CMY-16 (Asn346His), SHV-1, TEM-1, OXA-10 | >1024 | 1024 | 512 | 4 | 4 | >256 | 16 |
| Kp202_128B | OXA-48, CTX-M-15, CMY-16 (Tyr150Ser), SHV-1, TEM-1, OXA-10, SCO-1 | >1024 | 1024 | 1024 | 4 | 4 | 128 | 32 |
| E. coli DH10B CMY-16 constructs | ||||||||
| Ec202-WT | CMY-16 WT | 512 | 256 | >256 | 0.5 | ≤0.06 | 1 | ≤0.015 |
| Ec202-R150 | CMY-16 (Tyr150Ser) | 64 | 256 | 8 | 0.25 | ≤0.06 | 64 | 1 |
| Ec202-R346 | CMY-16 (Asn346His) | 256 | 128 | 64 | 0.125 | ≤0.06 | 16 | ≤0.015 |
| DH10B | — | 4 | ≤0.125 | ≤0.125 | 0.125 | ≤0.06 | 0.06 | ≤0.015 |
AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; IPM, imipenem; MEM, meropenem; AVI, avibactam; —, negative.
Genomic sequencing of Kp202 and its mutants
The parental strain Kp202 had a circular chromosome of 5.46 Mbp and seven plasmids, belonging to IncA/C (pKp202_1: 179 254 bp), IncFIBkpn (pKp202_2: 158 438 bp), IncL/M (pKp202_3: 63 499 bp), IncFIA-IncR (pKp202_4: 61 718 bp) and three ColE-like groups (pKp202_5, pKp202_6 and pKp202_4: 5359, 4052 and 3541 bp, respectively). Acquired resistance gene analysis showed the parental Kp202 harboured eight different types of β-lactamase gene from the four Ambler classes, including blaNDM-1 (on pKp202_1), blaOXA-48 (pKp202_3), blaCTX-M-15 (three copies: chromosome, pKp202_1 and pKp202_4), blaCMY-16 (pKp202_1), blaSHV-1 (chromosome), blaTEM-1 (two copies: pKp202_1 and pKp202_4), blaOXA-10 (pKp202_1) and blaSCO-1 (pKp202_2). In addition, Kp202 carried a mutant outer membrane protein OmpK35, with a premature stop codon at amino acid 63 due to a G deletion at nt 184, and a mutant OmpK36, with glycine and aspartic acid insertions at amino acid positions 134 and 135. We suspect the combination of diverse β-lactamases and the outer membrane protein defects may have contributed to the elevated aztreonam/avibactam MIC observed for Kp202.
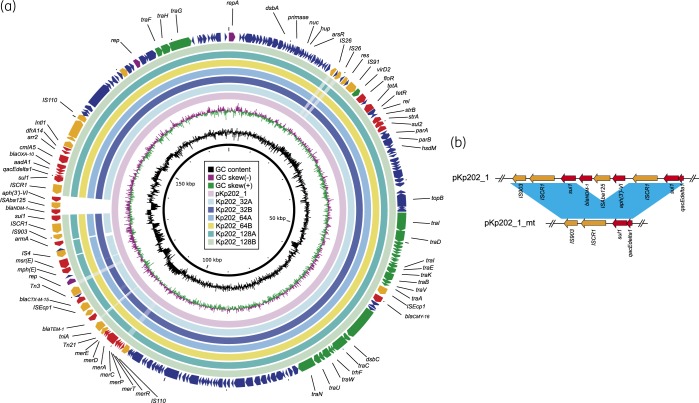
A core SNP genomic analysis showed that the genomes from the six resistant colonies were nearly identical to the parental Kp202 (<2 SNPs). Further sequence inspection showed that the six mutants were all positive for blaOXA-48, blaCTX-M-15, blaSHV-1, blaTEM-1 and blaOXA-10, while Kp202_64A and Kp202_128A were negative for blaSCO-1, due to the loss of the pKp202_2 plasmid in the two strains (data not shown). In addition, blaNDM-1 was lost in all six mutants. Examination of the blaNDM-1 neighbouring region in pKp202_1 showed that blaNDM-1 was located at a 9.3 kb region with the element aph(3′)-VI-ISAba125-blaNDM-1 flanked by two copies of sul1-ISCR1 (Figure 1a). A 6.45 kb fragment, encompassing aph(3′)-VI-ISAba125-blaNDM-1 and a copy of sul1-ISCR1, was deleted in all of the six mutants from the same site, likely through DR-mediated slippage (Figure 1b). The results indicated the elevated aztreonam/avibactam MIC level for the Kp202 selected mutants was unrelated to blaNDM. Moreover, our results also indicate that the blaNDM-1 in pKp202-1 was not stable under increased aztreonam challenge, as the gene was deleted in all selected mutants.
Figure 1.
(a) Plasmid structure comparison between blaNDM-1-harbouring pKp202_1 and the selected mutants. (b) The blaNDM-1 deletion in Kp202 selected mutants. Coloured arrows indicate ORFs, with purple, orange, green, red and blue arrows representing replication genes, mobile elements, plasmid transfer genes, the antimicrobial and heavy metal resistance gene, and plasmid backbone genes, respectively. Blue shading denotes regions of shared homology among different elements.
Interestingly, examination of the blaCMY-16 sequence among the six mutants found that isolates Kp202_64A and Kp202_128A carried an asparagine-to-histidine substitution at Ambler amino acid position 346 (Asn346His), while the remaining four isolates harboured a tyrosine-to-serine substitution at position 150 (Tyr150Ser). We suspect that the two amino acid substitutions might have contributed to aztreonam/avibactam resistance among these mutant strains. Real-time quantitative RT–PCR did not show any significant differential expression of blaCMY-16 between the parental Kp202 and the derived mutant strains (data not shown).
MICs for recombinant strains
The WT blaCMY-16 and the two mutant blaCMY-16 (Tyr150Ser and Asn346His) genes were cloned in plasmid vector pET28a, followed by electroporation into E. coli DH10B to generate constructs Ec202-WT, Ec202-R150 (Tyr150Ser) and Ec202-R346 (Asn346His), respectively. The MIC of aztreonam/avibactam increased from 1 to 64 mg/L for the CMY-16 Tyr150Ser construct (strain Ec202-R150) and from 1 to 16 mg/L for the CMY-16 Asn346His construct (strain Ec202-R346) (Table 2), indicating that the CMY-16 amino acid substitutions are responsible for the increased aztreonam/avibactam resistance. The MICs of aztreonam were not significantly affected and showed a <2-fold change in comparison with the WT CMY-16 construct (strain Ec202), suggesting that the increased aztreonam/avibactam resistance was mainly due to the reduced inhibition efficacy of avibactam against the CMY mutants. Along with the increase in aztreonam/avibactam resistance, the CMY-16 Tyr150Ser and Asn346His mutants also demonstrated lower ceftazidime MICs (>32- and >4-fold, respectively), suggesting the amino acid substitutions also reduce the ability of CMY-16 to degrade ceftazidime (Table 2).
To further evaluate the role of the CMY-16 mutations, we electroporated the CMY-16 mutant plasmid vectors into five clinical strains harbouring different carbapenemases and outer membrane porin defects. Among these transformants, the MICs of aztreonam/avibactam all increased by 16–128-fold (from ≤0.25 mg/L to 2–32 mg/L), which is consistent with the results from the E. coli DH10B constructs (Table 2). Interestingly, the MIC of aztreonam/ceftazidime/avibactam also increased by 256-fold for two dual-carbapenemase-producing mutant K. pneumoniae strains (NDM-5 and OXA-181, and NDM-1 and OXA-48) with OmpK defects (Table 3).
Table 3.
In vitro activity of aztreonam/avibactam and ceftazidime/aztreonam/avibactam against recombinant K. pneumoniae strains (mg/L)
| Strain | β-Lactamases | MLST ST | OmpK35/36 | MIC (mg/L) |
|
|---|---|---|---|---|---|
| ATM/AVI | CAZ/ATM/AVI | ||||
| Kp040 | IMP-4, DHA-1 | new | WT/WT | ≤0.25 | ≤0.125 |
| Kp040-CMY | IMP-4, DHA-1, CMY-16 | ≤0.25 | ≤0.125 | ||
| Kp040-R150 | IMP-4, DHA-1, CMY-16 (Tyr150Ser) | 4 | ≤0.125 | ||
| Kp040-R346 | IMP-4, DHA-1, CMY-16 (Asn346His) | 2 | ≤0.125 | ||
| Kp189 | NDM-7, CTX-M-15, SHV-11, TEM-1, OXA-1 | ST16 | WT/WT | ≤0.25 | ≤0.125 |
| Kp189-CMY | NDM-7, CTX-M-15, SHV-11, TEM-1, OXA-1, CMY-16 | ≤0.25 | ≤0.125 | ||
| Kp189-R150 | NDM-7, CTX-M-15, SHV-11, TEM-1, OXA-1, CMY-16 (Tyr150Ser) | 4 | ≤0.125 | ||
| Kp189-R346 | NDM-7, CTX-M-15, SHV-11, TEM-1, OXA-1, CMY-16 (Asn346His) | 4 | ≤0.125 | ||
| Kp214 | NDM-5, OXA-181, CTX-M-15, SHV-11, TEM-1 | ST147 | MT (ISEcp1 ins)/MT (135 DT ins) | ≤0.25 | ≤0.125 |
| Kp214-CMY | NDM-5, OXA-181, CTX-M-15, SHV-11, TEM-1, CMY-16 | ≤0.25 | ≤0.125 | ||
| Kp214-R150 | NDM-5, OXA-181, CTX-M-15, SHV-11, TEM-1, CMY-16 (Tyr150Ser) | 128 | 32 | ||
| Kp214-R346 | NDM-5, OXA-181, CTX-M-15, SHV-11, TEM-1, CMY-16 (Asn346His) | 64 | 32 | ||
| Kp231 | NDM-1, OXA-48, CTX-M-15, SHV-11, OXA-1 | ST377 | MT (IS1 ins)/WT | ≤0.25 | ≤0.125 |
| Kp231-CMY | NDM-1, OXA-48, CTX-M-15, SHV-11, OXA-1, CMY-16 | ≤0.25 | ≤0.125 | ||
| Kp231-R150 | NDM-1, OXA-48, CTX-M-15, SHV-11, OXA-1, CMY-16 (Tyr150Ser) | 128 | 32 | ||
| Kp231-R346 | NDM-1, OXA-48, CTX-M-15, SHV-11, OXA-1, CMY-16 (Asn346His) | 32 | 32 | ||
| Kp518 | KPC-2, SHV-11 | ST258 | MT (stop)/WT | ≤0.25 | ≤0.125 |
| Kp518-CMY | KPC-2, SHV-11, CMY-16 | ≤0.25 | ≤0.125 | ||
| Kp518-R150 | KPC-2, SHV-11, CMY-16 (Tyr150Ser) | 32 | ≤0.125 | ||
| Kp518-R346 | KPC-2, SHV-11, CMY-16 (Asn346His) | 16 | ≤0.125 | ||
ATM, aztreonam; AVI, avibactam; CAZ, ceftazidime; MT, mutant type; ins, insertion; DT ins, aspartate and threonine insertion.
Discussion
As a novel non-β-lactam β-lactamase inhibitor, avibactam offers a broader β-lactamase inhibition profile than traditional β-lactamase inhibitors, such as clavulanic acid, tazobactam and sulbactam, which inactivate only specific class A enzymes. Avibactam protects β-lactams from hydrolysis by class A (e.g. KPCs), class C (e.g. CMY) and some class D (e.g. OXA-48) enzymes. The combination of aztreonam/avibactam presents a novel approach to the treatment of infections caused by pathogens containing multiple β-lactamases, including isolates carrying MBLs. Our in vitro susceptibility testing also indicated that aztreonam/avibactam and ceftazidime/avibactam/aztreonam are potent against MBL-producing K. pneumoniae, including isolates co-harbouring more than one carbapenemase.
Our results demonstrated that selection of aztreonam/avibactam resistance is associated with mutations in blaCMY (Tyr150Ser or Asn346His) during in vitro selection. In a previous study on the binding analysis of avibactam to P. aeruginosa AmpC β-lactamase, Lahiri et al.18 identified eight key conserved amino acid residues (Ser64, Lys67, Gln120, Tyr150, Asn152, Lys315, Thr316 and Asn346) in chromosomal and plasmid AmpC β-lactamase enzymes that contribute to the binding interaction of avibactam with AmpC β-lactamase and other β-lactamases. The carboxamide group of avibactam interacted with the side chains of Asn152 and Gln120, and the sulphate moiety was positioned by Thr316, Lys315 and Asn346, whereas Tyr150 and Lys67 were positioned to participate in catalytic roles to enable formation of the covalent bond with Ser64.18 The authors identified two amino acids substitutions (Asn346Tyr and Tyr150Ser) in Citrobacter freundii chromosomal AmpC and E. coli plasmid-encoded CMY-6 proteins through in vitro selection experiments.18 The Tyr150Ser mutant significantly lowered the ability of avibactam against the hydrolysis of aztreonam, increasing the MIC value by 16-fold for the selected mutant strain, which is consistent with our results, except that we identified the same Tyr150Ser substitution in a different CMY-16 variant (Table 2). In addition, the authors suggested that the Asn346Tyr substitution, because of the increased size of the Tyr residue, could result in a steric clash with the sulphate group of avibactam, thus influencing the binding affinity of this inhibitor.18 We suspect the Asn346His substitution described in this study may use a similar mechanism to reduce the binding to avibactam. Moreover, the Asn346His substitution also changed the charge from neutral to positive, which may interfere with the binding of avibactam.
Recent studies showed that ceftazidime/avibactam in combination with aztreonam demonstrated excellent in vitro synergistic activity against and bactericidal effect on MBL-producing Enterobacteriaceae and P. aeruginosa strains.19,20 The success of treatment by ceftazidime/avibactam/aztreonam against MBL-producing strains has also been described in several clinical cases.11,21,22 Our in vitro study also showed that the combination of ceftazidime/avibactam/aztreonam had great activity against MBL-producing K. pneumoniae, including strains co-producing more than one carbapenemase (Table 1). However, the Kp202 CMY-16 mutants showed more than 128-fold increases in ceftazidime/avibactam/aztreonam MIC (Table 2). In addition, the transformants of CMY-16 Asn346Tyr and Tyr150Ser of two clinical carbapenem-resistant K. pneumoniae also showed increased resistance to ceftazidime/avibactam/aztreonam, indicating that this triple combination may lose the activity in carbapenem-resistant K. pneumoniae strains with AmpC mutations and outer membrane porin defects. However, our results also suggested that the usage of the triple combination of ceftazidime/avibactam/aztreonam may lower the chance of blaCMY-16 mutations arising.
Taken together, our study showed that the aztreonam/avibactam combination is active against MBL-producing K. pneumoniae, including strains co-producing more than one carbapenemase. However, carbapenemase- and AmpC-co-producing strains showed selected resistance to aztreonam/avibactam as well as ceftazidime/avibactam/aztreonam. In order to prevent selection resistance in clinical practice, rational strategies for employing different combinational agents and dosing regimens that optimize pharmacokinetic/pharmacodynamic target attainment should be considered.
Funding
This work was in part supported by grants from the National Institutes of Health (R01AI090155 to B.N.K. and R01AI100560, R01AI063517 and R01AI072219 to R.A.B.). This work was in part supported by the Science and Technology Research Program of Chongqing Municipal Education Commission (KJ1702022) and the Medical Research Program of Chongqing Health and Family Planning Commission in 2016 (2016MSXM001).
Transparency declarations
None to declare.
References
- 1. Kumar N, Singh VA, Pottathil S.. Metallo-β-lactamase- and serine carbapenemase-producing Klebsiella spp.: a global challenge. J Glob Antimicrob Resist 2018; 12: 185–6. [DOI] [PubMed] [Google Scholar]
- 2. Sader HS, Castanheira M, Shortridge D. et al. Antimicrobial activity of ceftazidime-avibactam tested against multidrug-resistant Enterobacteriaceae and Pseudomonas aeruginosa isolates from U.S. medical centers, 2013 to 2016. Antimicrob Agents Chemother 2017; 61: e01045–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lomovskaya O, Sun D, Rubio-Aparicio D. et al. Vaborbactam: spectrum of β-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriaceae. Antimicrob Agents Chemother 2017; 61: e01443–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wong D, van Duin D.. Novel β-lactamase inhibitors: unlocking their potential in therapy. Drugs 2017; 77: 615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kazmierczak KM, Biedenbach DJ, Hackel M. et al. Global dissemination of blaKPC into bacterial species beyond Klebsiella pneumoniae and in vitro susceptibility to ceftazidime-avibactam and aztreonam-avibactam. Antimicrob Agents Chemother 2016; 60: 4490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sader HS, Mendes RE, Pfaller MA. et al. Antimicrobial activities of aztreonam-avibactam and comparator agents against contemporary (2016) clinical Enterobacteriaceae isolates. Antimicrob Agents Chemother 2018; 62: e01856–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Karlowsky JA, Kazmierczak KM, de Jonge BLM. et al. In vitro activity of aztreonam-avibactam against Enterobacteriaceae and Pseudomonas aeruginosa isolated by clinical laboratories in 40 countries from 2012 to 2015. Antimicrob Agents Chemother 2017; 61: e00472–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chew KL, Tay MKL, Cheng B. et al. Aztreonam-avibactam combination restores susceptibility of aztreonam in dual-carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 2018; 62: e00414–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Endimiani A, Choudhary Y, Bonomo RA.. In vitro activity of NXL104 in combination with β-lactams against Klebsiella pneumoniae isolates producing KPC carbapenemases. Antimicrob Agents Chemother 2009; 53: 3599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Eighth Edition: M100 2018.
- 11. Marshall S, Hujer AM, Rojas LJ. et al. Can ceftazidime-avibactam and aztreonam overcome β-lactam resistance conferred by metallo-β-lactamases in Enterobacteriaceae? Antimicrob Agents Chemother 2017; 61: e02243–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Livermore DM, Warner M, Jamrozy D. et al. In vitro selection of ceftazidime-avibactam resistance in Enterobacteriaceae with KPC-3 carbapenemase. Antimicrob Agents Chemother 2015; 59: 5324–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mushtaq S, Warner M, Williams G. et al. Activity of chequerboard combinations of ceftaroline and NXL104 versus β-lactamase-producing Enterobacteriaceae. J Antimicrob Chemother 2010; 65: 1428–32. [DOI] [PubMed] [Google Scholar]
- 14. Bankevich A, Nurk S, Antipov D. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19: 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zankari E, Hasman H, Cosentino S. et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67: 2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lowe M, Kock MM, Coetzee J. et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014-2016. Emerg Infect Dis 2019; 25: 739–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wick RR, Judd LM, Gorrie CL. et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017; 13: e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lahiri SD, Johnstone MR, Ross PL. et al. Avibactam and class C β-lactamases: mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrob Agents Chemother 2014; 58: 5704–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davido B, Fellous L, Lawrence C. et al. Ceftazidime-avibactam and aztreonam, an interesting strategy to overcome β-lactam resistance conferred by metallo-β-lactamases in Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 2017; 61: e01008–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emeraud C, Escaut L, Boucly A. et al. Aztreonam plus clavulanate, tazobactam, or avibactam for treatment of infections caused by metallo-β-lactamase-producing Gram-negative bacteria. Antimicrob Agents Chemother 2019; 63: e00010–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mittal J, Szymczak WA, Guo Y. et al. Two for the price of one: emerging carbapenemases in a returning traveller to New York City. BMJ Case Rep 2018; 2018: bcr-2018–225440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shaw E, Rombauts A, Tubau F. et al. Clinical outcomes after combination treatment with ceftazidime/avibactam and aztreonam for NDM-1/OXA-48/CTX-M-15-producing Klebsiella pneumoniae infection. J Antimicrob Chemother 2018; 73: 1104–6. [DOI] [PubMed] [Google Scholar]