Hepatocellular carcinoma (HCC) is a common cancer that affects people worldwide with high morbidity and mortality, and its resistance to current chemotherapeutic drugs is a serious concern.
Hepatocellular carcinoma (HCC) is a common cancer that affects people worldwide with high morbidity and mortality, and its resistance to current chemotherapeutic drugs is a serious concern.
Abstract
Hepatocellular carcinoma (HCC) is a common cancer that affects people worldwide with high morbidity and mortality, and its resistance to current chemotherapeutic drugs is a serious concern. Cytotoxicity of silica nanoparticles (Nano-SiO2) towards cancer cells has been reported previously, but the specific mechanism is not fully clear. In this study, Nano-SiO2 showed a remarkable cytotoxic effect against HCC cells, regardless of whether the cells were drug resistant or not. Further study showed that Nano-SiO2 treatment leads to cell cycle arrest, apoptosis enhancement and necroptosis induction in the HCC cells. RNA-seq data, together with bioinformatics analysis, revealed that a series of genes involved in cancer cell death could be regulated by Nano-SiO2, among which ZBP-1 was up-regulated the most by Nano-SiO2 treatment. The siRNA based experiments demonstrated that ZBP-1 might play a key role in mediating Nano-SiO2 cytotoxic functions against HCC cells. These results have evidently signified the anti-tumor potential of Nano-SiO2 in the treatment of HCC.
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide and the second leading cause of cancer-related deaths in China.1 Traditional treatment includes resection and subsequent chemotherapy.2 However, during treatment, cancer cells develop resistance toward functionally and structurally different anticancer drugs via either acquired (due to host factors) or intrinsic (due to genetic or epigenetic) mechanisms.3 Effective novel agents that can counteract drug resistance in HCC need to be developed.
Nanotechnology has broadened the development of new anticancer drugs in recent years. Numerous substances with anticancer activity have expanded their efficacy after they were fabricated into nanoparticles, such as nanoselenium,4 nanogold,5 nanocopper,6 and silica nanoparticles (Nano-SiO2).7
Nano-SiO2 is a SiO2 particle with a diameter of 1–100 nm.8 Nano-SiO2 has a large surface area, which can promote its cellular absorption.9 Reports have demonstrated that Nano-SiO2 can exert toxicity and thus destroy normal cells and tissues, leading to conditions such as pulmonary toxicity;10 irrespective of this, Nano-SiO2 has been widely used in disease diagnosis, biological analysis and imaging, drug carriers, and other research studies.8
In the present study, we evaluated the efficacy of Nano-SiO2 against HCC and investigated its molecular mechanism. We found that Nano-SiO2 exerted potent cytotoxic effects on various HCC cell lines and primary isolated tumor cells but less effects on normal cells. RNA sequencing (RNA-seq), combined with molecular biological experiments, has shown that this inhibition can be achieved by ZBP1 enhancement.
Materials and methods
Preparation of nanoparticles
Nano-SiO2 with 30 nm diameter was purchased from Kisker Biotech (Steinfurt, Germany). The stock solution was prepared by diluting the particles in phosphate-buffered saline (PBS) to a final volume of 5 mg ml–1 and sonicated at 60 kHz for 20 min. Before immediate use, the stock solution was subjected to sonication under the same conditions.
Cell line culture
HCC cell lines HepG, SMMC7721, PLC, BEL-7402, and QGY7703, as well as the human normal hepatocyte line L-O2, were included in this study. RPMI-1640 Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum was used to culture the cells, together with a 1% antibiotic mixture of penicillin–streptomycin. The cells were grown in the desired culture medium and then incubated.
Clinical samples
A total of 79 HCC patients were included in this study. All HCC patients underwent liver resection at Linyi Central Hospital. HCC tissue samples were obtained from all included patients. The experiments involving the use of samples from HCC patients in this study were approved by the Ethics Committee of Linyi Central Hospital. Informed consent was obtained from all patients. All experiments were conducted in accordance with the Declaration of Helsinki.
Primary culture of HCC cells
HCC was identified by histological testing according to Edmondson's grading criteria. Tumor-adjacent tissues were obtained 3 cm away from HCC tissues. Isolation the HCC or tumor-adjacent primary cells was performed according to the previous description.11 Briefly, after hepatectomy the tumor tissues were immediately immersed in Hanks balanced salt solution (HBSS; Gibco) and transported to the laboratory. After removal of blood, the liver sample was cut into small fragments, gently dispersed, and placed in HBSS containing 0.03% pronase (Gibco), 0.05% type IV collagenase (Gibco), and 0.01% deoxyribonuclease (DNase, from bovine pancreas, Gibco) for 20 min at 37 °C. The resultant suspension was filtered through a 100 μm nylon filter (BD Falcon, Franklin Lakes, NJ, USA) and centrifuged at 50g for 2 min at 4 °C to obtain hepatocytes. The pellets were washed twice with HBSS containing 0.005% DNase. The final cell suspensions were cultured in collagen-coated T25 flasks (BD Falcon) in hepatocyte basal medium (Lonza, Basel, Switzerland) supplemented with 10% heat-inactivated FBS, 1 ng ml–1 hepatocyte growth factor (HGF, Prospec, Rehovot, Israel), and 1× antibiotic–antimycotic (Gibco) as HBM medium at 37 °C in a humidified incubator with 5% CO2. The medium was changed 24 h after seeding to remove dead cells and debris. When the cells reached 70–80% confluence, they were re-plated in HBM medium with supplements. Confluent cells were trypsinized, counted, and diluted 1 : 3–1 : 5 at every passage. Once the cells were maintained for more than 30 passages, the cells were collected and stored in liquid nitrogen.
Cell counting
Cells were counted using a cell counting plate. The space between the cover sheet and the counting plate was filled with cell suspension. The number of cells in the four large grids of the plate was determined manually. The number of cells was calculated using the following formula: Cell concentration (number per mL) = [(Number of cells in the grid)/4] × 104 mL–1
MTT assay
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was employed to determine the antitumor activities of Nano-SiO2 and antihepatoma drugs on all types of tumor cells. Each well of a 96-well plate was seeded with about 10 000 cells and then the cells were incubated for 24 h. Test agents at specific concentrations were used for cell treatment and then the cells were incubated for 24 h. The cells were ultimately treated with MTT solution and incubated for 3 h, followed by the addition of DMSO and incubation for 15 min. An ELISA microplate reader (DYNEX, USA) was used to measure the absorbance at 570 nm [optical density (OD) value]. The cytotoxic effect is expressed by the cell viability reduction rate (CVRR) and calculated using the following equation: CVRR = [(ODcontrol – ODdrug)/ODcontrol] × 100%.
Determination of the drug resistance patterns of HCC cells
Four first-line antihepatoma drugs, namely, cisplatin, fluorouracil (5FU), Taxol, and sorafenib, were included in this study. The peak plasma concentrations (PPCs) of these four drugs were determined, and drug resistance was considered to be CVRR < 50% at the PPC as recommended in a previous study.12
Cell cycle analysis
Cells were synchronized in G0 by serum starvation for 3 days followed by stimulation in DMEM supplemented with 10% FBS. Progression through the cell cycle was monitored by determination of the DNA content as previously described.13
Cell apoptosis analysis
An annexin-V- FLUOS staining kit (Roche-Boehringer) was used to detect the early stages of apoptosis, as represented by a flip of phosphatidylinositol to the outer membrane. The cells were washed with PBS and stained according to the manufacturer's protocol. The slides were mounted with Permafluor mounting medium (Immunotech, Marseille, France) and viewed under a fluorescence microscope (Axiophot Olympus).
RT-qPCR assay
Total RNA was extracted from the cells using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Medium without cells served as a negative control for this experiment. Subsequently, RT-qPCR was carried out with the PrimeScript RT-PCR kit (Takara Bio, Inc., Shiga, Japan), using β-actin as an internal control, in an Eppendorf Realplex4 machine (cat. no. X222687G; Hamburg, Germany). Reverse transcription reactions were performed using the following parameters: 16 °C for 30 min, 42 °C for 30 min and 84 °C for 5 min. The 2–ΔΔCq method was used for normalization.
RNA-sequencing assay
We fragmented the mRNA of the sample into 200 bp. Subsequently, a TruSeq RNA LT/HT sample preparation kit (Illumina, San Diego, CA, USA) was used to construct the cDNA library of the collected RNA and to synthesize the first and second strands of the collected RNA. Agilent 2200 TapeStation and Qubit 2.0 (Life Technologies, Carlsbad, CA, USA) were used to evaluate the purity of the DNA, and then the cDNA was diluted to 10 pM and sequenced using a HiSeq instrument (Illumina, San Diego, CA, USA).
Western blot assay
We washed the cells three times with cold phosphate buffer saline (PBS) and then lysed them with protein lysate (Pierce, Rockford, IL, USA). The supernatant of the mixture of the lysate and cell components was centrifuged at 4 °C for 15 minutes at 5000g. Then the protein concentration was measured using a Pierce kit for BCA protein determination (Pierce, Rockford, IL, USA). The purified protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene difluoride membrane. The membrane was sealed with 5% skimmed milk powder containing 0.05% Tween 20 Tris buffer saline (pH 7.4) and incubated with the primary antibody (Santa Cruz, Delaware Avenue, CA, USA) at a 1 : 200 ratio followed by incubation with the secondary antibody (Santa Cruz, Delaware Avenue, CA, USA) at a 1 : 5000 ratio. The target protein was detected by enhanced chemiluminescence (ECL) and film exposure.
Statistical analyses
Data are presented as the mean ± standard deviation from ≥3 separate experiments performed in triplicate. The differences between the groups were determined using two-tailed Student's t-test using SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA). P < 0.05 was considered to indicate a statistically significant difference.
Results
In vitro cytotoxic effect of Nano-SiO2 on HCC cells
HCC tissues were collected, and adjacent tissues from 79 patients were matched. Primary cells were then isolated. Cyclin D1 and GPC3, which can differentiate the primary HCC cells from the primary tumor-adjacent cells, exhibited significantly higher expression in the former than in the latter. These results are in accordance with previous studies.14 An in vitro study demonstrated that HCC cells treated with 100 μg mL–1 Nano-SiO2 exhibited higher reduced cell viability, compared with the adjacent cells (Fig. 1A). Subsequently, we found that 11 of the 79 primary cultured HCCs were resistant to cisplatin, 6 to 5-FU, 13 to Taxol, and 9 to sorafenib; there was no significant difference in the cytotoxic effects of Nano-SiO2 on these drug-resistant HCC cells and drug-sensitive HCC cells (Fig. 1B). Meanwhile, Nano-SiO2 exerted potent cytotoxic effects on all tested HCC cell lines but exerted significantly less inhibitory effects on human normal hepatocytes (L-O2) (Fig. 1C). These cytotoxic effects of Nano-SiO2 on HCC cells indicated that Nano-SiO2 treatment showed potential as an antitumor strategy against HCC.
Fig. 1. In vitro cytotoxic effect of Nano-SiO2 against HCC cells. (A) Cyclin D1 and GPC3 expressions were determined to identify HCC and tumor adjacent primary cells. (A and B) Cultured primary HCC cells and their matched tumor-adjacent cells were treated with or without 100 μg ml–1 Nano-SiO2 for 24 h, and the cell viability reduction rate (CVRR) was determined by MTT assay. (A) Comparison of the CVRR of HCC cells and the matched tumor adjacent group; (B) comparison of the CVRR of different drug-resistant groups of HCC cells. (C) Cultured human HCC cell lines and the normal liver tissue cell line were treated with or without 100 μg ml–1 Nano-SiO2 for 24 h, and the CVRR was determined by MTT assay.
Nano-SiO2 treatment leads to cell cycle arrest and apoptosis of HCC cells
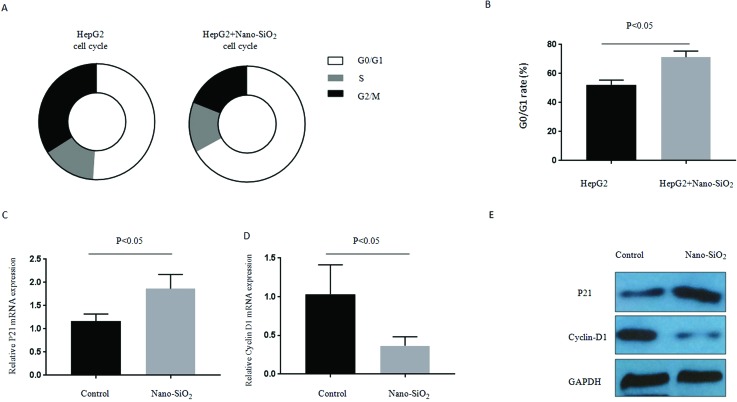
We found that Nano-SiO2 treatment could induce significant G1 cell cycle arrest relative to the controls in HepG2 cell lines (Fig. 2A). The G1 checkpoint is strictly regulated by cyclin-dependent kinase inhibitors, such as p21. The mRNA expression of p21 significantly increased in HepG2 treated with Nano-SiO2 (Fig. 2B). By contrast, the mRNA expression of cyclin D1, a cyclin that associates with and functions as a regulatory subunit of CDK4/6, decreased in HepG2 cells treated with Nano-SiO2 (Fig. 2C). This trend of expression of p21 and cyclin D1 was also demonstrated at the protein level by western blot assay (Fig. 2D).
Fig. 2. Nano-SiO2 treatment leads to cell cycle arrest in HCC cells. (A and B) The cultured human HCC cell line HepG2 was treated with or without 100 μg ml–1 Nano-SiO2 for 24 h, and the cell cycle was determined: (A) representive graphics of the cell cycle in the control group and the Nano-SiO2 group; (B) comparison of the G0/G1 rate of the control group and the Nano-SiO2 group. (C and D) Detection of mRNA expression of (C) p21 and (D) cyclin D1 by RT-PCR in cells described in (A) and (B). (E) Detection of protein expression of p21 and cyclin D1 by western blot assay in cells described in (A) and (B).
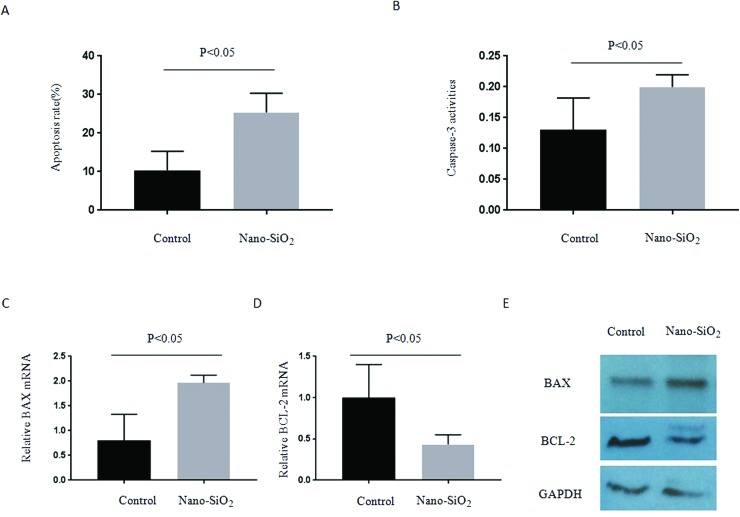
Cell apoptosis rates were subsequently determined using the annexin-V-FLUOS kit. Treatment with Nano-SiO2 significantly increased the HepG2 apoptosis rate (Fig. 3A). Similarly, caspase 3 activity was also induced by Nano-SiO2 (Fig. 3B). Treatment with Nano-SiO2 resulted in extremely higher Bax (pro-apoptotic, Fig. 3C) and lower Bcl-2 (antiapoptotic, Fig. 3D) mRNA expression levels after 24 h. Similar trends in protein expression were observed (Fig. 3E). These results demonstrated that Nano-SiO2 treatment can prevent HCC proliferation partly by cell cycle arrest and enhanced apoptosis.
Fig. 3. Nano-SiO2 treatment leads to cell apoptosis enhancement in HCC cells. (A and B) The cultured human HCC cell line HepG2 was treated with or without 100 μg ml–1 Nano-SiO2 for 24 h, and the (A) apoptosis rate and (B) caspase-3 activities were determined. (C and D) Detection of mRNA expression of (C) BAX and (D) cyclin D1 by RT-PCR in cells described in (A) and (B). (E) Detection of protein expression of BAX and BCL-2 by western blot assay in cells described in (A) and (B).
Nano-SiO2 treatment induces necroptosis of HCC cells
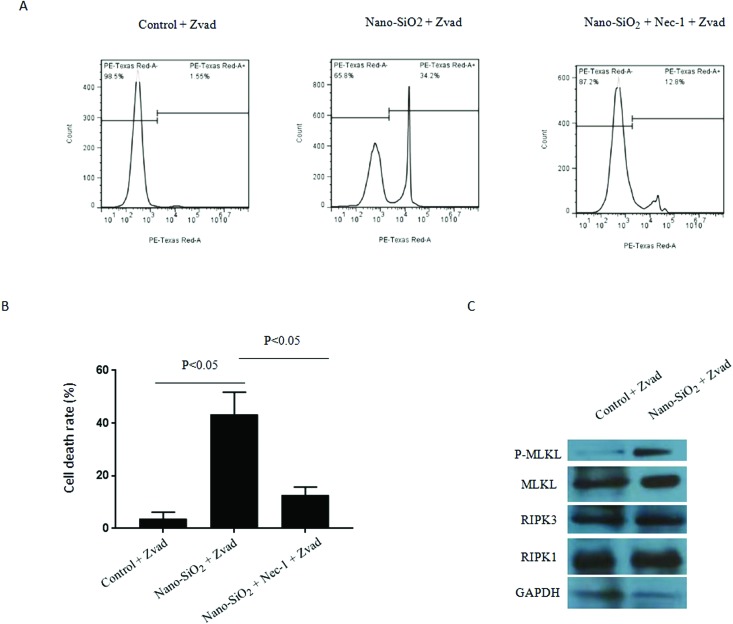
In addition to apoptosis, other modes of cell death, usually proactive programmed cell death, can inhibit the proliferation of cancer cells. Necrosis used to be considered a non-programmed mode of cell death; however, recent studies have shown that a new type of programmed necrosis, referred to as necroptosis, is closely associated with the cancer process.15 Necroptosis is optimally induced when the apoptotic machinery is interrupted.16 To delineate the regulation of necroptosis by Nano-SiO2, HepG2 cells were treated with Nano-SiO2 in combination with Z-VAD-FMK, an apoptosis inhibitor. In this cell model, Nano-SiO2 promoted cell death significantly, and this cell death process was inhibited in the presence of the RIPK1 kinase inhibitor Nec-1 (Fig. 4A), indicating that the induced cell death was necroptotic. Meanwhile, Nano-SiO2 activated the RIPK1/RIPK2/MLKL pathway, which was a signal pathway promoting necroptosis (Fig. 4B). The combined findings suggest that Nano-SiO2 induced dominant necroptosis in HepG2 cells.
Fig. 4. Nano-SiO2 treatment induces necroptosis in HCC cells. (A and B) The cultured human HCC cell line HepG2 was treated with or without 100 μg ml–1 Nano-SiO2, in the presence of Z-VAD-FMK (20 μM), together with or without Nec-1 (50 μM) for 24 hours. Nec-1 and Z-VAD-FMK were pretreated for 3 hours prior to Nano-SiO2 treatment. (A) Representative flow cytometry graphics of PI staining in the control group, Nano-SiO2 group and Nano-SiO2 + Nec-1 group; each group of cells was subjected to PI staining and subsequent flow cytometry, and the number of cells tested was 5000. (B) The percentages of necrotic cells (stained with PI) increased significantly in the Nano-SiO2 group and were significantly inhibited by pretreatment with Nec-1. (C) Detection of the phosphorylation level or protein expression of MLKL, RIPK1 and RIPK3 by western blot assay in the cells described in (A) and (B).
Global changes in the expression of genes in HCC cells upon Nano-SiO2 treatment
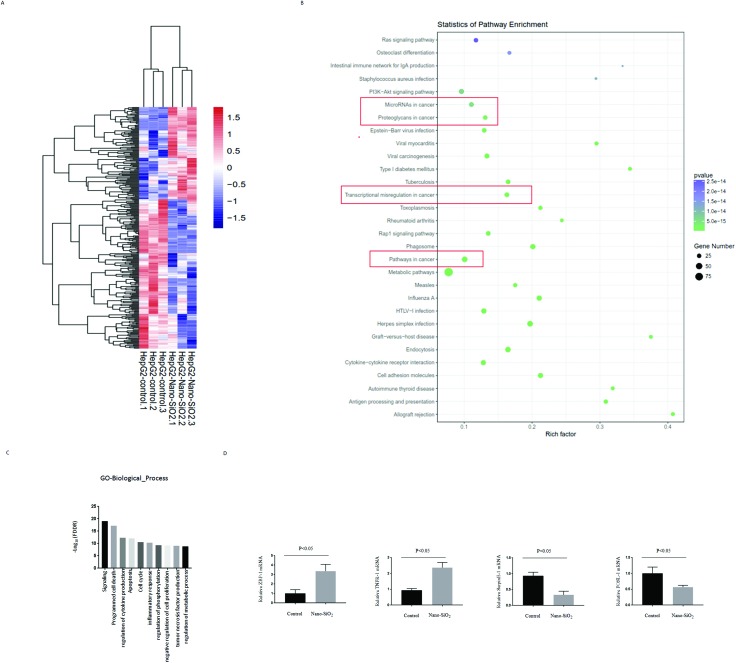
We used HepG2 cells treated with Nano-SiO2 to investigate global changes in gene expression. With a cutoff >1.5 fold and p ≤ 0.05, 384 genes were differentially expressed upon Nano-SiO2 treatment (Fig. 5A). KEGG pathway enrichment analyses revealed that numerous genes were involved in the cancer process (Fig. 5B). Gene Ontology (GO) enrichment demonstrated that Nano-SiO2 treatment could regulate many genes related to various types of cell death (Fig. 5C). RT-PCR confirmed that ZBP1, TNFR-1, SerpinE-1, and FOSL-1 could be regulated by Nano-SiO2; ZBP1 was upregulated most significantly.
Fig. 5. Genome-wide sequencing together with qRT-PCR analysis identifies global gene changes in Nano-SiO2 treated HepG2 cells. (A) The total RNA samples prepared in Fig. 2C and D were used to perform the RNA-seq experiment. Heatmap analysis displaying the deregulated targets in Nano-SiO2 HepG2 cells, compared with that in the control group. (B) KEGG bioinformatics analysis determined the pathways related to the differentially expressed genes identified in (A). Red-box labeling of cancer-related pathways. (C) GO bioinformatics analysis enriched the biological functions related to the differentially expressed genes identified in (A). (D) Of the deregulated genes found in (A), 4 genes closely related to cell death were selected for qRT-PCR verification.
Nano-SiO2 treatment induces necroptosis and plays a cytotoxic role in HCC cells by increasing ZBP1 expression
ZBP1, which reportedly leads to necroptosis,17 was significantly upregulated by Nano-SiO2 and could be downregulated by its siRNA, as verified by western blot assay (Fig. 6A). MLKL phosphorylation induced by Nano-SiO2 was reversed by ZBP1 knockdown. Furthermore, we found that the ZBP1 knockdown could reverse the necroptosis induced by Nano-SiO2 (Fig. 5B and C). The aforementioned results demonstrated that Nano-SiO2 treatment could induce necroptosis in HCC cells via ZBP1 enhancement. ZBP1 knockdown could also reduce the CVRR caused by Nano-SiO2. Thus, ZBP1 is an important mediator of the cytotoxic effect of Nano-SiO2 on HCC cells.
Fig. 6. Nano-SiO2 treatment induces necroptosis and exhibits a cytotoxic effect in a ZBP-1 dependent manner. (A) The cultured human HCC cell line HepG2 was treated with or without 100 μg ml–1 Nano-SiO2, in the presence in Z-VAD-FMK (20 μM), together with or without ZBP-1 siRNA for 24 hours. The phosphorylation level or protein expression of ZBP-1 and MLKL was detected by western blot assay. (B and C) The cultured human HCC cell line HepG2 was treated with or without 100 μg ml–1 Nano-SiO2, in the presence of Z-VAD-FMK (20 μM), together with or without ZBP-1 siRNA for 24 hours. Z-VAD-FMK was pretreated for 3 hours prior to Nano-SiO2 treatment. (B) Representative flow cytometry graphics of PI staining in the control group, Nano-SiO2 group and Nano-SiO2 + ZBP-1 siRNA group; (C) the percentages of necrotic cells (stained with PI) increased significantly in the Nano-SiO2 group and were significantly inhibited by pretreatment with ZBP-1 siRNA. (D) Cultured human HCC cell lines were treated with 100 μg ml–1 Nano-SiO2 together with or without ZBP-1 siRNA for 24 h, and the cell viability reduction rate (CVRR) was determined by MTT assay.
Discussion
The drug resistance of HCC cells has become increasingly serious in recent years, which is one of the important factors for the low five-year survival rate of HCC patients after surgery.18 Many mechanisms underlying chemotherapeutic drug resistance in hepatocellular carcinoma have been identified. However, apoptosis, originally an important anticancer mechanism was reported to regulate a large number of genes, disturbing the balance between apoptotic and antiapoptotic cells. This imbalance induces resistance to various chemotherapeutic drugs, and hence the emergence of multidrug-resistant HCC cases.19 This special mechanism hinders the development of drugs targeting apoptosis and requires the development of improved anticancer agents.
Nano-SiO2 is considered to be an effective antineoplastic agent and has been extensively studied.7,20,21 The results of the current study, together with previous reports, demonstrate that Nano-SiO2 exhibits a potential cytotoxic effect on HCC cells, regardless of whether the cancer cells are resistant to cisplatin, 5FU, Taxol, and sorafenib or not.
Various articles have reported on the pathway of the cytotoxic effect of Nano-SiO2. Jeon et al. showed the efficacy of Nano-SiO2 against breast cancer cells via modulation of EGFR signaling cascades.7 Ye et al. demonstrated that Nano-SiO2 induced apoptosis via activation of p53 and Bax mediated by oxidative stress in human hepatic cells.20 Promoting apoptosis is an important means for Nano-SiO2 function. The results of this study first demonstrated that Nano-SiO2 can cause cell cycle arrest and promote apoptosis, which is consistent with previous reports.21,22 Moreover, Nano-SiO2 can promote necroptosis, which acts differently from apoptosis. Necroptosis is a recently identified antitumor cell death mechanism that is independent of the caspase pathway.15 The functions of necroptosis in HCC development were identified in various studies;16 however, the regulation these diseases by necroptosis has rarely been reported. Genome-wide sequencing was conducted to explore why Nano-SiO2 stimulation could induce necroptosis. RNA-seq analysis, followed by KEGG and GO bioinformatics analysis, was conducted to identify cell death-related genes regulated by Nano-SiO2. An important finding was that Nano-SiO2 could upregulate ZBP1 expression, as determined by RNA-seq assay and verified by western blot analysis. Using ZBP1 siRNA, we found that Nano-SiO2 could enhance the necroptosis, together with cell viability reduction, via the ZBP1 involved pathway. The RNA-seq results also indicated that in addition to ZBP1, many genes closely related to necroptosis were significantly upregulated, such as TNFR1, an important factor promoting cell necroptosis.23 Further experiments are needed to examine the relationship between Nano-SiO2 and these molecules.
Numerous publications have shown that Nano-SiO2 produces toxicity both in vitro and in vivo.10,24,25 Sergent et al. demonstrated that Nano-SiO2 exerts potent cytotoxic effects on normal human epithelial intestinal cells.10 Moreover, increases in intracellular reactive oxygen species and catalase activity were among the common effects of Nano-SiO2, even at noncytotoxic concentrations.24 These toxicities of Nano-SiO2 mainly depend on size, dose, and cell type.25 Our results indicated that the proposed Nano-SiO2 treatment exerted significantly less inhibition effects on human normal hepatocytes than on HCC cells at the same concentration. This result indicated that Nano-SiO2 can potentially become an antitumor therapy agent in the liver. Additional data from animal experiments are needed to prove this finding.
Conflicts of interest
There are no conflicts of interest to declare.
References
- Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet-Tieulent J., Jemal A. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- Forner A., Llovet J. M., Bruix J. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- Awan F. M., Naz A., Obaid A., Ikram A., Ali A., Ahmad J. Sci. Rep. 2017;7:11448. doi: 10.1038/s41598-017-11943-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz L. Y., Wang D., Liu J. J. Photochem. Photobiol., B. 2019;191:123–127. doi: 10.1016/j.jphotobiol.2018.12.008. [DOI] [PubMed] [Google Scholar]
- Agarwalla P., Mukherjee S., Sreedhar B., Banerjee R. Nanomedicine. 2016;11:2529–2546. doi: 10.2217/nnm-2016-0224. [DOI] [PubMed] [Google Scholar]
- Chen W., Yang W., Chen P., Huang Y., Li F. ACS Appl. Mater. Interfaces. 2018;10:41118–41128. doi: 10.1021/acsami.8b14940. [DOI] [PubMed] [Google Scholar]
- Jeon D., Kim H., Nam K., Oh S., Son S. H., Shin I. Anticancer Res. 2017;37:6189–6197. doi: 10.21873/anticanres.12068. [DOI] [PubMed] [Google Scholar]
- Yang Y., Yu C. Nanomedicine. 2016;12:317–332. doi: 10.1016/j.nano.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Dey S., Bakthavatchalu V., Tseng M. T., Wu P., Florence R. L., Grulke E. A. Carcinogenesis. 2008;29:1920–1929. doi: 10.1093/carcin/bgn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergent J. A., Paget V., Chevillard S. Ann. Occup. Hyg. 2012;56:622–630. doi: 10.1093/annhyg/mes005. [DOI] [PubMed] [Google Scholar]
- Song Y., Kim J. S., Kim S. H., Park Y. K., Yu E., Kim K. H. J. Exp. Clin. Cancer Res. 2018;37:109. doi: 10.1186/s13046-018-0752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselsberger K., Peterson D. C., Thomas D. G., Darling J. L. Anticancer Drugs. 1996;7:331–338. [PubMed] [Google Scholar]
- Hinz M., Krappmann D., Eichten A., Heder A., Scheidereit C., Strauss M. Mol. Cell. Biol. 1999;19:2690–2698. doi: 10.1128/mcb.19.4.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü Z. L., Luo D. Z., Wen J. M. World J. Gastroenterol. 2005;11:3850–3384. doi: 10.3748/wjg.v11.i25.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Kepp O., Chan F. K., Kroemer G. Annu. Rev. Pathol. 2017;12:103–130. doi: 10.1146/annurev-pathol-052016-100247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahsavari Z., Karami-Tehrani F., Salami S. Asian Pac. J. Cancer Prev. 2015;16:7261–7266. doi: 10.7314/apjcp.2015.16.16.7261. [DOI] [PubMed] [Google Scholar]
- Lin J., Kumari S., Kim C., Van T. M., Wachsmuth L., Polykratis A. Nature. 2016;540:124–128. doi: 10.1038/nature20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricoli L., Niture S., Chimeh U., Kumar D. Front. Biosci., Landmark Ed. 2019;24:545–554. doi: 10.2741/4734. [DOI] [PubMed] [Google Scholar]
- Li S., Zhang D., Sheng S., Sun H. Int. J. Nanomed. 2017;12:5993–6003. doi: 10.2147/IJN.S137335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Liu J., Xu J., Sun L., Chen M., Lan M. Toxicol. in Vitro. 2010;24:751–758. doi: 10.1016/j.tiv.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Min Y., Li J., Liu F., Yeow E. K., Xing B. Angew. Chem., Int. Ed. 2014;53:1012–1016. doi: 10.1002/anie.201308834. [DOI] [PubMed] [Google Scholar]
- Asweto C. O., Wu J., Hu H., Feng L., Yang X., Duan J. Int. J. Environ. Res. Public Health. 2017;14:E289. doi: 10.3390/ijerph14030289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Huang Y., Chen L., Gu J., Zhou X. Hum. Cell. 2014;27:162–171. doi: 10.1007/s13577-014-0093-z. [DOI] [PubMed] [Google Scholar]
- Wen T., Yang A., Piao L., Hao S., Du L., Meng J. Int. J. Nanomed. 2019;14:4475–4489. doi: 10.2147/IJN.S208225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I. Y., Joachim E., Choi H., Kim K. Nanomedicine. 2015;11:1407–1416. doi: 10.1016/j.nano.2015.03.004. [DOI] [PubMed] [Google Scholar]