 To answer questions about nanoparticle toxicity, translocation and biokinetics following inhalation exposure, in vitro models should have functional barrier properties to mimic the in vivo situation as closely as possible.
To answer questions about nanoparticle toxicity, translocation and biokinetics following inhalation exposure, in vitro models should have functional barrier properties to mimic the in vivo situation as closely as possible.
Abstract
Inhalation is the most relevant entry point for nanoparticles (NPs) into the human body. To date, toxicity testing of nanomaterials in respect to oral, dermal and inhalative application is mainly based on animal experiments. The development of alternative test methods is the subject of current research. In vitro models can help to investigate mechanistic aspects, as e.g. cellular uptake or genotoxicity and might help to reduce in vivo testing. Lung cell lines are proper in vitro tools to assess NP toxicity. In respect to this, various cell models have been developed during the recent years, but often lack in a proper intact barrier function. However, besides other important in vivo criteria which are still missing like e.g. circulation, this is one basic prerequisite to come closer to the in vivo situation in certain mechanistic aspects such as particle translocation which is an important task for risk assessment of nanomaterials. Novel developed in vitro models may help to investigate the translocation of nanomaterials from the lung. We investigated the barrier function of the recently developed human lung cell lines CI-hAELVi and CI-huAEC. The cells were further exposed to CeO2 NPs and ZnO NPs, and their suitability as in vitro models for toxicological investigations was proven. The obtained data were compared with data generated with the A549 cell line. Measurement of transepithelial resistance and immunohistochemical examination of tight junctions confirmed the formation of a functional barrier for both cell lines for submerged and air–liquid cultivation. For particle exposure, hAELVi and huAEC cells showed comparable results to A549 cells without losing the barrier function. CeO2 NP exposure revealed no toxicity for all cell lines. In contrast, ZnO NPs was toxic for all cell lines at a concentration between 10–50 μg ml–1. Due to the comparable results to A549 cells CI-hAELVi and CI-huAEC offer new opportunities to investigate nanoparticle cell interactions more realistic than recent 2D cell models.
Introduction
Due to the increased use of nanomaterials in consumer products, investigations into their safety and potential risks are key tasks.1 Despite interspecies variations,2–4 understanding any potential implications of nanoparticles (NPs) to human health are normally conducted in animal models.5–13 However, based on the 3R (refine, reduce and replace) principle, the development of alternative testing methods is an important task.14 For this, in vitro models can be helpful to answer mechanistic issues like e.g. cellular uptake15 or genotoxicity.16 Due to their small diameter NPs deposit deep into the lung.17 Therefore, NPs are mainly taken up via inhalation18 followed by a presumed deposition in the lower regions of the lung. Here, they come in contact with bronchial epithelia cells and pneumocyte type I & II cells. There are several human in vitro systems reported to assess adverse effects of NP cell-interactions in the lung. This includes bronchial cell lines, alveolar cell lines, different co-culture models as well as 3D models.15,19–24 For instance, an increased oxidative stress and apoptosis of BEAS-2B cells after cerium dioxide (CeO2) NPs exposure has been previously reported.25 Another group used the BEAS-2B cells line as well as the bronchial 3D system MucilAir™ to investigate the toxicity of CeO2 NPs. They found that the 3D model is more resistant to oxidative stress and DNA damage than simple cell cultures.23 In contrast, there are also reports demonstrating protective functions of CeO2 NPs which could be attributed to their antioxidant properties as studied in details in many published work.14,26–29 For the alveolar region, A549 is the most frequently used cell line to study particle cell interactions. These cells are used either as a single monolayer or as co-culture in combination with other cell lines. For example, cytotoxicity of gold NPs in A549 cells was recently reported by inducing cell cycle arrest, oxidative stress and apoptosis.30 A549 cells were also used to determine the toxicity of copper oxide NPs,31 CeO2 NPs21 and zinc oxide (ZnO) NPs.32 In addition to single cell lines that allow investigation of mechanistic aspects only, there are approaches to improve the used cell models to closely mimic the in vivo situation by using more sophisticated cell models such as co-cultures or 3D cell models. E.g. a co-culture system of A549, alveolar macrophages and dendritic cells was used to investigate the uptake of polystyrene particles. Most of the particles were found in macrophages but A549 and dendritic cells were also able to take up polystyrene particles.33 Another conducted study even went one step further and developed a 3D co-culture model composed of A549, THP-1, mast cells (HMC-1) and endothelia cells (EA·hy 926). This tetraculture model was subsequently exposed to 50 nm SiO2 rhodamin labeled NPs. SiO2 NPs were only found in the macrophage like THP-1 cell line but not in A549 cells.34 Despite the improved complexity of these models, a decisive disadvantage about barrier function still remains. The epithelial cells used in all alveolar models were A549 cells, a cell line which do not possess an intact barrier function.35–37 Thus, they are not fully suited for studying the translocation of NPs. The NP translocation from the lung to secondary organs and tissues was previously described in the literature.11,18,38 There are hints that NPs reach extrapulmonary structures via the blood stream circulation.13,39 In 2006 rats were exposed to gold NPs. An uptake into epithelia cells and a translocation into the circulation occurred. However, an uptake by the endothelium has not been reported.40 This raises the question how the NPs reached the blood stream. A translocation of NPs loaded macrophages into the lymph nodes was recently shown which could be one further mechanism.8,41 In 2010 real-time intraoperative near-infrared fluorescence imaging was used to show that both mechanisms mentioned above may take place simultaneously.42 However, the exact mechanism is not yet fully understood and subject of current research. Human alveolar in vitro models with intact barrier function would allow a closer estimation of the in vivo situation in terms of translocation of NPs. Hence, the aim of this work is to determine a cell model that reflects the in vivo situation more realistic than current used models and allows studying the translocation of NP under more realistic conditions. For this purpose, we investigated the recently developed human alveolar type I cell line CI-hAELVi (human Alveolar Epithelial Lentivirus immortalized) hereinafter stated as hAELVi.43 hAELVi cells were characterized regarding their barrier function and the influence of CeO2 NPs and ZnO NPs. Furthermore, the recently developed airway epithelia cell line CI-huAEC (human Airway Epithelial Cells),44 a model of the lower respiratory tract, was examined for the same endpoints. The CI-huAEC cell line is hereinafter stated as huAEC. In addition, we evaluated the alveolar 3D model EpiAlveolar in terms of barrier function. The obtained data were compared to results achieved with A549 cells.
Experimental
Cell culture
A549 cells (ATCC cat. no.: CCL-185) were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum (FCS) (PAN-Biotech GmbH, Germany), 1% penicillin/streptomycin (PAN-Biotech GmbH, Germany) and 1% l-glutamine (PAN-Biotech GmbH, Germany). Cells were passaged two times per week.
CI-hAELVi (cat. no.: INS-CI-1015) and CI-huAEC (cat. no.: INS-CI-1011) cells were purchased from InSCREENeX GmbH (InSCREENeX GmbH, Germany). Both cell lines were cultured in CI-huAEC media supplemented with 1% penicillin/streptomycin (PAN-Biotech GmbH, Germany) and the CI-huAEC basal supplement provided by the manufacture (InSCREENeX GmbH, Germany). Cells were passaged two times per week.
EpiAlveolar
EpiAlveolar is a three dimensional human alveolar model and consists of lung epithelia cells, fibroblasts and endothelia cells.45 EpiAlveolar was purchased from MatTek (MatTek Corporation, USA). Cultivation was conducted in accordance to the manufacturer's protocol. The medium was supplemented with 1% penicillin/streptomycin (PAN-Biotech GmbH, Germany).
Air–liquid cultivation
For trans-epithelial electrical resistance measurements, cells were seeded on transwell membranes and cultured for two days in submerse culture conditions (cat. no. 353180, Coring B.V., Netherland; 0.4 μm pore size, 1.12 cm2) prior transferring them to the air–liquid phase. Therefore, the apical medium was removed and cells were washed once with PBS. Basal medium was changed every two days.
Trans-epithelial electrical resistance (TEER)
To determine the barrier properties of all lung models we conducted TEER measurements. Cell lines were seeded onto transwell membranes and cultured for two days under submerse conditions. Subsequently, the cells were divided in two groups and cultured for further 15 days: five membranes were further cultivated under submerge conditions (LL = liquid–liquid), six under air–liquid conditions (ALI). One insert without cells was used as background control. The background control was subtracted from the measured data. TEER measurements were performed each day with a Millicell-ERS system (Merk, Darmstadt, Germany) (STX2 electrode). Before measuring, cells were washed once with PBS. 1 ml fresh medium was added into the apical compartment and the cells were placed in the incubator for 1 h before measuring. In order to prevent the electrode being in contact with the plate wall, the membranes were transferred into a 6 well plate before starting the measurement. The 6-well plate was filled with 5 ml PBS per well. After measuring, apical medium was removed from ALI cultured inserts. The basolateral medium was changed every two days. For huAEC and hAELVi cells, membranes were coated with huAEC coating solution three hours before seeding. For A549, medium was added in the apical part of the membranes three hours before seeding. For EpiAlveolar, TEER measurement was performed as mentioned above. Due to the shortened life span of EpiAlveolar, TEER was only monitored for eight days.
ZO-1 staining
For the optical characterization of tight junctions cells were grown on microscopic dishes (cat. no. D35-20-1-N, IBL Baustoff + Labor GmbH, Austria) or on transwell membranes (cat. no. 353180, Coring B.V., Netherland; 0.4 μm pore size). Seeding density was 50 000 cells. ZO-1 staining was performed after 14 days as described below. Cells were washed three times with PBS and fixed with 4% paraformaldehyde for 15 minutes at room temperature (RT). Afterwards, the samples were permeabilized with 0.2% Triton X-100 (Merck, KGaA, Darmstadt, Germany) for 10 minutes at RT. Subsequently, a blocking step with PBS containing 10% FCS was performed. The primary anti-ZO-1 antibody (cat. no. 402200, Fisher Scientific, Germany) was diluted 1 : 200 in PBS containing 1% FCS and incubated at 4 °C overnight. The secondary antibody (rabbit IgG, Alexa 488, cat. no. A-11034, Fisher Scientific, Germany) was diluted 1 : 400 in PBS containing 1% FCS and incubated for 1 h at RT. Then cells were washed with PBS three times and counterstained with Hoechst or DAPI (1 μg ml–1). Samples were analyzed by a confocal laser scanning microscopy (LSM 700, Zeiss).
Growth curve and population doubling time
To assess the growth behavior of the different lung cells, 50 000 cells per well were seeded into a 6 well plate. Cells were harvested by trypsinization and counted in a haemocytometer by trypan blue dye exclusion after 24 h, 48 h, 72 h and 96 h. For each time point three wells were counted. The population doubling (PDT) time was determined based on the following equation:PDT = t/((Log (C1) – Log(C2)/Log(2))
With PDT = population doubling time (h), t = time point of harvesting (h), Log = 10 based Log, C1 = 1 cell number counted at harvesting time point, C2 = cell number initially seeded. PDT was calculated from the exponential growth phase (harvesting time points: 48, 72 and 96 h).
Particle characterization
Transmission electron microscope (TEM)
In situ TEM observation of NPs was performed by a JEM-2100HR transmission electron microscopy (JEOL, Japan) operated at 100 kV equipped with an energy-dispersive X-ray (EDX) spectrum. For TEM analysis, the sample solution was drop coated on TEM copper grids (Agar Scientific, United Kingdom) from a 10 μg ml–1 particle solution and allowed to dry overnight under RT.
Dynamic light scattering (DLS)/zeta potential
Determination of the hydrodynamic diameter and the zeta potential were performed with a Zetasizer Nano ZS from Malvern (Malvern Inc., UK) in MilliQ water and in both cell culture media. For analysis, particle concentration for both materials was set to 50 μg ml–1.
Nanoparticle tracking analysis (NTA)
NTA was performed with a NanoSight LM20 (NanoSight, Amesbury, UK), equipped with a 632 nm laser, in MilliQ water and in both cell culture media. For analysis, particle concentration was set to 250 ng ml–1 and 10 μg ml–1 for CeO2 and ZnO NPs, respectively. All measurements were performed at RT. The software used for recording and analyzing the data was NTA 2.3. All samples were measured for 60 seconds at five positions.
Particle toxicity
CeO2 NPs (NM-212) was chosen as a well characterized granular biopersistent particle (GBP).46 As zinc oxide is known as cytotoxic, it was chosen as positive particle control as well as soluble particle model. To determine adverse effects after submerged NP exposure, A549, huAEC and hAELVi cells were exposed to CeO2 NPs and ZnO NPs for 24 h. CeO2 NPs (JRC) and ZnO NPs were weighed and the particles were dispersed in MilliQ water to a final stock concentration of 2.5 mg ml–1. Subsequently, the particle dispersion was sonicated for 5 minutes and 9 seconds (Sonoplus HD 220/UW 2200, Bandelin, Germany) to avoid particle aggregation. For all experiments, particles were freshly prepared. For cell exposure, particles were diluted in media to reach the final concentration. After exposure, cell viability, cytotoxicity and ROS production was determined using a WST-1 (water soluble tetrazolium-1), a lactatdehydrogenase (LDH) assay and a 2′,7′-dichlorofluorescin diacetate (DCFDA) assay, respectively.
Cell viability
After particle exposure, the supernatant was transferred in a new 96 well plate and subsequently used for LDH analysis (see below). Cells were rinsed with PBS and fresh medium containing 10% WST-1 reagent (Roche Diagnostics GmbH, Germany) was added into the well (100 μl). After 1 h incubation at 37 °C, 90 μl was transferred in a new 96 well plate and the absorbance was measured with a Tecan plate reader using wavelengths of 450 nm and 562 nm (reference wavelength). Six technical replicates were performed.
Cytotoxicity
After particle exposure a LDH assay was conducted to check for membrane damage after particle exposure. The assay was performed according to the manufactures instructions (Roche Diagnostics GmbH, Germany). In brief, LDH reagent was added to the supernatant and incubated for 15 minutes in dark at RT. Afterwards the absorbance was measured with a Tecan plate reader at 450 nm. Six technical replicates were performed.
Reactive oxygen species
The level of intracellular reactive oxygen species (ROS) generation was determined by using a DCFDA assay. After particle exposure, the cells were rinsed with PBS and DCFDA (80 μM in Medium) (Merck KGaA, Darmstadt, Germany) was added to the cells and incubated for 45 minutes at 37 °C. Afterwards, DCFDA was aspirated and the cells were rinsed again once with PBS. New medium and the positive control (tert-butyl hydroperoxide (TBHP 1/20 000 from stock solution), Merck KGaA, Darmstadt, Germany) was added to the cells and further incubated for 2 h. Subsequently, DCFDA fluorescence intensity was measured within a plate reader (Biotek Synergy™ HTX multi detection reader, BioTek Instruments, Inc., Winooski, USA) at excitation and emission wavelengths of 485 and 528 nm, respectively. Three technical replicates were performed.
Statistical analysis
Data are shown as mean ± standard deviation. If not stated otherwise data represents three independent experiments. For statistical analysis a Mann–Whitney-U-Test was performed using Origin 9.1 software. *P > 0.05 was considered as significant; **P > 0.01; ***P > 0.001.
Results and discussion
Most studies investigating the interactions of NPs and airway epithelia were carried out with bronchial and alveolar cells.5,20,22,23,35,47,48 Unfortunately, the most commonly used alveolar model, the A549 cell line, possess a carcinogenic phenotype49 and lacks in a proper barrier function.35–37 Due to the regulation of paracellular substance transport, the barrier function is important for the systemic distribution of inhaled NPs.50 Therefore, we investigated the recently developed cell line hAELVi, a model for type I pneumocytes,43 as well as the new developed bronchial cell line huAEC44 in respect of their capability to form an intact and functional cell–cell-barrier. In addition, we also analyzed the more complex 3D human alveolar model EpiAlveolar in respect of ongoing experiments regarding particle uptake and location/translocation. All received data were compared to the frequently used alveolar cell line A549.
Particle characterization
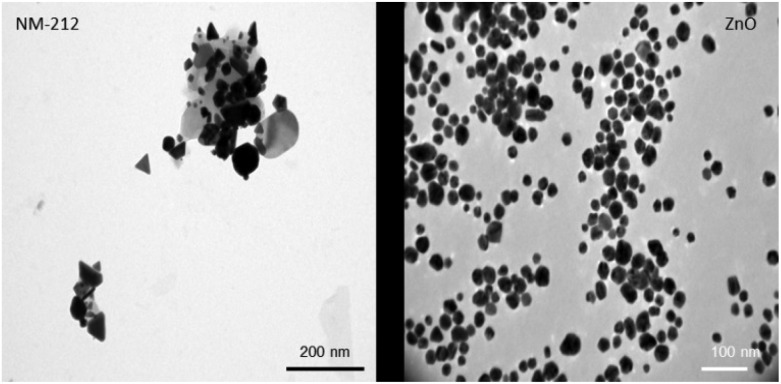
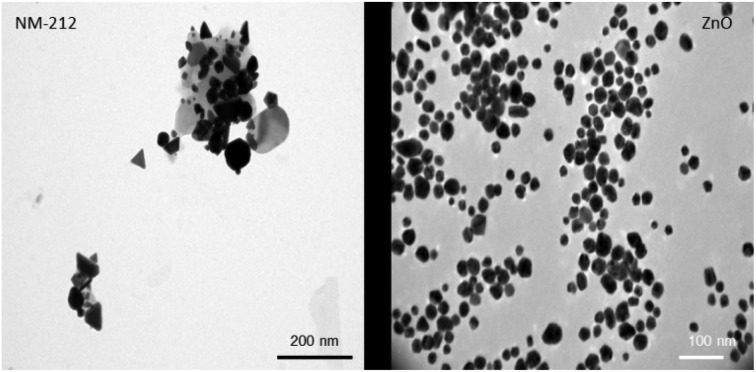
Compared to the widely used NANOGENOTOX protocol, we slightly modified the particle generation procedure (no bovine serum albumin, 10 ml dispersion volume instead of 6 ml and final concentration of 2.5 mg ml–1 instead of 2.56 mg ml–1). Therefore, the NPs were again thoroughly characterized.51 The particle size of CeO2 and ZnO NPs were characterized using TEM, NTA and DLS measurements. In addition the zeta potential was determined. Particle size distributions can be found in ESI.† As shown in Table 1, both particle types exhibit a comparable size and zeta potential. As depicted in Fig. 1, ZnO particles were spherical whereas CeO2 particles displayed a rather platelet shape. Furthermore, CeO2 NPs showed strong agglomeration behavior compared to ZnO NPs. DLS and NTA were used to determine the hydrodynamic diameter of both materials. For CeO2 NPs DLS revealed a slightly higher hydrodynamic diameter as NTA. This is mainly due to the fact that during the DLS measurements large particles contribute more to the diameter determination as NTA analysis. Nevertheless, our DLS results are consistent with data published by the manufacture JRC.52 For ZnO NPs, DLS and NTA analysis displayed a similar size of about 250 nm. Electron microscopy analysis revealed no agglomeration for ZnO NPs whereas CeO2 NPs showed a strong agglomeration behavior. Taken this into account, this explains the differences between the DLS and NTA data for CeO2 NPs.
Table 1. NPs characterization of CeO2 and ZnO.
| NTA [nm] | DLS [nm] | Zeta potential [mV] | |
| CeO2 in MilliQ | 164.1 ± 33.4 | 212.9 ± 20.6 | 22.6 ± 0.9 |
| ZnO in MilliQ | 265.6 ± 78.7 | 244.5 ± 4.6 | 25.5 ± 1.6 |
| CeO2 in DMEM | 86.0 ± 43.7 | 1550.8 ± 157.1 | –11.3 ± 0.8 |
| ZnO in DMEM | 227.9 ± 41.8 | 189.6 ± 10.3 | –10.8 ± 0.6 |
| CeO2 in huAEC | 146.5 ± 72.4 | 2159.7 ± 104.5 | –10.8 ± 0.7 |
| ZnO in huAEC | 231.3 ± 57.0 | 337.1 ± 28.7 | –9.5. ± 0.7 |
Fig. 1. NP characterization: Representative TEM pictures of CeO2 (left) and ZnO (right) NPs. ZnO NPs show a spherical morphology and less agglomeration whereas CeO2 NPs were more clustered and displayed a rather platelet like shape.
Characterization of lung cells: growth behavior
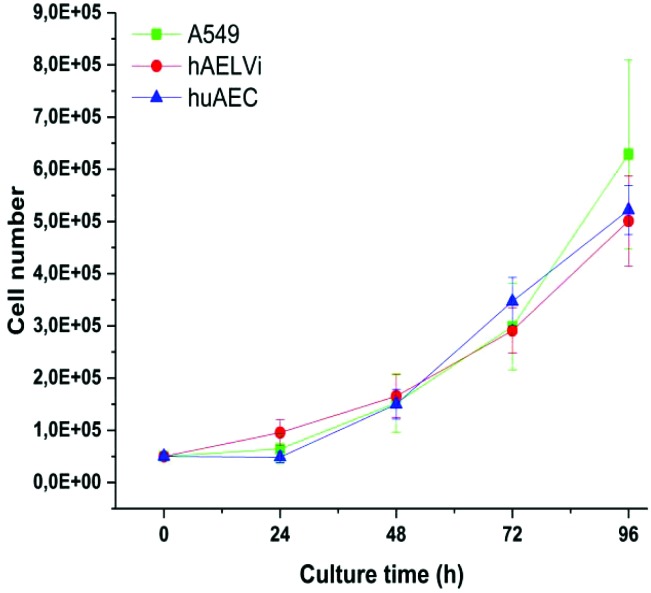
A549 cells are a well-established cell line in particle toxicity studies, whereas hAELVi and huAEC are relatively new cell lines. To the best of our knowledge, there are no data for huAEC cells published so far except the technique used to create them.44 Therefore, we firstly investigated the growth behavior of the different cell lines to basically understand their growth behavior. Fig. 2 illustrates the growth curve of all lung epithelia cell lines we used. As expected, all of them showed an exponential growth pattern.43,53 The population doubling time for all cell lines was 28 hours and is in accordance with previous A549 studies53,54 indicating a similar growth behavior than standard cell lines used in this field.
Fig. 2. Characterization of growth behavior: Growth curves of A549, hAELVi and huAEC cells show similar growth behavior for all cell types.
Characterization of barrier function: transepithelial resistance measurement and tight junction staining
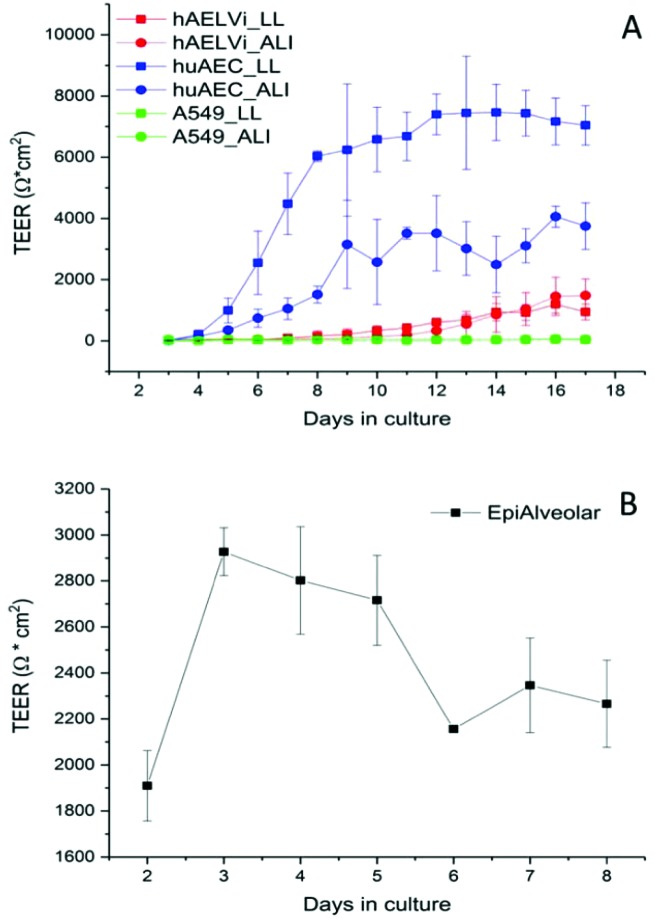
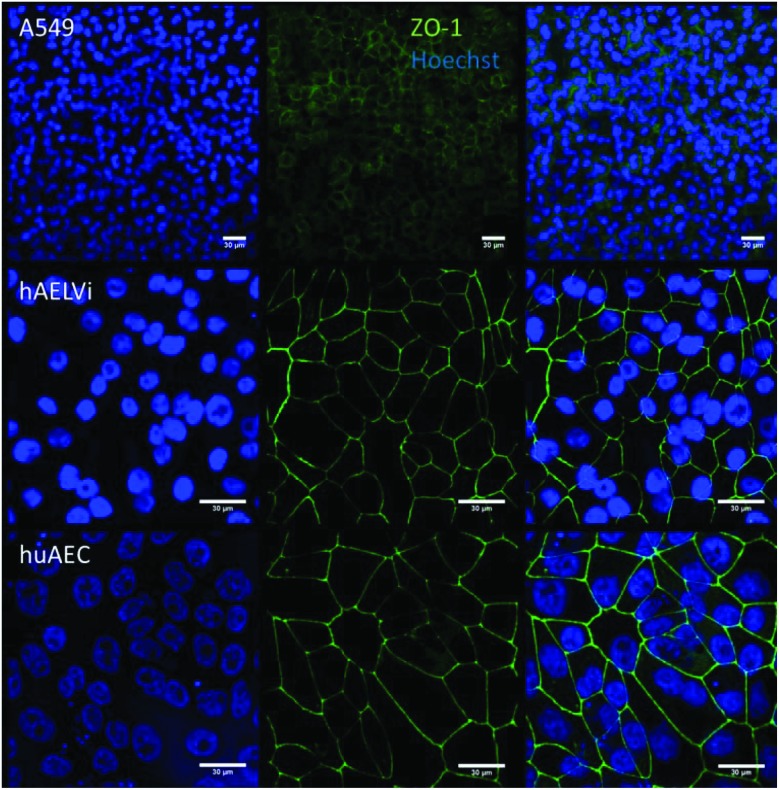
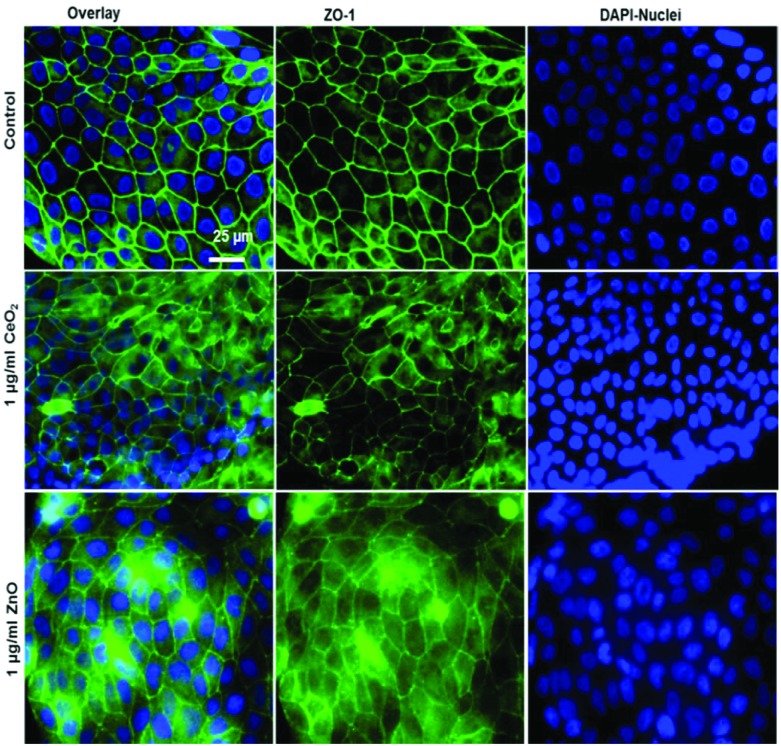
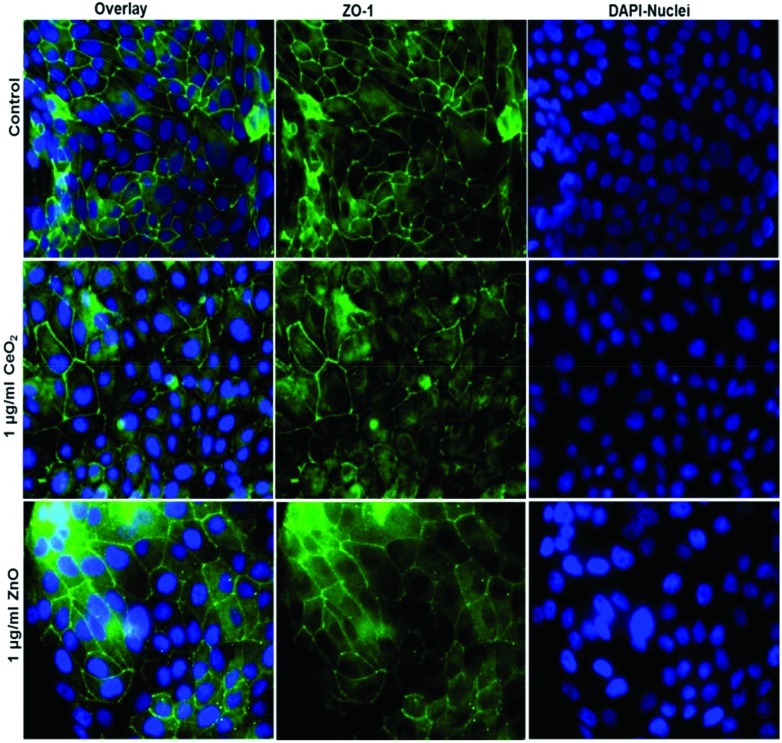
To characterize the barrier function of the different lung epithelia models, we performed TEER measurements and immunohistochemical analysis of the tight junction protein zonula occludens-1 (ZO-1).55 Air–liquid cultivation resembles the in vivo situation closer than standard liquid–liquid cultivation.56 Therefore, daily TEER experiments were performed under both culture conditions. As shown in Fig. 3 A hAELVi cells reached stable TEER values of about 1200–1500 Ω cm2. A549 showed no barrier formation with resistance values between 30–50 Ω cm2 which was expected as this cell line is known to lack functional tight junctions.35–37 TEER data of the novel cell lines revealed a distinct difference between huAEC and hAELVi. hAELVi cells evolved a resistance of about 1500 Ω cm2 which is slightly less as previously described.43 This might be due to the fact that Kuehn and co-workers used corning transwell membranes and SAGM medium instead of huAEC medium and falcon transwell membranes.36,43 In contrast to hAELVi cells, huAEC cells reached TEER values up to 3000–7000 Ω cm2, dependent on the culture conditions. An influence of the culture conditions on the barrier function of hAELVi and A549 cells was not observed. Notably, huAEC cells developed barrier properties two fold higher in submerged culture compared with air–liquid interface, which we assume on account of enormous nutrient resource available in submerged culture.57 Aside from the differences in the resistance values, the time to achieve high TEER values was also different between huAEC and hAELVi. For huAEC cells a strong barrier formation was detected at about day six to day eight, whereas hAELVi cell starts do display a tight barrier at about day 12. This is in consistent with the findings from Kuehn et al.43 where hAELVi cells start to develop TEER values of approximately 1000 Ω cm2 at day 12. While performing manually TEER measurements with an EVOM the position of the electrode is of crucial importance for the resistance value. This is one reason which may explain the large standard deviation for all cell lines achieved in our experiments. A study recently reported a lung on the chip system with integrated electrodes to investigate the resistance of primary humane airway epithelia cells for more than 60 days.58 Using such devices might help to overcome such kind of handling issues. Furthermore with a chip design a direct influence of NPs on the barrier function could be studied over a long period of time. In addition to the 2D models the 3D model EpiAlveolar was analyzed over eight days (Fig. 3B). During this time, a strong increase in TEER data was observed during the first two days. Subsequently, a daily decrease in TEER values was seen. This behavior fits with the short life span of primary cells.43 The achieved standard deviation was clearly smaller compared with the cell lines, which suggests good cell homogeneity in the model. Taken together, the measured TEER values of the new developed models are similar to primary bronchial and primary alveolar cells.36 Thus, hAELVi, huAEC as well as EpiAlveolar resembles the in vivo situation vastly better than the common used A549 cell line regarding a functional cell barrier as well as a potential in vitro model to investigate particle translocation. hAELVi cells are known to express the tight junction protein ZO-1.43,59 Due to the minimal amount of data about hAELVi, we decided to characterize them again in terms of growth and cell–cell-connections. Immunohistochemical staining of the tight junction protein ZO-1 was performed after 14 days. For huAEC cells there were no data reported so far about the barrier formation (TEER and tight junctions). To close this gap we analyzed huAEC cells concerning their barrier properties. As indicated on the TEER values we expected a ZO-1 expression in this cell line as well. hAELVi and huAEC cells developed a complete tight junction network (see Fig. 4). As already shown in the TEER data above, our immunofluorescence staining of ZO-1 confirmed the data of Kuehn et al.43 ZO-1 staining for A549 cells as comparison was negative as diffused signal can be viewed in Fig. 4 and verified the absence of a barrier function in this cell line as it was found in the resistance measurement which is in agreement with the literature.35–37 Unfortunately, direct ZO-1 staining of EpiAlveolar was not successful due to strong backscattering from the membrane. However the exact mechanism of particle translocation is still not fully understand. In 2005 a possible mechanism was published by Rothen-Rutishauser and colleagues.33 The authors exposed a co-culture model of A549 cells, macrophages and dendritic to polystyrene particles. The particles were added on top of the cell model without having contact to the dendritic cell layer. Particle localization revealed an uptake in all cell types, even in dendritic cells which have never been in direct contact to the particles. Further investigations showed particle localization in the pseudopods of the A549 cells. This might suggests a particle transfer between A549 and dendritic cells via the pseudopodia as possible translocation mechanism in vitro.33 Nevertheless other mechanisms, for example an influence on the tight junction formation are also conceivable, as has been recently reported for some materials e.g. CeO2 60 or multi-walled carbon nanotubes.48 Despite the increased permeability, a cytotoxic effect has not been observed for these materials.48,60 This suggests that the absence of cytotoxicity is not an indication of an intact barrier function. In respect to particle translocation and a possible altered permeability, we next stained huAEC cells for ZO-1 expression in routine culture exposed to CeO2 and ZnO NPs, under LL and ALI to ensure no damage on the tight junctions during particle exposure. As shown in Fig. 5 and 6, we did not observe any significant change in ZO-1 expression profile irrespective of exposure conditions. This means that the exposure conditions we used did not lead to an alteration of the tight junction barrier.
Fig. 3. Barrier function assessment of different lung epithelia cells via TEER measurement. (A) Evolution of TEER has been measured under liquid–liquid (LL) and air liquid (ALI) conditions in different cell models; n = 3. (B) Barrier function in the 3D model EpiAlveolar, n = 1 with 3 technical replicates. Data are shown as mean ± SD.
Fig. 4. Representative images of ZO-1 staining of lung cells after 14 days in culture. The first line represents nuclei staining in blue, 2nd line shows the tight junction protein ZO-1 in green and 3rd line displays the overlay of ZO-1 and nuclei.
Fig. 5. Tight junction staining reveals no major effect of NPs exposure at submerged conditions to huAEC cells. The control group shown in upper row contains the overlay image of fluorescently labelled tight junction protein ZO-1 in green and nuclei in blue (DAPI) as first image, 2nd and 3rd images show ZO-1 and nuclei staining respectively. The middle row displays the overlay of ZO-1 in green and nuclei in blue after 24 h exposure to 1 μg ml–1 CeO2 NPs as first image, 2nd and 3rd images show ZO-1 and nuclei staining respectively. The lower row shows the overlay of ZO-1 in green and nuclei in blue after 24 h exposure to 1 μg ml–1 ZnO NPs as first image, 2nd and 3rd images show ZO-1 and nuclei staining respectively.
Fig. 6. Tight junction staining reveals no major effect of NPs exposure at liquid–liquid-interface (ALI) to huAEC cells The control group shown in upper row contains the overlay image of fluorescently labelled tight junction protein ZO-1 in green and nuclei in blue (DAPI) as first image, 2nd and 3rd images show ZO-1 and nuclei staining respectively. The middle row displays the overlay of ZO-1 in green and nuclei in blue after 24 h exposure to 1 μg ml–1 CeO2 NPs as first image, 2nd and 3rd images show ZO-1 and nuclei staining respectively. The lower row shows the overlay of ZO-1 in green and nuclei in blue after 24 h exposure to 1 μg ml–1 ZnO NPs as first image, 2nd and 3rd images show ZO-1 and nuclei staining respectively.
This allows for future investigations to deepen the understanding of the exact mechanism of particle translocation. Taken together, all new developed cell models we investigated showed a distinct barrier formation. Therefore, all examined models exhibit the potential to examine NP translocation in vitro more realistically than current models.
Particle toxicity and metabolic activity analysis
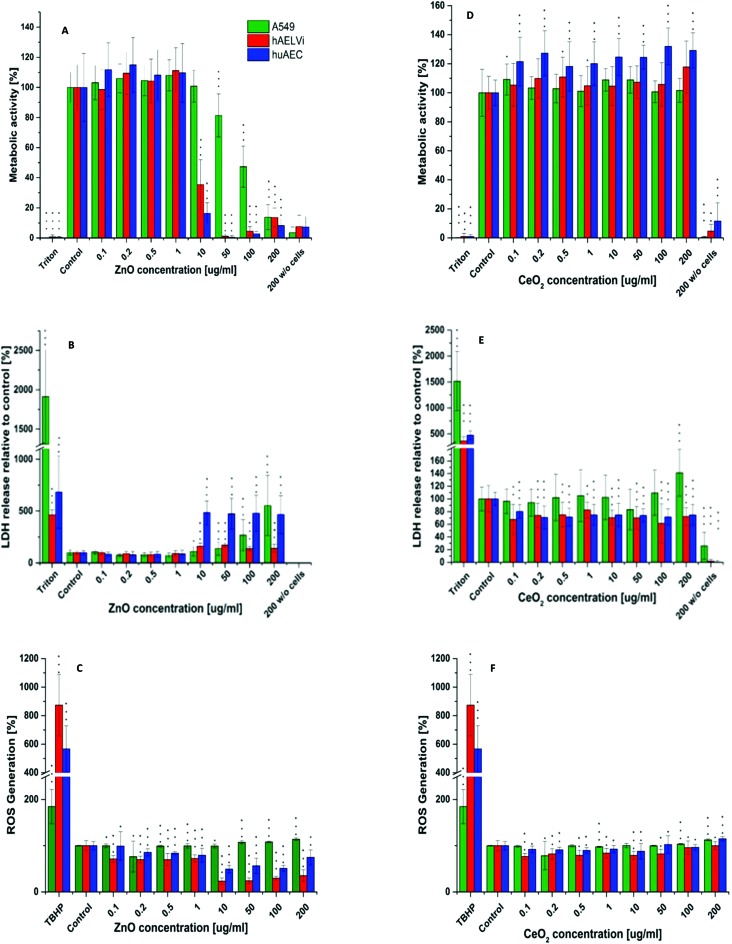
After characterization of the barrier properties, submerged cells were exposed to CeO2 NPs and ZnO NPs. For both particles the influence on metabolic processes (WST-1) as well as the cytotoxicity (LDH) and the generation of ROS was investigated. As depicted in Fig. 7, CeO2 NPs showed no adverse effect, neither in metabolic activity, nor in cytotoxicity for all three cell lines. In addition ROS production after CeO2 NP exposure was either equal or slightly decreased compared to the control in all cell lines used. Ce is known to change its oxidation state.27–29 Therefore, the decrease in ROS production could be due to the antioxidative properties of CeO2 as it is known for other cell types.26,28 Concerning the in vitro toxicity of CeO2 NPs, contradicting studies have been reported. For example, Sauer and colleagues examined the toxicity of CeO2 NPs to rat precision-cut lung slices. They reported no cytotoxicity between 10–100 μg ml–1. 1000 μg ml–1 was needed to reach a cytotoxic effect. Nevertheless, inflammation already occurred at 100 μg ml–1.61 Similar results were reported from another study where different CeO2 NPs were tested on several different cell lines in submerse conditions. In a concentration range of 0.1–10 μg cm–2 neither a cytotoxic effect nor cell death was seen but an increase in oxidative stress occurred.21 The generation of oxidative stress in BEAS-2B cells after CeO2 NP exposure was also described.19,25 Another study even revealed a protective function against oxidative stress of CeO2 NPs for A549 after 24 hours exposure62 which correlates with our findings (Fig. 7). These points to the fact that particle toxicity is cell type specific which was also reported by others.19,30,63,64 Despite the broad applied concentration range in our study, the absence of an adverse/toxic effect from CeO2 NPs exposure is not unexpected. Furthermore, our data are consistent with the findings of Shi et al. (2012) where the exposure of CeO2 NPs up to 200 μg ml–1 showed no cytotoxicity on epithelia cells.65 Zinc oxide NPs revealed a cytotoxicity at 10 μg ml–1 for all cells. For A549 cells a significant decrease in the metabolic activity was seen at 50 μg ml–1 whereas 10 μg ml–1 was sufficient to significantly decrease the metabolic activity of huAEC and hAELVi cells. Analysis of ROS production after ZnO exposure showed a strong decrease in ROS formation for hAELVi and huAEC cells at 10 μg ml–1. For A549 cells such a decrease in ROS formation could not be observed. This difference might be due to the above mentioned cell line depended toxicity which needs further investigations to make a final conclusion. In contrast to CeO2 NPs, ZnO NPs are known to be toxic to many different cell types as e.g. breast cancer cells65 fibroblasts66 and lung epithelia cells.67 In more sophisticated systems like precision-cut lung slices, ZnO NPs induced strong toxicity based on tissue destruction as early as 10 μg ml–1.61 Therefore, our data fit with the literature and confirms a cytotoxicity of ZnO NP also for the two new cell lines hAELVi and huAEC. However, if ZnO NPs show a toxic effect on cells an increase in ROS production should be expected. This was not the case in our study. We assume that the cytotoxicity and the decrease in metabolic activity at 10 μg ml–1 might lead to a decrease in DCFDA uptake. This could explain the low ROS detection which especially takes place at 10 μg ml–1 which is similar to the detected cytotoxicity level. To sum up, WST-1, LDH and ROS assay showed similar results for all cell lines which indicates that the new developed cell lines have a similar behavior under particle exposure as the frequently used A549 cells without the disadvantages such as a carcinogenic phenotype44,49 or the lack of a proper barrier function.35–37 Therefore, they mimic the in vivo situation closer than previously used cell lines.
Fig. 7. Cytotoxicity, metabolic activity and ROS assay. Metabolic activity, LDH release and ROS generation after ZnO NP exposure (A–C) and CeO2 NP exposure (C–D) in different lung epithelia cells. n = 3. Data are shown as mean ± SD. *p < 0.05, **p < 0.01 and ***p > 0.001 is compared to the respective control group.
Model of choice
Here we reported several different lung epithelia models and showed their ability to form functional tight junction networks which is a prerequisite for a realistic in vitro model particularly when particle translocation is one of the challenged tasks. So, we were able to show that all new models tested here exhibit potential as pulmonary in vitro model to study NP cell interactions. After inhalation, most of the NPs deposit in the alveolar region.17,68 Here they come in contact with pneumocytes type I, pneumocytes type II and alveolar macrophages; where type I cells cover about 95% of the alveolar surface.69,70 Consequently, type I pneumocytes are the cell type which comes into the majority of contact with NPs after inhalation. Taking this into account, hAELVi are supposed to be the model of choice aside from primary cells as currently, they are the only model representing the type I human pneumocytes.43 The bronchial epithelium is covered with a mucus layer. This respiratory mucus can promote to an agglomeration of NPs.24 Since inhaled NPs follow the whole airway down to the alveolar region there is the possibility that some particles can deposit in the bronchial region. Thus, the huAEC cell line also represents a relevant model to study the toxicity of NPs. Moreover, huAEC cells reflect the human airway epithelium which enables microparticle studies with this cell line as well. Due to the fact that the culture conditions for huAEC and hAELVi cells are identical, further NP studies will include a co-culture model covering both cell types as well as the exposure of airborne NPs at the air–liquid interface with the aim to resemble the in vivo situation even closer.56 In addition, the combination with macrophages will further increase the complexity of these models to allow more accurate in vitro particle translocation studies. To ensure a better representation of the in vivo situation different 3D models have been developed to study NP cell interaction and toxicological behavior. For instance, for the bronchial 3D model MucilAir™ a higher toxicity of CeO2 NPs was reported compared to the bronchial cell line BEAS-2B in terms of oxidative stress and DNA damage.23 The group of Brandenburger and colleagues used a co-culture model consisting of A549 cells, human blood monocyte derived macrophages and dendritic cells to investigate the effects of gold NPs.15 Another group exposed A549 cells, BEAS-2B cells and the MucilAir™ model to CeO2 NPs. They also reported a lower toxicity for the 3D model compared to a cellular monolayer.24 In 2013, a tetraculture composed of A549, THP-1, HMC-1 and EA·hy 926 cells was developed to study the particle uptake of 50 nm SiO2 NPs. The authors found that a particle uptake by macrophages34 which is in contrast to the findings from another group where 1 μm polystyrene latex particles were found in A549 cells, macrophages and dendritic cells.33 To the best of our knowledge EpiAlveolar is the only commercial 3D human alveolar model so far that includes different cell types45 which is not based on A549 cells (see above). Therefore we performed preliminary experiments to determine the barrier function with this newly developed model. Investigations regarding particle uptake and translocation will be tasks in the future. Our results showed that EpiAlveolar evolves a transepithelial resistances corresponding to primary alveolar cells in vivo.
Conclusions
Here we used the recently developed cell line hAELVi as an alveolar type I model43 to investigate the effect of CeO2 NPs and ZnO NPs. A549 cells were also exposed as they represent a human type II pneumocyte cell model. In addition, we examined the effect of these two nanomaterials on the novel developed airway epithelia cell line huAEC44 and characterized them for the first time regarding their barrier function and applicability as in vitro model for NP toxicity investigations. Cultivation of lung epithelia cells at the air–liquid interface resembles the in vivo situation closer as submerge conditions.56 Therefore, the barrier function was investigated under both conditions. Our data showed that both new cell lines evolve a proper tight junction network independent of if they are cultured under standard submerge conditions or at the air liquid interface. Submerged exposure to CeO2 NPs and ZnO NPs revealed a strong toxicity for ZnO at 10 μg ml–1 for huAEC and hAELVi cells where A549 were only significantly affected at 50 μg ml–1. CeO2 NPs showed no toxicity in any of cell lines used. These results indicate that both new cell lines respond similarly to NP exposure as the frequently used A549 cell line. The tight junctions were not affected by the NPs. As hAELVi and huAEC cells developed tight junctions under submerge and air–liquid culture conditions they can also be used to examine the effect of airborne NPs. Taken together, these two new cell lines behave similar like the A549 cell line which is the most frequently used cell line in terms of pulmonary toxicity testing of NPs. Moreover, they can be cultivated at the air–liquid interface without losing their barrier function which makes them interesting for the future and might be helpful for various issues such as particle translocation of airborne nanomaterials or the development of respirable drugs.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We thank Max Planck institute for solid state research, Stuttgart for transmission electron microscopy images.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c9tx00179d
References
- Ouyang Z., Mainali M. K., Sinha N., Strack G., Altundal Y., Hao Y., Winningham T. A., Sajo E., Celli J., Ngwa W. Phys. Med. 2016;32:631–635. doi: 10.1016/j.ejmp.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgharian B., Price O. T., Oldham M., Chen L. C., Saunders E. L., Gordon T., Mikheev V. B., Minard K. R., Teeguarden J. G. Inhalation Toxicol. 2014;26:829–842. doi: 10.3109/08958378.2014.935535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. S., Wilson W. E., Grant L. D. Inhalation Toxicol. 2005;17:355–385. doi: 10.1080/08958370590929475. [DOI] [PubMed] [Google Scholar]
- Kreyling W. G., Andre S., Collier C. G., Ferron G. A., Metivier H., Schumann G. J. Aerosol Sci. 1991;22:509–535. [Google Scholar]
- Demokritou P., Gass S., Pyrgiotakis G., Cohen J. M., Goldsmith W., McKinney W., Frazer D., Ma J., Schwegler-Berry D., Brain J., Castranova V. Nanotoxicology. 2013;7:1338–1350. doi: 10.3109/17435390.2012.739665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto Y., Izumi H., Yoshiura Y., Tomonaga T., Oyabu T., Myojo T., Kawai K., Yatera K., Shimada M., Kubo M., Yamamoto K., Kitajima S., Kuroda E., Kawaguchi K., Sasaki T. J. Nanopart. Res. 2015;17:46. doi: 10.1007/s11051-015-3249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreyling W. G., Hirn S., Moller W., Schleh C., Wenk A., Celik G., Lipka J., Schaffler M., Haberl N., Johnston B. D., Sperling R., Schmid G., Simon U., Parak W. J., Semmler-Behnke M. ACS Nano. 2014;8:222–233. doi: 10.1021/nn403256v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwotzer D., Ernst H., Schaudien D., Kock H., Pohlmann G., Dasenbrock C., Creutzenberg O. Part. Fibre Toxicol. 2017;14:23. doi: 10.1186/s12989-017-0204-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konduru N. V., Murdaugh K. M., Swami A., Jimenez R. J., Donaghey T. C., Demokritou P., Brain J. D., Molina R. M. Nanotoxicology. 2016;10:720–727. doi: 10.3109/17435390.2015.1113322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina R. M., Konduru N. V., Hirano H., Donaghey T. C., Adamo B., Laurenzi B., Pyrgiotakis G., Brain J. D. Inhalation Toxicol. 2016;28:550–560. doi: 10.1080/08958378.2016.1226449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller J., Wohlleben W., Ma-Hock L., Strauss V., Groters S., Kuttler K., Wiench K., Herden C., Oberdorster G., van Ravenzwaay B., Landsiedel R. Arch. Toxicol. 2014;88:2033–2059. doi: 10.1007/s00204-014-1349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeguarden J. G., Mikheev V. B., Minard K. R., Forsythe W. C., Wang W., Sharma G., Karin N., Tilton S. C., Waters K. M., Asgharian B., Price O. R., Pounds J. G., Thrall B. D. Part. Fibre Toxicol. 2014;11:46. doi: 10.1186/s12989-014-0046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. S., Morishita M., Wagner J. G., Fatouraie M., Wooldridge M., Eagle W. E., Barres J., Carlander U., Emond C., Jolliet O. Part. Fibre Toxicol. 2016;13:45. doi: 10.1186/s12989-016-0156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann M., Vennemann A., Sauer U. G., Wiench K., Ma-Hock L., Landsiedel R. J. Nanobiotechnol. 2016;14:16. doi: 10.1186/s12951-016-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberger C., Rothen-Rutishauser B., Muhlfeld C., Schmid O., Ferron G. A., Maier K. L., Gehr P., Lenz A. G. Toxicol. Appl. Pharmacol. 2010;242:56–65. doi: 10.1016/j.taap.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Barraud C., Corbiere C., Pottier I., Estace E., Blanchard K., Logie C., Lagadu S., Keravec V., Pottier D., Dionnet F., Morin J. P., Preterre D., Andre V., Monteil C., Sichel F. Toxicol. in Vitro. 2017;45:426–433. doi: 10.1016/j.tiv.2017.04.025. [DOI] [PubMed] [Google Scholar]
- Oberdorster G., Oberdorster E., Oberdorster J. Environ. Health Perspect. 2005;113:823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina R. M., Konduru N. V., Jimenez R. J., Pyrgiotakis G., Demokritou P., Wohlleben W., Brain J. D. Environ. Sci.: Nano. 2014;1:561–573. [Google Scholar]
- Park E. J., Choi J., Park Y. K., Park K. Toxicology. 2008;245:90–100. doi: 10.1016/j.tox.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Fytianos K., Chortarea S., Rodriguez-Lorenzo L., Blank F., von Garnier C., Petri-Fink A., Rothen-Rutishauser B. ACS Nano. 2017;11:375–383. doi: 10.1021/acsnano.6b06061. [DOI] [PubMed] [Google Scholar]
- Kroll A., Dierker C., Rommel C., Hahn D., Wohlleben W., Schulze-Isfort C., Gobbert C., Voetz M., Hardinghaus F., Schnekenburger J. Part. Fibre Toxicol. 2011;8:9. doi: 10.1186/1743-8977-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loret T., Peyret E., Dubreuil M., Aguerre-Chariol O., Bressot C., le Bihan O., Amodeo T., Trouiller B., Braun A., Egles C., Lacroix G. Part. Fibre Toxicol. 2016;13:58. doi: 10.1186/s12989-016-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooter I. M., Gröllers-Mulderij M., Steenhof M., Duistermaat E., van Acker F. A. A., Staal Y. C. M., Tromp P. C., Schoen E., Kuper C. F., van Someren E. Appl. In Vitro Toxicol. 2016;2:56–66. [Google Scholar]
- Kuper C. F., Grollers-Mulderij M., Maarschalkerweerd T., Meulendijks N. M. M., Reus A., van Acker F., Zondervan-van den Seuken E. K., Wouters M. E. L., Bijlsma S., Kooter I. M. Toxicol. in Vitro. 2015;29:389–397. doi: 10.1016/j.tiv.2014.10.017. [DOI] [PubMed] [Google Scholar]
- Eom H. J., Choi J. Toxicol. Lett. 2009;187:77–83. doi: 10.1016/j.toxlet.2009.01.028. [DOI] [PubMed] [Google Scholar]
- Singh R., Singh S. Colloids Surf., B. 2019;175:625–635. doi: 10.1016/j.colsurfb.2018.12.042. [DOI] [PubMed] [Google Scholar]
- Singh S. Biointerphases. 2016;11:4. doi: 10.1116/1.4966535. [DOI] [PubMed] [Google Scholar]
- Singh R., Karakoti A. S., Self W., Seal S., Singh S. Langmuir. 2016;32:12202–12211. doi: 10.1021/acs.langmuir.6b03022. [DOI] [PubMed] [Google Scholar]
- Pirmohamed T., Dowding J. M., Singh S., Wasserman B., Heckert E., Karakoti A. S., King J. E. S., Seal S., Self W. T. Chem. Commun. 2010;46:2736–2738. doi: 10.1039/b922024k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam V., Revathidevi S., Shanmuganayagam T., Muthulakshmi L., Rajaram R. RSC Adv. 2016;6:20598–20608. [Google Scholar]
- Jing X. F., Park J. H., Peters T. M., Thorne P. S. Toxicol. in Vitro. 2015;29:502–511. doi: 10.1016/j.tiv.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz A. G., Karg E., Brendel E., Hinze-Heyn H., Maier K. L., Eickelberg O., Stoeger T., Schmid O. BioMed Res. Int. 2013;2013:652632. doi: 10.1155/2013/652632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothen-Rutishauser B. M., Kiama S. G., Gehr P. Am. J. Respir. Cell Mol. Biol. 2005;32:281–289. doi: 10.1165/rcmb.2004-0187OC. [DOI] [PubMed] [Google Scholar]
- Klein S. G., Serchi T., Hoffmann L., Blomeke B., Gutleb A. C. Part. Fibre Toxicol. 2013;10:31. doi: 10.1186/1743-8977-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George I., Vranic S., Boland S., Courtois A., Baeza-Squiban A. Toxicol. in Vitro. 2015;29:51–58. doi: 10.1016/j.tiv.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Srinivasan B., Kolli A. R., Esch M. B., Abaci H. E., Shuler M. L., Hickman J. J. JALA. 2015;20:107–126. doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton H. L., Wan H., Cannell M. B., Gruenert D. C., Thompson P. J., Garrod D. R., Stewart G. A., Robinson C. Clin. Exp. Allergy. 1998;28:1273–1285. doi: 10.1046/j.1365-2222.1998.00354.x. [DOI] [PubMed] [Google Scholar]
- Konduru N. V., Murdaugh K. M., Sotiriou G. A., Donaghey T. C., Demokritou P., Brain J. D., Molina R. M. Part. Fibre Toxicol. 2014;11:44. doi: 10.1186/s12989-014-0044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleh C., Holzwarth U., Hirn S., Wenk A., Simonelli F., Schaffler M., Moller W., Gibson N., Kreyling W. G. J. Aerosol Med. Pulm. Drug Delivery. 2013;26:24–30. doi: 10.1089/jamp.2011.0951. [DOI] [PubMed] [Google Scholar]
- Takenaka S., Karg E., Kreyling W. G., Lentner B., Moller W., Behnke-Semmler M., Jennen L., Walch A., Michalke B., Schramel P., Heyder J., Schulz H. Inhalation Toxicol. 2006;18:733–740. doi: 10.1080/08958370600748281. [DOI] [PubMed] [Google Scholar]
- Konduru N. V., Molina R. M., Swami A., Damiani F., Pyrgiotakis G., Lin P., Andreozzi P., Donaghey T. C., Demokritou P., Krol S., Kreyling W., Brain J. D. Part. Fibre Toxicol. 2017;14:42. doi: 10.1186/s12989-017-0223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. S., Ashitate Y., Lee J. H., Kim S. H., Matsui A., Insin N., Bawendi M. G., Semmler-Behnke M., Frangioni J. V., Tsuda A. Nat. Biotechnol. 2010;28:1300–1303. doi: 10.1038/nbt.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn A., Kletting S., Carvalho-Wodarz C. D., Repnik U., Griffiths G., Fischer U., Meese E., Huwer H., Wirth D., May T., Schneider-Daum N., Lehr C. M. Altex-Alternatives to Animal Experimentation. 2016;33:251–260. doi: 10.14573/altex.1511131. [DOI] [PubMed] [Google Scholar]
- Lipps C., Klein F., Wahlicht T., Seiffert V., Butueva M., Zauers J., Truschel T., Luckner M., Koster M., MacLeod R., Pezoldt J., Huhn J., Yuan Q. G., Muller P. P., Kempf H., Zweigerdt R., Dittrich-Breiholz O., Pufe T., Beckmann R., Drescher W., Riancho J., Sanudo C., Korff T., Opalka B., Rebmann V., Gothert J. R., Alves P. M., Ott M., Schucht R., Hauser H., Wirth D., May T. Nat. Commun. 2018;9:994. doi: 10.1038/s41467-018-03408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson G., Mankus C., Oldach J., Child M., Spratt M., Kandarova H., Ayehunie S., Hayden P. Toxicol. Lett. 2013;221:S138. [Google Scholar]
- Laux P., Riebeling C., Booth A. M., Brain J. D., Brunner J., Cerrillo C., Creutzenberg O., Estrela-Lopis I., Gebel T., Johanson G., Jungnickel H., Kock H., Tentschert J., Tlili A., Schaffer A., Sips A., Yokel R. A., Luch A. NanoImpact. 2017;6:69–80. doi: 10.1016/j.impact.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson C. R., Ameer S. S., Ludvigsson L., Ali N., Alhamdow A., Messing M. E., Pagels J., Gudmundsson A., Bohgard M., Sanfins E., Karedal M., Broberg K., Rissler J. J. Nanopart. Res. 2016;18:86. doi: 10.1007/s11051-016-3389-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derk R., Davidson D. C., Manke A., Stueckle T. A., Rojanasakul Y., Wang L. Sens. Biosensing Res. 2015;3:38–45. doi: 10.1016/j.sbsr.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giard D. J., Aaronson S. A., Todaro G. J., Arnstein P., Kersey J. H., Dosik H., Parks W. P. J. Natl. Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- Powell D. W. Am. J. Physiol. 1981;241:G275–G288. doi: 10.1152/ajpgi.1981.241.4.G275. [DOI] [PubMed] [Google Scholar]
- Hartmann N. B., Jensen K. A., Baun A., Rasmussen K., Rauscher H., Tantra R., Cupi D., Gilliland D., Pianella F., Riego Sintes J. M. J. Toxicol. Environ. Health, Part B. 2015;18:299–326. doi: 10.1080/10937404.2015.1074969. [DOI] [PubMed] [Google Scholar]
- Singh C., Friedrichs S., Ceccone G., Gibson N., Jensen K. A., Levin M., Infante H. G., Carlander D. and Rasmussen K., Cerium Dioxide, NM-211, NM-212, NM-213. Characterisation and test item preparation 2014. [Google Scholar]
- Assanga-Iloki S. B., Gil-Salido A. A., Lewis-Luján L. M., Rosas-Durazo A., Acosta-Silva A. L., Rivera-Castañeda E. G., Rubio-Pino J. L. Int. J. Biotechnol. Mol. Biol. Res. 2013;4:60–70. [Google Scholar]
- Limame R., Wouters A., Pauwels B., Fransen E., Peeters M., Lardon F., De Wever O., Pauwels P. PLoS One. 2012;7:10. doi: 10.1371/journal.pone.0046536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. Y., Xue X. D., You K., Fu J. H. Respir. Res. 2016;17:50. doi: 10.1186/s12931-016-0364-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzulo A. A., Starner T. D., Scheetz T. E., Traver G. L., Tilley A. E., Harvey B. G., Crystal R. G., McCray P. B., Zabner J. Am. J. Physiol.: Lung Cell. Mol. Physiol. 2011;300:L25–L31. doi: 10.1152/ajplung.00256.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikram Singh A., Gharat T., Batuwangala M., Park B. W., Endlein T., Sitti M. J. Biomed. Mater. Res., Part B. 2018;106:1369–1382. doi: 10.1002/jbm.b.33922. [DOI] [PubMed] [Google Scholar]
- Henry O. Y. F., Villenave R., Cronce M. J., Leineweber W. D., Benz M. A., Ingber D. E. Lab Chip. 2017;17:2264–2271. doi: 10.1039/c7lc00155j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletting S., Barthold S., Repnik U., Griffiths G., Loretz B., Schneider-Daum N., de Souza Carvalho-Wodarz C., Lehr C. M. ALTEX. 2018;35:211–222. doi: 10.14573/altex.1607191. [DOI] [PubMed] [Google Scholar]
- Rothen-Rutishauser B., Grass R. N., Blank F., Limbach L. K., Muehlfeld C., Brandenberger C., Raemy D. O., Gehr P., Stark W. J. Environ. Sci. Technol. 2009;43:2634–2640. doi: 10.1021/es8029347. [DOI] [PubMed] [Google Scholar]
- Sauer U. G., Vogel S., Aumann A., Hess A., Kolle S. N., Ma-Hock L., Wohlleben W., Dammann M., Strauss V., Treumann S., Groters S., Wiench K., van Ravenzwaay B., Landsiedel R. Toxicol. Appl. Pharmacol. 2014;276:1–20. doi: 10.1016/j.taap.2013.12.017. [DOI] [PubMed] [Google Scholar]
- De Marzi L., Monaco A., De Lapuente J., Ramos D., Borras M., Di Gioacchino M., Santucci S., Poma A. Int. J. Mol. Sci. 2013;14:3065–3077. doi: 10.3390/ijms14023065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A. M., Limbach L. K., Van Duc L., Krumeich F., Athanassiou E. K., Gerber L. C., Moch H., Stark W. J. Toxicol. Lett. 2010;197:169–174. doi: 10.1016/j.toxlet.2010.05.012. [DOI] [PubMed] [Google Scholar]
- L'Azou B., Jorly J., On D., Sellier E., Moisan F., Fleury-Feith J., Cambar J., Brochard P., Ohayon-Courtes C. Part. Fibre Toxicol. 2008;5:22. doi: 10.1186/1743-8977-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. W., Karlsson H. L., Johansson K., Gogvadze V., Xiao L. S., Li J. T., Burks T., Garcia-Bennett A., Uheida A., Muhammed M., Mathur S., Morgenstern R., Kagan V. E., Fadeel B. ACS Nano. 2012;6:1925–1938. doi: 10.1021/nn2021056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Q., Yin L. H., Tang M., Pu Y. P. Biomed. Environ. Sci. 2011;24:661–669. doi: 10.3967/0895-3988.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Kim I. S., Baek M., Choi S. J. J. Nanosci. Nanotechnol. 2010;10:3453–3458. doi: 10.1166/jnn.2010.2340. [DOI] [PubMed] [Google Scholar]
- Geiser M., Kreyling W. G. Part. Fibre Toxicol. 2010;7:2. doi: 10.1186/1743-8977-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyts K., Napierska D., Dinsdale D., Klein S. G., Serchi T., Hoet P. H. M. Toxicol. in Vitro. 2015;29:234–241. doi: 10.1016/j.tiv.2014.10.024. [DOI] [PubMed] [Google Scholar]
- Crapo J. D., Barry B. E., Gehr P., Bachofen M., Weibel E. R. Am. Rev. Respir. Dis. 1982;126:332–337. doi: 10.1164/arrd.1982.126.2.332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.