Abstract
Increased levels of serum antibody titers against human adenovirus 36 (HAdV-D36) are associated with human obesity and experimental obesity in laboratory animals. While HAdV-D36 has been studied as an infectious agent implicated in obesity for over a decade, the complete genome sequence and its analysis have yet to be reported. A detailed analysis of the genome sequence of HAdV-D36 may be important to understand its role in obesity. Genomic and bioinformatic comparisons with other HAdVs identified differences that suggested unique functions. Global pairwise genome alignment with all sequenced human adenovirus D (HAdV-D) genomes revealed areas of nonconserved sequences in the hexon, E3 CR1β, E3 CR1γ, and fiber genes. Phylogenetic analysis of all HAdV-D36 proteins confirmed that this virus belongs to species Human adenovirus D. This genomic analysis of HAdV-D36 provides an important tool for comprehending the role that this unique adenovirus may play in human obesity. Low amino acid sequence identity in the E3 CR1β and CR1γ genes may suggest distinctive roles for these proteins. Furthermore, the predicted molecular models of the HAdV-D36 fiber protein seem to implicate a unique tissue tropism for HAdV-D36.
Keywords: Human adenovirus, Bioinformatics, Genomics
1. Introduction
As many as 66% of adults in the United States are overweight or obese (Brown et al., 2009) and in the final three decades of the 20th century, the numbers of children with obesity tripled (Troiano and Flegal, 1998). Although the number of obese adults doubled between 1980 and 2004, the dramatic increase in obesity may have stabilized, with no significant change in adult obesity between 2003–2004 and 2005–2006 (Ogden et al., 2006, 2007). Adverse health consequences associated with obesity include cardiovascular disease, diabetes, hepatic dysfunction, osteoarthritis, and reproductive disorders (Brown et al., 2009). Calorie excess is the most logical cause of obesity in America. While mutations in single genes such as those of leptin and leptin receptor are rare causes of obesity (Clement et al., 1998; Montague et al., 1997), an “obesigenic” genetic profile may occur through the accumulation of many different obesity single nucleotide polymorphisms (Bouchard, 2009). Recently, adenoviruses have been added to the list of potential causes of obesity (Atkinson et al., 2005).
Human adenoviruses (HAdVs) were first isolated from civilians and military trainees with respiratory disease in the early 1950s (Hilleman and Werner, 1954; Rowe et al., 1953). They were the first respiratory viruses to be isolated and characterized. Epidemiological studies confirmed that adenoviruses are the primary cause of acute febrile respiratory disease among military recruits (Dingle and Langmuir, 1968; Ginsberg et al., 1955) and have been persistent in the global population. The diverse human pathogens in this family are categorized into 54 types of adenoviruses in the genus Mastadenovirus (Ishiko and Aoki, 2009). The 54 known types are grouped into seven species, based on their immunochemical responses, nucleic acid characteristics, hexon and fiber protein characteristics, biological properties, and phylogenetic analysis (Echavarria, 2009; Ishiko and Aoki, 2009). HAdVs are transmitted by direct and indirect contact, through fomites, and by fecal-oral and respiratory routes, and are associated with acute respiratory disease, epidemic ocular (Walsh et al., 2009), genitourinary, and gastrointestinal infections (Jones et al., 2007). It has recently been suggested that human adenovirus type 36 (HAdV-D36) may also play a role in obesity.
HAdV-D36 was first described in 1980 (Wigand et al., 1980) from a child with diabetes and enteritis and subsequently has been shown to increase visceral adipose tissue in chickens, mice, and monkeys (Dhurandhar et al., 1992, 2000, 2002). In these experiments, the animals were noted to be viremic and HAdV-D36 DNA was detected in adipose tissue at 13 weeks after inoculation (Dhurandhar et al., 2000). Blood transfusion from obese HAdV-D36 infected chickens to non-obese, uninfected chickens produced infection and subsequent development of obesity (Dhurandhar et al., 2001). Emerging evidence supports HAdV-D36 as a cause of human obesity. Obese adults were observed to have a 30% prevalence of HAdV-D36 antibodies compared with only 11% of non-obese adults (Atkinson et al., 2005). In vitro adipogenesis is accelerated by infection of preadipocytes with HAdV-D36 (Vangipuram et al., 2004), possibly related to activity of the E4 gene dUTPase (ORF1) (Rogers et al., 2008). Understanding specific mechanisms involved in the adipogenic effect of HAdV-D36 will be augmented by a detailed description of the HAdV-D36 genome and a comparison of its similarities to and divergences from other human adenoviruses.
2. Materials and methods
2.1. Preparation of HAdV-D36 genome for sequencing
HAdV-D36p stock was acquired from Dr. David Schnurr at the California Department of Public Health. Virus was passed once onto monolayers of A549 cells in 25cm2 flasks to verify cytopathic effect and subsequently amplified in monolayers of A549 cells in 75cm2 flasks for intracellular viral DNA extraction by the method developed by Shinagawa and colleagues with modifications as previously described (Kajon and Erdman, 2007). For quality control of both identity and purity, 1 μg of viral DNA was digested with restriction endonuclease BamHI.
2.2. Amplification and sequencing of the HAdV-D36 genome
To amplify regions of HAdV-D36, we designed primers based on conserved adenovirus sequences in species – HAdV-D from GenBank. Amplicons of the predicted size were sequenced on an Applied Biosystems 3130x Genetic analyzer using the Big Dye Terminator kit (Applied Biosystems).
2.3. Bioinformatics
The proteins and genes of HAdV-D36 were compared to homologs in other HAdV-D genomes. Percent identities between proteins of HAdV-D36 and other HAdVs were determined using Fasta3 [EBI] and Blastp software (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi). Sequence alignments was performed using the mVISTA Limited Area Global Alignment of Nucleotides (LAGAN) program (http://genome.lbl.gov/vista/index.shtml) (Brudno et al., 2003) to compare all fully sequenced genomes in species HAdV-D to HAdV-D36.
2.4. Sequence analysis and genome annotation
Sequence data was filtered and assembled using SeqMan (Lasergene 8, Madison, WI). The genome assembly contained 957 high quality reads with an average length of 524 bp. Eight-fold was achieved for both strands of the genome. The average quality score was 52.7. Genome annotation was partially performed manually as well as by ORFinder (http://www.ncbi.nlm.nih.gov/projects/gorf/). An online sequence alignment program, mVISTA LAGAN [VISTA] was used for global pairwise sequence alignment (Brudno et al., 2003). Splice sites were predicted manually.
2.5. Nucleotide sequence accession numbers
The HAdV-D36 genome and annotation have been deposited in GenBank prior to manuscript submission: accession number GQ384080. The following HAdV genomes (GenBank accession numbers) were used for comparative analysis: HAdV-D8 (AB448767), HAdV-D9 (AJ854486), HAdV-D17 (AC_000006), HAdV-D19 (EF121005), HAdV-D22 (FJ404771), HAdV-D26 (EF153474), HAdV-D28 (FJ824826), HAdV-D37 (DQ900900), HAdV-D46 (AY875648), HAdV-D48 (EF153473), HAdV-D49 (DQ393829), HAdV-D53 (FJ169625), and HAdV-D54 (AB333801). The following accession numbers were used for HAdV fiber genes analysis: HAdV-D8 (BAD11146), HAdV-D9, (BAD24678), HAdV-D17 (AAD20325), HAdV-D19, (EF121005), HAdV-D22, (BAG69149), HAdV-D37 (ABK59080) HAdV-D53 (ACI04198), HAdV-A12 (NP_040933), HAdV-B3 (YP_002213796), HAdV-B14 (AAW33140), HAdV-C5 (AP_000226), HAdV-G52 (ABK35058), and HAdV-G52 (ABK35059). The following accession numbers were used for the HAdV-E3 CR1β genes: HAdV-D53 (ACI04194), HAdV-D22 (ACR81620), HAdV-D37 (BAH19189), HAdV-D46 (AAX70938), HAdV-D9 (AAL01123), HAdV-D19 (BAH18937), HAdV-D8 (BAC78833), and HAdV-D28 (ACQ91166).
2.6. Phylogenetic analysis of HAdV-D36
The protein sequences were aligned with the program ClustalW2, available on the homepage of the European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/clustalw2/). Nucleotide alignment of the whole genomes of HAdV-D was made with the program MAFFT (Multiple Alignment using Fast Fourier Transform, http://www.ebi.ac.uk/Tools/mafft/). Phylogenetic calculations were performed using the programs of the PHYLIP package. The calculations of the protein data were made by the distance matrix analysis programs named Protdist (by applying the categories model) and Fitch (by global rearrangements option) (Felsenstein, 1989). Whole nucleotide genome analysis was performed with the programs Dnadist (by F84 model) and Fitch (by global rearrangements). To confirm the results of the above calculations, bootstrap datasets (created new datasets from the original alignment) were made with the program Seqboot, analyzed with the above-described programs, and the program Consense was used to calculate the consensus tree (Felsenstein, 1989). The analyses were repeated with maximum likelihood method, too (data not shown), using the program PhyML (Guindon and Gascuel, 2003). This analysis resulted in similar tree topology as the distance matrix method. The phylogenetic trees were visualized using the program MEGA 4 (Tamura et al., 2007). In the cases of some HAdV-D types, we had to identify the E3 CR1 genes by reannotating this part of the genome (HAdV-D22 (CR1γ only), -D26, -D48, and -D49) from the available whole genome sequences (GenBank accession numbers: FJ404771, EF153474, EF153473, and DQ393829).
2.7. Molecular modeling
The molecular modeling of the fiber knob was performed with the SWISS-MODEL server (http://swissmodel.expasy.org/) and visualized using DeepView (http://www.expasy.org/spdbv/) (Guex and Peitsch, 1997).
3. Results
3.1. Physical features of the HAdV-D36 genome
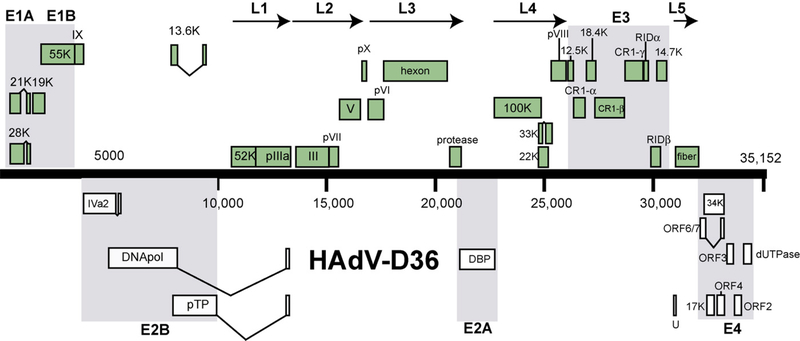
The genome length of HAdV-D36 is 35,152 bp, with a base composition of 22.5% A, 20.3% T, 28.7% G, 28.5% C, with the GC content at 57.2%, consistent with members of species HAdV-D (57.0% mean). The organization of the 39 open reading frames (ORFs) was similar to that of other mastadenoviruses (Fig. 1). The inverted terminal repeat (ITR) sequences for HAdV-D36 were determined to be 86 bp in length. Thus HAdV-D36 has the smallest ITR amongst all sequenced HAdVs.
Fig. 1.
Map of predicted open reading frames and their identities in the genome of HAdV-D36.
3.2. Genetic analysis of the HAdV-D36 genome
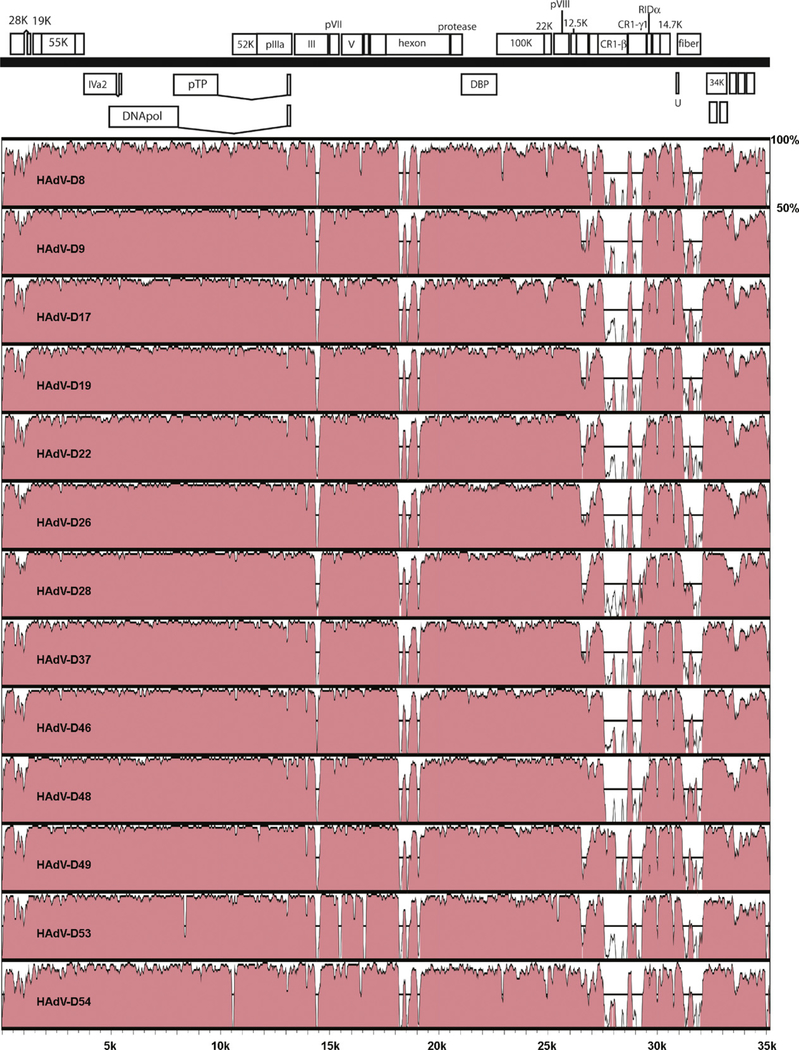
Comparison of HAdV-D36 with HAdV-D8, -D9, -D17, -D19, -D22, -D26, -D28, -D37, -D46, -D48, -D49, -D53, and -D54 using global sequence alignment revealed significant sequence divergence in the hexon, CR1β, CR1γ, and fiber coding sequences (Fig. 2).
Fig. 2.
Pairwise comparison of adenovirus genomes from species HAdV-D to HAdV-D36.
3.2.1. Penton
The Arg-Gly-Asp (RGD) motif binds to specific integrins to facilitate viral internalization into the host cell (Wickham et al., 1993). The RGD motif in HAdV-D36 was located at amino acids 310–312 of the HAdV-D36 penton gene. Recently it was demonstrated that recombination is common in the penton base gene in species HAdV-D (Robinson et al., 2009). By Bootscan and Simplot analysis, we found no evidence of recombination in the HAdV-D36 penton base gene (data not shown). The penton base protein from HAdV-D19 was the nearest relative of the HAdV-D36 penton, with 93.9% amino acid identity (Table 1).
Table 1.
Percent identities of the amino acid sequences of selected HAdV-D36 proteins and their homologs. Penton base, hexon, and fiber were chosen because they are important determinants of tropism. All other proteins in the table are shown here, because they had less than 92% amino acid identity to the corresponding proteins in HAdV-D36.
| HAdV-D8 | HAdV-D9 | HAdV-D19 | HAdV-D22 | HAdV-D28 | HAdV-D37 | HAdV-D49 | HAdV-D53 | |
|---|---|---|---|---|---|---|---|---|
| Penton base | 91.4 | 92.1 | 93.3 | 92.3 | 92.3 | 90.6 | 91.7 | 90.6 |
| Hexon | 91 | 90.5 | 90.1 | 91 | 90.5 | 90.7 | 91.8 | 91 |
| CR1β | 46.5 | 48.6 | 48.3 | 46 | 51 | 45.8 | 85 | 47.1 |
| CR1γ | 69.9 | 64.3 | 87 | 69 | 61 | 70.1 | 66.9 | 69.6 |
| Fiber | 69 | 64.3 | 62.4 | 68.7 | 65 | 62.4 | 66 | 63.5 |
| ORF4 | 90.8 | 91.7 | 89.2 | 91 | 83.5 | 90 | 91.7 | 91.7 |
| ORF3 | 88.9 | 88 | 88 | 86 | 87.2 | 88 | 88 | 88.9 |
| ORF2 | 86.2 | 86.9 | 86.9 | 86 | 93.8 | 86.9 | 87.7 | 86.2 |
| ORF1 | 84 | 92 | 47.2a | 91.2 | 91.2 | 47.2a | 91.2 | 84.8 |
Shorter proteins due to frameshift.
3.2.2. Hexon
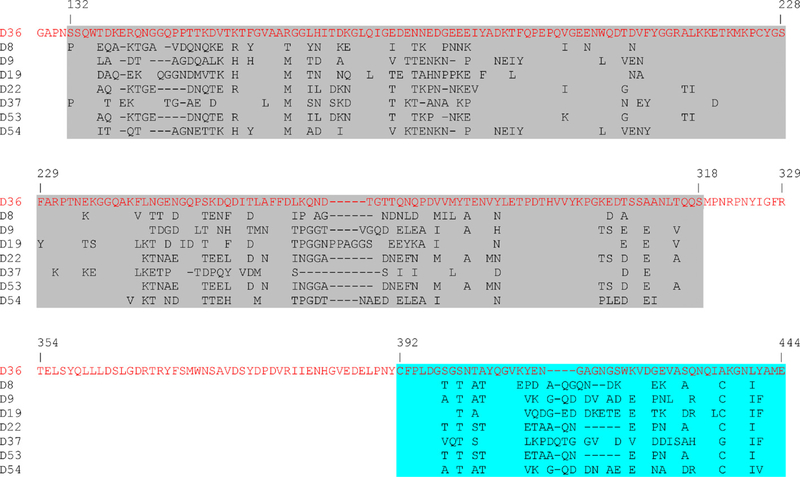
Recombination in the hexon gene of viruses in species HAdV-D is a well-documented phenomenon (Walsh et al., 2009). Recombination analysis did not reveal any evidence of recombination in the hexon gene of HAdV-D36 (data not shown). Overall, the HAdV-D36 hexon protein was on average 90.8% similar to other HAdV-Ds (Fig. 3 and Table 2). However, diversity was seen in the protruding regions of the hexon (loops 1 and 2) as evidenced by the low amino acid identity to other HAdVs in those regions (Table 2). This was expected since the primary serum neutralization epitopes of HAdVs are concentrated in loops 1 and 2 (Rux et al., 2003), both of which are recognized as being hypervariable.
Fig. 3.
Multiple-sequence alignment of hexon loops 1 and 2 from species HAdV-D. Loop 1 is highlighted in grey and loop 2 is highlighted in aqua. Parts of the conserved regions were deleted to save space. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Table 2.
Percent identities of the HAdV-D36 amino acids in hexon loops 1 and 2 to other HAdVs.
| Loop 1 | Loop 2 | |
|---|---|---|
| HAdV-D8 | 66.9 | 65.4 |
| HAdV-D9 | 64.3 | 59.3 |
| HAdV-D19 | 64.7 | 57.4 |
| HAdV-D22 | 63.5 | 68.6 |
| HAdV-D37 | 67.4 | 56.4 |
| HAdV-D53 | 64.6 | 68.6 |
| HAdV-A18 | 43 | 47.1 |
| HAdV-B7 | 51.3 | 51 |
| HAdV-C2 | 48.3 | 49.1 |
| HAdV-E4 | 52.4 | 51.9 |
| HAdV-F40 | 43 | 50 |
| HAdV-G52 | 41 | 41 |
3.2.3. E3
It has been established that E3 proteins gp 19, RIDα, RIDβ, and 14.7K interact with host cell proteins to disrupt the immune system (Elsing and Burgert, 1998; Horwitz, 2004). Three E3 genes encoding membrane proteins called CR1 (conserved region) genes have relatives in primate cytomegaloviruses (subfamily Betaherpesvirinae in the Herpesviridae) (Davison et al., 2003). CR1α functions together with RID protein complex to downregulate a set of TRAIL receptors thereby preventing the action of these important immune mediators (Lichtenstein et al., 2004). The exact functions of the E3 proteins CR1β and CR1γ are not known. The nearest relative to the HAdV-D36 CR1γ coding sequence was the HAdV-D19 CR1γ gene with 87% amino acid identity (Table 1). In contrast, the CR1γ homologs of HAdV-D37 and HAdV-D53 CR1γ proteins were 99 and 100% identical to HAdV-D19 and HAdV-D8, respectively (data not shown). Interestingly, only the HAdV-D49 CR1β coding sequence was similar to the HAdV-D36 CR1β coding sequence, with 85% amino acid identity (Table 1). All other CR1β coding sequences had very low amino acid identity with the HAdV-D36 CR1β protein (Table 1). In contrast, HAdV-D37 and HAdV-D53 CR1β proteins were identical to HAdV-D19 and HAdV-D8, respectively (data not shown). Our analysis of the HAdV-E3 proteins CR1β and CR1γ suggests they are uniquely divergent in HAdV-D36
3.2.4. Fiber
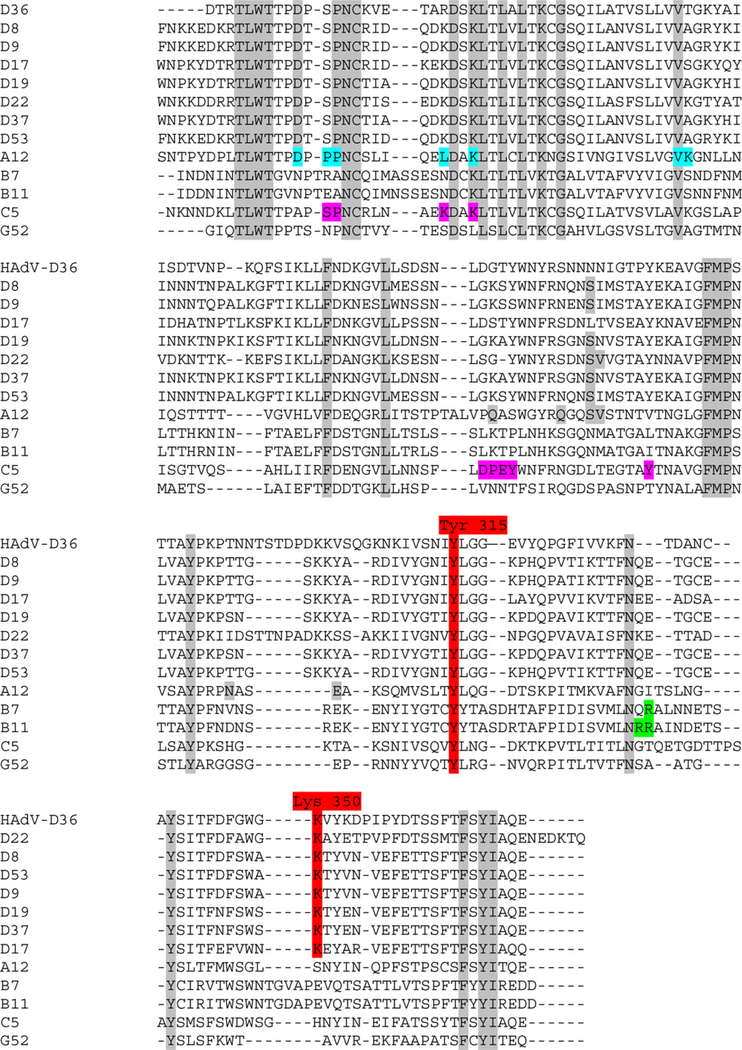
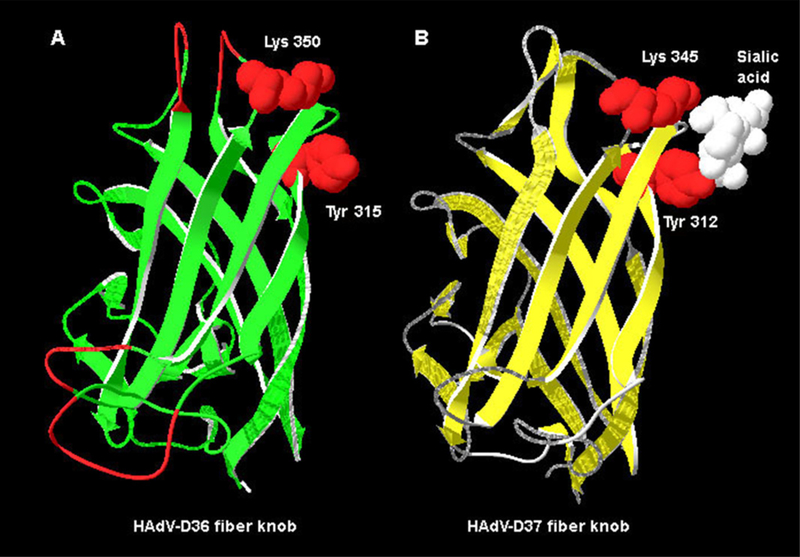
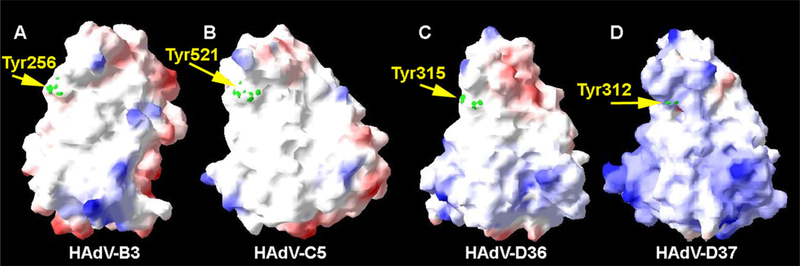
The fiber gene with the highest amino acid identity to HAdV-D36 was that of HAdV-D8 with 69% amino acid identity (Table 1). The HAdV-D36 fiber protein contains Tyr315 and Lys350 (present in all species HAdV-D fiber proteins) which are critical amino acids for binding sialic acid (Figs. 4 and 5A). The HAdV-D36 fiber protein contained 4 of the 13 residues known to contact the coxsackie-adenovirus-receptor (CAR) in CAR-binding HAdVs. The HAdV-D36 fiber does not contain Arg279 or Arg280 (Fig. 4), which are known to interact with CD46, the receptor for several viruses in species HAdV-B (Nemerow et al., 2009). The electrostatic surface potential, generated from a molecular model of the HAdV-D36 fiber knob, suggests that the HAdV-D36 fiber knob has a less positive surface charge in the area near the sialic acid binding site (Fig. 6).
Fig. 4.
Multiple alignment of adenovirus fiber knob sequences from species HAdV-D. Alignment contains adenovirus fiber knob from sequences from species HAdV-D (−8, −9, −17, −19, −22, −37, and −53), HAdV-A, HAdV-B, HAdV-C, and HAdV-G. Tyr315 and Lys350, the vital amino acids that form the sialic acid binding site, are highlighted in red. The conserved residues are highlighted in grey. Amino acids contacting CAR in the HAdV-A12–CAR complex crystal structure are shown on an aqua background. The residues in the HAdV-C5 fiber protein that were found to interact with CAR by mutagenesis (Kirby et al., 1999, 2000) are highlighted in magenta. Residues that interact with CD46 are highlighted in green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Fig. 5.
Molecular model of the HAdV-D36 fiber knob. (A) Molecular model of HAdV-D36 knob with Tyr315 and Lys350 highlighted in red. (B) Structure of the HAdV-D37 fiber knob bound to sialic acid (white). Lys345 and Tyr312, which bind sialic acid are highlighted in red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Fig. 6.
Electrostatic surface potential of HAdV-D36 fiber knob. Side view of fiber knobs from (A) HAdV-B3, (B) HAdV-C5, (C) HAdV-D36, and (D) HAdV-D37. The figure was prepared with DeepView (http://www.expasy.org/spdbv/) (Guex and Peitsch, 1997). The green dots demarcate the location of the conserved Tyr residues which are associated with binding sialic acid. Coloring of the surface potential ranges from +5 kT (blue) to −5 kT (red). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.2.5. E4
Previous studies showed that seven amino acids in domain 2 of the HAdV-D9 ORF1 dUTPase (G40, V41, D65, L89, F91, H93, and F97) are important for binding to a 70-kDa cellular phosphoprotein (Chung et al., 2007). Furthermore, the trimerization (TRI) and PDZ domain-binding motif (PBM) elements have also been shown to be important for tumor transformation (Chung et al., 2007). The dUTPase of HAdV-D36 contains an arginine at position 120 whereas position 120 is lysine in all other HAdVs for which we have sequence for in species HAdV-D (Fig. 7). For the three functional domains, this was the only unique feature of the HAdV-D36 dUTPase that we observed.
Fig. 7.
Multiple-sequence alignment of adenovirus dUTPase (E4-ORF1) proteins from species HAdV-D. Domain 2 is highlighted in grey, TRI is highlighted in green, and the PBM element is highlighted in aqua. Amino acids that are critical for the function of domain 2 (G40, V41, D65, L89, F91, H93, and F97) are in bold and in black boxes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
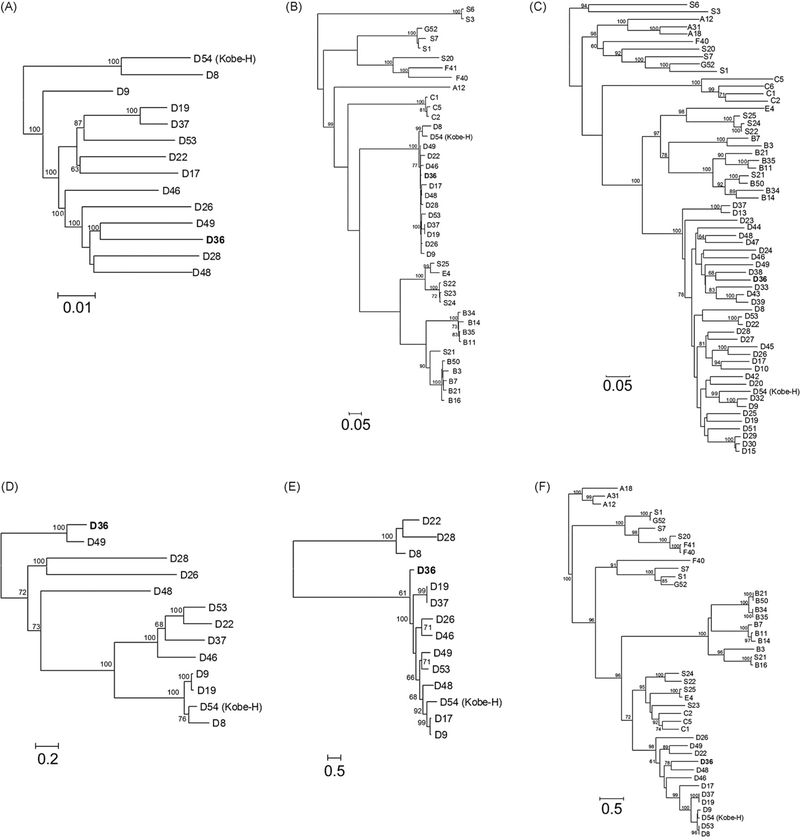
3.3. Phylogenetic analysis
Phylogenetic analysis using the full genomes of all sequenced HAdV-D types as well as the amino acid sequences of selected proteins clearly showed HAdV-D36 to be a member of species HAdV-D (Fig. 8). This was confirmed by the high bootstrap values for all proteins greater than 400 amino acids in length. Full fiber sequences are generally not used for phylogenetic calculations partly because there is a recombination hot spot at the fiber shaft/fiber knob border of species HAdV-D (Darr et al., 2009). Notably, Darr et al. indicated that HAdV-D36 itself was recombinant at this hot spot because the fiber knob sequence of HAdV-D36 was unique whereas its fiber shaft sequence was identical with three other HAdV-D types. We applied the knob region in these calculations (even if its length yielded only limited reliability as mirrored in the bootstrap values).
Fig. 8.
Phylogenetic analysis of HAdV-D36. Analysis of HAdV-D36 is based on the nucleic acid sequence of (A) complete genomes, as well as the predicted amino acid sequences of (B) DNA polymerase, (C) hexon, (D) CR1β, (E) CR1γ, and (F) fiber.
4. Discussion
The presence of anti-HAdV-D36 antibodies is associated with obesity in humans (Atkinson et al., 2005). We set out to sequence this virus to shed light on potentially interesting genes in the HAdV-D36 genome. Complete genome sequencing of HAdV-D36 revealed that the CR1β, CR1γ, and fiber genes were quite divergent from their respective HAdV homologs.
The tropism for HAdV-D36 has not been determined. However, it is known that HAdV-D36 is able to enter preadipocytes, adipocytes, primary human adipose-derived stem/stromal cells, and several animal tissues (Dhurandhar et al., 2000, 2002; Pasarica et al., 2008). Since the key amino acids for binding CD46, a receptor for several adenoviruses in species HAdV-B, are not present in the HAdV-D36 fiber knob, it is unlikely that CD46 is used by HAdV-D36. The HAdV-D36 knob contains 4 of 13 residues that are known to contact CAR, (utilized by many HAdVs as a primary attachment receptor). If HAdV-D36 does attach to CAR, it would be the only virus from species HAdV-D with that capability (Nemerow et al., 2009). Moreover, CAR is not expressed on 3T3-L1 cells (Orlicky et al., 2001), which HAdV-D36 is known to infect (Rogers et al., 2008), further decreasing the probability that HAdV-D36 uses CAR. Among HAdV-D fiber knobs, Tyr315 and Lys350, also found in HAdV-D36 (Fig. 4), are well conserved. This suggests that HAdV-D36 may use sialic acid for attachment to a host cell. We generated a molecular model of the HAdV-D36 fiber knob, based on a crystal structure of HAdV-D37, to predict where the important amino acids for sialic acid binding are located in three-dimensional space. Tyr315 and Lys350 were located in similar locations as the corresponding amino acids of HAdV-D37 (Fig. 5). In comparison to HAdV-D37, the electrostatic surface potential model of the HAdV-D36 fiber knob predicts that it has a less positive and more negative surface charges (Fig. 6). This implicates that HAdV-D36 may use a different receptor than HAdV-D37, a common cause of epidemic keratoconjunctivitis. However, this does not prove or disprove that HAdV-D36 binds sialic acid nor does it rule out unknown receptor(s) for this virus. HAdV-D36 is known to infect adipose, liver, and brain cells in rats (Pasarica et al., 2008), and it will be important in the future to determine its attachment and internalization receptors.
It is noteworthy that the coding sequence for the HAdV-D36 CR1β gene is 85% identical to the homologous coding sequence in HAdV-D49, yet unlike other CR1β coding sequences in species HAdV-D. Since little is known about the function of CR1β, we analyzed it using the Uniprot database to theorize a potential function. Proteins suggested by Uniprot as distant homologs did not have similar functions to one another and had identities equal to or lower than 20% (data not shown). Since CR1γ has low amino acid identity to homologous coding sequences, its roles in adipogenesis should be explored. Transcriptome analysis of adenoviral and host genes in adipocytes infected with HAdV-D36 would help elucidate which genes, human as well as viral, are involved with adipogenesis.
Phylogenetic analyses confirmed the earlier classification of HAdV-D36 into species HAdV-D either by using the full genome sequence, or the amino acid sequence of any proteins in species HAdV-D. On the other hand, it is not possible to conclude a more detailed relationship between types inside species HAdV-D because there are 20 prototype genomes which remain to be sequenced. Therefore, many proteins are not available for comparison. Secondly, adenoviruses in species HAdV-D are closely related to each other in such a way that the mathematical analysis methods cannot resolve their phylogeny with adequate reliability. The latter fact is mirrored in the low bootstrap values when short and highly variable proteins were used. The E3 proteins were so variable that only types within the same species could be applied in reliable alignments. Therefore we decided to calculate a phylogenetic tree based only on proteins from HAdV-D while the other trees reflect the evolutionary distance of all sequenced human, ape, and monkey adenovirus types (SAdV-1, 3, 6, 7, and 20–25; HAdV-1 to 54). The calculation problems may be better solved by completing the full genome sequencing of all known HAdV types. The available sequences (the calculated phylogenetic trees and the similarity plotting or bootstrap analyses) have not suggested any recombination events in the genome of HAdV-D36 (beside the one in the fiber at the shaft/knob border), which is noteworthy considering that recombination occurs frequently within species HAdV-D (Robinson et al., 2009; Walsh et al., 2009).
In this study we analyzed the complete genomic sequence of HAdV-D36. Although the organization of the HAdV-D36 genome is similar to other viruses in species HAdV-D, it contains great diversity in the CR1β, CR1γ, and fiber genes. Phylogenetic analysis of HAdV-D36 proteins confirmed HAdV-D36 as a member of species HAdV-D but did not reveal any further recombination events beside the one identified in the fiber.
Acknowledgements
The work reported herein was performed under United States Air Force Surgeon General-approved Clinical Investigation No. FDG20040024E. This work was approved by the David Grant Institutional Review Board.
BH was supported by Hungarian Research Fund grant K72484. JC was supported in part by U.S. Public Health Service NIH grants EY013124 and P30EY013104, Massachusetts Lions Eye Research Fund, Inc., and an unrestricted grant to the Department of Ophthalmology, Harvard Medical School from Research to Prevent Blindness, Inc.
Footnotes
The views expressed in this work are those of the authors and do not reflect the official policy or position of the Department of the Air Force, Department of the Navy, Department of the Army, the Department of Defense, or the US government.
References
- Atkinson RL, Dhurandhar NV, Allison DB, Bowen RL, Israel BA, Albu JB, Augustus AS, 2005. Human adenovirus-36 is associated with increased body weight and paradoxical reduction of serum lipids. Int. J. Obes. (Lond.) 29 (3), 281–286. [DOI] [PubMed] [Google Scholar]
- Bouchard C, 2009. Childhood obesity: are genetic differences involved? Am. J. Clin. Nutr 89 (5), 1494S–1501S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WV, Fujioka K, Wilson PW, Woodworth KA, 2009. Obesity: why be concerned? Am. J. Med 122 (4 (Suppl. 1)), S4–S11. [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S, 2003. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13 (4), 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SH, Frese KK, Weiss RS, Prasad BV, Javier RT, 2007. A new crucial protein interaction element that targets the adenovirus E4-ORF1 oncoprotein to membrane vesicles. J. Virol 81 (9), 4787–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement K, Vaisse C, Lahlou N, Cabrol S, Pelloux V, Cassuto D, Gourmelen M, Dina C, Chambaz J, Lacorte JM, Basdevant A, Bougneres P, Lebouc Y, Froguel P, Guy-Grand B, 1998. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature 392 (6674), 398–401. [DOI] [PubMed] [Google Scholar]
- Darr S, Madisch I, Hofmayer S, Rehren F, Heim A, 2009. Phylogeny and primary structure analysis of fiber shafts of all human adenovirus types for rational design of adenoviral gene-therapy vectors. J. Gen. Virol 90 (Pt 12), 2849–2854. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Akter P, Cunningham C, Dolan A, Addison C, Dargan DJ, Hassan-Walker AF, Emery VC, Griffiths PD, Wilkinson GW, 2003. Homology between the human cytomegalovirus RL11 gene family and human adenovirus E3 genes. J. Gen. Virol 84 (Pt 3), 657–663. [DOI] [PubMed] [Google Scholar]
- Dhurandhar NV, Israel BA, Kolesar JM, Mayhew G, Cook ME, Atkinson RL, 2001. Transmissibility of adenovirus-induced adiposity in a chicken model. Int. J. Obes. Relat. Metab. Disord 25 (7), 990–996. [DOI] [PubMed] [Google Scholar]
- Dhurandhar NV, Israel BA, Kolesar JM, Mayhew GF, Cook ME, Atkinson RL, 2000. Increased adiposity in animals due to a human virus. Int. J. Obes. Relat. Metab. Disord 24 (8), 989–996. [DOI] [PubMed] [Google Scholar]
- Dhurandhar NV, Kulkarni P, Ajinkya SM, Sherikar A, 1992. Effect of adenovirus infection on adiposity in chicken. Vet. Microbiol 31 (2–3), 101–107. [DOI] [PubMed] [Google Scholar]
- Dhurandhar NV, Whigham LD, Abbott DH, Schultz-Darken NJ, Israel BA, Bradley SM, Kemnitz JW, Allison DB, Atkinson RL, 2002. Human adenovirus Ad-36 promotes weight gain in male rhesus and marmoset monkeys. J. Nutr 132 (10), 3155–3160. [DOI] [PubMed] [Google Scholar]
- Dingle JH, Langmuir AD, 1968. Epidemiology of acute, respiratory disease in military recruits. Am. Rev. Respir. Dis 97 (6 (Suppl.)), 1–65. [DOI] [PubMed] [Google Scholar]
- Echavarria M, 2009. Adenovirus In: Zuckerman AJ, Banatvala JE, Schoub BD, Griffiths PD, Mortimer P (Eds.), Principles and Practice of Clinical Virology, 6th ed. John Wiley and Sons, San Diego, CA, pp. 463–488. [Google Scholar]
- Elsing A, Burgert HG, 1998. The adenovirus E3/10.4K-14.5K proteins downmodulate the apoptosis receptor Fas/Apo-1 by inducing its internalization. Proc. Natl. Acad. Sci. U.S.A 95 (17), 10072–10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J, 1989. Phylogeny Inference Package (Version 3.2). Cladistics 5, 164–166. [Google Scholar]
- Ginsberg HS, Gold E, Jordan WS Jr., Katz S, Badger GF, Dingle JH, 1955. Relation of the new respiratory agents to acute respiratory diseases. Am. J. Public Health Nations Health 45 (7), 915–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC, 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18 (15), 2714–2723. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O, 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol 52 (5), 696–704. [DOI] [PubMed] [Google Scholar]
- Hilleman MR, Werner JH, 1954. Recovery of new agent from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med 85 (1), 183–188. [DOI] [PubMed] [Google Scholar]
- Horwitz MS, 2004. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J. Gene Med 6 (Suppl. 1), S172–S183. [DOI] [PubMed] [Google Scholar]
- Ishiko H, Aoki K, 2009. Spread of epidemic keratoconjunctivitis due to a novel serotype of human adenovirus in Japan. J. Clin. Microbiol 47 (8), 2678–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MS 2nd, Harrach B, Ganac RD, Gozum MM, Dela Cruz WP, Riedel B, Pan C, Delwart EL, Schnurr DP, 2007. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol 81 (11), 5978–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajon AE, Erdman DD, 2007. Assessment of genetic variability among subspecies b1 human adenoviruses for molecular epidemiology studies. Methods Mol. Med 131, 335–355. [DOI] [PubMed] [Google Scholar]
- Kirby I, Davison E, Beavil AJ, Soh CP, Wickham TJ, Roelvink PW, Kovesdi I, Sutton BJ, Santis G, 1999. Mutations in the DG loop of adenovirus type 5 fiber knob protein abolish high-affinity binding to its cellular receptor CAR. J. Virol 73 (11), 9508–9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby I, Davison E, Beavil AJ, Soh CP, Wickham TJ, Roelvink PW, Kovesdi I, Sutton BJ, Santis G, 2000. Identification of contact residues and definition of the CAR-binding site of adenovirus type 5 fiber protein. J. Virol 74 (6), 2804–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein DL, Doronin K, Toth K, Kuppuswamy M, Wold WS, Tollefson AE, 2004. Adenovirus E3–6.7K protein is required in conjunction with the E3-RID protein complex for the internalization and degradation of TRAIL receptor 2. J. Virol 78 (22), 12297–12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague CT, Farooqi IS, Whitehead JP, Soos MA, Rau H, Wareham NJ, Sewter CP, Digby JE, Mohammed SN, Hurst JA, Cheetham CH, Earley AR, Barnett AH, Prins JB, O’Rahilly S, 1997. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature 387 (6636), 903–908. [DOI] [PubMed] [Google Scholar]
- Nemerow GR, Pache L, Reddy V, Stewart PL, 2009. Insights into adenovirus host cell interactions from structural studies. Virology 384 (2), 380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM, 2006. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA 295 (13), 1549–1555. [DOI] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, McDowell MA, Flegal KM, 2007. In: National Center for Health Statistics (Ed.), Obesity Among Adults in the United States—No Change Since 2003–2004. NCHS Data Brief No 1. United States Department of Health and Human Services, Hyattsville, MD. [Google Scholar]
- Orlicky DJ, DeGregori J, Schaack J, 2001. Construction of stable coxsackievirus and adenovirus receptor-expressing 3T3-L1 cells. J. Lipid Res 42 (6), 910–915. [PubMed] [Google Scholar]
- Pasarica M, Mashtalir N, McAllister EJ, Kilroy GE, Koska J, Permana P, de Courten B,Yu M,Ravussin E,Gimble JM,Dhurandhar NV,2008Adipogenic human adenovirus Ad-36 induces commitment, differentiation, and lipid accumulation in human adipose-derived stem cells. Stem Cells 26 (4), 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CM, Rajaiya J, Walsh MP, Seto D, Dyer DW, Jones MS, Chodosh J, 2009. Computational analysis of human adenovirus type 22 provides evidence for recombination between human adenoviruses species D in the penton base gene. J. Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers PM, Fusinski KA, Rathod MA, Loiler SA, Pasarica M, Shaw MK, Kilroy G, Sutton GM, McAllister EJ, Mashtalir N, Gimble JM, Holland TC, Dhurandhar NV, 2008. Human adenovirus Ad-36 induces adipogenesis via its E4 orf-1 gene. Int. J. Obes. (Lond.) 32 (3), 397–406. [DOI] [PubMed] [Google Scholar]
- Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG, 1953. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med 84 (3), 570–573. [DOI] [PubMed] [Google Scholar]
- Rux JJ, Kuser PR, Burnett RM, 2003. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol 77 (17), 9553–9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S, 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol 24 (8), 1596–1599. [DOI] [PubMed] [Google Scholar]
- Troiano RP, Flegal KM, 1998. Overweight children and adolescents: description, epidemiology, and demographics. Pediatrics 101 (3 (Pt 2)), 497–504. [PubMed] [Google Scholar]
- Vangipuram SD, Sheele J, Atkinson RL, Holland TC, Dhurandhar NV, 2004. A human adenovirus enhances preadipocyte differentiation. Obes. Res 12 (5), 770–777. [DOI] [PubMed] [Google Scholar]
- Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D, Jones MS, 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4 (6), e5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham TJ, Mathias P, Cheresh DA, Nemerow GR, 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73 (2), 309–319. [DOI] [PubMed] [Google Scholar]
- Wigand R, Gelderblom H, Wadell G, 1980. New human adenovirus (candidate adenovirus 36), a novel member of subgroup D. Arch. Virol 64 (3), 225–233. [DOI] [PubMed] [Google Scholar]