Abstract
Filamentous bacteriophage (Pf phage) contribute to the virulence of Pseudomonas aeruginosa infections in animal models, but their relevance to human disease is unclear. We sought to interrogate the prevalence and clinical relevance of Pf phage in patients with cystic fibrosis (CF) using sputum samples from two well-characterized patient cohorts. Bacterial genomic analysis in a Danish longitudinal cohort of 34 patients with CF revealed that 26.5% (n = 9) were consistently Pf phage positive. In the second cohort, a prospective cross-sectional cohort of 58 patients with CF at Stanford, sputum qPCR analysis showed that 36.2% (n = 21) of patients were Pf phage positive. In both cohorts, patients positive for Pf phage were older, and in the Stanford CF cohort, patients positive for Pf phage were more likely to have chronic P. aeruginosa infection and had greater declines in pulmonary function during exacerbations than patients negative for Pf phage presence in the sputum. Last, P. aeruginosa strains carrying Pf phage exhibited increased resistance to antipseudomonal antibiotics. Mechanistically, in vitro analysis showed that Pf phage sequesters these same antibiotics, suggesting that this mechanism may thereby contribute to the selection of antibiotic resistance over time. These data provide evidence that Pf phage may contribute to clinical outcomes in P. aeruginosa infection in CF.
INTRODUCTION
Cystic fibrosis (CF) is an autosomal recessive disease caused by mutations in the gene that encodes the cystic fibrosis transmembrane conductance regulator (CFTR), an anion transport protein present in mucosal surfaces (1). In the CF lung, disrupted chloride and bicarbonate transport result in the accumulation of tenacious secretions in the airways (2, 3). This, in turn, sets off a vicious cycle of infection, inflammation, and tissue damage resulting in severe and progressive obstructive pulmonary disease with substantial morbidity and mortality (4, 5). It is now well established that lung disease starts very early in the life of patients with CF, primarily as a consequence of mucus obstruction and bacterial infection (6, 7). Despite substantial therapeutic advancements in the medical management of CF, progressive airway obstruction, measured as a decline in forced expiratory volume in 1 s (FEV1), still predicts mortality (8).
Pseudomonas aeruginosa is a common pathogen recovered from CF airway secretions (9). Infection with P. aeruginosa likely starts as an intermittent infection progressing to a chronic stage with eventual conversion to a biofilm-entrenched state (10). These communities of bacteria encased in extracellular matrix allow P. aeruginosa to effectively adapt to the airway environment and persist despite strong immune response and antibiotic therapy (11, 12). Infection with P. aeruginosa typically begins in childhood. By age 3 years, 95% of children with CF have serologic evidence of intermittent P. aeruginosa infection (13). Antibiotic eradication protocols are now the standard of care when P. aeruginosa is first detected; however, successful eradication is variable and not sustained (14–16). As a result, nearly 60% of patients with CF have chronic P. aeruginosa infection when reaching adulthood (17, 18).
Chronic endobronchial infection with P. aeruginosa in patients with CF is associated with worsening lung function and increased mortality (19–22). Despite prolonged suppressive antibiotic therapy for those patients with CF and chronic P. aeruginosa infection, episodic exacerbations with increased symptomatology, decompensation in lung function, and progression of lung disease are still commonly observed (15, 19, 20, 23).
Filamentous bacteriophage, Pf phage, has previously been described in P. aeruginosa biofilms (24–26), and we recently reported that Pf phage promotes the formation of P. aeruginosa biofilms (27). In particular, we have shown that polymers present in the dense, viscous sputum that is characteristic of CF drive the self-assembly of Pf phage into a highly ordered liquid crystal via depletion attraction, a cohesive force that operates between crowded, like-charged elements in environments where sufficient ionic strength exists to screen their repulsive forces (27, 28). The capsid of Pf phage has a dense negative charge, as do most host and bacterial polymers, such as DNA, mucin, actin, and glycosaminoglycans (29–31). This crystalline architecture promotes the adhesiveness and viscosity of P. aeruginosa biofilms (27). Moreover, antibiotics have difficultly penetrating these lattice-like networks, leading to antibiotic tolerance (27, 28). These effects are highly concentration dependent, with higher Pf phage concentrations promoting increasingly pathogenic P. aeruginosa phenotypes (27).
In light of these effects, we have proposed that Pf phage could contribute to the pathogenesis of P. aeruginosa biofilm infections (28). Unlike lytic phage used in phage therapy, Pf phage are lysogenic and typically do not lyse (kill) their bacterial hosts. Consistent with a pathogenic role, Pf phage has been shown to contribute to the virulence of P. aeruginosa infections in animal models of acute lung infection (32, 33). Further, we reported that Pf phage was abundant in a small cohort of 10 patients with CF with advanced lung disease, being found at concentrations averaging 107 copies of Pf phage/ml of sputum (27). Pf phage is produced by P. aeruginosa isolates from CF sputum (34–36), and the Pf prophage (phage DNA integrated into the P. aeruginosa genome) has been found in P. aeruginosa isolates collected from patients with CF and in other clinical settings (25, 37, 38). However, much remains unknown regarding the prevalence of Pf phage within the CF population and the clinical implications of its presence.
We hypothesized that Pf phage is widespread in the CF patient population and that higher Pf phage concentrations are associated with features of advanced lung disease in CF. To test this hypothesis, we explored the prevalence of Pf prophage in P. aeruginosa isolates in an online dataset and in a previously described cohort of Danish patients with CF. We then performed a prospective cross-sectional cohort study at the Stanford CF Center to evaluate Pf phage concentrations in sputum and clinical outcomes in patients with CF.
RESULTS
Pf phage is present in more than half the strains cataloged within the Pseudomonas Genome Database
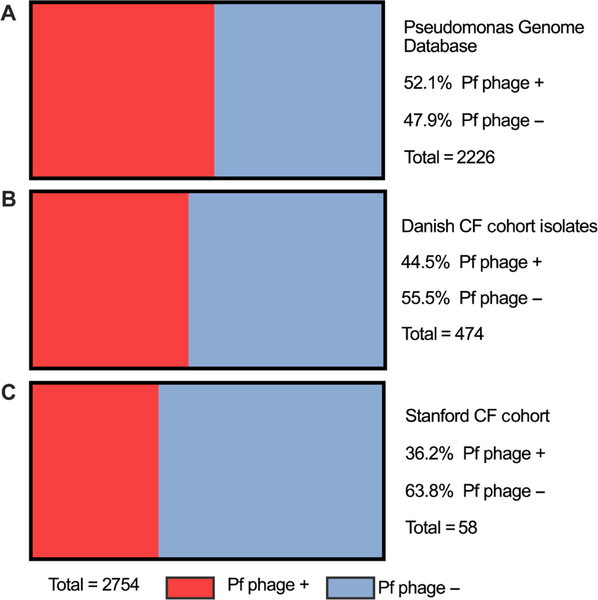
Because Pf is incorporated as a prophage, its carriage can be detected within genomic sequences of P. aeruginosa (37, 39). Exploiting this, we first interrogated the prevalence of Pf phage in a large collection of P. aeruginosa genetic sequences, the Pseudomonas Genome Database (www.pseudomonas.com). This is an internet- and community-based genome project that contains Pseudomonas genome sequences and their annotations. We queried the database for the presence of the Pf phage coaB coat protein gene as a marker for carriage of Pf phage. We found evidence of Pf phage in 1159 of 2226 P. aeruginosa sequences (52.1%) (Fig. 1A). Together, these data indicate that Pf phage is widespread within P. aeruginosa but that not all isolates harbor this bacteriophage.
Fig. 1. Pf phage is prevalent in the Pseudomonas Genome Database, a Danish CF cohort and the Stanford CF cohort.
(A) The prevalence of the detection of the Pf prophage in P. aeruginosa logged in the Pseudomonas Genome Database, an internet- and community-based genome project containing the genome sequences and annotations of both clinical and environmental isolates of P. aeruginosa. (B) The prevalence of the detection of Pf prophage in P. aeruginosa isolates from the Danish CF cohort previously described by Marvig et al. (40). Four hundred seventy-four P. aeruginosa whole-genome sequences were collected from a cohort of 34 Danish patients with CF who were followed longitudinally. There are multiple isolates from each patient represented in this figure. (C) The prevalence of Pf phage detected by quantitative polymerase chain reaction (qPCR) in the sputum of patients also with P. aeruginosa in their sputum from the Stanford CF cohort. Each patient contributed only one sputum sample.
Pf phage is present in a Danish CF cohort and is associated with older patient age
We then evaluated the prevalence of Pf phage within 474 P. aeruginosa whole-genome sequences collected from a cohort of 34 Danish patients with CF previously described (40). These patients were enrolled upon their first respiratory culture that grew P. aeruginosa and followed longitudinally, with one P. aeruginosa isolate collected and sequenced at each visit. We interrogated these whole-genome sequences for the presence of Pf phage coaB coat protein.
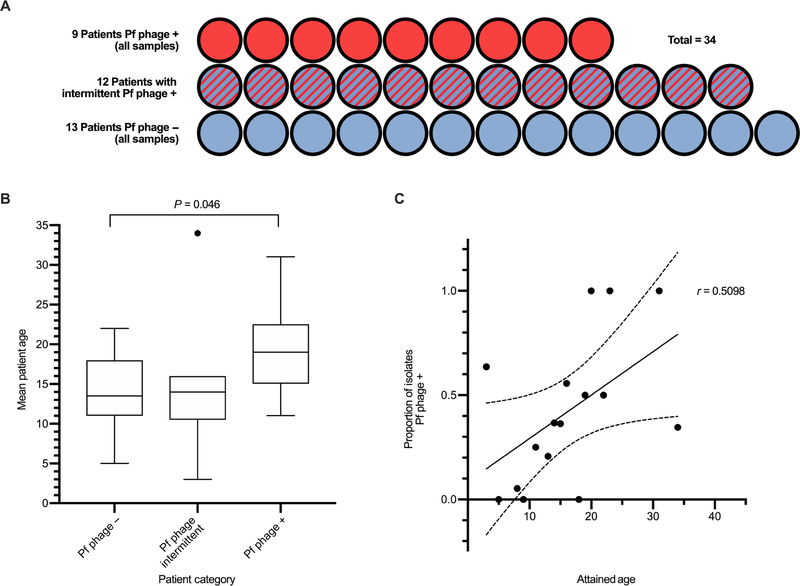
We found that 211 of the total 474 P. aeruginosa isolates (44.5%) were positive for Pf phage (Fig. 1B). Twenty-one of the 34 patients [61.8%; 95% confidence interval (CI), 43.6 to 77.8%] had at least one isolate containing Pf phage at any time during the study. Nine of the 34 patients (26.5%; 95% CI, 12.9 to 44.4%) were consistently Pf phage positive in all samples collected. Twelve (35%) had a mixture of isolates with and without Pf phage over the study period. Thirteen of the 34 patients (38.2%; 95% CI, 22.2 to 56.4%) were consistently Pf phage negative over the course of the study period (Fig. 2A).
Fig. 2. In a Danish CF cohort, the prevalence of Pf phage rises with increasing patient age.
(A) Patients were categorized as having all isolates with the Pf prophage detected (Pf phage +), intermittently having the Pf prophage detected (Pf phage intermittent), or all isolates with no Pf prophage detected (Pf phage −). (B) Mean age of patients with CF in each category. Age was unavailable for three subjects (n = 12, n = 9, n = 9). Boxes represent interquartile range (IQR) with whiskers showing 1.5 × IQR and dots as outliers. Comparison was made by Mann-Whitney test. The outliers on graph were included in analysis. Data are available in table S1. (C) Correlation between subject attained age at the date of the last isolate versus the proportion of total isolates in which Pf prophage was detected(r = 0.5098, P = 0.048). Data for n = 31 patients are included with each dot representing a specific age category and assessed by Pearson correlation.
Patients who were consistently Pf phage positive were significantly older than patients who never had Pf phage detected in their isolates (19.1 years versus 13.9 years; P = 0.046) (Fig. 2B). Moreover, subject age was directly correlated to the proportion of isolates with Pf phage detected for each patient (r = 0.5098, P = 0.048) (Fig. 2C).
These data confirm the finding that Pf phage is widespread within P. aeruginosa sequences and suggest there may be a tendency for P. aeruginosa strains that produce Pf phage to dominate in the sputum of individual patients with CF over time.
Pf phage are abundant in P. aeruginosa lung infections in a Stanford CF cohort
To further interrogate the role of Pf phage in P. aeruginosa infections in CF, we collected sputum samples from a total of 76 patients with CF followed at the Stanford CF center from December 2016 to February 2018. Patient characteristics are described in Table 1. The methodology involved in screening and enrolling these patients is detailed in Materials and Methods.
Table 1. Patient characteristics and microbiologic data in the Stanford CF cohort.
Wilcoxon ranked sum was used for analysis of continuous values, and Pearson’s χ2 was used for categorical values. P values for comparison of the two right columns, P. aeruginosa-positive Pf phage – patients versus P. aeruginosa-positive Pf phage-positive.
|
P. aeruginosa-negative by qPCR |
P. aeruginosa-positive by qPCR | P | ||
|---|---|---|---|---|
| Pf phage-negative | Pf phage-positive | |||
| Subjects, n | 18 | 37 | 21 | — |
| Sex, % male | 72.2% | 51.4% | 52.4% | 0.94 |
| Age, years (±SD) | 23.2 ± 13.5 | 25.9 ± 11.0 | 33.4 ± 10.9 | 0.006 |
| FEV1, % predicted (±SD) | 50.3 ± 24.0 | 54.1 ± 20.1 | 58.7 ± 26.5 | 0.57 |
| BMI, Z score (±SD) | −0.16 ± 1.0 | 0.8 ± 0.85 | 0.09 ± 0.9 | 0.37 |
| CFTR genotype | ||||
| Homozygous F508del, n (%) | 5 (27.8%) | 17 (46.0%) | 11 (54.5%) | 0.52 |
| Heterozygous F508del, n (%) | 8 (44.4%) | 14 (37.8%) | 5 (23.8%) | |
|
P. aeruginosa, copies/ml (range) |
0 | 2.2 × 108 (2.8 × 102–5.1 × 109) | 3.2 × 108 (1.5 × 105–1.9 × 109) | 0.01 |
| Pf phage, copies/ml (range) | 0 | 0 | 4.4 × 109 (1.3 × 106–3.1 × 1010) | — |
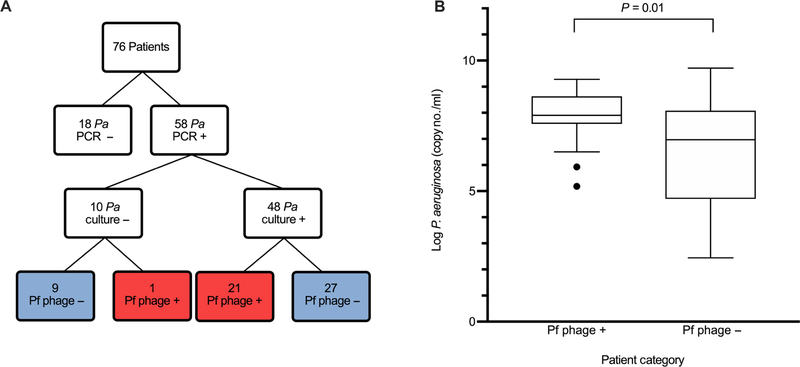
We first assessed the prevalence of P. aeruginosa infection in this cohort. We found that in our 76 patients, 58 had P. aeruginosa detected by qPCR analysis of the sputum. Forty-eight of those grew P. aeruginosa on concurrent respiratory culture. Of the remaining 10 patients that had P. aeruginosa detected by qPCR but did not grow P. aeruginosa on their respiratory culture, 9 had remote history of P. aeruginosa on previous respiratory cultures and 1 had transferred care to Stanford University, and previous records were unavailable for review. Of the samples with no P. aeruginosa detected by qPCR, 10 patients had a history of P. aeruginosa but were not culture positive at present and 8 had never grown P. aeruginosa from their sputum (Table 1 and Fig. 3A).
Fig. 3. P. aeruginosa concentration is higher in patients with Pf phage detected in the sputum within the Stanford CF cohort.
(A) Patients enrolled at the Stanford CF Center with detection of P. aeruginosa by both qPCR and respiratory culture and detection of Pf phage by qPCR. Of the 10 patients with P. aeruginosa detected by PCR but not grown on respiratory culture, 9 had history of P. aeruginosa on previous respiratory culture and 1 patient had no previous records available. Pa, P. aeruginosa. (B) P. aeruginosa concentrations in patients with Pf phage–positive sputum and Pf phage–negative sputum (P = 0.01). Boxes represent IQR with whiskers showing 1.5 × IQR and dots as outliers. Comparison was done by Wilcoxon ranked sum. Outliers on graph were included in analysis.
We then examined the presence or absence of Pf phage in these individuals using our previously published qPCR-based assay (27). The prevalence of Pf phage was 36.2% (21 of 58; 95% CI, 24.0 to 49.9%) in patients with P. aeruginosa infection and 27.6% (21 of 76; 95% CI, 18.0 to 39.1%) in all patients (Table 1). There was no Pf phage detected in any P. aeruginosa–negative samples. Pf phage concentrations averaged 4.4 × 109 copies/ml with a range from 1.3 × 106 to 3.1 × 1010 copies/ml (Fig. 4A). The presence of Pf phage in sputum was not associated with clinical characteristics such as FEV1, body mass index (BMI), or copy number of the CFTR mutation F508del (Table 1 and fig. S1).
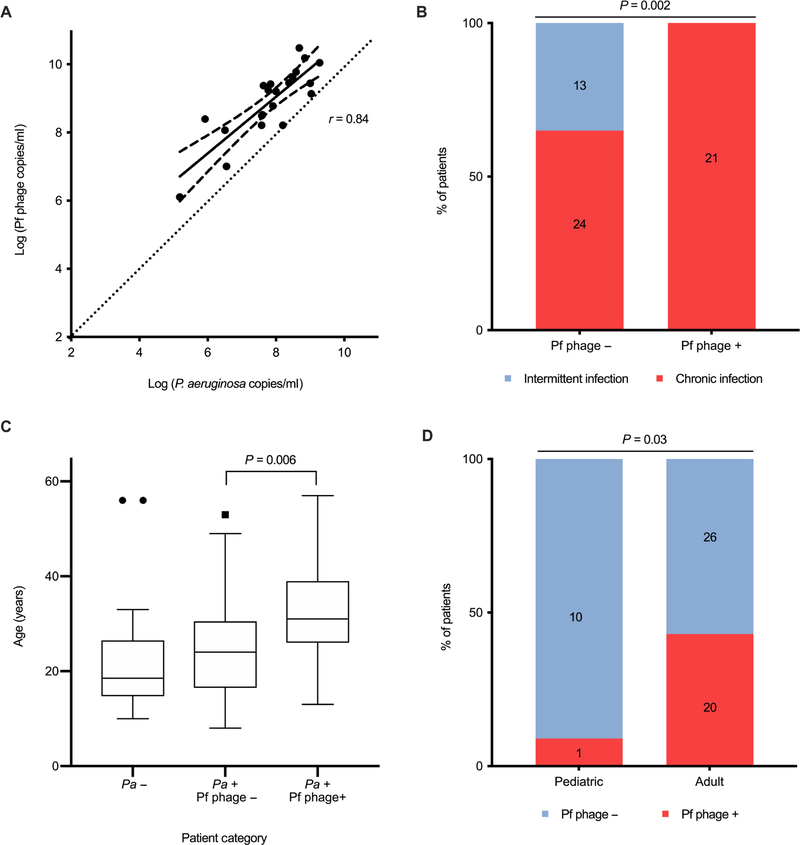
Fig. 4. Pf phage concentration correlates with P. aeruginosa burden, chronic infection and patient age in the Stanford CF cohort.
(A) Correlation between the concentrations of Pf phage and P. aeruginosa in patients positive for Pf phage (r = 0.84, P < 0.001). Dotted line, line for 1:1 correlation for reference. All samples were run in duplicate. The area between dashed lines represents the 95% CI. Correlation calculated by Kendall correlation. (B) Percentage of patients meeting the Leeds criteria for chronic P. aeruginosa infection in Pf phage–positive and Pf phage–negative samples (P = 0.002). Comparison by Pearson’s χ2 was used. (C) Age of patients with P. aeruginosa–negative sputum, P. aeruginosa–positive and Pf phage–negative sputum, and P. aeruginosa–positive and Pf phage–positive sputum. Boxes represent IQR with whiskers showing 1.5 × IQR and dots as outliers. Comparison was done by Wilcoxon ranked sum. (D) Prevalence of Pf phage in adult versus pediatric patients with CF infected with P. aeruginosa (P = 0.03). Comparison by Pearson’s χ2. Outliers on graph were included in analysis.
Pf phage is highly correlated with both the bacterial burden and chronicity of P. aeruginosa lung infection in the Stanford CF cohort
We next asked whether Pf phage correlated with bacterial burden in CF lung infections in the Stanford CF cohort. The sputum burden of P. aeruginosa was greater in patients positive for Pf phage (7.8 ± 1.1 log copies/ml) than patients negative for Pf phage (6.4 ± 2.0 log copies/ml) (P = 0.01) (Table 1 and Fig. 3B). Across individual patients, the amount of Pf phage and the amount of P. aeruginosa were closely correlated; however, the copy numbers of Pf phage DNA consistently averaged 1 to 2 logs higher than P. aeruginosa DNA copies (Fig. 4A). P. aeruginosa carry only a single integrated copy of Pf phage within their genomes (a prophage). The concentration of Pf phage reported in Fig. 4A, fig. S2, and Table 1 reflect the deduction of prophage from the total phage burden (raw Pf phage concentration by qPCR with the raw P. aeruginosa concentration by qPCR subtracted). Together, these data indicate that Pf phage are highly abundant in the sputum of infected patients and that these phage outnumber their bacterial hosts by 10 to 100×.
We next asked whether the presence of Pf phage was correlated with the chronicity of P. aeruginosa lung infections as defined by Leeds criteria, a well-validated and clinically useful set of criteria used to identify patients with CF who have progressed to a chronic infected stage with P. aeruginosa (41). A patient is categorized as having chronic infection if >50% of the respiratory cultures in the past 12 months have growth of P. aeruginosa. Patients positive for Pf phage were more likely to meet Leeds criteria (21 of 21) than patients negative for Pf phage (13 of 37) (P = 0.002) (Fig. 4B).
Together, these data indicate that, among patients with CF, the presence of Pf phage is highly correlated with both the bacterial burden and chronicity of P. aeruginosa lung infection.
Pf phage is more prevalent in older patients in the Stanford CF cohort
We observed that patients positive for Pf phage in the Stanford CF cohort were, on average, older (33.4 ± 10.9 years) than patients negative for Pf phage (25.9 ± 11.0 years) (P = 0.006) (Fig. 4C). Consistent with this, only 1 of 11 pediatric patients (9.1%; 95% CI, 0.2 to 41.3%) were Pf phage positive, whereas 20 of 46 (43.5%; 95% CI, 28.9 to 58.9%) adult patients were Pf positive (P = 0.03) (Fig. 4D). Moreover, in our recently published study, samples from 10 of 10 (100%; 95% CI, 69.2 to 100%) patients with CF who were undergoing lung transplant evaluation were Pf phage positive (27). Consistent with the results shown for the Danish CF cohort, these data confirm in a second independent cohort that the presence of Pf phage is associated with older patient age.
Pf phage is associated with decreased FEV1 in the Stanford CF cohort
We next asked whether the absence or presence of Pf phage was associated with worse pulmonary disease in the Stanford CF cohort. Overall, we did not observe significant differences in FEV1% predicted on the day of sputum collection between patients positive for Pf phage and patients negative for Pf phage infected with P. aeruginosa (P = 0.57) (Table 1 and fig. S1A). However, within patients positive for Pf phage, there was a modest inverse correlation between the concentration of Pf phage present in their sputum and their FEV1 value at the time of sampling (r = − 0.35, P = 0.01) (fig. S2).
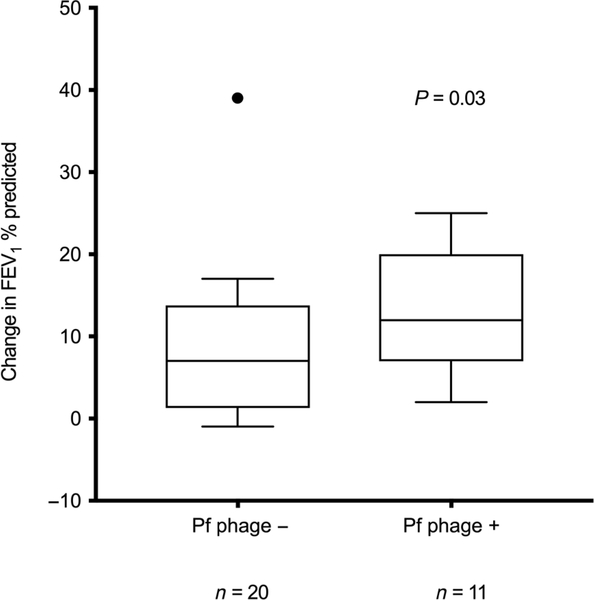
We also asked whether pulmonary exacerbations had a more detrimental effect in FEV1 in the setting of Pf phage presence by comparing the pulmonary function of patients during a pulmonary exacerbation (defined as a combination of a flare in symptomatology, treatment with increased airway clearance, and antibiotic therapy) to the pulmonary function when not in exacerbation. We found that the drop in FEV1 during exacerbation was significantly greater in patients positive for Pf phage (Fig. 5, P = 0.03). Together, these data suggest that Pf phage burden in the airways is associated with a decrease in FEV1 during exacerbations.
Fig. 5. Patients positive for Pf phage have reduced lung function during exacerbations in the Stanford CF cohort.
The difference between FEV1% predicted at time of sampling and at presentation of subsequent exacerbation in patients with Pf phage–positive sputum and in those with Pf phage–negative sputum (P = 0.03). Only patients who were enrolled while at baseline health and who had exacerbation after enrollment were included in this analysis (n = 11 patients positive for Pf phage, n = 20 patients negative for Pf phage). These data include only patients with P. aeruginosa detected in their sputum. Boxes represent IQR with whiskers showing 1.5 × IQR and dots as outliers. Comparisons were made by Mann-Whitney test. Outlier on graph was included in analysis. Data are available in table S2.
Clinical P. aeruginosa isolates from patients positive for Pf phage exhibit increased antibiotic resistance in the Stanford CF cohort
Given that older patients were more likely to be Pf phage positive, we suspected that strains producing Pf phage may have a selective advantage over strains that do not. We therefore examined the phenotypes and antibiotic resistance patterns in clinical isolates from patients positive and negative for Pf phage, as described in Table 2. These data are for the predominant clinical isolate in the same sputum samples used to assess Pf phage status.
Table 2. Phenotype and resistance patterns from respiratory culture from patients in the Stanford CF cohort.
Antibiotic resistance was assessed in clinical P. aeruginosa isolates cultured from the same sputum samples in which Pf phage status was assessed in our cohort. N = 28 Pf phage–negative samples and 20 Pf phage–positive samples. Comparison by Pearson’s χ2 test.
| P. aeruginosa + by qPCR | P | ||
|---|---|---|---|
| Pf phage − | Pf phage + | ||
| Mucoid strain | 58.3% | 85.71% | 0.03 |
| Ciprofloxacin | 12 (33%) | 6 (29%) | 0.71 |
| Tobramycin | 7 (19%) | 7 (33%) | 0.24 |
| Aztreonam | 5 (14%) | 7 (33%) | 0.0499 |
| Amikacin | 8 (22%) | 14 (67%) | 0.0009 |
| Cefepime | 7 (19%) | 8 (38%) | 0.12 |
| Ceftazidime | 7 (19%) | 5 (23%) | 0.70 |
| Meropenem | 4 (11%) | 10 (48%) | 0.002 |
| Piperacillin-tazobactam | 7 (19%) | 5 (24%) | 0.70 |
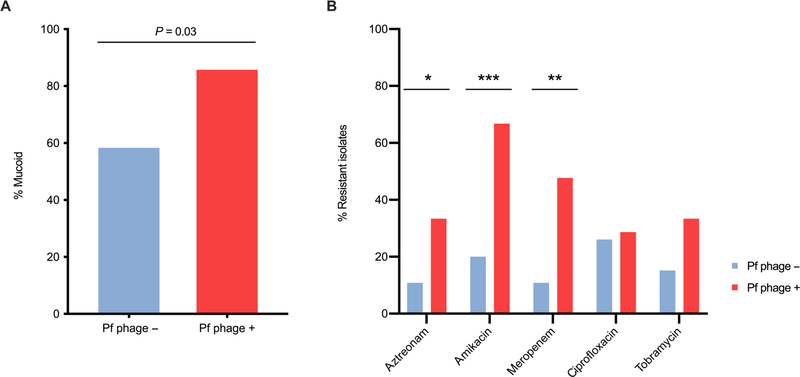
We found that more patients who are Pf phage positive have mucoid strains of P. aeruginosa, a marker of adaptation to the airway environment (58.3% versus 85.7%) (P = 0.03) (Fig. 6A). Further, patients who are Pf phage positive have P. aeruginosa strains that exhibit increased resistance to the antibiotics aztreonam (P = 0.0499), amikacin (P = 0.0009), and meropenem (P = 0.002), as determined by the Kirby Bauer disk diffusion method in our clinical laboratory (Table 2 and Fig. 6B). When comparing mucoid versus nonmucoid isolates, there were no differences in antibiotic resistance patterns (table S3). Because antibiotic therapy is an important component of CF care, these data suggest that an association with heightened antibiotic resistance may contribute to the link between Pf phage status and increased lung disease pathogenicity in CF.
Fig. 6. Clinical isolates of P. aeruginosa from patients with Pf phage–positive sputum exhibit increased antibiotic resistance and are more likely to be mucoid.
(A) Proportion of isolates that display mucoid phenotype in isolates from Pf phage–negative sputum and Pf phage–positive sputum (P = 0.03). Comparison by Pearson χ2 test. (B) The proportion of isolates that were resistant to antibiotics listed. n = 28 Pf phage–negative samples, n = 20 Pf phage–positive samples. Comparison was made by Pearson’s χ2 test. *P = 0.0499, **P = 0.002, ***P = 0.0009.
Pf phage sequesters antimicrobials
We previously reported that Pf phage spontaneously assemble into a liquid crystalline structure in the presence of polymers abundant in CF sputum and that these liquid crystals sequester antibiotics and prevent their diffusion through biofilms, thereby contributing to increased antibiotic tolerance (bacterial survival despite the presence of antibiotics) (27, 28). This may over time lead to selection for fixed antibiotic resistance (the ability to grow at elevated concentrations of antimicrobial drugs due to heritable resistance mechanisms).
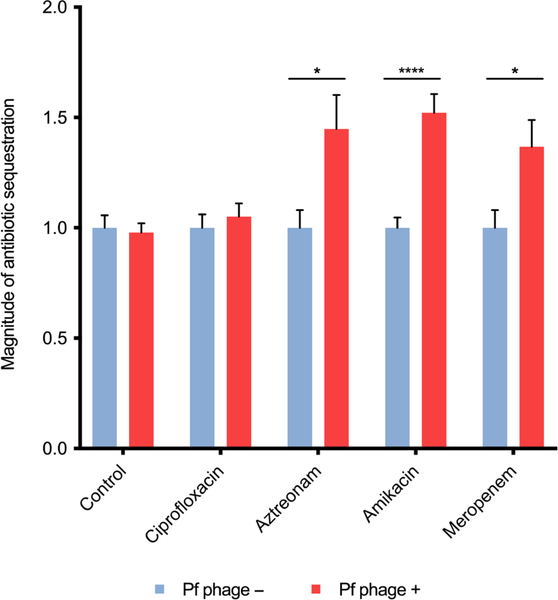
To test specifically whether this crystalline organization prevents diffusion of the antibiotics identified in Table 2, we used our previously described flow chamber system (27). Briefly, DNA, Pf phage, and an antibiotic were combined on one side of a microfilter apparatus, and then, the effluent was collected on the other side of the filter. This effluent was then added to antibiotic susceptible Escherichia coli to functionally assess antibiotic concentrations. Using this system, we observed that aztreonam (P < 0.05), amikacin (P < 0.0001), and meropenem (P < 0.05) were all sequestered by Pf phage–DNA complexes, whereas ciprofloxacin was not (Fig. 7).
Fig. 7. Antibiotics aztreonam, amikacin, and meropenem can be sequestered by Pf phage.
The magnitude of sequestered antibiotic (bacterial proliferation normalized to control conditions) is displayed for each antibiotic when run through dialysis cassette containing either Pf phage and DNA or DNA alone (control). Samples were run in triplicate (N = 2 experiments). Error bars indicate mean ± SEM. Comparisons were made by Student’s t test with an α level of 0.05. ****P < 0.0001, *P < 0.05.
DISCUSSION
Pf bacteriophage are prevalent in 36.2 to 52.1% of P. aeruginosa isolates, including in two independent cohorts of patients with CF, which is similar to that reported by Knezevic et al. (25), who reported that 56% of 241 clinical isolates carried Pf prophage. In the prospective Stanford CF cohort reported here, Pf phage was associated with chronic P. aeruginosa infection, worse lower pulmonary function during exacerbation, antibiotic resistance, and, in both Stanford and Danish CF cohorts, older patient age. Together, these observations suggest that Pf phage might contribute to poor clinical outcomes in patients with CF. Although bacteriophage are known to encode genes for toxins and other factors involved in acute infections (42), these findings suggest that a bacteriophage might also play a role in the pathogenicity of CF in humans.
Our data from both the Stanford and Danish CF cohorts together suggest that either patients with CF acquire Pf phage–producing strains of P. aeruginosa or the Pf phage–negative P. aeruginosa become infected with Pf phage as patients age and their disease progresses. This suggests that Pf carrying P. aeruginosa strains have a selective advantage over those that do not—an observation that may be explained by our observations of increased antibiotic resistance.
In light of the data presented here and our previously published work (27), we hypothesize that Pf phage promote the structure and function of P. aeruginosa biofilms in ways that drive the development of antibiotic resistance and chronic infection (fig. S3). This model is consistent with work by ourselves and other groups identifying Pf phage as a virulence factor and implicating Pf phage in the pathogenesis of P. aeruginosa lung infections in animal models (24, 25, 27, 28, 32, 33). Although lytic phage parasitize their bacterial hosts (43–46), our data suggest that Pf phage, a lysogenic phage, instead contributes to chronic P. aeruginosa infections.
We report that clinical isolates of P. aeruginosa collected from patients positive for Pf phage exhibit increased resistance to amikacin, meropenem, and aztreonam (Table 2 and Fig. 6B). Of note, in these studies, P. aeruginosa was grown in planktonic form (not as a biofilm). Therefore, our findings are indicative of resistance and not tolerance because there would be no biofilm or liquid crystal present in the assay, and the survival is related to genetic mutation conferring resistance. The association between sputum Pf phage and antibiotic resistance may be of high relevance to CF, given that the current standard of care in the United States for chronic lung infection is the use of inhaled topical antibiotics. Amikacin and aztreonam are all antibiotics commonly used to control P. aeruginosa infections in CF. Specifically, inhaled aztreonam, which reaches high concentrations in the airway, is one of the antibiotics that are routinely used as long-term therapy to control chronic P. aeruginosa infection. We propose that the tolerance conferred by the presence of Pf phage has allowed for selection of heritable antibiotic resistance genes as is reflected in the antibiotic susceptibilities measured in our isolates. These data provide a plausible mechanism for how P. aeruginosa strains that produce Pf phage might have a selective advantage and be disproportionately represented in the lungs of older patients with CF.
There was no difference in resistance to ciprofloxacin between Pf phage–positive and Pf phage–negative isolates. This was expected given our previous work showing ciprofloxacin was not sequestered by Pf phage (27), perhaps because of its neutral molecular structure. Whereas we saw that resistance to the aminoglycoside antibiotic amikacin was associated with the presence of Pf phage, there was no association with resistance to tobramycin, which contrasts to our previous report of sequestration of this antibiotic in vitro (27). This difference indicates to us that there may be additional mechanisms driving the development of tobramycin resistance beyond Pf phage–mediated sequestration (47), such as selective pressure from the high incidence of its chronic use among patients with CF. Alternatively, in addition to providing tolerance, bacteriophage can carry antibiotic resistance genes, as demonstrated in CF sputum (48), and this could provide an additional mechanism for influencing antibiotic resistance patterns. If the findings reported here are validated in larger studies and independent cohorts, a diagnostic test for the presence of Pf phage may have future implications for clinical care.
These studies together with the available animal data (32, 33) provide evidence of a role for Pf phage in the pathogenesis of P. aeruginosa chronic lung infections. Although in our cohort the overall FEV1 values, BMI, and other metrics of CF disease are not different in patients positive for versus patients negative for Pf phage, we did find a strong interactive effect with pulmonary exacerbations because the largest impact in FEV1 was among patients positive for Pf phage. Unfortunately, there is currently no ideal animal model for studying chronic pulmonary infection, making these associations difficult to evaluate experimentally.
In addition to promoting chronic infection by promoting antibiotic resistance, bacteriophage has been described as interacting with the mammalian immune system (49). Although this could have different effects on P. aeruginosa infection either favoring chronic infection or clearance, we recently reported that Pf phage suppresses local immunity in association with mouse and human wound infections. Studies are underway to examine the impact of Pf phage on immunity in patients with CF.
This study has several limitations. The methods used to collect and sequence samples in the datasets included in these analyses were highly heterogeneous. The Danish cohort and the Stanford cohort are CF specific, whereas the Pseudomonas Genome Database is not. Moreover, in these CF cohorts, only a single, dominant strain was sampled for each time point, whereas in patients with CF, there are often multiple P. aeruginosa lineages present (50–52). Our prevalence figures therefore may capture only a small part of the ecology of Pf phage–positive and Pf phage–negative P. aeruginosa strains. The cross-sectional design of our Stanford CF cohort does not permit us to definitively establish the role of Pf phage in the progression of CF lung disease. Future, prospective longitudinal studies will be necessary to tease apart the contribution of Pf phage to disease progression in individuals. It will also be critical to evaluate the relationships between Pf phage and specific antibiotic resistance mechanisms, host genetic modifiers, and associations with other microorganisms typical of polymicrobial lung infections in CF. Our ongoing work aims to further investigate Pf phage pathogenicity through a larger prospective longitudinal study with repeated sampling, including the investigation of changes at the individual level in association with episodes of exacerbation.
In conclusion, we report that Pf phage is abundant in P. aeruginosa in multiple settings and that higher concentrations of Pf phage are associated with increased patient age, chronic P. aeruginosa infection, lower FEV1 during pulmonary exacerbations, and increased antibiotic resistance profiles in a cohort of patients with CF. Together with the available in vitro and animal model data, these findings implicate Pf phage in the pathogenesis of CF lung disease and specifically in establishing chronic infection with P. aeruginosa. Our study identifies carriage of Pf phage as a potential biomarker for risk of antibiotic resistance and a potential therapeutic target for treatment of chronic P. aeruginosa infections in CF.
MATERIALS AND METHODS
Study design
We investigated the association between Pf phage and P. aeruginosa infections in CF in several sample sets. First, we investigated the prevalence of Pf phage in a large, public database of P. aeruginosa genomic sequences. Second, we determined the incidence of Pf phage in a population of well-characterized Danish patients with CF as to their temporal acquisition of P. aeruginosa. Third, we performed a prospective, cross-sectional cohort study in patients seen at the Stanford CF Center. In this group, we examined the associations between Pf status and patient demographics, patient clinical data, and microbiological strain attributes, as detailed below. In addition, we performed in vitro studies of Pf phage–mediated antibiotic sequestration.
Analysis of Pseudomonas Genome Database
Using the BLAST function on the Pseudomonas Genome Database website, we interrogated all P. aeruginosa strains for the sequence of the mature coaB major coat protein. We considered a sequence positive for Pf phage if there was a >80% identical match for the mature coaB sequence. This sequence was chosen as is necessary for mature phage production and contributes the anionic charge responsible for liquid crystal formation (27).
Analysis of Danish CF cohort
The whole-genome sequences from 474 isolates from 34 patients from the Copenhagen Cystic Fibrosis Center at the University Hospital, Rigshospitalet, Denmark described by Marvig et al. (40) were provided by the authors. In addition, they provided demographic information. We used Geneious Pro (Biomatters Limited) to interrogate the sequences for the presence of the coaB major coat protein.
The age of the patient at the last isolate of P. aeruginosa collected was used. Three patients were excluded from analysis of age because the ages were not available.
Subjects in Stanford CF cohort
This prospective, cross-sectional cohort study was conducted at the Stanford Cystic Fibrosis Center in Palo Alto, California. Patients were screened for study participation in a consecutive and unbiased manner. Eligible patients were defined as patients with confirmed diagnosis of CF, ability to expectorate, and ability to reliably perform pulmonary function testing. Patients who had received lung transplantation were excluded. Patients were identified either during hospital admission for treatment of a pulmonary exacerbation or at a routine clinic visit. Both pediatric and adult patients were recruited for participation. Patient data are reported in Table 1 and fig. S1.
Clinical data from the Stanford CF cohort
Clinical and laboratory data were extracted from the electronic medical record for each patient encounter. Pediatric age was defined as <18 years of age. Pulmonary exacerbation was defined as a change in clinical status that led to the patient being treated with antibiotics outside of the patient’s baseline regimen (such as chronic azithromycin therapy or chronic inhaled antibiotics) and an increase in airway clearance therapy regimen. Leeds criteria were used to define chronic P. aeruginosa infection (>50% of sputum cultures with P. aeruginosa in the preceding 12 months) (41, 53). FEV1 data recorded were from spirometry collected on the day of sputum sample collection (±7 days).
Sample acquisition and processing from the Stanford CF cohort
Expectorated sputum was collected from subjects in sterile specimen cups and frozen at −80°C. At a later date, samples were thawed and 200 μl of aliquot underwent mechanical homogenization by bead beating (MagNA Lyser Instrument, Roche) for 60 s at 6500 rpm. DNA extraction was performed with the QIAamp DNA Mini Kit by Qiagen as per the manufacturer’s protocol for tissue. DNA was eluted by adding 200 μl of elution buffer AL used.
Characterization of P. aeruginosa isolates in the Stanford CF cohort
On the day of sample collection, an additional sputum sample was sent to the clinical laboratory as per routine CF clinical care. In the clinical laboratory, respiratory cultures were performed as per Cystic Fibrosis Foundation recommendations (54). Briefly, respiratory samples from patients with CF are cultured on routine media [blood, chocolate, MacConkey, and colistin and nalidixic acid (CNA) agar] and Burkholderia cepacia Selective Agar (Remel) and BD BBL CHROMagar Staph aureus (Becton Dickinson). CF pathogens are identified by matrix-assisted laser desorption/ionization–time-of-flight, and antibiotic susceptibility testing is performed per Clinical & Laboratory Standards Institute recommendations. As per Stanford University Clinical Microbiology Laboratory protocol, a single representative clone is chosen for antibiotic susceptibility profiling. If multiple clonal phenotypes are observed, then a representative sample of each is tested. Antibiotic resistance is determined by the Kirby Bauer disc diffusion method. If there was not a sample sent to the clinical laboratory on day of sample collection, then susceptibilities were included from sputum sample within ±1 week. Isolates of P. aeruginosa are determined to be mucoid by visual identification based on colony morphology by the microbiology laboratory technicians.
Quantification of sputum P. aeruginosa and Pf phage by qPCR in the Stanford CF cohort
Pf phage and P. aeruginosa in sputum were quantified by qPCR. Two microliters of the DNA extracted from sputum samples was used as a template in 20-μl qPCR reactions containing 1× SensiFAST Probe Hi-ROX (catalog no. BIO-82020, Bioline), 200 nM probe, and 2 nM forward and reverse primers. Cycling conditions were as follows: 95°C for 2 min, (95°C for 15 s, 60°C for 20 s) × 40 cycles on a StepOnePlus Real-Time PCR system (Applied Biosystems). To quantify Pf, the primers [Pf-Conserve-F (TTCCCGCGTGGAATGC) and Pf-Conserve-R (CGGGAAGACAGCCACCAA)] targeting PA0717, a gene that is conserved across diverse strains of Pf phage (Pf1, Pf4, and Pf5), were used together with a PA0717 probe (AACGCTGGGTCGAAG). For P. aeruginosa quantification, primers targeting the 50S ribosomal subunit gene rpIU [rpIU-F (CAAGGTCCGCATCATCAAGTT) and rpIU-R (GGCCCTGACGCTTCATGT)] were used together with an rpIU probe (CGCCGTCGTAAGC). For the standard curves, the sequence targeted by the primers and probe was inserted into a pUC57 plasmid (Genewiz), and 10-fold serial dilutions of the plasmid were used in the qPCR reactions. Using these methods, we determined that the lower limit of detection in our qPCR assay for the RPIU gene of P. aeruginosa is 102 copies/ml and for the Pf phage conserved gene is 103 copies/ml. All samples were run in duplicate. The reported Pf phage concentrations represent the measured Pf phage with the P. aeruginosa concentration subtracted to account for detection of prophage DNA contained in the genome of P. aeruginosa.
Antibiotic sequestration assay
Sequestration experiments were performed by placing indicated concentrations of antibiotics with or without the mixture of Pf phage and DNA (Pf4, 1011 plaque-forming units/ml; DNA, 1.6 mg/ml) in a 2-kDa (0.5-ml volume) slide-A-Lyzer dialysis cassette (Thermo Fisher Scientific). Cassettes were dialyzed against 30-ml LB broth at 4°C overnight. Antibiotic concentrations are consistent with measured concentrations in the lung for both intravenous and inhaled dosing of antibiotics (55–58). LB broth outside the cassette was collected and used to quantify the amount of antibiotics by measuring the bactericidal effect. Briefly, log-phase E. coli DH5a culture was obtained by growing single colony in LB broth for 2 to 4 hours. The bacteria from 500 μl of the log-phase culture were spin down and mixed with 500 μl of the broth collected outside the cassette. The mixture was incubated at 37°C for 1 to 2 hours, and the number of viable bacteria was quantified by counting colony forming units (CFU) after plating serial dilutions. The CFUs for the groups with Pf phage were normalized to corresponding groups without Pf phage to obtain the degree of antibiotic sequestration.
Study approval
This study was approved by the Stanford University Institutional Review Board. Before inclusion in the study, informed consent was obtained from each subject for sputum collection and biobanking.
Statistics
Wilcoxon ranked sum was for analysis used to evaluate differences in concentration of Pf phage across continuous variables and Pearson’s χ2 or Fisher’s exact test across categorical variables. Mann-Whitney test was used for comparison of FEV1 values when well versus during exacerbation and comparison of patient age and phage status. For correlations with the concentration of Pf phage, Kendall correlation coefficients with log Pf phage were used. For correlations of patient age with proportion of isolates being Pf phage positive, Pearson correlation was used. For comparison of proportions of antibiotic resistance across phage and mucoid status, Pearson’s χ2 was used. For comparison of degree of antibiotic sequestration, Student’s t test was used. For all tests, an alpha level of 0.05 was used.
Supplementary Material
Acknowledgments:
We thank the CF research team at Stanford, specifically C. Dunn, D. Alvarez, S. Ryan, and W. Valencia for assistance with patient recruitment, obtaining consent and collection of samples. We would also like to thank D. Bassily, A. Yacob, and K. Engel for assistance in banking and processing sputum.
Funding:
E.B.B. was supported by the Stanford Child Health Research Institute, the Stanford Training Program in Pulmonary via the NIHLBI of Health under the award T32HL129970, the Translational Research and Applied Medicine (TRAM) Program at Stanford University under a pilot grant, and the Ross Mosier Laboratories Gift Fund. P.L.B. is supported by R21AI137432, R01 AI138981, and grants from the Cystic Fibrosis Foundation and the Dr. Ralph and Marian Falk Medical Research Trust Bank of America, N.A., Trustee. P.R.S. is supported by RO1 AI138981. H.K.J. was supported by The Novo Nordisk Foundation as a clinical research stipend (NNF12OC1015920), by Rigshospitalets Rammebevilling 2015–17 (R88-A3537), by Lundbeckfonden (R167-2013-15229), by Novo Nordisk Fonden (NNF15OC0017444), and by RegionH Rammebevilling (R144-A5287).
C.E.M. reports grants from Cystic Fibrosis Foundation, clinical trial support from Vertex Pharmaceuticals, Proteostasis, and Parion and consulting fees from Vertex Pharmaceuticals, Abbvie, and Gilead Sciences, all outside the submitted work. P.L.B. and P.R.S. are inventors on U.S. patent application no. 15/219,073 submitted on 22 July 2014 by Stanford University that covers the development of a vaccine that targets Pf bacteriophage.
Footnotes
Competing interests: The remainder of the authors have declared that no conflict of interest exists.
Data materials and availability: The genomic data analyzed in this study are deposited in the Sequence Read Archive under accession ERP004853. Further detail about accession codes for individual isolates are included in the Supplementary Materials of the original publication by Marvig et al. (40). All data associated with this study are present in the paper or in the Supplementary Materials.
REFERENCES AND NOTES
- 1.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, Drumm ML, Iannuzzi MC, Collins FS, Tsui L-C, Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science 245, 1066–1073 (1989). [DOI] [PubMed] [Google Scholar]
- 2.Welsh MJ, Ramsey BW, Accurso F, Cutting GR, in The Online Metabolic and Molecular Bases of Inherited Disease, Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, Gibson KM, Mitchell G, Eds. (The McGraw-Hill Companies Inc, 2014). [Google Scholar]
- 3.Poulsen JH, Fischer H, Illek B, Machen TE, Bicarbonate conductance and pH regulatory capability of cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. U.S.A 91, 5340–5344 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan TZ, Wagener JS, Bost T, Martinez J, Accurso FJ, Riches DW, Early pulmonary inflammation in infants with cystic fibrosis. Am. J. Respir. Crit. Care Med 151, 1075–1082 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Davis PB, Drumm M, Konstan MW, Cystic fibrosis. Am. J. Respir. Crit. Care Med 154, 1229–1256 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Hoegger MJ, Fischer AJ, McMenimen JD, Ostedgaard LS, Tucker AJ, Awadalla MA, Moninger TO, Michalski AS, Hoffman EA, Zabner J, Stoltz DA, Welsh MJ, Impaired mucus detachment disrupts mucociliary transport in a piglet model of cystic fibrosis. Science 345, 818–822 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoltz DA, Meyerholz DK, Pezzulo AA, Ramachandran S, Rogan MP, Davis GJ, Hanfland RA, Wohlford-Lenane C, Dohrn CL, Bartlett JA, Nelson IV GA, Chang EH, Taft PJ, Ludwig PS, Estin M, Hornick EE, Launspach JL, Samuel M, Rokhlina T, Karp PH, Ostedgaard LS, Uc A, Starner TD, Horswill AR, Brogden KA, Prather RS, Richter SS, Shilyansky J, McCray PB Jr., Zabner J, Welsh MJ, Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth. Sci. Transl. Med 2, 29ra31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corey M, Edwards L, Levison H, Knowles M, Longitudinal analysis of pulmonary function decline in patients with cystic fibrosis. J. Pediatr 131, 809–814 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Armstrong DS, Grimwood K, Carlin JB, Carzino R, Olinsky A, Phelan PD, Bronchoalveolar lavage or oropharyngeal cultures to identify lower respiratory pathogens in infants with cystic fibrosis. Pediatr. Pulmonol 21, 267–275 (1996). [DOI] [PubMed] [Google Scholar]
- 10.FitzSimmons SC, The changing epidemiology of cystic fibrosis. J. Pediatr 122, 1–9 (1993). [DOI] [PubMed] [Google Scholar]
- 11.Costerton JW, Stewart PS, Greenberg EP, Bacterial biofilms: A common cause of persistent infections. Science 284, 1318–1322 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Parsek MR, Singh PK, Bacterial biofilms: An emerging link to disease pathogenesis. Annu. Rev. Microbiol 57, 677–701 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Burns JL, Gibson RL, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith AL, Ramsey BW, Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis 183, 444–452 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Döring G, Flume P, Heijerman H, Elborn JS, Treatment of lung infection in patients with cystic fibrosis: Current and future strategies. J. Cyst. Fibros 11, 461–479 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Mogayzel PJ Jr., Naureckas ET, Robinson KA, Brady C, Guill M, Lahiri T, Lubsch L, Matsui J, Oermann CM, Ratjen F, Rosenfeld M, Simon RH, Hazle L, Sabadosa K, Marshall BC, Cystic fibrosis foundation pulmonary guideline. Pharmacologic approaches to prevention and eradication of initial Pseudomonas aeruginosa infection. Ann. Am. Thorac. Soc 11, 1640–1650 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Vidya P, Smith L, Beaudoin T, Yau YCW, Clark S, Coburn B, Guttman DS, Hwang DM, Waters V, Chronic infection phenotypes of Pseudomonas aeruginosa are associated with failure of eradication in children with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis 35, 67–74 (2016). [DOI] [PubMed] [Google Scholar]
- 17.2016. Patient Registry Annual Data Report; https://www.cff.org/research/researcherresources/patient-registry/2016-patient-registry-annual-data-report.pdf.
- 18.UK Cystic Fibrosis Registry Annual Data Report 2016; https://www.cysticfibrosis.org.uk/~/media/documents/the-work-we-do/uk-cf-registry/2016-registry-annual-data-report.
- 19.Rosenfeld M, Gibson RL, McNamara S, Emerson J, Burns JL, Castile R, Hiatt P, McCoy K, Wilson CB, Inglis A, Smith A, Martin TR, Ramsey BW, Early pulmonary infection, inflammation, and clinical outcomes in infants with cystic fibrosis. Pediatr. Pulmonol 32, 356–366 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Konstan MW, Morgan WJ, Butler SM, Pasta DJ, Craib ML, Silva SJ, Stokes DC, Wohl MEB, Wagener JS, Regelmann WE, Johnson CA; Scientific Advisory Group and the Investigators and Coordinators of the Epidemiologic Study of Cystic Fibrosis, Risk factors for rate of decline in forced expiratory volume in one second in children and adolescents with cystic fibrosis. J. Pediatr 151, 134–139.e1 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Nixon GM, Armstrong DS, Carzino R, Carlin JB, Olinsky A, Robertson CF, Grimwood K, Clinical outcome after early Pseudomonas aeruginosa infection in cystic fibrosis. J. Pediatr 138, 699–704 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Emerson J, Rosenfeld M, McNamara S, Ramsey B, Gibson RL, Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr. Pulmonol 34, 91–100 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Henry RL, Mellis CM, Petrovic L, Mucoid Pseudomonas aeruginosa is a marker of poor survival in cystic fibrosis. Pediatr. Pulmonol 12, 158–161 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Webb JS, Lau M, Kjelleberg S, Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol 186, 8066–8073 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knezevic P, Voet M, Lavigne R, Prevalence of Pf1-like (pro)phage genetic elements among Pseudomonas aeruginosa isolates. Virology 483, 64–71 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP, Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Secor PR, Sweere JM, Michaels LA, Malkovskiy AV, Lazzareschi D, Katznelson E, Rajadas J, Birnbaum ME, Arrigoni A, Braun KR, Evanko SP, Stevens DA, Kaminsky W, Singh PK, Parks WC, Bollyky PL, Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe 18, 549–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Secor PR, Jennings LK, Michaels LA, Sweere JM, Singh PK, Parks WC, Bollyky PL, Biofilm assembly becomes crystal clear - filamentous bacteriophage organize the Pseudomonas aeruginosa biofilm matrix into a liquid crystal. Microb. Cell 3, 49–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieleg O, Vladescu I, Ribbeck K, Characterization of particle translocation through mucin hydrogels. Biophys. J 98, 1782–1789 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park SH, Marassi FM, Black D, Opella SJ, Structure and dynamics of the membrane-bound form of Pf1 coat protein: Implications of structural rearrangement for virus assembly. Biophys. J 99, 1465–1474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janmey PA, Slochower DR, Wang Y-H, Wen Q, Cēbers A, Polyelectrolyte properties of filamentous biopolymers and their consequences in biological fluids. Soft Matter 10, 1439–1449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice SA, Tan CH, Mikkelsen PJ, Kung V, Woo J, Tay M, Hauser A, McDougald D, Webb JS, Kjelleberg S, The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 3, 271–282 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Secor PR, Michaels LA, Smigiel KS, Rohani MG, Jennings LK, Hisert KB, Arrigoni KR Braun TP. Birkland Y Lai TS Hallstrand PL Bollyky PK Singh WC Parks, Filamentous bacteriophage produced by Pseudomonas aeruginosa alters the inflammatory response and promotes noninvasive infection in vivo. Infect. Immun 85, e00648–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Platt MD, Schurr MJ, Sauer K, Vazquez G, Kukavica-Ibrulj I, Potvin E, Levesque RC, Fedynak FSL Brinkman J Schurr S-H Hwang GW Lau PA Limbach JJ Rowe MA Lieberman N Barraud J Webb S Kjelleberg DF Hunt DJ Hassett, Proteomic, microarray, and signature-tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late-stage cystic fibrosis airway conditions. J. Bacteriol 190, 2739–2758 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirov SM, Webb JS, O’May CY, Reid DW, Woo JKK, Rice SA, Kjelleberg S, Biofilm differentiation and dispersal in mucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 153, 3264–3274 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, Givskov M, Kjelleberg S, Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol 185, 4585–4592 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finnan S, Morrissey JP, O’Gara F, Boyd EF, Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J. Clin. Microbiol 42, 5783–5792 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, Olavarietta R, Doud M, Smith RS, Montgomery P, White JR, Godfrey PA, Kodira C, Birren B, Galagan JE, Lory S, Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. U.S.A 105, 3100–3105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winstanley C, Langille MGI, Fothergill JL, Kukavica-Ibrulj I, Paradis-Bleau C, Sanschagrin F, Thomson NR, Winsor GL, Quail MA, Lennard N, Bignell A, Clarke L, Seeger K, Saunders D, Harris D, Parkhill J, Hancock REW, Brinkman FSL, Levesque RC, Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool epidemic strain of Pseudomonas aeruginosa. Genome Res. 19, 12–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marvig RL, Sommer LM, Molin S, Johansen HK, Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet 47, 57–64 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Proesmans M, Balinska-Miskiewicz W, Dupont L, Bossuyt X, Verhaegen J, Høiby N, de Boeck K, Evaluating the “Leeds criteria” for Pseudomonas aeruginosa infection in a cystic fibrosis Centre. Eur. Respir. J 27, 937–943 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Boyd EF, Bacteriophage-encoded bacterial virulence factors and phage–pathogenicity island interactions. Adv. Virus Res 82, 91–118 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Friman V-P, Ghoul M, Molin S, Johansen HK, Buckling A, Pseudomonas aeruginosa adaptation to lungs of cystic fibrosis patients leads to lowered resistance to phage and protist enemies. PLOS ONE 8, e75380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters EM, Neill DR, Kaman B, Sahota JS, Clokie MRJ, Winstanley C, Kadioglu A, Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax 72, 666–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glonti T, Chanishvili N, Taylor PW, Bacteriophage-derived enzyme that depolymerizes the alginic acid capsule associated with cystic fibrosis isolates of Pseudomonas aeruginosa. J. Appl. Microbiol 108, 695–702 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Alemayehu D, Casey PG, McAuliffe O, Guinane CM, Martin JG, Shanahan F, Coffey RP Ross, C. Hill, Bacteriophages ϕMR299–2 and ϕNH-4 can eliminate Pseudomonas aeruginosa in the murine lung and on cystic fibrosis lung airway cells. MBio 3, e00029–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frimodt-Møller J, Rossi E, Haagensen JAJ, Falcone M, Molin S, Johansen HK, Mutations causing low level antibiotic resistance ensure bacterial survival in antibiotic-treated hosts. Sci. Rep 8, 12512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown-Jaque M, Rodriguez Oyarzun L, Cornejo-Sánchez T, Martín-Gómez MT, Gartner S, de Gracia J, Rovira S, Alvarez A, Jofre J, González-López JJ, Muniesa M, Detection of bacteriophage particles containing antibiotic resistance genes in the sputum of cystic fibrosis patients. Front. Microbiol 9, 856 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Belleghem JD, Dąbrowska K, Vaneechoutte M, Barr JJ, Bollyky PL, Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses 11, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darch SE, McNally A, Harrison F, Corander J, Barr HL, Paszkiewicz K, Holden S, Fogarty A, Crusz SA, Diggle SP, Recombination is a key driver of genomic and phenotypic diversity in a Pseudomonas aeruginosa population during cystic fibrosis infection. Sci. Rep 5, 7649 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mowat E, Paterson S, Fothergill JL, Wright EA, Ledson MJ, Walshaw MJ, Brockhurst MA, Winstanley C, Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am. J. Respir. Crit. Care Med 183, 1674–1679 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Williams D, Fothergill JL, Evans B, Caples J, Haldenby S, Walshaw MJ, Brockhurst MA, Winstanley C, Paterson S, Transmission and lineage displacement drive rapid population genomic flux in cystic fibrosis airway infections of a Pseudomonas aeruginosa epidemic strain. Microb. Genomics 4, doi: 10.1099/mgen.0.000167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee TWR, Brownlee KG, Conway SP, Denton M, Littlewood JM, Evaluation of a new definition for chronic Pseudomonas aeruginosa infection in cystic fibrosis patients. J. Cyst. Fibros 2, 29–34 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Saiman L, Siegel J, Infection control recommendations for patients with cystic fibrosis: Microbiology, important pathogens, and infection control practices to prevent patient-to-patient transmission. Am. J. Infect. Control 31, S6–S62 (2003). [PubMed] [Google Scholar]
- 55.Byl B, Jacobs F, Roucloux I, de Franquen P, Cappello M, Thys J-P, Penetration of meropenem in lung, bronchial mucosa, and pleural tissues. Antimicrob. Agents Chemother 43, 681–682 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dalhoff A, Pharmacokinetics and pharmacodynamics of aerosolized antibacterial agents in chronically infected cystic fibrosis patients. Clin. Microbiol. Rev 27, 753–782 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goldstein I, Wallet F, Robert J, Becquemin M-H, Marquette C-H, Rouby J-J, Lung tissue concentrations of nebulized amikacin during mechanical ventilation in piglets with healthy lungs. Am. J. Respir. Crit. Care Med 165, 171–175 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Beam TR Jr., R. P. Galask, L. T. Friedhoff, T. B. Platt, M. A. Leitz, Aztreonam concentrations in human tissues obtained during thoracic and gynecologic surgery. Antimicrob. Agents Chemother 30, 505–507 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.