Abstract
Objectives
Lipolysis, hydrolysis of triglycerides to fatty acids in adipocytes, is tightly regulated, poorly understood, and, if perturbed, can lead to metabolic diseases including obesity and type 2 diabetes. The goal of this study was to identify the genetic regulators of lipolysis and elucidate their molecular mechanisms.
Methods
Adipocytes from abdominal subcutaneous adipose tissue biopsies were isolated and were incubated without (spontaneous lipolysis) or with a catecholamine (stimulated lipolysis) to analyze lipolysis. DNA was extracted and genome-wide genotyping and imputation conducted. After quality control, 939 samples with genetic and lipolysis data were available. Genome-wide association studies of spontaneous and stimulated lipolysis were conducted. Subsequent in vitro gene expression analyses were used to identify candidate genes and explore their regulation of adipose tissue biology.
Results
One locus on chromosome 19 demonstrated genome-wide significance with spontaneous lipolysis. 60 loci showed suggestive associations with spontaneous or stimulated lipolysis, of which many influenced both traits. In the chromosome 19 locus, only HIF3A was expressed in the adipocytes and displayed genotype-dependent gene expression. HIF3A knockdown in vitro increased lipolysis and the expression of key lipolysis-regulating genes.
Conclusions
In conclusion, we identified a genetic regulator of spontaneous lipolysis and provided evidence of HIF3A as a novel key regulator of lipolysis in subcutaneous adipocytes as the mechanism through which the locus influences adipose tissue biology.
Keywords: Genetic variants, Lipolysis, Subcutaneous, Adipocytes, Gene expression
Highlights
-
•
A genome-wide association study identified a chromosome 19 locus that was associated with lipolysis in adipose tissue.
-
•
Of the genes in the locus, only HIF3A was strongly expressed during adipocyte differentiation.
-
•
In vitro analyses demonstrated that HIF3A plays a key role in the regulation of adipocyte lipolysis.
1. Introduction
Release of fatty acids through hydrolysis (lipolysis) of triglycerides in fat cells plays a major role in energy homeostasis. Lipolysis is highly regulated in a species-specific fashion. In humans, only catecholamines and natriuretic peptides have a strong lipolytic action among hormones as reviewed [[1], [2], [3]]. Human fat cells also have spontaneous lipolytic activity [1]. Inter-individual variations in lipolysis have important clinical impact. Large cross-sectional studies show significant relationships between high spontaneous or low catecholamine-stimulated lipolysis and a variety of metabolic conditions such as obesity and insulin resistance [[4], [5], [6]]. A recent prospective study demonstrated that subjects with high spontaneous lipolysis activity and low catecholamine-stimulated lipolysis had a markedly increased risk of future development of excess body fat, insulin resistance, and type 2 diabetes/glucose intolerance [7].
There is a strong genetic component to clinical traits of relevance to lipolysis, including body fat (measured by body mass index, BMI) [8], the location of stored lipids (as measured by waist-to-hip ratio (WHR) adjusted for BMI, WHRadjBMI) [9], and circulating triglyceride levels [10]. It is therefore likely that lipolysis is also regulated, at least in part, by genetic variations. Indeed, studies of monozygotic twins demonstrated a strong within-pair resemblance of spontaneous and catecholamine-stimulated lipolysis [11,12] and we reported that family history of type 2 diabetes is associated with increased spontaneous adipocyte lipolysis [13]. A recent large study on expression quantitative trait loci (eQTLs) in subcutaneous adipose tissue (SAT), the largest fat depot in the body, strongly suggests a genetic regulation of fat cell function [14].
To shed new light on the genetic influence on lipolysis, we conducted a genome-wide association study (GWAS) on 939 subjects from the GENiAL (GENetics of Adipocyte Lipolysis) cohort with phenotyping of spontaneous and catecholamine-stimulated lipolysis in SAT.
2. Materials and methods
2.1. Participants
The GENiAL cohort includes 273 men and 718 women whose characteristics are presented in Table 1. The participants were recruited between 1986 and 2016 by local advertisements for studies to examine the regulation of fat cell function with a focus on lipolysis. The recruitment period was extensive due to the labor-intensive methodology used for adipocyte analysis (only a few samples a week are possible); however, a single laboratory and consistent selection and analysis protocols were employed throughout.
Table 1.
Demographics of GENiAL cohort.
| Men |
Women |
|||||
|---|---|---|---|---|---|---|
| nonobese | obese | P | nonobese | obese | P | |
| lean/obese (n) | 177 | 96 | 269 | 449 | ||
| age (years) | 46(15) | 43(12) | 0.049 | 40(13) | 41(10 | 0.4 |
| height (cm) | 178(10) | 181(7) | 0.0089 | 167(6) | 166(6) | 0.0038 |
| body weight (kg) | 81(10) | 125(18) | <0.0001 | 67(9) | 108(17) | <0.0001 |
| BMI (kg/m2) | 25(2) | 38(5) | <0.0001 | 24(3) | 39(5) | <0.0001 |
| waist (cm) | 92(9) | 126(13) | <0.0001 | 84(10) | 119(13) | <0.0001 |
| WHR | 0.95(0.06) | 1.05(0.05) | <0.0001 | 0.86(0.07) | 0.96(0.07) | <0.0001 |
| systolic blood pressure (mm Hg) | 129(15) | 140(18) | <0.0001 | 120(16) | 130(16) | <0.0001 |
| diastolic blood pressure (mm Hg) | 78(10) | 85(12) | <0.0001 | 74(9) | 79(10) | <0.0001 |
| plasma total cholesterol (mmol/l) | 5.1(1.3) | 5.3(1.4) | 0.33 | 4.9(1.1) | 5.0(1.0) | 0.15 |
| plasma HDL cholesterol (mmol/l) | 1.2(0.4) | 1.0(0.2) | <0.0001 | 1.6(0.4) | 1.2(0.3) | <0.0001 |
| plasma triacylglycerides (mmol/l) | 1.70(1.98) | 2.33(2.49) | 0.038 | 1.06(0.64) | 1.52(0.86) | <0.0001 |
| plasma nonesterified fatty acids (mmol/l) | 0.49(0.17) | 0.62(0.21) | <0.0001 | 0.62(0.21) | 0.72(0.24) | <0.0001 |
| plasma glycerol (μmol/l) | 59.9(27.2) | 78.4(29.9) | <0.0001 | 81.1(42.6) | 116.9(54.1) | <0.0001 |
| fasting plasma glucose (mmol/l) | 5.4(1.2) | 6.3(2.3) | 0.0005 | 5.0(0.7) | 5.6(1.4) | <0.0001 |
| fasting serum insulin (mU/l) | 7.8(5.2) | 20.5(11.8) | <0.0001 | 6.2(3.3) | 14.7(7.9) | <0.0001 |
| HOMA-IR | 2.01(1.95) | 6.02(4.31) | <0.0001 | 1.40(0.87) | 3.82(2.88) | <0.0001 |
| Cell volume (pl) | 483(157) | 821(193) | <0.0001 | 473(177) | 860(180) | <0.0001 |
| Spontaneous lipolysis (mmol glycerol/2hrs/ESAT) | 0.8(0.7) | 3.8(2.8) | <0.0001 | 1.0(0.9) | 3.3(2.9) | <0.0001 |
| Stimulated lipolysis (mmol glycerol/2hrs/ESAT) | 6.6(5.2) | 3.6(1.6) | <0.0001 | 9.1(6.4) | 5.8(7.0) | <0.0001 |
Where: obese is defined as BMI>30 kg/m2; Spontaneous lipolysis rate was calculated as glycerol release divided by the lipid weight of the incubated fat cells; Stimulated lipolysis was calculated as the quotient of glycerol release at the maximum effective isoprenaline concentration divided by the spontaneous rate (no hormone present) of glycerol release from the isolated fat cells. continuous variables are presented as mean (sd), groups were compared with Student's t-test (unpaired). ESAT is estimated amount of abdominal subcutaneous adipose tissue corresponding to the region for needle biopsy.
The GENiAL cohort was used to analyze multiple adipose phenotypes, but all of the participants in this study had undergone baseline examination in our laboratory and their lipolysis and DNA measurements were available (Supplementary Fig. 1). A total of 939 study subjects with adipocyte lipolysis measurements were included. Descriptions of the study subjects are presented in Table 2. Overall, 57% were obese (defined as BMI ≥ 30 kg/m2). All lived in Stockholm, Sweden. A total of 194 of the participants had type 2 diabetes, hypertension, or dyslipidemia alone or in different combinations. None were treated with insulin, glitazones, or glucagon-like peptide analogues. Data on clinical variables and rates of fat cell lipolysis were published previously [5,6]. The current study was approved by the local committee on ethics and explained in detail to each subject. Informed consent was obtained from all of the participants; this was in written form since 1996.
Table 2.
Descriptions of the GENiAL participants included in this study.
| Men |
Women |
|||||
|---|---|---|---|---|---|---|
| nonobese | obese | P | nonobese | obese | P | |
| lean/obese (n) | 89 | 163 | 245 | 442 | ||
| age (years) | 47(15) | 43(13) | 0.044 | 40(13) | 41(10) | 0.7 |
| height (cm) | 178(6) | 181(7) | 0.0049 | 167(6) | 166(6) | 0.0076 |
| body weight (kg) | 80(10) | 125(18) | <0.0001 | 67(9) | 108(17) | <0.0001 |
| BMI (kg/m2) | 25(2) | 38(5) | <0.0001 | 24(3) | 39(5) | <0.0001 |
| waist (cm) | 92(9) | 126(13) | <0.0001 | 84(10) | 119(13) | <0.0001 |
| WHR | 0.95(0.06) | 1.05(0.05) | <0.0001 | 0.86(0.07) | 0.96(0.07) | <0.0001 |
| systolic blood pressure (mm Hg) | 129(15) | 140(19) | <0.0001 | 120(16) | 130(16) | <0.0001 |
| diastolic blood pressure (mm Hg) | 79(10) | 85(12) | <0.0001 | 74(10) | 80(10) | <0.0001 |
| plasma total cholesterol (mmol/l) | 5.1(1.3) | 5.3(1.4) | 0.43 | 4.9(1.1) | 5.0(1.0) | 0.29 |
| plasma HDL cholesterol (mmol/l) | 1.2(0.4) | 1.0(0.2) | <0.0001 | 1.6(0.4) | 1.2(0.3) | <0.0001 |
| plasma triacylglycerides (mmol/l) | 1.70(2.03) | 2.32(2.53) | 0.047 | 1.07(0.64) | 1.52(0.87) | <0.0001 |
| plasma nonesterified fatty acids (mmol/l) | 0.49(0.17) | 0.61(0.21) | 0.0002 | 0.63(0.21) | 0.72(0.24) | <0.0001 |
| plasma glycerol (μmol/l) | 60.0(27.0) | 76.8(27.3) | <0.0001 | 81.9(43.2) | 116.8(53.8) | <0.0001 |
| fasting plasma glucose (mmol/l) | 5.4(1.2) | 6.4(2.3) | 0.0006 | 5.0(0.7) | 5.6(1.4) | <0.0001 |
| fasting serum insulin (mIE/l) | 7.8(5.3) | 20.5(12.0) | <0.0001 | 6.2(3.4) | 14.7(7.9) | <0.0001 |
| HOMA-IR | 2.01(1.98) | 6.05(4.41) | <0.0001 | 1.40(0.88) | 3.80(2.88) | <0.0001 |
| cell volume (pl) | 495(154) | 812(193) | <0.0001 | 475(177) | 861(179) | <0.0001 |
| Spontaneous lipolysis (mmol glycerol/2hrs/ESAT) | 0.8(0.7) | 3.9(2.8) | <0.0001 | 1.0(0.9) | 3.4(2.9) | <0.0001 |
| Stimulated lipolysis (mmol glycerol/2hrs/ESAT) | 6.6(5.2) | 3.6(1.6) | <0.0001 | 9.1(6.4) | 5.8(7.0) | <0.0001 |
Where: obese is defined as BMI>30 kg/m2; Spontaneous lipolysis rate was calculated as glycerol release divided by the lipid weight of the incubated fat cells; Stimulated lipolysis was calculated as the quotient of glycerol release at the maximum effective isoprenaline concentration divided by the spontaneous rate (no hormone present) of glycerol release from the isolated fat cellscontinuous variables are presented as mean (sd), groups were compared with Student's t-test (unpaired).
SAT gene expression was studied in 114 women, a subset of the cohort. This cohort was previously described [15] and contained the same type of lipolysis data as presented herein. Gene expression of HIF3A was measured in 75 subjects from the GENiAL cohort with stored frozen abdominal subcutaneous adipocytes isolated as described below.
2.2. Clinical examination
The participants came to the Karolinska University Hospital's clinical research center in the morning after an overnight fast. Height, weight, and waist-to-hip ratio (WHR) were measured. Body fat content was measured via bioimpedance. A venous blood sample was obtained for extraction of DNA and clinical chemistry, which was performed by the hospital's accredited routine clinical chemistry laboratory. HOMA-IR as measure of systemic insulin resistance was calculated from the fasting levels of glucose and insulin as previously described [16]. SAT was obtained via needle aspiration biopsy lateral to the umbilicus as previously described [17]. The estimated abdominal subcutaneous adipose tissue (ESAT) area was calculated using a formula based on WHR, sex, age, waist circumference, and body fat as previously described and validated [18].
2.3. Adipose tissue phenotyping
The SAT samples were rapidly rinsed in sodium chloride (9 mg/ml) before removal of visual blood vessels and cell debris and subsequently subjected to collagenase treatment to obtain isolated adipocytes as previously described [19]. Fat cells were incubated as previously described [19]. In brief, cell suspensions (diluted to 2% volume/volume) were incubated for 2 h at 37 °C with air as the gas phase in Krebs–Ringer phosphate buffer (pH 7.4) supplemented with glucose (8.6 mmol/l), ascorbic acid (0.1 mg/ml), and bovine serum albumin (20 mg/ml) either without (spontaneous lipolysis) or with supplementation with synthetic non-selective β-adrenoreceptor agonist isoprenaline (Hässle, Mölndal, Sweden) at increasing concentrations (10−9-10−5 mol/l; stimulated lipolysis). The amount of glycerol, as a measure of lipolysis, was evaluated in an aliquot of medium at the end of the incubation [20]. This end product of lipolysis, unlike the other final fatty acid metabolites, is not re-utilized by fat cells. The spontaneous lipolysis rate was calculated as the glycerol release to the incubation medium divided by the lipid weight of the incubated fat cells. There was no consensus how to express the lipolysis rates (absolute terms, relative terms, per cell number, or per lipid weight). We expressed isoprenaline-stimulated lipolysis as the quotient of glycerol release at the maximum effective isoprenaline concentration divided by the spontaneous rate (no hormones present) of glycerol release from the isolated fat cells. Spontaneous lipolysis was expressed as glycerol release/cell weight multiplied by the weight of ESAT, that is, an estimate of the total release of glycerol from the ESAT area. The values were log10 transformed to improve normality (required for linear regression analysis). These modes of expression were preferred as they in linear regression showed better correlations with clinical parameters in the cohort than other ways of expressing lipolysis (results not shown). Previous in vitro studies demonstrated that spontaneous and isoprenaline-stimulated lipolysis were linear for at least 4 h in human subcutaneous fat cells [21].
2.4. Genetic analysis
DNA was extracted from whole blood using standard protocols. The samples were genotyped using the UK Biobank Axiom Array r3 platform and called using the Axiom analysis suite (https://assets.thermofisher.com/TFS-Assets/LSG/manuals/axiom_genotyping_solution_analysis_guide.pdf). Samples were excluded for cryptic relatedness, ambiguous sex, or low call rates (<95%). Multi-dimensional scaling, which essentially measures how genetically similar any two individuals are, was conducted in PLINK (http://zzz.bwh.harvard.edu/plink/download.shtml) [22] and the principal components (PCs) calculated for all individuals. PCs 1–4 were plotted and population outliers were excluded by visual inspection. SNPs were excluded for low call rates (<95%), failing Hardy–Weinberg equilibrium (P < 5 × 10−6), or low minor allele frequency (MAF < 1%). After quality control, imputation was performed using the haplotype reference consortium panel and, when variants were not available, the 1000G phase 3 reference panel [23]. Post-imputation quality control excluded SNPs with minor allele counts <3 and INFO <0.4 as well as related individuals (one of each pair of first- or second-degree relatives). After quality control, 885 samples and 9,260,588 SNPs were available for phenotypic analysis.
As previously noted, the spontaneous and stimulated lipolysis measurements were log10 transformed prior to statistical analysis. A GWAS was conducted in PLINK [22], using linear regression assuming an additive genetic model and adjusting for population structure (PCs1-3), age, sex, and BMI. BMI was adjusted for as the sample size precluded separate analysis in obese and non-obese individuals. Genome-wide significance was set at P < 5 × 10−8 and suggestive significance was set at P < 1 × 10−5. Only SNPs with MAF >1% were included in the results. SNPs were assigned to loci using the independent-pairwise function in PLINK [22] with default settings. Heritability was calculated using LD score regression (LDSR) [24].
2.5. Data mining
To identify SNPs with potentially functional effects on nearby genes, SNPs meeting the threshold for suggestive significance were assessed using the variant effect predictor (VEP) [25] and ANNOVAR [26] software. To identify genotype-specific gene expression patterns or eQTLs, SNPs reaching GWAS significance or those predicted by VEP or ANNOVAR to have functional effects were explored using GTEx [27].
2.6. Adipocyte cell culture and transfection with small interfering RNA
Isolation, growth, and differentiation of SAT-derived human mesenchymal stem cells (hMSCs) were previously described [28]. Marker gene expression (Cap analysis gene expression [CAGE] and qPCR data) and lipid accumulation images for these cells were previously published [29].
hMSCs were transfected using a Neon electroporator (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Briefly, 1 million of hMSCs at day 1 of differentiation were mixed with 40 nM ON-TARGETplus SMARTpool small interfering RNAs (siRNAs) targeting HIF3A or non-targeting siRNA pool (Dharmacon, Lafayette, CO, USA) and electroporated using a 100 μl Neon electroporation tip. Electroporation conditions were 1600 V, 20 ms width, and 1 pulse. Electroporation was repeated until the required amount of cells were collected for the experimental design. Following electroporation, the cells were plated in antibiotic-free medium at a density of 120,000 cells/well in 24-well plates or 12,000 cells/well in 96-well plates and cultured until days 6–13 of differentiation. The medium was collected at days 10 and 13 of differentiation for glycerol measurements as previously described [30]. The amounts of glycerol were normalized to the number of nuclei in each well (see “Quantification of neutral lipids and cell numbers during adipogenesis”).
For HIF3A protein analysis, hMSCs were reverse transfected 24 h before induction of adipogenesis using ON-TARGETplus SMARTpool siRNAs targeting HIF3A or non-targeting siRNA control #1 (Dharmacon) as previously described [31]. In this experimental design, the RNA and medium were collected at days 2, 6, and 9 after the induction of differentiation. Lipid accumulation was evaluated at day 9. For proteins, cells were collected 72 h post-transfection.
2.7. Quantification of neutral lipids and cell numbers during adipogenesis
Lipid accumulation was quantified at differentiation days 10 and 13. hMSCs differentiated in vitro were washed with PBS and fixed with 4% paraformaldehyde solution for 10 min at room temperature. The fixed cells were subsequently washed with PBS and stained with BODIPY 493/503 (0.2 μg/mL; Molecular Probes, Thermo Fisher Scientific) and Hoechst 33,342 (2 μg/mL; Molecular Probes) for 20 min at room temperature. Accumulation of intracellular lipids (BODIPY) and cell numbers (Hoechst) were quantified with a CellInsight CX5 High Content Screening (HCS) Platform (Thermo Fischer Scientific) using integrated protocols. Total BODIPY fluorescence (lipid droplets) was normalized to the number of nuclei in each well.
2.8. Isolation of RNA and analysis of gene expression
The hMSCs were collected at days 6, 10, and 13 of differentiation for isolation of RNA. Extraction of total RNA, measurement of concentration and purity, and reverse transcription were performed as previously described [30]. Isolation of RNA from frozen abdominal subcutaneous adipocytes was conducted as previously described [15]. Quantitative RT-PCR of the coding genes was performed using commercial TaqMan probes (Applied Biosystems, Foster City, CA, USA). Gene expression was normalized to the internal reference gene 18s. Relative expression was calculated using the 2 (−ΔΔ threshold cycle) method [32].
2.9. Analysis of protein expression
The hMSCs were transfected with siRNA against HIF3A 24 h prior to the induction of differentiation as previously described. At 72 h post-transfection, the cells were collected to isolate cytosolic and nuclear fractions as previously described [33]. Nuclei obtained from approximately 500,000 cells were analyzed in 150 μL of radioimmunoprecipitation assay buffer (Perce/Thermo Fisher) supplemented with 5 mM of NaF, 1 mM of Na3VO4, protease inhibitor cocktail set V (Calbiochem), and benzonase (Sigma–Aldrich) on ice for 30 min. Cytosolic proteins were clarified by centrifugation at 14,000 rpm for 30 min. The nuclear and cytosolic proteins were separated by 10% Mini-PROTEAN TGX Stain-Free Precast Gels (Bio-Rad) allowing direct protein visualization in the gels and after transfer onto the membranes. Protein transfer to the PVDF membranes was performed using a Trans-Blot Turbo Transfer System (Bio-Rad). The membranes were blocked in 3% ECL Advance Blocking Agent (GE Healthcare, Little Chalfont, UK). Antibodies against HIF3A were obtained from Proteintech (Rosemont, IL, USA) and Prosci (Poway, CA, USA), both used at 1:1000 dilution. Secondary rabbit IgG-horseradish peroxidase antibodies were obtained from Sigma–Aldrich. Antibody antigen complexes were detected by chemiluminescence using an ECL Select Western Blotting Detection Kit (GE Healthcare) in Chemidoc MP (Bio-Rad) and quantified using Image Lab software (Bio-Rad). Total lysate from Hep2D cells (Novus Biologicals, Abingdon, UK) was used as a positive control for HIF3A antibodies.
2.10. Statistical analysis of clinical and in vitro data
The values of clinical variables and results of the in vitro experiments are mean ± standard deviation (SD). Lipolysis, glycerol, and triglyceride values were normalized by log10 transformation prior to analysis. Standard statistical tests were used including the t-test and single or multiple regression as indicated in the figure/table legends using StatView software (Abacus Concepts Inc, Berkley, CA, USA) or JMP 13 (SAS, Cary, NC, USA).
3. Results
Demographic characteristics of all of the participants in the GENiAL cohort are presented in Table 1 and those analyzed in the GWAS are presented in Table 2. The GWAS cohort consisted predominantly of women who were younger and had a higher frequency of obesity than the men. Obesity was associated with higher systemic levels of fasting lipids, glucose, and insulin levels in both genders. Spontaneous lipolysis, measured as the rate of glycerol release from a defined segment of abdominal subcutaneous adipose tissue, was higher in the obese subjects. The stimulated lipolysis, measured as catecholamine-induced divided by basal glycerol release in isolated fat cells, was lower among the obese subjects. As expected, spontaneous and stimulated lipolysis were inversely correlated to one another (R2 = 0.49, standardized beta −0.70, p = 1 × 10−134).
To assess in vivo relevance, spontaneous and stimulated lipolysis measures were assessed for association with systemic metabolic variables (Table 3). Spontaneous lipolysis was positively correlated with the fasting plasma levels of triglycerides, free fatty acids, insulin, and HOMA-IR as measures of systemic insulin resistance after adjusting for age and sex (Table 3). For the triglycerides, insulin, and HOMA-IR, the results remained significant after adjusting to BMI (results not shown).
Table 3.
Associations between adipocyte spontaneous or stimulated lipolysis and systemic metabolic variables.
| Spontaneous lipolysis |
Stimulated lipolysis |
|||
|---|---|---|---|---|
| Standardized beta | P | Standardized beta | P | |
| plasma triglycerides (mmol/l) | 0.30 | 9.0 × 10−19 | −0.24 | 3.0 × 10−14 |
| plasma nonesterified fatty acids (mmol/l) | 0.22 | 5.3 × 10−9 | −0.07 | 0.049 |
| plasma glycerol (μmol/l) | 0.22 | 2.0 × 10−12 | −0.12 | 1.0 × 10−4 |
| fasting serum insulin (mU/l) | 0.57 | 4.0 × 10−71 | −0.38 | 2 × 10−31 |
| HOMAIR | 0.56 | 3.0 × 10−67 | −0.38 | 1.0 × 10−31 |
Spontaneous lipolysis rate was calculated as glycerol release divided by the lipid weight of the incubated fat cells; Stimulated lipolysis was calculated as the quotient of glycerol release at the maximum effective isoprenaline concentration divided by the spontaneous rate (no hormone present) of glycerol release from the isolated fat cells; Analysis was performed using multiple regression adjusting for age, and sex.
3.1. GWAS of spontaneous and stimulated lipolysis
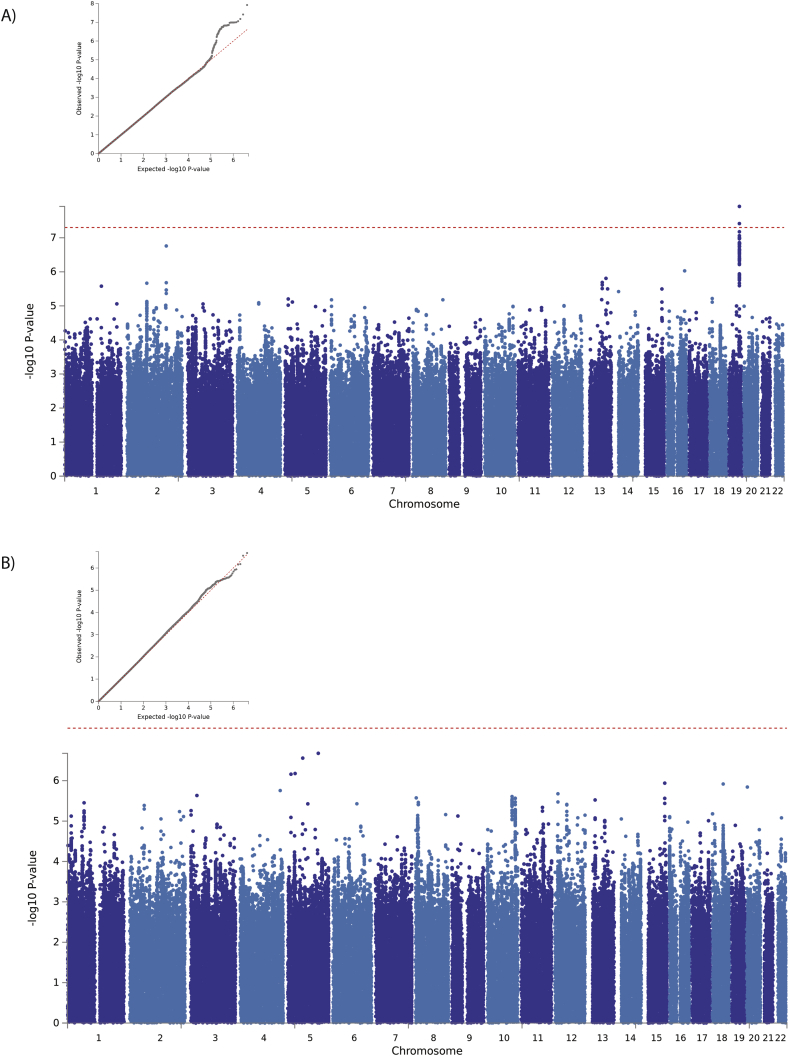
The results are presented in Figure 1 and Supplementary Tables 1 and 2 A total of 112 SNPs in 25 loci demonstrated suggestive associations (P < 1 × 10−5) with spontaneous lipolysis (Figure 1A, Table 4, and Supplementary Table 1), including one that demonstrated GWAS-significant (P < 5 × 10−8) associated SNPs on chromosome 19. Many genes were potentially impacted by the SNPs in this region of chromosome 19 (Figure 2A). For stimulated lipolysis (Figure 1B), 156 SNPs in 40 loci reached the threshold for suggestive association (Table 4 and Supplementary Table 2); however, no GWAS-significant signals were identified. The sample size precluded analysis stratifying by obesity status. However sensitivity analysis of the GWAS-significant signals demonstrated consistent directions, but weaker effects in the obese vs non-obese individuals (rs73048030: beta = −0.26 Se 0.08 P = 2.40 × 10−7 vs beta = −0.55 Se = 0.10 P = 7.46 × 10−4, respectively).
Figure 1.
Results of the GWAS of spontaneous and stimulated lipolysis. QQ and Manhattan plots for A) spontaneous lipolysis and B) isoprenaline-stimulated lipolysis. In the QQ plot, the red dotted line indicates the null distribution. In the Manhattan plot, the horizontal red line represents the threshold for GWAS significance (P < 5 × 10−8).
Table 4.
GWAS-significant and suggestive loci for spontaneous lipolysis and suggestive loci for stimulated lipolysis.
| Spontaneous lipolysis |
Stimulated lipolysis |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Pos | SNP | Ref | Alt | Freq | N | Beta | Se | L95 | U95 | P | N | Beta | Se | L95 | U95 | P |
| 1 | 44208924 | 1:44208924_D_I | I | D | 0.85 | 856 | 0.144 | 0.029 | 0.088 | 0.200 | 5.48E-07 | 874 | −0.076 | 0.022 | −0.118 | −0.033 | 0.0006 |
| 1 | 161567916 | rs371656041 | C | T | 0.99 | 856 | 0.449 | 0.095 | 0.263 | 0.636 | 2.64E-06 | 874 | −0.300 | 0.069 | −0.434 | −0.165 | 1.44E-05 |
| 1 | 229572764 | rs145251747 | D | I | 0.98 | 856 | 0.429 | 0.091 | 0.250 | 0.608 | 3.13E-06 | 874 | −0.257 | 0.070 | −0.394 | −0.120 | 0.0003 |
| 2 | 87271929 | rs71226525 | T | G | 0.54 | 856 | 0.097 | 0.020 | 0.057 | 0.137 | 2.16E-06 | 874 | −0.046 | 0.015 | −0.076 | −0.016 | 0.0027 |
| 2 | 170578879 | rs369684581 | A | G | 0.83 | 856 | 0.108 | 0.024 | 0.061 | 0.156 | 9.60E-06 | 874 | −0.056 | 0.018 | −0.092 | −0.020 | 0.0026 |
| 2 | 172907753 | rs60231166 | T | C | 0.74 | 856 | 0.121 | 0.023 | 0.076 | 0.166 | 1.74E-07 | 874 | −0.055 | 0.017 | −0.090 | −0.021 | 0.0016 |
| 3 | 67430871 | rs73098354 | G | A | 0.98 | 856 | −0.266 | 0.060 | −0.383 | −0.150 | 8.80E-06 | 874 | 0.192 | 0.045 | 0.103 | 0.280 | 2.63E-05 |
| 4 | 94856317 | rs2918080 | G | T | 0.78 | 856 | −0.096 | 0.021 | −0.138 | −0.054 | 8.80E-06 | 874 | 0.039 | 0.016 | 0.007 | 0.071 | 0.0164 |
| 5 | 14637275 | rs138114267 | A | C | 0.98 | 856 | 0.285 | 0.063 | 0.162 | 0.408 | 6.27E-06 | 874 | −0.204 | 0.047 | −0.297 | −0.111 | 1.92E-05 |
| 5 | 32955093 | rs114601412 | T | C | 0.95 | 856 | 0.176 | 0.039 | 0.100 | 0.253 | 7.70E-06 | 874 | −0.148 | 0.030 | −0.206 | −0.090 | 6.64E-07 |
| 6 | 6326042 | rs9502434 | A | G | 0.96 | 856 | 0.194 | 0.043 | 0.110 | 0.278 | 6.63E-06 | 874 | −0.129 | 0.033 | −0.192 | −0.065 | 8.72E-05 |
| 6 | 44053018 | rs554214639 | D | I | 0.85 | 856 | −0.111 | 0.024 | −0.158 | −0.063 | 6.38E-06 | 874 | 0.065 | 0.018 | 0.028 | 0.101 | 0.0005 |
| 8 | 133340555 | rs111301880 | T | C | 0.97 | 856 | 0.278 | 0.061 | 0.158 | 0.398 | 6.63E-06 | 874 | −0.206 | 0.045 | −0.295 | −0.116 | 6.90E-06 |
| 12 | 53131676 | rs11170254 | T | C | 0.09 | 856 | −0.141 | 0.032 | −0.203 | −0.079 | 9.83E-06 | 874 | 0.112 | 0.024 | 0.065 | 0.160 | 3.86E-06 |
| 13 | 71658551 | rs144654648 | T | C | 0.99 | 856 | 0.389 | 0.086 | 0.221 | 0.556 | 6.58E-06 | 874 | −0.214 | 0.065 | −0.342 | −0.087 | 0.0010 |
| 13 | 74083355 | rs117619390 | G | A | 0.98 | 856 | −0.293 | 0.061 | −0.413 | −0.173 | 2.05E-06 | 874 | 0.152 | 0.046 | 0.063 | 0.242 | 0.0009 |
| 13 | 90513818 | rs9588787 | T | C | 0.42 | 856 | −0.086 | 0.018 | −0.121 | −0.051 | 1.55E-06 | 874 | 0.051 | 0.014 | 0.024 | 0.077 | 0.0002 |
| 13 | 97727951 | rs149528824 | G | C | 0.99 | 856 | 0.417 | 0.089 | 0.243 | 0.591 | 3.17E-06 | 874 | −0.228 | 0.068 | −0.361 | −0.094 | 0.0008 |
| 14 | 19462974 | rs28701881 | T | C | 0.99 | 856 | 0.478 | 0.103 | 0.277 | 0.680 | 3.80E-06 | 874 | −0.366 | 0.082 | −0.526 | −0.205 | 8.87E-06 |
| 15 | 68691171 | rs199600197 | I | D | 0.11 | 856 | −0.171 | 0.035 | −0.240 | −0.102 | 1.34E-06 | 874 | 0.102 | 0.027 | 0.050 | 0.154 | 0.0001 |
| 15 | 92595793 | rs28660218 | T | C | 0.99 | 856 | 0.384 | 0.082 | 0.223 | 0.544 | 3.19E-06 | 874 | −0.305 | 0.062 | −0.426 | −0.183 | 1.15E-06 |
| 16 | 79960721 | rs143486974 | A | G | 0.98 | 856 | 0.360 | 0.073 | 0.217 | 0.503 | 9.35E-07 | 874 | −0.166 | 0.056 | −0.276 | −0.057 | 0.0030 |
| 18 | 12923043 | rs16939974 | C | T | 0.84 | 856 | 0.104 | 0.023 | 0.059 | 0.149 | 6.08E-06 | 874 | −0.075 | 0.017 | −0.109 | −0.041 | 1.40E-05 |
| 19 | 46633622 | rs73048030 | A | G | 0.98 | 856 | 0.377 | 0.066 | 0.249 | 0.506 | 1.20E-08 | 874 | −0.197 | 0.049 | −0.293 | −0.101 | 5.90E-05 |
| 19 | 46635857 | rs73048031 | C | T | 0.98 | 856 | 0.372 | 0.067 | 0.241 | 0.504 | 3.82E-08 | 874 | −0.197 | 0.050 | −0.294 | −0.099 | 8.74E-05 |
| 1 | 16291470 | rs3979178 | A | G | 0.73 | 856 | 0.065 | 0.019 | 0.028 | 0.103 | 0.0007 | 874 | -0.065 | 0.014 | -0.093 | -0.037 | 7.54E-06 |
| 1 | 72905881 | rs7524249 | C | A | 0.80 | 856 | 0.078 | 0.022 | 0.035 | 0.120 | 0.0003 | 874 | -0.076 | 0.016 | -0.107 | -0.044 | 3.39E-06 |
| 2 | 63587155 | rs148533396 | T | C | 0.98 | 856 | 0.245 | 0.062 | 0.124 | 0.366 | 8.17E-05 | 874 | -0.218 | 0.047 | -0.310 | -0.126 | 4.10E-06 |
| 2 | 87414589 | rs540268296 | D | I | 0.98 | 856 | 0.287 | 0.083 | 0.124 | 0.450 | 0.0006 | 874 | -0.280 | 0.062 | -0.403 | -0.158 | 8.15E-06 |
| 2 | 137306522 | rs116525497 | G | A | 0.98 | 856 | 0.249 | 0.060 | 0.131 | 0.367 | 4.08E-05 | 874 | -0.205 | 0.046 | -0.294 | -0.115 | 8.84E-06 |
| 2 | 218934359 | rs55703230 | T | C | 0.99 | 856 | 0.440 | 0.100 | 0.243 | 0.636 | 1.32E-05 | 874 | -0.347 | 0.076 | -0.496 | -0.198 | 5.82E-06 |
| 2 | 236618480 | rs560405754 | A | C | 0.99 | 856 | -0.282 | 0.094 | -0.466 | -0.098 | 0.0028 | 874 | 0.314 | 0.070 | 0.177 | 0.450 | 7.71E-06 |
| 3 | 2428151 | rs1032784 | A | G | 0.75 | 856 | -0.078 | 0.020 | -0.118 | -0.038 | 0.0001 | 874 | 0.070 | 0.015 | 0.040 | 0.100 | 5.51E-06 |
| 3 | 28133397 | rs1506686 | A | C | 0.99 | 856 | 0.321 | 0.087 | 0.150 | 0.492 | 0.0003 | 874 | -0.314 | 0.066 | -0.443 | -0.184 | 2.32E-06 |
| 3 | 116686382 | rs590225 | T | C | 0.82 | 856 | 0.078 | 0.023 | 0.033 | 0.124 | 0.0008 | 874 | -0.078 | 0.017 | -0.112 | -0.044 | 8.21E-06 |
| 4 | 177130984 | rs73007116 | G | T | 0.91 | 856 | 0.105 | 0.033 | 0.040 | 0.171 | 0.0016 | 874 | -0.118 | 0.025 | -0.166 | -0.070 | 1.75E-06 |
| 5 | 14799937 | rs114867704 | T | C | 0.97 | 856 | 0.246 | 0.060 | 0.128 | 0.363 | 4.69E-05 | 874 | -0.222 | 0.044 | -0.309 | -0.135 | 6.91E-07 |
| 5 | 66547938 | 5:66547938_G_A | A | G | 0.96 | 856 | 0.157 | 0.049 | 0.061 | 0.253 | 0.0015 | 874 | -0.190 | 0.037 | -0.262 | -0.118 | 2.76E-07 |
| 5 | 88866702 | rs74574939 | G | T | 0.99 | 856 | 0.250 | 0.076 | 0.101 | 0.398 | 0.0010 | 874 | -0.267 | 0.057 | -0.379 | -0.154 | 3.74E-06 |
| 5 | 134881956 | rs72787161 | T | G | 0.98 | 856 | 0.328 | 0.074 | 0.183 | 0.474 | 1.04E-05 | 874 | -0.288 | 0.055 | -0.395 | -0.180 | 2.10E-07 |
| 6 | 105831370 | rs117269303 | T | C | 0.99 | 856 | 0.270 | 0.076 | 0.121 | 0.420 | 0.0004 | 874 | -0.263 | 0.056 | -0.373 | -0.152 | 3.73E-06 |
| 6 | 123139459 | rs35141697 | D | I | 0.75 | 856 | -0.067 | 0.020 | -0.107 | -0.028 | 0.0009 | 874 | 0.070 | 0.015 | 0.040 | 0.100 | 4.94E-06 |
| 7 | 41024130 | rs200674393 | I | D | 0.98 | 856 | -0.329 | 0.077 | -0.481 | -0.177 | 2.45E-05 | 874 | 0.305 | 0.059 | 0.191 | 0.420 | 2.29E-07 |
| 8 | 3902830 | rs79055631 | T | G | 0.95 | 856 | 0.138 | 0.042 | 0.057 | 0.220 | 0.0009 | 874 | -0.148 | 0.031 | -0.209 | -0.086 | 2.66E-06 |
| 8 | 10375973 | rs111818085 | I | D | 0.89 | 856 | 0.064 | 0.027 | 0.012 | 0.117 | 0.0163 | 874 | -0.094 | 0.020 | -0.133 | -0.055 | 2.78E-06 |
| 8 | 13095207 | rs7007799 | G | A | 0.87 | 856 | 0.094 | 0.027 | 0.041 | 0.146 | 0.0005 | 874 | -0.094 | 0.020 | -0.133 | -0.054 | 3.45E-06 |
| 9 | 26337144 | rs10967318 | A | G | 0.99 | 856 | 0.311 | 0.101 | 0.112 | 0.509 | 0.0023 | 874 | -0.321 | 0.071 | -0.460 | -0.181 | 7.50E-06 |
| 10 | 109418980 | rs1902740 | T | C | 0.28 | 856 | 0.082 | 0.019 | 0.045 | 0.119 | 1.49E-05 | 874 | -0.069 | 0.014 | -0.097 | -0.042 | 1.21E-06 |
| 11 | 94458034 | rs117762377 | A | G | 0.99 | 856 | -0.174 | 0.079 | -0.328 | -0.019 | 0.0277 | 874 | 0.262 | 0.057 | 0.150 | 0.373 | 4.58E-06 |
| 12 | 14094162 | rs148192549 | C | T | 0.99 | 856 | 0.238 | 0.087 | 0.068 | 0.408 | 0.0062 | 874 | -0.304 | 0.064 | -0.429 | -0.179 | 2.11E-06 |
| 12 | 101524071 | 12:101524071_G_A | A | G | 0.98 | 856 | 0.158 | 0.064 | 0.032 | 0.285 | 0.0143 | 874 | -0.209 | 0.047 | -0.300 | -0.117 | 8.29E-06 |
| 12 | 133262001 | rs11147005 | T | C | 0.32 | 856 | 0.045 | 0.019 | 0.009 | 0.082 | 0.0148 | 874 | -0.062 | 0.014 | -0.089 | -0.035 | 7.14E-06 |
| 13 | 30567601 | rs142268497 | C | T | 0.99 | 856 | -0.246 | 0.085 | -0.413 | -0.080 | 0.0038 | 874 | 0.301 | 0.064 | 0.176 | 0.427 | 3.00E-06 |
| 13 | 73025966 | rs9318075 | A | G | 0.64 | 856 | 0.051 | 0.018 | 0.016 | 0.086 | 0.0041 | 874 | -0.059 | 0.013 | -0.085 | -0.033 | 9.92E-06 |
| 16 | 1595600 | rs56342298 | T | C | 0.95 | 856 | 0.108 | 0.039 | 0.031 | 0.185 | 0.0064 | 874 | -0.132 | 0.029 | -0.189 | -0.074 | 7.79E-06 |
| 17 | 73299631 | rs9892812 | G | A | 0.47 | 856 | -0.044 | 0.018 | -0.079 | -0.009 | 0.0146 | 874 | 0.059 | 0.013 | 0.033 | 0.085 | 9.79E-06 |
| 18 | 1772503 | rs541861443 | A | T | 0.42 | 856 | -0.078 | 0.019 | -0.116 | -0.041 | 4.75E-05 | 874 | 0.065 | 0.014 | 0.037 | 0.094 | 6.62E-06 |
| 18 | 48657387 | 18:48657387_C_T | T | C | 0.73 | 856 | 0.084 | 0.020 | 0.045 | 0.124 | 3.66E-05 | 874 | -0.074 | 0.015 | -0.104 | -0.045 | 1.21E-06 |
| 20 | 4213175 | rs79601367 | A | G | 0.96 | 856 | 0.160 | 0.045 | 0.071 | 0.249 | 0.0005 | 874 | -0.167 | 0.034 | -0.234 | -0.099 | 1.44E-06 |
| 22 | 32335786 | rs138027893 | A | C | 0.95 | 856 | 0.131 | 0.040 | 0.052 | 0.210 | 0.0012 | 874 | -0.135 | 0.030 | -0.194 | -0.076 | 8.31E-06 |
Beta, effect of Alt allele; Freq, frequency of Alt allele; Spontaneous lipolysis rate was calculated as glycerol release divided by the lipid weight of the incubated fat cells multiplied by the weight of ESAT; Stimulated lipolysis was calculated as the quotient of glycerol release at the maximum effective isoprenaline concentration divided by the spontaneous rate (no hormone present) of glycerol release from the isolated fat cells. Where SNPs were suggestive for both traits, they are listed under spontaneous lipolysis. The most significant SNP in each locus i listed in the table.
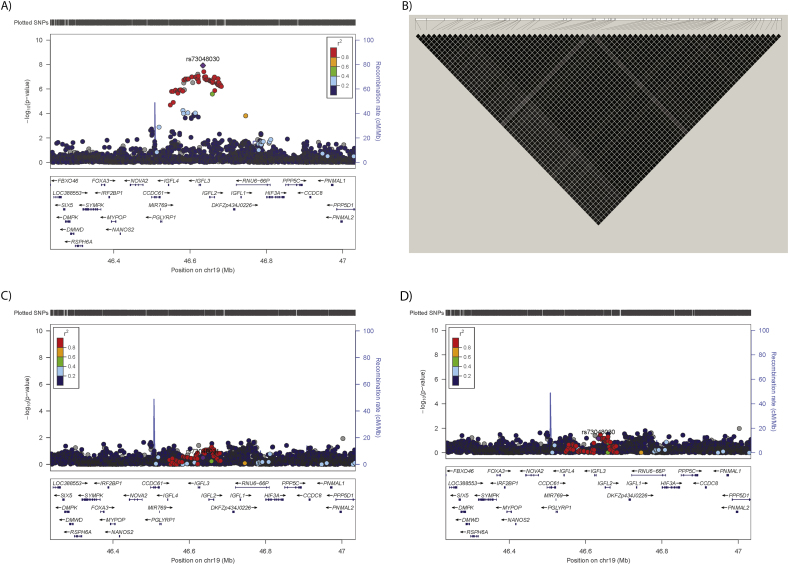
Figure 2.
Regional plot of the GWAS-significant spontaneous lipolysis-associated locus on chromosome 19. A) Regional plot of the GWAS-significant locus, B) linkage disequilibrium (LD) of SNPs meeting the threshold for suggestive significance, and C) regional plot after conditional analyses of the GWAS-significant rs73048030 or D) rs73048031. LD colors and values given in R2.
There was a strong overlap between SNPs associated with spontaneous and stimulated lipolysis; all of the SNPs displaying suggestive associations with spontaneous lipolysis were nominally associated with stimulated lipolysis (Supplementary Table 1), whereas 149 of the 156 SNPs displaying suggestive associations with stimulated lipolysis were nominally associated with spontaneous lipolysis (Supplementary Table 2). This was expected as spontaneous and stimulated lipolysis are correlated (inversely) and share several regulatory steps in signaling to the lipases that ultimately hydrolyze triglycerides [[1], [2], [3]].
Using LDSR, SNP heritability (observed scale) was estimated as 14.2% (h2SNP = 0.1426; SE = 0.468) for spontaneous lipolysis and 26.4% (h2SNP = 0.264; SE = 0.454) for stimulated lipolysis. Of note, the summary statistics meta-data for both spontaneous and stimulated lipolysis suggested that the files were not suitable for genetic correlation analyses (the mean chi-square was too low). Given that the sample size was <1,000 individuals, this was unsurprising.
3.2. Chromosome 19 locus was significantly associated with spontaneous lipolysis
The genome-wide significant spontaneous lipolysis-associated chromosome 19 locus (Figure 2A) encompasses a region of ∼130 Kb that includes two GWAS-significant SNPs and 68 suggestive SNPs (Supplementary Table 3). The linkage disequilibrium (LD) between SNPs meeting the threshold for suggestive evidence of association (Figure 2B) and the non-significant association of these SNPs with spontaneous lipolysis analyses when conditioning on the GWAS-significant SNPs, rs73048030 (Figure 2C), or rs73048031 (Figure 2D) demonstrated that this locus contained only one association signal (Supplementary Table 3). All of the SNPs in the chromosome 19 locus demonstrating GWAS-significant evidence of association with spontaneous lipolysis were assessed for potential functional effects using VEP and ANNOVAR. None of the SNPs were predicted to have significant or deleterious effects on nearby genes. The most severe consequence for each SNP is reported in Supplemental Table 4. The chromosome 19 locus was previously associated with inflammatory and hematological traits (Supplementary Table 5) [[34], [35], [36], [37], [38], [39], [40]]. This locus was also previously associated with lipoprotein disorders in UK Biobank (albeit in a case–control analysis with only 51 cases, http://pheweb.sph.umich.edu/pheno/277.51).
3.3. Expression analysis for chromosome 19 locus candidate gene identification
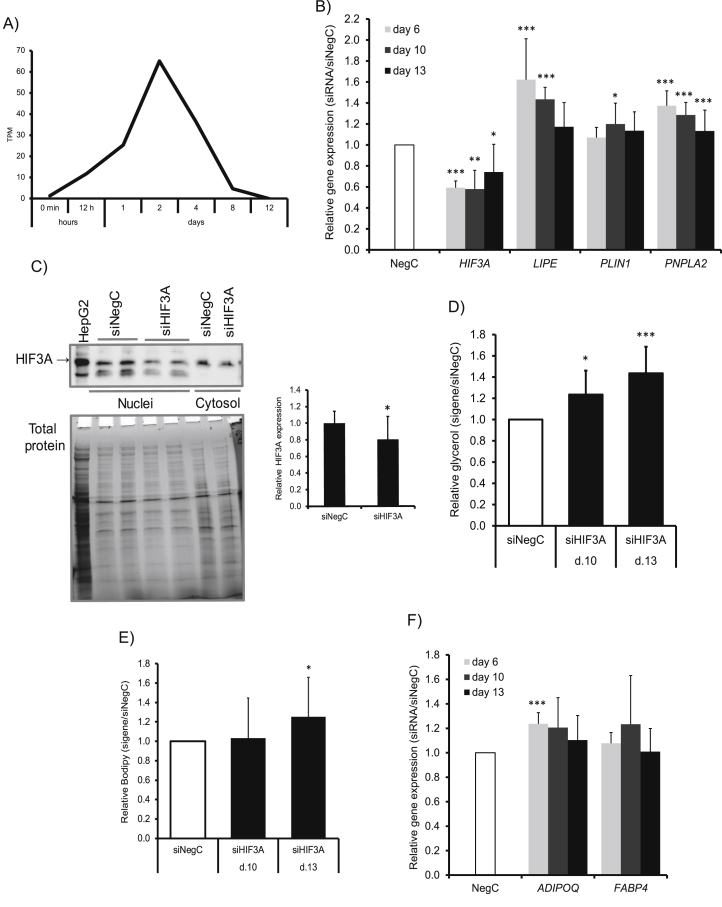
All of the transcripts in the chromosome 19 region bordered by a recombination rate of 25% from the lead SNPs were examined for expression in publicly available transcriptome data of our human adipocyte cultures and in isolated mature abdominal adipocytes from our clinical samples available in the FANTOM5 dataset (http://fantom.gsc.riken.jp/5/) and [29]. Only HIF3A was expressed in the isolated mature adipocytes (Supplementary Table 6) and precursor cells, that is the hMSCs undergoing differentiation to adipocytes (Figure 3A). PPP5C was expressed in the mature adipocytes but not precursors, so was not included in further analysis.
Figure 3.
Functional validation of HIF3A function in human adipocytes. A) HIF3A gene expression during hMSC differentiation in vitro. Data retrieved from FANTOM5 dataset (http://fantom.gsc.riken.jp/5/). B) and D-E) Expression of HIF3A was knocked down using siRNA in hMSCs 24 h after induction of differentiation and followed until differentiation days 6, 10, and 13, upon which B) the expression of HIF3A, LIPE, PLIN1, and PNPLA2 was monitored, D) glycerol amount in medium was measured, E) accumulation of neutral lipids was evaluated, and F) the expression of ADIPOQ and FABP4 was measured. C) The expression of HIF3A was knocked down using siRNA in hMSCs 24 h before induction of adipogenic differentiation of hMSCs; 72 h post-transfection, the cells were collected, fractionated to the cytosolic and nuclear fraction, and the proteins were analyzed by Western blotting. The expression of HIF3A was normalized to the total protein amount. Results in B-E are based on four biological/independent experiments. The expression of genes was normalized to the reference gene 18s. The results were analyzed using t-tests and presented as fold change ± SD relative to negative control of a corresponding time point (Neg C). Results in C) were analyzed using one-sided t-tests and presented as fold change ± SD relative to negative control (Neg C). ∗∗∗P < 0.005, ∗∗P < 0.01, ∗P < 0.05.
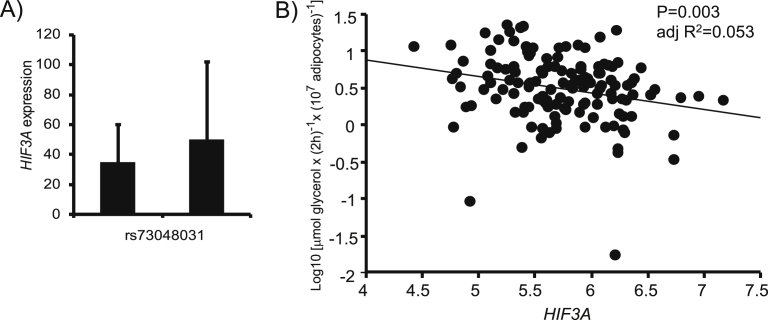
We also examined the genotype-dependent gene expression in the chromosome 19 locus. According to GTEx [27], the genome-wide significant SNPs on chromosome 19, rs73048030, and rs73048031 were eQTLs for IGL3 in the skin, but not for HIF3A in any investigated organ including the white adipose tissue (Supplementary Table 4). Adipose tissue comprises different cell types that can mask an eQTL effect on a single cell type. To test an adipocyte-specific effect of rs73048030 and rs73048031, we measured HIF3A in the isolated frozen adipocytes from GENiAL (N = 75). The common allele of rs73048031-C was marginally associated with higher gene expression (P = 0.076 two-sided; P = 0.038 one-sided) (Figure 4A). Due to the low MAF of rs73048031, no homozygous subject for the rare allele was included in the eQTL analysis. The results did not remain significant if adjusted for age, sex, and BMI, which was not surprising given the limited size of our cohort. The results for rs73048030 were similar although non-significant due to the lower genotype call rate (results not shown). No genotype-specific expression patterns were identified for PPP5C.
Figure 4.
A) Genotype-specific gene expression for HIF3A, with higher levels observed with 2 copies of rs73048031-C. Gene expression expressed relative to 18 S. Genotype groups were compared with Student's t-test. P = 0.076 (two-sided), P = 0.038 (one-sided). B) Relationship between abdominal SAT HIF3A expression and spontaneous lipolysis.
In addition, HIF3A in abdominal SAT in relation to lipolysis was assessed in a subset of the study cohort with lipolysis and gene expression measurements (114 women with a wide variation in BMI). HIF3A expression was inversely correlated with spontaneous lipolysis (Figure 4B). Based on these findings, HIF3A was further functionally evaluated.
3.4. Effect of candidate gene manipulation on adipose tissue biology
The expression of HIF3A during differentiation of hMSCs increased at the very beginning of differentiation, reaching peak expression at day 2 of differentiation and decreasing thereafter to almost the same level as in the beginning of differentiation (Figure 3A). Based on the high expression early in differentiation, we knocked down HIF3A using siRNA in the hMSCs at day 1 and followed the cells to differentiation days 10 and 13. This resulted in 45–50% decreased HIF3A expression at days 5, 9, and 12 post-transfection (that is, days 6, 10, and 13 of differentiation, Figure 3B); thus, approximately 50% of knockdown was left by the end of differentiation. In contrast, the expression levels of the genes central to lipolysis (hormone-sensitive lipase (LIPE ), perilipin 1 (PLIN1), and adipose triglyceride lipase (PNPLA2)) increased by HIF3A knockdown. The expression of LIPE increased by at least 60-40% at days 6 and 10. The expression of PNPLA2 increased by 40, 30, and 15%, respectively, at days 6, 10, and 13. The expression of PLIN1 increased by ∼20% at day 10 only (Figure 3B). The effect of gene silencing naturally gradually diminished due to the long post-transfection time of almost two weeks. We analyzed the effects of siRNA on HIF3A protein at day 2 of differentiation due to its highest expression at the beginning of differentiation. We detected a small but significant downregulation of HIF3A protein in the nuclear fraction but not in the cytosolic fraction (Figure 3C).
We further evaluated the glycerol levels in the conditioned medium as a marker of lipolysis. The glycerol levels increased significantly by approximately 20-50% at days 10 and 13, respectively (Figure 3D). We also evaluated the effects on lipid accumulation of HIF3A knockdown. The amount of neutral lipids increased by 30% at day 13 but not day 10 of differentiation (Figure 3E). Finally, HIF3A knockdown had only minor effects on adipocyte-specific genes. The expression of ADIPOQ increased by 25% at day 6 while the expression of FABP4 was not significantly affected (Figure 3F).
3.5. Analysis of previous clinically relevant associations for lipolysis-associated genetic loci
We next assessed whether lipolysis-associated SNPs in the present study were associated with obesesity-related clinical traits. None of the suggestive lipolysis-associated SNPs were previously reported to be associated with obesity (BMI, WHR, and % body fat) or related metabolic traits (type 2 diabetes, insulin resistance, and plasma lipids) according to the GWAS catalogue (https://www.ebi.ac.uk/gwas/, 20190410). However, a few SNPs were located in genes or within +/−100 Kbps of SNPs reported to be associated with these traits, for example, a locus on chromosome 2 harboring METAP1D was found to be associated with % body fat [41] and loci on chromosome 15 (SLCO3A1) and 18 with BMI [8,42] (Supplementary Table 7).
3.6. Data mining of spontaneous or stimulated lipolysis loci
The SNPs displaying suggestive associations with spontaneous or stimulated lipolysis were next investigated for genotype-specific gene expression (or eQTLs). Among the SNPs represented in GTEx, the SNPs associated with spontaneous lipolysis also demonstrated genotype-specific expression patterns on chromosome 2 for PLGLB1 in the SAT as well as on chromosome 19 for PPP5D1 in the brain and IGFL3 in the skin (Supplementary Table 4 and Supplementary Table 8). SNPs in six loci associated with stimulated lipolysis demonstrated eQTLs. The most interesting was rs9892812 on chromosome 17, which influenced gene expression in the SAT of NUP85, GGA3, and MRPS7 (Supplementary Table 4 and Supplementary Table 8). Other notable findings include rs11147005 on chromosome 12, the P2RX2 levels in the visceral adipose tissue, and many SNPs on chromosome 10 influencing ATE1 levels in the skeletal muscle (Supplementary Table 4 and Supplementary Table 8). Brief descriptions of genes highlighted by these analyses are presented in Supplementary Table 9. None of the eQTL-associated genes have established roles in lipolysis regulation. There are many caveats to consider when interpreting eQTL data (for example, the sample size, data collection time and method, and tissue selection); however, these data are included to be as comprehensive as possible.
4. Discussion
In this first GWAS of human fat cell lipolysis, we identified one genetic locus significantly associated with spontaneous lipolysis and multiple additional loci with suggestive associations with spontaneous and stimulated lipolysis. Furthermore, we provided evidence of HIF3A as a candidate gene with a key role in regulating the expression of genes central to lipolysis.
Regulation of SAT lipolysis is not well understood. Prior genetic studies implicated coding variants in or near genes encoding proteins with established and specific roles in lipolysis and glycerol release [43]. In contrast, in this unique cohort, we identified the SNPs associated with lipolysis that are not located in or near genes encoding proteins with established roles in lipolysis. This is in line with studies of other complex traits in which a GWAS of a well-defined phenotype can highlight novel biology. That prior studies were not validated herein suggests that they may represent spurious associations due to underpowered study designs.
While several nominal loci were identified, for robustness, we focused on the genome-wide significant locus on chromosome 19. This is a gene-dense region with high LD. Among the genes in this locus, only HIF3A was expressed in mature and precursor adipocytes and constituted an eQTL and was thus the focus of further evaluation. We provided functional support for transcription factor HIF3A explaining the association between the chromosome 19 locus and lipolysis: knockdown of HIF3A in hMSCs increased glycerol release and, consistent with this, increased the expression of genes encoding the major lipolysis-regulating lipases, that is, LIPE and PLNPLA2. Furthermore, the major lipid droplet-coating protein in fat cells, PLIN1, which is also critical for lipolysis, was influenced by HIF3A knockdown. In agreement with an inhibitory effect of HIF3A on lipolysis, we observed an inverse correlation between HIF3A expression and SAT spontaneous lipolysis in women and borderline nominally significant (P < 0.05) association with the rs73048031 genotype, whereby the spontaneous lipolysis-decreasing allele (C) was associated with increased HIF3A gene expression levels. We were unable to exclude the possibility that genes encoding secreted proteins could control lipolysis through distant-acting systemic effects, for example, IGFL3, which is primarily expressed in the skin and comprises a cis-eQTL in this organ for the lipolysis-associated SNPs on chromosome 19. However, there was no evidence that IGFL3 was present in the general circulation, making this scenario less likely.
The lipolysis-associated SNPs on chromosome 19 were not previously reported to be associated with clinical traits involving altered lipolysis, for example, adiposity, insulin resistance, or dyslipidemia, according to the GWAS catalogue. Furthermore, mice with a targeted disruption of the HIF3A locus displayed cardiac and pulmonary remodeling [44], but to the best of our knowledge, no phenotypes related to altered lipid or glucose metabolism have been reported. However, the HIF3A locus more generally has previously been associated with lipoprotein subclasses in Finns [45], which might be related to dysfunctional lipolysis. Furthermore, the influence of HIF3A on lipolysis could go beyond the genetic effects. In addition to hypoxia, HIF3A expression has also been reported to be regulated by the anti-lipolytic hormone insulin [46]. Furthermore, SAT CpG methylation of HIF3A has been associated with gene expression and BMI [47]. Also, reported results supported the hypothesis that HIF3A methylation is secondary to, rather than causal of, obesity [48]. We cannot exclude the possibility that the SNPs in the lipolysis-associated locus influenced DNA methylation. HIF3A could possibly represent a mechanistic link between obesity and metabolic complications since SAT gene expression is inversely correlated with systemic insulin resistance [49].
Hypoxia inducible factors (HIFs) are heterodimeric transcription factors that regulate adaptive responses to low oxygen tension [50]. Ectopic expression of HIF3A has been shown to enhance the adipogenic potential in mouse fat cell line 3T3-L1 cells [51]. In this study, knockdown of HIF3A did not suppress markers of adipocytes (FABP4 and ADIPOQ) and increased lipid accumulation, suggesting that the effects are species dependent. HIF3A binding motifs are not sufficiently well described to determine via bioinformatics analyses whether HIF3A directly represses LIPE, PNPLA2, and PLIN1. Experiments to address this were beyond the scope of the present study.
How regulation of lipolysis is related to traits such as obesity and insulin resistance is an area of obvious interest. However, the many loci influencing metabolic traits likely act through a variety of different mechanisms, with only a handful acting through lipolysis. Therefore, the gold standard methods (genetic correlation and polygenic risk scores) that consider the whole genome are inappropriate and could be misleading. Consideration of specific loci could provide more accurate estimates; however, defining the choice of loci is complex. For example, only the loci with evidence of genotype-specific gene expression patterns in adipose tissue could be considered, or with supportive experimental data, but these methods are fraught with issues. Expression data are limited in sample size, collection time points, and tissue selection, to name only a few concerns. Similarly, only a fraction of loci has been thoroughly studied experimentally. While this information provides additional evidence, there is a significant amount of bias that should be considered in their interpretation (while the observed associations could be true, lack of associations cannot be taken as true findings).
To date, lipolysis has not been extensively studied, and consensus in methodology is lacking. The lipolysis measurements used in this study were those most reflective of (best correlations with) clinical parameters. The limitations of this study include lack of replication, as we are unaware of any other datasets with comparable phenotypes of sufficiently large size to provide reasonable power for replication. The size of the cohort might be the reason for only finding one genome-wide significant locus for the lipolysis phenotypes. Nevertheless, the validity of our findings is supported by first, consistent effects of suggestive findings on the two phenotypes. That is, the effects on spontaneous lipolysis were consistently the inverse of the effects on stimulated lipolysis. Second, many SNPs showed effects on gene expression in adult adipose tissues. Finally, manipulation of the candidate gene expression levels in the GWAS-significant locus demonstrated clear effects on lipolysis. HIF3A has multiple splice variants, the expression of which are epigenetically regulated [52]. Whether there are also transcript-specific genetic effects on HIF3A expression remains to be determined. Finally, this study was underpowered for sex-specific analysis.
5. Conclusions
In conclusion, this first GWAS of adipocyte lipolysis identified a locus on chromosome 19 that was significantly associated with spontaneous lipolysis, demonstrated that genetic regulation of spontaneous and stimulated lipolysis overlaps, and provided evidence for HIF3A as a strong candidate gene with a novel and key regulatory function in SAT lipolysis in humans. We also highlighted multiple additional genetic loci with suggestive associations with lipolysis, including some that were previously linked to body fat storage and distribution.
6. Data availability
Summary statistics for the spontaneous and stimulated lipolysis GWAS are available upon request.
Acknowledgments
We thank all of the participants and staff involved in the GENiAL study. RJS is supported by a UKRI Innovation at HDR-UK Fellowship (MR/S003061/1). CML is supported by the Li Ka Shing Foundation; the NIHR Biomedical Research Center, Oxford; Widenlife; the NIH (5P50HD028138-27); and has collaborated with Novo Nordisk and Bayer in research, and in accordance with the policy of the University of Oxford, did not accept any personal payment. This study was supported by the Strategic Research Program in Diabetes at Karolinska Institute, Swedish Research Council, Novo Nordisk Foundation, and the Swedish Diabetes Association.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.01.009.
Author contributions
RJS designed the study, conducted the analysis, interpreted the data, and drafted the manuscript. AK and VL conducted the experiments, interpreted the data, and drafted the manuscript. JAL and CL oversaw imputation and edited the manuscript. CML edited the manuscript. ID and PA acquired the data, conceived and designed the study, interpreted the data, and drafted the manuscript. All of the authors agreed to be accountable for their contributions and approved the final manuscript.
Conflict of interest
None declared.
Declaration of interests
None.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Arner P. Human fat cell lipolysis: biochemistry, regulation and clinical role. Best Practice & Research Clinical Endocrinology & Metabolism. 2005;19:471–482. doi: 10.1016/j.beem.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Fruhbeck G., Mendez-Gimenez L., Fernandez-Formoso J.A., Fernandez S., Rodriguez A. Regulation of adipocyte lipolysis. Nutrition Research Reviews. 2014;27:63–93. doi: 10.1017/S095442241400002X. [DOI] [PubMed] [Google Scholar]
- 3.Lafontan M., Langin D. Lipolysis and lipid mobilization in human adipose tissue. Progress in Lipid Research. 2009;48:275–297. doi: 10.1016/j.plipres.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Girousse A., Tavernier G., Valle C., Moro C., Mejhert N., Dinel A.L. Partial inhibition of adipose tissue lipolysis improves glucose metabolism and insulin sensitivity without alteration of fat mass. PLoS Biology. 2013;11 doi: 10.1371/journal.pbio.1001485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryden M., Arner P. Subcutaneous adipocyte lipolysis contributes to circulating lipid levels. Arteriosclerosis, Thrombosis, and Vascular Biology. 2017;37:1782–1787. doi: 10.1161/ATVBAHA.117.309759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryden M., Arner P. Cardiovascular risk score is linked to subcutaneous adipocyte size and lipid metabolism. Journal of Internal Medicine. 2017;282:220–228. doi: 10.1111/joim.12641. [DOI] [PubMed] [Google Scholar]
- 7.Arner P., Andersson D.P., Backdahl J., Dahlman I., Ryden M. Weight gain and impaired glucose metabolism in women are predicted by inefficient subcutaneous fat cell lipolysis. Cell Metabolism. 2018;28:45–54 e3. doi: 10.1016/j.cmet.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S. Discovery and refinement of loci associated with lipid levels. Nature Genetics. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Despres J.P., Bouchard C. Monozygotic twin resemblance in fatness and fat cell lipolysis. Acta Geneticae Medicae et Gemellologiae. 1984;33:475–480. doi: 10.1017/s0001566000005924. [DOI] [PubMed] [Google Scholar]
- 12.Stich V., Harant I., De Glisezinski I., Crampes F., Berlan M., Kunesova M. Adipose tissue lipolysis and hormone-sensitive lipase expression during very-low-calorie diet in obese female identical twins. The Journal of Cinical Endocrinology and Metabolism. 1997;82:739–744. doi: 10.1210/jcem.82.3.3793. [DOI] [PubMed] [Google Scholar]
- 13.Dahlman I., Ryden M., Arner P. Family history of diabetes is associated with enhanced adipose lipolysis: evidence for the implication of epigenetic factors. Diabetes & Metabolism. 2018;44:155–159. doi: 10.1016/j.diabet.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Civelek M., Wu Y., Pan C., Raulerson C.K., Ko A., He A. Genetic regulation of adipose gene expression and cardio-metabolic traits. The American Journal of Human Genetics. 2017;100:428–443. doi: 10.1016/j.ajhg.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahlman I., Ryden M., Brodin D., Grallert H., Strawbridge R.J., Arner P. Numerous genes in loci associated with body fat distribution are linked to adipose function. Diabetes. 2016;65:433–437. doi: 10.2337/db15-0828. [DOI] [PubMed] [Google Scholar]
- 16.Matthews D.R., Hosker J.P., Rudenski A.S., Naylor B.A., Treacher D.F., Turner R.C. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 17.Kolaczynski J.W., Morales L.M., Moore J.H., Jr., Considine R.V., Pietrzkowski Z., Noto P.F. A new technique for biopsy of human abdominal fat under local anaesthesia with Lidocaine. International Journal of Obesity and Related Metabolic Disorders. 1994;18:161–166. [PubMed] [Google Scholar]
- 18.Andersson D.P., Arner E., Hogling D.E., Ryden M., Arner P. Abdominal subcutaneous adipose tissue cellularity in men and women. International Journal of Obesity. 2017;41:1564–1569. doi: 10.1038/ijo.2017.148. [DOI] [PubMed] [Google Scholar]
- 19.Lofgren P., Hoffstedt J., Naslund E., Wiren M., Arner P. Prospective and controlled studies of the actions of insulin and catecholamine in fat cells of obese women following weight reduction. Diabetologia. 2005;48:2334–2342. doi: 10.1007/s00125-005-1961-6. [DOI] [PubMed] [Google Scholar]
- 20.Hellmer J., Arner P., Lundin A. Automatic luminometric kinetic assay of glycerol for lipolysis studies. Analytical Biochemistry. 1989;177:132–137. doi: 10.1016/0003-2697(89)90027-4. [DOI] [PubMed] [Google Scholar]
- 21.Arner P. Relationship between intracellular cyclic AMP and lipolysis in human adipose tissue. Acta Medica Scandinavica. 1976;200:179–186. doi: 10.1111/j.0954-6820.1976.tb08217.x. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loh P.R., Danecek P., Palamara P.F., Fuchsberger C., A Reshef Y., K Finucane H. Reference-based phasing using the haplotype reference consortium panel. Nature Genetics. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R. An atlas of genetic correlations across human diseases and traits. Nature Genetics. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A. The ensembl variant effect predictor. Genome Biology. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium G.T. The genotype-tissue expression (GTEx) project. Nature Genetics. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettersson A.M., Stenson B.M., Lorente-Cebrián S., Andersson D.P., Mejhert N., Krätzel J. LXR is a negative regulator of glucose uptake in human adipocytes. Diabetologia. 2013;56:2044–2054. doi: 10.1007/s00125-013-2954-5. [DOI] [PubMed] [Google Scholar]
- 29.Arner E., Daub C.O., Vitting-Seerup K., Andersson R., Lilje B., Drabløs F. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundback V., Kulyte A., Strawbridge R.J., Ryden M., Arner P., Marcus C. FAM13A and POM121C are candidate genes for fasting insulin: functional follow-up analysis of a genome-wide association study. Diabetologia. 2018;61:1112–1123. doi: 10.1007/s00125-018-4572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiao H., Kulyté A., Näslund E., Thorell A., Gerdhem P., Kere J. Whole-exome sequencing suggests LAMB3 as a susceptibility gene for morbid obesity. Diabetes. 2016;65:2980–2989. doi: 10.2337/db16-0522. [DOI] [PubMed] [Google Scholar]
- 32.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Lorente-Cebrián S., Mejhert N., Kulyté A., Laurencikiene J., Åström G., Hedén P. MicroRNAs regulate human adipocyte lipolysis: effects of miR-145 are linked to TNF-alpha. PloS One. 2014;9 doi: 10.1371/journal.pone.0086800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429 e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christophersen I.E., Magnani J.W., Yin X., Barnard J., Weng L.C., Arking D.E. Vol. 10. Circ Cardiovasc Genet; 2017. Fifteen genetic loci associated with the electrocardiographic P wave. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law P.J., Berndt S.I., Speedy H.E., Camp N.J., Sava G.P., Skibola C.F. Genome-wide association analysis implicates dysregulation of immunity genes in chronic lymphocytic leukaemia. Nature Communications. 2017;8:14175. doi: 10.1038/ncomms14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nature Genetics. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mez J., Chung J., Jun G., Kriegel J., Bourlas A.P., Sherva R. Two novel loci, COBL and SLC10A2, for Alzheimer's disease in African Americans. Alzheimers Dement. 2017;13:119–129. doi: 10.1016/j.jalz.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vishal M., Sharma A., Kaurani L., Alfano G., Mookherjee S., Narta K. Genetic association and stress mediated down-regulation in trabecular meshwork implicates MPP7 as a novel candidate gene in primary open angle glaucoma. BMC Medical Genomics. 2016;9:15. doi: 10.1186/s12920-016-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hübel C., Gaspar H.A., Coleman J.R.I., Finucane H., Purves K.L., Hanscombe K.B. Genomics of body fat percentage may contribute to sex bias in anorexia nervosa. Am J Med Genet B Neuropsychiatr Genet. 2018 doi: 10.1002/ajmg.b.32709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kichaev G., Bhatia G., Loh P.R., Gazal S., Burch K., Freund M.K. Leveraging polygenic functional enrichment to improve GWAS power. The American Journal of Human Genetics. 2019;104:65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dahlman I., Arner P. Genetics of adipose tissue biology. Prog Mol Biol Transl Sci. 2010;94:39–74. doi: 10.1016/B978-0-12-375003-7.00003-0. [DOI] [PubMed] [Google Scholar]
- 44.Yamashita T., Ohneda O., Nagano M., Iemitsu M., Makino Y., Tanaka H. Abnormal heart development and lung remodeling in mice lacking the hypoxia-inducible factor-related basic helix-loop-helix PAS protein NEPAS. Molecular and Cellular Biology. 2008;28:1285–1297. doi: 10.1128/MCB.01332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis J.P., Huyghe J.R., Locke A.E., Jackson A.U., Sim X., Stringham H.M. Common, low-frequency, and rare genetic variants associated with lipoprotein subclasses and triglyceride measures in Finnish men from the METSIM study. PLoS Genetics. 2017;13 doi: 10.1371/journal.pgen.1007079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heidbreder M., Qadri F., Jöhren O., Dendorfer A., Depping R., Fröhlich F. Non-hypoxic induction of HIF-3alpha by 2-deoxy-D-glucose and insulin. Biochemical and Biophysical Research Communications. 2007;352:437–443. doi: 10.1016/j.bbrc.2006.11.027. [DOI] [PubMed] [Google Scholar]
- 47.Dick K.J., Nelson C.P., Tsaprouni L., Sandling J.K., Aïssi D., Wahl S. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 48.Potabattula R., Dittrich M., Schorsch M., Hahn T., Haaf T., El Hajj N. Male obesity effects on sperm and next-generation cord blood DNA methylation. PloS One. 2019;14 doi: 10.1371/journal.pone.0218615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Main A.M., Gillberg L., Jacobsen A.L., Nilsson E., Gjesing A.P., Hansen T. DNA methylation and gene expression of HIF3A: cross-tissue validation and associations with BMI and insulin resistance. Clinical Epigenetics. 2016;8:89. doi: 10.1186/s13148-016-0258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greer S.N., Metcalf J.L., Wang Y., Ohh M. The updated biology of hypoxia-inducible factor. The EMBO Journal. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hatanaka M., Shimba S., Sakaue M., Kondo Y., Kagechika H., Kokame K. Hypoxia-inducible factor-3alpha functions as an accelerator of 3T3-L1 adipose differentiation. Biological and Pharmaceutical Bulletin. 2009;32:1166–1172. doi: 10.1248/bpb.32.1166. [DOI] [PubMed] [Google Scholar]
- 52.Heikkila M., Pasanen A., Kivirikko K.I., Myllyharju J. Roles of the human hypoxia-inducible factor (HIF)-3alpha variants in the hypoxia response. Cellular and Molecular Life Sciences. 2011;68:3885–3901. doi: 10.1007/s00018-011-0679-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics for the spontaneous and stimulated lipolysis GWAS are available upon request.