Abstract
Medulloblastoma (MB) is the most common and deadliest brain tumor in children. Proline-, glutamic acid-, and leucine-rich protein 1 (PELP1) is a scaffolding protein and its oncogenic signaling is implicated in the progression of several cancers. However, the role of PELP1 in the progression of MB remains unknown. The objective of this study is to examine the role of PELP1 in the progression of MB. Immunohistochemical analysis of MB tissue microarrays revealed that PELP1 is overexpressed in the MB specimens compared to normal brain. Knockdown of PELP1 reduced cell proliferation, cell survival, and cell invasion of MB cell lines. RNA-Seq analysis revealed that PELP1 knockdown significantly downregulated the pathways related to inflammation, and extracellular matrix. Gene set enrichment analysis (GSEA) confirmed that the PELP1-regulated genes were negatively correlated with NF-κB, extracellular matrix, and angiogenesis gene sets. Interestingly, PELP1 knockdown reduced the expression of NF-κB target genes, NF-κB reporter activity, and inhibited the nuclear translocation of p65. Importantly, knockdown of PELP1 significantly reduced in vivo MB progression in orthotopic models and improved the overall mice survival. Collectively, these results suggest that PELP1 could be a novel target for therapeutic intervention in MB.
Keywords: Medulloblastoma, oncogene, PELP1, NF-κB, coactivator
INTRODUCTION
Medulloblastoma (MB) is the most common and deadliest primary brain tumor in children, and accounts for approximately 20% of all brain tumors in children under 19 years of age1,2. The cerebellum is the primary initiation site for MB and more than 80% of cases are diagnosed in children under the age of 15 years with the peak age between 6–8 years3. The annual incidence of MB is ~5 cases per 1 million individuals4,5. MB is heterogeneous with distinct histopathological and molecular sub groups and arise from distinct neuronal stem or progenitor cell populations during early life3.
The standard of care consists of surgical resection followed by craniospinal irradiation and adjuvant chemotherapy6. Despite recent advances in multimodal treatment, the 5-year overall survival of MB patients is approximately 60–70%. Unfortunately, improved outcome has been associated with significant long-term toxicities3. New therapies targeting aberrant signaling pathways in MB such as Sonic Hedgehog, Wnt, Myc, and growth factor signaling pathways are currently being investigated, however, they showed limited efficacy and are associated with significant toxic effects4,7. New therapies are urgently needed to improve outcomes and quality of life for MB patients. Understanding the molecular mechanisms and identifying the novel targets that drive the MB progression are necessary to target these tumors.
Proline glutamic acid and leucine rich protein 1 (PELP1) is a scaffolding protein that regulates several signaling pathways and many biological processes8. Structurally, PELP1 has 10 nuclear receptor interacting boxes LXXLL motifs that facilitate its interaction with nuclear receptors (NRs), proline rich motifs that facilitate interactions with SH3 domain-containing proteins, and C-terminus glutamic acid containing region which assists in interactions with histone proteins 8–10. PELP1 regulates gene expression by functioning as a coregulator of many transcriptional factors and NRs including estrogen receptor (ER), androgen receptor (AR), progesterone receptor (PR), and glucocorticoid receptor (GR) and signal transducer and activator of transcription 3 (STAT3)12,13. PELP1 is shown to play a key role in several biological functions such as transcriptional regulation by NRs, chromatin remodeling, cell cycle, DNA damage response, ribosome biogenesis, and RNA splicing8.
PELP1 is expressed in several tissues with highest expression noted in ovary, testis, mammary gland, uterus, and brain9. PELP1 is essential for estrogen mediated neuroprotective and cognitive functions in the brain via modulation of cellular survival, antioxidant, and inflammatory pathways14–16. Several recent studies have identified that PELP1 is overexpressed in multiple cancer types including breast17,18, prostate19, ovarian20, colon21, pancreas22, lung23, endometrial24, and salivary25. However, the role and significance of PELP1 in MB remains unknown.
In this study, we examined the role of PELP1 in the progression of MB. Using immunohistochemical analysis of MB tissue microarrays, we showed that PELP1 is highly expressed in MB compared to normal tissues. Functional assays demonstrated that PELP1 knockdown attenuated proliferation, survival, and invasiveness of MB cells. Mechanistically, PELP1 mediates NF-κB transactivation functions. Importantly knockdown of PELP1 improved the mice overall survival in orthotopic MB xenograft models.
2. MATERIALS AND METHODS
2.1. Cell lines and reagents
Human MB cell lines Daoy, and D556 were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and were maintained as per ATCC guidelines. All the MB cell lines were passaged in the user’s laboratory for fewer than 15 passages after receipt or resuscitation. All the cells were devoid of mycoplasma contamination and were confirmed by using mycoplasma PCR Detection Kit (Sigma-Aldrich St. Louis, MO). Cells identity was confirmed with Short Tandem Repeat (STR) polymorphism analysis using UT Health San Antonio Core facilities. Non-targeted siRNA and PELP1 siRNA were obtained from Thermo Fisher Scientific (Waltham, MA). The PELP1 antibody was purchased from Bethyl Laboratories (Montgomery, TX) and p-p65 (Ser536), p65, p-IKKα/β, IKKα, IKKβ, p-IκB and GAPDH antibodies were obtained from Cell Signaling Technology (Beverly, MA). PELP1 specific shRNA lentivirus, β-actin antibody and secondary antibodies were procured from Sigma-Aldrich (St. Louis, MO). Ki67 antibody was obtained from Abcam (Cambridge, MA) and MMP9 antibody was purchased from Millipore (Burlington, MA). Corning® BioCoat™ Growth Factor Reduced Matrigel Invasion Chamber assay was purchased from Thermo Fisher Scientific. CellTiter-Glo® Luminescent Cell Viability Assay kit and Dual Luciferase Assay system was obtained from Promega (Madison, WI). MB cells stably expressing control shRNA- and PELP1-shRNA were generated using human specific lentiviral non-targeted shRNA and PELP1-shRNA particles respectively and stable cells were selected with puromycin (1 μg/mL).
2.2. Tissue microarrays and immunohistochemistry
The MB tissue microarrays (TMAs) were obtained from US BioMax (Rockville, MD). Each TMA comprised 0.6-mm cores taken from paraffin-embedded specimens that represent a total of 20 cases/60 cores of MB of the brain, along with 3 normal brain tissues. Immunohistochemical staining of PELP1 was performed as described previously28. Briefly, tissue arrays were incubated in xylene and passed through a series of graded alcohols. Tissues were then subjected to antigen retrieval using the antigen retrieval solution (Vector Labs, Burlingame, CA) followed by incubation with 3% H2O2 solution for 20 min. Next, tissues were subjected to blocking using the Vector Lab Blocking Kit (Vector Labs) and incubated overnight with PELP1 primary antibody followed by incubation with secondary antibody for 30 minutes. The immunopositivity was developed by using the DAB substrate and counterstained with hematoxylin (Vector Labs). PELP1 immunoreactivity on TMA was scored using Allred scoring system38. Briefly, the PELP1 staining intensity was scored on a scale between zero and three and the proportion of positive stained cells was rated as one between 0 and 1%, two between 1 and 10%, three between 10 and 33%, four between 33 and 66%, and five between 66 and 100%. For mice experiments, orthotopic xenograft tumor sections were incubated overnight with Ki67 and MMP9 primary antibodies followed by secondary antibody incubation for 45 minutes. The immunopositivity was developed by using the DAB substrate and counterstained with hematoxylin.
2.3. Cell viability, proliferation, and clonogenic assays
The cell viability rates of MB cells that were transfected with control siRNA or PELP1 siRNA or transduced with either control shRNA or PELP1 shRNA were measured by using Cell Titer-Glo Luminescent Cell Viability Assay. Cells (2 × 103 cells/well) were seeded in 96-well, flat, clear-bottom, opaque-wall microplates and after various time intervals total ATP content as an estimate of total number of viable cells was measured on automatic Fluoroskan Luminometer according to manufacturer’s instructions. The cell proliferation rates of control or PELP1 knockdown cells were measured using IncuCyte Label-Free Cell Proliferation Assay with IncuCyte® Live-Cell Analysis System as per the manufacturer’s instructions. For the clonogenic assays, Daoy and D556 cells stably expressing either control shRNA or PELP1 shRNA (500 cells/well) were seeded in 6-well plates and after 14 days cells were then fixed with ice cold methanol followed by staining with 0.5% crystal violet solution. Colonies which contain ≥ 50 cells were counted and used in the analysis.
2.4. Cell invasion assays
The invasion ability of MB cells that were transduced with either control shRNA or PELP1 shRNA was determined using Corning® BioCoat™ Growth Factor Reduced Matrigel Invasion Chamber assay according to the manufacturer’s instructions. Briefly, 2 × 104 cells were seeded in the upper chamber in serum free medium and with 10% FBS containing media as a chemo attractant in bottom well. After 12 hours, the invaded cells on the bottom side of the membrane were fixed in methanol and stained with 0.5% crystal violet. The number of invaded cells in 5 random fields were counted and used for quantitative analysis.
2.5. Cell lysis and Western blotting
Whole cell lysates were prepared from MB cells using RIPA buffer containing protease and phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO). Lysates were mixed with 4X SDS sample buffer to run on SDS-PAGE gels. The resolved proteins were then transferred onto nitrocellulose membranes and the blots were blocked with 5% non-fat dry milk powder for 1 hour at room temperature. Primary antibody incubation was carried out at 4 °C for overnight followed by incubation with secondary antibodies for 1 hour at room temperature. Blots were developed using the ECL kit (Thermo Fisher Scientific, Waltham, MA).
2.6. RNA-Sequencing and RT-qPCR analysis
Daoy cells stably expressing either control shRNA or PELP1 shRNA were subjected to RNA isolation using the RNeasy mini kit according to the manufacturer’s protocol (Qiagen, Valencia, CA). The RNA purity was determined by using Agilent 2100 BioAnalyzer. Illumina TruSeq RNA Sample preparation was performed following manufacturer’s protocol, and the samples were run on an Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA) in duplicates as described previously26. Differential gene expression analysis was conducted using DESeq, and significant genes with at least 2.0-fold change with adjusted p<0.05 were chosen for analysis. The expression of selected genes was determined using quantitative real-time-PCR (RT-qPCR) using gene-specific primer sequences obtained from Harvard Primer Bank (http://pga.mgh.harvard.edu/primerbank/). cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) and RT-qPCR was performed using SYBR Green (Thermo Fisher Scientific) on an Illumina Real-Time PCR system. The gene expression levels were normalized to the house keeping gene β-actin transcript levels and the difference in fold expression was calculated by using delta-delta-CT method.
2.7. Reporter gene assays
Reporter gene assays were performed as described earlier27. Daoy, D556 and HEK293T model cells were seeded in 24 well plates and after overnight incubation, transiently transfected with 250 ng of NF-κB-Luc reporter vector along with PELP1 expressing or control vector using Turbofect transfection reagent (Thermo Fisher Scientific). Renilla reporter (50 ng) plasmid was co-transfected and used for normalization of transfection efficiency. For some assays, after a 48 hour transfection, cells were stimulated with 10 ng of TNF-α for 24 hours. Cells were lysed in Luciferase Lysis Buffer, and the luciferase activity was measured by using the dual luciferase assay system (Promega, Madison, WI) with a luminometer.
2.8. Immunocytochemistry
Daoy cells that stably express control shRNA or PELP1 shRNA were seeded onto 8 well chamber slides in serum free medium and after 12 hours cells were stimulated with TNF-α (20ng/ml) for 1 h. Cells were then washed with PBS followed by fixation with 4% paraformaldehyde and permeabilization with 0.2% Triton X-100. Cells were blocked in 5% normal goat serum followed by incubation with p65 primary antibody and fluorochrome-conjugated secondary antibody (Alexa Fluor 488). DAPI was used to visualize the nuclei and images were captured using confocal microscopy.
2.9. In vivo orthotopic tumor model
All mice experiments were performed after obtaining UT Health San Antonio Institutional Animal Care and Use Committee approval. Male and female athymic nude mice of approximately 8 to 10 weeks old were purchased from Charles River Laboratories (Wilmington, MA). Daoy cells expressing either control shRNA or PELP1 shRNA were labeled with GFP-Luciferase reporter and 1 × 106 cells were injected orthotopically into the cerebellum of a mouse with the coordinates of 1.5mm lateral, 1.5mm posterior from lambda and 3.0mm deep. Tumor progression was monitored weekly using the Xenogen in vivo Imaging System. Mice were euthanized and brains collected when mice become moribund or neurological deficits appeared. Mice survival was determined using Kaplan–Meier survival curves and log-rank test using GraphPad Prism 6 software.
2.10. Statistical analysis
Statistical differences between the groups were analyzed with unpaired Student’s t-test and one-way ANOVA using GraphPad Prism 6 software. All the data represented in the graphs are shown as means ± SE. A value of P < 0.05 was considered as statistically significant.
3. Results
3.1. PELP1 is overexpressed in MB
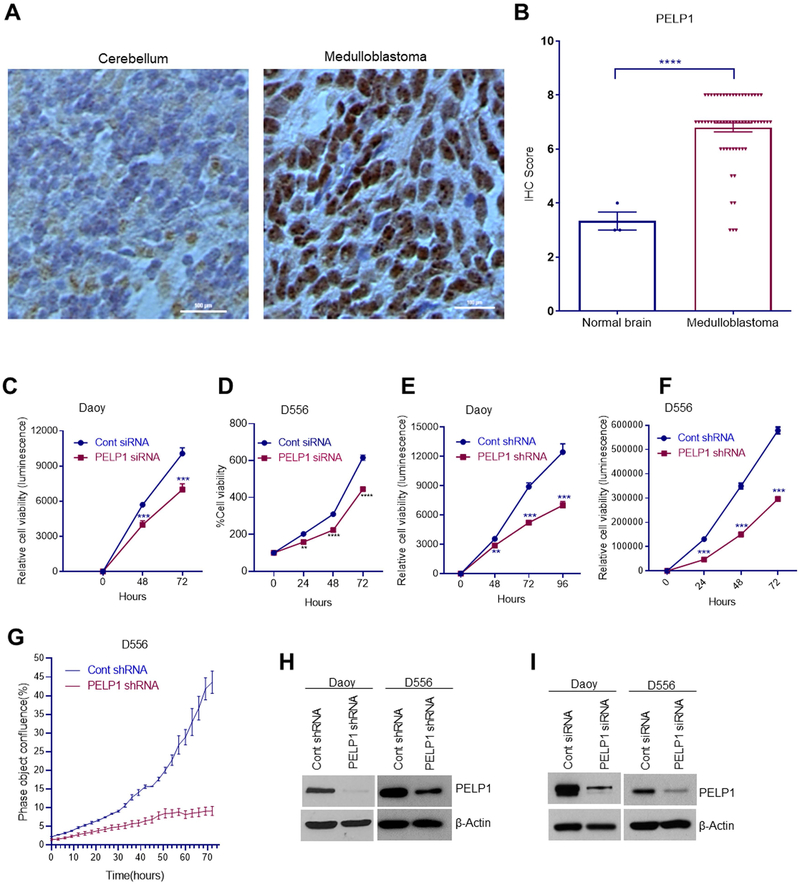
To examine the expression of PELP1 in MB, we performed immunohistochemistry (IHC) using tissue microarrays that consist of various MB and normal brain samples. IHC score was calculated as described previously29. Representative images of PELP1 staining in control and MB was shown in Fig. 1A and the quantitation of IHC was represented in Fig. 1B. MB samples exhibited high levels of PELP1 expression compared to normal brain which exhibited weaker PELP1 staining suggesting that PELP1 expression is upregulated in MB.
Figure 1.
PELP1 is highly expressed in MB and PELP1 knockdown reduced the cell viability of MB cells. A-B, MB tissue micro array that consists of normal control brain (n=3), and MB (n=60) samples were subjected to immunohistochemical staining with PELP1 antibody. B, The intensity and positivity of PELP1 staining was quantitated as described in methods section. Daoy and D556 cells were transfected with control siRNA or PELP1 siRNA (C,D) or stably expressing control shRNA or PELP1 shRNA (E,F) were examined for cell viability using Cell Titer-Glo assay. G, The cell proliferation rates of D556 cells stably expressing control shRNA or PELP1 shRNA were examined using IncuCyte Label-Free Cell Proliferation Assay. H,I, Knockdown of PELP1 and silencing of PELP1 in Daoy and D556 cells was confirmed by western blotting. Data are represented as mean ± SE of three independent experiments. ** p<0.01; *** p<0.001. ****p<0.0001.
3.2. PELP1 knockdown reduces the cell viability, proliferation, survival, and invasion of MB cells
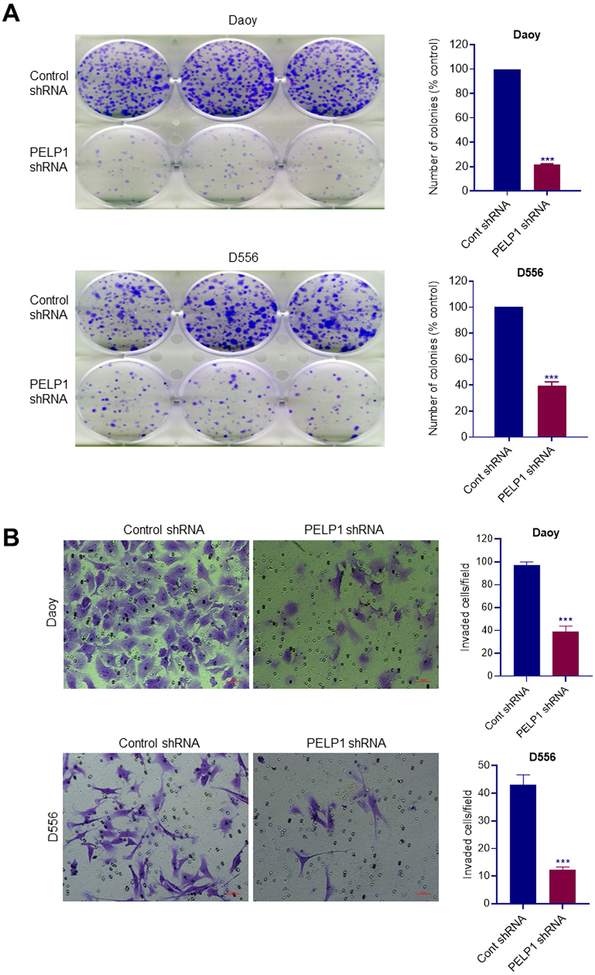
To examine the significance of PELP1 in MB we performed a knockdown of PELP1 expression. MB cells were transfected either with PELP1 siRNA or shRNA that target distinct sites on PELP1, and their viability rates were examined using Cell Titer-Glo assay. The silencing of PELP1 in MB cells using PELP1-siRNA (Fig. 1C, D) or PELP1-shRNA (Fig. 1E, F) resulted in significant reduction in cell viability compared to controls. To determine whether PELP1 knockdown modulates the proliferation of MB cells, we performed IncuCyte Label-Free Cell Proliferation Assay with IncuCyte® Live-Cell Analysis System. As shown in Fig. 1G, the knockdown of PELP1 significantly reduced the proliferation of MB cells compared to controls. Knockdown of PELP1 in these model cells was confirmed using western blotting (Fig. 1H, I). Next, we examined the effect of PELP1 knockdown on cell survival using colony formation assays. Knockdown of PELP1 significantly reduced the colony forming ability of MB cells compared to control shRNA cells (Fig. 2A). Since MB are highly invasive in nature, we next determined the effect of PELP1 knockdown on the invasion of MB cells. Invasion assay of control and PELP1 knockdown cells revealed that PELP1 knockdown significantly reduced the invasion of MB cells compared to control cells (Fig. 2B). These results suggest that PELP1 is essential for cell viability, proliferation, survival, and invasion of MB cells.
Figure 2.
PELP1 knockdown reduced the cell survival and invasion of MB cells. A, Daoy and D556 cells stably expressing control shRNA or PELP1 shRNA were subjected to colony formation assay and the number of colonies in each group was quantitated. B, The invasion ability of Daoy and D556 cells stably expressing control shRNA or PELP1 shRNA were examined using BioCoat Matrigel invasion chamber assays. Data are represented as mean ± SE of three independent experiments. *** p<0.001..
3.3. Transcriptomic analysis of PELP1 regulated genes in MB
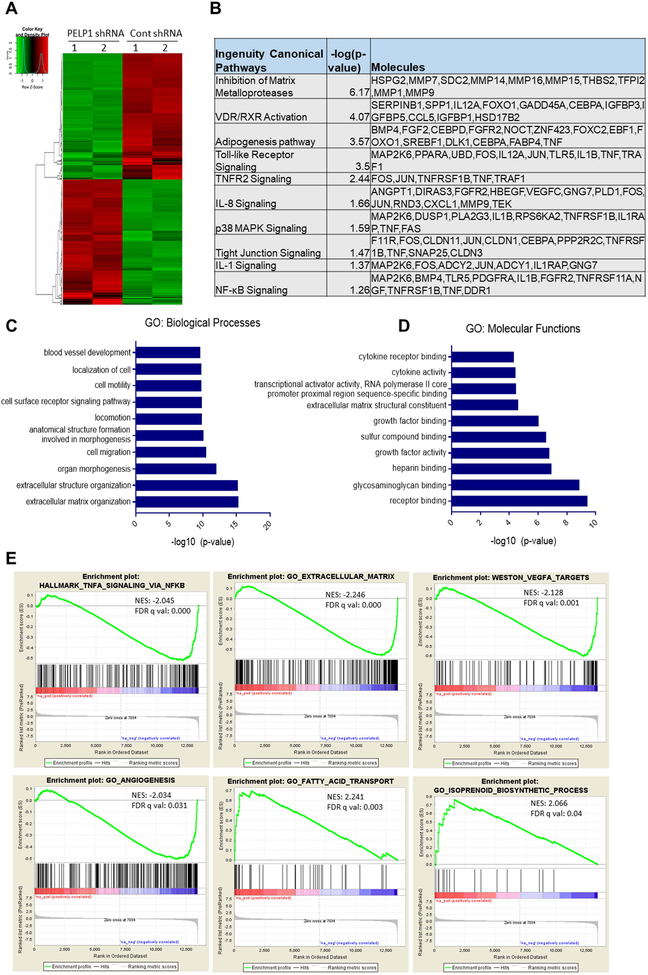
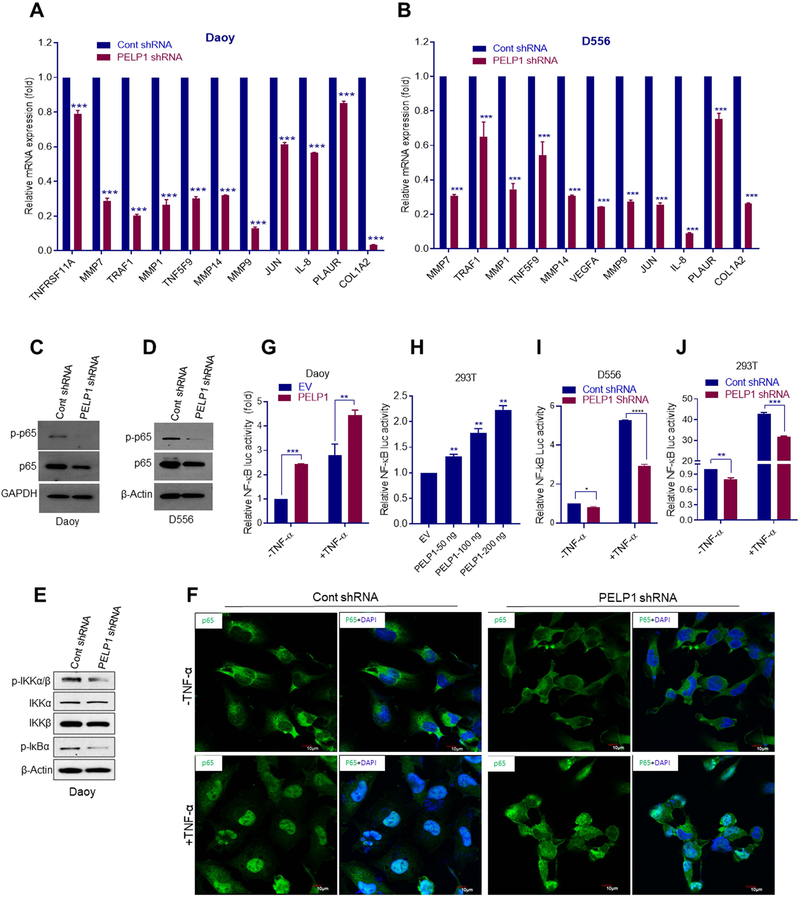
To find out the mechanism of PELP1 mediated functions in MB, we performed global transcriptomic analysis using control and PELP1 knockdown cells. Differential gene expression analysis identified that 845 genes were significantly altered between control and PELP1 knockdown cells with fold change difference >2.0 and adjusted p-value 0.05 of which 421were upregulated and 424 were down regulated. The complete list of sequenced genes is available in the GEO database under accession number GSE131347. The differentially expressed genes between control and PELP1 knockdown were shown in the heat map (Fig. 3A). The biological significance of differentially expressed genes was studied using ingenuity pathway analysis (IPA). The IPA of differentially expressed genes demonstrated that PELP1 regulated genes were related to inflammatory processes and extra cellular matrix: such as inhibition of matrix metalloproteases, toll like receptor signaling, TNFR2 signaling, IL-8 signaling, and NF-kB signaling (Fig. 3B). To further understand the significance of differentially expressed genes enrichment, further analysis was performed using Gene Ontology (GO). The results showed that the differentially expressed genes were enriched in biological processes such as extracellular matrix organization, cell migration and cell motility (Fig. 3C). In terms of molecular function, the differentially expressed genes were mainly enriched in receptor binding, extracellular matrix structural constituent, cytokine activity, and cytokine receptor binding (Fig. 3D). Importantly, GSEA demonstrated that PELP1 regulated genes showed negative correlation with the gene sets of TNF signaling via NF-κB, extracellular matrix, angiogenesis and VEGF targets and positive correlation with the gene sets of fatty acid transport, and isoprenoid biosynthetic process (Fig. 3E). Next, we validated the expression of selected genes related to these pathways using RT-qPCR in Daoy and D556 cell lines. As shown in Fig. 4A, B the expression of several NF-κB targets that are involved in extracellular matrix and invasion such as MMP7, MMP14, MMP9, PLAUR, MMP1 and other NF-κB targets such as IL-8, TNFRSF11A, TRAF1, JUN and COL1A2 were significantly downregulated in PELP1 knockdown MB cells compared to control cells. Collectively, these results suggest that PELP1 knockdown reduced the expression of NF-κB pathways genes involved on cell invasion, extracellular matrix, and angiogenesis.
Figure 3.
Analysis of global transcriptional changes modulated by PELP1 in MB cells. RNA isolated from Daoy cells stably expressing control shRNA or PELP1 shRNA was subjected to RNA-sequencing as described in methods. A, Heat map of differentially expressed genes between control shRNA and PELP1 shRNA groups was shown. B, Differentially expressed genes between the groups were subjected to pathway analysis employing IPA software, and the selected top canonical pathways were shown. Gene ontology (GO) analysis of differentially expressed genes in terms of either biological processes (C) or molecular functions (D) was examined using DAVID software. E, Gene set enrichment analysis (GSEA) testing correlation of PELP1-regulated genes with signatures of NF-κB signaling, extracellular matrix, angiogenesis, VEGF targets, fatty acid transport and isoprenoid biosynthetic process gene sets.
Figure 4.
PELP1 modulate NF-κB signaling. NF-κB signaling regulated genes were validated using RNA isolated from Daoy (A) and D556 (B) cells stably expressing control shRNA or PELP1 shRNA using RTqPCR. C-D, The expression of p-p65 and p65 levels was examined in Daoy and D556 cells stably expressing control shRNA or PELP1 shRNA using western blotting. E. The expression of phosphorylated IKKα/β and IκB were examined in Daoy cells stably expressing control shRNA or PELP1 shRNA using western blotting. F. Daoy cells that stably express control shRNA or PELP1 shRNA were serum starved for 12 h and stimulated with TNF-α (20 ng/ml) for 1 h and subjected to immunofluorescence staining using p65 antibody. Fluorescence was captured using confocal microscope. Daoy (G) and HEK293T (H) model cells were transfected with NF-κB-luciferase and renilla vectors along with either empty vector or PELP1 expression vector and after 48 hours luciferase reporter activity was measured with or without TNF-α stimulation. I,J, D556 and HEK293T cells that stably express control shRNA or PELP1 shRNA were transfected with NF-κB-luciferase and renilla vectors and after 48 hours, luciferase reporter activity was measured with or without TNF-α stimulation. Data are represented as mean ± SE of three independent experiments. ** p<0.01; *** p<0.001.
3.4. PELP1 activates NF-κB pathway
Previous studies have demonstrated that PELP1 functions as coactivator for several transcriptional factors13. To determine whether PELP1 modulates NF-κB activation, we examined the activation status of the NF-κB subunit p65. As shown in Fig. 4C, D, the levels of phosphorylated p65 which is the active form was significantly reduced in PELP1 knockdown MB cells compared to controls. The activation of NF-κB is tightly regulated by IκB which inhibits NF-κB nuclear translocation and activation. In response to stimuli such as TNF-α, IκB is phosphorylated by IκB kinase (IKK) complex resulting in its degradation and subsequent nuclear translocation of NF-κB. To study whether PELP1 knockdown affects the phosphorylation of IKKα/β and IκB, we determined their phosphorylation levels in control and PELP1 knockdown Daoy cells. As shown in Fig. 4E, PELP1 knockdown significantly reduced the phosphorylation of IKKα/β and IκB compared to control cells. To further confirm the inhibition of nuclear translocation of NF-κB subunit p65, Daoy cells that stably express control shRNA or PELP1 shRNA were stimulated with TNF-α and subjected to immunofluorescence analysis. As shown in Fig. 4F, we observed prominent nuclear localization of p65 with TNF-α stimulation in control cells, however in PELP1 shRNA cells p65 nuclear translocation was substantially compromised. These results suggest that PELP1 knockdown attenuated the activation and nuclear translocation of p65 via modulation of upstream kinases.
Next, we examined whether PELP1 regulates the transactivation functions of NF-κB using NF-κB luciferase reporter assays. As shown in Fig. 4G, co-expression of PELP1 and the reporter gene in Daoy cells significantly increased the NF-κB reporter activity. In addition, we observed that stimulation with TNF-α further potentiated the NF-κB reporter activity in PELP1 transfected cells compared to control (Fig. 4G). Similarly, we observed increased NF-κB reporter activity in 293T cells following PELP1 transfection (Fig. 4H). Further we observed that the NF-κB reporter activity is significantly attenuated in D556 and 293T cells that stably express PELP1 shRNA compared to control cells (Fig. 4I, J). Collectively, these results suggested that PELP1 has potential to activate the NF-κB pathway in MB cells.
3.5. PELP1 knockdown reduces MB progression in vivo and improved mice survival
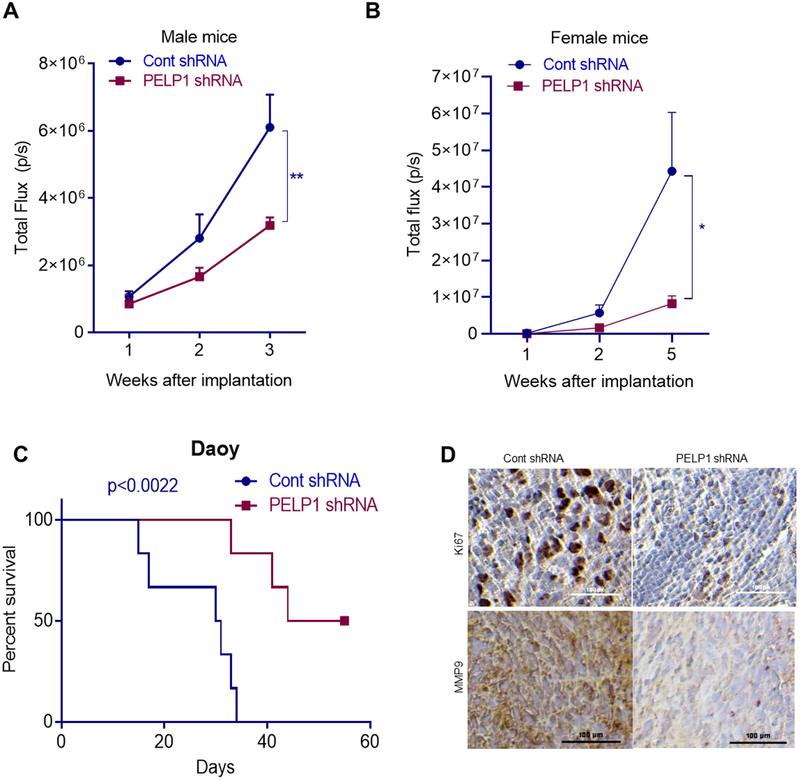
We next examined whether PELP1 knockdown decreases in vivo MB growth using mouse orthotopic models. Daoy cells stably expressing control shRNA or PELP1 shRNA were labelled with luciferase and orthotopically injected into the cerebellum of male and female mice. The progression of MB was examined using Xenogen in vivo Imaging System. PELP1 knockdown significantly decreased tumor progression in both male and female tumor bearing mice when compared to controls (Fig. 5A. B). Further, we recorded the survival for male tumor bearing mice and analyzed the data using Kaplan-Meier curves. Survival analysis of tumor bearing mice demonstrated the increased survival in PELP1 shRNA group compared to the mice injected with control shRNA cells (Fig. 5C). Next, we studied the expression levels of proliferation marker Ki67 in the tumors of control and PELP1 shRNA groups using immunohistochemistry. The number of Ki67 positive proliferating cells were significantly lower in PELP1 shRNA tumors compared to control shRNA tumors (Fig. 5D). In addition, we examined the expression of NF-κB target gene MMP9 in tumors immunohistochemically. As shown in Fig. 5D, the expression of MMP9 was significantly downregulated in PELP1 knockdown Daoy tumors compared to control shRNA-expressing tumors. Altogether, these results suggest that PELP1 knockdown reduced in vivo MB progression and improved the mice survival via attenuation of NF-κB pathway.
Figure 5.
PELP1 knockdown reduced MB progression and enhanced mice survival in orthotopic MB model. Daoy cells that stably express control shRNA or PELP1 sRNA were labeled with luciferase reporter and implanted orthotopically into the right cerebellum of male (A) and female (B) athymic nude mice. The tumor growth in terms of luminescence was examined using Xenogen in vivo imaging system. C, The survival of the control shRNA and PELP1 shRNA tumor bearing male mice was recorded and plotted using Kaplan-Meier survival curve (n=6). D, Mice brains collected from Daoy control shRNA and PELP1 shRNA groups, were fixed in formalin and processed for immunohistochemical staining for Ki67 and MMP9.
4. Discussion
PELP1 is a coregulator protein that regulates transactivation functions of many transcriptional factors and provides cancer cells with a distinct growth and survival advantage in many neoplasms. Importantly PELP1 is expressed in the brain and is essential for neuroprotective and cognitive functions. Although, PELP1 plays a vital role in many human malignancies, little is known about its role in MB. In this study, we provided evidence that PELP1 expression is upregulated in MB and is essential for MB cell survival and invasion. Importantly, PELP1 knockdown reduced the expression of several genes involved in cell adhesion, extracellular matrix, and invasion particularly those regulated by NF-κB. Finally, PELP1 knockdown significantly improved the overall survival of tumor bearing mice.
It has been shown that PELP1 expression is negatively correlated with patient survival in several cancer types. In ER positive breast cancer PELP1 is an independent prognostic predictor of shorter breast cancer specific survival30. In TNBC, dual expression of PELP1/Ki-67 is an independent prognostic factor for predicting poorer outcome31. PELP1 expression levels were upregulated in non-small cell lung cancer and correlated with degree of malignancy23. Furthermore, ESR2-PELP1 axis has shown to play an essential role in colorectal tumorigenesis and might have prognostic significance32. PELP1 is overexpressed in astrocytic tumors in small cohort of patients and serve as a prognostic biomarker33. Our results demonstrate that PELP1 is highly expressed in MB tissues compared to normal brain specimens are corroborated with findings observed in other cancers.
PELP1 functions are vital in oncogenic processes such as cell cycle, survival, metastasis, hormone therapy resistance, and autophagy8. Several reports have shown that PELP1 downregulation significantly reduced the proliferation, survival, migration and invasion of cancer cells in multiple cancer types12,34,35. Recently, we demonstrated that PELP1 overexpression contributes to mammary gland carcinoma using a tissue specific transgenic mouse model36. PELP1 plays a role in ER positive metastases via extranuclear ER-Src-PELP1-ILK1 pathway and knockdown of PELP1 reduced the invasion and metastasis of TNBC cells37,38. In this study we observed that knockdown of PELP1 reduced the cell proliferation, survival, and invasion of MB cells, supporting the previously observed effects of PELP1 knockdown in other cancer types. Further, PELP1 knockdown significantly enhanced the survival of MB xenograft bearing mice.
Our global transcriptomic analysis results demonstrated that knockdown of PELP1 modulates several pathways of inflammation and extracellular matrix regulation which are mediated by NF-κB signaling. GSEA results further demonstrated that PELP1 modulated gene sets were negatively correlated with NF-κB signaling, extracellular matrix, and angiogenesis gene sets. Recent studies demonstrated that NF-κB is overexpressed in MB, plays a vital role in MB growth and its inhibition reduced MB cell viability and growth both in vitro and in vivo models39. NF-κB pathway is a key mediator of many of inflammatory processes40,41. NF-κB is a family of dimeric transcription factors that play an important role in regulation of diverse biological processes such as immune responses, inflammation, cell proliferation and apoptosis42. The NF-κB pathway activates various genes involved in cell proliferation, apoptosis, adhesion, and angiogenesis (http://www.bu.edu/nf-kb/target-genes/)43. In our study, we observed downregulation of various NF-κB targets that are involved in cell invasion, adhesion, and cell proliferation such as MMP1, MMP7, MMP9, VEGFA, PLAUR, IL-8 JUN, and COL1A2 in PELP1 knockdown cells. These results suggested that PELP1 plays an essential role in the activation of NF-κB target genes.
A recent study by Girard et al. demonstrated that PELP1 regulates the NF-κB pathway in immortalized human mammary epithelial cells44. In this study, the authors demonstrated that cytoplasmic expression of PELP1 promotes breast cancer initiation via induction of non-canonical NF-κB signaling pathway via the up-regulation of inhibitor of κB kinase ϵ (IKKϵ). Our global RNA-Seq results demonstrated that the target genes of canonical NF-κB and phosphorylation of p65 were significantly reduced following PELP1 knockdown. Furthermore, we also observed that PELP1 knockdown resulted in reduction in phosphorylation of IKKα/β and IκB. In support of these observations, we observed that PELP1 co-expression enhances the NF-κB reporter activity, while PELP1 knockdown reduced NF-κB target genes in MB cells. Additionally, we observed that the expression of NF-κB target genes, such as MMP9, is significantly downregulated in PELP1 knockdown tumors compared to controls. These results suggest that PELP plays a role in canonical NF-κB signaling. However, future studies are needed to further dissect the mechanism of PELP1 regulation of NF-κB pathway and are beyond the scope of the current investigation.
To the best of our knowledge, this represents the first study demonstrating PELP1 expression upregulated in MB and contributes to MB progression via the modulation of NF-κB signaling. Collectively, the results of this study significantly advance our understanding of PELP1 mediated oncogenic functions in MB and suggest PELP1 may represent a novel therapeutic target in MB.
Funding sources
This study was supported by National Institute of Health grants (CA178499, P30CA054174–17; RKV); Max and Minnie Tomerlin Voelcker Fund (G.R.S) and CPRIT Predoctoral Fellowship RP170345 (KAA).
Footnotes
Data accessibility
All the data generated and/or analyzed during the current study are included in this article and are available from the corresponding author on reasonable request. RNA-Seq data has been deposited in the GEO database under a GEO accession number GSE131347.
Ethics statement
All animal experiments were performed in accordance to IACUC standards and by using approved protocols at UT Health San Antonio.
Conflict of Interest: The authors declare no potential conflicts of interest
References
- 1.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(2):83–103. [DOI] [PubMed] [Google Scholar]
- 2.Smoll NR, Drummond KJ. The incidence of medulloblastomas and primitive neurectodermal tumours in adults and children. Journal of clinical neuroscience : official journal of the Neurosurgical Society of Australasia. 2012;19(11):1541–1544. [DOI] [PubMed] [Google Scholar]
- 3.Northcott PA, Robinson GW, Kratz CP, et al. Medulloblastoma. Nature reviews Disease primers. 2019;5(1):11. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Garancher A, Ramaswamy V, Wechsler-Reya RJ. Medulloblastoma: From Molecular Subgroups to Molecular Targeted Therapies. Annual review of neuroscience. 2018;41:207–232. [DOI] [PubMed] [Google Scholar]
- 5.Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro-oncology. 2018;20(suppl_4):iv1–iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramaswamy V, Taylor MD. Medulloblastoma: From Myth to Molecular . Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2017;35(21):2355–2363. [DOI] [PubMed] [Google Scholar]
- 7.Ramaswamy V, Remke M, Bouffet E, et al. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta neuropathologica. 2016;131(6):821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sareddy GR, Vadlamudi RK. PELP1: Structure, biological function and clinical significance. Gene. 2016;585(1):128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vadlamudi RK, Wang RA, Mazumdar A, et al. Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor alpha. J Biol Chem. 2001;276(41):38272–38279. [DOI] [PubMed] [Google Scholar]
- 10.Nair SS, Mishra SK, Yang Z, Balasenthil S, Kumar R, Vadlamudi RK. Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res. 2004;64(18):6416–6423. [DOI] [PubMed] [Google Scholar]
- 11.Gonugunta VK, Nair BC, Rajhans R, Sareddy GR, Nair SS, Vadlamudi RK. Regulation of rDNA transcription by proto-oncogene PELP1. PLoS One. 2011;6(6):e21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girard BJ, Daniel AR, Lange CA, Ostrander JH. PELP1: a review of PELP1 interactions, signaling, and biology. Mol Cell Endocrinol. 2014;382(1):642–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vadlamudi RK, Kumar R. Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl Recept Signal. 2007;5:e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sareddy GR, Zhang Q, Wang R, et al. Proline-, glutamic acid-, and leucine-rich protein 1 mediates estrogen rapid signaling and neuroprotection in the brain. Proc Natl Acad Sci U S A. 2015;112(48):E6673–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thakkar R, Sareddy GR, Zhang Q, Wang R, Vadlamudi RK, Brann D. PELP1: a key mediator of oestrogen signalling and actions in the brain. Journal of neuroendocrinology. 2018;30(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thakkar R, Wang R, Sareddy G, et al. NLRP3 Inflammasome Activation in the Brain after Global Cerebral Ischemia and Regulation by 17beta-Estradiol. Oxidative medicine and cellular longevity. 2016;2016:8309031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajhans R, Nair S, Holden AH, Kumar R, Tekmal RR, Vadlamudi RK. Oncogenic potential of the nuclear receptor coregulator proline-, glutamic acid-, leucine-rich protein 1/modulator of the nongenomic actions of the estrogen receptor. Cancer Res. 2007;67(11):5505–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnan SR, Nair BC, Sareddy GR, et al. Novel role of PELP1 in regulating chemotherapy response in mutant p53-expressing triple negative breast cancer cells. Breast Cancer Res Treat. 2015;150(3):487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, Ravindranathan P, Ramanan M, et al. Central role for PELP1 in nonandrogenic activation of the androgen receptor in prostate cancer. Mol Endocrinol. 2012;26(4):550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dimple C, Nair SS, Rajhans R, et al. Role of PELP1/MNAR signaling in ovarian tumorigenesis. Cancer Res. 2008;68(12):4902–4909. [DOI] [PubMed] [Google Scholar]
- 21.Ning Z, Zhang Y, Chen H, et al. PELP1 suppression inhibits colorectal cancer through c-Src downregulation. Oxid Med Cell Longev. 2014;2014:193523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kashiwaya K, Nakagawa H, Hosokawa M, et al. Involvement of the tubulin tyrosine ligase-like family member 4 polyglutamylase in PELP1 polyglutamylation and chromatin remodeling in pancreatic cancer cells. Cancer Res. 2010;70(10):4024–4033. [DOI] [PubMed] [Google Scholar]
- 23.Slowikowski BK, Galecki B, Dyszkiewicz W, Jagodzinski PP. Increased expression of proline-, glutamic acid- and leucine-rich protein PELP1 in non-small cell lung cancer. Biomed Pharmacother. 2015;73:97–101. [DOI] [PubMed] [Google Scholar]
- 24.Vadlamudi RK, Balasenthil S, Broaddus RR, Gustafsson JA, Kumar R. Deregulation of estrogen receptor coactivator proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor in human endometrial tumors. J Clin Endocrinol Metab. 2004;89(12):6130–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vadlamudi RK, Balasenthil S, Sahin AA, et al. Novel estrogen receptor coactivator PELP1/MNAR gene and ERbeta expression in salivary duct adenocarcinoma: potential therapeutic targets. Hum Pathol. 2005;36(6):670–675. [DOI] [PubMed] [Google Scholar]
- 26.Sareddy GR, Viswanadhapalli S, Surapaneni P, Suzuki T, Brenner A, Vadlamudi RK. Novel KDM1A inhibitors induce differentiation and apoptosis of glioma stem cells via unfolded protein response pathway. Oncogene. 2017;36(17):2423–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Viswanadhapalli S, Garcia L, et al. Therapeutic utility of natural estrogen receptor beta agonists on ovarian cancer. Oncotarget. 2017;8(30):50002–50014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Sareddy GR, Zhou M, et al. Differential Effects of Estrogen Receptor beta Isoforms on Glioblastoma Progression. Cancer Res. 2018;78(12):3176–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sareddy GR, Nair BC, Krishnan SK, et al. KDM1 is a novel therapeutic target for the treatment of gliomas. Oncotarget. 2013;4(1):18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Habashy HO, Powe DG, Rakha EA, et al. The prognostic significance of PELP1 expression in invasive breast cancer with emphasis on the ER-positive luminal-like subtype. Breast Cancer Res Treat. 2010;120(3):603–612. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Dai J, McNamara KM, et al. Prognostic significance of proline, glutamic acid, leucine rich protein 1 (PELP1) in triple-negative breast cancer: a retrospective study on 129 cases. BMC Cancer. 2015;15:699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grivas PD, Tzelepi V, Sotiropoulou-Bonikou G, Kefalopoulou Z, Papavassiliou AG, Kalofonos H. Expression of ERalpha, ERbeta and co-regulator PELP1/MNAR in colorectal cancer: prognostic significance and clinicopathologic correlations. Cellular oncology : the official journal of the International Society for Cellular Oncology. 2009;31(3):235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kefalopoulou Z, Tzelepi V, Zolota V, et al. Prognostic value of novel biomarkers in astrocytic brain tumors: nuclear receptor co-regulators AIB1, TIF2, and PELP1 are associated with high tumor grade and worse patient prognosis. Journal of neuro-oncology. 2012;106(1):23–31. [DOI] [PubMed] [Google Scholar]
- 34.Gonugunta VK, Miao L, Sareddy GR, Ravindranathan P, Vadlamudi R, Raj GV. The social network of PELP1 and its implications in breast and prostate cancers. Endocr Relat Cancer. 2014;21(4):T79–86. [DOI] [PubMed] [Google Scholar]
- 35.Ravindranathan P, Lange CA, Raj GV. Minireview: Deciphering the Cellular Functions of PELP1. Mol Endocrinol. 2015;29(9):1222–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cortez V, Samayoa C, Zamora A, Martinez L, Tekmal RR, Vadlamudi RK. PELP1 overexpression in the mouse mammary gland results in the development of hyperplasia and carcinoma. Cancer Res. 2014;74(24):7395–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakravarty D, Nair SS, Santhamma B, et al. Extranuclear functions of ER impact invasive migration and metastasis by breast cancer cells. Cancer Res. 2010;70(10):4092–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy S, Chakravarty D, Cortez V, et al. Significance of PELP1 in ER-negative breast cancer metastasis. Mol Cancer Res. 2012;10(1):25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spiller SE, Logsdon NJ, Deckard LA, Sontheimer H. Inhibition of nuclear factor kappa-B signaling reduces growth in medulloblastoma in vivo. BMC cancer. 2011;11:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capece D, Verzella D, Tessitore A, Alesse E, Capalbo C, Zazzeroni F. Cancer secretome and inflammation: The bright and the dark sides of NF-kappaB. Seminars in cell & developmental biology. 2018;78:51–61. [DOI] [PubMed] [Google Scholar]
- 41.Liu T, Zhang L, Joo D, Sun SC. NF-kappaB signaling in inflammation. Signal transduction and targeted therapy. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harbor perspectives in biology. 2009;1(4):a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Molecular cancer. 2013;12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girard BJ, Knutson TP, Kuker B, McDowell L, Schwertfeger KL, Ostrander JH. Cytoplasmic Localization of Proline, Glutamic Acid, Leucine-rich Protein 1 (PELP1) Induces Breast Epithelial Cell Migration through Up-regulation of Inhibitor of kappaB Kinase and Inflammatory Cross-talk with Macrophages. The Journal of biological chemistry. 2017;292(1):339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]