Abstract
The translocator protein (TSPO), an 18-kDa transmembrane protein primarily found in the outer mitochondrial membrane, is evolutionarily conserved and widely distributed across species. In mammals, TSPO has been described as a key member of a multiprotein complex involved in many putative functions and over the years several classes of ligands have been developed to modulate these functions. This review considers the currently available atomic structures of mouse and bacterial TSPO and proposes a rationale for the development of new ligands for the protein. A review of TSPO monomeric and oligomeric states and their conformational flexibility, together with ligand binding site and interaction mechanisms, is provided. These data are expected to help the development of high-affinity ligands for TSPO-based therapies or diagnostics considerably.
Keywords: positron emission tomography, nuclear magnetic resonance, X-ray crystallography, protein ligand interactions, protein flexibility
TSPO: a pharmacological target
The translocator protein (TSPO), originally discovered in 1977 as a second target of the benzodiazepine diazepam [1], is an 18-kDa transmembrane protein. TSPO is an evolutionarily conserved protein widely distributed in most Eukarya, Archae and Bacteria, which can be traced back to 3.5 billion years ago [2]. In humans, under stress or inflammatory conditions, TSPO is overexpressed both in the central nervous system (CNS) [3–4] and in the peripheral nervous system (PNS) [5]. Therefore, TSPO appears as a diagnostic target for many brain diseases. A similar relationship between TSPO expression and stress regulation has been observed in plants under abiotic stress [6] and bacteria under oxidative stress/redox imbalance [7], suggesting a conserved function along evolution [8].
In mammals, TSPO has been described as a key member of a multiprotein complex involved in many putative functions (such as the synthesis of steroid hormones and heme, apoptosis, cell proliferation [1]), and several classes of ligands (see Glossary) have been developed to modulate these functions [1, 9]. TSPO was also shown to be involved in cell signalling and has been related to apoptosis and autophagy process [10]. TSPO levels are usually constitutively high in several organs, with an over-expression in glial cells and cancer which makes it suitable as diagnostic marker and drug target [11–12]. In healthy human brain, TSPO level is low, but is up-regulated under various neuropathological conditions including injury, stroke and neurodegenerative disorders [11,13]. However, it is paradoxically decreased in some psychiatric disorders. [3,14] Therefore, while TSPO has become an important diagnostic and therapeutic target, mostly in brain [3–5,14,15], via the identification and development of several classes of chemical entities that bind TSPO, it presents therapeutic challenges.
The structure of TSPO is formed by five transmembrane α helices tightly assembled with a pocket accepting ligand in between the bundle [16–19]. Although a number of studies tried to identify the specific domain of TSPO where the ligands bind, a number of amino acid sequences spread across the five transmembrane (TM) domains and their connecting loops were found to contribute to drug-ligand binding [20]. Thus, the true target sequence within TSPO for these ligands remains difficult to characterize and the ligand binding mechanism to TSPO itself remains unclear. Moreover, the discovery of a cholesterol-recognition amino acid consensus (CRAC) domain, binding cholesterol [21] with high affinity [22–23] in the C-terminus of the TM5 helix of TSPO defined a second ligand binding domain, which was also used to identify chemical entities binding and blocking cholesterol binding [24].
Developing ligands for TSPO
Currently known TSPO ligands have neuroprotective and regenerative properties [9,25]. TSPO exo- and endogeneous ligands stimulate neurosteroids [26–27], for example, allopregnenolone production, active in stress adaptation and treatment of posttraumatic stress disorders [28]. TSPO exogenous ligands enhance cholesterol efflux in choroidal endothelial cells, reduce reactive oxygen species (ROS) production and suppress inflammation and, thus, may have potential benefits for aged-related macular degeneration (AMD) patients [29].
Since the discovery of endogeneous molecules (such as cholesterol, porphyrins and endozepines) that interact with TSPO, various classes of synthetic ligands have been developed to improve the binding specificity or genotype sensitivity of ligands used as therapeutic drugs or to improve their labelling for imaging (ie PBR28, new carboxamide analogs, metal complexes [1,5,9,15]. While they belong to different structural families, all are heterocyclic with at least one nitrogen atom, and all have one or more carbonyl (C=O) group. For example, the prototypical TSPO ligand, PK 11195 [30] is part of the isoquinoline-carboxamide family (Box1, Figure IA).
The successful development of TSPO ligands for therapeutic and diagnostic purposes requires the answers to several questions: (1) What is the basal expression of TSPO versus pathologic overexpression? We know that there is elevated expression of TSPO in peripheral tissues, whereas protein expression is low in healthy brain and restricted to glial cells [31], but increases with age and brain diseases [31–33]. TSPO is also lowly expressed during homeostasis in immune cells but benzodiazepines, another class of TSPO ligands, modulate oxidative burst by neutrophils and macrophages [34].
(2) When imaging with positron emission tomography (PET), what is the accessibility of the target protein to the TSPO ligand-based PET probe, as well as the ratio of specific to non-specific probe binding [35]? An example is Ro5-4864, a well-characterized benzodiazepine TSPO ligand that failed to demonstrate PET imaging in brain [36], probably because of low affinity and high non-specific binding. However, Ro5-4864 has numerous physiological effects such as brain injuries [9] and can be docked to specific TSPO atomic structures [37] and hence it has been kept as a potential therapeutic but not as a diagnostic using PET. Another example to consider would be the a circumstance where TSPO might be in the plasma membrane of astrocytes in CNS [38] or mitochondrial membrane in PNS [5] and thus has different accessibility. Therefore, future TSPO ligands developments should correlate in vivo and in vitro binding to both the accessibility and the time that the ligands spend in contact with TSPO [39].
(3) How stable are the PET probes and the TSPO ligands themselves, and what is the influence of radiometabolites? TSPO ligands show different metabolic profiles when tested in vivo and in vitro [40]; the metabolic activities can influence the diagnostic and therapeutic efficiency of the ligands.
Apart from these factors, successful ligand binding also raises several questions concerning molecular level interactions of the various TSPO ligands with different affinities that have been tested over the last decade for PET imaging [35, 41]. Does in vivo ligand binding involve TSPO alone or the interface of TSPO in complex with one or more other proteins? Indeed, TSPO has been described as part of a complex with different protein partners [42–45]. If a multiprotein complex is active, TSPO ligand selectivity may be governed by the protein-complex composition and not by the interaction with TSPO alone and, thus, specific ligand binding to TSPO might be reduced.
Moreover, it has to be taken into account that overexpression of proteins other than TSPO and its partners in neuroinflammation, for example [4], could occur. These observations raise various binding-site related questions: what “makes” the binding site, which amino acids of the TSPO protein are involved in the binding site and which are involved in interactions driving ligand affinity and selectivity?
Hence, the successful development of TSPO ligands as drugs for diagnostics and therapeutics may gain from deep analysis of ligand interaction to its protein binding site using available atomic structures [16–19] that we will review below. This will help to optimize molecular docking for the analysis either of a series of ligands [46] or of different classes of ligands [37,47] and, thus, generate more efficient ligands. Such new ligands may help to characterize the pathologies in which TSPO is overexpressed, as well as to assess new drugs for therapies.
Factors to consider for development of new ligands for TSPO
Since the identification of TSPO by means of benzodiazepine diazepam binding to peripheral tissue [1], many ligands have been synthesized to optimize their biological properties [9,46]. The structure-affinity relationships were rationalized in light of binding affinities and pharmacophore interactions with a TSPO topological model initially designed with pockets fitting different parts of the ligands [47]. The determination of the first atomic structure [16] made possible the study of the interactions between ligands and the protein cavity by docking [37,46,48].
Several TSPO atomic structures have since been solved (Table 1), for example mouse TSPO by nuclear magnetic resonance (NMR) [16–17] and bacterial TSPO by X-ray diffraction [18–19]. The structures reveal similar folding with five TM helices but different oligomeric states, and one active site that can bind both the high affinity TSPO drug ligand PK 11195 (Box 1, Figure I) and protoporphyrin IX (PPIX, Figure 1A). It is interesting to note that even though sequences of TSPO from Rhodobacter sphaeroides (RsTSPO), Bacillus cereus (BcTSPO) and mouse (mTSPO) are relatively well conserved (25-35% identity), there is variability in amino acid composition for the active site between mammalian and bacterial TSPO [20]. The analysis of ligand interaction in mammalian and bacterial binding sites is the starting point for understanding what controls selectivity. This selectivity depends both on ligand molecular formula and structure and on ligand access to the TSPO binding cavity. It also depends on protein polymorphism. For example, the murine TSPO A147T mutation (Table 1), which is not in the actual binding cavity increases the flexibility and generates different binding properties for different ligands [42, 49–50]. The change in TSPO flexibility which has been recently described to decrease half-life for two human TSPO polymorphisms (A147T and R162H) [51] might alter ligand binding.
Table 1.
Table 1 shows the different experimentally determined structures of mammalian and bacterial TSPO gained using three different biophysical techniques NMR, EM and X-ray diffraction.
| TSPO species | Genotype | Moleculea in the ligand cavity | ID | Mediums of extraction | Method | Resolution (Å) | Oligomeric state | Ref | |
|---|---|---|---|---|---|---|---|---|---|
| Purification | Experimental | ||||||||
| mTSPO | WT | PK 11195 | 2MGYb 19608c |
DPC | DPC | Solution NMR 315K | - | monomer | [16] |
| A147Td | PK 11195 | 2N02b 25513c |
DPC | DPC | Solution NMR 315K | - | monomer | [69] | |
| WT | DAA1106 | - | DPC | DMPC | solid state NMR ~278K | - | dimer | [54] | |
| RsTSPO | WT | - | 1698e | DDM | E. coli lipidsf | electron microscopy | 10.0 | dimer | [55] |
| A139Tg | PPIX | 4UC1b | DM | LCP monolein | X-ray diff. 100K | 1.8 | dimer | [19] | |
| A139T | 4UC2b | DM | LCP monolein | X-ray diff. 100K | 2.4 | dimer | [19] | ||
| WT | - | 4UC3b | DM | LCP monolein | X-ray diff. 100K | 2.5 | dimer | [19] | |
| A139T | PPIX | 5DUOb | DM | LCP monolein | X-ray diff. 100K | 2.4 | dimer | [19] | |
| BcTSPO | WT | PK 11195 | 4RYIb | DDM | LCP monolein | X-ray diff. 100K | 3.49 | dimer | [18] |
| WT | - | 4RYJb | DDM | PEG | X-ray diff. 100K | 4.1 | dimer | [18] | |
| WT | Iodine | 4RYMb | DDM | LCP monolein | X-ray diff. 100K | 2.8 | monomer | [18] | |
| WT | PEG & 3 water | 4RYNb | DDM | LCP monolein, DDM | X-ray diff. 100K | 2.01 | monomer | [18] | |
| WT | 2 DMSO & 7 water | 4RYOb | DDM | LCP monolein | X-ray diff. 100K | 1.6 | monomer | [18] | |
| WT | 7 water | 4RYQb | DDM | LCP monolein | X-ray diff. 100K | 1.7 | monomer | [18] | |
| WT | 2 DMSO & 8 water | 4RYRb | DDM | LCP monolein | X-ray diff. 100K | 1.7 | monomer | [18] | |
Note:
corresponds to molecules found in the ligand cavity;
depicts TSPO structures deposited in PDB (Protein Data Bank);
depicts structures deposited in BRMB (Biological Resonance Magnetic Bank);
corresponds to the A147T mutation in mammals;
depicts structures in EMDB (Electron Microscopy Data Bank);
represents lipids extracted from E. coli ;
corresponds to the A139T mutation in bacteria (equivalent to the mammalian A147T mutation).
Abbreviations used: mTSPO: mouse TSPO; RsTSPO: Rhodobacter sphaeroides TSPO; BcTSPO: Bacillus cereus TSPO; DDM: DoDecylMaltoside; DM: DecylMaltoside DMSO: dimethyl sulfoxide DPC: DodecylPhosphoCholine LCP: Lipid Cubic Phase PEG: PolyEthylene Glycol WT: wild type.
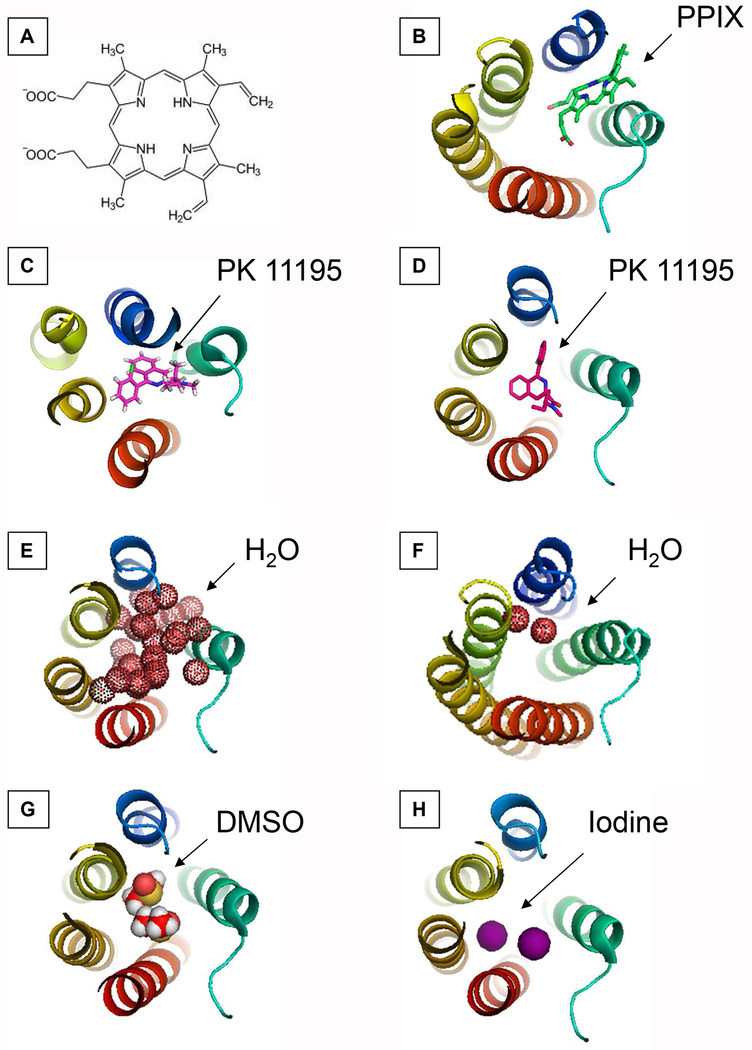
Figure 1. Overlap of molecules in the cavity of TSPO.
(A) Scheme of PPIX ligand structure.
(B) PPIX (carbons in green, oxygens in red) in RsTSPO (PDB entry 4UC1).
(C) PK 11195 (carbons in magenta, hydrogens in white, nitrogens in blue, chlorine in green) in BcTSPO (PDB entry 4RYI).
(D) PK 11195 (carbons in magenta, nitrogens in blue, chlorine in green) in mTSPO (PDB entry 2MGY).
(E) Water (doted red spheres) in BcTSPO (PDB entry 4RYQ). Dark red spheres correspond to water molecules in the selected slice whereas shadowed red spheres correspond to water molecules located underneath.
(F) Water (doted red spheres) in RsTSPO (PDB entry 4UC1).
(G) DMSO (carbons in red, hydrogens in white and sulfur in yellow) in BcTSPO (PDB entry 4RYO).
(H) Iodine (magenta spheres) in BcTSPO (PDB entry 4RYM)
In each case, the TSPO atomic structure is shown as rainbow cartoon colored as follows: TM1, blue; TM2, green; TM3, light green; TM4, yellow; TM5, red using PyMol (https://pymol.org/2/) [93].
The mode of action of the available TSPO ligands, especially in vivo, remains unclear. Several questions remain open, for example the oligomeric state of TSPO, the flexibility of the protein, the links between ligand and protein within the binding cavity, and the actual binding mechanism(s). The following sections will review what is currently known and the opportunities that can be used for future TSPO drug development.
Oligomeric states of TSPO
Several oligomeric states for mammalian TSPO have been reported in the literature ranging from monomers to high homo-oligomers in vivo [1, 52]. These reported oligomeric states depend on various factors such as the medium and conditions in which the structure is obtained, the method which the structure is studied [(NMR, electron microscopy (EM), X-ray crystallography)] and whether TSPO is interacting with other proteins in the experimental process. We give brief examples of these here.
Purified recombinant mTSPO in solution, solubilized by detergent, is usually in a monomeric state [53] and has permitted the elucidation of the first atomic structure (Table 1) [16]. Reconstituted in a membrane and studied by solid state NMR (ssNMR, Table 1), the mTSPO dimer interface has been found to include the G83xxxG87 motif of TM3 [54]. A highly stable dimer has been obtained from bacterial membrane by solubilization with a mild detergent, such as dodecylmaltoside (DDM), and was used to form tubular crystals upon detergent removal studied by EM (Table 1) [55]. However, monomers to dimers have been observed for bacterial TSPO (BcTSPO [18] and RsTSPO [19]) in X-ray structures obtained using crystals grown in lipidic cubic phase (LCP) (Table 1)). While the BcTSPO dimer interface includes the G42xxxG46 motif of TM2, the RsTSPO dimer interface reveals another type of motif, AxxxA (one in TM1 and 2 in TM3) involved in the interface of three different crystal packing arrangements [19]. Observation of several interfaces (TM2-TM2 for BcTSPO [18], TM3-TM3 for RsTSPO [19]), raises the question of either potential oligomer state-function relationships or the effect of crystallographic constraints. Moreover, water molecules have been resolved between the TM3 helices in the two monomers of RsTSPO, raising the question of a putative external transport pathway [19].
Electron microscopy (EM) of RsTSPO dimers [55] fitted with atomic models [19, 56] suggested that different interfaces depend on the model used: a TM3-TM3 interface was obtained using crystallographic structure, whereas a TM4-TM4 interface was obtained using a mTSPO derived model [57]. It is thus impossible to conclude what is the functional state of RsTSPO. However, it is interesting to note that AxxxA motifs (present in TM3 and TM4) have been suggested to be a common α helical interaction motif that provide stability of several proteins [58]. Further, interhelical axial distances might be greater for AxxxA motifs than for GxxxG ones [58], as observed comparing GxxxG motif interactions in mTSPO and BcTSPO versus AxxxA motif interactions in RsTSPO raising the question of the stability of the different oligomers.
Actually no atomic structure of human TSPO (hTSPO) is available. However, when overexpressed in E. coli and purified by its polyhistidine tag on a Ni-NTA column followed by size exclusion chromatography (SEC), hTSPO was suggested to form a hexameric structure, whereas RsTSPO with the same protocol, generated only dimers [59]. It might however be that TSPO is being misfolded when expressed in heterologous conditions. Moreover, TSPO might be dynamic and adopting different organizations depending on its environment such as the medium in which it is expressed/purified. Further, as noted above, other proteins form complex with TSPO and thus could affect ligand binding. For example, TSPO has been described to interact with various membrane partners [42–45] such as the voltage dependent ion channel (VDAC) [60], and TSPO exhibit higher affinity for benzodiazepine in protein complex than alone [22]. Moreover, ligand binding could affect oligomeric TSPO structure. Indeed, binding of cholesterol to the CRAC motif shifts the dynamic equilibrium of mTSPO dimer toward the monomer [54] and thus destabilizes the dimer. It has to be noted that this effect of cholesterol might be part of the potential transport process of cholesterol by TSPO activated by ligand binding such as PK 11195 to another site. This could occur through gliding of cholesterol from CRAC to a specific amino acid motif (LAF) in the middle of the TM5 [61] and another cholesterol recognition motif (CARC) located at the N-terminus of the TM5 [62]. It must be noted that bacteria and plants do not have cholesterol. Thus, the conserved function of TSPO among species remains unclear, as well as the effect or need of oligomeric states of TSPO for its function in different kingdoms. However, it was shown that covalent polymer formation observed upon UV or ROS exposure [52] reduces cholesterol binding whereas it increases PK 11195 one [52] suggesting that TSPO function involves a dynamic process. Moreover, the description by molecular modeling of at least two types of interfaces for mTSPO [37] involving different TM interfaces previously described in the literature [58,63–65] motifs suggests that the same protein can contain two motifs within the TM domain, one for homodimerization and another one for hetero-dimerization [66], leading to the formation of homo or heteropolymers between TSPO and other membrane proteins.
Finally, TSPO might be implicated in various dynamic oligomers, but in cellular studies have also suggested that formation of covalent oligomers might be part of TSPO turnover, the covalent polymers being degraded and new protein being synthetized [51,67], making the situation even more complicated.
TSPO flexibility and stability
Ligand binding to TSPO depends on its accessibility to the binding site, which itself depends on protein flexibility as illustrated by recent data from NMR and crystallography. [17–19] TSPO stability can be affected by single nucleotide polymorphisms (SNPs). A way to investigate such SNPs that can affect TSPO stability has been to search for deleterious SNPs in human TSPO in silico [51]. Most of the detected SNPs had low frequencies, except SNPs R162H and A147T. Both R162H and A147T mutations have been shown to decrease the half-life of the mutant TSPOs by about 25 percent, corresponding to a decrease of stability and an increase of flexibility [51].
Effect of R162H:
R162 is located in the C-terminal domain of TSPO and is outside of the binding site of PK 11195 [16]. Since it is known that C-terminus deletions of TSPO impacts ligand affinity [21,68], this mutation may be involved in the binding mechanism of TSPO with PK 11195, perhaps by its location on the access path to the binding site [3].
Effect of A147T:
A147 is located in the TM5 and is part of the binding site [16]. Comparison of WT and mutant of hTSPOs showed that the A147T mutation significantly modified the flexibility (in silico) and the stability (in cellulo) of the protein [51]. Solution NMR of hTSPO and mTSPO shows highly dynamic structure in the absence of PK 11195 [17] and detailed analysis of mTSPO revealed that A147 belongs to a highly flexible part of the protein [17]. This may suggest that ligand binding occurs differently for WT and mutant as observed in vivo with different affinities for WT and A147T mutant [49]. However, solution NMR atomic structures of WT and A147T mutant of mTSPO in complex with PK 11195 show the same structural and dynamic profile [69] suggesting that A147T mutation is mainly involved in the binding mechanism.
Moreover, bacterial TSPO (RsTSPO) in LCP 3-D crystals at cryogenic temperatures in the absence of ligand also shows structural changes between WT and the A139T mutant (equivalent to mammalian A147T and located in the same TM5) [19]. The WT structure shows a higher degree of flexibility than the mutant, in particular for the loop connecting the TM1 and TM2 that is not resolved due to the various conformations that avoid the determination of its structure. Interestingly, this loop has been described as important for ligand binding and protein stabilization [17,20–21,68].
In the X-ray structure of the A139T mutant of RsTSPO, a single PPIX, another TSPO ligand, binds only one of the two monomers and no substantial structural differences (Root Mean Square Deviation (RMSD) of 0.3Å) are observed between the TSPO apo and holo forms [19]. Particularly, the loop connecting TM1 and TM2 that caps the PPIX is similarly positioned in all monomers and closes the binding cavity, thus raising the question of the binding site accessibility [19]. This is also the case for BcTSPO where atomic structures with and without PK 11195 are highly superimposable (RMSD of 0.7Å) [18]. The lack of differences between TSPO structures with or without ligands, may be due to cryo-cooling penalties, which could hide transient conformational states favouring ligand accessibility to its binding site [70].
Ways to access protein flexibility and stability:
Characterization of protein flexibility can be obtained by looking at X-ray B-factor distribution throughout the amino acid sequence in PDB files. B-factors model thermal motion and are directly related to conformational heterogeneity; their calculation requires highly-resolved structures that still remain challenging for membrane proteins such as TSPO. Valuable information on protein flexibility can also be obtained by molecular dynamics (MD) simulations. For example, MD simulations of mTSPO in lipid membranes suggest that dimer formation is unstable [37] and contradicts experimental data previously described [54]. Furthermore, simulations with and without PK 11195 reveal rearrangement of TM helices [37, 71]. Moreover, MD simulation have also shown additional structural changes such as (i) the bending of TM2 and TM4 helices increases mainly in the presence of PK 11195, very likely related to ligand-protein constraints [37], and (ii) TM1, TM3 and TM5 helices show the largest rotation fluctuation, perhaps related to the reduced number of ligand contacts compared to TM2 and TM4 in mTSPO in presence of PK 11195 [71].
Ultimately, it is critical that the known characteristics of flexibility and stability of TSPO be taken into account when a new ligand is being designed.
TSPO ligand binding site
Structures obtained by NMR and X-ray crystallography show that bound PK 11195 and PPIX ligands are buried in the same cavity in between the five TM helices in mammalian and bacterial TSPOs (Figure 1) [18–19,46]. Ligand stabilization involves 10 to 20 amino acids depending on the complex, but only a few are highly conserved between species [16, 19–20]. In order to fully evaluate how the ligands fit into a the binding cavity, it is important to evaluate the volume of the cavity (between species, with and without ligand, whether there are water or other molecules within the cavity, WT versus mutant, etc.) and determine whether the TSPO binding cavity changes to adapt to the ligand. Indeed, molecules of various sizes such as PK 11195, PPIX and dimethylsulfoxide (DMSO) have been observed in the cavity of TSPO atomic structures (Figure 1) and have also been supported by molecular docking studies [37,46–48]. PK 11195 and PPIX, the common TSPO ligands, both fit within the lipophilic binding cavity of TSPO [11–19]. While PPIX is a rather soluble compound and protrudes outside between TM1 and TM2, PK 11195 is mostly hydrophobic and is almost inaccessible from the bulk, raising the question of the hydrophobicity-hydrophilicity of the TSPO binding cavity [16–19].
Indeed, the TSPO ligand binding site contains both hydrophobic and polar residues [16–20] that surprisingly accepts various molecules, such as water, iodine and DMSO (Table 1) [19]. It has to be noted that only high-resolution cryogenic X-ray crystallographic structures permit to localize small molecules. Thus, the different structures of BcTSPO [18] reveal the presence of many (95) water molecules (PDB ID-4RYQ), 2 DMSO molecules (PDB ID-4RYR), or 2 iodine molecules (PDB ID-4RYM) in the binding cavity in the absence of ligand (Figure 1). Both DMSO molecules form hydrogen bonds with highly conserved amino acids [18]. RsTSPO was crystallized in the presence of PK 11195 [19] but surprisingly was not visible in any structure.
These observations suggest that ordered water molecules may be involved in the interaction events (such as water displacement upon ligand binding) and energetics minima as previously described for trypsin [72]. High-resolution cryogenic X-ray crystallographic structures of RsTSPO [19] only resolved one or two water molecules (PDB ID-5DUO and 4UC1 respectively) in the ligand binding sites, in absence of ligand [19]. The water molecules form hydrogen bonds with some residues involved in PPIX binding (i.e. Y54, N84, T88, W135 and T139) [19]. The hydrogen bond mediated by one water molecule is present in almost all apo monomers [19]. The clear involvement of hydrogen bonds of the water molecules needs to be confirmed for instance by comparing cryogenic and room temperature crystallographic structures [72], as well as at low and high pH ones [73]. More generally, characterization of the hydrogen-bond network involving water molecules could help for drug development.
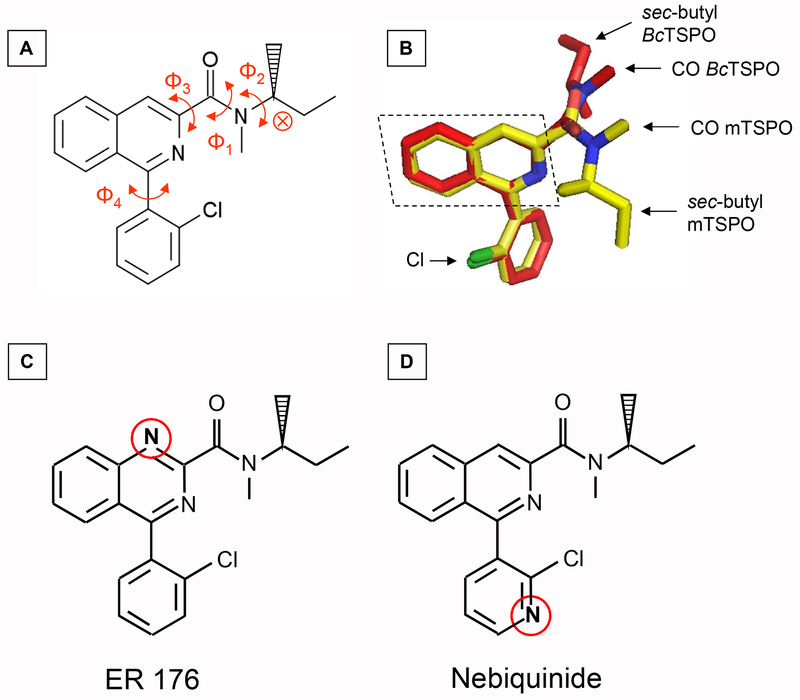
The number of amino acids involved in the binding pocket changes with the ligand type and also for the same ligand with the bacterial and mTSPO as previously described [20]. This might be attributed either to the different orientations that the same ligand could adopt within the cavity or to the change in orientation induced by atom substitutions on the heterocycle, such as observed on PK 11195 analogues (Box 1, Figure IC and D) [74–76]. Ligand ER 176 [74–75], which differs from the PK 11195 by only one carbon substituted by one nitrogen on the isoquinoline scaffold (Box 1, Figure IC)), has higher affinity for WT TSPO than PK 11195, but is sensitive to the human A147T mutation [76]. This might be due to reduced stabilisation by interactions with TM5 that contains the A147T mutation, thus inducing different stabilisation by residues of other TM, such as TM2 for example. The presence of chloride on the phenyl ring of ER 176 also seems important, since its absence decreases the binding affinity, but a change of its position on the phenyl ring has a smaller effect [75]. On the other hand, nebiquinide, which differs from the PK 11195 with one carbon substituted by one nitrogen on the phenyl ring (Box 1, Figure ID), has similar affinities to PK 11195 and is insensitive to the A147T mutation [76]. This indicates that neither the mutation nor structural changes induced by the mutation are involved in interactions in the ligand binding site.
TSPO ligand binding mechanism
A crucial element to improve selectivity and specificity of ligand is to understand what the mechanism of ligand binding is and, the protein conformational changes involved in permitting the fitting of ligand within the cavity. The accessibility of the binding site in TSPO is not completely known. Atomic structures of TSPO-ligand complexes suggest a potential gating access between TM1 and TM2 as evidenced by the PPIX protruding outside from the RsTSPO [18]. Atomic structure of the WT BcTSPO which loop linking TM1 and TM2 is not resolved, shows increased access to the ligand binding cavity [19]. The role of various loops in the ligand binding mechanism was proposed early on, based on affinity measurements on point mutants, as well as on deletions mutants of mammalian TSPO [21,68]. Structure analysis has confirmed the role of the loop connecting TM1 and TM2 that shows interaction with TM5 [77]. Implication of the loop linking TM3 and TM4, as well as the C-terminus, has been proposed recently [20]. It seems that these two loops and the C-terminus might contribute to driving the ligand into the cavity.
The role of water molecules during ligand recognition, as well as ligand stabilization within the cavity, has been described as a key parameter for protein-ligand complexes in solution [72]. When bound with different ligands, TSPO in the high-resolution atomic structures has been found to be associated with a different number of water molecules [18–19] raising the question of the contribution of water molecules in ligand binding. The stabilization of the ligand within the binding cavity of TSPO seems to involve exclusion of some ordered water molecules, while others remain involved in hydrogen bonds.
Concluding Remarks and Future Perspectives
The importance of TSPO in cell-specific functions in inflammation and repair has led to a large interest in developing ligands for its visualization and quantification. However, TSPO is not the only protein that is a marker of inflammation. Hence, the specificity of the ligands that bind to TSPO becomes an important criterion of their successful design and utility. Further, the development of new molecules or the optimization of existing ones to improve imaging remains an important goal. Moreover, it is anticipated that functional characterization of these molecules could lead to novel therapeutics. In this review we have reviewed the various TSPO structures and discussed the different aspects TSPO-ligand interactions that would be important in developing successful ligands for diagnostic and therapeutic purposes.
TSPO exist in different oligomeric states (see Outstanding Questions) and exhibit some flexibility in conformation. To access TSPO stability and flexibility in presence of different ligands, several approaches have been developed that are complementary to conventional methodologies reviewed earlier. These are mass spectrometry (MS), small angle X-ray scattering or small angle neutron scattering, methods that can allow to explore protein flexibility along with help characterize different oligomeric states of TSPO [78]. Indeed, recent advances in MS have given information on several membrane protein, dynamics, solvent accessibility, lipid/ligand interaction and ligand binding induced conformational perturbations [79]. Likewise, tools developed for the analysis of data from small angle scattering coupled with chromatographic set-up has permitted the characterization of oligomeric membrane protein such as aquaporin and Fhac protein transporter [80]. Hence, these techniques might be useful to study homo or hetero oligomers of TSPO.
Outstanding Questions.
What is the oligomeric state of TSPO in vivo inside the cell?
What is the reason for the loss of binding properties of human TSPO2 compared to TSPO1?
How can we optimize TSPO ligands to generate better diagnostics (for PET) and therapeutics (for example for allopregnenolone synthesis)?
Will ligand screening coupled with conformational dynamic studies based on molecular dynamic simulations lead to the generation of new TSPO ligands?
In some cases, extensive details on the flexible nature of the TSPO has not been entirely possible. Cryo-cooling penalties are probably responsible for missing the conformational states that show such details about flexibility and stability of TSPO. These can be avoided using recent technologies which exploit free-electron lasers and room temperature X-ray data collection to reduce the irradiation damage and should, therefore, allow sampling functionally relevant conformations as NMR experiments in solution [81].
Optimization of TSPO ligand to improve the affinity for the various TSPO sequences (see Outstanding Questions) remains to be realised since recent comparisons of PET efficiency of the various compounds designed to bind to human TSPO revealed large non-specific binding [35] or polymorphisms variability [41],. Ultimately, given that the TSPO endogenous ligand is a peptide [1], it might be interesting to develop a peptidic ligand. To overcome the peptide instability, a pseudopeptide or peptidomimetic could be designed [82] and the addition of cargo or cell penetrating peptide moiety could help to pass the hematoencephalic barrier to reach the brain [83].
Ligand binding kinetics, and its residence time in particular, are rarely studied despite their crucial role in ligand-protein complex formation [39]. Further it is known that water displacement increases the affinity for the ligand, whereas water that remained trapped represented an entropic disadvantage [72]. Thus, it is expected that TSPO ligands that fully displace water molecules may exhibit higher affinities. Hence, it is critical to gain primarily high-resolution atomic structures with and without different ligands and if possible precise location of water molecules to help design successful ligands. It might be helpful to perform experiments using neutron diffraction and low/high temperature X-ray diffraction to determine water molecule orientation and fully understand their contribution. However, ligand stabilization may involve a different set of amino acids with different types of interactions contributing to the stabilization.
A couple of in-silico studies involving TSPO have been reported since atomic structure determination: ligand-TSPO docking studies [19,37,84], dimer structural prediction [37], unbinding of TSPO chemical modulators in order to correlate the in vitro residence time to the in vivo efficacy [85]. Development of MD simulation up to microseconds should help to analyze the evolution of both loops and TM domains to understand ligand accessibility to the binding cavity, as well as water movements. Analysis of binding cavity dynamics would also be useful both in the absence and in the presence of ligand to characterize the involvement of specific/conserved amino acids [86].
Interestingly, in search of TSPO-like gene, a paralog gene TSPO2, has been identified in mammals that have different ligand binding properties than that of TSPO1 (referred to as TSPO in the text above) [2]. The cholesterol binding is conserved between the two proteins whereas binding to PK 11195 is lost in TSPO2 (see Outstanding Questions) [2]. If homology models and further experimental data are provided for TSPO2, one can learn from the differences between these two TSPO paralogs to better characterize the ligand binding site. Moreover, the unified structural model in membrane bilayers [87], recently obtained by comparative modelling from the mouse and bacterial TSPO structures, could be used as a starting condition for structural studies on human TSPO and help the structure-based design of high-affinity TSPO ligands. Ligand screening fragment libraries can be done to characterize new drugs, using for example surface plasmon resonance (SPR), as applied to many membrane proteins [88]. Combining cryogenic and room temperature X-ray data would also help to guide ligand design in order to optimize the affinity between the ligand and the binding site [89]. In-depth analysis and visualization of protein flexibility in interactions with ligands will be needed to push the limits of structural investigations [78] and to generate or optimize TSPO ligands.
Thus, the combination of all available and new structural information (X-ray, NMR, molecular dynamics simulations, role of water in ligand affinity, role of partners in vivo, etc.) will lead to an increased understanding of TSPO-ligand interactions that will be valuable for the development of new therapeutic and diagnostic TSPO ligands.
Box 1. Interactions of PK 11195 and PPIX with TSPO.
PK 11195 (Box 1, Figure IA) is a flexible ligand (low energy transition between isomers) with several rotamers [90] and one asymmetric carbon (red cross) giving two enantiomers (R) and (S), the former having a 2-fold greater affinity for TSPO than the latter [91]. Overlay of the alignment of (R)-PK 11195 bound to mTSPO (PDB ID-2MGY) and BcTSPO(PDB ID-4RYI) exhibits a rotation of the carboxamide group (Φ3) that places the CO and the sec-butyl group on opposite sides of the isoquinoline plane for the two TSPO (Box 1, Figure IB). In addition, the CO and the Cl of PK 11195 are placed on the same side of the isoquinoline plane for BcTSPO and on opposite side for mTSPO (Box 1, Figure IB). It is worth noting that the mTSPO structure is only stabilized in its holo form, and the presence of detergents may distort the positions of protein residues interacting with the ligand [92] whereas all the structures of BcTSPO have been solved in a lipid environment.
PPIX (Figure 1A) is rather a rigid ligand that binds to RsTSPO (Figure 1B) and has also been shown to fit the cavity of BcTSPO [19–20]. Six residues belonging to the TM2, TM3 and the loop connecting TM1 and TM2 of RsTSPO are within 3 Å distance of the ligand. The two COOHs of the PPIX fit inside the TSPO cavity, probably stabilized by hydrogen bonds and by unordered water molecules.
Box 1, Figure I. Interaction of PK 11195 with mTSPO and BcTSPO.
(A) Structure of (R) PK 11195 ligand. Φ1, Φ2, Φ3 and Φ4 are the respective dihedral angles for CH3-N-C=O, CH3-CH-N-CH3, N-CH-C=O and chlorophenyl-isoquinoline ring, respectively. Red cross shows the asymmetric carbon and the red arrows show the rotation of the bond corresponding to the various Φ angles.
(B) Overlay of aligned (R) PK 11195 bound in the binding cavity of mTSPO (yellow) and BcTSPO (red) (PDB ID-2MGY and 4RYI respectively). The isoquinoline plane is shown as dotted parallelogram.
(C) and (D) General structures of ER 176 and Nebiquinide, respectively. Red circles emphasize the chemical substitutions introduced compared to PK 11195 (A).
HIGHLIGHTS.
The transmembrane protein TSPO has been described as a diagnostic and therapeutic target for inflammation and, in particular, brain diseases.
Various classes of TSPO ligands have been developed since its discovery in 1977, and some of them are routinely used for diagnosis and treatment.
The design of new ligands is needed to improve cellular specificity, for instance to distinguish abnormal cells, but also for targeting to TSPO as part of a multi protein complex of various compositions depending on the cells.
Drug design will gain from TSPO structure-function analysis and computer simulation methods such as molecular dynamics.
ACKNOWLEDGENTS
J.J.L. and S.F. acknowledges financial support from CNRS. L.D. acknowledges financial support from CNRS, University of Technology of Compiègne and Hauts-de-France Region and the European Regional Development Fund (ERDF) 2014/2020 (project BESTMIP)., VP is supported by a grant from the National Institutes of Health (R01 AG21092) and the John Stauffer Dean’s Chair in Pharmaceutical Sciences (University of Southern California). MK is supported by the National Health and Medical Research Council of Australia (NHMRC) (APP1132524) and is an NHMRC Principal Research Fellow (APP1154692).
GLOSSARY
- Apo/halo
Protein without/with ligand bound.
- Astrocytes
Star-shaped glial cells from the brain.
- B-factor
the factor originated from thermal motion that is applied to the X-ray data for each atom (or groups of atoms). A high B-factor usually corresponds to a large flexibility.
- Cryo-cooling penalties
increase in random errors of atom positioning in the structure due to cryogenic cooling of the crystals used to avoid irradiation damage, and that could perturb protein conformation equilibrium.
- Detergents
reagents used for the solubilization of membrane proteins. Some can be denaturing to some extent like the ionic one, sodium dodecyl sulfate (SDS), some other non-ionic maintain the tertiary structure and are used in NMR or crystallography. Some of the commonly used are detergents are DPC, DM, DDM. Detergents can be removed, and protein transferred to a lipid environment (reconstitution).
- Heterocycle
compound that has at least one ring structure with at least one atom in the ring that is not carbon.
- Ligands
Ligands are molecules that target proteins to initiate or modulate the target protein’s function or functions by binding to them. Ligands can be more or less specific, and target proteins can have one or more binding sites for different ligands.
- LCP
Lipidic cubic phase is obtained by mixing aqueous and surfactant components that form a lattice of aqueous channels within lipid phase that permits the growth of membrane protein crystal.
- MD
computer simulation method for analyzing the physical movements of atoms and molecules. Molecular dynamics simulations permit various dynamic process such as protein folding or conformational changes, protein-protein association.
- Paralogs
genes that derive from the same ancestral gene
- ROS
Reactive Oxygen Species are chemically reactive chemical species containing oxygen, including peroxides, superoxide, hydroxyl radicals etc.
- Radiometabolites
the various derivative products (generated by cytochromes for example) from a PET probe when injected in vivo.
- RMSD
Root Mean Square Deviation can be used to compare two atomic structures. It measures of the average distance between the atoms of two superimposed proteins.
- Rotamers
conformers that arise from restricted rotation around a single bond.
- SEC
Size exclusion chromatography permit to separate molecules such as protein in solution according to their size.
- SNP
Single Nucleotide Polymorphism is a single nucleotide mutation occurring to some degree within a population. These may or may not be linked to diseases or changes in protein functioning.
- SPR
Surface Plasmon Resonance is a spectroscopic method that permits to measure a ligand binding to protein adsorbed on a surface.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLAIMER STATEMENT
The authors declare no conflict of interest
REFERENCES
- 1.Papadopoulos V et al. (2006) Translocator protein (18kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci 27(8), 402–409. [DOI] [PubMed] [Google Scholar]
- 2.Fan J et al. (2012) Structural and functional evolution of the translocator protein (18kDa) Curr. Mol. Med 12, 36–386. [DOI] [PubMed] [Google Scholar]
- 3.Guilarte TR (2019) TSPO in diverse CNS pathologies and psychiatric disease: a critical review and a way forward. Pharm. Therap 194, 44–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narayanaswami V et al. (2018) Emerging PET radiotracers and targets for imaging of neuroinflammation in neurodegenerative diseases: outlook beyond TSPO. Mol. Imag 17, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Largeau B et al. (2017) TSPO PET imaging: from microglial activation to peripheral sterile inflammation diseases? Contrast Media Mol. Imaging 6592139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillaumot D et al. (2009) The Arabidopsis TSPO-related protein is a stress and abscisic acid-regulated, endoplasmic reticulum-Golgi-localized membrane protein. Plant J. 60(2), 242–256. [DOI] [PubMed] [Google Scholar]
- 7.Zeng X and Kaplan S (2001) TspO as a modulator of the repressor/antirepressor (PsR/AppA) regulatory system in Rhodobacter sphaeroides. J. Bacteriol 183, 6355–6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batoko H et al. (2015) Enigmatic translocator protein (TSPO) and cellular stress regulation. Trends Biochem. Sci 40(9), 497–503. [DOI] [PubMed] [Google Scholar]
- 9.Ruppecht R et al. (2010) Translocator protein (18kDa) (TSPO) as a therapeutic target for neurological and psychiatric disorders. Nat. Rev. Drug. Discov 9, 971–988. [DOI] [PubMed] [Google Scholar]
- 10.Gatliff J and Campanella M (2015) TSPO is a redox regulator of cell mitophagy. Biochem. Soc. Trans 43, 543–552. [DOI] [PubMed] [Google Scholar]
- 11.Batarseh A and Papadopoulos V (2010) Regulation of translocator protein 18kDa (TSPO) expression in health and disease states. Mol. Cell. Endo 327, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhoola NH et al. (2018) Translocator protein (TSPO) as a potential biomarker in human cancers. Int. J. Mol. Sci 19, 2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrero-Rivera M et al. (2018) Translocator protein and new targets for neuroinflammation. Clin. Transl. Imaging 3, 391–402. [Google Scholar]
- 14.Barichello T et al. (2017) The translocator protein (18kDa) and its role in neuropsychiatric disorders. Neurosci. Biobehav. Rev 83, 183–199. [DOI] [PubMed] [Google Scholar]
- 15.Denora N and Natile G (2017) An update view of the translocator protein (TSPO) Int. J. Mol. Sci 18, 2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaremko L et al. (2014) Structure of the mitochondrial translocator protein in complex with a diagnostic ligand. Science 343, 1363–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaremko L et al. (2015) Conformational flexibility in the transmembrane protein TSPO. Chem. Eur. J 21, 16555–16563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y et al. (2015) Structure and activity of tryptophan-rich TSPO proteins. Science 347, 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F et al. (2015) Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism. Science 347, 555–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iatmanen-Harbi S et al. (2019) Characterization of the high-affinity drug ligand binding site of mouse recombinant TSPO. Int. J. Mol. Sci 20(6), 1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H and Papadopoulos V (1998) Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology 139(12), 4991–4997. [DOI] [PubMed] [Google Scholar]
- 22.Lacapere J-J et al. (2001) Structural and functional study of reconstituted peripheral benzodiazepine receptor. Biochem. Biophys. Res. Commun 284, 536–541 [DOI] [PubMed] [Google Scholar]
- 23.Jamin N et al. (2005) Characterization of the cholesterol recognition amino acid consensus sequence of the peripheral-type benzodiazepine receptor. Mol. Endocrinol 19, 588–594. [DOI] [PubMed] [Google Scholar]
- 24.Midzak A et al. (2011) Novel androstenetriol interacts with the mitochondrial translocator protein and controls steroidogenesis. J. Biol. Chem 286(11), 9875–9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werry EL et al. (2015) TSPO as a target for glioblastoma therapeutics. Biochem. Soc. Trans 43(4), 531–536. [DOI] [PubMed] [Google Scholar]
- 26.Do Rego JL et al. (2012) Regulation of neurosteroid biosynthesis by neurotransmitters and neuropeptides. Front. Endocrinol 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tonon M-C et al. (2019) Endozepines and their receptors: Structure, functions and pathophysiological significance, Accepted in Pharmacology and Therapeutics [DOI] [PubMed] [Google Scholar]
- 28.Rasmusson AM et al. (2017) Neuroactive steroids and PTSD treatment. Neurosci. Lett 10, 156–163. [DOI] [PubMed] [Google Scholar]
- 29.Biswas L et al. (2018) TSPO ligands promote cholesterol efflux and suppress oxidative stress and inflammation in choroidal endothelial cells. Int. J. Mol. Sci 19, 3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Fur G et al. (1983) Peripheral benzodiazepine binding sites: effect of PK 11195, 1-(2-chlorophenyl)-N-methyl-N-(1-methylpropyl)-3-isoquinolinecarboxamide: I. In vitro studies. Life Sci. 32(16) 1839–1847. [DOI] [PubMed] [Google Scholar]
- 31.Tong J et al. (2019) Concentration, distribution, and influence of aging on the 18kDa translocator protein in human brain: implications for brain imaging studies. J. Cereb. Blood Flow Metab doi: 10.1177/0271678X19858003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar A et al. (2012) Evaluation of age related changes in translocator protein (TSPO) in human brain using 11C-[R]-PK 11195 PET. J. Neuroinflam 9, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coughlin JM et al. (2014) Regional brain distribution of translocator protein using [11C]DPA-713 PET in individuals infected with HIV. J. Neurovirol 20, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zavala F (1997) Benzodiazepines, Anxiety and Immunity. Pharmacol. Ther 75(3) 199–216. [DOI] [PubMed] [Google Scholar]
- 35.Fujita M et al. (2017) Comparison of four 11C-labeled PET ligands to quantify translocator protein 18kDa (TSPO) in human brain: (R)-PK 11195, PBR28, DPA-713, and ER176 - based on recent publications that measured specific-to-non-displaceable ratios. EJNMMI research 7, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Junck L et al. (1989) PET imaging of human gliomas with ligands for the peripheral benzodiazepine binding site. Annals of neurology 26(6), 752–758. [DOI] [PubMed] [Google Scholar]
- 37.Zeng J et al. (2018) Structural prediction of the dimeric form of the mammalian translocator membrane protein TSPO: a key target for brain diagnostics. Int. J. Mol. Sci 19, 2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dupont AC et al. (2017) Translocator protein-18kDa (TSPO) positron emission tomography (PET) imaging and its clinical impact in neurodegenerative diseases. Int. J. Mol. Sci 18, 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costa B et al. (2016) TSPO ligand residence time: a new parameter to predict compound neurosteroidogenic efficacy. Sci. Rep 6, 18164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eberl S et al. (2017) Preclinical in vivo and in vitro comparison of the translocator protein PET ligands [18F]PBR102 and [18F]PBR111. Eur. J. Nucl. Med. Mol. Imaging 44, 296–307. [DOI] [PubMed] [Google Scholar]
- 41.Cumming P et al. (2018) Sifting through the surfeit of neuroinflammation tracers. J. Cereb. Blood Flow Metab 38(2), 204–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McEnery MW et al. (1992) Isolation of the mitochondrial benzodiazepine receptor: association with the voltage dependent anionic channel and the adenine nucleotide carrier. Proc. Natl. Acad. Sci. USA 89(8), 3170–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papadopoulos V et al. (2007) Is there a mitochondrial signaling complex facilitating cholesterol import? Mol. Cell. Endo 265-266, 59–64. [DOI] [PubMed] [Google Scholar]
- 44.Issop L et al. (2013) Organelle plasticity and interactions in cholesterol transport and steroid biosynthesis. Mol. Cell. Endo 371(1–2), 34–46. [DOI] [PubMed] [Google Scholar]
- 45.Guilarte TR et al. (2016) TSPO finds NOX2 in microglia for redox homeostasis. Trends Pharmacol. Sci 37(5), 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barresi E et al. (2015) Deepening the topology of the translocator protein binding site by novel N,N-dialkyl-2-arylindol-3-ylglyoxylamides. J. Med. Chem 58, 6081–6092. [DOI] [PubMed] [Google Scholar]
- 47.Cinone N et al. (2000) Development of a unique 3D interaction model of endogenous and synthetic peripheral benzodiazepine receptor ligands J. Comput. Aided Mol. Des 14, 753–768. [DOI] [PubMed] [Google Scholar]
- 48.Deeva OA et al. (2019) A novel dipeptide ligand for TSPO. Dokl. Biochem. Biophys 484(2), 17–20. [DOI] [PubMed] [Google Scholar]
- 49.Owen DR et al. (2012) An 18-kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J. Cereb. Blood Flow Metab 32, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Owen DR et al. (2017) TSPO mutations in rats and human polymorphism impair the rate of steroid synthesis. Biochem. J 474, 3985–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milenkovic VM et al. (2018) Effects of genetic variants in the TSPO gene on protein structure and stability. PLoS One. 13(4), e0195627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delavoie F et al. (2003) In vivo and in vitro peripheral-type benzodiazepine receptor polymerization: functional significance in drug ligand and cholesterol binding. Biochim. Biophys. Acta 42(15), 4506–4519. [DOI] [PubMed] [Google Scholar]
- 53.Lacapere J-J et al. (2014) Structural studies of TSPO, a mitochondrial membrane protein In Membrane proteins production for structural studies. Editor Muss-Veteau I; Springer; New York Heidelberg Dordrecht London, pp 393–421; ISBN 978-1-4939-0661-1. [Google Scholar]
- 54.Jaipuria G et al. (2017) Cholesterol-mediated allosteric regulation of the mitochondrial translocator protein structure. Nat. Commun 8, 14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korkhov V et al. (2010) Three-dimensional structure of Tspo by electron cryomicroscopy of helical crystals, Structure 18(6), 677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li F et al. (2013) Characterization and modeling of the oligomeric state and ligand binding behavior of the purified translocator protein 18kDa (TSPO) from Rhodobacter sphaeroides. Biochemistry 52, 5884–5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hinsen K et al. (2015) Construction and validation of an atomic model for bacterial TSPO from electron microscopy density, evolutionary constraints, and biochemical and biophysical data Biochim. Biophys. Acta 1848(2), 568–580. [DOI] [PubMed] [Google Scholar]
- 58.Kleiger G et al. (2002) GxxxG and AxxxA: common alpha-helical interaction motifs in proteins, particularly in extremophiles. Biochemistry 41, 5990–5997. [DOI] [PubMed] [Google Scholar]
- 59.Li L et al. (2012) Expression, purification and characterization of bacterial and human translocator protein 18kDa (TSPO) Biophys. J 247a-248a, 1252-Pos. [Google Scholar]
- 60.Shoshan-Barmatz V et al. (2019) VDAC1 and the TSPO: expression, interactions, and associated functions in health and disease states. Int. J. Mol. Sci 20, 3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li F et al. (2015) Identification of a key cholesterol binding enhancement motif in translocator protein 18kDa. Biochemistry 54, 1441–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fantini J et al. (2016) A mirror code for protein-cholesterol interactions in the two leaflets of biological membranes. Sci. Rep 6, 21907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Russ WP and Engelman DM (1999) The GxxxG motif: a framework for transmembrane helix-helix association. Biophys. J 296, 911–919. [DOI] [PubMed] [Google Scholar]
- 64.Eilers M et al. (2002) Comparison of helix interactions in membrane and soluble alpha-bundle protein. Biophys. J 82, 2720–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ridder A et al. (2005) Tryptophan supports interaction of transmembrane helices. J. Mol. Biol 354, 894–902. [DOI] [PubMed] [Google Scholar]
- 66.Gerber D et al. (2004) Two motifs within transmembrane domain, one for homodimerization and the other for heterodimerization. J. Biol. Chem 279, 2117–21182. [DOI] [PubMed] [Google Scholar]
- 67.Issop L et al. (2016) Translocator protein-mediated stabilization of mitochondrial architecture during inflammation stress in colonic cells. PLoS One 11(4), e0152919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farges R et al. (1994) Site-directed mutagenesis of the peripheral benzodiazepine receptor: identification of amino acids implicated in the binding site of Ro5–4864. Mol. Pharmacol 46, 1160–1167. [PubMed] [Google Scholar]
- 69.Jaremko L et al. (2015) Structural integrity of the A147T polymorph of mammalian TSPO. Chem. Bio. Chem 16(10), 1483–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fischer M et al. (2015) One crystal, two temperatures: cryocooling penalties alter ligand binding to transient protein sites. Chem. Bio. Chem 16, 1560–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rao RM (2019) Study of structural and functional dynamics of the translocator protein, a new target for treatment of malaria. PhD 2019.
- 72.Schiebel J et al. (2018) Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes. Nat. Commun 9, 3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomaston JL et al. (2015) High-resolution structures of the M2 channel from influenza A virus reveal dynamic pathways for proton stabilization and transduction. Proc. Natl. Acad. Sci. USA 112(46), 14260–14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ikawa M et al. (2017) The Biomarkers Consortium Radioligand Project Team. 11C-ER176, a radioligand for 18kDa translocator protein, has adequate sensitivity to robustly image all three affinity genotypes in human brain. J. Nucl. Med 58(2), 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zanotti-Fregonara P et al. (2014) Synthesis and evaluation of translocator 18kDa protein (TSPO) positron emission tomography (PET) radioligands with low binding sensitivity to human single nucleotide polymorphism rs6971. ACS Chem. Neurosci 5(10), 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kalina T et al. (2019) Synthesis and in vitro evaluation of new translocator protein ligands designed for positron emission tomography. Future Med. Chem 11(6), 539–550. [DOI] [PubMed] [Google Scholar]
- 77.Li F et al. (2016) Translocator protein 18kDa (TSPO): an old protein with new functions? Biochemistry 55, 2821–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palamini M et al. (2016) Identifying and visualizing macromolecular flexibility in structural biology. Front Mol. Biosci 3, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calabrese AN and Radford SE (2018) Mass spectrometry-enabled structural biology of membrane proteins. Methods 147, 187–205. [DOI] [PubMed] [Google Scholar]
- 80.Koutsioubas A (2017) Low-Resolution Structure of Detergent-Solubilized Membrane Proteins from Small-Angle Scattering Data. Biophys. J 113, 2373–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Woldeyes RA et al. (2014) E pluribus unum, no more: from one crystal, many conformations. Curr. Opin. Struct. Biol 28, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pernot M et al. (2011) Stability of peptides and therapeutic success in cancer. Expert Opin. Drug. Metab. Toxicol 7(7), 793–802. [DOI] [PubMed] [Google Scholar]
- 83.Kristensen M and Brodin B (2017) Routes for drug translocation across the blood-brain barrier: exploiting peptides as delivery vectors. J. Pharm. Sci 106, 2326–2334. [DOI] [PubMed] [Google Scholar]
- 84.Elkamhawy A et al. (2017) Design, synthesis, biological evaluation and molecular modelling of 2-(2-aryloxyphenyl)-1,4-dihydroisoquinolin-3(2H)-ones: A novel class of TSPOligands modulating amyloid-β-induced mPTP opening. Eur. J. Pharmac. Sc 104, 366–381. [DOI] [PubMed] [Google Scholar]
- 85.Bruno A et al. (2019) Unbinding of Translocator Protein 18kDa (TSPO) Ligands: From in Vitro Residence Time to in Vivo Efficacy via in Silico Simulations. ACS Chem. Neurosci. 10 (8), 3805–3814. [DOI] [PubMed] [Google Scholar]
- 86.Laurent B et al. (2015) Epock: rapid analysis of protein pocket dynamics. Bioinformatics 31(9), 1478–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xia Y et al. (2019) A unified structural model of the mammalian translocator protein (TSPO). J. Biomol. NMR, doi: 10.1007/s10858-019-00257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patching SG (2014) Surface plasmon resonance spectroscopy for characterization of membrane protein-ligand interactions and its potential for drug discovery. Biochim. Biophys. Acta 1838, 43–55. [DOI] [PubMed] [Google Scholar]
- 89.Fischer M et al. (2011) One crystal, two temperatures: cryocooling penalties alter ligand binding to transient protein sites. Chem. Bio. Chem 5 16, 1560–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee YS et al. (2012) Solution structures of the prototypical 18kDa translocator protein ligand, PK 11195, elucidated with 1H/13C NMR spectroscopy and quantum chemistry. ACS Chem. Neurosci 3, 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shah F et al. (1994) Synthesis of the enantiomers of [N-methyl-11C]PK 11195 and comparison of their behaviors as radioligands for PK binding sites in rats. Nucl. Med. Biol 21(3), 573–581. [DOI] [PubMed] [Google Scholar]
- 92.Chipot C et al. (2018) Perturbations of native membrane protein structure in alkyl phosphocholine detergents: A critical assessment of NMR and biophysical studies. Chem. Rev 118, 3559–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.DeLano WL (2008) The Pymol molecular graphics system. DeLano Scientific LLC: Palo Alto, CA, USA, Available online: https://www.pymol.org. [Google Scholar]