Abstract
Transcriptional dysregulation induced by disease-defining genetic alterations of proteins in transcriptional machinery is a key feature of cancers. MED12 is the central architectural subunit in the kinase module of Mediator, a large transcriptional regulatory complex that controls essential steps of transcription. Emerging evidence links deregulated MED12 to human cancers. MED12 is frequently mutated in benign tumors and cancers. While the missense mutations of MED12 in benign tumors disrupt the kinase activity of Mediator, MED12 mutations in cancers could eliminate the interaction between Mediator complex and RNA polymerase II, leading to severe transcriptional misregulation. Aberrant expression of MED12 is associated with the prognosis of various types of human cancers. Loss of MED12 function has been associated with the development of resistance to chemotherapeutics. Moreover, MED12 is modified by post-transcriptional regulations. Arginine methylation of MED12 has been shown to regulate MED12-mediated transcriptional regulation and response to chemotherapeutics in human cancer cell lines. In this mini-review, we will provide an overview of the roles of MED12 in the development of benign and malignant tumors as well as its roles in chemo-resistance. The studies of MED12 exemplifies that aberrant transcriptional programming is a therapeutic vulnerability for certain types of cancer.
Keywords: Mediator, MED12, mutation, methylation, drug resistance
In this mini-review, we will provide an overview of the roles of MED12 in the development of benign and malignant tumors as well as its roles in chemo-resistance. The studies of MED12 exemplifies that aberrant transcriptional programming is a therapeutic vulnerability for certain types of cancer.
MEDIATOR COMPOSITION AND FUNCTIONS
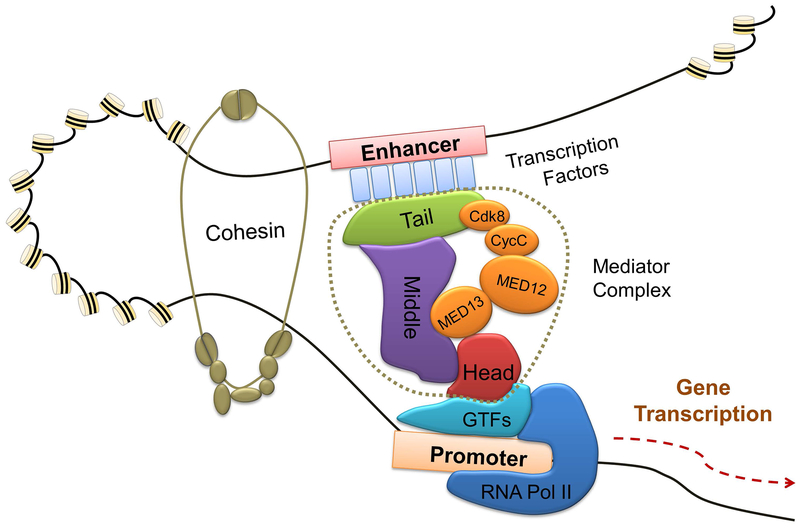
Using genetic and biochemical approaches, the Mediator of RNA polymerase II (RNAPII) was first identified in 1990s as a central integrator for transcription through interaction with gene-specific transcription factors as well as RNAPII, both in yeast and mammalian cells. Mediator is a multi-subunit complex, comprised of 25 subunits in budding yeast and approximately 30 subunits in humans. Because Mediator interacts with over 3000 transcription factors (TFs), it controls a vast array of the gene transcription in an evolutionarily conserved manner.1 Mediator is assembled in four distinct modules, termed “head”, “middle”, “tail”, and “kinase”.2 Structural and biochemical data obtained from yeast and humans revealed that the head and middle modules are functionally essential for transcription regulation, whereas the tail and CDK kinase modules play regulatory roles.1 Mediator interacts with RNAPII primarily through head module contacts. Gene-specific TFs generally bind Mediator through its tail and kinase domains, then transduce the regulatory information through the middle and head modules of Mediator to RNAPII (Figure 1). Thus, all Mediator modules are involved in gene-specific transcriptional regulation and the architecture of Mediator enables long-range gene activation.
Figure 1. Mediator is a central integrator and processor of RNA polymerase II transcription.
Mediator transduces regulatory information conveyed by signal-activated transcription factors to elicit gene expression changes in diverse biological processes, including development, differentiation, and homeostasis. Mediator is structurally organized into four functional modules: “head”, “middle”, “tail”, and “kinase”. The Mediator kinase module comprises of four subunits: MED13, MED12, CycC, and CDK8, which dynamically interact with the core modules during transcription elongation. The schematics show the relative position of Mediator components in transcription complex. Gene-specific transcription factors generally bind Mediator through its tail and kinase domains. Cohesion stabilizes the enhancer-promoter loop to enable the middle and head modules of Mediator to interact with RNA Pol II to conduct signaling dependent transcription.
FUNCTIONS OF MED12 IN MEDIATOR COMPLEX AND PHYSIOLOGICAL PROCESSES
MED12, along with MED13, Cyclin C (CycC), and either CDK8 or CDK19, comprise the “kinase” module that reversibly associates with the core Mediator. MED12 activates the kinase activity of CDK8 by bridging the interaction between MED13 and CycC-CDK8 (Figure 1). The kinase-stimulating activity of MED12 depends on its direct interaction with CycC.3 CycC, a highly conserved cyclin family member, consists of a negatively charged surface groove mediating its CDK8-binding as well as a CycC-specific surface for MED12 binding. MED12 binds to CycC via its N-terminus encoded largely by exons 1 and 2, where the hotspot mutations most frequently reside in hormone-dependent tumors. 3 Notably, reciprocal mutation of residues at the interface of MED12 and CycC on either protein uncouples CycC-CDK8 from core Mediator and severely impairs CycC-dependent CDK8 kinase activity.4 Moreover, MED13 interacts with the C terminus of MED12 and plays an allosteric role in regulating the interaction between a mutant form of MED12 and CycC-CDK8/CDK19. Thus, when MED13 is present, mutant MED12 can bind, but is unable to activate CycC-CDK8/CDK19.5 These studies implicate MED12-CycC interface as a putative target in CDK8-driven cancers.
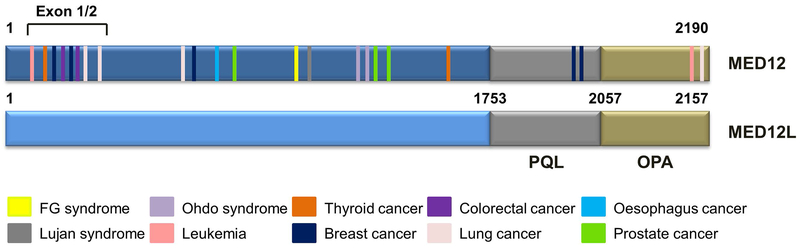
Because MED12 regulates essential physiological processes such as development and cell fate determination, deregulation of MED12 is often linked to human cancers. MED12 knockout mice elicit full developmental arrest at E7.5, demonstrating that MED12 plays critical roles in early development.6 MED12 (also called TRAP230 or HOPA) maps to Xq 13.1, a region highly mutated (~70%) in uterine leiomyomas. Missense mutations of MED12 can cause X-linked mental retardation, notably Opitz-Kaveggia syndrome (also known as FG syndrome) and Lujan-Fryns syndrome.7,8 The protein encoded by MED12L shares 67% amino acid sequence similarity to MED12 and contains MED12-like PQL and OPA domains,4 thus MED12L may compensate MED12 function in a context-dependent manner (Figure 2). This review will provide an overview of MED12 in human carcinogenesis and its regulation of therapeutic drug response.
Figure 2. Somatic mutations of MED12 in human cancers.
Pathogenic mutations and their approximate locations in MED12 are colored and annotated in the legend. The CycC/CDK8 binding interface on MED12, corresponding to its N-terminal 100 amino acids, is disrupted by exon1/2 mutations, leading to loss of Mediator-associated CDK activity. Besides the hot spot of exon 1/2, the distribution of MED12 mutations is dispersed in human cancers. Mutations shown here are drawn based on data from COSMIC database.
MED12 MUTATIONS IN HUMAN CANCERS
With the advance of next generation sequencing, the incidence of MED12 missense mutations in human tumors have been continuously growing. MED12 mutations occur at a high frequency (59-80%) in estrogen-dependent benign tumors, including uterine leiomyomas and fibroadenomas and phyllodes tumors of the breast. These mutations are clustered on highly conserved amino acids residues (L36, Q43 and G44) mapped to exon 2.9–11 As described above, MED12 activates CycC-CDK8 through a direct interaction between MED12 amino acids 1-100 (encoded by exons 1 and 2) and a conserved surface groove on CycC. Therefore, the benign tumor-linked mutations at the MED12 CycC-binding interface substantially impair CycC binding and CDK8 activation, leading to global gene expression changes in MED12 mutant expressing tumors. Because the kinase module is the regulatory unit in the assembly of transcription complex engaging signal-activated TFs, alteration of genes in transforming growth factor-β (TGF-β), Wnt/β-catenin and estrogen receptor α (ERα) signaling pathways have been reported, which may lead to initiation of tumorigenesis. Moreover, uterine leiomyomas associated MED12 mutation, Med12 c.131G>A, was reported as a “gain-of-function” mutation, which causes genomic instability and drives tumor formation.12
Large-scale genomic analyses have identified highly recurrent MED12 somatic mutations, albeit at a lower frequency, in hormone-dependent cancers, such as breast cancer, prostate cancer, and ovarian cancer, as well as in other cancers, such as lung cancer, colon cancer, and leukemia. Notably, the spectrum of MED12 mutations and cell types harboring MED12 mutations are different between benign tumors in estrogen-responsive tissues and in cancers. Barbieri et. al. reported recurrent MED12 missense mutations in 5.4% of prostate cancer by exome sequencing.13 In contrast to MED12 exon 2 mutations found in the stromal stem cells in uterine leiomyomas, the MED12 mutation in prostatic carcinoma resides in exon 26. The prominent mutation found in epithelial cells is the substitution of leucine 1224 by phenylalanine (L1224F). MED12 mutations in prostate cancer has been proposed to interfere with the androgen signaling pathway and the CDK8-dependent transcriptional regulation of p53.13 However, this notion was challenged by the finding of Kampjarvi and colleagues that the L1224F mutation affects neither the interaction between MED12 and Cyclin C-CDK8/19 in the kinase module nor the Mediator-associated CDK activity. Rather, the L1224F mutation on MED12 effected its binding to other Mediator subunits (MED1, MED13, MED13L, MED14, MED15, MED17, and MED24).14 In order to validate the prevalence of MED12 p.L1224F mutation, Stoehr et. al. analyzed a cohort of Caucasian patients with prostate cancer (n=223) and could not detect MED12 p.L1224F mutation in any cases.15 Similarly, Yoon and colleagues investigated the mutation sites in exons 2 and 26 of MED12 among 102 Korean patients who underwent radical prostatectomy for prostate cancer and could not identify MED12 mutations.16 Thus, the prevalence of MED12 somatic mutations in prostate cancer, and functional effects of the mutation in prostate carcinogenesis requires further study.
The distribution of MED12 mutations in leukemia is diverse. The first identified MED12 mutation (c.97G>T, p.E33X) in T-cell acute lymphoblastic leukemia (T-ALL) is a nonsense mutation.17 The mutation affects the last codon of exon 1 and results in the usage of alternative translation start site. As the result, an N-terminal truncated protein was generated which failed to localize to the nucleus due to the absence of nuclear localization signal (NLS).17 The mis-localization of MED12 abrogates its interaction with other Mediator components.17 Subsequent genome-sequencing identified additional MED12 mutations including a frame shift mutation p.V167fs, a missense mutation p.R1989H, and a splice site mutation g.chrX:70339329T>C.18 Interestingly, nearly 60% of MED12 mutations were identified from immature T-ALL cases, implying that pediatric T-ALL harbor genomic complexity. Identifying early disease driven mutations will help to stratify patients for personalized treatments.18,19 In Chronic Lymphocytic Leukemia (CLL), N-terminal MED12 mutations were identified with a frequency of 5.2% (37/709), 6.9% (12/188), and 8.8% (10/110) in three independent studies.20,21 NOTCH signaling is known to regulate differentiation and tumorigenesis in multiple cancers, such as CLL.22 Although the occurrence of MED12 and NOTCH1 mutations in CLL are mutually exclusive, NOTCH1 intracellular domain (NICD), the active form of NOTCH1, were found elevated in CLL samples harboring MED12 mutations. Because NICD is a substrate of Cyclin C-CDK8 kinase, this finding is congruent with the N-terminal MED12 mutations affecting CDK8 kinase activity and NOTCH1 activation. Therefore, mutating MED12 appears to be an alternative route to activate NOTCH signaling in CLL pathogenesis.19,21
MED12 mutations have also been identified in colorectal cancer (CRC), gastric cancer, and non-anaplastic thyroid cancer by next-generation sequencing. A MED12 exon 2 mutation was observed in 389 patients with colon cancer (0.3%).23 Subsequent reports by Kampjarvi et. al. and The Cancer Genome Atlas (TCGA) Network identified two MED12 exon 2 mutations in 392 (0.5%) and 224 (0.4%) CRC samples, respectively, indicating that mutations of MED12 are rare and may not contribute to CRC tumorigenesis.23–25 The frequency of MED12 mutations is much higher in hereditary diffuse gastric cancer and non-anaplastic thyroid cancer. MED12 mutations occur in 48% (14/30) of sporadic gastric cancers (ILe1115Thr) and in 14% (8/57) of non-anaplastic thyroid cancers (Gly44Cys).26,27 However, the number of patients in these series were small and these findings require validation. To date mutations of MED12 do not appear to correlate with prognosis, and the clinical relevance of MED12 mutations in pathogenesis of these cancer types remains to be determined. It is unclear if MED12 mutations are passenger or driver mutations and if MED12 mutations alter essential signaling pathways that cancer cells become addicted to.
Collectively, MED12 mutations are frequently found in different human cancer types, meeting the criteria of so-called “cancer driver genes” identified through large-scale genomic analyses.28 Coincidentally, a functionally related MED12L was implicated as a putative cancer driver gene in oral squamous cell carcinomas.29 The spectrum of MED12 mutations in human cancers is diverse, and distinct from mutations noted in benign tumors, in which mutations are largely clustered in exon 2. Table 1 and Figure 2 illustrate the MED12 mutations in different types of cancers in the COSMIC database (the Catalogue of Somatic Mutations in Cancer, http://cancer.sanger.ac.uk). Additional studies are needed to determine mutation prevalence using large cohorts of samples, and interrogate the functional significance of MED12 mutations in human carcinogenesis.
Table 1.
Alterations of MED12 in human cancers.
| Alteration | Point Mutation | Copy Number Variation | Gene Expression | |||||
|---|---|---|---|---|---|---|---|---|
| Tissue Type | sample | % | Gain/Loss | sample | % | Over/Under | sample | % |
| Adrenal gland | 9/735 | 1.22 | Over | 13/79 | 16.46 | |||
| Loss | 2/268 | 0.75 | Under | 1/79 | 1.27 | |||
| Autonomic ganglia | 1/1231 | 0.08 | ||||||
| Biliary tract | 13/814 | 1.6 | ||||||
| Bone | 8/796 | 1.01 | Loss | 1/80 | 1.25 | |||
| Breast | 888/6016 | 14.76 | Gain | 1/1544 | 0.06 | Over | 64/1104 | 5.8 |
| Loss | 9/1544 | 0.58 | Under | 28/1104 | 2.54 | |||
| Cervix | 14/386 | 3.63 | Gain | 3/313 | 0.96 | Over | 45/307 | 14.66 |
| Under | 4/307 | 1.3 | ||||||
| Central nervous | 36/3066 | 1.17 | Over | 27/697 | 3.87 | |||
| system | Loss | 7/1093 | 0.64 | Under | 16/697 | 2.3 | ||
| Endometrium | 67/1097 | 6.11 | Loss | 6/598 | 1 | Over | 47/602 | 7.81 |
| Genital tract | 1/126 | 0.79 | ||||||
| Haematopoieticand lymphoid | 110/6089 | 1.81 | Loss | 1/835 | 0.12 | Over | 7/221 | 3.17 |
| Kidney | 23/2668 | 0.86 | Over | 17/600 | 2.83 | |||
| Loss | 5/1027 | 0.49 | Under | 20/600 | 3.33 | |||
| Large intestine | 180/4609 | 3.91 | Gain | 1/773 | 0.13 | Over | 107/610 | 17.54 |
| Loss | 4/773 | 0.52 | Under | 12/610 | 1.97 | |||
| Liver | 38/2280 | 1.67 | Loss | 4/692 | 0.58 | Over | 20/373 | 5.36 |
| Lung | 135/4153 | 3.25 | Gain | 3/1185 | 0.25 | Over | 44/1019 | 4.32 |
| Loss | 11/1185 | 0.93 | Under | 25/1019 | 2.45 | |||
| NS | 28/356 | 7.87 | ||||||
| Oesophagus | 17/1633 | 1.04 | Over | 28/125 | 22.4 | |||
| Loss | 3/546 | 0.55 | Under | 1/125 | 0.8 | |||
| Ovary | 10/1431 | 0.7 | Over | 30/266 | 11.28 | |||
| Loss | 5/729 | 0.69 | Under | 5/266 | 1.88 | |||
| Pancreas | 21/2407 | 0.87 | Over | 6/179 | 3.35 | |||
| Loss | 1/929 | 0.11 | Under | 8/179 | 4.47 | |||
| Placenta | 1/5 | 20 | ||||||
| Pleura | 2/358 | 0.56 | ||||||
| Prostate | 55/3288 | 1.67 | ||||||
| Salivary gland | 5/359 | 1.39 | ||||||
| Skin | 78/1739 | 4.49 | Loss | 2/650 | 0.31 | Over | 19/473 | 4.02 |
| Small intestine | 3/111 | 2.7 | ||||||
| Soft tissue | 1875/5900 | 31.78 | Over | 13/263 | 4.94 | |||
| Loss | 1/286 | 0.35 | Under | 1/263 | 0.38 | |||
| Stomach | 47/1104 | 3.35 | Gain | 1/501 | 0.2 | Over | 37/285 | 12.98 |
| Loss | 12/501 | 2.4 | Under | 5/285 | 1.75 | |||
| Testis | 2/398 | 0.5 | Loss | 2/152 | 1.32 | |||
| Thyroid | 51/1868 | 2.73 | Over | 17/513 | 3.31 | |||
| Loss | 1/506 | 0.2 | Under | 17/513 | 3.31 | |||
| Upper aerodigestive tract | 23/1674 | 1.37 | Over | 52/522 | 9.96 | |||
| Loss | 5/563 | 0.89 | Under | 18/522 | 3.45 | |||
| Urinary tract | 41/1114 | 3.68 | Gain | 1/419 | 0.24 | Over | 63/408 | 15.44 |
| Loss | 7/419 | 1.67 | ||||||
DEREGULATION OF MED12 EXPRESSION IN HUMAN CANCERS
In addition to mutations, MED12 is often overexpressed in human cancers (Table 1). Nuclear accumulation of MED12 can be detected in 40% of castrate-resistant prostate cancers (CRPC) and 20% of locally recurrent prostate cancers (LOC), as compared to less than 11% in the androgen-dependent prostate cancer, and non-detectable level in the benign prostatic tissues.30 Because knockdown of MED12 decreases proliferation, reduces G1- to S-phase transition, and increases cell cycle inhibitor p27 level, MED12 expression levels have been associated with a high proliferative index. Positive feedback regulation between TGF-β signaling and MED12 transcriptional function has been reported. The activation of TGF-β signaling stimulates the nuclear accumulation of MED12 in cell lines, while knocking down MED12 reduced the expression of TGF-β target genes, such as vimentin. Moreover, genomic and epigenomic dysregulation of the MED12/MED12L axis also occurs frequently in CRPC. These studies indicate that MED12 may promote the proliferation of prostate cancer cells and contribute to anti-androgen resistance.30
In contrast to prostate cancer, MED12 expression shows the opposite prognostic association in breast cancer patients. Higher MED12 expression is associated with longer relapse-free survival (RFS) in patients treated with chemotherapy.31,32 The expression levels of CDK8 positively correlate with MYC, as well as CDK19, CycC, and MED13 in breast tumors but are inversely correlated with MED12. Moreover, p53 mutant expressing breast tumors typically have higher levels of expression of CDK8, CDK19 and CycC but lower levels of expression of MED12, indicating that MED12 may have distinct roles in regulating the CDK8 kinase module, or eliciting kinase module-independent function in breast cancers.33
The level of MED12 expression is important for hematopoietic stem cell (HSC) homeostasis, as in vivo knockout of MED12 results in rapid bone marrow aplasia, leading to acute lethality. Aranda-Orgilles and colleagues showed that MED12 maintains HSC function in a cell-autonomous manner and is independent of CycC/CDK8, as deletion of mediator kinase module subunits does not affect HSC survival. MED12, together with p300, preserves the enhancers in an active state. Loss of MED12 causes depletion of H3K27Ac, an active histone modification marked by p300, at the enhancers of essential HSC genes and abrogates hematopoietic-specific transcriptional programs. This MED12-dependent enhancer regulation may be essential for maintaining the normal physiological functions of HSC, and as a result, aberrant regulation leads to malignant hematopoiesis.34 MED12 expression is also strongly associated with the sensitivity of leukemia to chemotherapeutics. In Jurkat leukemia cells, loss of MED12 prevented cells from entering apoptosis following T chemotherapy18.
The oncogenic function of MED12 may involve crosstalk with oncogenic signaling pathways in ovarian and lung cancer. Loss of MED12 decreases the expression of EGFR in epithelial ovarian cancer (EOC). This coincides with the positive correlation of MED12 with EGFR expression in EOC patients. MED12 knockout in ovarian cancer cell lines decreases EGFR protein levels. Thus, MED12 may regulate the dormancy of EOC through regulation of EGFR.35 Besides the function of MED12 in genomic signaling, MED12 was reported to activate TGF-beta receptor 2 (TGFβR2) in the cytoplasm, constituting a non-genomic signaling pathway. MED12 has been detected in both the nucleus and cytoplasm in lung cancer cells. It is highly plausible that MED12 in different cellular compartments elicits different functions: nuclear MED12 may be involved in modulating the function of CDK8 kinase module to regulate transcription, whereas the cytoplasmic MED12 might interact with cell surface growth factors such as TGFβR2, both of which may contribute to therapeutic drug resistance. How MED12 cellular localization is regulated and the contribution of nuclear vs. cytoplasmic MED12 to the development of cancers are questions that remain to be elucidated.
FUNCTIONS OF MED12 METHYLATION IN HUMAN CARCINOGENESIS
Transcriptional abnormalities caused by aberrant epigenetic events are an emerging theme of the cancer epigenome.36 TFs, co-factors, and epigenetic enzymes are often subjected to post-transcriptional modifications (PTMs) that regulate their respective activities. Protein arginine methylation catalyzed by protein arginine methyltransferases (PRMTs) in mammalian cells share many of the attributes with other covalent modifications.37 We have identified MED12 as a substrate for coactivator-associated arginine methyltransferase 1 (CARM1), which asymmetrically dimethylates protein substrates on the arginine residues.38 We have shown that the expression levels of CARM1 and MED12 in cancers are positively correlated. In addition, their high expression often predicts better prognosis in breast cancer patients treated with chemotherapy. We identified two MED12 methylation sites, arginine 1862 (R1862), and arginine 1912 (R1912).38 By searching the COSMIC database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) for mutations on R1862 and R1912, we found that R1862 is mutated in a case of lung carcinoma, and a somatic, homozygous mutation at R1912 was identified in a melanoma from a patient who developed resistance to combined treatment with RAF and MEK (mitogen-activated protein kinase) inhibitors. Most recently, Mark Bedford’s group reported a third CARM1-catalyzed MED12 methylation site, arginine 1899 (R1899), is involved in recruitment of the Tudor domain-containing effector molecule (TDRD3), and potentiates the interaction of MED12 with activating noncoding RNAs (ncRNAs).39 These studies suggest that MED12 is modulated by methylation and possibly other PTMs, and distinct methylation sites may engage different functions of MED12 in cancer cells.
MED12 IN DRUG RESISTANCE
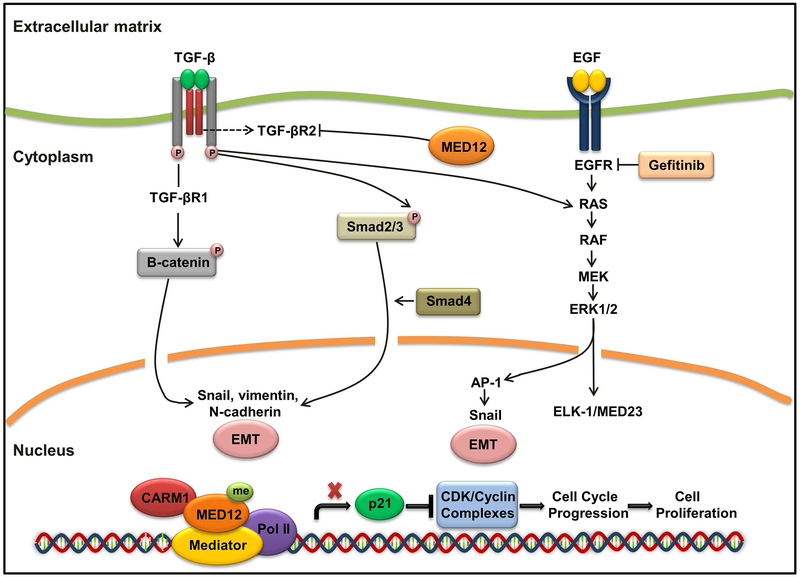
The finding of and mechanism by which MED12 is involved in mediating therapeutic drug response were described in two studies. It is known that mutations of oncogenes could be acquired during the course of treatment and lead to failure of response to therapeutic drugs. Lung cancers harboring activating EGFR mutations develop resistance to EGFR inhibitors. Similarly, melanomas with activating BRAF mutations develop resistance to BRAF inhibitors. Using a large-scale RNAi screen, Huang and his colleagues identified MED12 as a predictor of response to ALK and EGFR inhibitors in non-small-cell lung cancer (NSCLC) cells.40 They noted that a portion of MED12 is localized in cytoplasm, where it physically interacts with TGFβR2 and negatively regulates TGFβR2 signaling. MED12 suppression is accompanied by activation of TGF-βR signaling through the RAS-RAF-MEK-ERK pathway, causing resistance to EGFR, ALK, and BRAF inhibitors. In MED12-deficient cells, the inhibition of TGF-βR signaling could restore drug responsiveness, indicating that TGF-βR signaling is downstream of MED12 and ablation of both induces synthetic lethality in MED12-deficient, drug-resistant tumors.40 The findings by Huang and colleagues highlight the causal effects of MED12 deficiency in tumor resistance to tyrosine kinase inhibitor treatment (Figure 3).40,41 Reduced MED12 function was later confirmed by Rosell and colleagues as a hallmark of resistance to tyrosine kinase inhibitors in EGFR-mutant NSCLC.42 In contrast to NSCLC, MED12 function seems to be intact in small-cell lung cancer (SCLC), in which, no association has been found between the expression of MED12 and TGF-βR2 or with clinical variables such as overall survival. Thus, the functional link of MED12 and TGF-βR2 only exists in NSCLC subtype of lung cancer, and MED12 and TGFβR2 should not be taken as universal biomarkers for all types of lung cancer.43
Figure 3. MED12 and Cancer Drug Resistance.
In the canonical TGF-β pathway, TGF-βR2 activates TGF-βR1, which in turn phosphorylates Smad2 and Smad3. Smad2 and Smad3 then translocate to the nucleus and regulate the expression of TGF-β target genes. TGF-β signaling induces EMT through the phosphorylation of β-catenin, leading to the up-regulation of Snail, vimentin, and N-cadherin. Loss of MED12 causes drug resistance by increasing the level of TGFβR2, which activates the ERK and SMAD pathways. This leads to cell proliferation and features of EMT. Additionally, methylation of MED12 by CARM1 renders cancer cells sensitive to chemotherapy drugs through suppression of p21 transcription.
We have shown that the higher levels of MED12 and CARM1 predict better response to chemotherapeutics in breast cancer.31 Methylation of MED12 by CARM1 renders cells sensitive to 5-fluorouracil, but not to the RTK (Receptor Tyrosine Kinases) inhibitors in vitro and in vivo, suggesting that the mechanism of mediating drug response in breast cancer cells is different from TGF-βR2-mediated pathways in NSCLC. Moreover, p21/WARF1 levels are up-regulated in MED12 methylation defective cells. In breast cancer, high levels of p21/WAF1 are associated with a poor prognosis in patients with breast cancer treated with chemotherapy. Thus, MED12 methylation may serve as a predictive biomarker for patients with breast cancer who receive chemotherapy (Figure 3).31 This is in keeping with the study of Huang et. al. where loss of MED12 not only resulted in TKI resistance, but also rendered lung cancer cells resistant to 5-fluorouracil and cisplatin.40 Thus, these findings demonstrate that alterations of MED12, both transcriptionally and post-translationally, modulate chemotherapeutic response, suggesting that interruption of MED12 signaling pathway could be a new strategy for treating drug-resistant cancers.
THE ROLES OF MED12 IN REGULATION OF SUPERENHANCER-ASSOCIATED GENES
Besides gene amplification, high levels of oncogene expression in cancer cells is often attributed to transcriptional regulation by highly active enhancers, so called super-enhancers. Inhibiting cancer-acquired, super-enhancer-addicted transcription has emerged as a new means to target historically non-druggable oncogenic proteins, such as c-Myc, in highly proliferative cancer cells. Because transcriptional regulators, such as Mediator and BRD4, are enriched in super-enhancers, super-enhancer-associated genes are often highly sensitive to the inhibition of BRD4. BET (bromodomain and extraterminal motif) inhibitors have been extensively investigated for treating hematopoietic cancers.44 Tumors can develop resistance to BET inhibitors with prolonged treatment. Thus, there is sustained interest in identifying combinatory therapies to selectively decrease the expression of super-enhancer-associated oncogenes, such as MYC. A functional shRNA screen conducted in AML cells identified MED12, MED13, MED23, and MED24 subunits as sharing functional similarity with BRD4, i.e., restrain myeloid maturation.45 These findings indicate that BRD4 and Mediator functionally coordinate in gene-specific transcriptional activation that are essential for AML maintenance. Therefore, targeting Mediator subunits MED12, MED13, MED23, and MED24 can potentially inhibit cell proliferation in AML cells.45
Recently, Kuuluvainen and colleagues showed that depletion of MED12 or MED13/MED13L drastically reduced the expression of super-enhancer addicted oncogenes (e.g., MYC) in colon cancer cells, leading to growth inhibition. The transcription factor involved is β-catenin, a known TF that interacts with MED12. β-catenin depletion causes reduction of MED12 binding to the super-enhancer of MYC. Although both Mediator components and BRD4 regulate the expression of oncogenes in super-enhancer dependent fashion in cancer cells, the set of genes effected by MED12 or MED13/MED13L depletion and inhibition of BRD4 do not completely overlap, suggesting that more efficient inhibition may be achieved by co-targeting MED12 and BRD4. At least in colon cancers, the oncogenic gene expression shows dependency of three Mediator subunits, MED12 and MED13/MED13L. Therefore, targeting the Mediator subunits alone, or in combination with BRD4 inhibition, may provide new treatment strategy for colon cancer.46
RELEVANCE OF MED12 TO ESTROGEN PATHWAY
Although, in breast cancer cell line models, MED12 is in the activator complex regulating estrogen receptor (ER) transcriptional activity, the mechanism by which MED12 affects estrogen receptor (ER) signaling remains unclear. One model is that Mediator and cohesin cooperate to enforce the long-range chromosomal interactions (via looping) that are essential for enhancer-driven RNAP II transcription (Figure 1). Mediator and cohesin co-occupy different promoters to generate cell-type-specific DNA loops to control gene expression program.47 Recently, MED12 was found to co-localize on the estrogen receptor-alpha (ESR1) gene with the cohesion subunit SMC3. The occupancy of either subunit across the ESR1 gene depends on the other, and both are required for RNAP II mediated transcriptional initiation, suggesting that high order chromatin architecture controlled by MED12 and cohesin may be exploited for regulation of ER expression and treatment of estrogen-dependent breast cancer.48 However, control of ER expression and ER-regulated transcription in ER-driven malignancies is far more complex and has been elegantly reviewed by others.49,50 Dr. Wen Liu’s group recently reported that MED12 interacts with the JmjC-domain-containing protein (JMJD6) and recruits JMJD6 to the active enhancers bound by ERα, releasing paused RNA polymerase on cognate estrogen target genes. These studies identified JMJD6 as a critical regulator for productive transcription of estrogen/ERα-bound enhancer coding genes. JMJD6 is a multi-functional protein: it is an iron (Fe2+) and 2-oxoglutarate (2-OG)-dependent dioxygenase as well as protein arginine demethylase. Because JMJD6 plays an essential role in ER transcription and can hydroxylate lysines on both histones and non-histone proteins,51,52 it may be exploited as a molecular target for developing epigenetic therapies for ERα-positive breast cancers.53 Although MED12 functionally regulates ERα signaling, mutations of MED12 were more common in triple negative breast cancer (TNBC) than ERα-positive breast cancer, indicating that MED12 might play different roles in the TNBC and estrogen-dependent breast cancer,54 as in the cases for lung cancer.
CONCLUSIONS
The reason for the dispersed mutation spectrum of MED12 in human cancers is intriguing. MED12 mutations are frequently found in estrogen-dependent benign tumors and in various cancer types, suggesting that mutation of MED12 could be a tumor initiating event. However, whether MED12 mutations in benign tumors in conjunction with other genetic mutations drive cancer progression await further investigation. The expression levels of MED12 are frequently altered in various types of human cancers, yet the prognostic association may be cancer type-specific. Attention should also be drawn to the subcellular localization of MED12 and post-transcriptional regulation of MED12, since both genomic and non-genomic functions of MED12 have been associated with therapeutic drug resistance. While the exact function of MED12 overexpression or mutations in the context of Mediator complex remain to be elucidated, MED12 is a therapeutic vulnerability in broad types of human cancers and synthetic lethal screens should be exploited for treatment of MED12 deregulated cancers in the future.
Despite the strong implication of MED12 in human carcinogenesis, many questions remain to be addressed. For example, why the frequency of MED12 mutations in malignant tumors is much lower than in benign tumors? How is MED12 protein nuclear localization controlled? Are PTMs of MED12 involved in the regulation of genomic and non-genomic functions of MED12? Although our search of the COSMIC database reveals copy-number variations of MED12 in various human cancers (Table 1), the alteration of copy number changes related to human carcinogenesis has not been reported. It is worth noting that MED12 is located on the X-chromosome, thus future mutation analyses should take into consideration whether mutation of MED12 occurs on the activated or inactivated X-chromosome in gynecological cancers. Finally, more effort is needed to delineate the cell- and tissue-type specific functions of MED12 and its mechanism of action in normal cells and cancer cells.
Acknowledgments:
The authors would like to thank members of Xu laboratory for helpful discussions, especially thank Kristine Donahue for editing.
Funding: This project is supported by National Natural Science Foundation of China (81502603), Natural Science Foundation of Zhejiang Province (LGF18H160016) and Medicine and Health Science Fund of Zhejiang Province (2018RC021) to Zhang, S., NIH R01 CA213293 and R01 CA236356 to W.X..
Footnotes
Competing interests: The authors declare that they have no competing interests.
REFERENCES
- 1.Soutourina J Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol. 2018;19: 262–274. [DOI] [PubMed] [Google Scholar]
- 2.Tsai KL, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat Struct Mol Biol. 2013;20: 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turunen M, Spaeth JM, Keskitalo S, et al. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep. 2014;7: 654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark AD, Oldenbroek M, Boyer TG. Mediator kinase module and human tumorigenesis. Crit Rev Biochem Mol Biol. 2015;50: 393–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park MJ, Shen H, Spaeth JM, et al. Oncogenic exon 2 mutations in Mediator subunit MED12 disrupt allosteric activation of cyclin C-CDK8/19. J Biol Chem. 2018;293: 4870–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocha PP, Scholze M, Bleiss W, Schrewe H. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development. 2010;137: 2723–2731. [DOI] [PubMed] [Google Scholar]
- 7.Risheg H, Graham JM Jr., Clark RD, et al. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat Genet. 2007;39: 451–453. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz CE, Tarpey PS, Lubs HA, et al. The original Lujan syndrome family has a novel missense mutation (p.N1007S) in the MED12 gene. J Med Genet. 2007;44: 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim WK, Ong CK, Tan J, et al. Exome sequencing identifies highly recurrent MED12 somatic mutations in breast fibroadenoma. Nat Genet. 2014;46: 877–880. [DOI] [PubMed] [Google Scholar]
- 10.Tan WJ, Chan JY, Thike AA, et al. MED12 protein expression in breast fibroepithelial lesions: correlation with mutation status and oestrogen receptor expression. J Clin Pathol. 2016;69: 858–865. [DOI] [PubMed] [Google Scholar]
- 11.Ng CC, Tan J, Ong CK, et al. MED12 is frequently mutated in breast phyllodes tumours: a study of 112 cases. J Clin Pathol. 2015;68: 685–691. [DOI] [PubMed] [Google Scholar]
- 12.Mittal P, Shin YH, Yatsenko SA, et al. Med12 gain-of-function mutation causes leiomyomas and genomic instability. J Clin Invest. 2015;125: 3280–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44: 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kampjarvi K, Kim NH, Keskitalo S, et al. Somatic MED12 mutations in prostate cancer and uterine leiomyomas promote tumorigenesis through distinct mechanisms. Prostate. 2016;76: 22–31. [DOI] [PubMed] [Google Scholar]
- 15.Stoehr R, Taubert H, Gaisa NT, et al. Lack of evidence for frequent MED12 p.L1224F mutation in prostate tumours from Caucasian patients. J Pathol. 2013;230: 453–456. [DOI] [PubMed] [Google Scholar]
- 16.Yoon N, Lim S, Kang SY, et al. Mutation of MED12 is not a frequent occurrence in prostate cancer of Korean patients. Asian J Androl. 2017;19: 346–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heikkinen T, Kampjarvi K, Keskitalo S, et al. Somatic MED12 nonsense mutation escapes mRNA decay and reveals a motif required for nuclear entry. Hum Mutat. 2017;38: 269–274. [DOI] [PubMed] [Google Scholar]
- 18.Spinella JF, Cassart P, Richer C, et al. Genomic characterization of pediatric T-cell acute lymphoblastic leukemia reveals novel recurrent driver mutations. Oncotarget. 2016;4: 65485–65503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Bie J, Demeyer S, Alberti-Servera L, et al. Single-cell sequencing reveals the origin and the order of mutation acquisition in T-cell acute lymphoblastic leukemia. Leukemia. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JA, Hwang B, Park SN, et al. Genomic Profile of Chronic Lymphocytic Leukemia in Korea Identified by Targeted Sequencing. PLoS One. 2016;11: e0167641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu B, Slabicki M, Sellner L, et al. MED12 mutations and NOTCH signalling in chronic lymphocytic leukaemia. Br J Haematol. 2017;179: 421–429. [DOI] [PubMed] [Google Scholar]
- 22.Arruga F, Gizdic B, Serra S, et al. Functional impact of NOTCH1 mutations in chronic lymphocytic leukemia. Leukemia. 2014;28: 1060–1070. [DOI] [PubMed] [Google Scholar]
- 23.Je EM, Kim MR, Min KO, Yoo NJ, Lee SH. Mutational analysis of MED12 exon 2 in uterine leiomyoma and other common tumors. Int J Cancer. 2012;131: E1044–1047. [DOI] [PubMed] [Google Scholar]
- 24.Kampjarvi K, Makinen N, Kilpivaara O, et al. Somatic MED12 mutations in uterine leiomyosarcoma and colorectal cancer. Br J Cancer. 2012;107: 1761–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majewski IJ, Kluijt I, Cats A, et al. An alpha-E-catenin (CTNNA1) mutation in hereditary diffuse gastric cancer. J Pathol. 2013;229: 621–629. [DOI] [PubMed] [Google Scholar]
- 27.Ibrahimpasic T, Xu B, Landa I, et al. Genomic alterations in fatal forms of non-anaplastic thyroid cancer: Identification of MED12 and RBM10 as novel thyroid cancer genes associated with tumor virulence. Clin Cancer Res. 2017;23: 5970–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onken MD, Winkler AE, Kanchi KL, et al. A surprising cross-species conservation in the genomic landscape of mouse and human oral cancer identifies a transcriptional signature predicting metastatic disease. Clin Cancer Res. 2014;20: 2873–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaikhibrahim Z, Offermann A, Braun M, et al. MED12 overexpression is a frequent event in castration-resistant prostate cancer. Endocr Relat Cancer. 2014;21: 663–675. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Zeng H, Wang Q, et al. MED12 methylation by CARM1 sensitizes human breast cancer cels to chemotherapy drugs. Sci Adv. 2015;9: e1500463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dos Anjos Pultz B, da Luz FA, de Faria PR, Oliveira AP, de Araujo RA, Silva MJ. Far beyond the usual biomarkers in breast cancer: a review. J Cancer. 2014;5: 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh M Multilayered prevention and treatment of chronic inflammation, organ fibrosis and cancer associated with canonical WNT/betacatenin signaling activation (Review). Int J Mol Med. 2018;42: 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aranda-Orgilles B, Saldana-Meyer R, Wang E, et al. MED12 regulates HSC-specific enhancers independently of Mediator kinase activity to control hematopoiesis. Cell Stem Cell. 2016;19: 784–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo XL, Deng CC, Su XD, et al. Loss of MED12 induces tumor dormancy in human epithelial ovarian cancer via downregulation of EGFR. Cancer Res. 2018;78: 3532–3543. [DOI] [PubMed] [Google Scholar]
- 36.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11: 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shishkova E, Zeng H, Liu F, et al. Global mapping of CARM1 substrates defines enzyme specificity and substrate recognition. Nat Commun. 2017;8: 15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng D, Vemulapalli V, Lu Y, et al. CARM1 methylates MED12 to regulate its RNA-binding ability. Life Sci Alliance. 2018;1: e201800117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang S, Holzel M, Knijnenburg T, et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151: 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosell R Mediating resistance in oncogene-driven cancers. N Engl J Med. 2013;368: 1551–1552. [DOI] [PubMed] [Google Scholar]
- 42.Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. The Lancet. 2013;382: 720–731. [DOI] [PubMed] [Google Scholar]
- 43.Yokouchi H, Nishihara H, Harada T, et al. Immunohistochemical profiling of receptor tyrosine kinases, MED12, and TGF-βRII of surgically resected small cell lung cancer, and the potential of c-kit as a prognostic marker. Oncotarget. 2017;8: 39711–39726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roe JS, Mercan F, Rivera K, et al. BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell. 2015;58: 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhagwat AS, Roe JS, Mok BYL, Hohmann AF, Shi J, Vakoc CR. BET bromodomain inhibition releases the Mediator complex from select cis-regulatory elements. Cell Rep. 2016;15: 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuuluvainen E, Domenech-Moreno E, Niemela EH, Makela TP. Depletion of Mediator kinase module subunits represses superenhancer-associated genes in colon cancer cells. Mol Cell Biol. 2018;38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kagey MH, Newman JJ, Bilodeau S, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prenzel T, Kramer F, Bedi U, Nagarajan S, Beissbarth T, Johnsen SA. Cohesin is required for expression of the estrogen receptor-alpha (ESR1) gene. Epigenetics Chromatin. 2012;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnal JF, Lenfant F, Metivier R, et al. Membrane and nuclear estrogen receptor alpha actions: from tissue specificity to medical implications. Physiol Rev. 2017;97: 1045–1087. [DOI] [PubMed] [Google Scholar]
- 50.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296 :1642–1644. [DOI] [PubMed] [Google Scholar]
- 51.Liu W, Ma Q, Wong K, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155: 1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. [DOI] [PubMed] [Google Scholar]
- 53.Gao WW, Xiao RQ, Zhang WJ, et al. JMJD6 licenses ERalpha-dependent enhancer and coding gene activation by modulating the recruitment of the CARM1/MED12 co-activator complex. Mol Cell. 2018;70: 340–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu ZY, Xie N, Tian C, et al. Identifying circulating tumor DNA mutation profiles in metastatic breast cancer patients with multiline resistance. EBioMedicine. 2018;32: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]