Abstract
Structural characterization of the HIV-1 Envelope (Env) glycoprotein has facilitated the development of Env probes to isolate HIV-specific monoclonal antibodies (mAbs). However, preclinical studies have largely evaluated these virus-specific mAbs against chimeric viruses, which do not naturally infect non-human primates, in contrast to the unconstrained simian immunodeficiency virus (SIV)mac239 clone. Given the paucity of native-like reagents for the isolation of SIV-specific B cells, we examined a method to isolate SIVmac239-specific mAbs without using Env probes. We first activated virus-specific B cells by inducing viral replication after the infusion of a CD8β-depleting mAb or withdrawal of antiretroviral therapy in SIVmac239-infected rhesus macaques. Following the rise in viremia, we observed 2- to 4-fold increases in the number of SIVmac239 Env-reactive plasmablasts in circulation. We then sorted these activated B cells and obtained 206 paired Ab sequences. After expressing 122 mAbs, we identified 14 Env-specific mAbs. While these Env-specific mAbs bound to both the SIVmac239 SOSIP.664 trimer and to infected primary rhesus CD4+ T cells, five also neutralized SIVmac316. Unfortunately, none of these mAbs neutralized SIVmac239. Our data show that this method can be used to isolate virus-specific mAbs without antigenic probes by inducing bursts of contemporary replicating viruses in vivo.
Keywords: SIV, monoclonal antibodies, rhesus macaques, CD8b depletion, antiretroviral therapy interruption
Introduction
Thirty-eight percent of HIV-infected people worldwide remain untreated despite the existence of antiretroviral therapy (ART).1,2 Currently, ART needs to be taken daily to control HIV replication. However, multiple strategies are being evaluated as alternatives to ART for prevention, therapy, and long-term remission, including virus-specific monoclonal antibodies (mAbs).3,4 Unlike ART, Abs have the ability to not only block viral infection, but to induce killing of infected cells by recruitment of cytolytic effector cells.5 Several studies have highlighted the importance of Ab-mediated viral suppression using Fc effector mechanisms, including Ab-dependent cellular cytotoxicity (ADCC).6, 7, 8, 9 Ab-mediated elimination of infected cells could therefore play a critical role not only in controlling HIV replication but also in reducing the size of the viral reservoir. Understanding the role of Abs and their in vivo mechanisms of action could greatly facilitate the development of HIV/AIDS cure strategies.
A single injection of neutralizing mAbs, administered either individually or as a cocktail, has been shown to be capable of preventing infection and suppressing simian-human immunodeficiency virus (SHIV) replication in Indian rhesus macaques.10, 11, 12, 13, 14, 15 While these results are promising, these chimeric SHIV strains are considered constrained viruses, which do not naturally infect non-human primates.16 In contrast, challenging rhesus macaques with simian immunodeficiency virus (SIV) results in robust infection with extremely fit and pathogenic viruses,17,18 especially when using the pathogenic SIVmac239 clone.19 In fact, the live attenuated SIV and the recombinant near full-length SIV rhesus rhadinovirus vaccines are the only vaccine strategies that have effectively provided protection from acquisition after challenge with this pathogenic clone.20, 21, 22 Additionally, the infusion of CD4-immunoglobulin (Ig)G2, eCD4 Ig, and 5L7 IgG1 also protected rhesus macaques against SIVmac239.23, 24, 25 Adeno-associated virus (AAV)-delivered 5L7 IgG1 prevented infection of a single macaque, presumably through ADCC activity, since 5L7 IgG1 does not detectably neutralize SIVmac239.25 To date, there are only three clonally related SIVmac239-specific neutralizing mAbs available.26 Unfortunately, the paucity of virus-specific Abs with therapeutic potential limits our ability to test Ab-based therapies with the tier 3 SIVmac239 challenge virus.
Methods for isolating virus-specific mAbs have been revolutionized in the past decade.27 Indeed, a better understanding of HIV Envelope (Env) structure has facilitated the design of stabilized soluble trimers that have been used as probes to sort HIV-specific B cells.28, 29, 30 While von Bredow et al.31 have recently generated a soluble SIVmac239 SOSIP.664 Env trimer, other soluble recombinant SIV probes were previously available, although they were not conformationally authentic.32 Unfortunately, these tools have not yet been successfully employed to isolate additional SIVmac239-specific mAbs with preventative or therapeutic capabilities in vivo.
In this study, we used an alternative strategy to isolate SIV-specific B cells that does not require prior knowledge of the SIVmac239 Env structure. We reasoned that rapid and transient viral replication, and the associated viral antigen production, would lead to the activation and recirculation of SIV-specific B cells. We therefore induced viral replication in three SIVmac239-infected rhesus macaques by treating them with a CD8β-depleting mAb or interrupting ART. We detected increased frequencies of SIV-specific plasmablasts in all three animals, and we isolated 206 paired Ab sequences from single B cells. Fourteen of the 122 screened mAbs were specific for SIVmac239 Env, targeting the different variable loops (V loops) and the gp41 fusion protein. Five of them neutralized the related SIVmac316 strain. Interestingly, all of these Env-specific mAbs also bound to the recently developed SIVmac239 SOSIP.664 trimer and to SIVmac239-infected primary rhesus CD4+ T cells, suggesting that these mAbs could possess antiviral activity in vivo.
Results
Selection of Animals with SIVmac239-Neutralizing Serum Titers
To identify animals with serum neutralizing reactivity against SIVmac239, we screened 34 SIVmac239-infected Indian rhesus macaques (Table 1). Twenty-five of these macaques belonged to our elite controller (EC) cohort, and nine were conventional progressors (CPs). The EC cohort was comprised of animals that had been controlling SIV replication to below 1,000 viral RNA (vRNA) copies/mL of plasma for up to 17 years (Table 1). We reasoned that long-term ECs might possess Abs with high levels of somatic hypermutation (SHM) given their prolonged exposure to low levels of antigen. CPs with high viral loads were also selected due to their high levels of circulating antigen, which may enhance SHM.
Table 1.
SIVmac239- and SIVsmE543-Specific Neutralizing Serum Titers of 34 SIVmac239-Infected Macaques
| Animal | Sex | MHC Class I Alleles | Weeks after Infection | SIVmac239 ID50 Titers | SIVsmE543 ID50 Titers | EC/CP |
|---|---|---|---|---|---|---|
| r01068 | M | Mamu-B*08+ | 728 | 1:14 | <1:10 | CP |
| r06029 | F | negative for relevant alleles | 41 | 1:23 | ND | EC |
| r08019 | F | Mamu-A*01+ | 136 | 1:23 | ND | EC |
| rh2358 | M | Mamu-A*01+, -B*08+ | 278 | 1:36 | <1:10 | EC |
| r08009 | M | Mamu-A*01+, -B*08+, -B*17+ | 70 | 1:36 | ND | EC |
| r08024 | M | Mamu-A*01+, -B*08+ | 82 | 1:40 | ND | EC |
| r09029 | M | Mamu-B*08+ | 728 | 1:42 | <1:10 | EC |
| r08046 | M | Mamu-B*17+ | 75 | 1:42 | ND | EC |
| r09062 | F | Mamu-A*02+, -B*17+ | 73 | 1:50 | ND | EC |
| r08054 | M | Mamu-B*08+ | 171 | 1:61 | <1:10 | EC |
| r08034 | M | Mamu-B*17+ | 75 | 1:65 | ND | CP |
| r05007 | F | Mamu-B*17+ | 79 | 1:83 | ND | EC |
| r02019 | F | Mamu-B*08+ | 291 | 1:85 | <1:10 | EC |
| r08062 | M | Mamu-B*17+ | 75 | 1:88 | ND | EC |
| r07032 | M | Mamu-B*08+ | 143 | 1:90 | ND | EC |
| r08047 | M | Mamu-A*02+, -B*17+ | 75 | 1:96 | ND | EC |
| r09083 | F | Mamu-B*17+ | 68 | 1:99 | ND | EC |
| r02075 | M | Mamu-B*08+ | 8 | 1:115 | ND | EC |
| r00008 | F | Mamu-A*02+, -B*08+ | 74 | 1:129 | ND | EC |
| r95061 | F | Mamu-A*01, -A*02+, -B*17+ | 886 | 1:131 | <1:10 | EC |
| r04132 | F | Mamu-B*08+ | 139 | 1:141 | ND | EC |
| r10072 | M | Mamu-B*08+ | 82 | 1:145 | ND | CP |
| r08016 | M | Mamu-B*08+ | 173 | 1:151 | <1:10 | CP |
| rh2365 | M | Mamu-A*02+, -B*08+ | 728 | 1:171 | <1:10 | EC |
| r09001 | M | Mamu-B*17+ | 77 | 1:192 | ND | EC |
| r08038 | M | Mamu-A*01+, -B*08+ | 80 | 1:193 | ND | EC |
| r11002 | F | Mamu-A*02+ | 88 | 1:263 | <1:10 | CP |
| rhBB35 | F | Mamu-B*08+ | 74 | 1:337 | ND | CP |
| r09089 | M | Mamu-B*08+ | 168 | 1:344 | <1:10 | EC |
| r11021 | M | Mamu-A*01+ | 85 | 1:411 | <1:10 | CP |
| r09037 | F | Mamu-B*08+ | 172 | 1:426 | 1:207 | EC |
| r11039 | M | Mamu-A*01+ | 84 | 1:1,141 | <1:10 | CP |
| r04135 | F | Mamu-A*02+, -B*08+ | 422 | 1:2,060 | 1:129 | EC |
| r10051 | F | Mamu-B*08+ | 80 | 1:2,157 | ND | CP |
Serum reactivity against SIVmac239 and SIVsmE543 was determined in 34 SIVmac239-infected macaques using the TZM-bl neutralization assay. Three of these CP macaques were treated with ART to control viral replication. Eight animals had moderate-to-high serum-neutralizing activity; five of these were CPs and three were ECs. Note that macaques r04135 and r09037 also made mAbs against the heterologous strain SIVsmE543. ID50, inhibitory plasma dilution at which 50% virus neutralization was attained; ND, not determined; EC, elite controller; CP, conventional progressor.
Although most animals exhibited marginal serum neutralizing activity, three EC and five CP macaques had moderate-to-high SIVmac239-neutralizing titers (>1:200; Table 1). Of these macaques, r04135 and r09037 also had low levels of neutralizing activity against the heterologous strain SIVsmE543, a divergent molecular clone from an independent SIV isolate.33 Macaque r10051, a CP macaque that experienced a chronic phase viral load set point of >106 vRNA copies/mL, generated the highest neutralizing titers against SIVmac239. We therefore selected r04135, r09037, and r10051 for subsequent experiments due to their potent serum neutralization activity against both SIVmac239 and SIVsmE543.
Induction of Viral Rebound by CD8β+ T Cell Depletion and ART Interruption
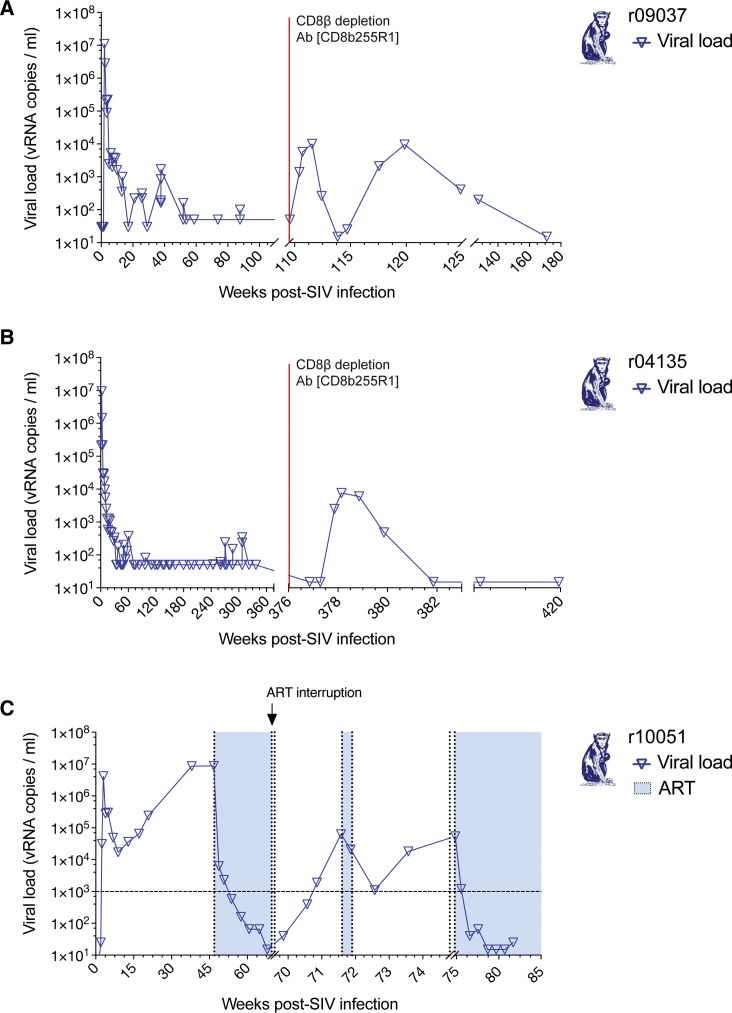
Previous studies have shown that CD8+ T cell responses are critical for suppression of viremia in EC macaques.34, 35, 36, 37, 38, 39 Our group, and others, have successfully induced SIV replication after the infusion of the anti-CD8β mAb CD8β255R1, which specifically targets rhesus CD8αβ+ T cells but not natural killer (NK), NKT, or γδ T cells.40 Replication of acute viruses is usually followed by a surge of viral antigen-specific plasmablasts in the peripheral blood of infected individuals.41, 42, 43 Abs produced by these antigen-specific plasmablasts are thought to be associated with the development of nAb titers for the cognate viruses.41 Since SIV-infected ECs control viral replication after the acute phase of infection, we reasoned that the depletion of CD8+ T cells using the anti-CD8β mAb CD8β255R1 would increase viral replication and antigen exposure and, in turn, reactivate SIV-specific B cells. Accordingly, we treated r04135 and r09037 with the anti-CD8β mAb, and then confirmed depletion of CD3+ CD8αβ+ T cells by flow cytometry. The absolute counts of CD3+ CD8αβ+ T cells were significantly reduced in r09037, while CD8αα+ T cell and NK cell counts remained near or slightly above baseline levels (Figure S1). Next, we assessed viral replication kinetics in these macaques (Figures 1A and B). Both animals had detectable viral loads at 1 week after depletion and reached peak viremia of 104 vRNA copies/mL on day 14 after CD8β+ T cell depletion. However, while r04135 required 35 days to control SIVmac239, r09037 did not establish control of viral replication to <1,000 vRNA copies/mL until more than 1 year after CD8β depletion.
Figure 1.
Viral Loads after CD8β T Cell Depletion and ART Interruption
We administered the CD8β-depleting Ab CD8β255R1 to the SIVmac239-infected ECs (A) r09037 and (B) r04135, which induced viral replication in vivo. (C) After the ART discontinuation, we detected SIVmac239 replication in r10051.
Of all the animals initially screened for neutralization activity, r10051 had the highest SIVmac239-nAb serum titers (Table 1). At 47 weeks post-infection, we administered ART for 5 months until plasma viremia reached <15 vRNA copies/mL to prevent progression to AIDS (Figure 1C). We reasoned that removal of ART would result in viral rebound and a consequent burst of recalled SIV-specific plasmablasts in circulation. Viral loads became detectable 2 days after ART discontinuation and slowly increased, reaching peak viremia of >104 vRNA copies/mL on day 14 after ART interruption (Figure 1C). ART was then resumed for 2 days. Viremia dropped from 104 to 103 vRNA copies/mL in 1 week (21 days after initial ART discontinuation), and then progressively increased until reaching >104 vRNA copies/mL again on day 35.
SIV-Specific Plasmablasts Recirculate following CD8β+ T Cell Depletion and ART Interruption-Induced Viral Rebound
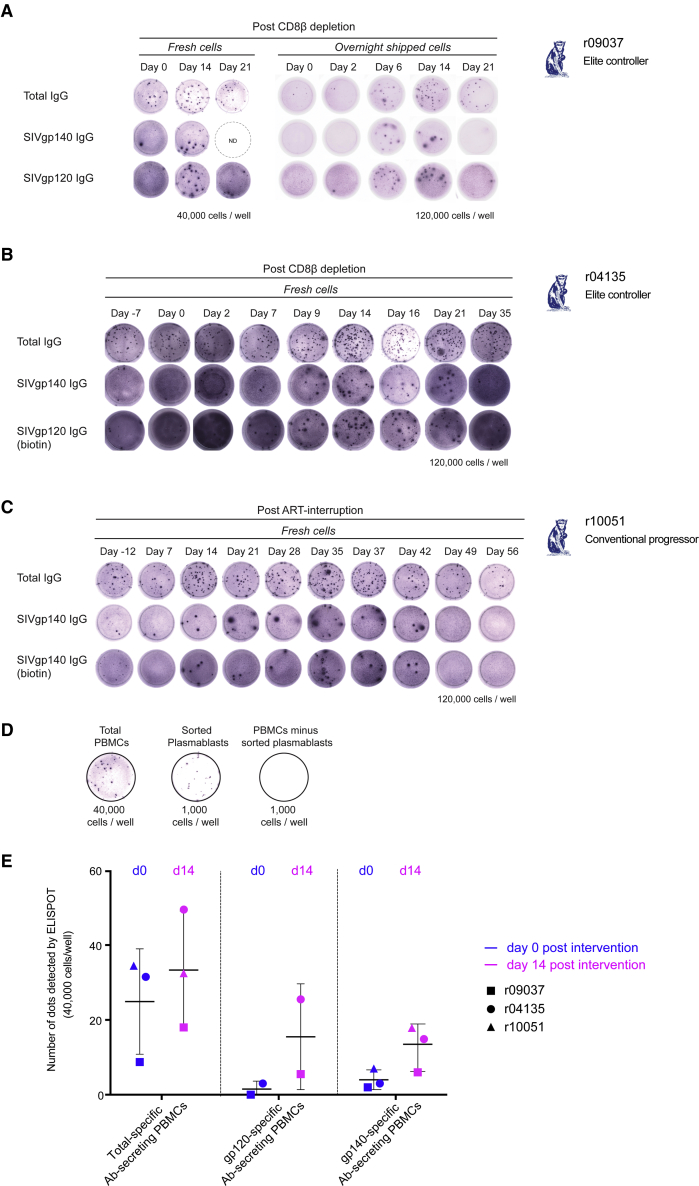
After CD8β depletion, we detected increased frequencies of SIVmac239-specific plasmablasts in peripheral blood mononuclear cells (PBMCs) by IgG enzyme-linked immunospot (ELISPOT). We observed this increase from day 6 to 21 after CD8β depletion in both ECs, and from day 14 to 42 after ART interruption in the CP (Figure 2A–2C). Plasmablast expansion peaked at day 14 after CD8β depletion and at day 35 after ART withdrawal. We confirmed the phenotype of the spot-forming population by sorting PBMCs based on a previously described plasmablast phenotype (CD3− CD20− CD14− CD16− human leukocyte antigen [HLA]-DR+ CD11c− CD123− CD80+ cells)44 and demonstrated that these sorted plasmablasts were actively secreting IgG by ELISPOT (Figure 2D).
Figure 2.
Recall of SIVmac239-Specific Plasmablasts after CD8β+ T Cell Depletion and ART Interruption
Of the total PBMCs, we detected an increased number of SIVmac239-specific plasmablasts by IgG ELISPOT in circulation in (A) r09037, (B) r04135, and (C) r10051. (A–C) Upper rows indicate the total Ab-secreting cells; middle and bottom rows show SIV-specific responses. (D) Sorting for plasmablast surface markers enriched for the Ab-secreting cell population as shown by ELISPOT comparison among unsorted total PBMCs, sorted cells, and the remaining cell fraction. (E) Frequency of total IgG-secreting cells and SIV-specific plasmablasts per 40,000 PBMCs (ELISPOT) from days 0 and 14 after CD8β+ T cell depletion or ART withdrawal from all three animals.
Of the total PBMCs in circulation, less than 0.2% of cells were actively secreting IgG in all three animals at the time of the interventions. At 40,000 cells/well, a small percentage of these IgG-secreting cells were SIVmac239-specific: 11.4% in r09037, 9.5% in r04135, and 20.3% in r10051 (Figure 2). During the peak of plasmablast expansion, the number of SIVmac239-specific B cells increased by 2- to 4-fold compared to baseline (29.2%, 31.9%, and 51.4% of the total IgG-secreting cells in r09037, r04135, and r10051, respectively) (Figure 2E).
Recalled Plasmablasts Expressed Mutated Abs, Some of Them with Long CDRH3s
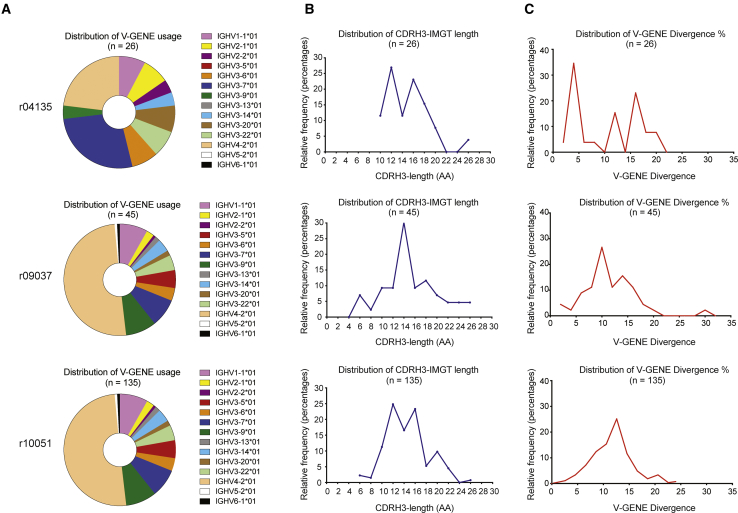
To understand the characteristics of the Ab repertoire induced by SIVmac239 infection in Indian rhesus macaques, we sorted single plasmablasts from r10051, r09037, and r04135 before and during peak plasmablast expansion. From these time points, we amplified a total of 304 variable heavy (VH) chains and 261 variable light (VL) chains, resulting in 206 plasmablast-derived VH/VL sequences: 26 mAbs from r04135, 45 mAbs from r09037, and 135 mAbs from r10051. We then examined the amplified sequences and identified one allele (IGHV4-2*01) present in the 62.2% and 50.4% of the mAbs isolated from r09037 and r10051, respectively. In r04135, this allele was the second most prevalent (23.1%), along with IGHV3-7*01, which was present in 26.9% of the isolated mAbs (Figure 3A).
Figure 3.
Characteristics of the VH from Recalled Plasmablasts from r04135, r09037, and r10051 after Induction of Viremia
(A) V-GENE distribution. (B) Histogram representation of VH CDR3 length distribution for all three macaques. (C) Histogram representation of SHM (% nucleotide difference from germline). Note that among 206 plasmablast-derived sequences from all three animals, we detected heavy chains with CDRH3 lengths of up to 26 aa and with >22% nucleotide divergence to their rhesus V-GENE germline sequences (IMGT). After CD8β depletion, plasmablasts were sorted at days 8 and 14 in r09037 and at days 14 and 16 in r04135. Following ART withdrawal, plasmablasts of r10051 were sorted at days 7, 9, 14, 16, and 21. The genetic analyses for each animal were performed with pooled samples from all time points after viremia induction.
HIV-specific human neutralizing mAbs usually share some common characteristics: atypically long VH CDR3 loops and greatly mutated sequences (as high as 39 aa long and 29.7% nucleotide divergence from germline).45,46 In contrast, typical human mAbs have a median VH CDR3 length of 14 aa and 5% nucleotide divergence from germline.47 Of the 206 plasmablast-derived VH/VL sequences, we identified 92 mAbs with VH CDR3s longer than 14 aa, with the highest being 26 aa long (Figure 3B), and 194 mAbs with a nucleotide divergence >5%. The most mutated mAbs reached 22% nucleotide divergence from germline (Figure 3C).
Abs Bind to Different Env Regions, SIVmac239 SOSIP.664, and Primary Infected CD4+ T Cells
Of the 206 plasmablast-derived VH/VL sequences, we expressed 122 mAb pairs and determined their specificity. Fourteen of these mAbs bound to the SIVmac239 gp140 foldon trimer (FT) protein.32 We then tested their binding to the following viral Env subunits: gp120, gp41, gp140 FT ΔV1/V2/V3, gp140 FT ΔV4, and the SIVmac239 SOSIP.664 (Table 2). 5L7 IgG1, which has previously been shown to bind the SIVmac239 Env and induce ADCC, was also included in our screening.25 While P1E09 and 5L7 IgG1 bound to the V4 loop, four other mAbs targeted the V1/V2/V3 loops. Binding specificity of one of the V1/V2/V3-specific mAbs, P5C08, was later verified by Pepscan, which targeted a linear epitope in the V1/V2 loops (Figure S2). We also identified one mAb, P2A12, with reactivity against gp140 FT that failed to bind when tested against both ΔV1/V2/V3 and ΔV4 constructs. Interestingly, all mAbs isolated from r10051, in addition to one from r09037, were gp41-specific. Lastly, all but one gp140 FT-specific mAb, P1G08, detectably bound SIVmac239 SOSIP.664 (Table 2). Since most of these mAbs bound SIVmac239 SOSIP.664, we assessed the neutralization activity of these mAbs. Surprisingly, none neutralized SIVmac239. However, five mAbs did neutralize SIVmac316, which differs from SIVmac239 by only 8 aa in Env.48 Interestingly, only the V1/V2/V3-specific mAbs and P2A12 neutralized the SIVmac316 variant. P2A12, whose epitope lies somewhere in the V loops, completely neutralized SIVmac316, whereas the other four neutralizing mAbs exhibited a maximum percentage neutralization (Vmax) of less than 80% at the highest concentrations tested (Table 2).
Table 2.
Functional Characteristics of the Env-Specific Abs Isolated from r04135, r09037, and r10051
| gp140 FT | gp140 FT ΔV1–V3 | gp140 FT ΔV4 | gp120 | gp41 | SIVmac239 SOSIP.664 | SIVmac239 IC50 (μg/mL) | SIVmac316 IC50 (μg/mL) | SIVmac316 Vmax | Binding to Infected CD4+ T Cells | |
|---|---|---|---|---|---|---|---|---|---|---|
| P2E04 | ++ | − | ++ | ++ | − | ++ | >50 | 1.33 | 72% | +++ |
| P2A12 | ++ | − | − | ++ | − | ++ | >50 | 0.0082 | 100% | +++ |
| P4D07 | ++ | ++ | ++ | − | ++ | ++ | >50 | >50 | ND | ++ |
| P5C08 | ++ | − | ++ | ++ | − | ++ | >50 | 0.129 | 85% | +++ |
| P1C08 | ++ | − | ++ | ++ | − | ++ | >50 | 0.0755 | 85% | +++ |
| P1E09 | ++ | ++ | − | ++ | + | ++ | >50 | >50 | ND | +++ |
| P5H02 | ++ | + | ++ | ++ | + | ++ | >50 | 8.078 | 54% | +++ |
| P2A04 | ++ | ++ | ++ | − | ++ | ++ | >50 | >50 | ND | + |
| P4A09 | ++ | ++ | ++ | − | ++ | ++ | >50 | >50 | ND | + |
| P2A11 | ++ | ++ | ++ | − | ++ | ++ | >50 | >50 | ND | + |
| P1G09 | ++ | ++ | ++ | − | ++ | ++ | >50 | >50 | ND | + |
| P1D12 | ++ | ++ | ++ | − | ++ | ++ | >50 | >50 | ND | + |
| P1G08 | ++ | ++ | ++ | − | + | − | >50 | >50 | ND | + |
| P2E05 | ++ | ++ | ++ | − | ++ | ++ | >50 | >50 | ND | + |
| 5L7 | ++ | ++ | + | ++ | − | ++ | >50 | 0.011 | 100% | +++ |
Of the 122 plasmablast-derived screened mAbs from all three macaques, 14 bound gp140 FT by ELISA. We determined the target region by using gp140 FT, gp140 FT ΔV1V2V3, gp140 FT ΔV4, gp120, gp41, and SIVmac239 SOSIP.664 in an ELISA format. None of the 14 SIV-specific mAbs exhibited neutralization activity against SIVmac239 at 50 μg/m, but 5 were capable of neutralizing SIVmac316 in a TZM-bl neutralization assay. We also assessed the ability of these Env-specific mAbs to bind SIVmac239-infected primary rhesus CD4+ T cells. IC50, inhibitory mAb concentration at which 50% virus neutralization was attained; Vmax, maximum percentage neutralization; ND, not determined. At 1 μg/mL: ++, optical density 450 (OD450) ≥ 0.5; +, 0.5 < OD450 ≥ 0.1, −, OD450 < 0.1. At 10 μg/mL: +++, binding ≥ 40%; ++, 40% < binding ≥ 20%; +, binding < 20%.
Ab effector functions that mediate antiviral immunity in vivo often depend on the ability of a mAb to bind Env on the infected cell surface. Rather than overexpressing recombinant Env in an immortalized cell line, we chose to study the interaction of mAbs with cell surface Env in the context of natural SIVmac239 infection—that is, in primary rhesus CD4+ T cell cultures—to provide a more accurate representation of what might occur when a mAb encounters an infected cell in vivo. In contrast to conventional cell-binding assays, our assay accounts for other variables that may affect mAb-Env interactions and mAb specificity for infected cells, including Env shedding and virus-induced cytopathic effects. Thus, we employed a flow cytometry-based assay to evaluate the affinity of the isolated mAbs for bona fide SIVmac239-infected cells. We assessed mAb binding to infected CD4+ T cells at 48 h post-spinoculation, using intracellular Gag p27 staining as a marker of bona fide infection. Because spinoculation results in the infection of only a portion of the total CD4+ T cells in culture (generally 20%–30% for rhesus CD4+ T cells), most cells are uninfected. All of the isolated mAbs exhibited very high specificity for Gag p27+ cells, as none of the mAbs bound the uninfected cells in the cultures. Interestingly, the mAbs with the strongest binding strength (i.e., highest affinity), P1E09 and P2A12, were the ones directed against either the V4 loop or a shared epitope that was disrupted in both ΔV1/V2/V3 and ΔV4 constructs (Figure S3). These two mAbs had nearly identical binding profiles to 5L7 IgG1, which binds the V4 loop (Figure S3), although they do not bind overlapping epitopes. We also identified two distinct Env-binding profiles in our group of isolated mAbs. gp120-specific mAbs bound a significantly higher proportion of infected cells than did gp41-specific mAbs (Table 2). Interestingly, the gp41-specific mAb that had not shown detectable reactivity against SIVmac239 SOSIP.664 by ELISA, P1G08, bound the lowest proportion of infected primary CD4+ T cells of all of the mAbs (Figure S3).
Discussion
In this study, we show that CD8β+ T cell depletion in SIV-infected ECs or ART discontinuation in CPs induces transient increases in viral replication that, in turn, is associated with recirculation of SIV-specific Ab-secreting B cells. Using this methodology, we isolated 206 plasmablast-derived VH/VL paired sequences from these recently activated B cells and generated 122 mAbs. Fourteen of these 122 mAbs were SIVmac239 Env-specific and targeted different parts of the Env glycoprotein, including the V loops and the gp41 fusion protein. Additionally, these mAbs bound SIVmac239 SOSIP.664 trimer, primary rhesus CD4+ T cells infected with SIVmac239, and five of them also neutralized SIVmac316, suggesting that these mAbs may contribute to control of viral replication in vivo.
Although both interventions successfully induced viral rebound and recirculation of SIV-specific plasmablasts, the plasmablast recall rate observed in the ART-treated animal was slower compared to that of the CD8β-depleted ECs. While rebound from these two ECs peaked at day 14, plasmablasts from r10051 took 35 days to reach peak expansion after ART interruption. One possibility for the delayed viral rebound and appearance of SIV-specific plasmablasts could be due to the presence of ART drugs and the time needed to eliminate them from the organism, including phosphorylated intracellular active metabolites. The slower viral replication kinetics observed in this animal is also consistent with this explanation. It is also possible that SIV-specific CD8 T+ cells could have been developed while this animal was receiving ART. These cells would have been able to eliminate the circulating wild-type virus, and, therefore, the delayed appearance of SIV-specific plasmablasts would have been due to the emergence of escape variants.
Of the SIV strains commonly used in the macaque model of AIDS, SIVmac239 is the most difficult to protect against. SIVmac239 replicates to high levels during the acute phase (up to 109 vRNA copies/mL) and reaches a set point of approximately 106 vRNA copies/mL during the chronic phase. Even though macaques mount high-frequency SIV-specific CD8+ T cell responses, the virus is still able to escape.40,49,50 Most animals develop AIDS-like symptoms and have to be euthanized within 1–2 years post-infection. Although most SIVmac239-infected macaques develop detectable serological neutralizing activity against this tier 3 virus, the titers are typically low (<1:100) and strain-specific, closely resembling a primary HIV isolate. Notably, Gorman et al.26 have recently reported the isolation of the first three SIVmac239-neutralizing mAbs. These mAbs bind a new epitope in the SIV gp120 core and are the first full-length mAbs capable of completely neutralizing SIVmac239 and other tier 2 SIV variants. Previously, CD4-IgG2 and eCD4-Ig were the only Ab-like molecules that could neutralize SIVmac239 and had provided protection against infection in vivo.23,24 Interestingly, CD4-IgG2-mediated protection against SIVmac239 was the first evidence of protection from acquisition ever achieved by a mAb or an Ab-like molecule against this virus. Unfortunately, there are currently no in vivo data available for the new SIVmac239-neutralizing mAbs. In summary, these examples show that the isolation of mAbs against SIVmac239 greatly expands the number of tools for use in prophylaxis and therapy studies in non-human primate models using pathogenic SIV, complementing studies previously conducted using SHIVs that may be subject to artificial constraints in vivo.
The majority of SIV-reactive mAbs described to date are not capable of neutralizing SIVmac239.25,32,51, 52, 53 Because these mAbs lacked neutralization, their antiviral potentials were often assessed by the ability to neutralize related viruses as a surrogate measurement. For example, the 5L7 immunoadhesin, which neutralizes SIVmac316, protected animals against challenges with this clone in vivo.54 Despite the inability to neutralize SIVmac239, 5L7 protected a single macaque from SIVmac239 infection when delivered as an authentic full-length IgG1, and it was present in plasma at high concentrations.25 It is possible, therefore, that the mAbs isolated in our study could similarly exert antiviral activity. In particular, we have identified two mAbs, P1E09 and P2A12, that have nearly identical binding profiles to 5L7 IgG1 in our infected CD4+ T cell binding assay. These Abs also bound to the native-like SIVmac239 SOSIP.664 trimer. Taken together, these reactivity data suggest that our SIV-specific mAbs recognize conformationally authentic SIVmac239 Env trimers. Thus, it is possible that some of our isolated mAbs, even though they do not neutralize SIVmac239, might have in vivo antiviral effects when present at sufficiently high concentrations.
Our ability to isolate only SIVmac239-specific binding mAbs is not completely unexpected based on the small number of screened mAbs reported herein. The frequency of B cells secreting HIV-specific neutralizing Abs is thought to be less than 1% of all HIV-specific B cells.55 In fact, Walker et al.56 used an antigen-agnostic approach and screened 30,300 memory B cells from 1,800 HIV-infected patients for neutralization activity. Only 2% of cultured cells bound to HIV-1 Env, and 0.6% neutralized one or both of the HIV-1 primary isolates utilized in this study (JR-CSF and SF162). Thus, a higher throughput screening might be required for the isolation of neutralizing mAbs against SIVmac239 from infected animals. Similarly, Mason et al.32 did not isolate SIVmac239 nmAbs when using SIVmac239 scaffolded probes and competitive probe-binding techniques. It is possible that the authors would have achieved this goal had they used animals with higher neutralization titers or screened a greater number of mAbs. In fact, this group has later isolated the first mAbs that fully neutralize SIVmac239 after screening a much larger number of cells.26 Alternatively, in our study, the contemporary rebound virus might be mostly represented by Env sequence variants that had escaped recognition of the SIVmac239 neutralizing Abs detected by our assays, particularly variations contained in the Env V loops. Thus, most of the rebound plasmablasts might be encoding Abs that recognize these contemporary sequence variants, but not the original SIVmac239. It has been shown that the isolation of virus-specific mAbs after infection from non-persistent acute viruses—which present little sequence variation in comparison to SIV—is relatively straightforward. For example, Wrammert et al.57 isolated mAbs from plasmablasts after influenza vaccination, 71% of which had high affinity to viral ligands. Similarly, our group isolated plasmablast-derived flavivirus-specific mAbs from a dengue vaccinee and a Zika virus-infected individual, where 90% of the mAbs neutralized the target virus.43,58 Our current data, consequently, highlight the difficulty of isolating the infrequent neutralizing mAbs elicited during HIV and SIVmac239 infections.
In conclusion, we have shown that CD8β+ T cell depletion and ART withdrawal strategies can be used to enrich for circulating SIVmac239-specific B cells. In contrast to memory or plasma B cell isolation approaches, we identified activated B cell clones responding to contemporary replicating viruses in vivo. These B cells encoded diverse mAbs that recognize both the native-like SIVmac239 SOSIP.664 trimer and infected primary CD4+ T cells. While the in vivo antiviral potential of these Abs remains undetermined, our data show that it is possible to isolate B cells encoding viral-specific Abs without using antigenic probes. In addition, this technique could provide new insights into the rhesus humoral responses against SIV, and allow the isolation of B cells from other persistent pathogens that lack structural characterization.
Materials and Methods
Research Animals and Ethics Statement
The Indian rhesus macaques (Macaca mulatta) utilized in this study were kept at the Wisconsin National Primate Research Center (WNPRC), cared for in strict accordance with the Weatherall report guidelines (i.e., housing, feeding, environmental enrichment, and steps to minimize suffering), and adhered to the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.59 All procedures were performed according to approved protocols by the University of Wisconsin Graduate School Animal Care and Use Committee (animal welfare assurance no. A3368-01; protocol numbers G005654, G00703, and G00729). Macaques from this study were housed in temperature-controlled facilities, maintained in a 12 h light/12 h dark schedule, and monitored daily for overall health.
Three macaques were selected for this study due to their high neutralizing titers against SIVmac239.40,60,61 During the length of this study, two macaques underwent CD8β+ T cell depletion, and the third macaque was discontinued from ART for 35 days. All procedures, including mAb infusions, were performed under anesthesia, and all efforts were made to minimize potential suffering. Additional animal information, including major histocompatibility complex (MHC) class I alleles, age, and sex, is shown in Table 1.
SIV Viral Loads
SIV viral load measurements were based on a previously published protocol.62 Total RNA was extracted from 0.5 mL of EDTA-anticoagulated Indian rhesus macaque plasma samples using QIAGEN DSP virus/pathogen Midi kits on a QIASymphony SP laboratory automation instrument platform. A two-step RT-PCR reaction was performed using random hexamers in a reverse transcription reaction, followed by 45 PCR cycles. Each sample had six replicates, and the limit of reliable quantification in a 0.5 mL vol of plasma was 15 vRNA copies/mL. The primers and probe utilized for this procedure are as follows: forward primer SGAG21, 5′-GTCTGCGTCAT(dP)TGGTGCATTC-3′; reverse primer SGAG22, 5′-CACTAG(dK)TGTCTCTGCACTAT(dP)TGTTTTG-3′; probe, PSGAG23, 5′-FAM-CTTC(dP)TCAGT(dK)TGTTTCACTTTCTCTTCTGCG-BHQ1-3′.
Pseudovirus Neutralization Assay
The replication-incompetent SIVmac239 and SIVmac316 pseudoviruses were produced by transfection of human embryonic kidney (HEK)293T cells with the SIVmac239 or SIVmac316 env plasmids with an env-deficient backbone plasmid (pSG3Δenv) at a ratio of 1:2, using the X-tremeGENE 9 transfection reagent (Roche).
The cell culture supernatants were harvested after 72 h and filter-sterilized (0.22 μm). Neutralization was tested later by incubating pseudovirus and mAbs at 37°C for 1 h before transferring them onto TZM-bl cells, as previously described.63 Each sample was tested in duplicate wells. SIVmac239 neutralization started at 100 μg/mL followed by six serial 2-fold dilutions, while SIVmac316 neutralization was set up at 12.5 μg/mL and subsequently 4-fold diluted seven times to ensure high sensitivity and range of detection. Neutralization IC50 (inhibitory mAb concentration at which 50% virus neutralization was attained) titers were calculated using non-linear fit of the transformed data in GraphPad Prism v7.0.
CD8β Depletion
CD8β chain-expressing lymphocytes were transiently depleted using rhesus IgG1 anti-CD8β mAb clone CD8β255R1 (NIH Nonhuman Primate Reagent Resource catalog no. PR-2557, RRID:AB_2716321). The Ab was administered intravenously at 50 mg/kg dose to animals r04135 and r09037.
ART
The ART cocktail consisted of tenofovir (20 mg/kg) and emtricitabine (40 mg/kg), which were given daily at 60 mg/kg via subcutaneous injection. To prepare 4 L of the formulation, 2 initial L of sterile water were added to a flask, containing a stir bar, and placed onto a stirrer/hot plate. Four hundred grams of tenofovir was slowly added to the flask and let stir with heat until the drug went into solution. An additional liter of sterile water was then added, along with 160 g of emtricitabine. To facilitate emtricitabine going into solution, NaOH was added. Five hundred mLs of sterile water was then added in the flask and the solution was let stir continuously until all powder was in solution. The pH was adjusted with NaOH or HCl to 7.0–7.2 if necessary. Under sterile conditions, the final solution was doubled filtered using Nalgene sterile filters, aliquoted, and stored at −20°C.
Plasmablast Sort
We sorted single plasmablasts at various time points after either depletion of CD8β+ T cells or interruption of ART. Plasmablasts were defined as CD3− CD20− CD14− CD16− HLA-DR+ CD11c− CD123− CD80+. Prior to staining, PBMCs were isolated using a Ficoll-Paque gradient (GE Healthcare). Fresh PBMC samples were sorted on a BD FACSJazz sorter (Becton Dickinson). Single plasmablasts were sorted into 96-well plates containing a lysis buffer to preserve the RNA (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2, 6.25 mM dithiothreitol [DTT], and 250 ng/well of yeast tRNA [Life Technologies]; 20 U of RNase inhibitor [New England Biolabs]; and 0.0625 L/well of IGEPAL CA-630 [Sigma]). The RNA plates were immediately frozen and stored at −80°C until subsequent cloning.
Env-Specific ELISPOT
ELISPOT assays were performed as previously described on PBMCs or sorted plasmablasts at the indicated time points to determine plasmablast frequencies.64 All total IgG and SIV Env-specific ELISPOT assays were performed with 120,000 or 40,000 cells per well, in duplicate, according to the manufacturer’s instructions (Mabtech). For the total IgG assay, we used a purified anti-human IgG mAb at 15 μg/mL. To determine frequencies of SIV-specific plasmablasts, the wells were coated with gp120 or gp140, each at a concentration of 1 μg/mL. Each animal was tested individually with each plate including at least two separate positive control wells and two or more negative control wells. Wells were imaged and spots were enumerated with an AID ELISPOT reader (Autoimmun Diagnostika).
mAb Amplification, Cloning, Expression, and Purification
Sorted plasmablasts were immediately frozen on dry ice for subsequent amplification of the reverse-transcribed RNA encoding the variable region of the heavy (H), lambda (L), and kappa (K) Ab chains, as previously described.27 First, cDNA was synthesized in a 25 μL reaction mixture using the original sorted plates. Each reaction had 1 μL of 150 ng random hexamers (IDT), 2 μL of 10 mM deoxynucleotide triphosphate (dNTP; Life Technologies), 1 μL of SuperScript III reverse transcriptase (Life Technologies), 1 μL of molecular biology-grade water, and 20 μL of single-sorted cell sample in lysis buffer (described above). The reaction was set up at 42°C for 10 min, 25°C for 10 min, 50°C for 60 min, and 94°C for 10 min. Upon completion, cDNA was stored at −20°C.
Three nested PCRs were performed using a mix of 5′ V gene-specific primers with matching 3′ primers, which were designed to specifically bind encoding the constant regions of the H, K, and L chains. PCRs were prepared using HotStarTaq Plus DNA polymerase (QIAGEN). The last round of PCR was carried out with primers adapted (and modified) from Sundling et. al,27 which contained ends compatible with Gibson Assembly subcloning and the rhesus IgG1 expression vectors (InvivoGen). After cloning, the plasmids encoding heavy and light chains of each mAb were co-transfected using the Expi293 expression system (Thermo Fisher Scientific) that includes suspension-adapted HEK293 cell lines. Sixteen to 18 h post-transfection, we boosted the reaction by adding enhancers and then harvested the secreted mAb-containing supernatants 5 days later. All mAbs were purified using protein A columns (GE Healthcare), and purified protein concentrations were determined by measuring absorbance at 280 nm (NanoDrop, Thermo Scientific).
ELISA
The specificity of the isolated mAbs was determined by ELISA. All mAbs were initially screened using purified SIVmac239 gp140 FT at 1 μg/mL. If reactive, the mAbs were then tested against gp120 (Immune Technologies), gp41 (ImmunoDX), SIVmac239 SOSIP.664, SIVmac239 ΔV1/V2/V3 gp140 FT, and SIVmac239 ΔV4 gp140 FT (based on Mason et al.32). ELISA plates were coated with 100 μL of the SIV Env protein at 5 μg/mL and incubated overnight at 4°C. However, the SOSIP.664 trimer plates were coated with 100 μL of streptavidin at 5 μg/mL. On the following day, the SOSIP.664 plates were first washed with 1× PBS with Tween 20, and then SOSIP.664 at 5 μg/mL was added to the wells for 1 h at 37°C. Subsequently, all plates were then washed with 1× PBS with Tween 20 and blocked with 300 μL of 5% powdered milk in PBS for 1 h at 37°C. The plates were washed again, and 100 μL of the isolated mAbs, at a final concentration of 1 μg/mL, was added to the corresponding wells prior to incubating for 1 h at 37°C. The plates were washed again and 100 μL of a 1:10,000 dilution of goat anti-human IgG horseradish peroxidase (HRP) (SouthernBiotech) was added to all wells prior to the final incubation for 1 h at 37°C. Lastly, the plates were washed and then developed using 100 μL of 3,3′,5,5′-tetramethylbenzidine (TMB) (MilliporeSigma). When the reaction was stopped after a short incubation with 100 μL of TMB Stop Solution (MilliporeSigma), the plates were read at 450 nm (Biotek Synergy 2). To determine the reactivity of the mAbs, the background wells were averaged and extracted from all samples.
Flow Cytometry Analysis of mAb Binding to Infected Cells
We assessed the ability of the isolated mAbs to bind native Env conformations on the surface of SIVmac239-infected rhesus CD4+ T cells using a flow cytometry-based assay. Our protocol for generation of rhesus CD4+ phytohemagglutinin (PHA) blasts permissive for SIV replication was adapted from that of Sacha and Watkins.65 Briefly, rhesus PBMCs were isolated from EDTA-anticoagulated blood by Ficoll-Paque Plus (GE Healthcare) density centrifugation. CD4+ T cells were isolated from PBMCs using a non-human primate CD4+ T cell isolation kit (Miltenyi Biotec) and magnetic-activated cell sorting (MACS). CD4+ T cells were stimulated for 48 h in a T cell activation cocktail containing 2.5 μg/mL anti-CD3 (clone 6G12, NIH Nonhuman Primate Reagent Resource), 2.5 μg/mL anti-CD28 (clone L293, BD Biosciences), 2.5 μg/mL anti-CD49d (clone 9F10, BD Biosciences), 5 μg/mL PHA-P (Sigma-Aldrich), and 100 U/mL IL-2 (Roche) in R15 medium. Following the 48 h stimulation period, CD4+ T cells were resuspended and cultured in R15-100 medium (R15 with 100 U/mL IL-2). CD4+ T cells were infected with SIVmac239 by spinoculation (2 h, 1,800 × g, 23°C, 250 ng of p27 antigen per 106 cells), washed twice in R15 medium, and then cultured in R15-100 medium for 48 h at 37°C. Prior to staining, dead cells and debris were removed by Ficoll-Paque Plus density centrifugation. Cells were first stained with the isolated mAbs at the indicated concentrations. After washing, cells were stained with a cocktail containing the following fluorophore-conjugated mAbs and amine-reactive dye: anti-monkey IgG Alexa Fluor 647 (clone SB108a, SouthernBiotech), anti-human CD4 Brilliant Violet (BV)605 (clone OKT4, BioLegend), anti-human CD8 BV785 (clone RPA-T8, BioLegend), aqua live/dead dye (Live/Dead fixable aqua dead cell stain kit, Invitrogen). Cells were washed and then fixed and permeabilized using Cytofix/Cytoperm (BD Biosciences) and Perm/Wash buffer (BD Biosciences), respectively. Finally, intracellular staining was conducted with anti-human CD3 peridinin chlorophyll protein (PerCP)-Cy5.5 (clone SP34-2, BD Biosciences) and anti-SIVmac Gag p27 fluorescein isothiocyanate (FITC) (clone 55-2F12, NIH AIDS Reagent Program, FITC conjugated at WNPRC) to permit identification of bona fide infected cells. Following a final wash in Perm/Wash buffer, at least 35,000 events per sample were acquired on a BD LSR II flow cytometer. Data analysis was conducted with FlowJo 9.9.6. To determine the relative binding affinities of the isolated mAbs, we compared the proportions of infected cells (live CD3+ Gag p27+ cells) bound by rhesus IgG (Alexa Fluor 647+ cells) for each sample.
Author Contributions
Conceptualization, D.I.W. and D.M.M.; Writing – Original Draft, N.P.-L. and D.M.M.; Writing – Review & Editing, N.P.-L., B.C.R., M.J.R., J.M.M.-N., M.A.M., J.D.L., E.G.R., D.R.B., D.I.W., and D.M.M.; Investigation, N.P.-L., C.M.D., B.C.R, M.J.R., V.K.B., M.J.G., L.G.-N., M.G.P., K.L., G.S., R.A., K.L.W., N.P., J.M.M.-N., S.P.F., and D.M.M., Resources, D.I.W. and D.M.M.; Methodology, N.P.-L., C.M.D., B.C.R., M.J.R., R.A., and D.M.M.; Supervision, J.W., J.D.L., M.A.M., E.G.R., D.R.B., D.I.W., and D.M.M.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the University of Miami Institute of AIDS and Emerging Infectious Diseases, made possible by the State of Florida contract awarded to the University of Miami (CODMR), the Miami Clinical and Translational Science Institute (CTSI), and by NIH Grants R37AI052056, P01AI094420, R01AI108421, 7R24OD011161-12, 7R24RR015371-13, 7R56AI049120-11, and 1F30AI147881-01, and under contracts HHSN261200800001E and 75N91019F00129. This project was supported in part by P30AI073961 to the Miami Center for AIDS Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. We are grateful to Gilead Sciences, Inc. for supplying the tenofovir and emtricitabine used in this study; the NIH Nonhuman Primate Reagent Resource for providing the depleting Ab reagent (CD8β255R1) and T cell-activating Ab (anti-CD3 clone 6G12); the Quantitative Molecular Diagnostics Core of the AIDS and Cancer Virus Program of the Frederick National Laboratory for Cancer Research for plasma viral load analysis; and to Leydi Machado for administrative assistance and the members of the Immunology Services Unit at WNPRC for their technical assistance. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.01.010.
Supplemental Information
References
- 1.Barré-Sinoussi F., Ross A.L., Delfraissy J.F. Past, present and future: 30 years of HIV research. Nat. Rev. Microbiol. 2013;11:877–883. doi: 10.1038/nrmicro3132. [DOI] [PubMed] [Google Scholar]
- 2.GBD 2017 HIV Collaborators Global, regional, and national incidence, prevalence, and mortality of HIV, 1980–2017, and forecasts to 2030, for 195 countries and territories: a systematic analysis for the Global Burden of Diseases, Injuries, and Risk Factors Study 2017. Lancet HIV. 2019;6:e831–e859. doi: 10.1016/S2352-3018(19)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hessell A.J., Malherbe D.C., Haigwood N.L. Passive and active antibody studies in primates to inform HIV vaccines. Expert Rev. Vaccines. 2018;17:127–144. doi: 10.1080/14760584.2018.1425619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caskey M., Klein F., Nussenzweig M.C. Broadly neutralizing antibodies for HIV-1 prevention or immunotherapy. N. Engl. J. Med. 2016;375:2019–2021. doi: 10.1056/NEJMp1613362. [DOI] [PubMed] [Google Scholar]
- 5.Kumar R., Qureshi H., Deshpande S., Bhattacharya J. Broadly neutralizing antibodies in HIV-1 treatment and prevention. Ther. Adv. Vaccines Immunother. 2018;6:61–68. doi: 10.1177/2515135518800689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker L.M., Burton D.R. Passive immunotherapy of viral infections: “super-antibodies” enter the fray. Nat. Rev. Immunol. 2018;18:297–308. doi: 10.1038/nri.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bournazos S., Ravetch J.V. Anti-retroviral antibody FcγR-mediated effector functions. Immunol. Rev. 2017;275:285–295. doi: 10.1111/imr.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bournazos S., Klein F., Pietzsch J., Seaman M.S., Nussenzweig M.C., Ravetch J.V. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazar G.A., Dang W., Karki S., Vafa O., Peng J.S., Hyun L., Chan C., Chung H.S., Eivazi A., Yoder S.C. Engineered antibody Fc variants with enhanced effector function. Proc. Natl. Acad. Sci. USA. 2006;103:4005–4010. doi: 10.1073/pnas.0508123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barouch D.H., Whitney J.B., Moldt B., Klein F., Oliveira T.Y., Liu J., Stephenson K.E., Chang H.W., Shekhar K., Gupta S. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shingai M., Nishimura Y., Klein F., Mouquet H., Donau O.K., Plishka R., Buckler-White A., Seaman M., Piatak M., Jr., Lifson J.D. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hessell A.J., Jaworski J.P., Epson E., Matsuda K., Pandey S., Kahl C., Reed J., Sutton W.F., Hammond K.B., Cheever T.A. Early short-term treatment with neutralizing human monoclonal antibodies halts SHIV infection in infant macaques. Nat. Med. 2016;22:362–368. doi: 10.1038/nm.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J., Ghneim K., Sok D., Bosche W.J., Li Y., Chipriano E., Berkemeier B., Oswald K., Borducchi E., Cabral C. Antibody-mediated protection against SHIV challenge includes systemic clearance of distal virus. Science. 2016;353:1045–1049. doi: 10.1126/science.aag0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julg B., Liu P.T., Wagh K., Fischer W.M., Abbink P., Mercado N.B., Whitney J.B., Nkolola J.P., McMahan K., Tartaglia L.J. Protection against a mixed SHIV challenge by a broadly neutralizing antibody cocktail. Sci. Transl. Med. 2017;9:eaao4235. doi: 10.1126/scitranslmed.aao4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautam R., Nishimura Y., Pegu A., Nason M.C., Klein F., Gazumyan A., Golijanin J., Buckler-White A., Sadjadpour R., Wang K. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature. 2016;533:105–109. doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatziioannou T., Evans D.T. Animal models for HIV/AIDS research. Nat. Rev. Microbiol. 2012;10:852–867. doi: 10.1038/nrmicro2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lifson J.D., Haigwood N.L. Lessons in nonhuman primate models for AIDS vaccine research: from minefields to milestones. Cold Spring Harb. Perspect. Med. 2012;2:a007310. doi: 10.1101/cshperspect.a007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith S.M., Holland B., Russo C., Dailey P.J., Marx P.A., Connor R.I. Retrospective analysis of viral load and SIV antibody responses in rhesus macaques infected with pathogenic SIV: predictive value for disease progression. AIDS Res. Hum. Retroviruses. 1999;15:1691–1701. doi: 10.1089/088922299309739. [DOI] [PubMed] [Google Scholar]
- 19.Daniel M.D., Letvin N.L., King N.W., Kannagi M., Sehgal P.K., Hunt R.D., Kanki P.J., Essex M., Desrosiers R.C. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 20.Daniel M.D., Kirchhoff F., Czajak S.C., Sehgal P.K., Desrosiers R.C. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 21.Martins M.A., Bischof G.F., Shin Y.C., Lauer W.A., Gonzalez-Nieto L., Watkins D.I., Rakasz E.G., Lifson J.D., Desrosiers R.C. Vaccine protection against SIVmac239 acquisition. Proc. Natl. Acad. Sci. USA. 2019;116:1739–1744. doi: 10.1073/pnas.1814584116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Nieto L., Castro I.M., Bischof G.F., Shin Y.C., Ricciardi M.J., Bailey V.K., Dang C.M., Pedreño-Lopez N., Magnani D.M., Ejima K. Vaccine protection against rectal acquisition of SIVmac239 in rhesus macaques. PLoS Pathog. 2019;15:e1008015. doi: 10.1371/journal.ppat.1008015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poignard P., Moldt B., Maloveste K., Campos N., Olson W.C., Rakasz E., Watkins D.I., Burton D.R. Protection against high-dose highly pathogenic mucosal SIV challenge at very low serum neutralizing titers of the antibody-like molecule CD4-IgG2. PLoS ONE. 2012;7:e42209. doi: 10.1371/journal.pone.0042209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner M.R., Fellinger C.H., Kattenhorn L.M., Davis-Gardner M.E., Weber J.A., Alfant B., Zhou A.S., Prasad N.R., Kondur H.R., Newton W.A. AAV-delivered eCD4-Ig protects rhesus macaques from high-dose SIVmac239 challenges. Sci. Transl. Med. 2019;11:eaau5409. doi: 10.1126/scitranslmed.aau5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuchs S.P., Martinez-Navio J.M., Piatak M., Jr., Lifson J.D., Gao G., Desrosiers R.C. AAV-delivered antibody mediates significant protective effects against SIVmac239 challenge in the absence of neutralizing activity. PLoS Pathog. 2015;11:e1005090. doi: 10.1371/journal.ppat.1005090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorman J., Mason R.D., Nettey L., Cavett N., Chuang G.Y., Peng D., Tsybovsky Y., Verardi R., Nguyen R., Ambrozak D. Isolation and structure of an antibody that fully neutralizes isolate SIVmac239 reveals functional similarity of SIV and HIV glycan shields. Immunity. 2019;51:724–734.e4. doi: 10.1016/j.immuni.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Sundling C., Phad G., Douagi I., Navis M., Karlsson Hedestam G.B. Isolation of antibody V(D)J sequences from single cell sorted rhesus macaque B cells. J. Immunol. Methods. 2012;386:85–93. doi: 10.1016/j.jim.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Sanders R.W., Derking R., Cupo A., Julien J.P., Yasmeen A., de Val N., Kim H.J., Blattner C., de la Peña A.T., Korzun J. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klasse P.J., Ketas T.J., Cottrell C.A., Ozorowski G., Debnath G., Camara D., Francomano E., Pugach P., Ringe R.P., LaBranche C.C. Epitopes for neutralizing antibodies induced by HIV-1 envelope glycoprotein BG505 SOSIP trimers in rabbits and macaques. PLoS Pathog. 2018;14:e1006913. doi: 10.1371/journal.ppat.1006913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georgiev I.S., Joyce M.G., Yang Y., Sastry M., Zhang B., Baxa U., Chen R.E., Druz A., Lees C.R., Narpala S. Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J. Virol. 2015;89:5318–5329. doi: 10.1128/JVI.03451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Bredow B., Andrabi R., Grunst M., Grandea A.G., 3rd, Le K., Song G., Berndsen Z.T., Porter K., Pallesen J., Ward A.B. Differences in the binding affinity of an HIV-1 V2 apex-specific antibody for the SIVsmm/mac envelope glycoprotein uncouple antibody-dependent cellular cytotoxicity from neutralization. MBio. 2019;10:e01255-19. doi: 10.1128/mBio.01255-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mason R.D., Welles H.C., Adams C., Chakrabarti B.K., Gorman J., Zhou T., Nguyen R., O’Dell S., Lusvarghi S., Bewley C.A. Targeted isolation of antibodies directed against major sites of SIV Env vulnerability. PLoS Pathog. 2016;12:e1005537. doi: 10.1371/journal.ppat.1005537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirsch V., Adger-Johnson D., Campbell B., Goldstein S., Brown C., Elkins W.R., Montefiori D.C. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J. Virol. 1997;71:1608–1620. doi: 10.1128/jvi.71.2.1608-1620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martins M.A., Tully D.C., Shin Y.C., Gonzalez-Nieto L., Weisgrau K., Bean D.J., Gadgil R., Gutman M.J., Domingues A., Maxwell H.S. Rare control of SIVmac239 infection in a vaccinated rhesus macaque. AIDS Res. Hum. Retroviruses. 2017;33:843–858. doi: 10.1089/aid.2017.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin X., Bauer D.E., Tuttleton S.E., Lewin S., Gettie A., Blanchard J., Irwin C.E., Safrit J.T., Mittler J., Weinberger L. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matano T., Shibata R., Siemon C., Connors M., Lane H.C., Martin M.A. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 1998;72:164–169. doi: 10.1128/jvi.72.1.164-169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitz J.E., Kuroda M.J., Santra S., Sasseville V.G., Simon M.A., Lifton M.A., Racz P., Tenner-Racz K., Dalesandro M., Scallon B.J. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 38.Mudd P.A., Martins M.A., Ericsen A.J., Tully D.C., Power K.A., Bean A.T., Piaskowski S.M., Duan L., Seese A., Gladden A.D. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature. 2012;491:129–133. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedrich T.C., Valentine L.E., Yant L.J., Rakasz E.G., Piaskowski S.M., Furlott J.R., Weisgrau K.L., Burwitz B., May G.E., León E.J. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 2007;81:3465–3476. doi: 10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martins M.A., Tully D.C., Cruz M.A., Power K.A., Veloso de Santana M.G., Bean D.J., Ogilvie C.B., Gadgil R., Lima N.S., Magnani D.M. Vaccine-induced simian immunodeficiency virus-specific CD8+ T-cell responses focused on a single Nef epitope select for escape variants shortly after infection. J. Virol. 2015;89:10802–10820. doi: 10.1128/JVI.01440-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wrammert J., Onlamoon N., Akondy R.S., Perng G.C., Polsrila K., Chandele A., Kwissa M., Pulendran B., Wilson P.C., Wittawatmongkol O. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J. Virol. 2012;86:2911–2918. doi: 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buckner C.M., Moir S., Ho J., Wang W., Posada J.G., Kardava L., Funk E.K., Nelson A.K., Li Y., Chun T.W., Fauci A.S. Characterization of plasmablasts in the blood of HIV-infected viremic individuals: evidence for nonspecific immune activation. J. Virol. 2013;87:5800–5811. doi: 10.1128/JVI.00094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magnani D.M., Silveira C.G.T., Ricciardi M.J., Gonzalez-Nieto L., Pedreño-Lopez N., Bailey V.K., Gutman M.J., Maxwell H.S., Domingues A., Costa P.R. Potent plasmablast-derived antibodies elicited by the National Institutes of Health dengue vaccine. J. Virol. 2017;91:e00867-17. doi: 10.1128/JVI.00867-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silveira E.L., Kasturi S.P., Kovalenkov Y., Rasheed A.U., Yeiser P., Jinnah Z.S., Legere T.H., Pulendran B., Villinger F., Wrammert J. Vaccine-induced plasmablast responses in rhesus macaques: phenotypic characterization and a source for generating antigen-specific monoclonal antibodies. J. Immunol. Methods. 2015;416:69–83. doi: 10.1016/j.jim.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sok D., Burton D.R. Recent progress in broadly neutralizing antibodies to HIV. Nat. Immunol. 2018;19:1179–1188. doi: 10.1038/s41590-018-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burton D.R., Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francica J.R., Sheng Z., Zhang Z., Nishimura Y., Shingai M., Ramesh A., Keele B.F., Schmidt S.D., Flynn B.J., Darko S., NISC Comparative Sequencing Program Analysis of immunoglobulin transcripts and hypermutation following SHIVAD8 infection and protein-plus-adjuvant immunization. Nat. Commun. 2015;6:6565. doi: 10.1038/ncomms7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mori K., Ringler D.J., Kodama T., Desrosiers R.C. Complex determinants of macrophage tropism in env of simian immunodeficiency virus. J. Virol. 1992;66:2067–2075. doi: 10.1128/jvi.66.4.2067-2075.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans D.T., O’Connor D.H., Jing P., Dzuris J.L., Sidney J., da Silva J., Allen T.M., Horton H., Venham J.E., Rudersdorf R.A. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 1999;5:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- 50.Allen T.M., O’Connor D.H., Jing P., Dzuris J.L., Mothé B.R., Vogel T.U., Dunphy E., Liebl M.E., Emerson C., Wilson N. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 51.Robinson J.E., Cole K.S., Elliott D.H., Lam H., Amedee A.M., Means R., Desrosiers R.C., Clements J., Montelaro R.C., Murphey-Corb M. Production and characterization of SIV envelope-specific rhesus monoclonal antibodies from a macaque asymptomatically infected with a live SIV vaccine. AIDS Res. Hum. Retroviruses. 1998;14:1253–1262. doi: 10.1089/aid.1998.14.1253. [DOI] [PubMed] [Google Scholar]
- 52.Cole K.S., Alvarez M., Elliott D.H., Lam H., Martin E., Chau T., Micken K., Rowles J.L., Clements J.E., Murphey-Corb M. Characterization of neutralization epitopes of simian immunodeficiency virus (SIV) recognized by rhesus monoclonal antibodies derived from monkeys infected with an attenuated SIV strain. Virology. 2001;290:59–73. doi: 10.1006/viro.2001.1144. [DOI] [PubMed] [Google Scholar]
- 53.Johnson W.E., Sanford H., Schwall L., Burton D.R., Parren P.W., Robinson J.E., Desrosiers R.C. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 2003;77:9993–10003. doi: 10.1128/JVI.77.18.9993-10003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson P.R., Schnepp B.C., Zhang J., Connell M.J., Greene S.M., Yuste E., Desrosiers R.C., Clark K.R. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 2009;15:901–906. doi: 10.1038/nm.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwong P.D., Mascola J.R. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker L.M., Phogat S.K., Chan-Hui P.Y., Wagner D., Phung P., Goss J.L., Wrin T., Simek M.D., Fling S., Mitcham J.L., Protocol G Principal Investigators Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wrammert J., Smith K., Miller J., Langley W.A., Kokko K., Larsen C., Zheng N.Y., Mays I., Garman L., Helms C. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magnani D.M., Rogers T.F., Beutler N., Ricciardi M.J., Bailey V.K., Gonzalez-Nieto L., Briney B., Sok D., Le K., Strubel A. Neutralizing human monoclonal antibodies prevent Zika virus infection in macaques. Sci. Transl. Med. 2017;9:eaan8184. doi: 10.1126/scitranslmed.aan8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weatherall D. 2006. The use of non-human primates in research. Report of the Academy of Medical Sciences/CRS.https://mrc.ukri.org/documents/pdf/the-use-of-non-human-primates-in-research/ [Google Scholar]
- 60.Martins M.A., Gonzalez-Nieto L., Shin Y.C., Domingues A., Gutman M.J., Maxwell H.S., Magnani D.M., Ricciardi M.J., Pedreño-Lopez N., Bailey V.K. The frequency of vaccine-induced T-cell responses does not predict the rate of acquisition after repeated intrarectal SIVmac239 challenges in Mamu-B*08+ rhesus macaques. J. Virol. 2019;93:e01626-18. doi: 10.1128/JVI.01626-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valentine L.E., Loffredo J.T., Bean A.T., León E.J., MacNair C.E., Beal D.R., Piaskowski S.M., Klimentidis Y.C., Lank S.M., Wiseman R.W. Infection with “escaped” virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques. J. Virol. 2009;83:11514–11527. doi: 10.1128/JVI.01298-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H., Wang S., Kong R., Ding W., Lee F.H., Parker Z., Kim E., Learn G.H., Hahn P., Policicchio B. Envelope residue 375 substitutions in simian-human immunodeficiency viruses enhance CD4 binding and replication in rhesus macaques. Proc. Natl. Acad. Sci. USA. 2016;113:E3413–E3422. doi: 10.1073/pnas.1606636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sok D., Pauthner M., Briney B., Lee J.H., Saye-Francisco K.L., Hsueh J., Ramos A., Le K.M., Jones M., Jardine J.G. A prominent site of antibody vulnerability on HIV envelope incorporates a motif associated with CCR5 binding and its camouflaging glycans. Immunity. 2016;45:31–45. doi: 10.1016/j.immuni.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saletti G., Çuburu N., Yang J.S., Dey A., Czerkinsky C. Enzyme-linked immunospot assays for direct ex vivo measurement of vaccine-induced human humoral immune responses in blood. Nat. Protoc. 2013;8:1073–1087. doi: 10.1038/nprot.2013.058. [DOI] [PubMed] [Google Scholar]
- 65.Sacha J.B., Watkins D.I. Synchronous infection of SIV and HIV in vitro for virology, immunology and vaccine-related studies. Nat. Protoc. 2010;5:239–246. doi: 10.1038/nprot.2009.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.