Abstract
Breast cancer is the most common malignancy and the leading cause of cancer-related death among women worldwide. The underlying mechanisms for breast cancer development, especially in young women, are not completely understood. Although there are several experimental models to understand the biology of breast cancer such as immortalized cell lines, many of these cell lines have been in culture for decades and most of them have been derived from Caucasians or African-Americans. So, it is required to establish a new cell line derived from primary tumors and Asian women. In this study Pari-Institute for Cancer Research (Pari-ICR) was derived from the primary breast tumor of a 36-years old patient with invasive ductal carcinoma. We characterized the cell line by examining morphology, expression of different markers, and functional profile. Immunocytochemistry showed that this cell line does not express estrogen and progesterone receptors as well as human epidermal growth factor receptor 2 (HER2). Pari-ICR cell line expresses high levels of Vimentin, Ezrin, and S100 but does not express EpCAM, Cytokeratin19, Pan-cytokeratin, Nestin, and Desmin. Its doubling time of Pari-ICR was about 22h and was able to grow as colonies in soft agar. It displayed a higher ability of migration and invasion in comparison with MCF-7 cell line. This breast cancer cell line can serve as a model for understanding the molecular mechanisms of breast carcinogenesis. Moreover, it can be used as an appropriate resource to find novel biomarkers or assess new drugs.
Key Words: Breast cancer, cell line establishment, Triple-negative-, Pari-ICR
Introduction
Breast cancer is the most common malignancy and the leading cause of cancer-related death among women worldwide (Banin Hirata et al., 2014; Zhang et al., 2015). In Iran, breast cancer accounts for 24.6% of total cancers with the crude incidence rate of about 22.6 per 100,000 females, which even is increasing annually. According to Iran National Cancer Registry (NCR), it affects the Iranian women at least 10 years earlier compared to women in developed countries and about 21% of women with breast cancer are under the age of 40 years (Jazayeri et al., 2015; Otaghvar et al., 2015). Among different subtypes, triple-negative and HER-2 positive breast cancers are more prevalent in young women and tend to have more aggressive behaviors (Anders et al., 2009; Carey et al., 2010; Assi et al., 2013).
Triple-negative breast cancers (TNBCs) are a heterogeneous group of tumors, both clinically and pathologically. These cancers are defined by the lack of express estrogen and progesterone receptors as well as human epidermal growth factor receptor 2 (HER2) expressions and account for approximately 15% of all breast cancers. TNBCs, which are significantly more frequent in women younger than 40 years, have a poor prognosis and reduced overall survival (Dent et al., 2007; Rapoport et al., 2014; Collignon et al., 2016).
Despite massive studies, the molecular pathways involved in breast cancer development, especially in young women, are not completely understood. Different experimental approaches can be applied to understand the biology of breast cancer such as those using immortalized cell lines. While cancer cell lines maintain many features of the tumor tissue (Neve et al., 2006; Vargo-Gogola and Rosen, 2007; Van Staveren et al., 2009), there are some difficulties with using them. For instance, the majority of them have been isolated from a metastatic tumor of older patients and many of these cell lines have been in culture for decades(Ku et al., 2013; Pandrangi et al., 2014). In addition, the genetic diversity of tumor cells may cause unpredictable tumor behavior and treatment failure. It has been recently evidenced that there is variability in the pharmacokinetics, pharmacodynamics, and tolerance of anticancer drugs among individuals with different ethnicities. In other words, ethnic variations have been found in anticancer drug response and in toxicity from anticancer drugs, especially among Asian and Caucasian cancer patients (Phan et al., 2009). Therefore, new cell lines derived from primary breast tumors of Asian young women are highly needed. The aim of this study is to describe the establishment and partial characterization of a triple negative breast cancer cell line derived from a primary tumor of 36-years old women with invasive ductal carcinoma of the breast.
Materials and Methods
Clinical history
The cancer cell line was obtained from the primary breast tumor of a 36 years old patient with grade II invasive ductal carcinoma. This study was approved by the Ethics Committee of Shiraz University of Medical Sciences and an informed consent was obtained from patient.
The patient underwent a right-side mastectomy and axillary lymph node dissection. Metastases were found in 8 out of 16 excised lymph nodes. Under sterile condition, the tumor specimen was washed twice with phosphate-buffered saline (PBS) and minced with a scalpel into small pieces of approximately 2 mm2 in size. These small tissue pieces were seeded into a Petri dish, cultured in a complete DMEM/F12 medium (Gibco, USA), supplemented with 10% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin-streptomycin (Bioidea, UK). After about 10 days, epithelial and mesenchymal cells were projected out of small tissue fragments. Next, the cells were enzymatically harvested (Trypsin 0.25%) and passaged with a 1:5 split ratio at the confluence of 80-90%. After 11 passages, tumor cells outcompeted the mesenchymal cells due to their faster growth and shorter doubling time. Pari-ICR were frozen periodically and stored in liquid nitrogen at various passages.
Calculation of growth rate
To calculate cell growth rate, the Pari-ICR cell line was plated at a concentration of 2.5× 104in a DMEM/F12 medium supplemented with 10% FBS and 1% penicillin-streptomycin. Triplicate wells were counted after staining with trypan blue dye at 24-h intervals. The growth curve was plotted and doubling time calculated using free online doubling time calculating software (http://www.doubling-time.com).
Colony-forming assay
A two-layer method was performed. After trypsinization, a single cell suspension of the established cell line from 100 to 1,000 cells per plate were prepared in a solution containing 0.36% agar in DMEM/F12 supplemented with 10% FBS and 1% penicillin-streptomycin. The mixture was deposited on base layers (containing 0.75% agar with DMEM/F12 plus 10% FBS and 1% penicillin-streptomycin) and incubated at 37oC under 5% CO2. After 10 days, the numbers of colonies containing more than 50 cells were counted by an inverted microscope.
Migration and invasion assays
Migration and invasion assays were performed using transwell permeable inserts with or without coated Matrigel. After 24h of serum starvation, 5 × 104 cells in 100µl serum-free DMEM/F12 with 1% penicillin-streptomycin were added to the upper chamber of the transwell, while the lower chamber of the transwell was filled with 600 μl DMEM supplemented with 10% FBS. After 24h of incubation at 37oC with 5% CO2, non-migratory cells on the upper surface of the membrane were removed with cotton swab. The migrated or invaded cells on the lower surface were fixed with ice-cold alcohol, stained with a solution containing 0.5% crystal violet in 10% ethanol, and counted under phase contrast microscope. The assay was performed in triplicate.
Chromosomal Analysis
To identify chromosomal abnormalities, karyotyping of Pari-ICR cell line was performed. Briefly, confluent cells were exposed to 0.2pg/ml Colcemid at 37oC for 2h and then detached by trypsinization. The harvested cells were treated with 0.5% potassium chloride solution for 30 min. After fixation with a fixative solution (methanol/glacial acetic acid 3:1), the cell suspension was spread on wet slides. Finally, the slides were colored in Gimsa staining solution for chromosome analysis.
Comparative genomic hybridization (CGH)
DNA amplification was performed using Sure PlexDNA amplification system according to the manufacturer’s protocol (Illumina, UK). After DNA amplification, control DNA [DNA from a normal male] and sample DNA were directly labeled with Cy3 and Cy5 Fluorophores, respectively. Sample DNA was hybridized overnight with a control DNA undercover slide. Finally, the array field was scanned by laser (Innopsys, France) and the microarray data on chromosome Loss/gain across 24 chromosomes were analyzed using Blue Fuse software (BlueGenome, UK).
Immunofluorescent assay
Pari-ICR cell line was seeded on coverslips at a density of 100 × 103 cells and grown for 24 h. The cells were then permeabilized with cold methanol for 15 min at room temperature. Blocking was done with 1% BSA (Biosera) in PBS containing 0.5% Tween 20 (PBST). Next, the slides were incubated overnight at 4oC with primary antibodies against Vimentin, Nestin, Ezrin, S100, Desmin, Pan-cytokeratin, and Cytokeratin19 [all from Santa Cruz]. After incubation, the cells were washed 3 times with PBST and incubated with corresponding secondary fluorescent antibodies (Goat anti-rabbit IgG- conjugated with FITC or goat anti-mouse IgG-conjugated with FITC, Santa Cruz) for 2h at room temperature. After washing with PBST, coverslips were mounted using amounting medium containing DAPI or PI and examined with a fluorescent microscope.
Flow cytometry
Expressions of cell surface markers were analyzed by flow cytometry. Cells were detached and washed with PBS twice, dissociated into single cell suspension, and incubated with fluorescent-conjugated monoclonal antibodies including anti CD73, CD90, CD166, CD29, CD24, CD44, CD10, CD45, CD34, CD105, CD133, and CD146(all from BD Bioscience, USA). After 30 min of incubation in the dark at 4oC, the cells were washed and resuspended in PBS and analyzed with FACS Calibur flow cytometer. Flow data were analyzed using FlowJo software version 7.6.2 (Tritar Inc., USA).
Results
Establishment of human breast cancer cell line
Pari-ICR was established from a 36-year-old female patient, diagnosed with invasive ductal carcinoma of the breast.
Culture dish was examined daily using a phase contrast microscope. An outgrowth of tumor-like cells with a spindle shape was seen from breast explants 10 days after culture initiation. After 11 passages, the epithelial cells were enriched and a homogenous population was seen. These spindle-shaped cells grow as an adherent monolayer. So far, Pari-ICR cell line has been grown for 150 passages (Figure 1).
Figure 1.
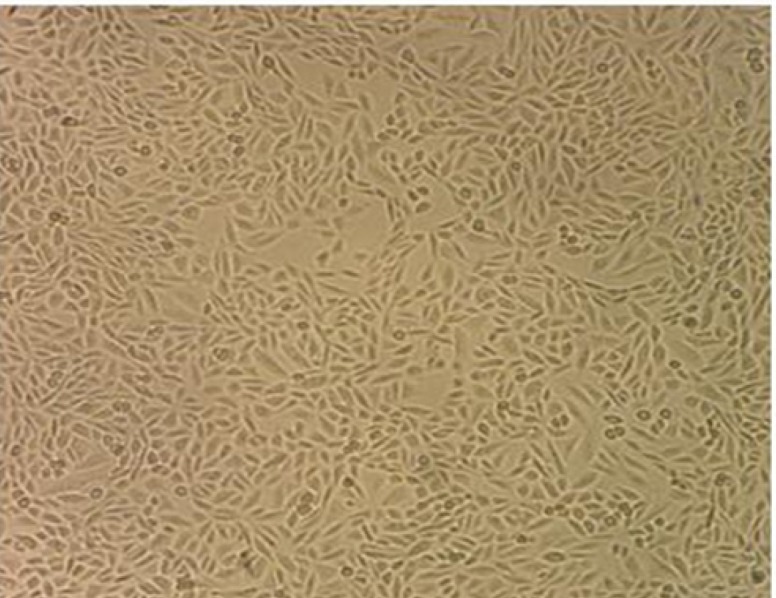
Cellular Morphology of Pari-ICR Cell Line in Cell Culture. The epithelial-like cells grew as a monolayer
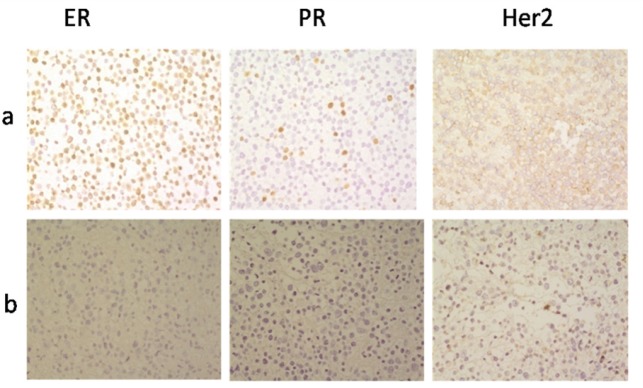
Immunocytotochemical study revealed that Pari-ICR cell line was negative for ER, PR and HER2, while in the control cell line (MCF-7), ER and PR were positive in about 90% and 10% of the nucleus, respectively. HER2/neu had faint and incomplete membranous staining on MCF-7 (score 1+) (Figure 2).
Figure 2.
Immunocytochemical Profiles of Estrogen Receptor (ER), Progesterone Receptor (PR) and HER2 Receptor. (a) MCF-7 cell line: positive for ER (90%) and PR (10%) receptors, weakly positive for HER2/neu receptor. (b) Pari-ICR cell line: Negative for ER, PR and Her2/neureseptors. Images were taken at 400× magnification
Characterization of the cell line
Growth characteristic and anchorage-independent growth in soft-agar
Doubling time of Pari-ICR was calculated at passages 50 during the exponential growth phase of the cell line by counting the number of cells for 6 days. The doubling time of Pari-ICR was about 22h (Figure 3).
Figure 3.
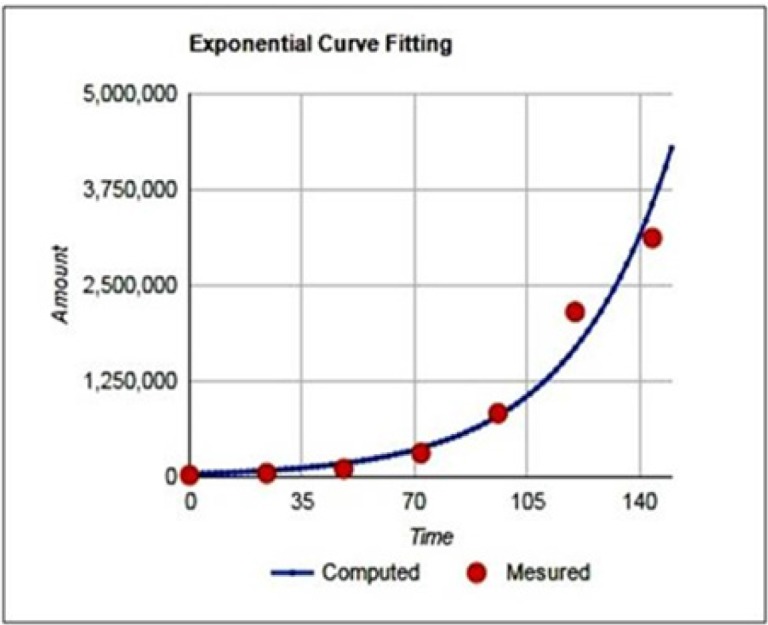
Growth Curve of Pari-ICR. Doubling time calculated using free online doubling time calculating software (http://www.doubling-time.com). The doubling time of Pari-ICR was about 22h
To evaluate the ability of the established cell line to grow into a colony, the colony formation assay was used. After 3 weeks, Pari-ICR formed large colonies on agar with mean efficiency of 30% (Figure 4).
Figure 4.
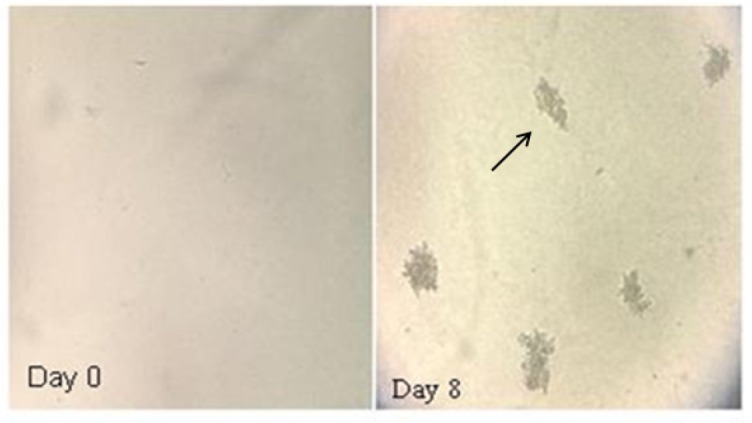
Representative Images of Colonies Formed in Soft Agar by Pari-ICR at day 0 and day 8. Single cell suspensions from 100 to 1000 cells per well were prepared. After 10 days, the number of colonies containing more than 50 cells was counted by an inverted microscope
Migration and invasion assay
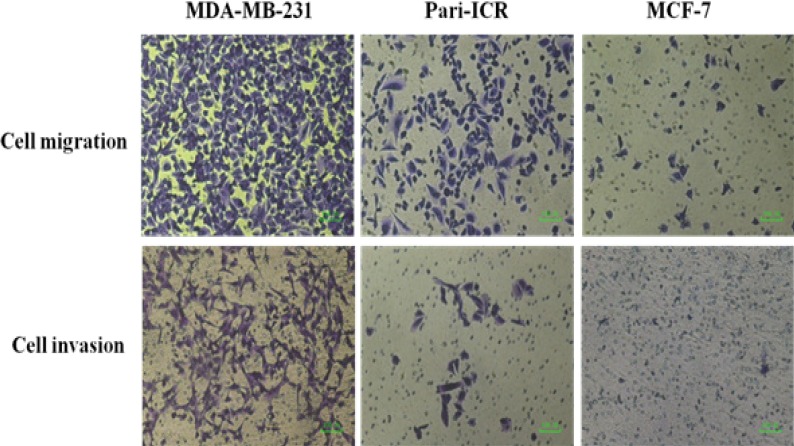
Cell migration and invasion assay was performed in order to investigate this ability of Pari- ICR cell line in comparison to MDA-MB231 and MCF-7 breast cancer cell lines. As shown in Figure 5, the migrated cell numbers of Pari-ICR cell line was higher than MCF-7 but lower than MDA-MB231.
Figure 5.
Migration and Invasion Assays of Pari-ICR Compared to MDA-MB-231 (Positive Control) and MCF-7 (Negative Control) Cell Lines
Phenotypic profiling of established cell line
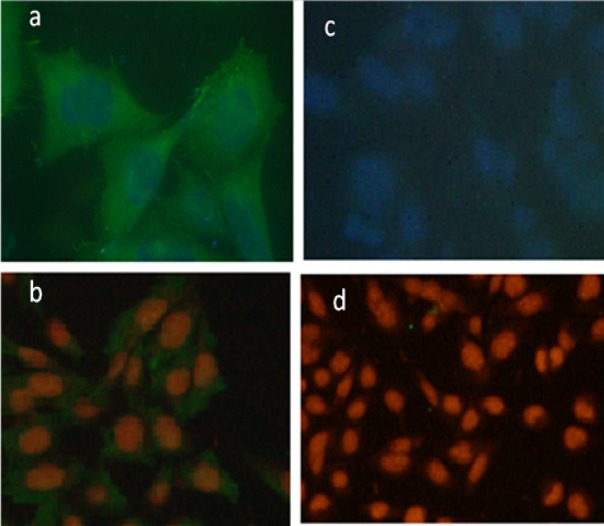
By immunofluorescent staining we observed that Pari-ICR cell line expressed a high level of Vimentin and Ezrin but not Cytokeratin19, Pan-cytokeratin, Nestin, and Desmin (Figure 6).
Figure 6.
Immunofluorescence Staining of Pari-ICR Cell Line. Expression of Ezrin and Vimentin in pari-ICR was investigated by imunofluorescence assay. (a) Immunofluorescence staining with anti-Ezrin antibody, (b) Immunofluorescence staining with anti-Vimentin antibody, (c,d) Negative controls. Nucleus is stained by DAPI in figures (a) & (c), and by PE in figures (b) & (d)
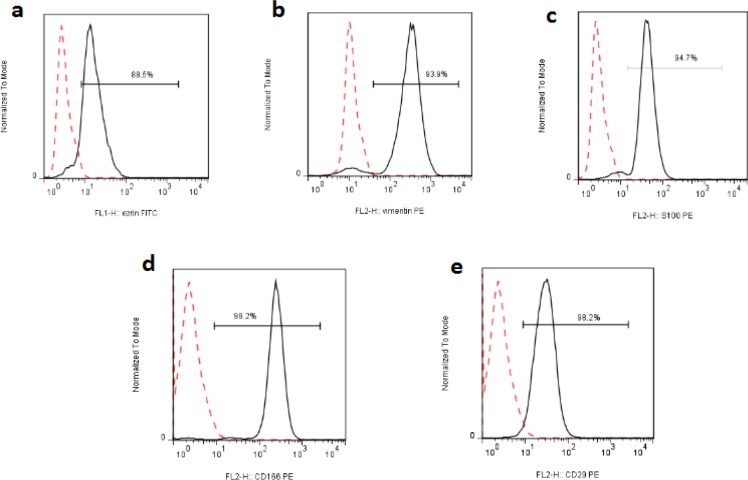
Flowcytometric analysis showed that Pari-ICR cell line was positive for Vimentin, Ezrin, S100, CD166, and CD29 but negative for EPCAM, Pan-cytokeratin, CD24, CD44, CD10, CD45, CD34, CD105, CD133, and CD146 (Figure 7).
Figure 7.
Expression of Different Markers by FACS in the Pari-ICR Cell Line. Pari-ICR cells were incubated with indicated antibodies, and analyzed by flow cytometery. a) Ezrin, b) Vimentin, c) S100, d) CD166, e) CD29. Dashed lines represent isotype control staining
Chromosomal analysis of the cell line
To detect gain and loss of chromosomes, comparative genomic hybridization array was performed and showed chromosomal losses on chromosomes 4 and 18 and gains on chromosomes 1, 12, and17 (Figure 8).
Figure 8.
Array CGH of Pari-ICR Cell Line Shows Loss on Chromosomes 4, 18, Y and Gain on Chromosomes 1, 12, 17 and X
Discussion
Breast cancer is a public health problem worldwide. It is a very heterogeneous disease, which has different biological features and clinical implications that lead to heterogeneous responses to therapy (Dai et al., 2015). Although, new approaches have been recently presented for the diagnosis and treatment, breast cancer accounts for 22% of new cancer cases each year (Pires et al., 2016). Moreover, although 5-year survival rate of breast cancer in developed countries is 85%, it is around 50-60% in developing countries (Movahedi et al., 2012).
In vitro and in vivo models are important for providing additional information in breast carcinogenesis mechanisms and testing therapeutic strategies. Among the various models, cancer cell lines are the most widely used models. The cell lines are a pure population of cells and an infinite source of material, which can be applied easily to provide reproducible results with the same protocol (Burdall et al., 2003; Van Staveren et al., 2009; Turin et al., 2014). The majority of cancer cell lines have been passaged thousands of times (Shen et al., 2009), the process that may change their characteristics. Therefore, newly established cancer cell lines derived from primary tumors are always in demand.
In this study, we established a new triple negative breast cancer cell line, Pari-ICR, derived from a primary tumor of a young patient with invasive ductal carcinoma. The newly established cells grew well as an adherent monolayer in DMEM/F12 medium without any extra essential components for growth. Pari-ICR has spindle-shaped morphology like MDA-MB-231. The doubling time of Pari-ICR was 22h that is faster than the proliferation rate of many other breast cancer cell lines (Engel and Young, 1978). The immunocytochemistry staining revealed that the established cell line was negative for ER, PR, and HER2. Pari-ICR was negative for epithelial markers such as cytokeratin 19, pan-cytokeratin and EpCAM but it was strongly positive for Vimentin.
Keratins are normally expressed in all types of epithelial cells and their neoplastic derivatives. Vimentin is an intermediate filament that is expressed in a wide variety of mesenchymal cell types and has important effect during invasion and migration. Several studies have reported that Vimentin is overexpressed in a large number of cancers including breast cancer. It has been shown that Vimentin expression had a significant correlation with the loss of one or more cytokeratins in malignant cells (Sommers et al., 1994; Satelli and Li, 2011; Vassilopoulos et al., 2014). Thomas et al. reported that Vimentin immunopositivity in breast tissues was inversely associated with keratin expression and directly associated with poor differentiation, negative estrogen/progesterone receptor status, and poor prognosis (Thompson et al., 1992; Choi et al., 2016). Although histogenesis of the Vimentin expressing tumors is unclear, myoepithelial histogenesis has been proposed. Moreover, it has been suggested that Vimentin expression can be considered as a sign of epithelial-mesenchymal transition (EMT), reflecting the final step of tumor dedifferentiation, which is generally associated with a high potential of tumor cell invasion (Korsching et al., 2005; Satelli and Li, 2011)
Sommers et al., (1994) have shown that human breast cancer cell lines were distributed along a spectrum of differentiation from well differentiated (epithelioid cell lines) to poorly differentiated (fibroblastic cell lines). They showed that fibroblastic cell lines such as MDA-MB-231 expressed Vimentin whereas cell lines such as MCF-7 and SKBR3 did not. The cell lines that were positive for Vimentin had reduced levels of keratins. In addition, Vimentin expression was associated with in-vitro invasiveness. Accordingly, overexpression of Vimentin in Pari-ICR may indicate the high metastatic potential of this cell line.
Ezrin, CD166 and CD29 are other markers that were expressed by Pari-ICR. Ezrin is a member of ERM family (Ezrin, Radixin, Moesin) that links the plasma membrane to the cytoskeleton of the cell. Erzin is detectable in normal and malignant cells and is involved in several functions such as cell migration, proliferation, survival/apoptosis, and adhesion (Meng et al., 2010; Gamei et al., 2014). Overexpression of this protein correlates with metastasis and poor prognosis in various types of human cancers including breast cancer (Ghaffari et al., 2014). A recent study demonstrated that inhibition of Ezrin with Ezrin shRNA in MCF-7 cell line decreased cell motility and invasiveness (Li et al., 2010).
Activated leukocyte cell adhesion molecule (ALCAM) also known as CD166 is a glycoprotein belonging to the immunoglobulin superfamily of adhesion molecules. CD166 functions as a cell-cell adhesion molecule in hemophilic (ALCAM-ALCAM) and heterophilic (ALCAM-CD6) interactions (Davies and Jiang, 2010). This glycoprotein, which is expressed in a wide variety of cell types, often shows increased expression in certain cancers including melanoma, colorectal, prostate, breast, ovarian, bladder, and esophageal cancer suggesting that CD166 is under restricting regulation during carcinogenesis (Davies and Jiang, 2010; Levin et al., 2010; Xiao et al., 2017). On the other hand, it has been shown that inhibition of CD166 promotes migration and invasion of tumor cells (Rosso et al., 2007; Kijima et al., 2012; Fujiwara et al., 2014). In breast cancer, the findings are controversial. While expression of ALCAM protects breast cancer cells against programmed cell death, its overexpression reduces the growth and migration of the tumor cells (Thompson et al., 1992; Jezierska et al., 2006; Rosso et al., 2007; Davies and Jiang, 2010). CD29, also known as Integrin β1, is a protein involved in cell motility and cancer metastasis. The malignant breast cancer cell line, MDA-MB-231, expresses high levels of β1 integrins, which promote invasive properties, whereas non-invasive MCF-7 cell line expresses low levels of β1 integrins (Hermann et al., 2016). It has been shown that the lack of CD29 results in a decreased cell migration (Vassilopoulos et al., 2014). Migration/invasion assays revealed that the established cell line had migration and invasiveness ability that was higher than MCF-7 but lower than MDA-MB231. This property of Pari-ICR cell line was in accordance with the expression of CD29.
Another marker expressed by Pari-ICR was S100. The S100 gene family, which is comprised of over 20 members, encodes low molecular weight calcium-binding proteins (Choi et al., 2016). They participate in a wide range of biological processes such as proliferation, migration and/or invasion, inflammation, and differentiation (Bresnick et al., 2015). Recent data suggest that specific S100 family members are engaged in cancer progression, especially metastasis (McKiernan et al., 2011). Recent studies indicated that abnormal expression of S100 proteins – mainly including S100A2, S100A4, and S100A7 – is related to metastasis of breast tumor(McKiernan et al., 2011; Li et al., 2014). The anchorage-independent growth in semisolid media is a property of most transformed cells that is correlated with tumorigenicity and metastatic potential. The established cell line was able to grow as colonies in the soft agar.
In conclusion, an immortalized triple negative breast cancer cell line was established from a primary breast tumor that exhibited high growth potential. Compared to other breast cancer cell lines that have been derived from a metastatic tumor, Pari-ICR seems to be a more plausible model. This breast cancer cell line can be served as a model for understanding molecular mechanisms of breast carcinogenesis in young patients. Moreover, it can be used as an appropriate source to find novel biomarkers and for assessment of new drugs.
Statement conflict of Interest
The authors have no conflict of interest to declare.
Acknowledgments
This work was financially supported by a grant from Shiraz university of Medical Sciences (grant number: 94-7652) and in part by Shiraz Institute for Cancer research (grant number: ICR-100-508).
References
- Anders CK, Johnson R, Litton J, et al. Breast cancer before age 40 years. Semin Oncol. 2009:237–49. doi: 10.1053/j.seminoncol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assi HA, Khoury KE, Dbouk H, et al. Epidemiology and prognosis of breast cancer in young women. J Thorac Dis. 2013;5:2–8. doi: 10.3978/j.issn.2072-1439.2013.05.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin Hirata BK, Oda JMM, Losi Guembarovski R, et al. Molecular markers for breast cancer: prediction on tumor behavior. Dis Markers. 2014;2014:513158. doi: 10.1155/2014/513158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer. 2015;15:96–109. doi: 10.1038/nrc3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdall SE, Hanby AM, Lansdown MR, et al. Breast cancer cell lines: friend or foe? Breast Cancer Res. 2003;5:89. doi: 10.1186/bcr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey L, Winer E, Viale G, et al. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683–92. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- Choi J, Kim D-I, Kim J, et al. Hornerin is involved in breast cancer progression. J Breast Cancer. 2016;19:142–7. doi: 10.4048/jbc.2016.19.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collignon J, Lousberg L, Schroeder H, et al. Triple-negative breast cancer: treatment challenges and solutions. Breast Cancer. 2016;8:93. doi: 10.2147/BCTT.S69488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Li T, Bai Z, et al. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5:2929. [PMC free article] [PubMed] [Google Scholar]
- Davies S, Jiang WG. ALCAM, activated leukocyte cell adhesion molecule, influences the aggressive nature of breast cancer cells, a potential connection to bone metastasis. Anticancer Res. 2010;30:1163–8. [PubMed] [Google Scholar]
- Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- Engel LW, Young NA. Human breast carcinoma cells in continuous culture: a review. Cancer Res. 1978;38:4327–39. [PubMed] [Google Scholar]
- Fujiwara K, Ohuchida K, Sada M, et al. CD166/ALCAM expression is characteristic of tumorigenicity and invasive and migratory activities of pancreatic cancer cells. PLoS One. 2014;9:e107247. doi: 10.1371/journal.pone.0107247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamei MM, el Naby NMA, El-Ashmawy AA, et al. Prognostic value of ezrin expression in common epithelial tumors: An immunohistochemical study. J Microsc Ultrastruct. 2014;2:67–76. [Google Scholar]
- Ghaffari A, Hoskin V, Szeto A, et al. A novel role for ezrin in breast cancer angio/lymphangiogenesis. Breast Cancer Res. 2014;16:438. doi: 10.1186/s13058-014-0438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann M-R, Jakobson M, Colo GP, et al. Integrins synergise to induce expression of the MRTF-A–SRF target gene ISG15 for promoting cancer cell invasion. J Cell Sci. 2016;129:1391–403. doi: 10.1242/jcs.177592. [DOI] [PubMed] [Google Scholar]
- Jazayeri SB, Saadat S, Ramezani R, et al. Incidence of primary breast cancer in Iran: Ten-year national cancer registry data report. Cancer Epidemiol. 2015;39:519–27. doi: 10.1016/j.canep.2015.04.016. [DOI] [PubMed] [Google Scholar]
- Jezierska A, Matysiak W, Motyl T. ALCAM/CD166 protects breast cancer cells against apoptosis and autophagy. Med Sci Monit. 2006;12:263–73. [PubMed] [Google Scholar]
- Kijima N, Hosen N, Kagawa N, et al. CD166/activated leukocyte cell adhesion molecule is expressed on glioblastoma progenitor cells and involved in the regulation of tumor cell invasion. Neurooncology. 2012;14:1254–64. doi: 10.1093/neuonc/nor202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsching E, Packeisen J, Liedtke C, et al. The origin of vimentin expression in invasive breast cancer: epithelial–mesenchymal transition, myoepithelial histogenesis or histogenesis from progenitor cells with bilinear differentiation potential? J Pathol. 2005;206:451–7. doi: 10.1002/path.1797. [DOI] [PubMed] [Google Scholar]
- Ku J-L, Park S-C, Kim K-H, et al. Establishment and characterization of seven human breast cancer cell lines including two triple-negative cell lines. Int J Oncol. 2013;43:2073–81. doi: 10.3892/ijo.2013.2144. [DOI] [PubMed] [Google Scholar]
- Levin TG, Powell AE, Davies PS, et al. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology. 2010;139:2072–82. doi: 10.1053/j.gastro.2010.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Men X, Zhang W. S100 protein in breast tumor. Indian J Cancer. 2014;51 doi: 10.4103/0019-509X.154046. [DOI] [PubMed] [Google Scholar]
- Li J, Tu Y, Wen J, et al. Role for ezrin in breast cancer cell chemotaxis to CCL5. Oncol Rep. 2010;24:965. doi: 10.3892/or.2010.965. [DOI] [PubMed] [Google Scholar]
- McKiernan E, McDermott EW, Evoy D, et al. The role of S100 genes in breast cancer progression. Tumor Biol. 2011;32:441–50. doi: 10.1007/s13277-010-0137-2. [DOI] [PubMed] [Google Scholar]
- Meng Y, Lu Z, Yu S, et al. Ezrin promotes invasion and metastasis of pancreatic cancer cells. J Transl Med. 2010;8:61. doi: 10.1186/1479-5876-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahedi M, Haghighat S, Khayamzadeh M, et al. Survival rate of breast cancer based on geographical variation in iran, a national study. Iran Red Crescent Med J. 2012;14:798–804. doi: 10.5812/ircmj.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaghvar HA, Hosseini M, Tizmaghz A, et al. A review on metastatic breast cancer in Iran. Asian Pac J Trop Biomed. 2015;5:429–33. [Google Scholar]
- Pandrangi SL, Bagadi SAR, Sinha NK, et al. Establishment and characterization of two primary breast cancer cell lines from young Indian breast cancer patients: mutation analysis. Cancer Cell Int. 2014;14:14. doi: 10.1186/1475-2867-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan VH, Moore MM, McLachlan AJ, et al. Ethnic differences in drug metabolism and toxicity from chemotherapy. Expert Opin Drug Metab Toxicol. 2009;5:243–57. doi: 10.1517/17425250902800153. [DOI] [PubMed] [Google Scholar]
- Pires BRB, De Amorim ÍSS, Souza LDE, et al. Targeting Cellular signaling pathways in breast cancer stem cells and its implication for cancer treatment. Anticancer Res. 2016;36:5681–91. doi: 10.21873/anticanres.11151. [DOI] [PubMed] [Google Scholar]
- Rapoport BL, Nayler S, Demetriou GS, et al. Triple negative breast cancer pathologic diagnosis and current chemotherapy treatment options. Oncol Hematol Rev. 2014;10:25–32. [Google Scholar]
- Rosso O, Piazza T, Bongarzone I, et al. The ALCAM shedding by the metalloprotease ADAM17/TACE is involved in motility of ovarian carcinoma cells. Mol Cancer Res. 2007;5:1246–53. doi: 10.1158/1541-7786.MCR-07-0060. [DOI] [PubMed] [Google Scholar]
- Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68:3033–46. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Gu M, Liang D, et al. Establishment and characterization of three new human breast cancer cell lines derived from Chinese breast cancer tissues. Cancer Cell Int. 2009;9:2. doi: 10.1186/1475-2867-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommers CL, Byers SW, Thompson EW, et al. Differentiation state and invasiveness of human breast cancer cell lines. Breast Cancer Res Treat. 1994;31:325–35. doi: 10.1007/BF00666165. [DOI] [PubMed] [Google Scholar]
- Thompson EW, Paik S, Brünner N, et al. Association of increased basement membrane invasiveness with absence of estrogen receptor and expression of vimentin in human breast cancer cell lines. J Cell Physiol. 1992;150:534–44. doi: 10.1002/jcp.1041500314. [DOI] [PubMed] [Google Scholar]
- Turin I, Schiavo R, Maestri M, et al. In vitro efficient expansion of tumor cells deriving from different types of human tumor samples. Med Sci. 2014;2:70–81. [Google Scholar]
- Van Staveren W, Solís DW, Hebrant A, et al. Human cancer cell lines: Experimental models for cancer cells in situ? For cancer stem cells? Biochim Biophys Acta. 2009;1795:92–103. doi: 10.1016/j.bbcan.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat Rev Cancer. 2007;7:659–72. doi: 10.1038/nrc2193. [DOI] [PubMed] [Google Scholar]
- Vassilopoulos A, Chisholm C, Lahusen T, et al. A critical role of CD29 and CD49f in mediating metastasis for cancer-initiating cells isolated from a Brca1-associated mouse model of breast cancer. Oncogene. 2014;33:5477–82. doi: 10.1038/onc.2013.516. [DOI] [PubMed] [Google Scholar]
- Xiao M, Yan M, Zhang J, et al. Cancer stem-like cell related protein CD166 degrades through E3 ubiquitin ligase CHIP in head and neck cancer. Exp Cell Res. 2017;353:46–53. doi: 10.1016/j.yexcr.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-Q, Han Y-Z, Nian Q, et al. Tumor invasiveness, not lymphangiogenesis, is correlated with lymph node metastasis and unfavorable prognosis in young breast cancer patients (≤ 35 Years) PLoS One. 2015;10:e0144376. doi: 10.1371/journal.pone.0144376. [DOI] [PMC free article] [PubMed] [Google Scholar]