Highlights
-
•
We conducted a GWAS to identify genetic loci linked to WMHV in non-demented elders.
-
•
Rs7220676 near HS3ST3A1 and MIR548H3 genes was significantly associated with WMHV.
-
•
Rs7220676 was also correlated with rates of cognitive decline.
Keywords: White matter hyperintensity volume, Cognition, Genome-wide association study
Abstract
Background
White matter hyperintensity has been correlated with cognitive disorders and its genetic predictors remain unclear. Here we conducted a genome-wide association study to identify novel genetic determinants that were correlated with white matter hyperintensity volume (WMHV) among non-demented elders.
Methods
Three hundred and fifty non-Hispanic Caucasian subjects aged 55–80 years were included from the Alzheimer's Disease Neuroimaging Initiative cohort. Associations of WMHV with genetic polymorphisms were explored using multiple linear regression under an additive genetic model. Further studies were conducted to explore the influence of genetic variants on cognition-related phenotypes.
Results
Rs7220676 near HS3ST3A1 and MIR548H3 genes was associated with WMHV levels at genome-wide significance (P = 2.96 × 10−8). Single nucleotide polymorphisms comprising rs9675262 (near HS3ST3A1 and MIR548H3 genes, P = 1.15 × 10−7), rs9820240 (in DCLK3 gene, P = 2.23 × 10−7), rs10916409 (near ISCA1P2 gene, P = 4.55 × 10−6), and rs540422 (in PICALM gene, P = 9.68 × 10−6) were identified as suggestive loci linked to WMHV levels. The minor allele of rs7220676 (C) showed association with lower log (WMHV) in a dose-dependent manner. Besides, rs7220676 was correlated with rates of cognitive decline assessed by Mini-mental State Examination and memory scores.
Conclusions
A novel locus near HS3ST3A1 and MIR548H3 genes was associated with WMHV levels and it may be involved in neurodegenerative diseases.
1. Introduction
White matter hyperintensities (WMH) are regions of increased signal on T2-weighted magnetic resonance imaging (MRI) sequences of the brain (Yoshita et al., 2006). The extent of WMH has been correlated with mild cognitive impairment (MCI) (DeCarli et al., 2001; Lopez et al., 2003), dementia (Elias et al., 2004; Wu et al., 2002), as well as cortical atrophy (Barnes et al., 2013). Besides, WMH may contribute to the progression from MCI to dementia (Wu et al., 2002; Wolf et al., 2000). The severity of WMH increased among patients diagnosed as clinical or preclinical Alzheimer's disease (AD) (Yoshita et al., 2006; Luchsinger et al., 2009), which reliably predicts who are at risk of developing AD (Prins et al., 2004), and predicts rate of cognitive decline among AD individuals (Provenzano et al., 2013). As for the interaction between WMH and AD pathology, WMH may not only be an independent process causing dementia in combination with coexisting yet unrelated AD pathology, but also lead directly to AD pathology by increasing amyloid accumulation rates due to ischemic disorders or by inducing ischemia because of amyloid accumulation in the vessels (cerebral amyloid angiopathy) (Cordonnier and van der Flier, 2011; Chen et al., 2006; Smith and Greenberg, 2009; Olichney et al., 2000). These lines of evidence together indicated that WMH might be involved in the pathogenesis of cognitive disorders and be promising in monitoring cognitive progression at early stage, and assessing treatment efficacy in clinical practice or drug trials.
Although previous articles have uncovered some loci that affect WMH volume (WMHV), the associations of these genetic modulators with disease risk remain poorly understood. A recent meta-analysis revealed that PLEKHG1 was linked to WMHV and ischemic stroke, motivating further identification of WMHV-associated loci (Traylor et al., 2019). The application of quantitative phenotypes in genome-wide association studies (GWAS) shed light on associations between genes and their correlated pathways (Ramanan et al., 2012). Moreover, MRI measurements have great strengths in monitoring changes in brain structure and function, such as adequate sensitivity, non-invasiveness, ease of access, and good tolerance (Duncan et al., 2013). And WMHV can be reliably measured in vivo. Herein, we performed a GWAS using WMHV as an endophenotype to explore genetic predictors associated with WMHV in non-demented elders. These genetic factors may be involved in cognition-related pathophysiological processes.
2. Methods
2.1. Alzheimer’s Disease Neuroimaging Initiative (ADNI)
ADNI, a large, multicenter, longitudinal neuroimaging study, was initiated in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies, and non-profit organizations (Mueller et al., 2005). ADNI was established to explore the effectiveness of integrating neuroimaging, genetic/biological markers, as well as clinical and neuropsychological assessments in measuring the early diagnosis and progression of AD. Regional ethical committees of all institutions approved the ADNI study. Written informed consent was obtained from all participants or authorized representatives.
2.2. Participants
Data used in preparation for this study were derived from the ADNI database (http://adni.loni.usc.edu/). Baseline and longitudinal clinical and neuropsychological test results, neuroimaging scan information, and data about clinical diagnosis from all participants were available. Detailed procedures for sample screening were presented in Appendix 1. The full cohort with both GWAS and WMHV data comprised 425 subjects aged 55–80 years. To reduce the likelihood of population stratification effects in the GWAS, all samples were restricted to non-Hispanic Caucasians (n = 385). To avoid the influence of dementia on results, only non-demented individuals at baseline (n == 355) were included in this study. To test cryptically related subjects and/or sample mix-ups, genomic identify-by-descent and multidimensional scaling (MDS) components were performed using PLINK (Chang et al., 2015). This step excluded 3 subjects who showed cryptically associated and clustering separately from the other participants (Appendix 2), leaving 352 valid samples. Finally, all samples presented close clustering with individuals of European descent using the HapMap cohort.
2.3. WMHV and QC
WMH regions were segmented via a multimodal segmentation method, white matter lesion segmentation (WMLS), using T1-weighted and fluid-attenuated inversion recovery (FLAIR) images (Zacharaki et al., 2008). FLAIR MRI was corrected for inhomogeneity and warped to T1 images to provide the segmentation. WMHs are seeded at points that are >3.5 standard deviations (SD) from the mean signal in white matter, and final segmentation is based on a Bayesian approach, combining spatial priors and tissue class constraints. The WMH segmentation also comprised segmentations of white matter, gray matter, and cerebrospinal fluid (CSF); the sum of the tissue volumes was applied as a surrogate of intracranial volume (ICV). For analysis, WMHV values were log transformed to achieve a normal distribution because the values were skewed (Shapiro-Wilk test P < 0.05). Log (WMHV) was then used as a quantitative outcome phenotype for the GWAS. QC was conducted to mitigate the influence of extreme values on statistical results. Mean and SD of baseline WMHV measures were calculated by observers blinded to clinical data, and the figures greater or smaller than 5.5-fold SD from the mean value were considered as extreme outliers and were excluded from this analysis. After removal of 2 outliers, there were 350 valid samples left.
2.4. Vascular information, neuroimaging, and cognition
The baseline vascular information, including history of heart disease and hypertension, were accessed by screening the medical information database. Body mass index (BMI) was utilized to determine obese conditions following the world health organization criteria: overweight/obese ≥25 kg/m2, normal weight <25 kg/m2.
Structural brain MRI was acquired using a Siemens Trio 3.0T scanner or Vision 1.5T scanner. Regional volume estimates were processed using Free-surfer software package version 4.3 and 5.1 image processing framework for the 1.5T and 3.0T MRI images, respectively. Regions of interests (ROIs) included the hippocampus and entorhinal cortex.
Cognition status was assessed by Mini-mental State Examination (MMSE) and memory (MEM) scores. The MEM was a weighted score based on memory items in Rey Auditory Verbal Learning Test (RAVLT), the Alzheimer's Disease Assessment Scale-Cognitive section (ADAS-cog), the MMSE and Logical Memory.
2.5. Genotyping and QC
All samples were genotyped using the Illumina Human Hap610-Quad BeadChips featuring 2379,855 single nucleotide polymorphisms (SNPs). Prior to association analysis, all samples and genotypes underwent stringent QC with the following criteria: call rates for SNPs > 98%, call rates for individuals > 95%, minor allele frequencies (MAF) > 0.20 and Hardy-Weinberg equilibrium test P > 0.001. We set the MAF value > 0.20 for SNPs to improve statistical power and reduce potentially false-positive results in the context of modest sample size. Finally, the filters based on QC produced a total of 695,203 imputed and genotyped SNPs for analyses. The overall genotyping rate for the remaining dataset was 99.7%.
2.6. Statistical analyses
Mann-Whitney U test was used to determine the difference in baseline WMHV measures of different diagnostic groups. We performed a GWAS of WMHV with genetic polymorphisms, using multiple linear regression under an additive genetic model in PLINK v1.9 software. Principle component analysis was performed in PLINK and the first three MDS components were applied as covariates in the GWAS. Age at baseline, gender, dosage of APOE ε4 allele, baseline diagnosis and ICV were also used as covariates. To account for multiple comparisons, we set the thresholds for genome-wide significant and suggestive associations at P < 5 × 10−8 and P < 1 × 10−5, respectively (Risch and Merikangas, 1996). Genome-wide associations were visualized using R package “qqman” and regional association plots were generated with the LocusZoom web tool (http://locuszoom.org/).
The difference in log (WMHV) of different genotype groups was determined using a multiple linear regression model in R software, after adjusting for age, gender and baseline diagnosis. To further explore the associations of genotypes with log (WMHV) under different vascular status, we conducted subgroup analyses by history of heart disease, history of hypertension, and obesity. As for subgroup analyses, age, gender and baseline diagnosis were also included as covariates in the multiple linear regression model. Considering the relatively low follow-up rate, we limited the maximum follow-up time to 6 years. We also calculated the relative MMSE change in percentage (yearly MMSE change/MMSE at baseline) and figures were calculated from the 1-, 2-, 3-, 4-, 5- and 6-year visits. Linear regression models were used to examine the associations of relative MMSE change in percentage with the top SNPs. Furthermore, the associations of top SNPs with MEM scores and brain structures of interest from baseline and longitudinal perspectives were also tested in R software using the multiple linear regression and linear mixed models, respectively. Age, gender, dosage of APOE ε4 allele, baseline diagnosis and ICV were applied as covariates.
2.7. Bioinformatics analyses
SNP annotations were conducted via the NCBI Database of SNPs (http://www.ncbi.nlm.nih.gov/SNP/). Expression quantitative trait loci (eQTL) analyses were performed using multiple publicly available datasets in human brain tissues (http://caprica.genetics.kcl.ac.uk/BRAINEAC; Allen Institute Human Brain Atlas; http://human.brain-map.org/) and the whole blood (http://www.genenetwork.nl/bloodeqtlbrowser/).
3. Results
3.1. Characteristics of included subjects
Demographic characteristics of the study population are listed in Table 1 and Appendix 3. Briefly, 240 MCI (105 women, 69.6 ± 5.9 years) and 110 HC (62 women, 72.2 ± 4.1 years) subjects were enrolled from the ADNI cohort. Linear correlations between age at baseline and log (WMHV) were found in both HC (P = 0.0374) and MCI (P < 0.0001) groups (Appendix 4). However, there was no significant difference in WMHV between males and females (MCI P = 0.105, HC P = 0.974, Total P = 0.153) among the diagnostic groups (Appendix 5). MCI group (45.4%) had a higher frequency of ε4 allele within APOE gene than HC group (29.1%) (P = 0.004). MCI group also had higher baseline WMHV compared to HC group (P < 0.001) (Appendix 6).
Table 1.
Demographic information.
| Baseline diagnosis | MCI | HC | Total |
|---|---|---|---|
| Sample size, n (%) | 240 (68.6) | 110 (31.4) | 350 |
| Age at baseline, mean (SD) | 69.6 (5.9) | 72.2 (4.1) | 70.4 (5.5) |
| Females, n (%) | 105 (43.8) | 62 (56.4) | 167 (47.7) |
| Education years, mean (SD) | 16.3 (2.6) | 16.4 (2.5) | 16.3 (2.6) |
| APOE ε4 carrier, n (%)a | 109 (45.4) | 32 (29.1) | 141 (40.3) |
| Hypertension, n (%) | 97 (40.4) | 47 (42.7) | 144 (41.1) |
| Heart disease, n (%) | 26 (10.8) | 14 (12.7) | 40 (11.4) |
| Obesity, n (%) | 165 (68.8) | 76 (69.1) | 241 (68.9) |
| MMSE score at baseline, mean (SD) | 28.2 (1.6) | 29.0 (1.3) | 28.5 (1.5) |
| MEM score at baseline, mean (SD) | 0.40 (0.66) | 1.09 (0.50) | 0.62 (0.69) |
| WMHV at baseline, mean (SD)b | 5.5 (6.6) | 5.3 (7.3) | 5.5 (6.8) |
Abbreviations: APOE, apolipoprotein E; HC, healthy controls; MCI, mild cognitive impairment; MEM, memory; MMSE, Mini-mental State Examination; SD, standard deviation; WMHV, white matter hyperintensity volume.
MCI group had a higher frequency of APOE ε4 than HC group (P = 0.004).
The MCI group had higher baseline WMHV compared to HC group (P < 0.001).
3.2. SNPs associated with WMHV
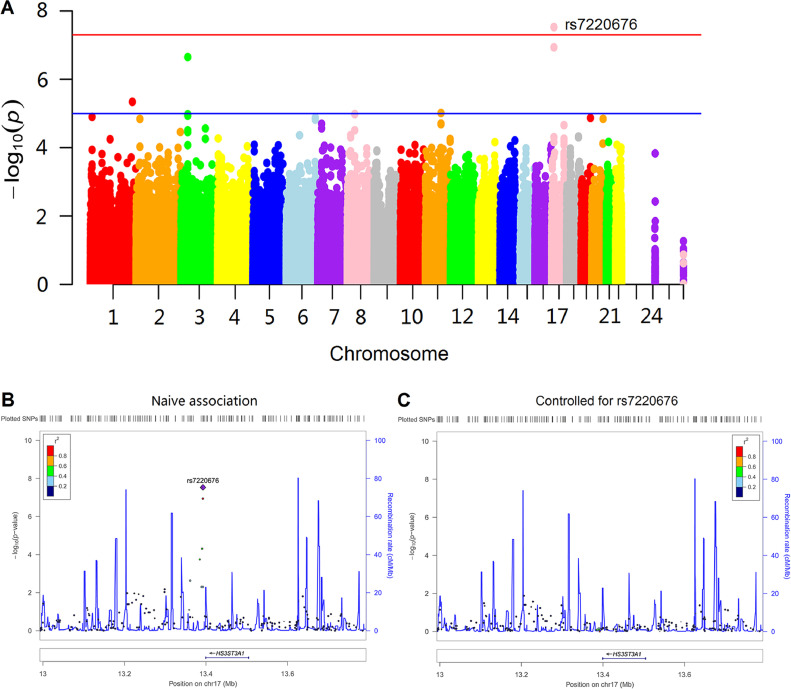
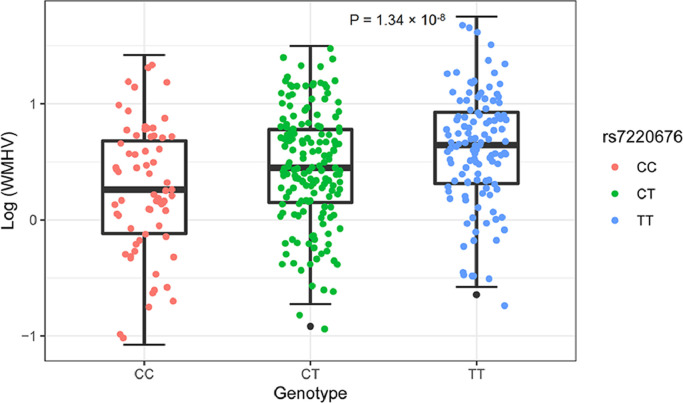
A total of 350 subjects were identified for GWAS. After adjustment for age, gender, dosage of APOE ε4 allele, baseline diagnosis, ICV and the first three MDS components, a genome-wide significant association of rs7220676 (near HS3ST3A1 and MIR548H3 genes, P = 2.96 × 10−8) with WMHV was detected (Fig. 1A). In a multiple linear regression model adjusted for age, gender and baseline diagnosis, the minor allele of rs7220676 (C) (CC: 0.2440 ± 0.5629, CT: 0.4555 ± 0.4878, TT: 0.6187 ± 0.4806; P = 1.34 × 10−8) was associated with lower log-WMHV measures in a dose-dependent manner among the whole samples (Fig. 2). Further analysis also found this linear association of genotypes and log (WMHV) levels in both MCI (CC: 0.3071 ± 0.5543, CT: 0.4638 ± 0.4958, TT: 0.5836 ± 0.4905; P = 2.48 × 10−4) and HC (CC: 0.1060 ± 0.5569, CT: 0.4385 ± 0.4704, TT: 0.7018 ± 0.4456; P = 4.35 × 10−6) groups (Appendix 6). Regarding subgroup analyses by vascular factors, the minor allele of rs7220676 (C) was associated with lower log-WMHV measures in a dose-dependent manner both among the population without (P = 1.03 × 10−5) and with hypertension (P = 1.80 × 10−4). Furthermore, the minor allele of rs7220676 (C) was correlated with lower log-WMHV measures in a dose-dependent manner among the population without heart disease (P = 9.46 × 10−10) or with obesity (P = 8.33 × 10−8). Nonetheless, the association was not significant among those with heart disease (P = 0.698) or normal BMI (P = 0.058) (Appendix 7).
Fig. 1.
Manhattan and regional plots for associations with white matter hyperintensity volumes. (A) Genome-wide signal intensity (Manhattan) plots showing the −log10 (p value) for individual single nucleotide polymorphisms. (B) Regional association results for the 12.9 Mb to 13.8 Mb region of chromosome 17. (C) Association results for the 12.9 Mb to 13.8 Mb region of chromosome 17 controlling for rs7220676.
Fig. 2.
Log (white matter hyperintensity volumes) of different genotypes. The minor allele of rs7220676 (C) showed association with log (white matter hyperintensity volumes) in a dose-dependent manner (P = 1.34 × 10−8). The P value was calculated using a multiple linear regression model after adjusting for age, gender and baseline diagnosis.
SNPs mapped closely to the top SNP (rs7220676) region were also analyzed (Fig. 1B). These nearby SNPs showed associations with WMHV levels at P < 0.01. After controlling for genotypes of rs7220676, the association signals for these nearby SNPs were dramatically attenuated (Fig. 1C). The Quantile-Quantile plot didn't show evidence of spurious inflation in test statistics (the genomic inflation factor= 1) due to population stratification or other confounders (Appendix 8).
Four SNPs met criteria for suggestive levels of genome-wide significance (P < 1 × 10−5) (Table 2), including rs9675262 (near HS3ST3A1 and MIR548H3 gene, P = 1.15 × 10−7), rs9820240 (in DCLK3 gene, P = 2.23 × 10−7), rs10916409 (near ISCA1P2 gene, P = 4.55 × 10−6), and rs540422 (in PICALM gene, P = 9.68 × 10−6). The minor allele of rs9675262 (T) (P = 3.77 × 10−8), the minor allele of rs9820240 (A) (P = 9.73 × 10−7), and the minor allele of rs10916409 (C) (P = 1.56 × 10−5) were associated with lower log-WMHV measures in a dose-dependent manner among the whole samples, whereas the minor allele of rs540422 (T) (P = 3.01 × 10−5) was associated with higher log-WMHV measures in a dose-dependent manner (Appendix 9).
Table 2.
Top SNPs associated with WMHV.
| CHR | BP | SNP | MAF | Closest Gene | SNP Type | P |
|---|---|---|---|---|---|---|
| 17 | 13,392,543 | rs7220676 | C = 0.4229 | HS3ST3A1, MIR548H3 | intergenic | 2.96E-08 |
| 17 | 13,395,795 | rs9675262 | T = 0.4373 | HS3ST3A1, MIR548H3 | intergenic | 1.15E-07 |
| 3 | 36,796,647 | rs9820240 | A = 0.2145 | DCLK3 | intron | 2.23E-07 |
| 1 | 229,156,248 | rs10916409 | C = 0.4411 | ISCA1P2 | intergenic | 4.55E-06 |
| 11 | 85,707,054 | rs540422 | T = 0.3401 | PICALM | intron | 9.68E-06 |
Abbreviations: BP, base pair (variant position); CHR, chromosome; MAF, minor allele frequency; SNP, single nucleotide polymorphism; WMHV, white matter hyperintensity volume.
3.3. Impact of the top SNPs on cognitive scores and brain structures
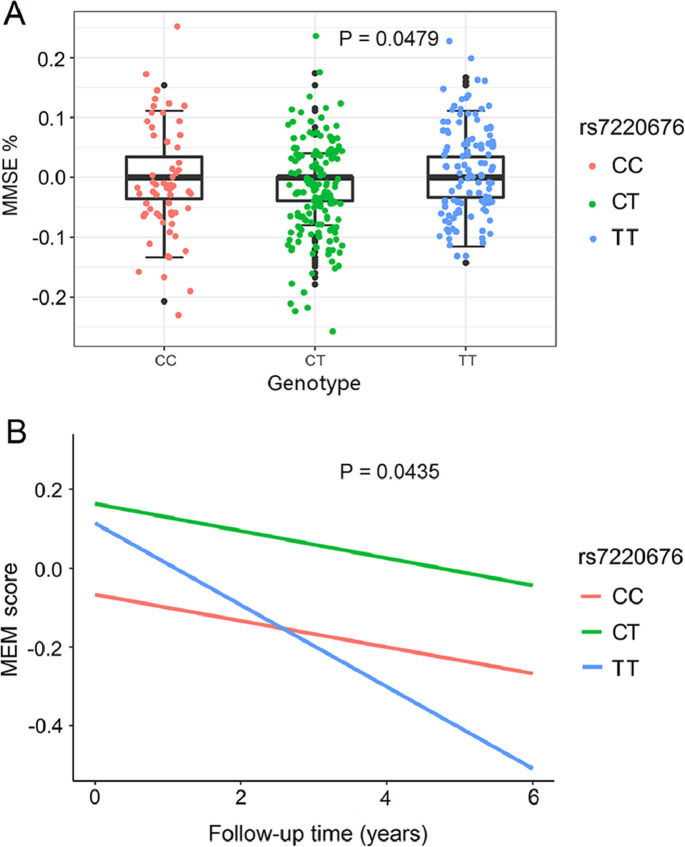
Some cognitive scores (MMSE and MEM scores) and specific brain structures (hippocampal volume and entorhinal volume) were chosen as cognition-related measures. Associations of rs7220676 with the above indexes from both cross-sectional and longitudinal perspectives were determined after adjusting for age, gender, APOE ε4 dosage, baseline diagnosis and ICV. In total samples, rs7220676 decreased the percentage of decline in MMSE score during 1 year of follow-up (CC: −0.0164 ± 0.0639; CT: −0.0188 ± 0.0697; TT: 2.0170 × 10−5 ± 0.0664; P = 0.0479) (Fig. 3A). MMSE scores decreased faster in subjects with TT genotypes than in those with CC genotypes (P = 0.0496). However, rs7220676 was not associated with MMSE% change during 2-, 3-, 4-, 5- or 6-year of follow-up. Also, rs7220676 was not correlated with MMSE scores at baseline. Besides, no significant influences of rs9675262 genotypes were revealed on MEM scores and brain structures of interest using both baseline and longitudinal measurements. In subgroup analysis, the minor allele of rs7220676 (C) was associated with slower cognitive decline assessed by MEM score in HC population (P = 0.0435) (Fig. 3B), whereas no other significant associations of rs9675262 genotypes were revealed with other phenotypes of interest in HC and MCI groups.
Fig. 3.
Associations of rs7220676 with cognitive scores. (A) Rs7220676 was associated with the percentage of decline in Mini-mental State Examination (MMSE) score during 1 year of follow-up (P = 0.0479). MMSE score decreased faster in subjects with TT genotypes than those with CC genotypes (P = 0.0496). (B) The minor allele of rs7220676 (C) was associated with slower cognitive decline assessed by memory (MEM) score among healthy controls (P = 0.0435).
3.4. Bioinformatics analyses
As for eQTL analyses, the minor allele of rs9820240 (A) (P = 0.0055) and the minor allele of rs540422 (T) (P = 0.04) were downregulated in putamen and frontal cortex, respectively. Besides, rs540422 has a cis-eQTL effect on PICALM (Z= −4.57, P = 4.83 × 10−6) in whole blood (Appendix 10) (Westra et al., 2013).
4. Discussion
This study conducted a GWAS of WMHV in non-demented elders. We identified genome-wide significant associations of a novel SNP (rs7220676) near HS3ST3A1 and MIR548H3 genes and 4 additional suggestive association loci (in DCLK3, PICALM gene, near ISCA1P2, HS3ST3A1 and MIR548H3 genes) with baseline WMHV levels. The minor alleles of rs7220676 (C), rs9675262 (T), rs9820240 (A) and rs10916409 (C) showed association with lower log- WMHV in a dose-dependent manner, whereas the minor allele of rs540422 (T) was associated with higher log-WMHV measures in a dose-dependent manner. Besides, rs7220676 was correlated with rates of cognitive decline assessed by MMSE and MEM scores.
Both rs7220676 and rs9675262 are located on chromosome 17 near HS3ST3A1 and MIR548H3 genes. HS3ST3A1 gene encodes the enzyme 3-Osulfotransferase, which catalyzes the biosynthesis of a specific subtype of heparan sulfate (HS), 3-O-sulfated heparan sulfate. HS3ST3A1 gene has been reported to be involved in human immunodeficiency virus (HIV) infection (Joubert et al., 2010), Plasmodium falciparum parasitaemia (Atkinson et al., 2012; Nguyen et al., 2018), lung cancer (Nakano et al., 2012), and respiratory papillomatosis (Wang et al., 2008). However, no research has explored the relationships between HS3ST3A1 gene and cognitive disorders. MIR548H3 gene is a member of micro Ribonucleic Acid (microRNA) family, which participates in the post-transcriptional modulation of gene expression in multicellular organisms through influencing the stability and translation of messenger Ribonucleic Acids (mRNAs) (Nilsen, 2007). Accumulating evidence showed that microRNAs might play an important role in the development of central nervous system and neuropsychiatric diseases (Mellios and Sur, 2012; Kim et al., 2017), and the potential associations of MIR548H3 gene with neuroticism (Kim et al., 2017), epilepsy and mild intellectual disability (Baglietto et al., 2014) have been discussed in previous studies. At present, the specific mechanisms through which MIR548H3 regulates cognition-related diseases remain poorly understood. There are several fundamental issues that need to be addressed, for example, whether microRNAs are particularly important in specific neurodegenerative diseases and which microRNA targets are linked to the disease, etc. Collectively, there is still a long way to go for research on the mechanism between the novel loci and cognitive function.
There was a suggestive SNP in the PICALM gene. The involvement of this gene in cognitive impairment has been highlighted. As one of the most highly validated AD risk factors, PICALM can be expressed in all tissues, and it shows prominent expression in neurons, where it is non-selectively distributed in the pre-and postsynaptic structures (Harold et al., 2009). Mutations in PICALM can cause synaptic perturbations, possibly through synaptic vesicle cycling, thus increasing the risk of AD. Alternatively, PICALM may influence AD risk through amyloid precursor protein (APP) trafficking via endocytic pathways, causing changes in amyloid beta (Aβ) levels (Harold et al., 2009). This evidence also got support from the biological aspect. One experiment in yeast, nematodes, and rat cortical neurons confirmed the role of PICALM in suppressing the toxicity of soluble Aβ oligomers and linking Aβ to the genetics of AD (Treusch et al., 2011). Moreover, the impact of PICALM on disproportionate frontal damage, executive dysfunction (Morgen et al., 2014), hippocampal degeneration (Yang et al., 2016), Pick disease, progressive supranuclear palsy (PSP), corticobasal degeneration (CBD) and frontotemporal lobar degeneration (FTLD) (Ando et al., 2016) provided further evidence support. However, the regions on chromosome 3 (DCLK3) and 1 (ISCA1P2) associated with WMHV have not been previously shown to have correlations with cognitive disorders. DCLK3 can modulate the expression of many genes participating in transcriptional regulation and nucleosome/chromatin remodeling in a kinase-dependent way. The kinase that DCLK3 expressed is preferentially distributed in neurons of the striatum and dentate gyrus, and DCLK3 may produce neuroprotection against Huntington's disease (Galvan et al., 2018). Also, DCLK3 may promote colorectal cancer progression (Liu et al., 2017), and involve in the development of glaucoma (Axenovich et al., 2011). But the function of ISCA1P2 has never been investigated. Further studies are especially warranted to explore how DCLK3 and ISCA1P2 genes mediate the progression of cognitive diseases.
Compared with previous articles similar to our topic (Fornage et al., 2011; Elliott et al., 2018; Verhaaren et al., 2015; Shen et al., 2010), this study was conducted in a different population and has found novel loci associated with WMHV in non-demented elders. Although some suggestive SNPs didn't reach the genome-wide significance level, they may have the potential significance in other studies with sample enrichment. When controlling for rs7220676 genotypes, the nearby association signals diminished, which indicated that the associations in nearby loci might be driven by rs7220676. Besides, the associations of rs7220676 genotypes with WMHV were not significant among the population with heart disease or normal BMI, which may be due to the insufficient sample. Given that the relationship between rs7220676 and vascular factors has never been reported, it's difficult to explain the specific mechanisms. After the GWAS scan, we assessed associations of rs7220676 with other cognition-related endophenotypes. Interestingly, the minor allele of rs7220676 (C) was associated with change rate of MEM score in HC population rather than in MCI patients. It could thus reasonably be implied that the modifying effects of rs7220676 might be more evident in subjects with normal cognition. Our research suggested that rs7220676 might be associated with cognitive disease among non-demented elders, and further studies are needed to confirm this.
Some limitations must be acknowledged. Firstly, the sample size for analysis was relatively small especially in subgroup analyses, leading to limited ability to identify variants with small effects. Secondly, only non-Hispanic Caucasians were included to avoid population stratification across ethnicities, which limited the generalizability of this study. Thirdly, we applied a stringent MAF threshold (MAF > 0.20) to improve statistical power. However, this restriction may lead to misses of less common SNPs. Fourthly, in our article, MCI patients included those who remained stable and those who developed into dementia at specific follow-up time, which may lower the specificity of the effect. However, since the conversion of MCI to dementia was complex and time-dependent, it's inappropriate to exclude those progressive MCI patients. The follow-up time was difficult to be defined and the follow-up data was limited. Fifthly, only the MMSE% change during 1 year of follow-up was found to be associated with rs7220676, which may be due to the higher missing rate during a longer follow-up. Replicated and independent studies with sample enrichment are warranted to confirm our findings.
In conclusion, we identified novel genome-wide significant associations of a SNP (rs7220676) near HS3ST3A1 and MIR548H3 genes and 4 additional suggestive association loci (in DCLK3, PICALM genes, near ISCA1P2, HS3ST3A1 and MIR548H3 genes) with baseline WMHV levels. The mechanisms underlying the associations between these novel genetic loci and cognitive disorders are worth exploring. Besides, further validation of these novel genetic associations in large samples and different populations is warranted.
CRediT authorship contribution statement
Yu Guo: Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing. Xue-Ning Shen: Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing. Xiao-He Hou: Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing. Ya-Nan Ou: Writing - original draft, Writing - review & editing. Yu-Yuan Huang: . Qiang Dong: . Lan Tan: Writing - original draft, Writing - review & editing. Jin-Tai Yu: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (91849126, 81571245, and 81771148), the National Key R&D Program of China (2018YFC1314700), Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX01) and Zhangjiang Lab, Tianqiao and Chrissy Chen Institute, and the State Key Laboratory of Neurobiology and Frontiers Center for Brain Science of Ministry of Education, Fudan University. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.nicl.2020.102209.
Appendix. Supplementary materials
References
- Ando K., Tomimura K., Sazdovitch V. Level of PICALM, a key component of clathrin-mediated endocytosis, is correlated with levels of phosphotau and autophagy-related proteins and is associated with tau inclusions in AD, PSP and Pick disease. Neurobiol. Dis. 2016;94:32–43. doi: 10.1016/j.nbd.2016.05.017. [DOI] [PubMed] [Google Scholar]
- Atkinson A., Garnier S., Afridi S., Fumoux F., Rihet P. Genetic variations in genes involved in heparan sulphate biosynthesis are associated with Plasmodium falciparum parasitaemia: a familial study in Burkina Faso. Malar. J. 2012;11:108. doi: 10.1186/1475-2875-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axenovich T., Zorkoltseva I., Belonogova N. Linkage and association analyses of glaucoma related traits in a large pedigree from a Dutch genetically isolated population. J. Med. Genet. 2011;48:802–809. doi: 10.1136/jmedgenet-2011-100436. [DOI] [PubMed] [Google Scholar]
- Baglietto M.G., Caridi G., Gimelli G. RORB gene and 9q21.13 microdeletion: report on a patient with epilepsy and mild intellectual disability. Eur. J. Med. Genet. 2014;57:44–46. doi: 10.1016/j.ejmg.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Barnes J., Carmichael O.T., Leung K.K. Vascular and Alzheimer’s disease markers independently predict brain atrophy rate in Alzheimer’s Disease Neuroimaging Initiative controls. Neurobiol. Aging. 2013;34:1996–2002. doi: 10.1016/j.neurobiolaging.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.W., Gurol M.E., Rosand J. Progression of white matter lesions and hemorrhages in cerebral amyloid angiopathy. Neurology. 2006;67:83–87. doi: 10.1212/01.wnl.0000223613.57229.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordonnier C., van der Flier W.M. Brain microbleeds and Alzheimer’s disease: innocent observation or key player? Brain. 2011;134:335–344. doi: 10.1093/brain/awq321. [DOI] [PubMed] [Google Scholar]
- DeCarli C., Miller B.L., Swan G.E., Reed T., Wolf P.A., Carmelli D. Cerebrovascular and brain morphologic correlates of mild cognitive impairment in the National Heart, Lung, and Blood Institute Twin Study. Arch. Neurol. 2001;58:643–647. doi: 10.1001/archneur.58.4.643. [DOI] [PubMed] [Google Scholar]
- Duncan G.W., Firbank M.J., O’Brien J.T., Burn D.J. Magnetic resonance imaging: a biomarker for cognitive impairment in Parkinson’s disease? Mov. Disord. 2013;28:425–438. doi: 10.1002/mds.25352. [DOI] [PubMed] [Google Scholar]
- Elias M.F., Sullivan L.M., D'Agostino R.B. Framingham stroke risk profile and lowered cognitive performance. Stroke. 2004;35:404–409. doi: 10.1161/01.STR.0000103141.82869.77. [DOI] [PubMed] [Google Scholar]
- Elliott L.T., Sharp K., Alfaro-Almagro F. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018;562:210–216. doi: 10.1038/s41586-018-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornage M., Debette S., Bis J.C. Genome-wide association studies of cerebral white matter lesion burden: the CHARGE consortium. Ann. Neurol. 2011;69:928–939. doi: 10.1002/ana.22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan L., Francelle L., Gaillard M.C. The striatal kinase DCLK3 produces neuroprotection against mutant huntingtin. Brain. 2018;141:1434–1454. doi: 10.1093/brain/awy057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D., Abraham R., Hollingworth P. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert B.R., Lange E.M., Franceschini N. A whole genome association study of mother-to-child transmission of HIV in Malawi. Genome Med. 2010;2:17. doi: 10.1186/gm138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.E., Kim H.N., Yun Y.J. Meta-analysis of genome-wide SNP- and pathway-based associations for facets of neuroticism. J. Hum. Genet. 2017;62:903–909. doi: 10.1038/jhg.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N.Q., Ter Huurne M., Nguyen L.N. The non-coding variant rs1800734 enhances DCLK3 expression through long-range interaction and promotes colorectal cancer progression. Nat. Commun. 2017;8:14418. doi: 10.1038/ncomms14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez O.L., Jagust W.J., Dulberg C. Risk factors for mild cognitive impairment in the Cardiovascular Health Study Cognition Study: part 2. Arch. Neurol. 2003;60:1394–1399. doi: 10.1001/archneur.60.10.1394. [DOI] [PubMed] [Google Scholar]
- Luchsinger J.A., Brickman A.M., Reitz C. Subclinical cerebrovascular disease in mild cognitive impairment. Neurology. 2009;73:450–456. doi: 10.1212/WNL.0b013e3181b1636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N., Sur M. The emerging role of microRNAs in schizophrenia and autism spectrum disorders. Front. Psychiatry. 2012;3:39. doi: 10.3389/fpsyt.2012.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgen K., Ramirez A., Frolich L. Genetic interaction of PICALM and APOE is associated with brain atrophy and cognitive impairment in Alzheimer’s disease. Alzheimers Dement. 2014;10:S269–S276. doi: 10.1016/j.jalz.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Mueller S.G., Weiner M.W., Thal L.J. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin. N. Am. 2005;15:869–877. doi: 10.1016/j.nic.2005.09.008. xi-xii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Shimizu K., Kawashima O. Establishment of a human lung cancer cell line with high metastatic potential to multiple organs: gene expression associated with metastatic potential in human lung cancer. Oncol. Rep. 2012;28:1727–1735. doi: 10.3892/or.2012.1972. [DOI] [PubMed] [Google Scholar]
- Nguyen N.T., Vives R.R., Torres M. Genetic and enzymatic characterization of 3-O-sulfotransferase SNPs associated with Plasmodium falciparum parasitaemia. Glycobiology. 2018;28:534–541. doi: 10.1093/glycob/cwy038. [DOI] [PubMed] [Google Scholar]
- Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Olichney J.M., Hansen L.A., Hofstetter C.R., Lee J.H., Katzman R., Thal L.J. Association between severe cerebral amyloid angiopathy and cerebrovascular lesions in Alzheimer disease is not a spurious one attributable to apolipoprotein E4. Arch. Neurol. 2000;57:869–874. doi: 10.1001/archneur.57.6.869. [DOI] [PubMed] [Google Scholar]
- Prins N.D., van Dijk E.J., den Heijer T. Cerebral white matter lesions and the risk of dementia. Arch. Neurol. 2004;61:1531–1534. doi: 10.1001/archneur.61.10.1531. [DOI] [PubMed] [Google Scholar]
- Provenzano F.A., Muraskin J., Tosto G. White matter hyperintensities and cerebral amyloidosis: necessary and sufficient for clinical expression of Alzheimer disease? JAMA Neurol. 2013;70:455–461. doi: 10.1001/jamaneurol.2013.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V.K., Shen L., Moore J.H., Saykin A.J. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet. 2012;28:323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Shen L., Kim S., Risacher S.L. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: a study of the ADNI cohort. Neuroimage. 2010;53:1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.E., Greenberg S.M. Beta-amyloid, blood vessels, and brain function. Stroke. 2009;40:2601–2606. doi: 10.1161/STROKEAHA.108.536839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traylor M., Tozer D.J., Croall I.D. Genetic variation in PLEKHG1 is associated with white matter hyperintensities (n = 11,226) Neurology. 2019;92:e749–e757. doi: 10.1212/WNL.0000000000006952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch S., Hamamichi S., Goodman J.L. Functional links between Abeta toxicity, endocytic trafficking, and Alzheimer’s disease risk factors in yeast. Science. 2011;334:1241–1245. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaaren B.F., Debette S., Bis J.C. Multiethnic genome-wide association study of cerebral white matter hyperintensities on MRI. Circ. Cardiovasc. Genet. 2015;8:398–409. doi: 10.1161/CIRCGENETICS.114.000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Han D.M., Kang H.W., Ma L.J., Ye J.Y., Xiao Y. [Primary study on glycan structure in pathopoiesis mechanism of recurrent respiratory papillomatosis] Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2008;43:355–359. [PubMed] [Google Scholar]
- Westra H.J., Peters M.J., Esko T. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H., Ecke G.M., Bettin S., Dietrich J., Gertz H.J. Do white matter changes contribute to the subsequent development of dementia in patients with mild cognitive impairment? A longitudinal study. Int. J. Geriatr. Psychiatry. 2000;15:803–812. doi: 10.1002/1099-1166(200009)15:9<803::aid-gps190>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Wu C.C., Mungas D., Petkov C.I. Brain structure and cognition in a community sample of elderly Latinos. Neurology. 2002;59:383–391. doi: 10.1212/wnl.59.3.383. [DOI] [PubMed] [Google Scholar]
- Yang X., Li J., Liu B., Li Y., Jiang T. Impact of PICALM and CLU on hippocampal degeneration. Hum. Brain Mapp. 2016;37:2419–2430. doi: 10.1002/hbm.23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshita M., Fletcher E., Harvey D. Extent and distribution of white matter hyperintensities in normal aging, MCI, and AD. Neurology. 2006;67:2192–2198. doi: 10.1212/01.wnl.0000249119.95747.1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharaki E.I., Kanterakis S., Bryan R.N., Davatzikos C. Measuring brain lesion progression with a supervised tissue classification system. Med. Image Comput. Comput. Assist. Interv. 2008;11:620–627. doi: 10.1007/978-3-540-85988-8_74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.