Abstract
HPV16 causes half of cervical cancers worldwide; for unknown reasons, most infections resolve within two years. Here, we analyze the viral genomes of 5,328 HPV16-positive case-control samples to investigate mutational signatures and the role of human APOBEC3-induced mutations in viral clearance and cervical carcinogenesis. We identify four de novo mutational signatures, one of which matches the COSMIC APOBEC-associated signature 2. The viral genomes of the precancer/cancer cases are less likely to contain within-host somatic HPV16 APOBEC3-induced mutations (Fisher’s exact test, P = 6.2 x 10−14), and have a 30% lower nonsynonymous APOBEC3 mutation burden compared to controls. We replicate the low prevalence of HPV16 APOBEC3-induced mutations in 1,749 additional cases. APOBEC3 mutations also historically contribute to the evolution of HPV16 lineages. We demonstrate that cervical infections with a greater burden of somatic HPV16 APOBEC3-induced mutations are more likely to be benign or subsequently clear, suggesting they may reduce persistence, and thus progression, within the host.
Subject terms: Cervical cancer, Human papilloma virus
The APOBEC mutational signature is prevalent in different tumour types. Here, using HPV16- positive cervical samples, the authors show that the signature is more prevalent in the viral genome of benign or clearing HPV16 infections compared to the viral genomes of the more advanced precancerous lesions or cervical cancer.
Introduction
High-risk human papillomaviruses (HR-HPVs) are small double-stranded DNA viruses that cause cervical cancer1 and a large proportion of other anogenital and oropharyngeal cancers2,3. HPV16 is the most potent of the 12 HR-HPV types; it accounts for >50% of the >500,000 incident cervical cancer cases worldwide annually4–6. It is unknown why HPV16 is more carcinogenic than other HPV types, or why the majority of HPV16 infections “clear” (are either eliminated or controlled) while others persist and lead to cervical precancer and cancer7–9. Within HPV16, genetic variation partly predicts risk of precancer and cancer. For example, the sublineages (A1–A4, B1–B4, C1–C4, D1–D4) of HPV16, defined by genetic variation, have been associated with substantial differences in cervical carcinogenicity10–22, and specific sublineages are linked to adenocarcinomas with an odds ratio (OR) of >10021. There is also much finer genetic variation among circulating HPV16 isolates23,24, which varies by viral gene region and infection outcome, with more variation observed in women with low-grade or benign HPV16 infections than in those with cancers24,25. Most notably, the E7 oncogene lacks nonsynonymous (amino acid changing) variants in cervical cancers from around the world compared with controls, illustrating that rigid E7 conservation is necessary for carcinogenicity24.
While cell-mediated immunity is thought to explain much of HPV clearance, innate immunity may also be important26. Specifically, the expression of human apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3 (APOBEC3) family of cytidine deaminases, APOBEC3A or APOBEC3B (hA3A/B), have been shown to be upregulated following HPV infection and act as a restriction factor, activated by the HPV16 E6 and E7 oncoproteins27–32. This elevated activity of hA3A/B enzymes can mediate mutations in both the host and viral genomes32–34. Large comprehensive human cancer genomic studies have characterized somatic mutations and discovered APOBEC3-mediated mutational signatures among multiple cancer types35–41, particularly enriched in HPV-positive cervical35 and HPV-positive head and neck cancers42–44. A recent study of oral squamous cell carcinomas (OSCCs) determined that the somatic APOBEC3 mutation burden was strongly linked to the total mutation burden in HPV-positive, but not HPV-negative OSCCs44. APOBEC3 mutagenesis is a source of oncogenic driver events35,43–45 (e.g., PIK3CA E545K hotspot mutations in cervical cancers35) and contributes to clonal evolution and intratumor heterogeneity46 (for more details, see refs. 32,47–49). At the same time, APOBEC3 cytidine deaminases have been shown to induce mutations in HPV genomes and act as a restriction factor in the early stages of HPV infection30,50,51.
A targeted sequencing study of 9 HPV16 precancers demonstrated that the HPV16 genome was hyperedited with G > A and C > T changes by human APOBEC3 cytidine deaminases52; subsequently, G > A and C > T hypermutations were verified in two small studies of the E2 gene and long control region (LCR) of the HPV16 genome53,54. A recent study showed that APOBEC3 cytidine deaminase was a driver of HPV mutations at the trinucleotide APOBEC3 target, TpCpN, across 151 HPV16/52/58 whole genomes25, particularly in low-grade lesions. It appears that host–pathogen coevolution has selected for HPV16 genomes with fewer APOBEC3 attackable motifs55. Thus, the remaining motifs are probably necessary or important for the full infectious viral life cycle. There have been no large-scale genomic studies to comprehensively characterize APOBEC3 mutagenesis and other mutational signatures across the HPV16 genome in cervical precancer/cancer cases and controls.
To investigate the viral genetic variation across the HPV16 genome potentially induced by human APOBEC3 cytidine deaminases and other mutational processes (signatures) and evaluate how these variants contribute to infection outcome (i.e., viral clearance or carcinogenesis), we analyze HPV16 whole-genome sequence data from 5328 case–control samples. Our primary analysis includes 3579 HPV16 genomes21,24 from 1265 controls (women with a benign HPV16 infection defined as causing ≤ cervical intraepithelial neoplasia [CIN] grade 1 [CIN1], and/or “clearing”) and 1032 CIN2, 1170 CIN3 precancer, and 112 cancer cases in the prospective NCI-Kaiser Permanente Northern California (PaP) Cohort56. We replicate case findings in 1749 HPV16 genomes24, including HPV16 from 444 CIN3 precancer and cancer cases (i.e., CIN3+) in a cross-sectional U.S. population17,57–59 from the Study to Understand Cervical Cancer Early Endpoints and Determinants (SUCCEED), and 1305 invasive cervical cancers collected internationally by the International Agency for Research on Cancer (IARC)60–63.
Results
Whole-genome sequencing of HPV16
Using a PCR based next-generation sequencing (NGS) assay64, we performed HPV16 whole-genome sequencing of 5328 HPV16-positive cervical precancer/cancer cases and controls (Table 1). The mean number of sequencing reads aligned across the HPV16 genome per sample was 520,127 (standard error 76,629), with a mean of 3696 sequencing reads per gene region.
Table 1.
Summary of HPV16-infected women from three studies by case status.
| Study | Status | No. of women |
|---|---|---|
| NCI-Kaiser PaP | Control | 1265 |
| CIN2 | 1032 | |
| CIN3 | 1170 | |
| Cancer | 112 | |
| SUCCEED | CIN3 | 314 |
| Cancer | 130 | |
| IARC | Cancer | 1305 |
| Total | 5328 |
CIN2 cervical intraepithelial neoplasia (CIN) grade 2, CIN3 CIN grade 3.
Mutational signature analysis identified four signatures
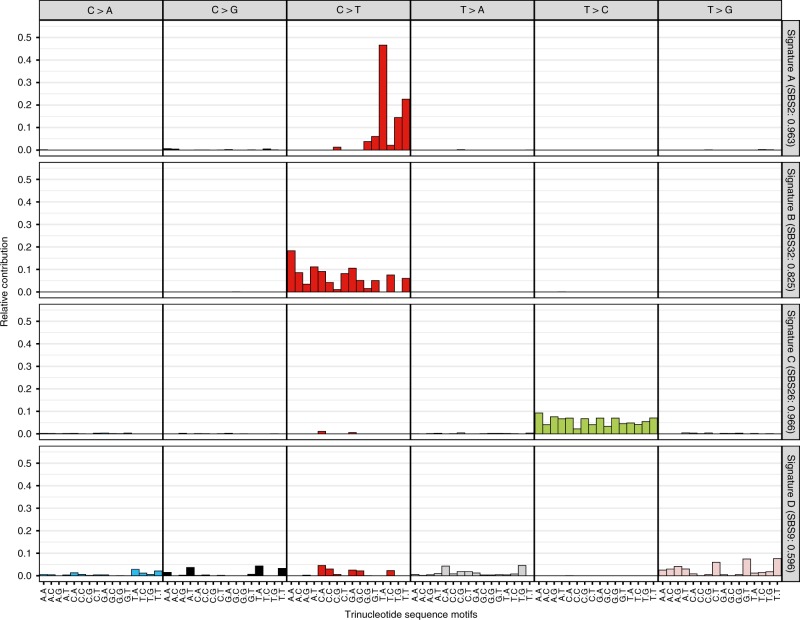
We first characterized the 96 trinucleotide mutation types taking into account the sequence context immediately 5′ and 3′ to each mutated base, and conducted a de novo mutational signature analysis of all variants across 3579 HPV16 genomes. We identified four mutational signatures present in the HPV16 genomes (Fig. 1). Signature A was characterized by C > T mutations enriched at the TCW motif, and it was highly similar to the known COSMIC (the Catalogue of Somatic Mutations in Cancer)65 single-base substitution (SBS) signature 2 (cosine similarity = 0.963), which has been associated with the activity of the APOBEC cytidine deaminases. Signature B was represented by C > T mutations outside the TCW motif (excluding C > T mutations in signature A), with some similarity to COSMIC65 SBS signature 32 (cosine similarity = 0.825), the etiology of this signature is less understood and suggested to be associated with damage to guanine and repair by transcription-coupled nucleotide excision repair. Signature C was characterized by T > C mutations, and highly similar to COSMIC65 SBS signature 26 (cosine similarity = 0.966), which has been associated with defective mismatch repair. The fourth signature D illustrated a relatively flat signature across many different mutation types with little similarity to a known COSMIC65 SBS signature.
Fig. 1. Four de novo mutational signatures identified using all variants across the HPV16 genome in women from the PaP cohort.
The x-axis indicates the 5′ and 3′ nucleotides for each of the top panel substitutions for the three base-pair motifs. The y-axis shows the single-base substitution (SBS) composition of each mutational signature by the 96 trinucleotide sequence motifs. For each identified signature, shown as A–D, the similarity was determined to the known COSMIC SBS signatures (https://cancer.sanger.ac.uk/cosmic/signatures/SBS/). The identified signature letter (A–D) and in parentheses the most similar COSMIC SBS signature number along with the cosine similarity are shown along the right y-axis. Cosine similarity ranges from 0 to 1, with a cosine of 1 indicating a perfect match.
We further evaluated the distribution of the mutations identified by each signature in our case–control samples and examined if they were linked to viral clearance or carcinogenicity with a primary focus on the HPV16 APOBEC3-induced mutations.
HPV16 APOBEC3 mutations are more prominent at a low variant allele fraction
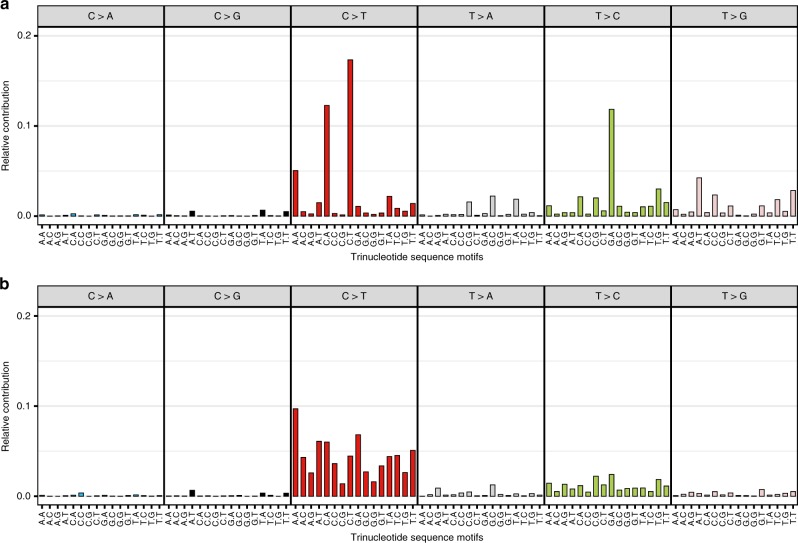
As expected, cytosine-to-thymine (C > T) changes were the most frequent variants observed (48.7% of total mutations; Supplementary Fig. 1). We observed that the distribution of mutation types differed by variant allele fraction (VAF; Fig. 2a, b). VAF refers to the fraction or percent of total viral sequence reads per woman containing the mutation at a given genomic position (see “Methods” for more details). The VAF is expected to be ~1.0 if the variant was present in the virus at acquisition (i.e., constitutive variant in a haploid HPV genome), and in a lower fraction of the reads if the viral mutation occurred de novo during the infection period (i.e., a within-host somatic mutation). We evaluated constitutive HPV16 variants at a high VAF in each woman, defined based on the distribution of mutations as a variant occurring in >60% of the sequence reads at that locus, and within-host HPV16 somatic mutations at a low VAF, defined as occurring in 10–60% of the sequence reads (variants in <10% of the reads were excluded), separately.
Fig. 2. Frequency of the 96 trinucleotide mutation types for variants across the HPV16 genome in women from the PaP cohort.
Illustrated for (a) high variant allele fraction (VAF) and (b) low VAF variants. The x-axis indicates the 5′ and 3′ nucleotides for each of the top panel substitutions for the three base-pair motifs.
The C > T substitutions were most prominent at low VAFs (Fig. 2a, b). The previously established APOBEC3 mutation types39, characterized by a C > T substitution with thymine on its 5′ side and adenine or thymine on its 3′ side (TCW motif [W is A or T]), identified as mutational signature A, and also C > T substitutions outside the TCW motif (signature B), were specifically more prevalent at a low VAF compared with high VAF (APOBEC3 mutations: 10.2% of low VAF, and 4.8% of high VAF; proportion test, P-value < 0.001; C > T outside the TCW motif: 59.8% of low VAF, and 40.7% of high VAF; proportion test, P-value < 0.001). However, APOBEC3-associated39 C > G substitutions at the TCW motif were rare across the HPV16 genome at both low and high VAFs (0.7% of low VAF, and 1.2% of high VAF). Supplementary Fig. 2 illustrates the distribution of APOBEC3-induced mutations across levels of VAF and shows that most mutations were in either very low or very high VAF levels. The within-host viral somatic mutations at low VAF were most pronounced in the controls, while the highest VAF constitutive variants, likely representing variants present in the HPV16 genome at acquisition, were equivalent among cases and controls.
We also observed mutation types characterized by T > C (signature C) and T > G (signature D) mutations across the HPV16 genomes at high VAF (Fig. 2a). T > G mutations are expected to be rare DNA changes66; the reason for their higher frequency in HPV16 genomes is unknown.
Within-host HPV16 APOBEC3 mutations are more frequent in controls
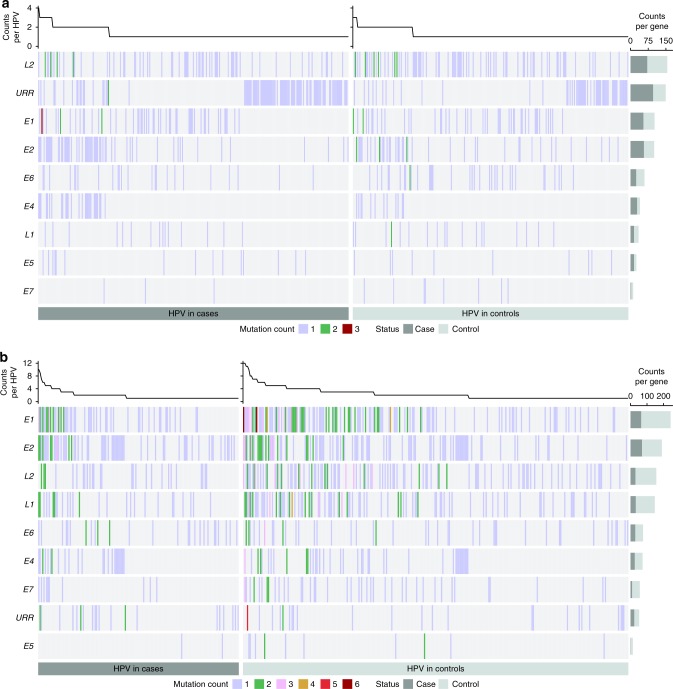
To test if APOBEC3-induced mutations, and the other mutational signature substitutions, were associated with case–control status, we compared the HPV16 APOBEC3-induced mutations (Fig. 3a, b) in the precancer/cancer cases and controls stratified by low/high VAF (see “Methods” for more details). Within-host viral somatic APOBEC3-induced mutations were present in significantly fewer CIN3 + cases (11.9%) compared with controls (23.2%; OR 0.45, 95% CI 0.36–0.56, Fisher’s exact test, P-value = 6.2 × 10−14; Table 2). The results were similar for CIN2 + cases compared with controls (OR 0.48, 95% CI 0.40–0.57, Fisher’s exact test, P-value = 5.8 × 10−16; Supplementary Table 1). In a subset analysis, we compared only the incident cases (N = 333) that developed CIN3 + during the follow-up study period (i.e., after baseline or enrollment) to the controls (i.e., women that cleared their HPV16 or did not progress to CIN2+), and also showed that the incident CIN3+ cases had significantly fewer somatic APOBEC3-induced mutations than controls (OR 0.57, 95% CI 0.41–0.79, Fisher’s exact test, P-value = 0.0003). We further evaluated if somatic APOBEC3-induced mutations were associated with case/control status for each viral gene region. L1 and L2 gene regions had significantly more somatic APOBEC3-induced nonsynonymous mutations in the controls compared with CIN3+ cases (Wald test, P-value = 0.01 and 6.7 × 10−4, respectively; Supplementary Table 2). There was no apparent clustering of mutations in these gene regions by functional domain (Supplementary Fig. 3a, b).
Fig. 3. The number of APOBEC3-induced mutations across the HPV16 genome by gene region in women from the PaP cohort.
The plots show only the cases and controls with one or more APOBEC3-induced mutations at a (a) high variant allele fraction (VAF) and (b) low VAF. Each vertical line represents a sample with at least one APOBEC3-induced mutation, colored by the number of mutations observed, as 1–3 (see legend), per viral gene region. Samples with no APOBEC3-induced mutations are not illustrated; the size of the case and control panels correspond to the number of individuals with at least one APOBEC3-induced mutation. The samples are organized along the x-axis by status (case vs. control). Cases are cervical intraepithelial neoplasia grade 3 and cancer cases (CIN3+). The right y-axis represents viral gene regions with the overall frequency of APOBEC3 mutations summarized, taking into account the sample sizes of the cases/controls and potential APOBEC3-mutable sites, for CIN3+ cases in dark gray and controls in light gray. The top panel histogram summarizes the total APOBEC3-induced mutations for the cases and controls across the HPV16 genomes. URR upstream regulatory region, E6 early gene 6, E7 early gene 7, E1 early gene 1, E2 early gene 2, E4 early gene 4, E5 early gene 5, L2 late gene 2, L1 late gene 1.
Table 2.
Cases and controls with and without APOBEC3-induced mutations by variant allele fraction in the NCI-Kaiser PaP cohort.
| VAF | Status | No APOBEC3 mutation | APOBEC3 mutation | % | OR | 95% CI | P-valuea |
|---|---|---|---|---|---|---|---|
| Lowb | CIN3+ | 1129 | 153 | 11.9% | 0.45 | (0.36–0.56) | 6.2 × 10−14 |
| Control | 971 | 294 | 23.2% | Ref. | |||
| Highc | CIN3 + | 997 | 285 | 22.2% | 1.14 | (0.94–1.39) | 0.17 |
| Control | 1012 | 253 | 20.0% | Ref. |
VAF variant allele fraction, CIN3+ cervical intraepithelial neoplasia grade 3 and cancer cases, OR odds ratio, CI confidence intervals, Ref. referent group.
aFisher’s exact test, two-sided, comparing the number of women with at least one APOBEC3-induced mutation and those without APOBEC3-induced mutations in the CIN3+ cases to controls.
bLow VAF is defined as VAF >10% and < = 60%.
cHigh VAF is defined as VAF >60%.
In contrast, there was no significant difference between the high VAF, constitutive HPV16 APOBEC3-induced variants in the CIN3+ cases compared with controls (OR 1.14, 95% CI 0.94–1.39, Fisher’s exact test, P-value = 0.17; Table 2). By gene region, there were more constitutive APOBEC3 nonsynonymous variants in the cases compared with controls in E4 (Mutation burden ratio 7.99, 95% CI 1.85–34.4; Wald test, P-value = 0.01; Supplementary Table 2).
We replicated the prevalence of APOBEC3-induced mutations throughout the HPV16 genome in precancer/cancer cases in two independent case study populations at both low and high VAFs (Supplementary Table 3). In particular, 12.4% and 12.5% of precancer/cancer cases had HPV16 somatic APOBEC3-induced mutations in SUCCEED and IARC, respectively, similar to the 11.9% observed in the PaP cohort cases and significantly fewer than the PaP controls (23.2%; OR 0.47, Fisher’s exact test, P-value = 5.1 × 10−7 and 1.1 × 10−12, respectively). The frequency of HPV16 APOBEC3-induced variants at high VAF among the SUCCEED and IARC CIN3 + cases was similar to the PaP CIN3 + cases, and slightly higher than the PaP controls (Supplementary Table 3).
Within-host viral somatic C > T substitutions outside the TCW motif (signature B) and the T > C substitutions (signature C) were also present in significantly fewer CIN3 + cases (36.1% and 18.5%, respectively) compared with controls (43.2% and 32.9%, respectively; Fisher’s exact test P-value < 0.001) in the PaP cohort (Supplementary Table 4). However, the frequency of these specific substitutions did not replicate in the precancer/cancer cases in our two independent case study populations (SUCCEED and IARC), so we did not evaluate their relationship to case–control status further. The substitutions characterized by mutational signature D were not associated with case–control status at low VAF (Supplementary Table 4).
Characteristics of HPV16 mutations induced by APOBEC3 enzymes
We first estimated all possible DNA changes and resultant amino acid changes across the HPV16 reference genome. For the HPV16 A1 sublineage reference genome (see Methods), there were a total of 263 APOBEC3 targetable sites, and fewer targetable sites for the more carcinogenic HPV16 D2/D3 sublineage10–22 genomes, 247/246 APOBEC3 targetable sites (Supplementary Table 5). Although HPV16 A1/A2 sublineages had only 14–17 more APOBEC3 targetable sites than the D2/D3 sublineages, the controls with A1/A2 sublineages had more APOBEC3 mutations than D2/D3 (31.2% vs. 15.8%; P = 0.04). Of the possible HPV16 APOBEC3 targetable sites (A1 reference genome), 96.2% of these APOBEC3 substitutions would result in a nonsynonymous change compared with 77.2% of non-APOBEC3 sites (proportion test, P-value < 0.001). Consequently, 95.2% of the APOBEC3-induced mutations we observed were nonsynonymous, which was significantly higher than the proportion observed for the non-APOBEC3 mutations, 71.2% (proportion test, P-value < 0.001; Supplementary Table 6).
For the HPV16 infections with at least one APOBEC3-induced mutation, we compared the APOBEC3-induced mutation burden per virus in cases vs. controls and between nonsynonymous vs. synonymous mutations, adjusting for the number of possible HPV16 APOBEC3-mutable sites (see “Methods” for more details). For within-host viral somatic mutations, CIN3 + cases had a significantly lower APOBEC3-induced mutation burden compared with controls (mutation burden ratio 0.71, 95% CI 0.61–0.82; Wald test, P-value = 6.42 × 10−6) for all mutations and stratified by both nonsynonymous (mutation burden ratio 0.71; Wald test, P-value = 1.2 × 10−5) and synonymous mutations (ratio 0.68; Wald test, P-value = 0.28) (Table 3; Supplementary Table 7). There was no evidence of selection against nonsynonymous relative to synonymous mutations (ratio of nonsynonymous-to-synonymous mutation rate 1.0; Wald test, P-value = 0.99; Table 3). Here, the nonsynonymous and synonymous mutation rates were evaluated based on the number expected relative to the number of APOBEC3 targetable sites that could result in a nonsynonymous or synonymous mutation. Most of these somatic HPV16 APOBEC3-induced mutations at low VAF were rare in the population with a minor allele frequency (MAF) < 0.01, and rare somatic APOBEC3 mutations were significantly more frequent in the controls compared to the cases (proportion test, P-value = 1.5 × 10−4).
Table 3.
Burden of APOBEC3-induced mutations in cases and controls from the NCI-Kaiser PaP cohort. Mutations are compared in CIN3+ cases vs. controls and for nonsynonymous (nonsyn) vs. synonymous (syn) mutations by variant allele fraction.
| VAF | Parameter | Interpretation | Mutation burdena | 95% CI | P-valued |
|---|---|---|---|---|---|
| Lowb | rsyn | Enrichment of synonymous mutations in cases vs. controls | 0.68 | (0.34–1.36) | 0.28 |
| rnonsyn | Enrichment of nonsynonymous mutations in cases vs. controls | 0.71 | (0.61–0.83) | 1.2 × 10−5 | |
| W | Selection of nonsynonymous mutations vs. synonymous mutations in controls | 1.00 | (0.70–1.44) | 0.99 | |
| Highc | rsyn | Enrichment of synonymous mutations in cases vs. controls | 1.27 | (0.73–2.21) | 0.40 |
| rnonsyn | Enrichment of nonsynonymous mutations in cases vs. controls | 0.82 | (0.68–1.00) | 0.05 | |
| W | Selection of nonsynonymous mutations vs. synonymous mutations in controls | 0.55 | (0.35–0.85) | 7.9 × 10−3 |
VAF variant allele fraction, CIN3+ cervical intraepithelial neoplasia grade 3 and cancer cases, CI confidence intervals.
aMutation burden ratio of APOBEC3-induced mutations was calculated using a Poisson regression model to compare the mutation burden or enrichment of APOBEC3-induced mutations per virus between cases and controls for nonsynonymous and synonymous mutations (r); selection of nonsynonymous mutations in the controls was estimated adjusting for the number of cases and controls and the potential APOBEC3-mutable bases that result in nonsynonymous and synonymous mutations (w).
bLow VAF is defined as VAF >10% and < = 60%.
cHigh VAF is defined as VAF >60%.
dP-values are generated by the Wald test of a Poisson regression model.
In contrast, at high VAF, there was no significant difference in the mutation burden of APOBEC3-induced nonsynonymous and synonymous mutations in cases and controls, and there was evidence of negative selection against nonsynonymous mutations relative to synonymous mutations (mutation burden ratio 0.55; Wald test, P-value = 7.9 × 10−3; Table 3).
Among the possible APOBEC3 targetable bases on each strand, the percent mutated on the positive strand (5.8/kb) was comparable with that estimated on the negative strand (6.2/kb, proportion test, P-value = 0.61; Supplementary Table 8). In addition, among the APOBEC3A (YTCA, Y is a pyrimidine base) and APOBEC3B (RTCA, R is a purine base) possible mutable bases67, we compared the rate of having at least one APOEBC3A or APOBEC3B mutation and found no significant difference (0.35 vs. 0.34/kb, proportion test, P-value = 0.86; Supplementary Table 9). When further separating APOBEC3-induced mutations by high/low VAF, there was no significant difference between the APOBEC3 mutation rates on the positive or negative strand, or for APOBEC3A or APOBEC3B mutations (Supplementary Tables 8 and 9).
APOBEC mutations contributed to the evolution of HPV16 lineages
We determined that APOBEC3 editing may have contributed to the evolution of HPV16 lineages using 239 HPV16 non-A1/2 sublineage sequences. We specifically evaluated the HPV16 nucleotide positions that are known to “define” each of the HPV16 main lineages68 (i.e., lineage/sublineage diagnostic SNPs; each SNP in the lineage haplotypes) compared with the derived ancestral sequence for each main lineage and sublineage at each node of the phylogenetic tree (Supplementary Fig. 4). We determined that there was a range of 6–41% of the lineage-defining SNPs for each of the different sublineages potentially induced by APOBEC3 (Supplementary Fig. 4). The D2/D3 sublineages, which are known to be the most carcinogenic of the HPV16 sublineages21, had the greatest number of lineage-defining SNPs potentially induced by APOBEC3 (35 and 41%, respectively).
Discussion
We report the largest HPV16 whole-genome sequencing study to date evaluating viral genome mutational signatures and identify viral APOBEC3 mutations likely induced during a woman’s infection linked to benign or clearing infections. We24 and others23 have previously shown that there is high genetic diversity among HPV16 isolates circulating in the population; these studies were focused on viral SNPs presumed to be “inherited” variants present at HPV acquisition and detected in all or nearly all viral sequence reads at a given locus (high VAF). In contrast, here we use deep NGS to more finely evaluate “acquired” somatic mutations presumed to be induced recently during a woman’s infection and detected in a minority of sequence reads (low VAF, but with sufficient read depth >10×). We have discovered additional somatic viral genetic diversity that is likely driven by APOBEC3 activity and associated with benign infections or subsequent viral clearance in our large prospective cohort. Our data suggest that these APOBEC3-induced mutations may constrain the viability, and by extension the oncogenic potential, of HPV16. We further determined that mutations induced by APOBEC3 cytidine deaminases contributed to HPV16 genetic diversity that shaped viral evolution of the important HPV16 lineages.
The combination of our large study population and deep NGS technology has revealed that HPV16 genomes, which have generally been considered stable during a persistent infection, can accumulate somatic mutations during infection, presenting in a fraction of the viruses within a host driven by APOBEC3 activity and other mutational processes. These somatic mutations, including APOBEC3-induced mutations, likely result in clearance or a reduced ability of the virus to persist. We observed four mutational signatures across HPV16 genomes, and the somatic substitutions related to the three main mutational signatures (A, B, and C) were all more prevalent in the controls compared with the precancer/cancer cases. However, the frequency of somatic substitutions related to signatures B and C did not replicate in two additional case populations; further follow-up is needed to determine the etiology and relevance of these substitutions. The APOBEC3-induced mutations (signature A) were enriched in the controls compared with cases in all three of our precancer/cancer case populations. This suggests that APOBEC3 is primarily inducing mutations during a woman’s infection when HPV16 is replicating, and ssDNA is exposed and targetable, during a productive infection69 in benign or low-grade (<CIN2) lesions. This APOBEC3 mutagenesis within the host prior to HPV clearance and transmission likely additionally contributes to the high diversity of HPV16 in the population previously reported23,24, and to viral evolution25,55,70.
There was no disparity between the mutation burden of nonsynonymous (i.e., missense or nonsense) and synonymous somatic mutations in controls, suggesting that these mutations are likely recent somatic mutations arising during a woman’s infection which would not have had time to be selectively removed from the viral population (i.e., ineffective purifying selection). These mutations were also most often rare in our population (i.e., observed in <1% of women) or singletons (i.e., observed in only one woman among 3500+), further suggesting that most of them arose recently within the host. It is possible that a minority of the rare low VAF variants could be due to PCR error or artifacts, although given our stringent quality control including the requirement that all variants be present in >10 sequence reads, our low assay error rate, and the specific enrichment of APOBEC3 signature mutations in controls at a low VAF instead of random mutations across all samples (as would be expected from errors), this is likely minimal. We observed a significantly higher proportion of viral nonsynonymous APOBEC3-induced mutations compared with that for other non-APOBEC3 mutations. This is consistent with the TpC dinucleotide depletion at the third codon position observed in the viral open-reading frame55,70, which would result in our observed viral APOBEC3-induced mutations primarily occurring at the first and second codon positions and thus causing the enrichment of nonsynonymous changes. Given that 95% of the APOBEC3-induced mutations were nonsynonymous and more frequent in the controls, we presume these mutations were deleterious to viral persistence and thus constrained the viability of HPV16. Alternatively, it is also possible that these APOBEC3-induced mutations are a biomarker of an innate immune response to the virus.
Earlier targeted HPV16 sequencing studies52–54 and an HPV whole-genome sequencing study of 151 HPV16, HPV52, or HPV58 samples25 also identify APOBEC3-induced mutations in HPV genomes. Our within-host viral somatic mutations are consistent with the study by Hirose et al.25, suggesting that the high levels of HPV genomic variation they observed, particularly in the low-grade CIN1 lesions, were likely the result of accumulating somatic mutations during infection. For a more direct comparison of our data with the HPV16 genome data from Hirose et al.25, we downloaded their 45 HPV16 genomes (GenBank accession numbers: LC368952 to LC368996) and created a 96 trinucleotide mutation-type plot after exclusion of the common evolutionary-derived HPV16 lineage-defining substitutions68,71, since inclusion of lineage-defining substitutions would skew the distribution for evaluations of recent or within-host mutations. The resulting distribution of mutation types looks similar to ours (Supplementary Fig. 5), including specific APOBEC3-associated variants and T > C and T > G substitutions. Although, mutational signature extraction was not possible for these 45 genomes25 due to the overall small number of variants.
Interestingly, we did not detect leading/lagging or transcribed DNA strand biases or APOBEC3-associated C > G changes at TCW motifs, which correspond to the APOBEC3 COSMIC65 SBS signature 13, as observed in human somatic genomes related to APOBEC mutagenesis36,39. Our observations are consistent with previous HPV data25,52 and suggest a difference between HPV and human genome APOBEC3 mutagenesis. It is possible that the antiviral APOBEC3 response to HPV infection is separate or slightly different from the role of APOBEC3 in host somatic genome mutagenesis.
The distribution of APOBEC3-induced mutations across the viral genome was different between the controls and precancer/cancer cases, suggesting that sites in specific regions of the viral genome may be “hit” more often by APOBEC3, or mutations at specific sites may have more deleterious effects to the virus if absent from the cases, such as mutations in L1 and L2. Alternatively, since APOBEC3 mutations occur when the viral DNA is single stranded, the mutations observed in the L1 and L2 gene regions more frequently in the controls may reflect that these regions of the viral genome are more likely to be transcribed and single stranded in the controls with a productive infection.
As noted, HPVs have evolved to limit the number of TpC dinucleotides in their genomes to avoid restriction55,70, yet even with a limited number of APOBEC3 targetable sites, somatic APOBEC3-induced mutations were still observed and enriched in benign infections, suggesting an antiviral effect. We note that the majority of viruses did not have APOBEC3-induced mutations. We may be underestimating the level of APOBEC3-induced mutations due to our stringent quality control and low VAF cut point of 10%, instead of >0.5%25, which would not detect the lowest VAF-induced mutations. It is also possible that some of the viruses with somatic APOBEC3-induced mutations were rapidly cleared, if they were less viable, and thus not part of our study.
In contrast, the high VAF constitutive APOBEC3 variants, which included a lower burden of nonsynonymous variants, were likely present in the HPV16 population for a longer period of time and/or present at viral acquisition and reflect the natural variants circulating in the population. Purifying selection would have prevented a disproportionate number of nonsynonymous variants from reaching high frequencies in HPV16 populations, since they are more likely than synonymous variants to be disadvantageous (e.g., leading to a reduced ability to persist in the host), as we observed. These high VAF APOBEC3 variants were overall equivalent in cases and controls, and possibly neutral with respect to carcinogenesis. However, we did observe that there were specifically more high VAF E4 variants in the cases, suggesting that these genetic variants could be slightly advantageous to viral persistence.
APOBEC3 upregulation occurs throughout disease progression to inhibit the HPV infection30. However, we still observed absent or reduced somatic APOBEC3-induced mutations in the precancer/cancer cases, which represent infections that have been successfully persisting for years, suggesting that these viruses may be evading host restriction by APOBEC3 and/or the more homogeneous viral genomes in the cases reflects outgrowth of a clone with a selective growth advantage. The somatic viral APOBEC3 mutations induced within the host that are not deleterious to the virus may instead aid in evasion of the host adaptive immune response by altering viral antigens, and this viral clone would then be selected for in that host. It is also possible that the precancer/cancer HPV16 viruses may be partially evading restriction through integration of HPV DNA into the host genome, where a portion of the viral genome and episomal HPV genomes are lost69,72 and therefore there is less viral DNA present in these infections to be targeted by APOBEC3 enzymes. In addition, increased viral DNA methylation of the cases73,74 may partially protect the viral genome from mutation by APOBEC3 enzymes. If APOBEC3 activity is upregulated and failed to clear HPV in the cases with advanced lesions, this may be contributing to the off-target host somatic mutations observed in cervical and other HPV-associated cancers. The difference in APOBEC3-induced mutations in the cases and controls may also reflect differences in activation and/or regulation of the innate immune response that leads to APOBEC3 expression and downstream viral mutations32, partially related to the activity of IFN-α75,76, inflammation and NF-κB signaling77,78, TP5331,79, or human genetic variation44,80,81.
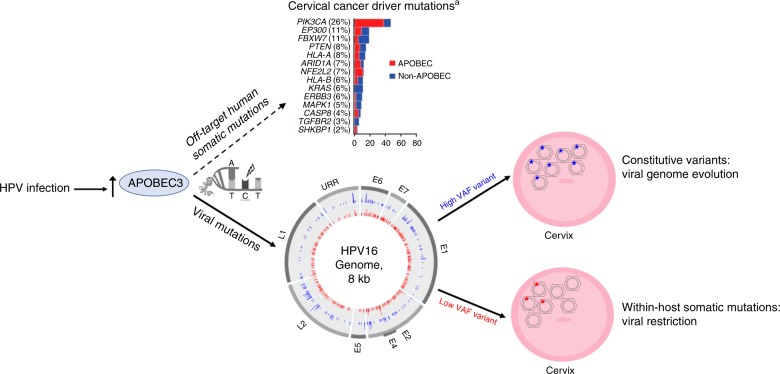
In summary (Fig. 4), we determined that APOBEC3 cytidine deaminases induce somatic mutations across the HPV16 genome, these mutations have contributed to the evolution of important viral lineages, and infections with somatic viral mutations induced during a woman’s infection were more likely to become benign infections or infections that subsequently cleared.
Fig. 4. Summary of the effects of mutations induced by the activity of APOBEC3.
Viral APOBEC3-induced mutations are illustrated in the circle plot by viral gene region in the inner ring for low variant allele fraction (VAF) somatic mutations in red, and the outer ring for high VAF constitutive variants in blue for all individuals in the PaP cohort (mutations in both cases and controls are illustrated). Modified from the Cancer Genome Atlas Research Network35.
Methods
Study populations
The cases and controls for the large discovery phase of our study were chosen from the Kaiser Permanente Northern California (KPNC)-NCI HPV Persistence and Progression (PaP) cohort21. This study population has been previously described56. The HPV Persistence and Progression (PaP) Cohort is a repository of residual cervical specimens stored in specimen transport medium (STM; Qiagen, Valencia, CA), from women who underwent cervical cancer screening from January 2007 to January 2011 at Kaiser Permanente Northern California (KPNC). Women could opt out of having their residual cervical specimens retained; only 8% of women with collected specimens opted out from having their specimen banked and tested. De-identified demographic and clinical information as well as all HPV and cytology test results and cervical histopathology were obtained on the cohort from electronic health records.
This cross-sectional study included 3579 exfoliated cervical cell specimens collected at enrollment previously determined to contain HPV16 DNA56, including 112 cancers, 1170 CIN3, 1032 CIN2, and 1265 controls (<CIN2) in follow-up through 2015. The precancer (CIN2 and CIN3) and cancer cases were diagnosed at baseline (i.e., at enrollment; prevalent cases) or during the study follow-up period after the baseline specimens were collected (i.e., incident cases). The controls were defined as women having enrollment specimens with HPV16 DNA, and no histologic evidence of equivocal precancer or worse (CIN2+) during the follow-up study period according to the coded data obtained from electronic health records. Therefore, controls were the HPV16-positive women that either cleared their infections or had not progressed to CIN2+ during the follow-up study period. Women are followed as long as possible, and only censored if they received treatment for a CIN2+ lesion, or until the last documented follow-up cytology or histology. The study protocol was reviewed and approved yearly by Kaiser Permanente and the National Cancer Institute Institutional Review Boards.
To confirm the findings from the PaP Cohort, we evaluated HPV16-positive women from the Study to Understand Cervical Cancer Early Endpoints and Determinants (SUCCEED). The details of the study design and specimen collection were previously described57–59. Briefly, a total of 2004 women were enrolled into SUCCEED between November 2003 and October 2009. We recruited women that were referred to colposcopy or treatment at the University of Oklahoma Dysplasia Clinic based at the University of Oklahoma Health Sciences Centre (OUHSC), with a recent abnormal Pap smear diagnosis or a biopsy diagnosis of CIN/cancer. Here, we included all CIN3+ exfoliated cervical cell specimens previously found to contain HPV16 DNA, including 444 women: 314 CIN3 and 130 cancer cases. Written informed consent was obtained from all women enrolled in the study, and Institutional Review Board approval was provided by OUHSC and the US National Cancer Institute.
In total, 645 additional HPV16-positive cervical cell or tissue (frozen biopsy or formalin-fixed paraffin-embedded [FFPE]) specimens from cervical cancer cases were studied to assess the worldwide generalizability of our main finding, from the biobank at IARC. These samples were part of the IARC-coordinated cervical cancer case series, cervical cancer case–control studies and population-based HPV prevalence surveys from 39 countries worldwide60–63. Both local and IARC ethical committees approved all studies. We sequenced all HPV16-positive histologically confirmed cervical cancers with adequate DNA left in the IARC biobank.
HPV16 detection and DNA isolation
DNA was extracted from the banked STM specimens as previously described82. Typing methods varied for different subsets of the cohort, many of the enrollment PaP samples were tested by the Burk laboratory (Bronx, NY) using MY09/M11 L1 degenerate primer PCR (MY09/11 PCR) and type-specific dot-blot hybridization methods82,83. Other specimens were tested with either the Linear Array® HPV Genotyping System (Roche Molecular Diagnostics, Pleasanton, CA) or typed by BD using Onclarity (BD, Sparks, MD).
Details of DNA isolation and HPV detection have been previously described57,84. Briefly, DNA was isolated from 1 mL aliquots of PreservCyt-fixed cells using the QIAamp DNA Blood Mini Kit (Qiagen) following a rinse in Hanks’ balanced salt solution (HBSS). The Linear Array® HPV Genotyping System (Roche Molecular Diagnostics) was used to detect HPV genotypes. Hybridization of PCR products to linear arrays and subsequent signal detection were performed using the Auto-LiPA automated staining system (Innogenetics N.V., Belgium). Hybridization to both β-globin probes was required to report genotyping results. A hybridization signal was called “positive” when an unambiguous, continuous band was observed on the array.
DNA was extracted from frozen biopsy specimens, cervical cells, or FFPE at IARC, as previously described85. Samples were genotyped for 37 HPV types using a GP5+/6+-based PCR system86 in one centralized laboratory (Department of Molecular Pathology, Vrije University, Amsterdam, The Netherlands).
Ion Torrent library preparation and sequencing
We used a custom Thermo Fisher Ion Torrent AmpliSeq HPV16 panel approach to amplify the entire 7906 bp HPV16 genome, as previously described64. In brief, the next-generation sequencing (NGS) assay used the Thermo Fisher Life Sciences’ Ion Torrent S5 and a custom HPV16 Ion Ampliseq panel of 47 multiplexed primer sets. Custom overlapping degenerate primers were designed to cover the entire viral genomes for all HPV16 variant lineages. After amplification, an Ion Torrent adapter-ligated library was generated following the manufacturer’s Ion AmpliSeq Library Preparation kit 2.0-96LV protocol with slight modifications (Life Technologies, Part #4480441). Raw sequencing reads were quality and adaptor trimmed using the Torrent Suite™ Software and aligned to the HPV16 reference sequence (7906 bp, NCBI accession number NC_001526) using the Torrent Mapping Alignment Program v5.0.13. SNP calls were made using the Torrent Variant Caller v.5.0.3, and variants were annotated with HPV gene/region using snpEff v.3.6c87. Pipeline settings and parameters can be found at https://github.com/cgrlab/cgrHPV16.
HPV16 variant lineage classification
HPV16 variant lineage assignment was based on the maximum likelihood (ML) tree topology constructed using RAxML MPI v7.2.8.2788 that included 16 HPV16 European and non-European variant lineage reference sequences. We excluded samples with overall poor coverage, per individual nucleotide site per sample with low reads (<5), as previously described24.
Statistical analyses
To identify the mutational processes generating the HPV variants, we carried out a de novo mutational signature analysis. There are a total of 12 possible single-nucleotide variants (SNV). The SNVs on the complementary DNA strands are considered the same, and we use pyrimidines (C and T) to annotate the SNV. Therefore, we have the following six basic types of SNVs: C > A, C > G, C > T, G > A, G > C, and G > T. We further considered the adjacent nucleotides in both 5′ and 3′ directions around the SNV as a three base-pair motif and obtained 96 (4 × 6 × 4) mutation types. We calculated the frequency of variants belonging to 96 mutation types for 3579 HPV16 genomes in a 96 × 3579 mutational catalog matrix. The mutational catalog matrix is regarded as a combination of mutational signatures induced by multiple mutational processes. To extract de novo mutational signatures, we applied the non-negative matrix factorization89 and compared the similarity of resulting mutational signatures with the COSMIC mutational signatures v3 measured by the cosine similarity.
We calculated the frequency of 96 mutation types, based on a three base-pair motif, across the HPV16 genome. Among these mutation types, APOBEC3-induced mutations are identified as C > T or C > G mutations specifically at the TCW motif (W is A or T), for which only T allowed in the 5′ end and G and C are excluded in the 3′ end40. This definition of APOBEC3-induced mutations has been well established and is more stringent than the motif defined (C > T mutations at motif YCN with Y a C or T and N being any nucleotide) by Vartanian et al.52.
Both rare (minor allele frequency (MAF) < 0.01) and common (MAF > = 0.01) APOBEC3-induced mutations were examined, and variants occurring in <10% of the sequence reads and the lineage-diagnostic sites68 were excluded. For each APOBEC3-induced mutation, we estimated the variant allele frequency (VAF) for HPV16 sequence reads per woman. Note, the difference between MAF and VAF: MAF quantifies the frequency of an APOBEC3-induced mutation across all samples in a given study or among women in a population (regardless of case and control status), while VAF measures the frequency of sequence reads containing the APOBEC3-induced mutation among all reads covering a specific APOBEC3 motif within a sample or per woman. Hence, the VAF reflects the percentage of HPV viruses, or sequence reads, with the specific APOBEC3-induced mutation in a sample. The VAF is expected to be ~1.0, if the APOBEC3-induced mutation was present in the virus at acquisition and being replicated subsequently; in contrast, the VAF would be much lower if the APOBEC3-induced mutation occurs de novo during the infection period, and the virus with this somatic mutation has not become dominant in the sample.
We defined “high VAF” as a variant occurring in >60% of the sequence reads, which we have previously published performs well for calling the predominant HPV variant for a haploid HPV genome24. To evaluate lower level within-host somatic changes, we defined “low VAF” as variants occurring in 10% to < = 60% of sequence reads. To minimize false positive mutation calls, we used a VAF lower cut point of 10% and required at least ten sequence reads for each variant call.
To count the number of APOBEC3 targetable sites, we first counted the TCW motifs across the HPV16 reference A1 genome, and for each main HPV16 sublineage. Since there are three possible changes at each nucleotide position (C > A, C > T and C > G), APOBEC3 targetable sites for the C > T and C > G changes only at TCW motif were counted as two-thirds of the number of TCW motifs.
To examine the relative contribution of APOBEC3A- and APOBEC3B-induced mutations across the HPV genome, we compared the proportion of APOBEC3A mutations and APOBEC3B mutations separately by their specific motifs. APOBEC3A and APOBEC3B specific motifs have been reported as distinguishable in the yeast genome as YTCA (for APOBEC3A) and RTCA (for APOBEC3B) (where Y is a pyrimidine base and R is a purine base)67.
We examined the association between the presence or absence of mutations with case and control status for all samples.
Logistic regression was used to obtain the odds ratio (OR) and 95% confidence intervals (CI) for precancer/cancer risk for the specified exposure groups using the controls (i.e., women with HPV16 and <CIN2) as the referent group. A chi-squared test was used to compare the distribution of women having an APOBEC3-induced mutation (per individual, coded as “yes” at least one APOBEC3 mutation or “no” APOBEC3 mutations) among HPV16 sublineages.
For a subset of samples with at least one APOBEC3-induced mutation, we further compared the mutation burden of APOBEC3-induced mutations per virus between nonsynonymous and synonymous variants and between cases and controls, which adjusts the sample size of the cases and controls and the potential mutable bases of APOBEC3-induced nonsynonymous and synonymous mutations, respectively. Let Yijk represent the number of synonymous (denoted by j = 1) or nonsynonymous (j = 2) APOBEC3-induced mutations in cases (i = 2) and controls (i = 1) for each type of APOBEC3-induced mutation (k = 1, 2…, 8). The Yijk’s are modeled by a Poisson distribution with expected count E(Yijk): (1) with the number of potential mutable bases and t the mutation burden for synonymous mutations in controls; (2) with the mutation burden ratio of nonsynonymous and synonymous mutations in controls, where and and similar notations hold for E(Y11) and N11; (3) E(Y21k) = N21k × rsyn with the enrichment of synonymous mutations in cases compared to controls, where and ; and (4) E(Y22k) = N22k × t × w rnonsyn with mutation burden ratio or the enrichment of nonsynonymous mutations in cases compared to controls, where and and similar notations hold for E(Y12) and N12. w is essentially the dN/dS ratio measuring the selection of nonsynonymous mutations compared to synonymous mutations in controls; if w = 1, it suggests that the mutations are neutral, while w < 1 suggests that nonsynonymous mutations are under negative or purifying selection. A Poisson regression model was fit to obtain the maximum likelihood estimation (MLE) of t, w, rsyn, and rnonsyn.
Statistical analyses were performed with R version 3.5.1; all statistical tests were two-sided.
To determine if APOBEC3-induced mutations contributed to the evolution of HPV16 lineages, we conducted the following analyses: (1) ancestral HPV16 sequences and our current day HPV16 genome sequences were used to create a phylogenetic tree that represents HPV16 ancestral states (Supplementary Fig. 4). Ancestral HPV16 sequences included nine HPV16 lineage/sublineages sequences (All, A, A1, A4, B1, C1, D, D2, and D3) that were inferred using the Maximum Likelihood method90 under the Tamura-Nei model91. The initial tree was inferred using a pre-computed tree file. The rates among sites were treated as being uniform among sites (Uniform rates option). The analysis included 63 nucleotide reference sequences from R.D.B. All positions containing gaps and missing data were eliminated. There were a total of 7697 HPV16 genome positions in the final data set. We also utilized 239 current day HPV16 sequences that represented A1, A4, B1, C1, D2, and D3 sublineages. All current day HPV16 sequences were controls from the PaP cohort. Evolutionary analyses were conducted in MEGA792. (2) For non-A1 lineages, we selected common HPV16 variant positions that are known lineage-defining positions. For the A1 sublineage, which is the reference sublineage (i.e., there are no lineage-defining positions), we evaluated all common variants (MAF > 1%) occuring within the A1 sublineage viruses. (3) We aligned each ancestral sequence to an ancestral sequence from the previous node in the phylogenetic tree. For example, ancestral A1 was compared with the ancestral A sequence, and ancestral D2 was compared with the ancestral D sequence. Then we calculated the percentage of mutations that were potentially induced by APOBEC3 among lineage-defining positions of that particular lineage or sublineage. (4) When comparing current day HPV16 sequences to the ancestral sequences, we looped over all available current day sequences of that particular sublineage and calculated the average percentage of APOBEC3-induced mutation among lineage-defining positions.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This study was funded by the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does neither necessarily reflect the views or policies of the Department of Health and Human Services nor mention trade names, commercial products, or organizations imply endorsement by the U.S. Government. Work at IARC was supported by a grant from the Institut National du Cancer (INCa), France (SHSESP 16-006). Work by R.D.B. was supported in part by the National Cancer Institute (CA78527) and the Einstein Cancer Research Center (P30CA013330) from the National Cancer Institute (to R.D.B.). C.W.N. was supported by a Gerstner Scholars Fellowship from the Gerstner Family Foundation at the American Museum of Natural History.
Author contributions
Study conceptualization and supervision were carried out by B.Z. and L.M. Sample collection, resources, and/or clinical characterization were performed by M.S., G.C., N.W., T.R.-B., T.L., S.F., P.E.C., J.W. and R.Z. HPV sequencing and data curation were performed by M.Y., M.C., J.F.B., S.B., M.K.S., R.D.B., L.B., D.R. and M.D. Sequence bioinformatics and assessment were performed by L.M., M.Y., M.P. and C.W.N. Formal statistical analyses were performed by B.Z., Y.X., D.L., K.Y., and L.S. The paper was drafted by L.M., B.Z. and Y.X., and reviewed by all co-authors.
Data availability
The HPV sequencing data have been deposited in the Genbank database under the accession codes MG847621-MG850835. All the other data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding author upon reasonable request. A reporting summary for this article is available as a Supplementary Information file.
Code availability
The software and algorithms used for our analyses are specified in the corresponding methods section. Pipeline settings and parameters used to call HPV16 variants can be found at https://github.com/cgrlab/cgrHPV16.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Youri Pavlov and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Bin Zhu, Yanzi Xiao.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-14730-1.
References
- 1.Walboomers JMM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 2.Ndiaye C, et al. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: a systematic review and meta-analysis. Lancet Oncol. 2014;15:1319–1331. doi: 10.1016/S1470-2045(14)70471-1. [DOI] [PubMed] [Google Scholar]
- 3.Hartwig S, et al. Estimation of the epidemiological burden of HPV-related anogenital cancers, precancerous lesions, and genital warts in women and men in Europe: potential additional benefit of a nine-valent second generation HPV vaccine compared to first generation HPV vaccines. Papillomavirus Res. 2015;1:90–100. doi: 10.1016/j.pvr.2015.06.003. [DOI] [Google Scholar]
- 4.Global Burden of Disease Cancer Collaboration. The global burden of cancer 2013. JAMA Oncol. 2015;1:505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Sanjose S, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 6.Guan P, et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J. Cancer. 2012;131:2349–2359. doi: 10.1002/ijc.27485. [DOI] [PubMed] [Google Scholar]
- 7.McCredie M, et al. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol. 2008;9:425–434. doi: 10.1016/S1470-2045(08)70103-7. [DOI] [PubMed] [Google Scholar]
- 8.Kjær SK, Frederiksen K, Munk C, Iftner T. Long-term absolute risk of cervical intraepithelial neoplasia grade 3 or worse following human papillomavirus infection: role of persistence. J. Natl Cancer Inst. 2010;102:1478–1488. doi: 10.1093/jnci/djq356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiffman M, et al. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Prim. 2016;2:16086. doi: 10.1038/nrdp.2016.86. [DOI] [PubMed] [Google Scholar]
- 10.Hildesheim A, et al. Human papillomavirus type 16 variants and risk of cervical cancer. J. Natl Cancer Inst. 2001;93:315–318. doi: 10.1093/jnci/93.4.315. [DOI] [PubMed] [Google Scholar]
- 11.Pientong C, et al. Association of human papillomavirus type 16 long control region mutation and cervical cancer. Virol. J. 2013;10:30. doi: 10.1186/1743-422X-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xi LF, et al. Risk for high-grade cervical intraepithelial neoplasia associated with variants of human papillomavirus types 16 and 18. Cancer Epidemiol. Biomark. Prev. 2007;16:4–10. doi: 10.1158/1055-9965.EPI-06-0670. [DOI] [PubMed] [Google Scholar]
- 13.Schiffman M, et al. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 2010;70:3159–3169. doi: 10.1158/0008-5472.CAN-09-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornet I, et al. HPV16 genetic variation and the development of cervical cancer worldwide. Br. J. Cancer. 2013;108:240–244. doi: 10.1038/bjc.2012.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gheit T, et al. Risks for persistence and progression by human papillomavirus type 16 variant lineages among a population-based sample of Danish women. Cancer Epidemiol. Biomark. Prev. 2011;20:1315–1321. doi: 10.1158/1055-9965.EPI-10-1187. [DOI] [PubMed] [Google Scholar]
- 16.Zehbe I, Voglino G, Delius H, Wilander E, Tommasino M. Risk of cervical cancer and geographical variations of human papillomavirus 16 E6 polymorphisms. Lancet. 1998;352:1441–1442. doi: 10.1016/S0140-6736(05)61263-9. [DOI] [PubMed] [Google Scholar]
- 17.Zuna RE, et al. Association of HPV16 E6 variants with diagnostic severity in cervical cytology samples of 354 women in a US population. Int. J. Cancer. 2009;125:2609–2613. doi: 10.1002/ijc.24706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sichero L, et al. High grade cervical lesions are caused preferentially by non-European variants of HPVs 16 and 18. Int. J. Cancer. 2007;120:1763–1768. doi: 10.1002/ijc.22481. [DOI] [PubMed] [Google Scholar]
- 19.Berumen J, et al. Asian-American variants of human papillomavirus 16 and risk for cervical cancer: a case–control study. J. Natl Cancer Inst. 2001;93:1325–1330. doi: 10.1093/jnci/93.17.1325. [DOI] [PubMed] [Google Scholar]
- 20.Freitas LB, et al. Human papillomavirus 16 non-european variants are preferentially associated with high-grade cervical lesions. PLoS ONE. 2014;9:e100746. doi: 10.1371/journal.pone.0100746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirabello L, et al. HPV16 sublineage associations with histology-specific cancer risk using HPV whole-genome sequences in 3200 women. J. Natl Cancer Inst. 2016;108:djw100. doi: 10.1093/jnci/djw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirabello L, et al. The intersection of HPV epidemiology, genomics and mechanistic studies of HPV-mediated carcinogenesis. Viruses. 2018;10:80. doi: 10.3390/v10020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Weele P, Meijer CJLM, King AJ. Whole-genome sequencing and variant analysis of human papillomavirus 16 infections. J. Virol. 2017;91:e00844-17. doi: 10.1128/JVI.00844-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirabello L, et al. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell. 2017;170:1164–1174.e6. doi: 10.1016/j.cell.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirose Y, et al. Within-host variations of human papillomavirus reveal APOBEC signature mutagenesis in the viral genome. J. Virol. 2018;92:e00017–e00018. doi: 10.1128/JVI.00017-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amador-Molina A, Hernández-Valencia JF, Lamoyi E, Contreras-Paredes A, Lizano M. Role of innate immunity against human papillomavirus (HPV) infections and effect of adjuvants in promoting specific immune response. Viruses. 2013;5:2624–2642. doi: 10.3390/v5112624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori S, Takeuchi T, Ishii Y, Kukimoto I. Identification of APOBEC3B promoter elements responsible for activation by human papillomavirus type 16 E6. Biochem. Biophys. Res. Commun. 2015;460:555–560. doi: 10.1016/j.bbrc.2015.03.068. [DOI] [PubMed] [Google Scholar]
- 28.Mori, S. et al. Human papillomavirus 16 E6 upregulates APOBEC3B via the TEAD transcription factor. J. Virol. 91, e02413-16 (2017). [DOI] [PMC free article] [PubMed]
- 29.Vieira, V. C. et al. Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B. MBio5, e02234-14 (2014). [DOI] [PMC free article] [PubMed]
- 30.Warren CJ, et al. APOBEC3A functions as a restriction factor of human papillomavirus. J. Virol. 2015;89:688–702. doi: 10.1128/JVI.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Periyasamy M, et al. p53 controls expression of the DNA deaminase APOBEC3B to limit its potential mutagenic activity in cancer cells. Nucleic Acids Res. 2017;45:11056–11069. doi: 10.1093/nar/gkx721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith NJ, Fenton TR. The APOBEC3 genes and their role in cancer: insights from human papillomavirus. J. Mol. Endocrinol. 2019;62:R269–R287. doi: 10.1530/JME-19-0011. [DOI] [PubMed] [Google Scholar]
- 33.Yang B, Li XS, Lei LQ, Chen J. APOBEC: from mutator to editor. J. Genet. Genomics. 2017;44:423–437. doi: 10.1016/j.jgg.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell. 2002;10:1247–1253. doi: 10.1016/S1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 35.The Cancer Genome Atlas Research N, et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng H, et al. Whole-exome sequencing identifies multiple loss-of-function mutations of NF-kappaB pathway regulators in nasopharyngeal carcinoma. Proc. Natl Acad. Sci. USA. 2016;113:11283–11288. doi: 10.1073/pnas.1607606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013;45:977. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The Cancer Genome Atlas, N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. 2014;7:1833–1841. doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Gillison ML, et al. Human papillomavirus and the landscape of secondary genetic alterations in oral cancers. Genome Res. 2019;29:1–17. doi: 10.1101/gr.241141.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buisson Rémi, Langenbucher Adam, Bowen Danae, Kwan Eugene E., Benes Cyril H., Zou Lee, Lawrence Michael S. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science. 2019;364(6447):eaaw2872. doi: 10.1126/science.aaw2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015;5:704–712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henderson S, Fenton T. APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol. Med. 2015;21:274–284. doi: 10.1016/j.molmed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 48.Burns MB, Leonard B, Harris RS. APOBEC3B: pathological consequences of an innate immune DNA mutator. Biomed. J. 2015;38:102–110. doi: 10.4103/2319-4170.148904. [DOI] [PubMed] [Google Scholar]
- 49.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat. Rev. Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warren CJ, Westrich JA, Van Doorslaer K, Pyeon D. Roles of APOBEC3A and APOBEC3B in human papillomavirus infection and disease progression. Viruses. 2017;9:233. doi: 10.3390/v9080233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mangeat B, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 52.Vartanian J, Guétard D, Henry M, Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–233. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- 53.Kukimoto I, et al. Hypermutation in the E2 gene of human papillomavirus type 16 in cervical intraepithelial neoplasia. J. Med. Virol. 2015;87:1754–1760. doi: 10.1002/jmv.24215. [DOI] [PubMed] [Google Scholar]
- 54.Wakae K, et al. Detection of hypermutated human papillomavirus type 16 genome by Next-Generation Sequencing. Virology. 2015;485:460–466. doi: 10.1016/j.virol.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 55.Warren, C. J., Pyeon, D., Van Doorslaer, K., Pandey, A. & Espinosa, J. M. Role of the host restriction factor APOBEC3 on papillomavirus evolution. Virus Evol.1, pii: vev015 (2015). [DOI] [PMC free article] [PubMed]
- 56.Castle P, et al. Human papillomavirus (HPV) genotypes in women with cervical precancer and cancer at Kaiser Permanente Northern California. Cancer Epidemiol. Biomark. Prev. 2011;20:946–953. doi: 10.1158/1055-9965.EPI-10-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wentzensen N, et al. Multiple human papillomavirus genotype infections in cervical cancer progression in the study to understand cervical cancer early endpoints and determinants. Int. J. Cancer. 2009;125:2151–2158. doi: 10.1002/ijc.24528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang SS, et al. Human papillomavirus (HPV) cofactors by disease progression and HPV types in the Study to Understand Cervical Cancer Early Endpoints and Determinants (SUCCEED) Cancer Epidemiol., Biomark. Prev. 2009;18:113–120. doi: 10.1158/1055-9965.EPI-08-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wentzensen N, et al. Grading the severity of cervical neoplasia based on combined histopathology, cytopathology, and HPV genotype distribution among 1,700 women referred to colposcopy in Oklahoma. Int. J. Cancer. 2009;124:964–969. doi: 10.1002/ijc.23969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bosch FX, et al. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 61.Muñoz N, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 62.Clifford GM, et al. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys: a pooled analysis. Lancet. 2005;366:991–998. doi: 10.1016/S0140-6736(05)67069-9. [DOI] [PubMed] [Google Scholar]
- 63.Crosbie, E. J., Einstein, M. H., Franceschi, S. & Kitchener, H.C. Human papillomavirus and cervical cancer. Lancet382, 889–899 (2013). [DOI] [PubMed]
- 64.Cullen M, et al. Deep sequencing of HPV16 genomes: a new high-throughput tool for exploring the carcinogenicity and natural history of HPV16 infection. Papillomavirus Res. 2015;1:3–11. doi: 10.1016/j.pvr.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tate JG, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2018;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graur D, Li W-H. Fundamentals of Molecular Evolution. MA: Sinauer Associates, Inc, Sanderland; 2000. [Google Scholar]
- 67.Chan K, et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 2015;47:1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith B, et al. Sequence imputation of HPV16 genomes for genetic association studies. PLoS ONE. 2011;6:e21375. doi: 10.1371/journal.pone.0021375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Doorbar J, et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30:F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 70.Warren CJ, Pyeon D. APOBEC3 in papillomavirus restriction, evolution and cancer progression. Oncotarget. 2015;6:39385–39386. doi: 10.18632/oncotarget.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology. 2013;445:232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McBride AA, Warburton A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 2017;13:e1006211. doi: 10.1371/journal.ppat.1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clarke MA, et al. Human papillomavirus DNA Methylation as a potential biomarker for cervical cancer. Cancer Epidemiol. Biomark. Prev. 2012;21:2125–2137. doi: 10.1158/1055-9965.EPI-12-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mirabello L, et al. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. Int J. Cancer. 2013;132:1412–1422. doi: 10.1002/ijc.27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peng G, Lei KJ, Jin W, Greenwell-Wild T, Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J. Exp. Med. 2006;203:41–46. doi: 10.1084/jem.20051512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koning FA, et al. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maruyama W, et al. Classical NF-κB pathway is responsible for APOBEC3B expression in cancer cells. Biochem. Biophys. Res. Commun. 2016;478:1466–1471. doi: 10.1016/j.bbrc.2016.08.148. [DOI] [PubMed] [Google Scholar]
- 78.Leonard B, et al. The PKC/NF-κB signaling pathway induces APOBEC3B expression in multiple human. Cancers. 2015;75:4538–4547. doi: 10.1158/0008-5472.CAN-15-2171-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Menendez D, Nguyen T-A, Snipe J, Resnick MA. The cytidine deaminase APOBEC3 family is subject to transcriptional regulation by p53. Mol. Cancer Res. 2017;15:735–743. doi: 10.1158/1541-7786.MCR-17-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Middlebrooks CD, et al. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat. Genet. 2016;48:1330–1338. doi: 10.1038/ng.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wittkopp CJ, Adolph MB, Wu LI, Chelico L, Emerman M. A single nucleotide polymorphism in human APOBEC3C enhances restriction of lentiviruses. PLoS Pathog. 2016;12:e1005865–e1005865. doi: 10.1371/journal.ppat.1005865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Burk RD, et al. Sexual behavior and partner characteristics are the predominant risk factors for genital human papillomavirus infection in young women. J. Infect. Dis. 1996;174:679–689. doi: 10.1093/infdis/174.4.679. [DOI] [PubMed] [Google Scholar]
- 83.Castle PE, et al. Comparisons of HPV DNA detection by MY09/11 PCR methods. J. Med. Virol. 2002;68:417–423. doi: 10.1002/jmv.10220. [DOI] [PubMed] [Google Scholar]
- 84.Dunn ST, Allen RA, Wang S, Walker J, Schiffman M. DNA extraction: an understudied and important aspect of HPV genotyping using PCR-based methods. J. Virol. Methods. 2007;143:45–54. doi: 10.1016/j.jviromet.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 85.Cornet I, et al. Human papillomavirus type 16 genetic variants: phylogeny and classification based on E6 and LCR. J. Virol. 2012;86:6855–6861. doi: 10.1128/JVI.00483-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jacobs MV, et al. Distribution of 37 mucosotropic HPV types in women with cytologically normal cervical smears: the age-related patterns for high-risk and low-risk types. Int. J. Cancer. 2000;87:221–227. doi: 10.1002/1097-0215(20000715)87:2<221::AID-IJC11>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 87.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 89.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- 91.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 92.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The HPV sequencing data have been deposited in the Genbank database under the accession codes MG847621-MG850835. All the other data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding author upon reasonable request. A reporting summary for this article is available as a Supplementary Information file.
The software and algorithms used for our analyses are specified in the corresponding methods section. Pipeline settings and parameters used to call HPV16 variants can be found at https://github.com/cgrlab/cgrHPV16.