Abstract
Recurrent and chronic respiratory tract infections in cystic fibrosis (CF) patients result in progressive lung damage and represent the primary cause of morbidity and mortality. Staphylococcus aureus (S. aureus) is one of the earliest bacteria in CF infants and children. Starting from early adolescence, patients become chronically infected with Gram-negative non-fermenting bacteria, and Pseudomonas aeruginosa (P. aeruginosa) is the most relevant and recurring. Intensive use of antimicrobial drugs to fight lung infections inevitably leads to the onset of antibiotic resistant bacterial strains. New antimicrobial compounds should be identified to overcome antibiotic resistance in these patients. Recently interesting data were reported in literature on the use of natural derived compounds that inhibited in vitro S. aureus and P. aeruginosa bacterial growth. Essential oils, among these, seemed to be the most promising. In this work is reported an extensive study on 61 essential oils (EOs) against a panel of 40 clinical strains isolated from CF patients. To reduce the in vitro procedure and render the investigation as convergent as possible, machine learning clusterization algorithms were firstly applied to pick-up a fewer number of representative strains among the panel of 40. This approach allowed us to easily identify three EOs able to strongly inhibit bacterial growth of all bacterial strains. Interestingly, the EOs antibacterial activity is completely unrelated to the antibiotic resistance profile of each strain. Taking into account the results obtained, a clinical use of EOs could be suggested.
Subject terms: Antimicrobial resistance, Clinical microbiology
Introduction
Cystic fibrosis (CF), one of the most common lethal genetic disorders in Caucasian population, is inherited as an autosomal recessive disease and affects 70.000 persons worldwide (Cystic Fibrosis Foundation, CFF). The defective gene, identified in 1989, is the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) that is carried by 4% of persons (among Caucasians). Since CFTR encodes for a chloride channel of the epithelial cell surface, CF patients manifest a variety of multi-organ problems due to the alteration of sodium and chloride secretion across cell membranes and the subsequent luminal dehydration1. The impairment of mucociliary clearance, which should remove all microbes entering the airways, leads to the production of a thick and dehydrated mucus in the CF lung, which promotes the airway chronic bacterial colonization2.
The microbiology of CF respiratory tract is peculiar. In the early stage of life, it is characterized by the prevalence of the Gram-positive bacterium Staphylococcus aureus (S. aureus). Overall, in 2017 more than half of affected individuals had at least one culture positive for methicillin sensitive S. aureus (MSSA). The highest prevalence of methicillin resistant S. aureus (MRSA) occurs in individuals between the ages of 10 and 30, while MSSA reaches the peak among patients younger than 10 (Cystic Fibrosis Foundation. 2017. Patient Registry Annual Data Report. https://www.cff.org/Research/Researcher-Resources/Patient-Registry/2017-Patient-Registry-Annual-Data-Report.pdf).
In early adolescence, CF patients’ lung becomes chronically infected with Gram-negative non-fermenting bacteria. Among these, Pseudomonas aeruginosa (P. aeruginosa) is the most relevant and recurring, so that 30% of CF children and up to 80% of CF adults (25 years old and older) have lungs chronically colonized by this pathogen3. P. aeruginosa isolated from respiratory secretions demonstrates great phenotypic diversity and develops genetic mutations over time to adapt and survive in the complex environment of the CF airway4. P. aeruginosa mucoid phenotype, defined by the exopolysaccharide alginate overproduction within lungs of CF patients, is a hallmark of chronic infection and predictive of poor prognosis. Indeed, mucoid P. aeruginosa has also been associated with failure of eradication and, compared to non-mucoid counterpart, exhibits enhanced resistance to multiple antibiotics and host immune effectors5.
Due to current therapeutic treatments, life expectancy for CF patients has consistently grown, reaching a median life of 40 years. Assuming a positive trend of clinical care improvements at the actual rate, CF patients born in 2010 are expected to live up to 50 years of age6.
The intensive use of antimicrobial drugs to fight lung infections inevitably leads to the onset of antibiotic resistant bacterial strains. New antimicrobial compounds should be identified to overcome antibiotic resistance during the treatment of CF lung infections.
Recent investigation has disclosed a few small molecules, such as peptides or mannosides, showing promising efficacy in prevention and treatment of both bacterial and fungal biofilm infection in vivo7. Nevertheless, due to their mechanism of action based on a specific binding to a main target, the use of small molecules is known to select more and more resistant strains8. Interestingly in the recent literature appeared some reports on the use of natural derived compounds that showed in vitro the potentiality to inhibit the development of CF associated infections9–12. In particular essential oils seemed to be the most promising agents among tested natural compounds10,11. In this study is reported an extensive study on 61 essential oils (EOs) against a panel of 40 bacterial strains isolated from CF patients (see Table 1).
Table 1.
Classification of bacterial strains based on their biofilm formation ability.
| Bacterial strains | Biofilm producer | Bacterial strains | Biofilm producer |
|---|---|---|---|
| 6538P* | STRONG | PAO1* | STRONG |
| 25923* | STRONG | PA14* | STRONG |
| 1S | WEAK | 21P | STRONG |
| 2S | MODERATE | 22P | NP |
| 3S | WEAK | 23P | MODERATE |
| 4S | WEAK | 24P | NP |
| 5S | WEAK | 25P | WEAK |
| 6S | WEAK | 26P | NP |
| 7S | MODERATE | 27P | NP |
| 8S | WEAK | 28P | NP |
| 9S | WEAK | 29P | NP |
| 10S | MODERATE | 30P | WEAK |
| 11S | WEAK | 31P | MODERATE |
| 12S | WEAK | 32P | WEAK |
| 13S | WEAK | 33P | NP |
| 14S | WEAK | 34P | WEAK |
| 15S | MODERATE | 35P | WEAK |
| 16S | WEAK | 36P | WEAK |
| 17S | MODERATE | 37P | STRONG |
| 18S | WEAK | 38P | MODERATE |
| 19S | WEAK | 39P | WEAK |
| 20S | MODERATE | 40P | WEAK |
To reduce the in vitro procedure and to render the investigation as convergent as possible the following workflow was followed. Unsupervised machine learning algorithms and techniques, as implemented in python language13, were firstly applied to pick-up a fewer number of representative strains (RS) among the panel of 40. To this aim, a number of categorical descriptors were collected and used to cluster the CF isolated strains. The clusters’ centroids indicated the RS to be investigated for their susceptibility to a list of commercial EOs at fixed doses. Three EOs showed a great efficacy to reduce the microrganisms growth and were therefore promptly assayed against all the available clinical isolates. The three EOs confirmed the initial assumption demonstrating their ability to inhibit bacterial growth. Gas chromatography coupled with mass spectrometry (GC/MS) was then performed on the three EOs to investigate on the likely chemical components mainly responsible for the antibacterial activity.
Results
Characterization of biofilm formation of clinical bacterial strains
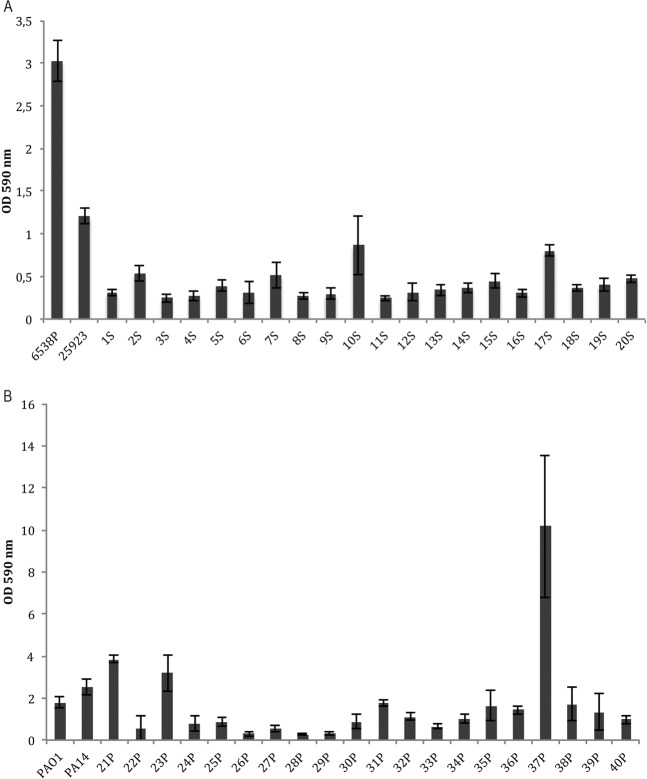
Clinical bacterial strains were investigated for their ability to produce biofilm (Fig. 1). Biofilm formation was evaluated at 37 °C in BHI for 18 h as described in Material and Methods section.
Figure 1.
Biofilm formation of S. aureus clinical and reference strains (A) and P. aeruginosa clinical and reference strains (B). The biofilm formation was evaluated after 18 h incubation in polystyrene plates at 37 °C. The data are reported as OD 590 nm after crystal violet staining. Each data point represents the mean ± SD of four independent samples.
Biofilm formation was also evaluated for four reference strains included in the experimental plan. Figure 1A reports biofilm formation of bacterial strains belonging to S. aureus species. Clinical strains, named from 1S to 20S, were classified as “weak” or “moderate” biofilm producers according to Cafiso and coworker, 200714. Both reference strains for S. aureus species are strong biofilm producers. Figure 1B reports biofilm formation of bacterial strains belonging to P. aeruginosa species. Clinical strains, named from 21P to 40P, were classified as: “non producer”, “weak”, “moderate” and “strong” biofilm producers according to Perez and Barth, 201115 (Table 1).
Selection of representative microorganisms by machine learning
The 40 selected strains were divided accordingly to the main strains families into S. aureus and P. aeruginosa dataset and imported into a python pandas dataframe. The principal components analysis (PCA) indicated that 90% of the variance is explained by the first 10th principal components (PCs) (Fig. 1S). Nevertheless graphical inspection of the PC1 versus PC2 scores and loadings plots indicated the PCs as potential new variables to cluster the datasets (Fig. 2S). As a matter of fact, application of the Silhouette Analysis16 coupled with the k-means clustering17 to the first 2 PCs indicated the optimal number of clusters to be 6 and 3 for the P. aeruginosa and S. aureus strains, respectively (Figs. 3S and 4S). For each cluster, the nearest datapoint to cluster centroid was selected yielding to a selection of representative strains to be screened with the commercial EOs. Analysis of data revealed the six samples, precisely 22P, 25P, 26P, 27P, 37P and 39P as the representatives for P. aeruginosa, whereas samples 4S, 5S and 19S were selected for S. aureus.
Antimicrobial activity of EOs on P. aeruginosa and S. aureus clinical strains from cystic fibrosis patients
Essential oils were tested for their ability to inhibit bacterial growth of P. aeruginosa and S. aureus clinical and reference strains. Analysis was performed on three representative S. aureus strains and six representative P. aeruginosa strains, previously selected by machine learning analysis. EOs were tested at a concentration of 1% v/v (Table 2). Several EOs have shown antimicrobial activity on many bacterial strains. It is worthy to note that the P. aeruginosa reference strain PAO1 is the most resistant to the action of EOs, since it was inhibited by only four EOs.
Table 2.
Antimicrobial activity of EOs listed in Table 2S on representative clinical strains and reference strains of S. aureus and P. aeruginosa.
| Eos ID | 6538P | 25923 | 4S | 5S | 19S | PaO1 | PA14 | 22P | 25P | 26P | 27P | 37P | 39P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1% | 1% | 1% | ||||||||||
| 2 | |||||||||||||
| 3 | 1% | 1% | |||||||||||
| 4 | 1% | 1% | 1% | 1% | 1% | 1% | 1% | ||||||
| 5 | |||||||||||||
| 6 | 1% | 1% | 1% | 1% | |||||||||
| 7 | |||||||||||||
| 8 | |||||||||||||
| 9 | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | ||
| 10 | |||||||||||||
| 11 | |||||||||||||
| 12 | |||||||||||||
| 13 | |||||||||||||
| 14 | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | ||
| 15 | |||||||||||||
| 16 | |||||||||||||
| 17 | |||||||||||||
| 18 | |||||||||||||
| 19 | |||||||||||||
| 20 | |||||||||||||
| 21 | |||||||||||||
| 22 | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% |
| 23 | |||||||||||||
| 24 | |||||||||||||
| 25 | |||||||||||||
| 26 | |||||||||||||
| 27 | |||||||||||||
| 28 | |||||||||||||
| 29 | |||||||||||||
| 30 | |||||||||||||
| 31 | |||||||||||||
| 32 | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% |
| 33 | |||||||||||||
| 34 | |||||||||||||
| 35 | |||||||||||||
| follows | |||||||||||||
| 36 | |||||||||||||
| 37 | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | |||||
| 38 | 1% | ||||||||||||
| 39 | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% | 1% |
| 40 | |||||||||||||
| 41 | |||||||||||||
| 42 | |||||||||||||
| 43 | |||||||||||||
| 44 | |||||||||||||
| 45 | |||||||||||||
| 46 | 1% | 1% | 1% | 1% | 1% | 1% | 1% | ||||||
| 47 | |||||||||||||
| 48 | 1% | 1% | 1% | 1% | 1% | 1% | 1% | ||||||
| 49 | |||||||||||||
| 50 | |||||||||||||
| 51 | |||||||||||||
| 52 | 1% | 1% | |||||||||||
| 53 | |||||||||||||
| 54 | 1% | 1% | |||||||||||
| 55 | |||||||||||||
| 56 | |||||||||||||
| 57 | |||||||||||||
| 58 | |||||||||||||
| 59 | 1% | 1% | 1% | 1% | 1% | 1% | 1% | ||||||
| 60 | 1% | 1% | 1% | 1% | |||||||||
| 61 | 1% | 1% | 1% |
This analysis allowed to identify three EOs active against all the representative strains used, namely cade essential oil (22 in Table 2S, CEO), birch essential oil (32 in Table 2S, BEO) and Ceylon cinnamon peel essential oil (39 in Table 2S, CCPEO). Thus, these 22, 32 and 39 EOs were tested on all clinical bacterial strains. Results summarized in Table 3 confirmed that BEO, CEO and CCPEO exerted a strong and effective bactericidal potency on all tested clinical strains.
Table 3.
Antimicrobial activity corresponding to minimal bactericidal concentration of previously selected EOs on all 40 clinical isolates.
| Bacterial strains | CEO | BEO | CCPEO | Bacterial strains | CEO | BEO | CCPEO |
|---|---|---|---|---|---|---|---|
| ATCC6538P | 1% | 1% | 1% | PA O1 | 1% | 1% | 1% |
| ATCC25923 | 1% | 1% | 1% | PA 14 | 1% | 1% | 1% |
| SA01 | 1% | 1% | 1% | PA21 | 1% | 1% | 1% |
| SA02 | 1% | 1% | 1% | PA22 | 1% | 1% | 1% |
| SA03 | 1% | 1% | 1% | PA23 | 1% | 1% | 1% |
| SA04 | 1% | 1% | 1% | PA24 | 1% | 1% | 1% |
| SA05 | 1% | 1% | 0.1% | PA25 | 1% | 1% | 1% |
| SA06 | 1% | 1% | 1% | PA26 | 1% | 1% | 1% |
| SA07 | 1% | 1% | 1% | PA27 | 1% | 1% | 1% |
| SA08 | 1% | 1% | 1% | PA28 | 1% | 1% | 1% |
| SA09 | 1% | 1% | 1% | PA29 | 1% | 1% | 1% |
| SA10 | 1% | 1% | 1% | PA30 | 1% | 1% | 1% |
| SA11 | 1% | 1% | 1% | PA31 | 1% | 1% | 1% |
| SA12 | 1% | 1% | 1% | PA32 | 1% | 1% | 1% |
| SA13 | 1% | 1% | 1% | PA33 | 1% | 1% | 1% |
| SA14 | 1% | 1% | 1% | PA34 | 1% | 1% | 1% |
| SA15 | 1% | 1% | 1% | PA35 | 1% | 1% | 1% |
| SA16 | 1% | 1% | 1% | PA36 | 1% | 1% | 1% |
| SA17 | 1% | 1% | 1% | PA37 | 1% | 1% | 1% |
| SA18 | 1% | 1% | 1% | PA38 | 1% | 1% | 1% |
| SA19 | 1% | 1% | 1% | PA39 | 1% | 1% | 1% |
| SA20 | 1% | 1% | 1% | PA40 | 1% | 1% | 1% |
Chemical composition analysis of active selected essential oils
The results of GC and GC-MS analyses of the essential oils are reported in Table 3S–5S. In the BEO, 21 components were identified and the major constituents were δ-cadinene, calamenene and creosol (22.2%, 15.2% and 12.8% respectively) (Table 3S). The chemical composition of CCPEO was characterized by the presence of 19 compounds and by a high amount of cinnamaldehyde (49.4%) followed by eugenol (21.2%) (Table 4S). The chemical composition of the CEO indicated 21 components and the most abundant were delta-cadinene (27.7%), calamenene (14.8%) and creosol (12.6%) (Table 5S). At first glance the chemical composition of the CEO seems very similar to that of BEO as the main compounds showed comparable percentages. Among the minor components of CEO, α-selinene (2.2%), aromadendrene (1.1%) and gleenol (1.1%) were found, whereas isoledene (5.7%) was found in BEO. At a deeper analysis the chemical qualitative profiles were compared and a 0.62 tanimoto index was calculated, thus indicating that although displaying a similar chromatogram the two EOs are indeed different. EOs producer was also inquired and their technical staff confirmed the two oils sharing high similarity quantitative profile in the main constituents.
Discussion
Long-term administration of antibiotics to prevent and treat airway infections in CF patients has been shown to be associated with the emergence of multi-drug (MDR) antimicrobial resistant microorganisms18.
In particular, mecA/mecC genes acquisition in S. aureus and accumulation of resistance mechanisms after antibiotic exposure in P. aeruginosa, both key pathogens in CF lung, are a concern in this context19,20.
Multidrug resistance significantly limits effective therapeutic options, affecting clinical outcome and prognosis of patients. For this reason, the identification and development of new antibacterial agents is fundamental to improve survival and quality of life of individuals with CF. Therefore the development of antimicrobial agents provided with novel molecular mechanisms that may allow to control bacterial infectious diseases without diffusing antibacterial resistance is desirable21.
Unsupervised Machine Learning algorithms13 applied to a panel of 40 strains of S. aureus and P. aeruginosa isolated from CF patients, led to select fewer representative strains using phenotypical and genotypical characteristics as categorical descriptors. Therefore, the antibacterial activity of all tested EOs was initially assessed on 9 selected bacterial strains: six representative strains for P. aeruginosa and 3 representative strains for S. aureus. The activity of all 61 EOs was also assessed on reference strains. Antimicrobial assays led to identify 3 EOs (CEO, BEO and CCPEO) out of the tested 61, that exhibited the highest antibacterial activity on the previously selected bacterial strains and reference ones. The antibacterial activity of the 3 selected EOs was then extended to all strains of both species. Interestingly all three EOs showed an utmost antimicrobial potency on all studied strains. Nothing can be yet ruled out on the chemical compounds’ role. Future studies involving machine learning application10,11, will be devoted to investigate the importance of chemical constituent either on biofilm modulation or in antibacterial potencies.
Several papers aimed at elucidating the antimicrobial mechanism of action of EOs. For example, cinnamaldehyde, the major component of cinnamon, is able to disrupt the transmembrane potential of P. aeruginosa22.
Furthermore, EOs of different origin (lavender, lemongrass, marjoram, peppermint, tea tree and rosewood) show antimicrobial activity against Burkholderia cepacia complex by inducing changes in membrane fatty acid composition, followed by membrane disruption23. Also, EO from Alluaudia procera was active against S. aureus ATCC25923, a multi-resistant strain24. Reported data confirmed the possibility to use EOs as therapeutic strategies in multi-resistant strains probably due to the heterogeneous composition of the oils themselves.
Notably, in this work we found EOs antibacterial activity unrelated to the antibiotic resistance profile of each strain.
This observation is of particular relevance as it suggests the EOs potential uses by topical administration without taking into account the complexity of drug resistance profile of the microbiota in each single patient.
In conclusion the approach herein applied allowed to minimize the experimental steps and it was possible to identify the most promising EOs on the basis of probabilistic evaluations that confirmed their wide spectra of antibacterial potency with a reduced set of experiments.
From a literature survey (www.scopus.com, accessed 2019 December 13, keywords: essential oil, antibacterial activity and resistance) no evidence of resistance to EOs antibacterial activity has yet been reported. This is a characteristic particularly relevant for antibacterial candidates to be administered for a chronic disease such as CF.
Indeed some papers report an increase of susceptibility to antibiotics after treatment with essential oils25,26.
Although a pletora of publications did not show development of resistance to EOs, a very recent publication suggested the induction of efflux pumps and multidrug resistance in P. aeruginosa by Cinnamaldehyde, the main component of cinnamon27. Therefore, in light of the recent reports, much still needs to be clarified on the effect of essential oils on bacterial multi-drug resistance.
Methods
Ethics approval and informed consent
The approval for this research was granted by the Ethics Committee of Children’s Hospital and Institute Research Bambino Gesù in Rome, Italy (No 1437_OPBG_2017 of July 2017), and it was performed according to the principles of the Helsinki Declaration. Informed consent was obtained from all individual participants and all parents/legal guardians included in the study.
Clinical isolates from CF patients
In this study were used 40 bacterial strains (20 S. aureus, 20 P. aeruginosa) obtained from respiratory specimens of 30 CF patients (13 males, 17 females; medium age 20.5) in follow-up at Pediatric Hospital Bambino Gesù (OPBG) of Rome, Italy. In particular, 27 bacterial strains were isolated from sputum, 11 from hypopharyngeal suction and 2 from throat swabs (Tables 4 and 5). As reference strains were used: S. aureus ATCC 6538P (6538P) and S. aureus ATCC 25923 (25923) commonly recognized as reference strains for antimicrobial testing; P. aeruginosa PAO1 (PAO1) and P. aeruginosa PA14 respectively recognized as moderately and highly virulent28.
Table 4.
The 20 Staphylococcus aureus clinical isolates and their characterization by several properties.
| ID pt | ID | SAM | Date | Str | Ph | QUIN | B | ER | CLI | LIN | RCLI | CF | CPA | GEN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1S | ESP | 10/11/2006 | MRSA | SCV | R | S | Nt | R | S | N | Cp | J | |
| 2 | 2S | ESP | 11/22/2007 | MRSA | SCV | R | S | Nt | R | S | N | Ca | X | N |
| 3 | 3S | ESP | 1/15/2009 | MRSA | SCV | S | S | Nt | S | S | N | X | E | |
| 4 | 4S | AT | 2/20/2009 | MRSA | — | S | S | Nt | S | S | P | A | ||
| 5 | 5S | ESP | 11/13/2009 | MRSA | — | R | S | Nt | R | S | N | Sp | C | |
| 6 | 6S | AT | 1/10/2011 | MRSA | — | R | S | Nt | R | S | P | K | ||
| 7 | 7S | ESP | 4/4/2011 | MRSA | — | R | S | Nt | R | S | N | Ca | X | D |
| 8 | 8S | AT | 7/22/2013 | MRSA | — | R | S | Nt | S | S | N | I | ||
| 9 | 9S | ESP | 1/15/2014 | MRSA | — | S | S | Nt | R | S | P | Ca | X | C |
| 10 | 10S | AT | 1/29/2015 | MRSA | — | S | S | Nt | R | S | N | Ca/Cd/Pb | G | |
| 11 | 11S | AT | 6/15/2017 | MSSA | — | S | S | R | R | S | P | C | ||
| 12 | 12S | AT | 6/15/2017 | MSSA | — | S | S | R | R | S | P | U | ||
| 13 | 13S | AT | 5/23/2017 | MSSA | — | I | S | I | I | S | N | Sa | B | |
| 14 | 14S | AT | 5/25/2017 | MSSA | — | S | S | S | S | S | N | C | ||
| 15 | 15S | AT | 5/24/2017 | MSSA | — | S | S | R | S | S | N | X | C | |
| 16 | 16S | AT | 5/26/2017 | MSSA | — | S | S | R | R | S | N | H | ||
| 17 | 17S | AT | 5/25/2017 | MSSA | — | S | S | R | R | S | N | Af | M | |
| 18 | 18S | ESP | 5/24/2017 | MSSA | — | S | S | R | R | S | P | Ca | X | C |
| 19 | 19S | ESP | 6/15/2017 | MSSA | — | S | S | R | R | S | P | X | L | |
| 20 | 20S | ESP | 5/19/2017 | MSSA | — | R | S | R | R | S | P | X | F |
ID pt: patient identification; ID: strain code; SAM:Sample; Date: Date of collection; Str:Strain; Ph: phenotype; QUIN: quinolones; B: Trimethoprim/Sulfamethoxazole; ER: Erythromycin; CLI: Clindamycin; LIN: linezolid; RCLI: Inducible Clindamycin resistance; CF: Fungal Co-infection; CPA: P. aeruginosa co-infection; GEN: pts genotype; Esp: sputum; AT: hypopharyngeal suction; MRSA: Methicillin Resistant S. aureus; MSSA: Methicillin Sensitive S. aureus; SCV: Small colony variant; R: Resistant;S: Susceptible; I: Intermediate; N: Negative; Nt: non-tested; Af: Aspergillus fumigatus; Ca: Candida albicans; Cp: Candida parapsilosis; Sp: Scedosporium prolificans; Cd: Candida dubliniensis; Pb: Pseudoallescheria boydii; Sa: Scedosporium apiospermum. X: denotes positive for this feature; -: denotes common phenotype. See Table 6 showing the correlation between letter code, CFTR gene mutation of the patient and bacterial strain isolated from the same patient.
Table 5.
The 20 Pseudomonas aeruginosa clinical isolates and their characterization by several properties.
| ID pt | ID | SAM | date | Str | Ph | CAR | PTC | AM | QUIN | MB | CEF | COL | 1St | E | L | CF | CSA | GEN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21 | 21P | ESP | 8/8/2006 | PA MDR | s | R | R | R | R | R | R | S | X | E | ||||
| 21 | 22P | ESP | 1/11/2017 | PA MDR | w | R | S | R | R | S | R | S | X | E | ||||
| 22 | 23P | ESP | 6/24/2005 | PA MDR MBL+ | sm | R | S | R | R | S | R | S | X | B | ||||
| 22 | 24P | ESP | 3/27/2017 | PA MDR MBL+ | s | R | S | R | R | S | R | S | X | Ca/Cl | B | |||
| 23 | 25P | AT | 9/3/2010 | PA | sm | S | S | I | S | I | S | S | X | B | ||||
| 24 | 26P | TF | 8/27/2008 | PA | i | S | S | S | S | I | S | S | X | G | ||||
| 24 | 27P | AT | 1/31/2017 | PA | sm | S | S | S | S | S | S | S | X | X | G | |||
| 25 | 28P | ESP | 5/24/2012 | PA | sm | S | S | S | S | S | S | S | X | U | ||||
| 25 | 29P | AT | 9/13/2017 | PA | m | S | S | S | S | S | S | S | X | U | ||||
| 9 | 30P | ESP | 9/6/2010 | PA | i | S | S | S | S | I | S | S | X | B | ||||
| 9 | 31P | ESP | 1/11/2017 | PA | m | S | S | S | R | S | S | S | X | X | B | |||
| 26 | 32P | AT | 12/5/2006 | PA | sm | S | S | R | S | I | S | S | X | F | ||||
| 26 | 33P | AT | 12/28/2016 | PA | m | S | S | S | S | I | S | S | X | Ca | X | F | ||
| 27 | 34P | ESP | 5/11/2005 | PA | i | MP I/IP R | S | S | CI S/LE R | I | S | S | X | Ca | D | |||
| 27 | 35P | ESP | 3/29/2017 | PA MDR | sm | R | R | R | R | I | R | S | X | D | ||||
| 28 | 36P | TF | 2/11/2008 | PA | sm | S | S | S | S | I | S | S | X | A | ||||
| 28 | 37P | AT | 2/22/2017 | PA | m | MP S/IP R | S | R | S | S | R | S | X | A | ||||
| 29 | 38P | ESP | 3/7/2006 | PA MDR | s | R | R | R | R | R | R | S | X | B | ||||
| 29 | 39P | ESP | 1/25/2017 | PA MDR | m | R | R | R | R | I | R | S | X | B | ||||
| 30 | 40P | AT | 7/1/2013 | PA | i | S | S | S | S | S | S | S | X | Ca | X | C |
ID pt: patient identification;ID: strain code;SAM: Sample; Date: Date of collection; Str: Strain; Ph: Phenotype; CAR: Carbapenems; MP: Meropenem; IP: Imipenem; PTC: Piperacillin/tazobactam; AM:
Aminoglycosides; QUIN: Quinolones; CI: Ciprofloxacin; LE: Levofloxacin; MB: Monobactam; CEF: Cephalosporins; COL: Colistin; 1 St: P. aeruginosa first isolate; E: P. aeruginosa early isolate; L: P. aeruginosa late isolate; CF: Fungal co-infection; CSA: S. aureus co-infection; Gen: pts genotype; BP: Biofilm Producer; Esp:sputum; AT: hypopharyngeal suction; TF: throat swabs; PA: P. aeruginosa; PA MDR: P. aeruginosa multi-drug resistant; PA MBL+: P. aeruginosa Metallo-Beta-Lactamases producing; s: small colony phenotype; w- wrinkled colony surface; m: mucoid colony; i: irregular colony edges; sm: smooth phenotype; R: Resistant; S: Susceptible; I: Intermediate; CA: Candida albicans; CL: Candida lusitaniae; X: denotes positive for the feature. See Table 6 showing the correlation between letter code, CFTR gene mutation of the patient and bacterial strain isolated from the same patient.
Patients were treated according to current standards of care29 with at least four microbiological controls per year. Informed consent was obtained from all subjects aged 18 years and older and from parents of all subjects under 18 years of age prior to enrolment.
Microbiological cultures have been performed according to approved Guidelines, using selective media, manual and automatic systems (API20NE, Vitek2, MALDI-TOF mass spectrometry) for isolates identification and 16S rRNA sequencing to assess ambiguous identifications.
The strains were selected from a local bacteria collection including about 10.000 CF bacterial isolates.
The species S. aureus and P. aeruginosa have been chosen for their clinical relevance in the natural history of CF disease, since they are related to a worst prognostic impact compared to other pathogens whose role is still under discussion.
In order to represent the complexity of CF lung microbiota population attending OPBG Center, a selection of specific strains with different phenotypic and biochemical features has been performed. The strains’ characteristics are described in Tables 4 and 5.
Qualitative description of the clinical isolates
Twenty S. aureus strains with a different susceptibility profile, belonging to 20 CF patients, were selected: 10 Methicillin-Sensitive (MSSA) and 10 Methicillin-Resistant (MRSA). Among the MRSA strains, three S. aureus with phenotypic “small colony variants” (SCVs) have been chosen, characterized by slow growth of small, unpigmented, non-haemolytic colonies.
Antimicrobial susceptibility profiles of MSSA and MRSA isolates were defined by automatic system Vitek2 (Biomerieux, France) or manual system E-test (Liofilchem, Italy). In particular, susceptibility to quinolones (ciprofloxacin, levofloxacin), trimethoprim/sulfamethoxazole, erythromycin, clindamycin, linezolid was assessed, according to EUCAST (www. EUCAST.org) criteria. Moreover, the clindamycin-inducing resistance test (40% positive test) was performed to classify S. aureus isolates that could develop acquired resistance to erythromycin or other macrolides during therapy with this antibiotic (Table 4)30.
Twenty P. aeruginosa isolates belonging to 11 CF patients were also selected (Table 5). The selected strains had been categorized as first, early and late isolates. In particular, seven strains have been associated to first acquisition of P. aeruginosa (first strains), 2 strains have been isolated 1 year after the first acquisition (early strains) and 11 strains have been isolated at least 5 years after the onset of chronic colonization (late strains).
Moreover, different phenotypes (mucoid, wrinkle surface, irregular edges or smooth) and strains with different antibiotic susceptibility patterns, e.g. P. aeruginosa producing Metallo-Beta-Lactamases (MBL)31 or P. aeruginosa multi-drug resistant (MDR), have been selected.
Susceptibility testing to carbapenems (imipenem, meropenem), piperacillin/tazobactam, aminoglicosides (tobramycin, amikacin), quinolones (ciprofloxacin, levofloxacin), monobactam (aztreonam), and cephalosporins (ceftazidime, cefepime) was carried out by Minimum Inhibitory Concentration (MIC) determined by E-test on Mueller Hinton (MH) agar plates, according to EUCAST criteria. The colistin MIC values were evaluated by Broth Microdilution (ComASP Colistin Liofilchem, Italy); 35% of P. aeruginosa isolates were MDR (i.e. resistant to three or more classes of antimicrobials)32 (Table 3). Table 1 of Supplementary Materials reports the percentage of bacterial strains resulted sensitive or resistant to different classes of antibiotics here tested (Table 1S).
Co-infection by bacterial (P. aeruginosa/S. aureus) and fungal agents (Aspergillus fumigatus/Candida albicans/Candida parapsilosis/Candida dubliniensis/Candida lusitaniae/Scedosporium prolificans/Scedosporium apiospermum//Pseudoallescheria boydii) was also evaluated for each patient (Tables 4 and 5).
Table 6 reports letters’ code correspondence for the strains associated genotype reported in Tables 4 and 5.
Table 6.
Table shows the correlation between letter code, CFTR gene mutation of the patient and bacterial strain isolated from the same patient.
| Code | Genotype | ID strain |
|---|---|---|
| A | 621+1G > T/R553X | 4S |
| B | F508del/1717-1G- > A | 13S |
| C | F508del/F508del | 5S |
| C | F508del/F508del | 9S |
| C | F508del/F508del | 11S |
| C | F508del/F508del | 14S |
| C | F508del/F508del | 15S |
| C | F508del/F508del | 18S |
| D | F508del/G1244E | 7S |
| E | F508del/G542X | 3S |
| F | F508del/L1077P | 20S |
| G | F508del/R1162X | 10S |
| H | F508del/R117L + L997F | 16S |
| I | F508del/R585X | 8S |
| J | F508del/W1282X | 1S |
| K | G542X/3271 + 42A/T | 6S |
| L | L636P/P499A | 19S |
| M | N1303K/2184insA | 17S |
| N | Q220X/A1006E | 2S |
| U | None | 12S |
| A | F508del/E193K | 36P |
| A | F508del/E193K | 37P |
| B | F508del/F508del | 23P |
| B | F508del/F508del | 24P |
| B | F508del/F508del | 25P |
| B | F508del/F508del | 30P |
| B | F508del/F508del | 31P |
| B | F508del/F508del | 38P |
| B | F508del/F508del | 39P |
| C | F508del/G542X | 40P |
| D | F508del/l1234V | 34P |
| D | F508del/l1234V | 35P |
| E | N1303K/3849 + 10kbC > T | 21P |
| E | N1303K/3849 + 10kbC > T | 22P |
| F | R347P/L571S | 32P |
| F | R347P/L571S | 33P |
| G | W1282X/2789 + 5G- > A | 26P |
| G | W1282X/2789 + 5G- > A | 27P |
| U | None | 28P |
| U | None | 29P |
Biofilm production assay
The quantification of biofilm production was based on microtiter plate biofilm assay (MTP) as reported in literature12. Briefly, the wells of a sterile 96-well flat-bottomed polystyrene plate were filled with 100 µL of the appropriate medium. 1/100 dilution of overnight bacterial cultures was added into each well (about 0.5 OD 600 nm). The plates were incubated aerobically for 18 h at 37 °C. Biofilm formation was measured using crystal violet staining. After incubation, planktonic cells were gently removed; each well was washed three times with double-distilled water and patted dry with a piece of paper towel in an inverted position. To quantify biofilm formation, each well was stained with 0.1% crystal violet and incubated for 15 min at room temperature, rinsed twice with double-distilled water, and thoroughly dried. The dye bound to adherent cells was solubilized with 20% (v/v) glacial acetic acid and 80% (v/v) ethanol. After 30 min of incubation at room temperature, OD590 was measured to quantify the total biomass of biofilm formed in each well. Each data point is composed of 4 independent experiments, each performed at least in 6-replicates.
Statistical analysis of biological evaluation
Data reported were statistically validated using Student’s t-test comparing mean absorbance of treated and untreated samples. The significance of differences between mean absorbance values was calculated using a two-tailed Student’s t-test. A p value of <0.05 was considered significant.
Chemical composition analysis of active selected essential oils
EOs were purchased from Farmalabor srl (Assago, Italy) and analyzed to characterize their composition as following.
Chemical analyses of EOs were performed by a Turbomass Clarus 500 GC-MS/GC-FID from Perkin Elmer instruments (Waltham, MA, USA) equipped with a Stabilwax fused-silica capillary column (Restek, Bellefonte, PA, USA) (60 m × 0.25 mm, 0.25 mm film thickness). The operating conditions used were as follows: GC oven temperature was kept at 40 °C for 5 min and programmed to 220 °C at a rate of 6 °C/min, and kept constant at 220 °C for 20 min. Helium was used as carrier gas (1.0 mL/min). Solvent delay 0–2 min and scan time 0.2 s. Mass range was from 30 to 350 m/z using electron-impact at 70 eV mode. 1 μL of each essential oil was diluted in 1 mL of methanol and 1 μL of the solution was injected into the GC injector at the temperature of 280 °C. Relative percentages for quantification of the components were calculated by electronic integration of the GC-FID peak areas. The identification of the constituents was achieved by comparing the obtained mass spectra for each component with those reported in mass spectra Nist and Wiley libraries. Linear retention indices (LRI) of each compound were calculated using a mixture of aliphatic hydrocarbons (C8-C30, Ultrasci) injected directly into GC injector at the same temperature program reported above.
Determination of EOs minimal inhibitory concentration (MIC)
The MIC was determined as the lowest concentration at which the observable bacterial growth was inhibited. MICs were determined according to the guidelines of Clinical Laboratory Standards Institute (CLSI33). Each EO was solubilized by adding DMSO, to generate a mother stock solutionof 1 g/mL. Appropriate dilution (106 cfu/mL) of bacterial culture in exponential phase was used. Antimicrobial activity of each EO was evaluated at a concentration of 1 mg/mL range. Experiments were performed in quadruplicate.
Unsupervised machine learning clusterization of clinical isolates
The cluster analysis was implemented in the Python (version 3.6) programming language13. The S. aureus and P. aeruginosa datasets were imported in a jupyter-notebook (version 5.7)34 and the categorical variables loaded into a Pandas35 dataframe were transformed into dummy indicator variables for the subsequent Principal Component Analysis (PCA) using the utilities available in the Pandas (version 0.23) library. The PCA analysis was performed using the scikit-learn library (version 0.20)36 to extract the first 20 principal components (PCs, Fig. 1S). The scores and loadings were graphically inspected on plots generated using the matplotlib library (version 3.0)37 (Fig. 2S). The PCs were used as features for the k-means clusterization. Silhouette analysis16 was performed to evaluate the separation distance between the resulting clusters and choose an optimal value for the number of clusters. Optimal number of clusters was identified by the maximum silhouette scores as graphically reported in Fig. 3S. Through k-means, the centroid of each cluster was calculated and the closest datapoint directly indicated the RS (Fig. 4S).
Supplementary information
Acknowledgements
This work was supported by PRIN 2017 (prot. 2017JL8SRX) (to RR), Ateneo 2019 (prot. RM11916B8876093E) (to RR) and Ateneo 2018 (prot. RM118164361B425B) (to RR).
Author contributions
R.R., R.P., A.P., G.V., S.G., V.T., E.V.F., L.S. and M.A. are responsible for the conception of the study, statistical analysis, interpretation of resultsand drafting the manuscript. All authors have seen and approved the final version of the manuscript. L.S. and R. R. are the guarantors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rino Ragno and Rosanna Papa.
Contributor Information
Rino Ragno, Email: rino.ragno@uniroma1.it.
Laura Selan, Email: laura.selan@uniroma1.it.
Supplementary information
is available for this paper at 10.1038/s41598-020-59553-8.
References
- 1.Harris A, Argent BE. The cystic fibrosis gene and its product CFTR. Semin. Cell. Biol. 1993;4:37–44. doi: 10.1006/scel.1993.1005. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, G. G. Pseudomonas aeruginosa Biofilm Formation in the CF Lung and Its Implications for Therapy. In Sriramulu, D. (ed), Cystic Fibrosis IntechOpen, Rijeka (2012).
- 3.Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2003;168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 4.Hauser AR, Jain M, Bar-Meir M, McColley SA. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin. Microbiol. Rev. 2011;24:29–70. doi: 10.1128/CMR.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malhotra S, Limoli DH, English AE, Parsek MR, Wozniak DJ. Mixed Communities of Mucoid and Nonmucoid Pseudomonas aeruginosa Exhibit Enhanced Resistance to Host Antimicrobials. MBio. 2018;9:e00275–18. doi: 10.1128/mBio.00275-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacKenzie T, et al. Longevity of patients with cystic fibrosis in 2000 to 2010 and beyond: survival analysis of the Cystic Fibrosis Foundation patient registry. Ann. Intern. Med. 2014;161:233–241. doi: 10.7326/M13-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017;15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molchanova N, Hansen PR, Franzyk H. Advances in Development of Antimicrobial Peptidomimetics as Potential. Drugs. Molecules. 2017;22:E1430. doi: 10.3390/molecules22091430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith, W. D. et al. Current and future therapies for Pseudomonas aeruginosa infection in patients with cystic fibrosis. FEMS Microbiol Lett364 (2017). [DOI] [PubMed]
- 10.Patsilinakos A, et al. Machine Learning Analyses on Data including Essential Oil Chemical Composition and In Vitro Experimental Antibiofilm Activities against Staphylococcus Species. Molecules. 2019;24:E890. doi: 10.3390/molecules24050890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Artini M, et al. Antimicrobial and Antibiofilm Activity and Machine Learning Classification Analysis of Essential Oils from Different Mediterranean Plants against Pseudomonas aeruginosa. Molecules. 2018;23:E482. doi: 10.3390/molecules23020482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papa R, et al. Anti-Biofilm Activities from Marine Cold Adapted Bacteria Against Staphylococci and Pseudomonas aeruginosa. Front. Microbiol. 2015;6:1333. doi: 10.3389/fmicb.2015.01333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perkel JM. Pick up Python. Nature. 2015;518:125–126. doi: 10.1038/518125a. [DOI] [PubMed] [Google Scholar]
- 14.Cafiso V, et al. Agr-Genotyping and transcriptional analysis of biofilm-producing Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2007;51:220–227. doi: 10.1111/j.1574-695X.2007.00298.x. [DOI] [PubMed] [Google Scholar]
- 15.Perez LRR, Barth AL. Biofilm production using distinct media and antimicrobial susceptibility profile of Pseudomonas aeruginosa. Braz. J. Infect. Dis. 2011;15:301–304. doi: 10.1016/s1413-8670(11)70196-9. [DOI] [PubMed] [Google Scholar]
- 16.Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 17.Celebi ME, Kingravi HA, Vela PA. A comparative study of efficient initialization methods for the k-means clustering algorithm. Expert. Syst. Appl. 2013;40:200–210. doi: 10.1016/j.eswa.2012.07.021. [DOI] [Google Scholar]
- 18.Lopez-Causape C, Rojo-Molinero E, Macia MD, Oliver A. The problems of antibiotic resistance in cystic fibrosis and solutions. Expert. Rev. Respir. Med. 2015;9:73–88. doi: 10.1586/17476348.2015.995640. [DOI] [PubMed] [Google Scholar]
- 19.Dodemont M, et al. Emergence of livestock-associated MRSA isolated from cystic fibrosis patients: Result of a Belgian national survey. J. Cyst. Fibros. 2019;18:86–93. doi: 10.1016/j.jcf.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Murray JL, Kwon T, Marcotte EM, Whiteley M. Intrinsic Antimicrobial Resistance Determinants in the Superbug Pseudomonas aeruginosa. MBio. 2015;6:e01603–01615. doi: 10.1128/mBio.01603-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev. Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Topa SH, et al. Cinnamaldehyde disrupts biofilm formation and swarming motility of Pseudomonas aeruginosa. Microbiology. 2018;164:1087–1097. doi: 10.1099/mic.0.000692. [DOI] [PubMed] [Google Scholar]
- 23.Vasireddy L, Bingle LEH, Davies MS. Antimicrobial activity of essential oils against multidrug-resistant clinical isolates of the Burkholderia cepacia complex. PLoS One. 2018;13:e0201835. doi: 10.1371/journal.pone.0201835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poma P, et al. Essential Oil Composition of Alluaudia procera and in Vitro Biological Activity on Two Drug-Resistant Models. Molecules. 2019;24:E2871. doi: 10.3390/molecules24162871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karumathil DP, Nair MS, Gaffney J, Kollanoor-Johny A, Venkitanarayanan K. Trans-Cinnamaldehyde and Eugenol Increase Acinetobacter baumannii Sensitivity to Beta-Lactam Antibiotics. Front. Microbiol. 2018;23:1011. doi: 10.3389/fmicb.2018.01011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosato A, et al. Elucidation of the synergistic action of Mentha Piperita essential oil with common antimicrobials. PLoS One. 2018;13:e0200902. doi: 10.1371/journal.pone.0200902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tetard A, Zedet A, Girard C, Plésiat P, Llanes C. Cinnamaldehyde Induces Expression of Efflux Pumps and Multidrug Resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2019;63:e01081–19. doi: 10.1128/AAC.01081-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikkelsen H, McMullan R, Filloux A. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One. 2011;6:e29113. doi: 10.1371/journal.pone.0029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerem E, Conway S, Elborn S, Heijerman H, Consensus C. Standards of care for patients with cystic fibrosis: a European consensus. J. Cyst. Fibros. 2005;4:7–26. doi: 10.1016/j.jcf.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Levin TP, Suh B, Axelrod P, Truant AL, Fekete T. Potential clindamycin resistance in clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus: report of a clinical failure. Antimicrob. Agents. Chemother. 2005;49:1222–1224. doi: 10.1128/AAC.49.3.1222-1224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palzkill T. Metallo-beta-lactamase structure and function. Ann N Y Acad Sci. 2013;1277:91–104. doi: 10.1111/j.1749-6632.2012.06796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meletis, G. & Bagkeri, M. Pseudomonas aeruginosa: Multi-Drug-Resistance Development and Treatment Options. In Basak, S. (ed), Cystic Fibrosis IntechOpen, Rijeka (2013).
- 33.Humphries RM, et al. CLSI Methods Development and Standardization Working Group Best Practices for Evaluation of Antimicrobial Susceptibility Tests. J. Clin. Microbiol. 2018;56:10. doi: 10.1128/JCM.01934-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kluyver, T. et al. Jupyter development t. Jupyter Notebooks? a publishing format for reproducible computational workflows, p 87–90. In Loizides, F., Scmidt, B. (ed), IOS Press (2016).
- 35.McKinney, W. Data Structures for Statistical Computing in Python, p 51–56. In Millman SvdWaJ (ed) (2010).
- 36.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J.Mach. Learn.Res. 2011;12:2825–2830. [Google Scholar]
- 37.Hunter JD. Matplotlib: A 2D graphics environment. Computing in Science & Engineering. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.