Significance
Celiac disease (CeD) is an autoimmune-like disorder that is driven by the ingestion of dietary gluten. There is a strong human leukocyte antigen (HLA) association with the disease, with the majority of CeD patients being HLA-DQ2.5 positive. While HLA-DQ2.2 is closely related to HLA-DQ2.5, it is less of a risk factor in CeD. Our understanding behind this differential disease association remained unclear. We give the molecular basis for the differential risks of the two HLA-DQ variants, and show that small differences in the peptide–HLA landscape cause profound differences in the responding T cell repertoires.
Keywords: celiac disease, human leukocyte antigen, X-ray crystallography, T cell receptor
Abstract
The highly homologous human leukocyte antigen (HLA)-DQ2 molecules, HLA-DQ2.5 and HLA-DQ2.2, are implicated in the pathogenesis of celiac disease (CeD) by presenting gluten peptides to CD4+ T cells. However, while HLA-DQ2.5 is strongly associated with disease, HLA-DQ2.2 is not, and the molecular basis underpinning this differential disease association is unresolved. We here provide structural evidence for how the single polymorphic residue (HLA-DQ2.5-Tyr22α and HLA-DQ2.2-Phe22α) accounts for HLA-DQ2.2 additionally requiring gluten epitopes possessing a serine at the P3 position of the peptide. In marked contrast to the biased T cell receptor (TCR) usage associated with HLA-DQ2.5–mediated CeD, we demonstrate with extensive single-cell sequencing that a diverse TCR repertoire enables recognition of the immunodominant HLA-DQ2.2-glut-L1 epitope. The crystal structure of two CeD patient-derived TCR in complex with HLA-DQ2.2 and DQ2.2-glut-L1 (PFSEQEQPV) revealed a docking strategy, and associated interatomic contacts, which was notably distinct from the structures of the TCR:HLA-DQ2.5:gliadin epitope complexes. Accordingly, while the molecular surfaces of the antigen-binding clefts of HLA-DQ2.5 and HLA-DQ2.2 are very similar, differences in the nature of the peptides presented translates to differences in responding T cell repertoires and the nature of engagement of the respective antigen-presenting molecules, which ultimately is associated with differing disease penetrance.
Celiac disease (CeD) is a heritable and chronic inflammatory disorder that is caused by a maladaptive immune response to cereal gluten proteins (1). While exhibiting features of food intolerance, the disease also has many autoimmune characteristics including highly disease-specific autoantibodies targeting the enzyme transglutaminase 2 (TG2). CeD patients, but not healthy subjects, have gluten-specific CD4+ T cells (2). These T cells, persisting for decades in patients (3), are considered drivers of the CeD pathophysiology that includes the formation of an inflammatory lesion in the proximal small intestine and the generation of autoantibodies (4). The gluten-specific T cells are uniquely restricted by disease-associated HLA-DQ allotypes, namely HLA-DQ2.5 (HLA-DQA1*05/HLA-DQB1*02:01), HLA-DQ2.2 (HLA-DQA1*02:01/HLA-DQB1*02:02), and HLA-DQ8 (HLA-DQA1*03:01/HLA-DQB1*03:02) (5–7). About 90% of patients affected by CeD are HLA-DQ2.5 positive, and those who do not carry HLA-DQ2.5 are equally often either HLA-DQ2.2 or HLA-DQ8. Interestingly, while the risk of the three disease-associated HLA-DQ allotypes are very different, the clinical presentations of patients with the different HLA-DQ allotypes are the same (7). This suggests that the differential risks relate to likelihood of generating an anti-gluten T cell response, and once the gluten-specific T cells are established and clonally expanded, exposure to gluten leads to the same pathophysiological reactions.
The gluten peptides recognized by T cells of CeD patients are posttranslationally modified. TG2 is essential in this process as the enzyme can convert glutamine residues in polypeptides to glutamate, mostly following a QXP recognition motif (8, 9). Gluten proteins are exceptionally rich in such sequence motifs but naturally sparse in negatively charged residues that is required for a high-affinity binding to peptide-binding pockets within the antigen-binding cleft of the disease-associated HLA-DQ allotypes (7). The TG2-mediated deamidation of gluten peptides leads to an increase in binding affinity and the formation of kinetically stable peptide–HLA-DQ complexes (10). Hence, this posttranslational modification is essential for the immunogenicity of these antigens (11–13), and the immune reactions inflicting damage that ensue T cell recognition of HLA-DQ gluten peptide complexes is likely the mechanism behind the absolute association of certain HLA-DQ allomorphs with CeD (7). Generally, T cell clones restricted by the three CeD-associated HLA allotypes recognize distinct and separate sets of gluten epitopes (4, 14, 15). This selectivity is based on the capacity of peptides to form kinetically stable peptide–HLA-DQ complexes as only peptides that form stable peptide complexes can elicit T cell clonal expansions in vivo (14, 16).
While HLA-DQ8 binds and presents gluten peptides with glutamate at anchor positions P1 and/or P9 (15, 17, 18), HLA-DQ2.5 and HLA-DQ2.2 binds and presents gluten peptides with glutamate residues at anchor positions P4, P6, or P7 (15, 19–23). Three known HLA-DQ2.2–restricted epitopes, namely DQ2.2-glut-L1 (PFSEQEQPV), DQ2.2-glia-α1 (QGSVQPQQL), and DQ2.2-glia-α2 (QYSQPEQPI), have sequences similar to HLA-DQ2.5–binding peptides, with the exception that they all carry serine at P3 (14, 24, 25). As seen for HLA-DQ2.5–restricted epitopes, the HLA-DQ2.2–restricted epitopes display a hierarchy with DQ2.2-glut-L1 being the epitope recognized by most T cells (24). In binding experiments with DQ2.2-glut-L1 variant peptides substituted at P3, HLA-DQ2.2 but not HLA-DQ2.5 showed preferential binding of the serine and threonine variants (14). An earlier study had indicated that HLA-DQ2.2, in contrast to HLA-DQ2.5, utilizes a P3 anchor (20), and a P3 anchor selectivity of HLA-DQ2.2 for serine or threonine was demonstrated when comparing the repertoires of endogenous peptide ligands eluted from purified HLA-DQ2.5, HLA-DQ2.2, and HLA-DQ7.5 molecules (25).
The HLA-DQ2 molecules, HLA-DQ2.5 and HLA-DQ2.2, are highly homologous. Previous studies have ascribed the differences in conferred risk associated with CeD as a result of the polymorphism in the DQα chain between HLA-DQ2.5 and HLA-DQ2.2 (14, 16). In particular, the Tyr22α to Phe22α polymorphism is located near the P3 peptide binding pockets of the HLA-DQ molecule. Here, we examined the role of Phe22α variant in HLA-DQ2.2 that influence peptide binding preferences and how the T cell receptors (TCRs) derived from DQ2.2 CeD patients engage the HLA-DQ2.2:DQ2.2-glut-L1 complex. We provide a molecular basis for these key interactions in HLA-DQ2.2–mediated CeD and demonstrate how this is different from HLA-DQ2.5– and HLA-DQ8–mediated epitope presentation and TCR recognition in CeD.
Results
DQ2.2-glut-L1–Specific TCR Repertoire.
We performed high-throughput DNA sequencing of rearranged TRAV and TRBV genes of the single HLA-DQ2.2:DQ2.2-glut-L1 tetramer binding CD4+ T cells isolated from six T cell lines (TCLs) of four CeD patients described in a previous study (14), specifically CD555 (TCL555.A.1.4 and TCL555.A.2.2), CD594 (TCL594.5.2 and TCL594.8.1), CD627 (TCL627.7.3), and CD1005 (TCL1005.1). Following processing of sequencing data, we obtained productive paired TCRαβ sequences of 549 cells that were categorized into 18 unique clonotypes (Table 1). Cells expressing paired identical nucleotide TRAV/TRBV genes are defined as a clonotype. We also obtained five unique clonotypes by sequencing TRAV/TRBV gene sequences of the T cell clones generated from the same four patients. Of those five unique clonotypes, cells expressing the same TCR as that of three unique T cell clones were observed as clonally expanded population in the single-cell sequencing data generated by tetramer-based sorting. In total, we generated 20 unique clonotypes of DQ2.2-glut-L1–specific T cells from tetramer-sorted single cells and DQ2.2-glut-L1–specific T cell clones.
Table 1.
DQ2.2-specific TCR sequences
| No. | Donor | TRAV | TRAJ | CDR3a | TRBV | TRBD | TRBJ | CDR3b | No. of cells | TCC |
| 1 | 1005 | 12-3 | 34 | CAMTLNTDKLIF | 4-2 | 1 | 1-2 | CASSLQGGDYGYTF | 1 | |
| 2 | 1005 | 38-1*01 | 32*02 | CAFMKEDGGATNKLIF | 20-1*01 | 1*02 | 2-5*01 | CSASPGNTGVAQYF | 5 | 1005.2.54 and 1005.2.60 |
| 3 | 1005 | 26-1 | 48 | CIVYLSNFGNEKLTF | 5-5 | 2 | 2-2 | CASSLIGGGELFF | 1 | — |
| 4 | 1005 | 29/DV5 | 42 | CAASAHYGGSQGNLIF | 12-5 | 1 | 1-1 | CASGPGTATEAFF | 1 | — |
| 5 | 1005 | 38-2/DV8 | 58 | CAYRSQETSGSRLTF | 20-1 | 1 | 2-5 | CSASWLGFGETQYF | 9 | — |
| 6 | 1005 | 38-2/DV8 | 58 | CAYTPLETSGSRLTF | 20-1 | 1 | 2-7 | CSARAPITGRFYEQYF | 4 | — |
| 7 | 1005 | 21 | 42 | CAVRPGGSQGNLIF | 4-2 | 2 | 2-5 | CASSLTTSEETQYF | 74 | — |
| 29/DV5 | 30 | CAASADLTNRDDKIIF | ||||||||
| 8 | 1005 | 38-1 | 42 | CAFMKGSYGGSQGNLIF | 7-8 | 1 | 2-7 | CASSRTANYEQYF | 28 | — |
| 40 | 40 | FLLGITSGTYKYIF | ||||||||
| 9 | 1005 | 26-1 | 54 | CIVRVAEGAQKLVF | 20-1 | 2-5 | CSATKETQYF | 1 | — | |
| 10 | 1005 | 26-1 | 54 | CIVRVAEGAQKLVF | 20-1 | 1 | 2-5 | CSATKETQYF | 3 | — |
| 11 | 1005 | 21*02 | 31*01 | CAVHTGARLMF | 7-3*01 | 1*01 | 2-3*01 | CASSHGASTDTQYF | NA | 1005.2.56, 1005.2.57, 1005.2.58, 1005.2.62, and 1005.2.65 |
| 12 | 555 | 26-1 | 54 | CIVRVAEGAQKLVF | 20-1 | — | 2-5 | CSATKETQYF | 3 | — |
| 13 | 555 | 6 | 26 | CALVRDYGQNFVF | 7-2 | 2 | 2-7 | CASSTSGGGARSYEQYF | 1 | — |
| 14 | 555 | 1-1*01 | 9*01 | CAWTGGGGFKTIF | 6-2*01 | 2*01 | 2-7*01 | CASRKPGGDYEQYF | 80 | 555A.1.4.38 |
| 15 | 555 | 21 | 58 | CAVIGTSGSRLTF | 7-3 | 1 | 2-3 | CASSSGAATDTQYF | 166 | — |
| 16 | 594 | 26-1 | 54 | CIVRPAQGAQKLVF | 29-1 | 2 | 1-2 | CSVSSGGEGGNGYTF | 1 | — |
| 17 | 594 | 9-2*01 | 57*01 | CASPQGGSEKLVF | 11-2*01 | 1*01 | 1-1*01 | CASSSGGWGGGTEAFF | 62 | 594.5.2.6† |
| 27 | 21 | CAGRDDFNKFYF | ||||||||
| 18 | 627 | 6 | 39 | CALGLNNNAGNMLTF | 10-3 | 1 | 2-5 | CAISPRQAETQYF | 1 | — |
| 19 | 627 | 21 | 11 | CAASGYSTLTF | 11-1 | 1 | 2-3 | CASTHGAGSDTQYF | 108 | — |
| 20 | 627 | 3*01 | 8*01 | CAVSGNTGFQKLVF | 28*01 | 2*02 | 1-5*01 | CASTSRGGNSNQPQHF | NA | 627.1.3.199 |
| DV1*01 | 12*01 | CALGATVMDSSYKLIF |
The table shows the total overview of unique TCRαβ paired sequences generated from high-throughput sequencing of HLA-DQ2.2:DQ2.2-glut-L1 tetramer-sorted single T cells and/or by Sanger sequencing of DQ2.2-glut-L1–specific TCCs. Due to uncertainty in the allele assignment in the TCRαβ sequences generated from single-cell TCR sequencing approach, the alleles for the V, D, and J genes are not shown for the TCRαβ sequences observed only in the single-cell TCR sequencing data. Three TCRαβ sequences generated from single-cell TCR sequencing (nos. 2, 14, and 17) were also found to be expressed by the DQ2.2-glut-L1–specific TCCs, which are mentioned in rightmost column. Two of the TCRαβ sequences (nos. 11 and 20) were observed only by Sanger sequencing of DQ2.2-glut-L1–specific TCCs (last column) and not observed in TCR sequencing data of HLA-DQ2.2:DQ2.2-glut-L1 tetramer sorted cells.
By performing Sanger sequencing, we observed that TCC594.5.2.6 expresses single TCRα chain (TRAV9-2*01/TRAJ57*01). However, by performing single-cell TCR sequencing, we were able to identify T cells that expressed an additional TCRα chain (TRAV27/TRAJ21).
We observed that few clonotypes dominate the DQ2.2-glut-L1–specific TCR repertoire in the TCLs derived from HLA-DQ2.2–positive CeD patients. In the majority of the patients, one or two expanded clonotypes dominated the T cell repertoire. However, we cannot exclude that some of this bias is the effect of in vitro culture of the TCLs. As the number of unique DQ2.2-glut-L1–specific TCRs is small and the dataset is obtained from TCLs generated by in vitro culture, caution should be exercised on generalizing features of the DQ2.2-glut-L1–specific TCR repertoire. In our fairly limited dataset, we observed that while some V genes like TRAV21, TRAV26-1, TRBV20-1, and TRBV7-2/3 genes were expressed in more than one CeD patient, there was no apparent V gene bias, preferential TRAV:TRBV pairing or conserved CDR3 features in DQ2.2-glut-L1–specific TCR repertoire. In two patients (CD1005, CD555), we observed T cells expressing TCRs with identical CDR3α and CDR3β amino acid sequences. The CDR3 of these public TCRs are encoded by different nucleotides, representing a phenomenon termed convergent recombination (26).
Among these HLA-DQ2.2:DQ2.2-glut-L1–specific T cell clones, four TCRs were expressed and successfully refolded: TCR 555 (TRAV1-1*01/TRBV6-2*01), TCR 594 (TRAV9-2*01/TRBV11-2*01), TCR 627 (TRAV3*01/TRBV28*01), and TCR 1005.2.56 (TRAV21*02/TRBV7-3*01) (Table 1 and SI Appendix, Table S1). These four HLA-DQ2.2-glut-L1 reactive TCRs do not contain a conserved arginine residue in the CDR loops that underpin the response to gluten antigens in HLA-DQ2.5:DQ2.5-glia-α2, HLA-DQ8:DQ8-glia-α1, and HLA-DQ8:DQ8.5-glia-γ1 reactive TCRs and displayed more diverse TRAV/TRBV gene usage (22).
Structural Overview of Two TCR:HLA-DQ2.2:DQ2.2-glut-L1 Complexes.
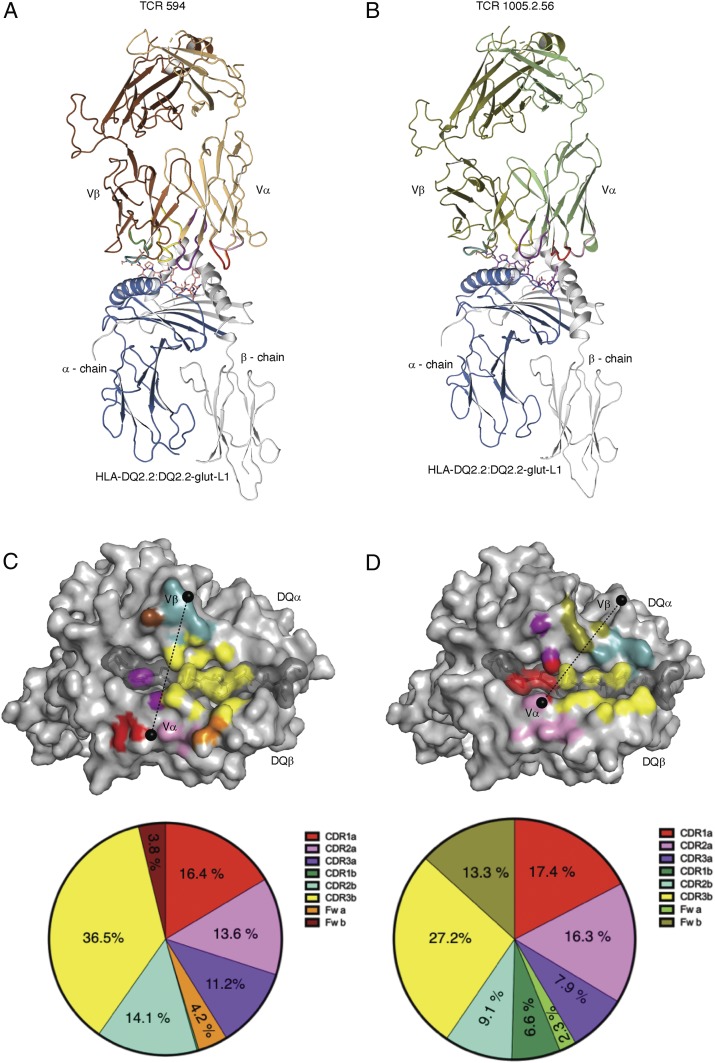
To establish how TCRs interacts with HLA-DQ2.2:DQ2.2-glut-L1, we determined the structure of the TCR 594:HLA-DQ2.2:DQ2.2-glut-L1 and TCR 1005.2.56:HLA-DQ2.2:DQ2.2-glut-L1 ternary complexes at 2.8 Å and 3.0 Å resolution, respectively (Fig. 1 and SI Appendix, Table S2). The TCR 594 docked centrally above HLA-DQ2.2:DQ2.2-glut-L1, ∼85° across the antigen-binding cleft, where the Vα and Vβ chains of the TCR lay above the β-helix and α-helix of HLA-DQ2.2, respectively (Fig. 1 A and C). The total buried surface area (BSA) upon ligation of TCR 594 with HLA-DQ2.2:DQ2.2-glut-L1 was ∼1,900 Å2. The Vα and Vβ domains of the TCR 594 contributed 45.4% and 54.6%, respectively, to the BSA at the interface (Fig. 1C). Notably 36.5% BSA arose from the CDR3β loop, which played a dominant role in contacting the epitope and HLA-DQ2.2 (Fig. 1C). There was a relatively even distribution of contacts among the CDR loops toward the HLA-DQ2.2 molecule, with the CDR1α, CDR2α, CDR3α, and CDR2β loops, contributing 16.4%, 13.6%, 11.2%, and 14.1% BSA to the interface, respectively.
Fig. 1.
Structures of TCR-HLA-DQ2.2:DQ2.2-glut-L1 ternary complexes. TCR 594 (A) and TCR 1005.2.56 (B). Footprint of the CDR loops of the TCR 594 (C) and TCR 1005.2.56 (D) on the HLA-DQ2.2:DQ2.2-glut-L1. CDR1α, CDR2α, and CDR3α are colored in red, pink, and purple, respectively, while the CDR1β, CDR2β, and CDR3β are colored in green, light teal, and yellow, respectively. The TCR framework for each Vα- and Vβ-chain is colored orange and brown, respectively, for TCR 594, and colored lime and deep-olive, respectively, for TCR 1005.2.56. The pie chart below show percentage of the CDR loops contributing to the buried surface area.
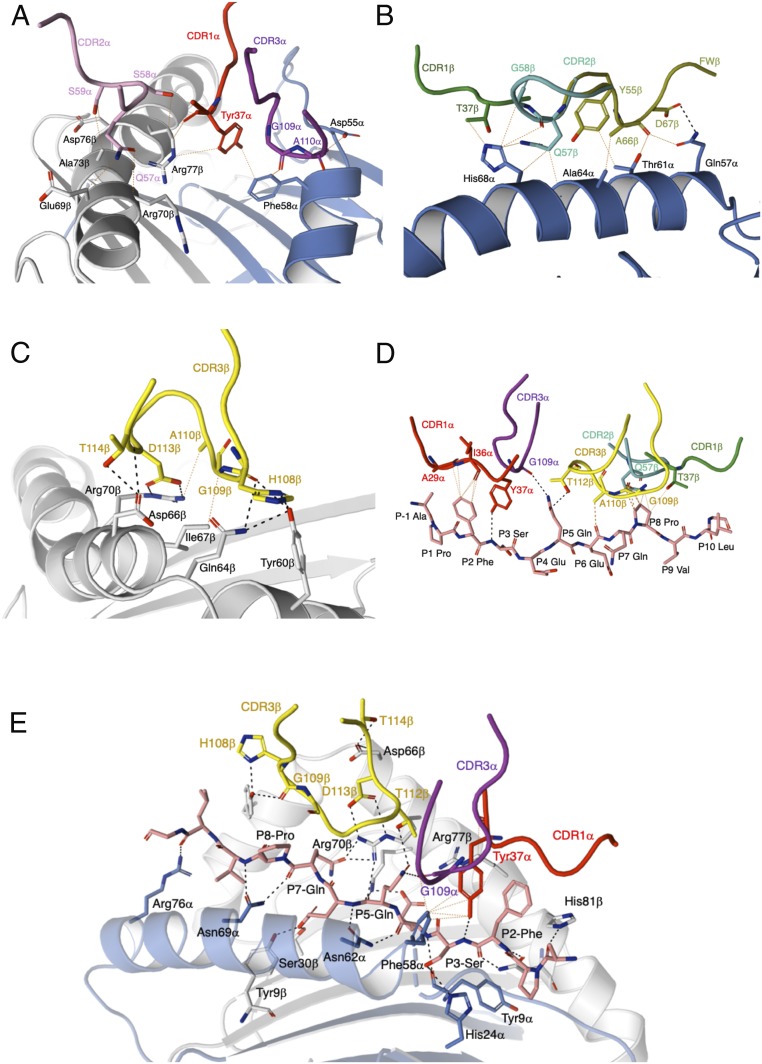
The TCR 594 α-chain interacted with the HLA-DQ2.2 β-chain, spanning from Asp66β to Arg80β (Fig. 2A and SI Appendix, Table S3). Here, Tyr30α from the CDR1α loop made van der Waals (vdw) contacts with Asp76β and Arg80β of HLA-DQ2.2, while Thr57α and Asn58α from the CDR2α loop made vdw contacts with Glu69β, Arg72β, and Ala73β (Fig. 2A). The neighboring Vα framework residue Lys55α further stabilizes the CDR2α–HLA-DQ2.2 interactions by forming a salt bridge with Asp66β and Glu69β. Regarding the TCR β-chain, the CDR2β loop made the majority of the contacts with the HLA-DQ2.2 α-chain (Fig. 2B). Namely, multiple vdw contacts between Gln57β, Asn58β, and Gly64β from the TCR and the HLA-DQ2.2 residues Lys39α, Gln57α, and Thr61α were observed. Moreover, additional vdw contacts were made between the Vβ framework residue Val66β and the HLA-DQ2.2 residue Asp55α (Fig. 2B). Accordingly, the TCR 594 engaged the HLA-DQ2.2 molecule in a standard docking mode, with varied contributions from each of the germ-line–encoded CDR loops.
Fig. 2.
TCR 594 CDR loops in contact with the HLA-DQ2.2:DQ2.2-glut-L1. (A) The CDR1α and CDR2α loop contacting HLA-DQ2.2 β-chain. (B) The CDR2β contacts with HLA-DQ2.2 α-chain. (C) CDR3α and CDR3β interactions with the DQ2.2-glut-L1 peptide. (D) Key interaction around the P5-Gln residue of the DQ2.2-glut-L1 epitope.
The TCR 1005.2.56 also adopted a canonical docking position at ∼60° across the antigen-binding cleft (Fig. 1 B and D). The total BSA upon ligation of TCR 1005.2.56 with HLA-DQ2.2:DQ2.2-glut-L1 was slightly higher than TCR 594 with ∼2,030 Å2, where the contribution of the Vα and Vβ domains of the TCR 1005.2.56 to the BSA at the interface were 43.8% and 56.2%, respectively. The TCR 1005.2.56 CDR loops made relatively even contact across the HLA-DQ2.2:DQ2.2-glut-L1 (Figs. 1D and 3). However, the TCR 1005.2.56 made contact with the DQ2.2-glut-L1 epitope with both germ-line–encoded CDR1α and nongerm-line–encoded CDR3β loop (Figs. 1D and 3D), rather than just the CDR3 residues as seen in the TCR594 ternary complex.
Fig. 3.
TCR 1005.2.56 CDR loops in contact with the HLA-DQ2.2:DQ2.2-glut-L1. (A) The CDR1α, CDR2α, and CDR3α loops contact the HLA-DQ2.2 α-chain and β-chain. The CDR1β, CDR2β (B), and CDR3β (C) interactions with the HLA-DQ2.2 α-chain and β-chain, respectively. (D) The CDR1α and CDR3β loops of the TCR 1005.2.56 that interacted the DQ2.2-glut-L1 peptide. The CDR3α, CDR1β, and CDR2β loop contacts with the peptide. (E) Key interaction around the P5-Gln and P7-Gln residue of the DQ2.2-glut-L1 epitope.
Compared to the TCR 594, the CDR1α loop of the TCR 1005.2.56 shifted closer to the peptide antigen (SI Appendix, Fig. S2A), where Tyr37α made contacts with Phe58α and Arg77β from HLA-DQ2.2-glut-L1, as well as forming a hydrogen bond with the main chain of the peptide P3-Ser and vdw interactions with the P2-Phe (Fig. 3 A and D and SI Appendix, Tables S4 and S5). This further allows the CDR3α loop to make vdw contact with the Asp55α and Phe58α from HLA-DQ2.2 (Fig. 3A). The CDR2α loop of the TCR 1005.2.56 made similar contacts with the Glu69β, Arg70β, and Ala73β from HLA-DQ2.2 through vdw interactions (Fig. 3A), comparable to the TCR 594 interactions (Fig. 2A). All three CDRβ loops from the TCR 1005.2.56 also interacted with the HLA-DQ2.2 molecule (Fig. 3 B and C). Similar vdw contacts between CDR1β and CDR2β with a stretch of the helix of the HLA-DQ2.2 α-chain were observed (Fig. 3B). The Vβ framework residue Tyr55β, Ala66β, and Asp67β further supported the interactions between CDR1β and CDR2β and the HLA-DQ2.2 by making hydrogen bond with HLA-DQ2.2 residue Gln57α and additional vdw contacts with Thr61α, Ala64α, and His68α (Fig. 3B). The TCR 1005.2.56 CDR3β also made extensive hydrogen bonds with the HLA-DQ2.2 β-chain (Fig. 3C). Overall, all CDR loops from both Vα and Vβ domains of the TCR 1005.2.56 made a fairly even number of contacts with both the HLA-DQ2.2 as well as the DQ2.2-glut-L1 epitope (Fig. 3E).
DQ2.2-glut-L1 Peptide-Mediated TCR Interactions.
The DQ2.2-glut-L1 epitope residues that were exposed for TCR contact were P2-Phe, P5-Gln, and P7-Gln (Fig. 2C and SI Appendix, Table S5). The nongerm-line–encoded CDR3 loops of the TCR 594 contacted all three positions, contributing to 22% BSA of the peptide HLA interface (SI Appendix, Table S5). Here, the CDR3α loop made limited contact, interacting only with the P2-Phe in the DQ2.2-glut-L1 peptide (Fig. 2C). However, there was an intricate network of polar interactions between the Glu112α residue of CDR3α loop, Arg70β of the HLA-DQ2.2, and the P5-Gln in the DQ2.2-glut-L1 peptide (Fig. 2D). This P5-Gln of the DQ2.2-glut-L1 peptide also formed a hydrogen bond with Gly109β of the CDR3β loop, and all together, appeared as a focal point for the CDR3 α/β-loops of the TCR 594 on to the HLA-DQ2.2:DQ2.2-glut-L1 (Fig. 2D).
The TCR 594 interacted with P7-Gln mostly via its CDR3β loop (Fig. 2C and SI Appendix, Table S5). The small glycine residues in the CDR3β loop, namely Gly109β, Gly110β, and Gly112β which flanked the Trp111β, permitted close contact with the peptide and formed hydrogen bond with the side chain of P5-Gln and P7-Gln (Fig. 2 C and D). The CDR3β loop of the TCR 594 also displayed extensive interactions with the surrounding residues, where Trp111β formed a hydrogen bond with the side chain of Asn62 on the HLA-DQ2.2 α-chain, while Glu116β formed a hydrogen bond with Asp66 on the HLA-DQ2.2 β-chain (Fig. 2D). Trp111β of the CDR3β loop made additional vdw contact with HLA-DQ2.2 α-chain at residues Phe58α, Thr61α, and Val65α. Further vdw interactions were made between the Gly112β, Gly113β, and Thr115β of the CDR3β loop and the HLA-DQ2.2 β-chain including residues Asp66β, Thr61β, and Ile67β. Interestingly, the positioning of the Trp111β in the CDR3β of the TCR 594 is analogous to that of the conserved Arg109 found in the CDR3β loop of the HLA-DQ2.5:DQ2.5-glia-α2 reactive TCRs (SI Appendix, Fig. S1), making similar contact to HLA-DQ2.2 residues Phe58α, Thr61α, and Asn62α, as well as gluten epitope residues at position P5 and P7 (Fig. 2 C and D).
Gly109α and Thr112β from the TCR 1005.2.56 also interacted with gluten epitope residues at position P5 (Fig. 3 D and E). However, the overall TCR:peptide interaction was further extended by contacts between the germ-line–encoded CDR loops and the DQ2.2-glut-L1 peptide residues P2-Phe, P3-Ser, P6-Glu, and P8-Pro, including Tyr37α from the CDR1α loop, Gln57α from the CDR2α loop, and Thr37α from the CDR1α loop (Fig. 3D). Notably, the P7-Gln residue in the TCR1005.2.56:HLA-DQ2.2:DQ2.2-glut-L1 ternary complex was orientated toward the antigen-binding cleft and formed a hydrogen bond with Arg70β from HLA-DQ2.2 (Fig. 3E) and was not involved in any direct TCR contact. In TCR 594:HLA-DQ2.2:DQ2.2-glut-L1, Arg70β forms a salt bridge with the Glu112α from the CDR3α loop and made polar contacts with the P5-Gln of the DQ2.2-glut-L1 (Fig. 2D). In the TCR1005.2.56:HLA-DQ2.2:DQ2.2-glut-L1 ternary complex, however, the HLA-DQ2.2 residue Arg70β had rotated toward the P7 peptide binding pocket, forming hydrogen bond with P7-Gln, pulling it inwards, while making hydrogen bond with Thr112β and Asp113β from the CDR3β loop of TCR 1005.2.56 (Fig. 3 D and E). Overall, the TCR 1005.2.56 contacts with the HLA-DQ2.2:DQ2.2-glut-L1 are more well distributed along the peptide compared to TCR 594 whose interactions focused on the central region of the peptide.
Comparison of HLA-DQ2.2:DQ2.2-glut-L1 and HLA-DQ2.5:DQ2.5-glia-α2.
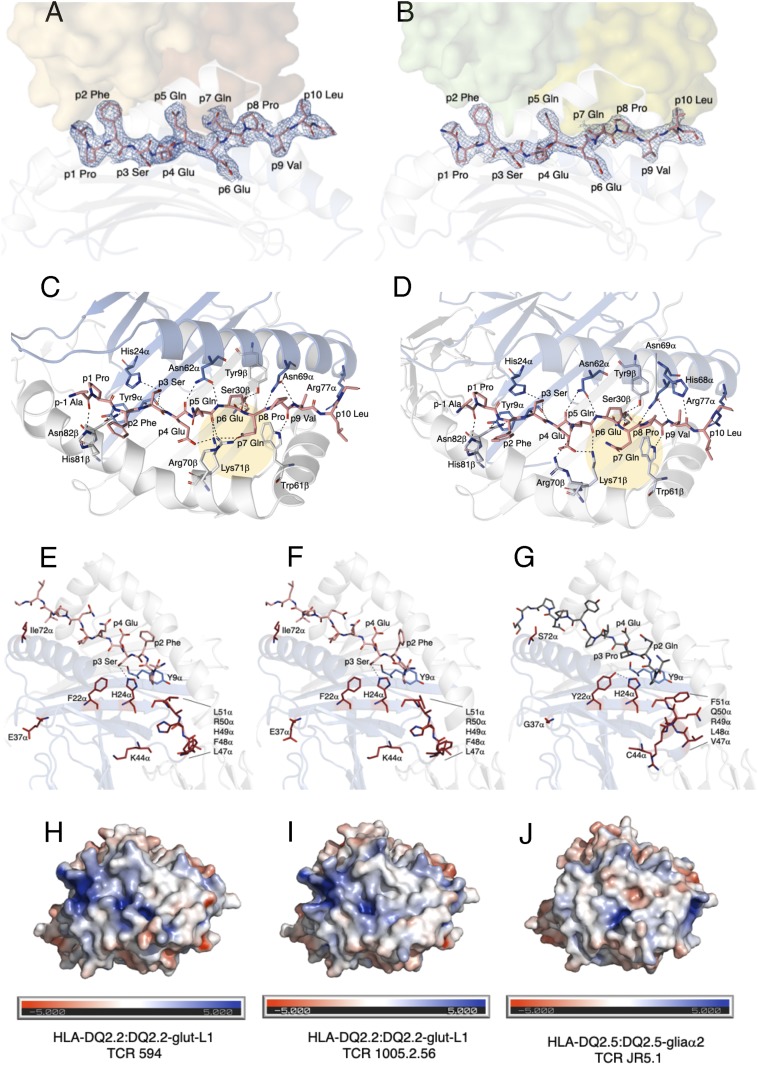
The DQ2.2-glut-L1 peptide (PFSEQEQPV) used in this study was derived from low molecular weight glutenin protein subsequent to TG2 mediated deamidation of the P4 and P6 glutamine residues in the native peptide sequence (PFSQQQQPV) to glutamate (14, 24). Crystal structures of HLA-DQ2.5:DQ2.5-glia-α1 and HLA-DQ2.5:DQ2.5-glia-α2 reveal that lysine at position 71 of the HLA-DQ2.5 β-chain is involved in binding of the P4-Glu and P6-Glu (22, 23, 27). The electron density maps of the DQ2.2-glut-L1 peptide recognized by TCR 594 (Fig. 4A) and TCR 1005.2.56 (Fig. 4B) were unambiguous. A more comprehensive hydrogen bonding network is shown from the view of a TCR for these two ternary complexes in Fig. 4 C and D, corresponding to P1 to P9 residues of the DQ2.2-glut-L1 peptide shown in Fig. 4 A and B. Similar HLA-peptide contacts were observed in HLA-DQ2.2:DQ2.2-glut-L1, where the P4-Glu and P6-Glu are buried deep into the binding pockets, acting as anchor residues, interacting with Tyr9β, Ser30β, and Lys71β of the HLA-DQ2.2 (Fig. 4 C and D).
Fig. 4.
Comparison of HLA-DQ2.2-glut-L1 and HLA-DQ2.5 in complexed with gluten epitopes. The DQ2.2-glut-L1 peptide is bound in the peptide-binding groove, with carbon colored in salmon, nitrogen colored in blue, and oxygen colored in red. The TCR 594 (A) and TCR 1002.56 (B) docked on top of the HLA-DQ2.2:DQ2.2-glut-L1 are shown as solid surface, and each of the TCR αβ pair were colored in brown/orange and green/lime, respectively. The peptide’s 2Fo-Fc electron density map is shown in blue and contoured to 1 σ. Hydrogen bond interactions between HLA-DQ2.2 and the DQ2.2-glut-L1 epitope in the TCR 594:HLA-DQ2.2:DQ2.2-glut-L1 ternary complex (C) and in the TCR 1005.2.56:HLA-DQ2.2:DQ2.2.-glut-L1 ternary complex (D) are shown. Comparison of the DQ2.2-glut-L1 peptide bound to HLA-DQ2.2 in the two ternary complexes solved showed minor movement in P7-Gln residues, highlighted in yellow circle. Polymorphic residues on the ectodomain of HLA-DQ differing between HLA-DQ2.2 (E and F) and HLA-DQ2.5 (G) are represented in stick and colored in fire-brick. The His24α interacts with P3-Ser in DQ2.2-glut-L1 (E and F) but with Tyr22α in HLA-DQ2.5 (G). The solvent-accessible electrostatic potential was calculated for HLA-DQ2.2:DQ2.2-glut-L1 in complexed with TCR 594 (H) or TCR 1005.2.56 (I) and HLA-DQ2.5:DQ2.5-glia-α-2 (J). Electrostatic calculations were performed using APBS (±5 kT/e).
Upon superposition, HLA-DQ2.2:DQ2.2-glut-L1 and HLA-DQ2.5.DQ2.5-glia-α2 showed no major structural differences, with an rmsd of 0.50 Å. In their membrane distal domains the HLA-DQ2.2 and HLA-DQ2.5 have identical DQβ chains and differ by 10 residues in their DQ α-chains (Fig. 4 E–G). None of these polymorphic residues of HLA-DQ2.2 made direct contact with the DQ2.2-glut-L1 epitope, or with the TCR in both of the TCR 594:HLA-DQ2.2:DQ2.2-glut-L1 and TCR 1005.2.56:HLA-DQ2.2:DQ2.2-glut-L1 complexes solved in this study (Fig. 4 E and F). The only key polymorphic residue that is in contact with the DQ2.2-glut-L1 peptide is the Phe/Tyr substitution in position 22 of the DQ α-chain. Here, we clearly show that the presence of P3-Ser in HLA-DQ2.2:DQ2.2-glut-L1 forms a hydrogen bond to side chain of His24α and main chain of Tyr9α of the HLA-DQ2.2 molecule (Fig. 4 E and F). By contrast, in HLA-DQ2.5, Tyr22α hydrogen bonds with the His24α (Fig. 4G) (22, 23). Moreover, these two HLA-DQ2.2:DQ2.2-glut-L1 complexes displayed relatively conserved electrostatic-potential surfaces for TCR interaction (Fig. 4 H and I). Hence, such selective P3-Ser peptide binding preferences is attributed to a hydrogen-bonding network whose functionality is differentially influenced by tyrosine and phenylalanine. While the polymorphic Phe22α is located at the base of the HLA peptide binding cleft and has no direct contact with the TCR, it provides a basis for understanding why P3-Ser binds more favorably to HLA-DQ2.2 compared to HLA-DQ2.5. These structural data are in keeping with a model previously presented (14).
Role of P3-Ser as an Anchor in Peptide Binding to HLA-DQ2.2.
To further investigate the role of P3-ser as an anchor residue for HLA-DQ2.2, we performed a competitive peptide binding assay and compared the binding of DQ2.2-glut-L1 and 10 different P3-Ser substituted peptide variants (SI Appendix, Table S6) to HLA-DQ2.2 (Fig. 5). We contrasted the results with testing the same peptides for binding to HLA-DQ2.5 (Fig. 5).
Fig. 5.
Binding of unsubstituted and P3-susbstituted variants of the DQ2.2-glut-L1 epitope to HLA-DQ2.2 and HLA-DQ2.5. Synthetic peptides containing the DQ2.2-glut-L1 epitope (Ac-QQPPFSEQEQPVLPQ, nine amino acid core sequence underlined), and variants substituted at P3 serine were tested as competitor peptides. The inhibitory effect of the competitor peptides is shown as IC50 values. One 10-fold titration experiment and three 4-fold titration experiments were performed. (A) Representative data showing results of one of the three fourfold titration experiments. (B) Results from all four independents experiments depicted as relative binding capacities (i.e., compared to the unsubstituted DQ2.2-glut-L1 epitope). The relative binding values from each experiment are shown as dots with bars representing mean values. A missing dot for some peptides is due to the titration curve not reaching its relevant IC50 value. Error bars, SD.
Among all of the 11 peptides with different amino acids at P3 of the DQ2.2-glut-L1 epitope, the peptide containing P3-Ser displayed the best binding to HLA-DQ2.2. Between the 10 P3-Ser–substituted peptides, threonine substitution had the least impact. The variant with cysteine also was a reasonable binder. A possible explanation why threonine and cysteine could serve as P3 anchors for HLA-DQ2.2 could be that their side chain can establish the above-mentioned hydrogen bond to the side chain of His24α and the main chain of Tyr9α. Further to this notion, α-aminobutyric acid, which is structurally similar to serine except for lacking the hydroxyl group, bound less well than the serine variant. Notwithstanding, α-aminobutyric acid was better than many of the other amino acids tested, indicating additional importance of size and hydrophobicity for optimal anchoring into the P3 pocket of HLA-DQ2.2. The patterns of binding of the 11 DQ2.2-glut-L1 variants to HLA-DQ2.5 was very different for that of HLA-DQ2.2 with P3-Ser not being preferred. Taken together, the binding experiments substantiate the notion of P3-Ser as a key anchor for peptide binding to HLA-DQ2.2 with the hydroxyl group of the serine side being functionally important.
TCR Affinity Measurements for the HLA-DQ2.2:DQ2.2-glut-L1 Complex.
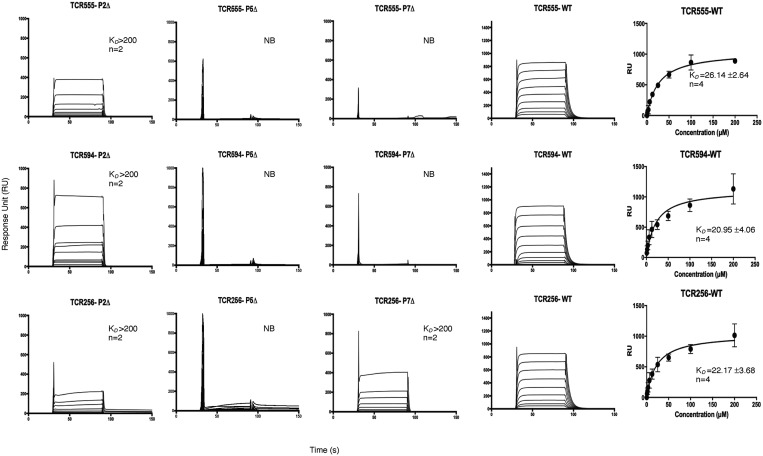
As determined by surface plasmon resonance (SPR), the steady-state affinity (KD) of the TCR 555, TCR 594, and TCR 1005.2.56 for the HLA-DQ2.2:DQ2.2-glut-L1 were 26.14 ± 2.64, 20.95 ± 4.06, and 22.17 ± 3.68 μM, respectively (Fig. 6). These values corroborate previous T cell proliferation studies (14). None of these TCRs tested showed any binding to HLA-DQ2.5:CLIP1 or HLA-DQ2.2:DQ2.2-glia-α2, thereby highlighting the lack of cross-reactivity of these TCRs.
Fig. 6.
Affinity measurement of TCR 555, 594, and 1005.2.56. Affinity for HLA-DQ2.2:DQ2.2-glut-L1 WT and the P2-Ala, P5-Ala, and P-7Ala epitope mutants were measure via SPR. Two to four independent experiments (with the number of replicates shown as n numbers) were carried out for each of the TCR against each of HLA-DQ2.2-glut-L1 WT and epitope mutants. Binding curve showed represent an independent experiment of TCR 555, TCR 594, TCR 1005.2.56 (labeled as TCR256) binding to HLA-DQ2.2:DQ2.2-glut-L1. All data were combined for each TCR and a one-site specific-binding model was used for curve fitting. HLA-DQ2.5-CLIP was used as negative control and acted as baseline reference value. Error bars, SD. NB, no binding.
To determine the extent of peptide sensitivity of these TCRs, we generated three peptide mutants corresponding to the solvent-exposed residues at position P2, P5, and P7 by replacing them with an alanine. The impact of these substitutions was measured by SPR. The PheP2Ala mutation reduced the affinity by at least 10-fold (KD > 200 μM), whereas the GlnP5Ala mutation abolished TCR binding to HLA-DQ2.2-glut-L1. This is consistent with the crystal structure where P5-Gln played a central role in mediating TCR 594 and TCR 1005.2.56 interaction in HLA-DQ2.2:DQ2.2-glut-L1 (Figs. 2 C and D and 3 D and E). The GlnP7Ala mutation abolished the binding of TCR 555 and TCR 594, whereas the TCR 1005.2.56 showed weak binding (KD > 200 μM) to this mutant, suggesting a different mode of interaction with this TCR. Indeed, as shown in the crystal structure, the P7-Gln in the TCR1005.2.56:HLA-DQ2.2:DQ2.2-glut-L1 ternary complex did not make any direct contact with the TCR, contrary to the TCR594:HLA-DQ2.2:DQ2.2-glut-L1 ternary complex (Fig. 3 D and E).
Discussion
HLA-DQ2.2 differs from HLA-DQ2.5 and HLA-DQ8 in gluten peptide-binding preferences and CeD risk association. Immunodominant gluten epitopes that binds stably to HLA-DQ2.5 were found to bind unstably to HLA-DQ2.2 and, furthermore, did not raise T cell responses in vivo in CeD patients carrying HLA-DQ2.2 (14, 16). Rather T cell clones from CeD patients expressing HLA-DQ2.2 recognized a unique set of gluten epitopes that commonly had a serine residue at position P3 (14, 16). Here, we provide a molecular basis for the T cell-mediated response to an HLA-DQ2.2–restricted immunodominant gluten epitope. We show that, in contrast to HLA-DQ2.5– and HLA-DQ8–mediated CeD, a diverse T cell repertoire underpins the response to HLA-DQ2.2 presenting the glut-L1 epitope. Moreover, we provide a structural basis underpinning this diverse TCR usage, by determining the crystal structures of two TCR:HLA-DQ2.2:DQ2.2-glut-L1 complexes.
Several studies have provided insight into TCR recognition of gluten epitopes presented by HLA-DQ2.5, HLA-DQ8, and HLA-DQ8.5. For instance, the HLA-DQ8:DQ8-glia-α1 reactive TCRs exhibit biased TRAV26-2/TRBV9-1 gene usage (18), and a nongerm-line–encoded arginine residue in either the CDR3α or CDR3β loop (23, 28). Moreover, TRAV20+/TRBV9+ TCRs reactive with HLA-DQ8:DQ8-glia-α1 and HLA-DQ8:DQ8.5-glia-γ1 also possessed a key arginine residue in their CDR1α loops (28, 29). All TRAV26-1/TRBV7-2+ TCRs specific for DQ2.5-glia-α2 had a nongerm-line–encoded arginine at position 109 of the CDR3β loop, which forms key interactions with the P5-Leu and P7-Tyr residues found within the gliadin epitopes (22). The HLA-DQ2.5:DQ2.5-glia-α1 reactive TCRs also shows biased TCR gene usage, TRAV4 and TRBV20-1 or TRBV29-1 (22, 30), but they do not carry a conserved arginine that is essential for recognition (31, 32). Ensuing structural studies have identified specific structural features that provided a basis for the selection of biased TCR usage and the preferential usage of the Arg residue (22, 23).
The TCR594:HLA-DQ2.2:DQ2.2-glut-L1 interactions resembled some features of both TCR S2 and TCR S16 that recognized HLA-DQ2.5:DQ2.5-glia-α1 and HLA-DQ2.5:DQ2.5-glia-α2, respectively. Like the S2 TCR, the germ-line–encoded CDRs on the TCR 594 Vα possesses a tyrosine that make similar interactions with the HLA-DQ2 β-chain helix. Contrary to TCR S2, the TCR 594 interaction with the P5-Gln and P7-Gln was reminiscent of TCR S16 and JR5.1 recognition of the HLA-DQ2.5:DQ2.5-glia-α2 epitope. Here, although the TCR594 did not possess a conserved arginine residue within the CDR3β loop, Trp111β in the CDR3β loop of the TCR 594 played an analogous role, forming extensive interactions with the HLA-DQ2.2:DQ2.2-glut-L1 complex (SI Appendix, Fig. S1). The nature of the contacts between the TCR 594 and the HLA-DQ2.2:DQ2.2-glut-L1 also provided a basis for understanding the lack of cross-reactivity toward HLA-DQ2.5:DQ2.5-glia-α2. Indeed, the HLA-DQ2.2:DQ2.2-glut-L1 reactive TCRs studied here appeared to be epitope-specific and epitope-driven.
However, the TCR1005.2.56-HLA-DQ2.2-glut-L1 interactions resembled TCR Bel502 and Bel602 that recognized HLA-DQ8-glia-α1 and HLA-DQ8-glia-γ1, respectively (28, 29) (SI Appendix, Fig. S2). Here, TCR Bel502 and Bel602 possess a conserved Arg37 in the CDR1α loop that interacts with the backbone of the gliadin peptide near the N termini. The TCR 1005.2.56, like TCR 594, did not bear an arginine in the CDR3β loop nor in the CDR1α loop. However, the Tyr37α in the CDR1α loop of the TCR1005.2.56 sat atop of the N terminus of the DQ2.2-glut-L1 epitope in the same manner as the conserved Arg37α in the CDR1α loop of TCR Bel502 and Bel602 (SI Appendix, Fig. S2B).
Despite HLA-DQ2.2 compensating for the substitution of Tyr22α by selectively binding to gluten epitopes with P3-Ser, it bears a lower risk of CD in comparison to HLA-DQ2.5. A higher chance of proteolytic degradation and stricter binding requirements are likely what limits the repertoire of potential HLA-DQ2.2 epitopes. In addition, the DQ2.2-glut-L1 has a chymotrypsin cleavage site between its P2 and P3 residues, making it more susceptible to proteolytic degradation. The requirement of the deamidation of glutamine residues at P4 and/or P6 for optimal binding to HLA-DQ2.2 further also limits the number of candidate antigenic epitopes in gluten for individuals with this allotype. These restrictions likely limit the bioavailability of epitopes that could bind to HLA-DQ2.2.
In summary, we have investigated the structural basis of how HLA-DQ2.2–restricted and CeD-derived TCRs bind to HLA-DQ2.2 complexed with the immunodominant DQ2.2-glut-L1 epitope. The single polymorphic residue in HLA-DQ2.2 (Phe22α) influences the hydrogen bond network within the peptide-binding groove of HLA-DQ2.2 and selectively bind to gluten epitopes that carry a P3-serine as anchor residue. What is remarkable is that while the TCR-accessible features of the antigen-binding clefts of HLA-DQ2.5 and HLA-DQ2.2 are very similar, the nature of the bound epitopes generate markedly different responding TCR repertoires. In HLA-DQ2.5–mediated CeD, the response is characterized by biased TCR usage, while in HLA-DQ2.2–mediated CeD, a diverse TCR response is generated. Thus, relatively subtle differences in the peptide-centric atomic landscape can have profound effects on the ensuing T cell response, which is correlated with disease pathogenesis.
Methods
Patient Material.
We characterized T cells isolated from gut biopsy-derived T cell lines of four DQ2.2+ subjects that have been described previously (14). According to the Oslo definitions of CeD and related terms (33), three of these subjects (CD555, CD594, and CD627) fulfilled the criteria of CeD while one subject (CD1005) fulfilled the criteria of potential CeD having normal gut histology but highly positive anti-TG2 serology. The subjects gave written informed consents for participation in the previous study, and updated written informed consent forms were obtained for this study. The responsible clinicians had access to sensitive patient data, whereas the research was done on coded material. The study protocol was approved by the Regional Ethical Committee (South East) for Medical and Health Research Ethics (present project ID: 6544). From the T cell lines, we sorted single T cells with HLA-DQ tetramers for TCR sequencing and TCRs of already-established T cell clones were sequenced (TCC627.1.3.199, TCC594.5.2.6, and TCC555A.1.4.38 from ref. 14 and TCC1005.2.54, TCC1005.2.56, TCC1005.2.57, TCC1005.2.58, TCC1005.2.60, TCC1005.2.62, and TCC1005.2.65 from ref. 34).
Tetramer Staining, Surface Antibody Staining, and Fluorescence-Activated Cell Sorting.
Tetramers of recombinant HLA-DQ2.2 covalently linked with DQ2.2-glut-L1 peptide (QPPFSEQEQPVL, underlined 9-mer core amino acid sequence) and CLIP1 (PVSKMRMATPLLMQA, negative control for nonspecific binding) were produced as per Dorum et al. (24) and conjugated with phycoerythrin (PE)-labeled streptavidin (Invitrogen) and allophycocyanin (APC)-labeled streptavidin (ProZyme), respectively. Cryopreserved T cell lines were thawed and stained with PE-conjugated HLA-DQ2.2:DQ2.2-glut-L1 (10 µg/mL) and APC-conjugated HLA-DQ2.2: CLIP1 (10 µg/mL) for 2 h at 37 °C. Following tetramer staining, the cells were stained with the mix of surface antibodies for fluorescence-activated cell sorting (FACS). We sorted live singlets that were CD3+, CD8−, CD4+, HLA-DQ2.2:DQ2.2-glut-L1 tetramer+, and HLA-DQ2.2:CLIP1 tetramer− (SI Appendix, Fig. S3). The cells were sorted in 96-well plate containing lysis buffer for single-cell TCR sequencing. The sorting was performed with a FACS Aria II instrument (BD Biosciences) at the Flow Cytometry Core Facility (Oslo University Hospital), and the flow cytometry data were analyzed with FlowJo software (FlowJo LLC). The following antibodies were used in the study: CD3-Superbright 600 (eBioscience), CD4-APC-H7 (BD Biosciences), and CD8-PerCP (eBioscience). LIVE/DEAD marker fixable violet stain (Thermo Fischer Invitrogen) was used to exclude dead cells.
TCR Sequencing of T Cell Clones.
Sequences of rearranged TCR-α and TCR-β genes of T cell clones were obtained by Sanger sequencing using a protocol as described (35).
Single-Cell TCR Gene Sequencing and Processing of TCR Gene Sequences.
We used a previously published protocol, based on nested PCR amplification using multiplex TRAV and TRBV primers, for single-cell TCRαβ sequencing (3, 36). The processing of the raw sequences generated from Illumina NGS was carried out as described (3). Sequencing was performed using Illumina MiSeq (250 bp PE) platform at the Norwegian Sequencing Centre (Oslo University Hospital).
Competitive Peptide Binding Assays.
The competitive peptide binding assay was performed as described previously (14, 37). In brief, the Epstein–Barr virus (EBV)-transformed B-cell lines VAVY (#9023) and MOU (#9050) were used as the source of HLA-DQ2.5 and HLA-DQ2.2 molecules. Microtiter plates (96 well, Nunc) were coated with an anti-DQ mAb (mAb and SPV-L3; 2 µg per well) as capture antibody for overnight at 4 °C. The wells were then blocked with phosphate buffered saline/gelatin (0.2%) at 37 °C for 5 h. The plates were washed, and cell lysate of EBV cells (equivalent of 4 million cells) was added to wells and incubated at 4 °C overnight. The plates were washed and biotinylated indicator peptide P418 (EPRAPWIEQEGPEYW-biotin, 0.5 µM) together with unlabeled competitor peptides (GL Biochem) were added in binding buffer with protease inhibitors (Complete ethylenediaminetetraacetic acid [EDTA]-free tablets, Roche Molecular Biochemicals) and 1 mM dithiothreitol followed by 48-h incubation at 37 °C. The plates were washed and incubated with streptavidin-europium diluted (1000× dilution) in DELFIA assay buffer (Wallac) for 45 min at room temperature. The plates were washed and incubated with DELFIA enhancement solution (Wallac) for 15 min at room temperature. Binding of the indicator peptide was then measured as Eu counts in a time-resolved fluorometer (Wallac 1420 multilabel counter). IC50 values (the concentration of the competitor peptide required for half-maximal inhibition of the binding of the indicator peptide) were established for each peptide. One 10-fold titration experiment and three 4-fold titration experiments were performed. The relative binding capacity was calculated as the ratio of the IC50 value of the DQ2.2-glut-L1 to the IC50 value of the P3-substituted peptide variants.
Protein Expression and Purification.
cDNAs (Genscript) encoding the individual extracellular domains of TCRα-chain and β-chain, with an engineered interchain disulfide linkage in the constant domains, were cloned into the pET-30 vector and expressed in Escherichia coli BL21DE3. The TCR proteins were purified from inclusion bodies, as described previously (38, 39). The TCR αβ heterodimers were refolded in the presence of 5 M Urea, followed by dialysis and further purification through anion exchange chromatography, size-exclusion chromatography, and hydrophobic-interaction chromatography as described previously (18).
For SPR and structural experiments, the extracellular domains of the HLA-DQ2.2 α-chain and HLA-DQ2.2-glut-L1 β-chain (with glut-L1 epitope covalently linked to the HLA-DQ2.2 β-chain) were coexpressed as soluble protein in High Five insect cells (Trichoplusia ni BTI-TN-5B1-4 cells; Invitrogen), via a baculovirus expression system as described previously (17). Briefly, baculovirus HLA-DQ2.2 expression constructs had enterokinase-cleavable Fos and Jun zippers at the C-terminal ends of the α-chain and β-chain, respectively, to promote dimerization. The C terminus of the β-chain also included a BirA recognition sequence for biotinylation as described previously (18), as well as a histidine tag for purification using immobilized metal ion affinity chromatography. Before crystallization, the Fos and Jun zippers were removed by enterokinase (Genscript) digestion, followed by separation using anion exchange chromatography (HiTrap Q HP, GE Healthcare). Purified HLA-DQ2.2:DQ2.2-glut-L1 was mixed in a 1:1 ratio with purified TCR 594 or TCR 1005.2.56, and the protein complex was then purified by size exclusion chromatography (Superdex 200; GE Healthcare). Three HLA-DQ2.2-glut-L1 epitope mutants were generated by replacing the glut-L1 WT core sequence (PFSEQEQPV) with glut-L1-P2Δ (PASEQEQPV), glut-L1-P5Δ (PFSEAEQPV), and glut-L1-P7Δ (PFSEQEAPV) separately in the HLA-DQ2.2 β-chain and coexpressed with HLA-DQ2.2 α-chain via a baculovirus expression system as described above.
Crystallization, Data Collection, and Processing.
The TCR 594/TCR 1005.2.56 and the HLA-DQ2.2:DQ2.2-glut-L1 complex, in 10 mM Tris (pH 8), 150 mM NaCl, were concentrated to 6 mg/mL. Crystallization was carried out using the hanging drop vapor diffusion method at 17 °C. For TCR594:HLADQ2.2:DQ2.2-glut-L1, diffraction quality crystal was obtained in conditions 0.1 M 2-(N-morpholino)ethanesulfonic acid pH 6.5, 5% vol/vol MPD, and 15% wt/vol PEG6K, and crystallization droplets were set up by adding 1 μL of the mother liquor to 1 μL of protein solution. Crystals typically appeared within 2–7 d. For TCR1005.2.56:HLA-DQ2.2:DQ2.2-glut-L1, diffraction quality crystal was obtained in an optimized Morpheus screen conditions 0.1 M Mops/Hepes pH 7.5, 12.5% vol/vol PEG1K, 12.5% vol/vol MPD, 12.5% wt/vol PEG 3350, and 0.04 M nitrate phosphate sulfate mixture (40), and crystallization droplets were set up by adding 1 μL of the mother liquor to 1 μL of protein solution and with the addition of 0.2 μL of seed stock. Crystals typically appeared within 2–3 wk. Protein crystals were exposed to cryoprotectant, 20% glycerol for 30 s before being flash-cooled in liquid nitrogen. Datasets were collected on the MX2 beamline of the Australian Synchrotron. Data were integrated with XDS and scaled and merged in Aimless. Phases were obtained using molecular replacement in PhaserMR, CCP4suite (41). Protein Data Bank (PDB) ID code 4OZH (chain A and B, without glycans or peptides for HLA; chain G and H for TCR) was used as search model for HLA-DQ2.2:DQ2.2-glut-L1 and TCR 594 for molecular replacement. The data collected for the TCR594:HLADQ2.2:DQ2.2-glut-L1was highly anisotropic, thus ellipsoidal truncations and anisotropic scaling were carried out at the Diffraction Anisotropic Server (42). The structure was built and refined in Coot and PHENIX (43, 44). Final model was validated in Molprobity (44).
Surface Plasmon Resonance Measurement and Analysis.
Surface plasmon resonance experiments were conducted on a BIAcore T200 system (GE Healthcare Life Sciences) instrument using HBS buffer (10 mM Hepes-HCl, pH 7.4, 150 mM NaCl, and 0.005% surfactant P20). The biotinylated HLA-DQ2.5:CLIP2, HLA-DQ2.2:DQ2.2-glut-L1, and respective peptide mutant DQ2.2-glut-L1-P2Δ, DQ2.2-glut-L1-P5Δ, DQ2.2-glut-L1-P7Δ were immobilized on a streptavidin-coated Biacore Series S Sensor Chip SA (SA Chip; GE Healthcare Life Sciences), creating a surface density of ∼800–1,000 response units (RU). The TCR 555, TCR 594, and TCR 1005.2.56 (one in two serial dilutions from the maximum concentration of 200 μM) were injected over the chip at 10 μL/min for 2 min. The TCR response to HLA-DQ2.5:CLIP2 alone was subtracted from the response to HLA-DQ2.2:DQ2.2-glut-L1 and respective epitope mutants. Data in the main text are representative of two or four independent experiments. The equilibrium dissociation constant, KD, was generated with nonlinear regression analysis of the combined data using Prism (GraphPad Software).
Data Availability.
The protein structural data that support the findings of this study have been deposited in the RCSB PDB with the ID codes 6PX6 and 6PY2. All of the single-cell TCR sequencing raw data generated in this study are available in the European Genome-phenome Archive (EGAS00001003673).
Supplementary Material
Acknowledgments
This research was supported by the National Health and Medical Research Council (Australia) and Australian Research Council (ARC) (J.R.); by the South-Eastern Norway Regional Health Authority Projects 2011050 and 2015009; Research Council of Norway Project 179573/V40 through the Centre of Excellence funding scheme, and Project 233885; and the Stiftelsen Kristian Gerhard Jebsen Project SKGJ-MED-017 (to L.M.S.). J.R. is supported by an ARC Australian Laureate Fellowship. We thank Khai Lee Loh, Mai Tran, and Bjørg Simonsen for their excellent technical assistance. The protein crystal X-ray diffraction data were collected on MX2 beamline at the Australian Synchrotron facility, Melbourne, Australia.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, http://www.wwpdb.org/ (PDB ID codes 6PX6 and 6PY2). Single-cell TCR sequencing raw data generated in this study are available in the European Genome-phenome Archive (EGAS00001003673).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914308117/-/DCSupplemental.
References
- 1.Abadie V., Sollid L. M., Barreiro L. B., Jabri B., Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu. Rev. Immunol. 29, 493–525 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Molberg O., et al. , Gliadin specific, HLA DQ2-restricted T cells are commonly found in small intestinal biopsies from coeliac disease patients, but not from controls. Scand. J. Immunol. 46, 103–109 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Risnes L. F., et al. , Disease-driving CD4+ T cell clonotypes persist for decades in celiac disease. J. Clin. Invest. 128, 2642–2650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sollid L. M., Jabri B., Triggers and drivers of autoimmunity: Lessons from coeliac disease. Nat. Rev. Immunol. 13, 294–302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jabri B., Sollid L. M., T cells in celiac disease. J. Immunol. 198, 3005–3014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sollid L. M., et al. , Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J. Exp. Med. 169, 345–350 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sollid L. M., Molecular basis of celiac disease. Annu. Rev. Immunol. 18, 53–81 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Fleckenstein B., et al. , Gliadin T cell epitope selection by tissue transglutaminase in celiac disease. Role of enzyme specificity and pH influence on the transamidation versus deamidation process. J. Biol. Chem. 277, 34109–34116 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Vader L. W., et al. , Specificity of tissue transglutaminase explains cereal toxicity in celiac disease. J. Exp. Med. 195, 643–649 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia J., Sollid L. M., Khosla C., Equilibrium and kinetic analysis of the unusual binding behavior of a highly immunogenic gluten peptide to HLA-DQ2. Biochemistry 44, 4442–4449 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Molberg O., et al. , Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 4, 713–717 (1998). [DOI] [PubMed] [Google Scholar]
- 12.van de Wal Y., et al. , Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J. Immunol. 161, 1585–1588 (1998). [PubMed] [Google Scholar]
- 13.Qiao S. W., Bergseng E., Molberg O., Jung G., Fleckenstein B., Sollid L. M., Refining the rules of gliadin T cell epitope binding to the disease-associated DQ2 molecule in celiac disease: importance of proline spacing and glutamine deamidation. J. Immunol. 175, 254–261 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Bodd M., Kim C. Y., Lundin K. E., Sollid L. M., T-cell response to gluten in patients with HLA-DQ2.2 reveals requirement of peptide-MHC stability in celiac disease. Gastroenterology 142, 552–561 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Tollefsen S., et al. , HLA-DQ2 and -DQ8 signatures of gluten T cell epitopes in celiac disease. J. Clin. Invest. 116, 2226–2236 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fallang L.-E., et al. , Differences in the risk of celiac disease associated with HLA-DQ2.5 or HLA-DQ2.2 are related to sustained gluten antigen presentation. Nat. Immunol. 10, 1096–1101 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Henderson K. N., et al. , A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity 27, 23–34 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Broughton S. E., et al. , Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity 37, 611–621 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Johansen B. H., et al. , Both alpha and beta chain polymorphisms determine the specificity of the disease-associated HLA-DQ2 molecules, with beta chain residues being most influential. Immunogenetics 45, 142–150 (1996). [DOI] [PubMed] [Google Scholar]
- 20.van de Wal Y., et al. , Unique peptide binding characteristics of the disease-associated DQ(α 1*0501, β 1*0201) vs the non-disease-associated DQ(α 1*0201, β 1*0202) molecule. Immunogenetics 46, 484–492 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Kim C.-Y., Quarsten H., Bergseng E., Khosla C., Sollid L. M., Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc. Natl. Acad. Sci. U.S.A. 101, 4175–4179 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen J., et al. , T-cell receptor recognition of HLA-DQ2-gliadin complexes associated with celiac disease. Nat. Struct. Mol. Biol. 21, 480–488 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Petersen J., et al. , Determinants of gliadin-specific T cell selection in celiac disease. J. Immunol. 194, 6112–6122 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Dørum S., et al. , HLA-DQ molecules as affinity matrix for identification of gluten T cell epitopes. J. Immunol. 193, 4497–4506 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Bergseng E., et al. , Different binding motifs of the celiac disease-associated HLA molecules DQ2.5, DQ2.2, and DQ7.5 revealed by relative quantitative proteomics of endogenous peptide repertoires. Immunogenetics 67, 73–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venturi V., Price D. A., Douek D. C., Davenport M. P., The molecular basis for public T-cell responses? Nat. Rev. Immunol. 8, 231–238 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Jabri B., Chen X., Sollid L. M., How T cells taste gluten in celiac disease. Nat. Struct. Mol. Biol. 21, 429–431 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Petersen J., Kooy-Winkelaar Y., Loh K. L., Tran M., Van Bergen J., Koning F., Rossjohn J., Reid H. H., Diverse T cell receptor gene usage in HLA-DQ8-associated celiac disease converges into a consensus binding solution. Structure 24, 1643–1657 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Rossjohn J., Koning F., A biased view toward celiac disease. Mucosal Immunol. 9, 583–586 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Dahal-Koirala S., et al. , Discriminative T-cell receptor recognition of highly homologous HLA-DQ2-bound gluten epitopes. J. Biol. Chem. 294, 941–952 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao S.-W., Christophersen A., Lundin K. E., Sollid L. M., Biased usage and preferred pairing of α- and β-chains of TCRs specific for an immunodominant gluten epitope in coeliac disease. Int. Immunol. 26, 13–19 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Qiao S.-W., et al. , Posttranslational modification of gluten shapes TCR usage in celiac disease. J. Immunol. 187, 3064–3071 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Ludvigsson J. F., et al. , The Oslo definitions for coeliac disease and related terms. Gut 62, 43–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorum S., et al. , HLA-DQ molecules as affinity matrix for identification of gluten T cell epitopes. J. Immunol. 193, 4497–4506 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Dahal-Koirala S., et al. , TCR sequencing of single cells reactive to DQ2.5-glia-α2 and DQ2.5-glia-ω2 reveals clonal expansion and epitope-specific V-gene usage. Mucosal Immunol. 9, 587–596 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Han A., Glanville J., Hansmann L., Davis M. M., Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 32, 684–692 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vader W., et al. , The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc. Natl. Acad. Sci. U.S.A. 100, 12390–12395 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulter J. M., et al. , Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein Eng. 16, 707–711 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Garboczi D. N., et al. , Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384, 134–141 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Gorrec F., The MORPHEUS II protein crystallization screen. Acta Crystallogr. F Struct. Biol. Commun. 71, 831–837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strong M., et al. , Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The protein structural data that support the findings of this study have been deposited in the RCSB PDB with the ID codes 6PX6 and 6PY2. All of the single-cell TCR sequencing raw data generated in this study are available in the European Genome-phenome Archive (EGAS00001003673).