Significance
Toll-like receptors (TLRs) sense and respond to foreign nucleic acids, but this specificity raises the risk of inappropriate responses to self-nucleic acids, leading to autoimmune diseases. Limiting TLR9 activation to endosomes is thought to reduce recognition of self-DNA. To test the importance of this compartmentalized activation for immune tolerance, we developed mice with inducible expression of a mutated TLR9 that bypasses the requirement for endosomal activation. While induction of expression in adults resulted in rather mild disease, expression during development resulted in neonatal fatality dependent on type II interferon (IFN) signaling and driven by macrophages and IFN-γ–producing natural killer cells. This study uses a new model to reveal unique aspects of TLR9-driven inflammatory disease, especially early in life.
Keywords: Toll-like receptor, inflammation, autoinflammation
Abstract
Recognition of self-nucleic acids by innate immune receptors can lead to the development of autoimmune and/or autoinflammatory diseases. Elucidating mechanisms associated with dysregulated activation of specific receptors may identify new disease correlates and enable more effective therapies. Here we describe an aggressive in vivo model of Toll-like receptor (TLR) 9 dysregulation, based on bypassing the compartmentalized activation of TLR9 in endosomes, and use it to uncover unique aspects of TLR9-driven disease. By inducing TLR9 dysregulation at different stages of life, we show that while dysregulation in adult mice causes a mild systemic autoinflammatory disease, dysregulation of TLR9 early in life drives a severe inflammatory disease resulting in neonatal fatality. The neonatal disease includes some hallmarks of macrophage activation syndrome but is much more severe than previously described models. Unlike TLR7-mediated disease, which requires type I interferon (IFN) receptor signaling, TLR9-driven fatality is dependent on IFN-γ receptor signaling. NK cells are likely key sources of IFN-γ in this model. We identify populations of macrophages and Ly6Chi monocytes in neonates that express high levels of TLR9 and low levels of TLR7, which may explain why TLR9 dysregulation is particularly consequential early in life, while symptoms of TLR7 dysregulation take longer to manifest. Overall, this study demonstrates that inappropriate TLR9 responses can drive a severe autoinflammatory disease under homeostatic conditions and highlights differences in the diseases resulting from inappropriate activation of TLR9 and TLR7.
Recognition of nucleic acids by Toll like receptors (TLRs) enables the immune system to sense and respond to diverse pathogens; however, this strategy raises the risk of initiating inappropriate immune responses to self-derived nucleic acids. Recognition of self-RNA and DNA by TLR7 or TLR9, respectively, has been implicated in the pathology of autoimmune and autoinflammatory diseases, such as systemic lupus erythematosus (SLE) and macrophage activation syndrome (MAS) (1–8). One important mechanism that reduces recognition of self-ligands is the localization of nucleic acid sensing TLRs in endosomes, which limits access to extracellular self-derived nucleic acids released from necrotic cells or apoptotic cells that undergo secondary necrosis (9). The requirement for proteolytic cleavage of the ectodomains of TLR9 and TLR7 within endosomes further reinforces this compartmentalized activation of TLRs (10–12).
When these regulatory mechanisms fail, inappropriate activation of TLR7 or TLR9 can result in autoimmune and autoinflammatory pathology (7, 8, 10, 13–15). Mice with additional copies of the Tlr7 gene (TLR7.1 transgenic mice, or TLR7tg) develop spontaneous disease that shares many of the hallmarks of SLE and MAS. Specifically, TLR7tg mice develop anti-nuclear antibodies, a type I IFN-dependent increase in myeloid precursor cells in the bone marrow, and cytopenia associated with increased inflammatory hemophagocytes in the spleen (8, 15). In addition, transgenic mice that express high levels of human TLR8 develop multiorgan inflammation (16). In contrast, overexpression of TLR9 does not induce overt disease (17), so it has been harder to determine whether there are detrimental consequences downstream of inappropriate TLR9 activation.
Chronic TLR9 stimulation through repeated CpG injections results in MAS-like disease, characterized by anemia, splenomegaly, and inflammatory myelopoiesis (18, 19), but it is unclear whether this pathology recapitulates the consequences of chronic activation of TLR9 by self-DNA. A recent report described two endolysosomal exonucleases, PLD3 and PLD4, capable of degrading TLR9 ligands (20). PLD4-deficient mice develop a relatively mild MAS-like disease that is rescued by TLR9 deficiency (20). Mice lacking both PLD3 and PLD4 develop a much more severe disease (20), but whether this disease depends on TLR9 activation has not been addressed. Collectively, these reports suggest that the disease outcomes associated with chronic dysregulation of TLR7 and TLR9 are distinct, but the lack of an animal model of disease clearly based on TLR9 dysregulation has precluded a close comparison of the diseases driven by these two nucleic acid sensors.
To overcome these limitations, we have built on our earlier studies of TLR9 regulation to generate a mouse model of TLR9 dysregulation. We previously described a mutant TLR9 receptor that no longer requires ectodomain processing (hereinafter called TLR9TransmembraneMutation, or TLR9TM) and showed that reconstitution of lethally irradiated mice with retrovirally transduced hematopoietic stem cells (HSCs) expressing TLR9TM led to a rapid and fatal disease (12). While these experiments formally demonstrated the importance of compartmentalized activation of TLR9, the ectopic overexpression of TLR9TM driven by a retroviral promoter and the increased levels of extracellular nucleic acids due to irradiation limited our ability to track the development of disease or draw any general conclusions about the consequences about TLR9 dysregulation under homeostatic conditions.
We have generated mice in which TLR9TM is expressed from within the endogenous Tlr9 locus in a Cre recombinase-dependent manner. This system allows us to examine the consequences of bypassing compartmentalized activation of TLR9 in cells that endogenously express TLR9 under homeostatic conditions, early or late in life. When TLR9TM expression was induced later in life, we observed mild inflammation with many aspects similar to TLR7-driven diseases. In contrast, induction of TLR9TM expression ab initio resulted in fatal disease, revealing a particular sensitivity to dysregulated TLR9 activation early in life. In contrast to TLR7-driven disease models, TLR9-driven disease required IFN-γ receptor signaling but not type I IFN receptor signaling. Disease was correlated with IFN-γ production by NK cells, suggesting a role for NK cells in promoting this autoinflammatory disease. These findings demonstrate that proper compartmentalization of TLR9 is necessary to prevent recognition of self-DNA under homeostatic conditions and establish a new model of TLR9 dysregulation.
Results
Dysregulation of TLR9 in Adult Mice Induces Systemic Inflammation.
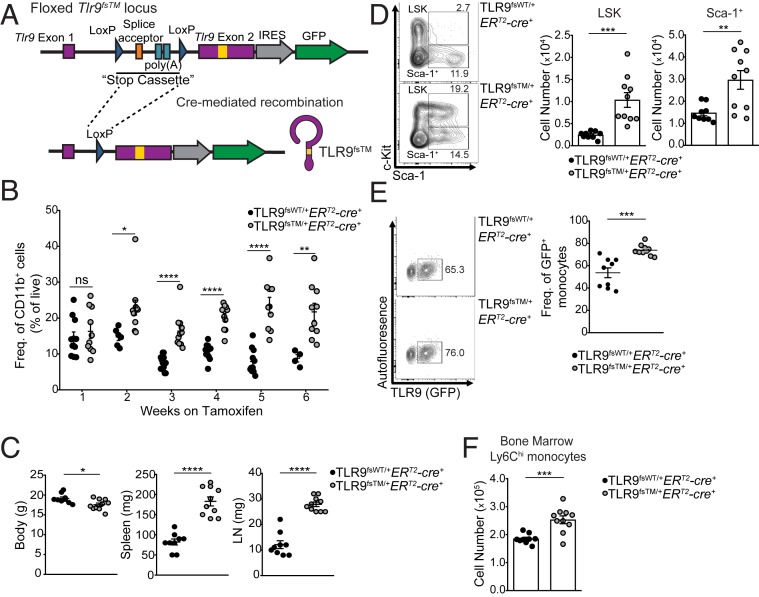
We generated mice that enabled inducible expression of TLR9TM from the endogenous Tlr9 promoter (TLR9flox-stop-TM, hereinafter TLR9fsTM). These mice had three key features: 1) the Tlr9 transmembrane mutation that negates the requirement for compartmentalized activation (12), 2) a loxP-flanked transcriptional STOP cassette upstream of exon 2 to prevent TLR9 expression in the absence of Cre recombinase, and 3) an IRES-GFP reporter gene downstream of the TLR9 coding sequence to allow tracking of TLR9-expressing cells via cytoplasmic fluorescence (Fig. 1A and SI Appendix, Fig. S1A). We also generated a line of Cre-inducible Tlr9 knockin mice without the transmembrane mutation, referred to as TLR9flox-stop-WT (hereinafter TLR9fsWT), to serve as controls for these studies (SI Appendix, Fig. S1B). As expected, we did not detect GFP+ cells in TLR9fsWT and TLR9fsTM mice in the absence of Cre, and cells from TLR9fsTM or TLR9fsWT mice without Cre did not respond to the TLR9 ligand (CpG-B) (SI Appendix, Fig. S1 C and D). Furthermore, TLR9-expressing bone marrow macrophages (BMMs) still produced TNF in response to CpG when pretreated with chloroquine, while BMMs expressing WT TLR9 were inhibited (SI Appendix, Fig. S1E), consistent with our previous work (12).
Fig. 1.
TLR9 dysregulation in adult mice results in systemic inflammation and expansion of Ly6Chi monocytes. (A) Diagram of TLR9fsTM knockin mice. The TLR9 mutation is shown as a yellow box. (B) On tamoxifen treatment, TLR9-expressing mice exhibit an increase in circulating myeloid cells compared with mice expressing TLR9WT. Mice were fed tamoxifen diet beginning at weaning and bled every week to monitor GFP expression. Shown is the frequency of live, single, CD11b+ cells in the blood. Data combined from independent experiments examining three or more mice per genotype per timepoint are shown as mean ± SEM and analyzed using the two-tailed Student’s t test. (C) TLR9TM-expressing mice exhibit weight loss, splenomegaly, and lymphomegaly compared with TLR9WT mice after 7 wk of tamoxifen diet. Data combined from independent experiments are shown as mean ± SEM and analyzed using the two-tailed Student’s t test. Mouse numbers: TLR9fsWT/+ERT2-cre+, n = 9; TLR9fsTM/+ERT2-cre+, n = 10. (D, Left) Representative flow cytometry plots for bone marrow of TLR9fsWT/+ERT2-cre+ and TLR9fsTM/+ERT2-cre+ mice. Gates for LSK and Sca-1+ progenitor cells are indicated. (D, Right) Quantification of LSK and Sca-1+ populations. Data combined from independent experiments are shown as mean ± SEM and analyzed using the two-tailed Student’s t test. Mouse numbers: TLR9fsWT/+ERT2-cre+, n = 9; TLR9fsTM/+ERT2-cre+, n = 10. (E, Left) Representative flow cytometry plots for bone marrow of TLR9fsWT/+ERT2-cre+ and TLR9fsTM/+ERT2-cre+ examining TLR9WT and TLR9TM expression in Ly6Chi monocytes (CD45+CD3e–B220–Ly6G–CD11b+F480midloLy6Chi) cells. (E, Right) Quantification of the frequency of GFP+ Ly6Chi cells from bone marrow. Data are compiled from independent experiments. Mouse numbers: TLR9fsWT/+ERT2-cre+, n = 9; TLR9fsTM/+ERT2-cre+, n = 10. (F) Quantification of total Ly6Chi monocytes from TLR9fsWT/+ERT2-cre+ and TLR9fsTM/+ERT2-cre+ bone marrow. Data combined from independent experiments are shown as mean ± SEM and analyzed using the two-tailed Student’s t test. Mouse numbers: TLR9fsWT/+ERT2-cre+, n = 9; TLR9fsTM/+ERT2-cre+, n = 10. In all panels, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To test whether bypassing compartmentalized activation of TLR9 is sufficient to break tolerance under steady-state conditions, we bred TLR9fsTM and TLR9fsWT mice to Rosa26-ERT2-cre mice to enable tamoxifen-inducible expression of each receptor. Beginning at weaning, TLR9fsTM/+ERT2-cre+ and TLR9fsWT/+ERT2-cre+ mice were placed on a tamoxifen-containing diet. GFP+CD11b+ myeloid cells were detectable in the peripheral blood of TLR9fsWT/+ERT2-cre+ and TLR9fsTM/+ERT2-cre+ mice after 1 wk of tamoxifen administration (SI Appendix, Fig. S2A). GFP expression was limited to CD45+ cells, as expected for expression driven by the endogenous Tlr9 promoter (SI Appendix, Fig. S2B). As early as 2 wk after tamoxifen treatment, TLR9fsTM/+ERT2-cre+ mice exhibited an increase in CD11b+ cells in the blood compared with TLR9fsWT/+ERT2-cre+ mice (Fig. 1B), suggesting an alteration in the circulating myeloid compartment due to TLR9TM expression. In addition, after 7 wk of tamoxifen administration, TLR9fsTM/+ERT2-cre+ mice demonstrated reduced body weight and enlarged spleens and lymph nodes compared with TLR9fsWT/+ERT2-cre+ mice (Fig. 1C).
Examination of bone marrow from TLR9fsTM/+ERT2-cre+ mice revealed an expansion in Lin–Sca-1+cKit+ (LSK) and Lin–Sca-1+cKit– (Sca-1+) cell populations (Fig. 1D). Similar abnormalities in hematopoiesis have been described in TLR7tg mice. The inflammation in these animals also leads to expansion of a population of Ly6Chi monocytes, and TLR7 and TLR9 signaling in these cells has been shown to induce the differentiation and development of inflammatory hemophagocytes (8). Furthermore, it was recently shown that stimulation of bone marrow progenitors via TLR9 can lead to the production of Ly6Chi monocytes (19, 21). Interestingly, TLR9fsTM/+ERT2-cre+ mice have increased frequencies of TLR9-expressing Ly6Chi monocytes (Lin–CD45+CD11b+F4/80neg-loLy6Chi), as well as increased numbers of total Ly6Chi monocytes in the bone marrow, compared with TLR9fsWT/+ERT2-cre+ mice (Fig. 1 E and F). Together, these data indicate that dysregulated TLR9 activation alters hematopoiesis in the bone marrow, which induces an expansion of Ly6Chi monocytes.
In contrast to the fatal inflammation observed in mice reconstituted with HSCs transduced with retroviruses encoding TLR9TM (12), the phenotype of tamoxifen-treated TLR9fsTM/+ERT2-cre+ was significantly less severe. Full body irradiation induces significant cell death and DNA release, so we reasoned that mice expressing TLR9TM in the ERT2-cre system might not succumb to fatal inflammation, because extracellular self-nucleic acids are not abundant under homeostatic conditions due to tightly regulated programmed cell death processes as well as the presence of DNA exonucleases (22–25). To induce cell death and increase the amount of self-DNA ligand, TLR9fsTM/+ERT2-cre+ mice were fed a tamoxifen diet for 10 wk and then administered a sublethal dose of irradiation (SI Appendix, Fig. S2C). Unexpectedly, irradiated TLR9fsTM/+ERT2-cre+ mice survived similarly to their littermate controls (SI Appendix, Fig. S2C). Furthermore, exposure to DNA ligand by repeated DNA injections also did not lead to fatal inflammation. Thus, excessive extracellular DNA arising as a result of irradiation does not appear to be sufficient to explain the previously observed lethal disease seen in mice overexpressing TLR9TM in hematopoietic cells.
Dysregulation of TLR9 Causes Neonatal Fatality.
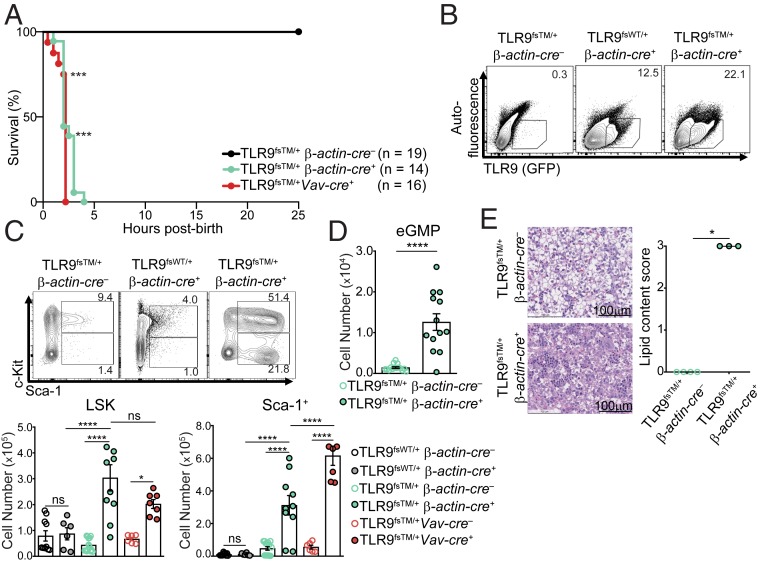
Based on the relatively mild disease observed in TLR9fsTM/+ERT2-cre+ mice, we next examined mice in which TLR9TM is expressed throughout development. We crossed TLR9fsTM/+ mice to Tmem163Tg(ACTB-cre)2Mrt (β-actin-cre) mice to generate mice in which TLR9TM is expressed ab initio. Surprisingly, no TLR9fsTM/+β-actin-cre+ progeny were present at the time of weaning, suggesting that expression of TLR9TM in this context may be fatal. Timed pregnancies revealed that TLR9fsTM/+β-actin-cre+ neonates were born at the expected Mendelian frequency but died shortly after birth, compared with TLR9fsWT/+β-actin-cre+ mice, which survived to adulthood (Table 1 and Fig. 2A).
Table 1.
Survival of TLR9fsWT and TLR9fsTM neonates
| Cre recombinase | TLR9 genotype | %Expected | %Born | %Survival 24 h postbirth | Mice analyzed |
| β-actin-cre– | TLR9fsWT/+ | 50 | 44.4 | 100 | 16 |
| β-actin-cre+ | TLR9fsWT/+ | 50 | 55.6 | 100 | 20 |
| β-actin-cre– | TLR9fsTM/+ | 50 | 60.3 | 100 | 47 |
| β-actin-cre+ | TLR9fsTM/+ | 50 | 39.7 | 0 | 31 |
| Vav-cre– | TLR9fsTM/+ | 50 | 51.2 | 100 | 21 |
| Vav-cre+ | TLR9fsTM/+ | 50 | 48.8 | 0 | 20 |
Fig. 2.
Dysregulation of TLR9 early in life is fatal. (A) TLR9fsTM/+ β-actin-cre–, TLR9fsTM/+ β-actin-cre+, and TLR9fsTM/+Vav-cre+ neonate survival curve. Results are combined from multiple litters and analyzed using the logrank (Mantel–Cox) test. (B) The frequency of TLR9 (GFP+)-expressing cells among live, single CD45+ cells in the neonatal liver of indicated genotypes. Data are representative of six independent experiments. (C) TLR9fsTM/+ β-actin-cre+ neonates exhibit expanded LSK and Sca-1+ cell populations. Representative flow cytometry plots of neonatal livers from the indicated genotypes are shown. Bar graphs show quantification of LSK and Sca-1+ populations in neonatal livers from indicated genotypes. Data from multiple litters are shown as mean ± SEM and analyzed using one-way ANOVA with Tukey’s multiple comparisons posttest. Mouse numbers: TLR9fsWT/+ β-actin-cre–, n = 10; TLR9fsWT/+ β-actin-cre+, n = 6; TLR9fsTM/+ β-actin-cre–, n = 14; TLR9fsTM/+ β-actin-cre+, n = 10; TLR9fsTM/+Vav-cre–, n = 6; TLR9fsTM/+Vav-cre+, n = 7. (D) TLR9fsTM/+ β-actin-cre+ neonates exhibit emergency myelopoiesis in the neonatal liver. Quantification of Sca-1+CD34+CD16/32+ myeloid cells found within the LSK gate. Data from multiple litters are shown as mean ± SEM and analyzed using one-way ANOVA with Tukey’s multiple comparisons posttest. Mouse numbers: TLR9fsTM/+ β-actin-cre–, n = 15; TLR9fsTM/+ β-actin-cre+, n = 13. (E, Left) Representative photomicrographs of H&E staining of neonatal livers from indicated genotypes. (E, Right) Pathology scoring of liver lipid content as a measure of inflammation. Pathology scoring scheme: 0 = hepatocytes contain diffuse cytoplasmic lipid vacuoles, 1 = hepatocytes contain many small cytoplasmic lipid vacuoles, 2 = hepatocytes contain a few small cytoplasmic lipid vacuoles, 3 = hepatocytes contain no lipid. Bar, 100 μm. Data are combined from multiple experiments and analyzed using Kruskal-Wallis analysis of variance and Dunn’s multiple comparisons test. Mouse numbers: TLR9fsTM/+ β-actin-cre– n = 4 and TLR9fsTM/+ β-actin-cre+ n = 3. In all panels, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To investigate the basis for this rapid death, we examined TLR9fsTM/+β-actin-cre+ neonates for signs of TLR9-driven immune activation. We detected increased frequencies of TLR9+(GFP+)CD45+ cells in the livers of TLR9fsTM/+β-actin-cre+ neonates compared with TLR9fsWT/+β-actin-cre+ neonates (Fig. 2B). Moreover, LSK, Sca-1+, and Ly6Chi monocyte populations were expanded in TLR9fsTM/+β-actin-cre+ neonatal livers (Fig. 2C and SI Appendix, Fig. S3A). We also observed an increase in emergency granulocyte/macrophage progenitor (eGMP) cells within the LSK gate (Lin–Sca-1+cKit+CD34+CD16/32+), indicating that hematopoiesis in TLR9fsTM/+β-actin-cre+ neonates had shifted to generate more myeloid progenitor cells, as has been described in adult TLR7tg mice (Fig. 2D and SI Appendix, Fig. S3B) (15). In addition, hematoxylin and eosin (H&E) staining revealed extensive inflammation in the livers of TLR9fsTM/+β-actin-cre+ neonates (Fig. 2E), but no phenotypic differences were observed in the lungs, heart, kidneys, or nasal passages.
Previous studies have reported that nonhematopoietic cells, such as cardiomyocytes, express TLR9 (26). To test whether the liver inflammation and neonatal death observed in TLR9fsTM/+β-actin-cre+ mice is driven by hematopoietic cell expression of TLR9TM, we bred TLR9fsTM mice to Commd10Tg(Vav1-icre)A2Kio (Vav-cre), which is expressed only in hematopoietic cells. TLR9fsTM/+Vav-cre+ neonates died shortly after birth and displayed the same signs of inflammation in the neonatal liver (Table 1 and Fig. 2 A and C). These data suggest that excessive TLR signaling by TLR9TM in hematopoietic cells in the neonatal liver drives an expansion of myeloid cells, presumably in response to an abundance of DNA ligand during development.
Dysregulated TLR9 Activation in Neonatal Macrophages and Monocytes Correlates with Severe Inflammatory Disease in Neonates.
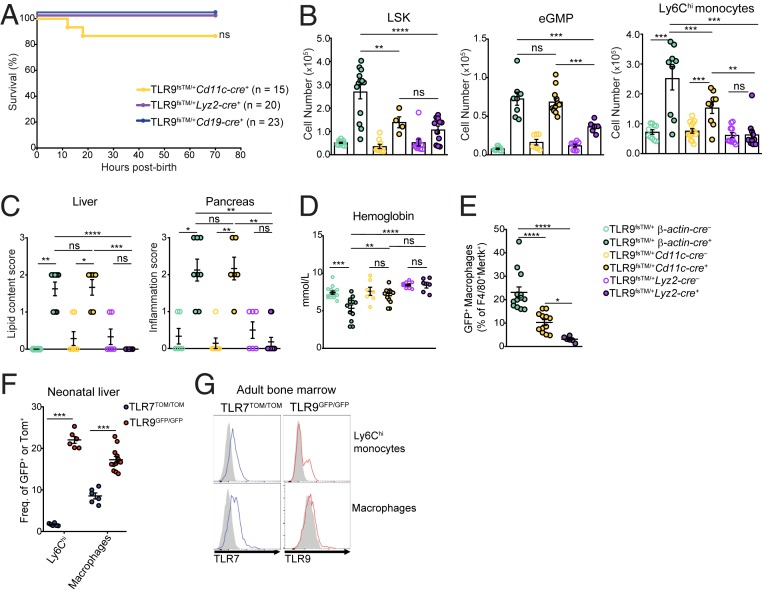
The Cre-inducible nature of our system allows us to test the importance of dysregulated TLR9 in specific immune cell types for disease. We bred TLR9fsTM mice to 1) Cd11c-cre (Tg(Itgax-cre)1–1Reiz/J) mice, to drive expression in CD11c+ cells (which include dendritic cells, some macrophages, and some B cells); 2) Lyz2-cre (Lyz2tm1(cre)Ifo) mice, to drive expression in macrophages, monocytes, and neutrophils; and 3) Cd19-cre (Cd19tm1(cre)Cgn) mice, to drive expression in B cells. Progeny from TLR9fsTM mice crossed to Lyz2-cre and Cd19-cre were born at expected Mendelian frequencies and survived to adulthood with no apparent disease (Fig. 3A). In contrast, approximately 15% of the TLR9fsTM/+Cd11c-cre+ neonates died within 24 h of birth, and the remaining mice were severely runted well into adulthood (Fig. 3A and SI Appendix, Fig. S4A). Collectively, these results indicate that dysregulation of TLR9 in B cells and Lyz2+ cells, such as monocytes and macrophages, is not sufficient to induce significant pathology, but TLR9TM expression in CD11c+ cells partially recapitulates the phenotype observed in TLR9fsTM/+β-actin-cre+ and TLR9fsTM/+Vav-cre+ neonates.
Fig. 3.
Expression of dysregulated TLR9 by neonatal macrophages and Ly6Chi monocytes correlates with liver inflammation and pancreatitis. (A) Survival curve of indicated genotypes. Results are combined from multiple litters and analyzed using Log-rank (Mantel-Cox) test. (B) TLR9fsTM/+ β-actin-cre+ and TLR9fsTM/+ Cd11c-cre+ neonates exhibit an increase in LSK, eGMP, and Ly6Chi monocyte cell populations in the liver compared with TLR9fsTM/+ Lyz2-cre+ neonates. Data combined from multiple litters are shown as mean ± SEM and analyzed using one-way ANOVA with Tukey’s multiple comparisons posttest. Mouse numbers: TLR9fsTM/+ β-actin-cre–, n = 15; TLR9fsTM/+ β-actin-cre+, n = 13; TLR9fsTM/+ Cd11c-cre–, n = 9; TLR9fsTM/+ Cd11c-cre+, n = 4 and n = 10; TLR9fsTM/+ Lyz2-cre–, n = 10; TLR9fsTM/+ Lyz2-cre+, n = 11. (C) Histology scores of liver (Left) and pancreas (Right) sections from the indicated mice. Pathology scoring scheme for liver: 0 = hepatocytes contain diffuse cytoplasmic lipid vacuoles; 1 = hepatocytes contain many small cytoplasmic lipid vacuoles; 2 = hepatocytes contain a few small cytoplasmic lipid vacuoles; 3 = hepatocytes contain no lipid. Pathology scoring scheme for pancreas: inflammation with exocrine cell necrosis or zymogen depletion or both: 0 = unaffected; 1 = minimal diffuse to mild multifocal; 2 = mild diffuse to moderate multifocal; 3 = moderate diffuse to marked multifocal. Data are combined from multiple experiments and analyzed using Kruskal–Wallis ANOVA and Dunn’s multiple comparisons test. Mouse numbers: TLR9fsTM/+ β-actin-cre–, n = 6; TLR9fsTM/+ β-actin-cre+, n = 8; TLR9fsTM/+ Cd11c-cre–, n = 7; TLR9fsTM/+ Cd11c-cre+, n = 6; TLR9fsTM/+ Lyz2-cre–, n = 6; TLR9fsTM/+ Lyz2-cre+, n = 11. (D) TLR9fsTM/+ β-actin-cre+ neonates suffer from anemia. Blood hemoglobin levels from the indicated mice are shown. Results combined from multiple experiments are shown as mean ± SEM and analyzed using one-way ANOVA with Tukey’s multiple comparisons posttest. Mouse numbers: TLR9fsTM/+ β-actin-cre–, n = 14, TLR9fsTM/+ β-actin-cre+, n = 12; TLR9fsTM/+ Cd11c-cre–, n = 7; TLR9fsTM/+ Cd11c-cre+, n = 7; TLR9fsTM/+ Lyz2-cre–, n = 8; TLR9fsTM/+ Lyz2-cre+, n = 6. (E) The frequency of TLR9-expressing neonatal macrophages correlates with disease severity. Quantification of CD45+CD3e–CD19–Ly6G–CD11b+F4/80+Mertk+ TLR9 (GFP+) macrophages from the neonatal livers of the indicated genotypes. Results combined from multiple experiments are shown as mean ± SEM and analyzed using one-way ANOVA with Tukey’s multiple comparisons posttest. Mouse numbers: TLR9fsTM/+ β-actin-cre+, n = 13; TLR9fsTM/+ Cd11c-cre+, n = 12; TLR9fsTM/+ Lyz2-cre+, n = 6. (F) The frequency of macrophages and Ly6Chi monocytes expressing TLR9 (GFP+) and TLR7 (TdTomato+). Results combined from multiple experiments are shown as mean ± SEM and analyzed using the two-tailed Student’s t test. Mouse numbers: TLR9GFP/GFP, n = 12; TLR7TOM/TOM, n = 6. (G) Representative histograms of TLR9 and TLR7 expression in Ly6Chi monocytes and macrophages (CD45+CD3e–CD19–Ly6G–Ly6C–CD11b+F4/80+) from adult bone marrow from TLR9GFP/GFP or TLR7TOM/TOM reporter mice compared with B6 WT controls (gray). In all panels, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To better define the determinants of the severity of the phenotype in TLR9fsTM mice crossed to each of the Cre drivers, we first compared the cellular composition of neonatal livers from TLR9fsTM/+β-actin-cre+, TLR9fsTM/+Cd11c-cre+, and TLR9fsTM/+Lyz2-cre+ neonates. Compared with the TLR9fsTM/+β-actin-cre+ neonates, the expansion of LSK cells, eGMP cells, Sca-1+ cells, and Ly6Chi monocytes was reduced in the TLR9fsTM/+Cd11c-cre+ and TLR9fsTM/+Lyz2-cre+ neonates (Fig. 3B and SI Appendix, Fig. S4B). There were no cellular abnormalities in TLR9fsTM/+Cd19-cre+ neonates (SI Appendix, Fig. S4C). Analyses of H&E-stained tissue sections of neonatal liver and pancreas revealed a similar correlation with disease severity (Fig. 3C).
Finally, we measured blood hemoglobin levels after birth to assess or anemia, a hallmark of MAS- and TLR7-driven autoinflammation (8, 19). TLR9fsTM/+β-actin-cre+ neonates had significantly lower levels of blood hemoglobin levels compared with TLR9fsTM/+β-actin-cre– littermate controls (Fig. 3D). TLR9fsTM/+Cd11c-cre+ neonates had slightly reduced levels, although the difference did not reach statistical significance, while there was no difference between TLR9fsTM/+Lyz2-cre+ and TLR9fsTM/+Lyz2-cre– neonates (Fig. 3D). Overall, these results indicate that inflammation of the liver and pancreas, as well as anemia, correlate with the fatality observed in TLR9fsTM/+β-actin-cre+ and TLR9fsTM/+Cd11c-cre+ neonates.
We next considered whether the varied disease severity in different Cre-driver lines could be linked to cell-intrinsic TLR9TM activation in specific cell populations in neonates. While the frequency of TLR9-expressing Ly6Chi cells was increased in TLR9fsTM/+β-actin-cre+ and TLR9fsTM/+Vav-cre+ neonates, these cells did not express TLR9TM in TLR9fsTM/+Cd11c-cre+ and TLR9fsTM/+Lyz2-cre+ neonates, suggesting that a different TLR9-expressing cell type may contribute to disease (SI Appendix, Fig. S4D). Consistent with this possibility, we found that TLR9TM expression in neonatal liver macrophages (CD45+CD11b+F4/80+Mertk+) across the different Cre lines correlated well with disease severity (Fig. 3E and SI Appendix, Fig. S4E) (27). These liver macrophages expressed CD64 but not CD115, Ly6C, or CCR2, distinguishing them from Ly6Chi monocytes (SI Appendix, Fig. S4E). The frequency of GFP+ neonatal liver macrophages was similar in TLR9fsTM/+β-actin-cre+ and TLR9fsWT/+β-actin-cre+ neonates, indicating that these cells express TLR9 in the absence of inflammation (SI Appendix, Fig. S4F). The frequency of TLR9TM-expressing neonatal macrophages was considerably lower in TLR9fsTM/+Cd11c-cre+ mice and hardly detectable in TLR9fsTM/+Lyz2-cre+ neonates (Fig. 3E). The difference in TLR9 expression in these cells across the different mouse Cre lines likely stems from the timing of expression of each of the Cre-drivers. While Cd11c-cre and Lyz2-cre would be expected to drive expression in macrophages, it has recently been demonstrated that Cd11c-cre expression during development is delayed relative to Vav expression (and presumably β-actin), and Lyz2 expression is delayed even further (28). Thus, these analyses reveal a critical window during development in which dysregulation of TLR9 activation in neonatal macrophages is particularly consequential.
In TLR7tg mice, dysregulation of TLR7 signaling by Ly6Chi cells drives hemophagocytic disease, but disease becomes apparent only later in life (8). One potential explanation for this delay relative to TLR9-expressing mice is that TLR7 and TLR9 expression differ in key cell populations during the developmental window in which dysregulated activation leads to severe disease. Analysis of TLR9GFP/GFP and TLR7TOM/TOM reporter mice, in which GFP and TdTomato are expressed from the endogenous TLR9 and TLR7 locus, respectively (17), revealed that approximately 20% of neonatal liver macrophages express TLR9, but a much lower proportion express TLR7 (Fig. 3F). Similarly, Ly6Chi monocytes in livers of neonates express TLR9 but not TLR7 (Fig. 3F). Interestingly, the analyses of macrophages and Ly6Chi monocytes from adult bone marrow revealed quite distinct expression levels; both populations express TLR7, while TLR9 expression is low in adult macrophages in the bone marrow (Fig. 3G). Based on these findings, we conclude that the expression of TLR9, but not of TLR7, by liver macrophages and Ly6Chi monocytes explains why dysregulation of TLR9 signaling early in life leads to a much more severe phenotype compared with dysregulated TLR7 signaling.
TLR9-Driven Disease Requires IFN-γ Receptor Signaling, Rather than Type I IFN Receptor Signaling.
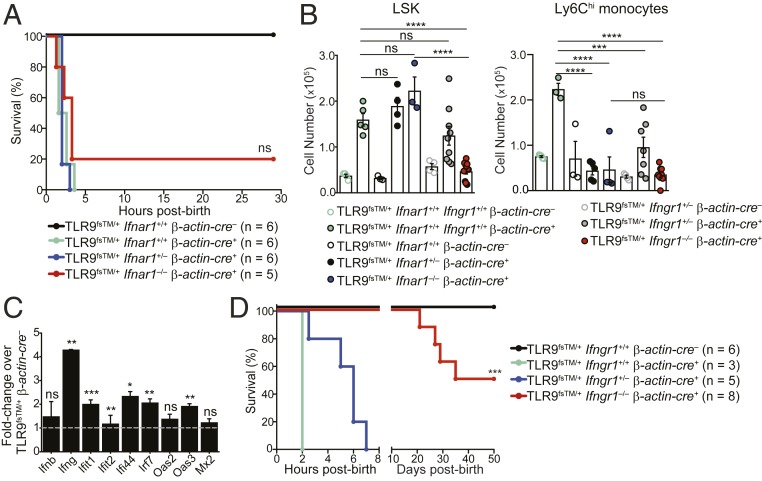
We next sought to identify the inflammatory signaling pathways underlying the disease observed in TLR9fsTM/+β-actin-cre+ neonates. The inflammatory disease in TLR7tg mice requires IFN-α receptor (IFNAR) signaling (15). To test whether type I IFN contributes to the inflammation in TLR9fsTM/+β-actin-cre+ neonates, we crossed TLR9fsTM/+ and β-actin-cre+ mice to Ifnar1−/− mice. Surprisingly, 80% of TLR9fsTM/+β-actin-cre+Ifnar1−/− neonates died within 5 h of birth (Fig. 4A). One TLR9fsTM/+β-actin-cre+Ifnar1−/− mouse survived after birth, but this animal was extremely runted. Furthermore, TLR9fsTM/+β-actin-cre+Ifnar1−/− neonates still exhibited increases in LSK, Sca-1+, and Ly6Chi cell populations (Fig. 4B and SI Appendix, Fig. S5A). Based on these analyses, we conclude that, in contrast to many other autoinflammatory disease models, IFNAR signaling is not required for the pathology observed in TLR9fsTM/+β-actin-cre+ neonates.
Fig. 4.
Fatal inflammation in TLR9TM-expressing neonates requires IFN-γ receptor signaling but not type I IFN signaling. (A) Survival curves of TLR9fsTM/+β-actin-cre+Ifnar1−/− neonates. Results are combined from multiple litters and analyzed using the logrank (Mantel–Cox) test. (B) LSK and Ly6Chi monocyte cell populations do not expand in TLR9fsTM/+β-actin-cre+Ifngr1−/− neonates. Quantification of LSK and Ly6Chi monocytes from neonatal livers of indicated genotypes (combined from multiple litters) is shown as mean ± SEM and analyzed using one-way ANOVA with Tukey’s multiple comparisons posttest. Mouse numbers: TLR9fsTM/+ β-actin-cre–, n = 4; TLR9fsTM/+ β-actin-cre+, n = 5; TLR9fsTM/+β-actin-cre–Ifnar1+/+, n = 4; TLR9fsTM/+β-actin-cre+Ifnar1+/−, n = 4; TLR9fsTM/+β-actin-cre+Ifnar1−/−, n = 3; TLR9fsTM/+β-actin-cre–Ifngr1+/−, n = 4; TLR9fsTM/+β-actin-cre+Ifngr1+/−, n = 7; TLR9fsTM/+β-actin-cre+Ifngr1−/−, n = 9. (C) Quantitative PCR analysis of inflammatory genes in TLR9fsTM/+β-actin-cre+ neonatal livers. Data are representative of two independent experiments and presented as fold change of expression in TLR9fsTM/+β-actin-cre+ vs. TLR9fsTM/+ β-actin-cre–. Bars show mean ± SD; statistics calculated using the two-tailed Student’s t test. (D) Survival curves of TLR9fsTM/+β-actin-cre+Ifngr1−/− neonates. Results are combined from multiple litters and analyzed using the logrank (Mantel-Cox) test. In all panels, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We searched for other inflammatory pathways induced when compartmentalized activation of TLR9 is bypassed early in life. Gene expression analysis of livers from TLR9fsTM/+β-actin-cre+ neonates revealed a significant induction of IFN-γ and IFN-stimulated genes (Fig. 4C), suggesting a potential role for type II IFN in TLR9-driven fatality. To test whether IFN-γ contributes to the inflammation in TLR9fsTM/+β-actin-cre+ neonates, we crossed TLR9fsTM/+ and β-actin-cre+ mice to Ifngr1−/− mice. All TLR9fsTM/+β-actin-cre+Ifngr1−/− neonates survived for 20 d postbirth, while none of the TLR9fsTM/+β-actin-cre+Ifngr1+/− and TLR9fsTM/+β-actin-cre+Ifngr1+/+ neonates survived past 7 h (Fig. 4D). We noted that TLR9fsTM/+β-actin-cre+Ifngr1−/− neonates were runted compared with TLR9fsTM/+β-actin-cre–Ifngr1−/− neonates, and a number of these mice died between 20 and 50 d postbirth, indicating that additional inflammatory pathway(s) can contribute to disease pathology later in life. Consistent with reduced disease in TLR9fsTM/+β-actin-cre+Ifngr1−/− neonates, there was no expansion of LSK and Ly6Chi cell populations in livers of these mice (Fig. 4B). The rescue of disease was not related to any change in TLR9 expression in neonatal macrophages (SI Appendix, Fig. S5B). Together, these data indicate that the pathology associated with TLR9 dysregulation is largely driven by IFN-γ receptor signaling.
Dysregulation of TLR9 Activation Leads to Expansion of IFN-γ–Producing NK Cells.
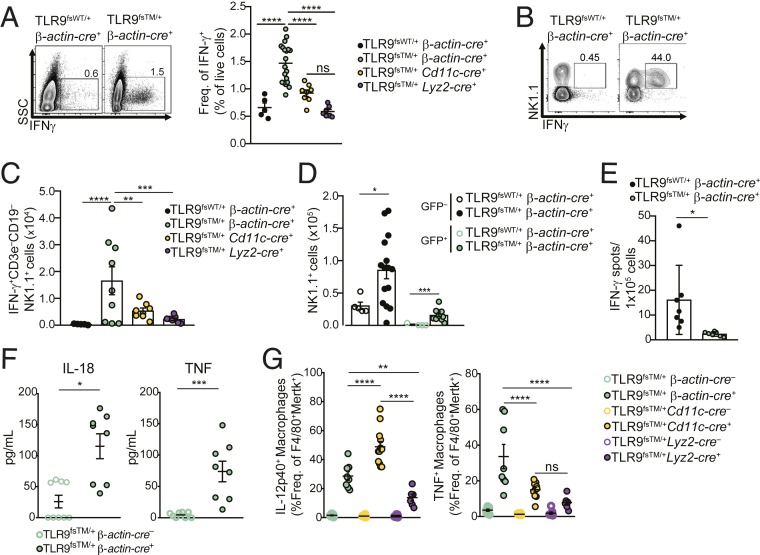
To identify the cells responsible for producing IFN-γ in TLR9fsTM/+β-actin-cre+ mice, we performed intracellular cytokine staining on neonatal liver cells. These analyses identified a clear population of IFN-γ–producing cells in TLR9fsTM/+β-actin-cre+ neonates that was absent in TLR9fsWT/+β-actin-cre+ neonates (Fig. 5A). The vast majority of these IFN-γ+ cells were NK1.1+ cells (CD3–CD19–NK1.1+) and expressed a number of cell markers indicative of NK cells, including NKG2D, NKp46, NKG2A/C/E, and CD226 (DNAM) (Fig. 5B and SI Appendix, Fig. S6 A and B) (29–32). The number of IFNγ+ NK cells was greatly expanded in the livers of TLR9fsTM/+β-actin-cre+ neonates (Fig. 5C), suggesting that they contribute to TLR9-driven disease.
Fig. 5.
Expansion of IFN-γ–producing NK cells correlates with neonatal inflammation. (A, Left) Representative flow cytometry plots of IFN-γ–producing cells in neonatal livers of TLR9fsWT/+β-actin-cre+ and TLR9fsTM/+β-actin-cre+ mice. (A, Right) Quantification of IFN-γ+ cells in neonatal livers of mice of the indicated genotypes. Data combined from multiple litters are shown as mean ± SEM and analyzed using one-way ANOVA with Tukey’s multiple comparisons posttest. Mouse numbers: TLR9fsWT/+ β-actin-cre+, n = 5; TLR9fsTM/+ β-actin-cre+, n = 19; TLR9fsTM/+ Cd11c-cre+, n = 8; TLR9fsTM/+ Lyz2-cre+, n = 7. (B) Expansion of IFN-γ+ NK1.1+ cells in TLR9fsTM/+β-actin-cre+ neonates. Shown are representative flow cytometry plots of NK1.1 and IFN-γ staining on CD3–CD19–NK1.1+ gated cells from neonatal livers of the indicated mice. (C) Quantification of IFN-γ+CD3–CD19–NK1.1+ subsets of indicated genotypes. Results from multiple litters are shown as mean ± SEM and analyzed using one-way ANOVA with Tukey’s multiple comparisons posttest. Mouse numbers: TLR9fsWT/+ β-actin-cre+, n = 5; TLR9fsTM/+ β-actin-cre+, n = 9; TLR9fsTM/+ Cd11c-cre+, n = 8; TLR9fsTM/+ Lyz2-cre+, n = 7. (D) Quantification of GFP+ and GFP– CD3–CD19–NK1.1+ cells in neonatal livers. Results are combined from multiple litters and shown as mean ± SEM and analyzed using the two-tailed Student’s t test. Mouse numbers: TLR9fsWT/+ β-actin-cre+, n = 5; TLR9fsTM/+ β-actin-cre+, n = 15. (E) Analysis of IFN-γ secretion by GFP+ and GFP– total cells by ELISpot. Quantification of IFN-γ secreting cells of indicated genotypes. Results combined from multiple litters are shown as mean ± SEM and analyzed using the two-tailed Student’s t test. Mouse numbers: TLR9fsWT/+ β-actin-cre+, n = 7; TLR9fsTM/+ β-actin-cre+, n = 7. (F) Neonatal liver cells from TLR9fsTM/+ B-actin-cre+ mice secrete more IL-18 and TNF compared to littermate controls. Shown are cytokine levels of adherent fetal liver cells cultured from TLR9fsTM/+ β-actin-cre+ and TLR9fsTM/+ β-actin-cre– neonates. Results are combined from two independent experiments and analyzed using the two-tailed Student’s t test. Mouse numbers: TLR9fsTM/+ β-actin-cre–, n = 9; TLR9fsTM/+ β-actin-cre+, n = 8. (G) Neonatal macrophages from TLR9fsTM/+ β-actin-cre+ and TLR9fsTM/+ Cd11c-cre+ neonates secrete significantly more TNF and IL-12p40 compared with TLR9fsTM/+ Lyz2-cre+ neonates. Results are combined from multiple experiments and analyzed using one-way ANOVA with Tukey’s multiple comparisons posttest. Mouse numbers: TLR9fsTM/+ β-actin-cre–, n = 10; TLR9fsTM/+ β-actin-cre+, n = 8; TLR9fsTM/+ Cd11c-cre–, n = 7; TLR9fsTM/+ Cd11c-cre+, n = 12; TLR9fsTM/+ Lyz2-cre–, n = 8; TLR9fsTM/+ Lyz2-cre+, n = 6. In all panels, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To more closely examine the link between IFN-γ producing NK cells and the disease observed in mice with dysregulated TLR9, we performed similar analyses in TLR9fsTM/+Cd11c-cre+ and TLR9fsTM/+Lyz2-cre+ mice. The frequency and number of IFN-γ–producing NK cells was increased in TLR9fsTM/+Cd11c-cre+ neonates but not to the extent observed in TLR9fsTM/+β-actin-cre+ neonates, while TLR9fsTM/+Lyz2-cre+ neonates showed no significant increase relative to control mice (Fig. 5 A and C). Thus, the presence of IFN-γ–producing NK cells correlated with disease severity.
Production of IFN-γ by NK cells could result from cell-intrinsic activation of TLR9 or from stimulation by cytokines, such as IL-18 and IL-12, produced by other TLR9-expressing cells (33, 34). To distinguish between these possibilities, we examined GFP expression in the expanded NK cells as a proxy for TLR9 expression. Interestingly, we observed that both GFP+ and GFP– NK1.1+ cells expanded in TLR9fsTM/+β-actin-cre+ neonates (Fig. 5D). These data suggest that the activation of NK cells does not require TLR9 expression and may occur via exposure to inflammatory cytokines.
To examine this possibility more directly, we sorted GFP– and GFP+ total cells from TLR9fsTM/+β-actin-cre+ neonatal livers and analyzed their IFN-γ production by ELISpot. The IFN-γ–producing cells were predominately cells that lacked expression of TLR9(GFP–) (Fig. 5E), indicating that inflammatory cytokines produced by another TLR9-expressing cell are most likely driving NK cell expansion and IFN-γ production. Consistent with this possibility, neonatal liver cells from TLR9fsTM/+β-actin-cre+ neonates produced IL-18 and TNF when cultured ex vivo (Fig. 5F). Using intracellular cytokine staining, we verified that neonatal macrophages and Ly6Chi monocytes produce IL-12p40 and TNF in TLR9fsTM/+β-actin-cre+ mice (Fig. 5G and SI Appendix, Fig. S6C). Moreover, the frequency of cytokine-producing cells across TLR9fsTM/+β-actin-cre+, TLR9fsTM/+Cd11c-cre+, and TLR9fsTM/+Lyz2-cre+ neonates generally correlated with disease severity (Fig. 5G and SI Appendix, Fig. S6C).
Together, these data suggest a model in which dysregulated TLR9 activation in neonatal macrophages induces expansion of Ly6Chi monocytes. In turn, macrophages and Ly6Chi monocytes secrete inflammatory cytokines that activate NK cells, leading them to expand and produce IFN-γ, ultimately driving neonatal fatality.
Discussion
Both TLR7 and TLR9 can respond to self-nucleic acids under certain circumstances, but whether dysregulated responses by each receptor result in similar mechanisms of disease has been difficult to address. In this study, we use a model of TLR9-driven disease to uncover that TLR9-driven IFN-γ production leads to detrimental autoinflammation with some similarities to MAS-like disease. Unlike models of TLR7-driven disease, the pathology in this TLR9-driven model was largely independent of type I IFN. The phenotype of TLR9fsTM/+β-actin-cre+ mice that we describe here is very severe, with mice living only a few hours after birth. This disease was characterized by an apparent activation of neonatal macrophages in the liver, an increase in Ly6Chi monocytes, altered hematopoiesis, and an increase in the number of IFN-γ producing-NK cells. Surprisingly, this fatal inflammation was dependent on IFN-γ receptor signaling, indicating different mechanisms downstream of TLR9- and TLR7-driven autoinflammatory diseases. Finally, our analysis of TLR9 and TLR7 reporter mice provides evidence that the expression of these nucleic acid sensing receptors is distinct in the key cell types implicated in TLR9-driven disease.
A unique aspect of our study is the ability to control the timing of TLR9TM, which revealed the severe sensitivity of neonates to dysregulated TLR9 activation relative to adult animals. Mice induced to express TLR9TM at the time of weaning develop signs of systemic inflammation that are similar to disease observed in TLR7-overexpressing mice. In contrast, mice that experience dysregulated TLR9 during development suffer severe liver inflammation and lethality. One potential explanation for the difference between neonates and adults is the abundance of nucleic acids early in life due to cell death associated with development. However, our inability to exacerbate disease in adults by providing additional nucleic acids, either through irradiation or injection of TLR9 ligands, argues against this possibility. Rather, the severe phenotype in TLR9TM-expressing neonates may reflect the particular sensitivity of the organism to inflammation during this critical developmental period. For example, apoptosis and necroptosis of cells during organ development may lead to release of nucleic acids, allowing for recognition of endogenous DNA by TLR9. It is also possible that commensal-derived nucleic acids can stimulate TLR9TM; however, the fact that neonates are born with severe liver and pancreas damage and die so quickly after birth argues against this possibility. Our findings that both macrophages in the neonatal liver and Ly6Chi monocytes express TLR9 but little or no TLR7 suggest that these cells may be responsive to DNA but not RNA ligands during embryogenesis, explaining the different timing and severity of TLR9-driven vs. TLR7-driven disease. It is also possible that regulatory mechanisms in adult mice not present in neonates ameliorate the consequences of TLR9 dysregulation; for example, IL-10 can suppress MAS-associated pathology in adult mice (18). Perhaps the differential induction of IL-10 in adult vs. neonatal mice is responsible for the different disease severity. In addition, the fact that neonatal fatality is dependent on IFN-γ receptor signaling suggests aberrant type II IFN can cause pathology early in life, whereas type I IFN may play a greater role in adult autoinflammatory disease. Finally, our observation that TLR9fsTM/+β-actin-cre+ neonates exhibit emergency myelopoiesis in the liver and cell infiltration in the pancreas, suggests that developing organs may be particularly sensitive to inflammatory signals early in life.
Recently, it has been shown that mice lacking exonuclease PLD4 also develop a TLR9- and IFN-γ–dependent inflammatory disease but survive to adulthood, while Pld3−/−Pld4−/−mice die 2 to 3 wk after birth. Whether the disease in Pld3−/−Pld4−/−mice is dependent on TLR9 and IFN-γ has not been determined, but these animals exhibit elevated levels of systemic IFN-γ and other inflammatory cytokines, suggestive of dysregulated TLR9 activation (20). The difference in disease severity between TLR9fsTM mice and mice lacking PLD3 and/or PLD4 implies that a hierarchy of mechanisms restrict TLR9 responses to self-nucleic acids. Endosomal exonucleases, such as PLD3 and PLD4, function to limit self-DNA accumulation within endosomes, but this activity is clearly not the sole mechanism preventing TLR9 responses to self-DNA. Additional nucleases have been implicated in the removal of extracellular self-DNA (23–25), and it is likely that these nucleases, acting in distinct cellular and extracellular compartments, prevent self-DNA from triggering properly localized TLR9. Our analysis of TLR9fsTM mice reveals the critical importance of compartmentalized activation; these mice develop severe disease despite the presence of functional nucleases.
Expansion of a population of atypical NK cells (NK1.1+CD11c+) has been described in TLR7tg mice (35, 36). These atypical NK cells express low levels of NKG2D and require type I IFN to induce inflammation (36). In contrast, in our model of TLR9-driven disease, we observe expansion of NK cells expressing classic NK cell markers, including NKG2D and NKp46, and show that these cells act as potent sources of IFN-γ in this IFN-γ–dependent disease. NK production of IFN-γ may promote the expansion and activation of macrophages; these activated macrophages have the potential to phagocytose red blood cells, thereby driving anemia and neonatal death (8). Thus, it remains unclear how similar the NK cell populations are in TLR7-driven vs. TLR9-driven disease. In studies examining patients with active SLE or juvenile rheumatoid arthritis, circulating NK cells are potent IFN-γ producers, but the frequency of these cells is low (37, 38), making it challenging to determine whether NK cells have a protective or disease-promoting role during different stages of autoinflammatory disease.
Our study suggests that inappropriately activated NK cells can contribute to autoinflammatory responses. Further investigation into the specific mechanisms leading to NK cell expansion and type II IFN-mediated inflammation in human patients may help identify the most opportune stage of disease at which to administer therapeutics.
Methods
The study methods are described in detail in SI Appendix. Mice were housed under specific pathogen-free conditions at the University of California (UC) Berkeley. All mouse experiments were approved by and performed in accordance with the guidelines of the Animal Care and Use Committee at UC Berkeley. Unless noted otherwise, mice were analyzed as neonates (e18.5 to 1 to 3 h postbirth) or as adults (6 to 12 wk). Timed pregnancies were estimated considering the day of vaginal plug formation as 0.5 d postcoitus. TLR9fsTM and TLR9fsWT knockin mice were generated by the UC Davis Mouse Biology Program. For cell staining, dead cells were excluded using DAPI (Thermo Fisher Scientific), and all stainings were performed in flow cytometry buffer including anti-CD16/32 Fc blocking antibody (2.4G2; UCSF Antibody Core) and normal mouse serum (Sigma-Aldrich). Cells were stained for 30 min at 4 °C with antibodies. All cells were analyzed on an LSRFortessa X-20 flow cytometer (BD Biosciences), and data were analyzed with FlowJo (TreeStar). For blood hemoglobin analysis, 10 μL of neonatal blood was collected immediately postsacrifice. Hemoglobin blood levels were measured using an AimStrip hemoglobin testing system (Germaine Laboratories; catalog no. 78200). For secreted cytokine analysis, fetal liver cells were collected, processed, and incubated in tissue culture-treated wells for 4 h. Suspension cells were removed, and adherent cells were incubated for an additional 14 h. Supernatants were collected and analyzed using the 36-plex Luminex Mouse Inflammation Kit (ProcartaPlex; catalog no. EPX360-26092-901).
Data Availability.
All data are available in the paper and SI Appendix.
Supplementary Material
Acknowledgments
We thank R. Vance and members of the G.M.B. and Vance laboratories for constructive discussions and critical reading of the manuscript; J. Tatah for assistance with analysis of the TLR9fsTM Cd19-cre mice; H. Nolla and A. Valeros for assistance with flow cytometry at the Flow Cytometry Facility of the Cancer Research Laboratory at UC Berkeley; and G. Reiner, S. McWhirter, and Aduro BioTech for Luminex reagents. This work was supported by the NIH (Grants A1072429 and AI063302, to G.M.B.), the Lupus Research Institute, and the NSF (Graduate Research Fellowship DGE1106400, to A.G.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.M.-R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911579117/-/DCSupplemental.
References
- 1.Barbalat R., Ewald S. E., Mouchess M. L., Barton G. M., Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 29, 185–214 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Marshak-Rothstein A., Toll-like receptors in systemic autoimmune disease. Nat. Rev. Immunol. 6, 823–835 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen S. R., et al. , Toll-like receptor 9 controls anti-DNA autoantibody production in murine lupus. J. Exp. Med. 202, 321–331 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen S. R., et al. , Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity 25, 417–428 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Lau C. M., et al. , RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J. Exp. Med. 202, 1171–1177 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leadbetter E. A., et al. , Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature 416, 603–607 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Pisitkun P., et al. , Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science 312, 1669–1672 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Akilesh H. M., et al. , Chronic TLR7 and TLR9 signaling drives anemia via differentiation of specialized hemophagocytes. Science 363, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton G. M., Kagan J. C., Medzhitov R., Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat. Immunol. 7, 49–56 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Ewald S. E., et al. , The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456, 658–662 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park B., et al. , Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat. Immunol. 9, 1407–1414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mouchess M. L., et al. , Transmembrane mutations in Toll-like receptor 9 bypass the requirement for ectodomain proteolysis and induce fatal inflammation. Immunity 35, 721–732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deane J. A., et al. , Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity 27, 801–810 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian S., et al. , A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc. Natl. Acad. Sci. U.S.A. 103, 9970–9975 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buechler M. B., Teal T. H., Elkon K. B., Hamerman J. A., Cutting edge: Type I IFN drives emergency myelopoiesis and peripheral myeloid expansion during chronic TLR7 signaling. J. Immunol. 190, 886–891 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guiducci C., et al. , RNA recognition by human TLR8 can lead to autoimmune inflammation. J. Exp. Med. 210, 2903–2919 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts A. W., et al. , Tissue-resident macrophages are locally programmed for silent clearance of apoptotic cells. Immunity 47, 913–927.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behrens E. M., et al. , Repeated TLR9 stimulation results in macrophage activation syndrome-like disease in mice. J. Clin. Invest. 121, 2264–2277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weaver L. K., Chu N., Behrens E. M., Brief report: Interferon-γ-mediated immunopathology potentiated by Toll-like receptor 9 activation in a murine model of Macrophage Activation Syndrome. Arthritis Rheumatol. 71, 161–168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavin A. L., et al. , PLD3 and PLD4 are single-stranded acid exonucleases that regulate endosomal nucleic-acid sensing. Nat. Immunol. 19, 942–953 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yáñez A., et al. , Granulocyte-monocyte progenitors and monocyte-dendritic cell progenitors independently produce functionally distinct monocytes. Immunity 47, 890–902.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashkenazi A., Salvesen G., Regulated cell death: Signaling and mechanisms. Annu. Rev. Cell Dev. Biol. 30, 337–356 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Sisirak V., et al. , Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell 166, 88–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apostolov E. O., et al. , Deoxyribonuclease I is essential for DNA fragmentation induced by gamma radiation in mice. Radiat. Res. 172, 481–492 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawane K., et al. , Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science 292, 1546–1549 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Boyd J. H., Mathur S., Wang Y., Bateman R. M., Walley K. R., Toll-like receptor stimulation in cardiomyoctes decreases contractility and initiates an NF-kappaB dependent inflammatory response. Cardiovasc. Res. 72, 384–393 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Hoeffel G., et al. , C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider C., et al. , Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 15, 1026–1037 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Jamieson A. M., et al. , The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity 17, 19–29 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Marcus A., et al. , Recognition of tumors by the innate immune system and natural killer cells. Adv. Immunol. 122, 91–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibuya A., et al. , DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity 4, 573–581 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Lanier L. L., NK cell recognition. Annu. Rev. Immunol. 23, 225–274 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Lauwerys B. R., Garot N., Renauld J. C., Houssiau F. A., Cytokine production and killer activity of NK/T-NK cells derived with IL-2, IL-15, or the combination of IL-12 and IL-18. J. Immunol. 165, 1847–1853 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Takeda K., et al. , Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity 8, 383–390 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Voynova E. N., Skinner J., Bolland S., Expansion of an atypical NK cell subset in mouse models of systemic lupus erythematosus. J. Immunol. 194, 1503–1513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voynova E., Qi C. F., Scott B., Bolland S., Cutting Edge: Induction of inflammatory disease by adoptive transfer of an atypical NK cell subset. J. Immunol. 195, 806–809 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schleinitz N., Vély F., Harlé J. R., Vivier E., Natural killer cells in human autoimmune diseases. Immunology 131, 451–458 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hervier B., et al. , Phenotype and function of natural killer cells in systemic lupus erythematosus: Excess interferon-γ production in patients with active disease. Arthritis Rheum. 63, 1698–1706 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the paper and SI Appendix.