Significance
Life depends on the propagation of heritable information across successive generations. An RNA polymerase ribozyme was obtained by in vitro evolution that has an unprecedented level of activity in copying complex RNA templates. The polymerase is able to synthesize its own evolutionary ancestor, an RNA ligase ribozyme, in the form of three fragments that assemble to give a functional complex, as well as to synthesize the complements of each of those three fragments. However, when pushed to the limits of its activity, the polymerase operates with lower fidelity, which is an impediment to maintaining functional information, as would be needed to provide an RNA-based living system.
Keywords: directed evolution, RNA enzyme, RNA replication
Abstract
The RNA-based organisms from which modern life is thought to have descended would have depended on an RNA polymerase ribozyme to copy functional RNA molecules, including copying the polymerase itself. Such a polymerase must have been capable of copying structured RNAs with high efficiency and high fidelity to maintain genetic information across successive generations. Here the class I RNA polymerase ribozyme was evolved in vitro for the ability to synthesize functional ribozymes, resulting in the markedly improved ability to synthesize complex RNAs using nucleoside 5′-triphosphate (NTP) substrates. The polymerase is descended from the class I ligase, which contains the same catalytic core as the polymerase. The class I ligase can be synthesized by the improved polymerase as three separate RNA strands that assemble to form a functional ligase. The polymerase also can synthesize the complement of each of these three strands. Despite this remarkable level of activity, only a very small fraction of the assembled ligases retain catalytic activity due to the presence of disabling mutations. Thus, the fidelity of RNA polymerization should be considered a major impediment to the construction of a self-sustained, RNA-based evolving system. The propagation of heritable information requires both efficient and accurate synthesis of genetic molecules, a requirement relevant to both laboratory systems and the early history of life on Earth.
In all known extant life, heritable information is maintained within the sequence of nucleic acid genomes that are copied by polymerase proteins. In the RNA world, which is thought to have preceded the invention of genetically encoded proteins, this task likely would have been performed by an RNA polymerase ribozyme that copies RNA molecules, including itself (1–3). For an RNA-based organism to self-replicate and undergo Darwinian evolution, the RNA polymerase ribozyme must have sufficient activity to amplify RNAs as complex as itself and must do so with sufficient accuracy to maintain heritable information against deleterious mutations (4, 5). A demonstration of these properties is essential both for understanding the nature of presumed RNA-based life and for constructing synthetic RNA life in the laboratory (6, 7).
Most examples of the RNA-catalyzed synthesis of complex RNA molecules have relied on polymerase ribozymes that are descendants of the class I ligase. These ribozymes were obtained by in vitro evolution, initially selecting for the RNA-templated joining of two RNA molecules and thereafter selecting for the templated polymerization of multiple successive nucleoside 5′-triphosphates (NTPs) (8–10). Numerous rounds of directed evolution have been carried out over the past two decades, leading to greatly improved polymerase activity and most recently yielding polymerase ribozymes that are capable of synthesizing other functional ribozymes (10–15). At the utmost limits of demonstrated activity, substantial portions of complex functional RNAs, including tRNA and the catalytic subunit of the polymerase itself, can be synthesized, albeit only at very low yields and by relying on preformed oligonucleotides to compose a substantial portion of the functional sequence (14, 15). The synthesis of a complex ribozyme in its entirety has remained elusive because complex structural features of the ribozyme generally correspond to the most difficult portions of the template for the polymerase to copy.
Although a ribozyme must adopt a folded structure to achieve catalysis, it need not be composed of a single, contiguous RNA strand. Complex ribozymes that exist in nature, such as the ribosome and spliceosome (16, 17), are formed by Watson–Crick pairing and other noncovalent interactions among multiple strands of RNA. Similarly, both natural and synthetic ribozymes can be engineered to assemble from smaller, more loosely structured fragments (18–21). Examples of these constructs include the class I ligase and some of its polymerase descendants, which can be divided into fragments that self-assemble and retain function (22–24). In an RNA-based organism, polymerase ribozymes composed of multiple fragments may have been replicated more easily, and these smaller fragments may have emerged more readily through the prebiotic synthesis of RNA (2, 3, 18).
Here directed evolution was used to obtain a highly improved class I polymerase ribozyme by selecting for its ability to synthesize a functional hammerhead ribozyme, which catalyzes the site-specific cleavage of RNA. An RNA polymerase that was isolated after 14 rounds of evolution has the ability to synthesize complex and highly structured RNAs. Most notably, the improved polymerase can synthesize a functional copy of its own evolutionary ancestor, the class I ligase, by generating three RNA fragments that can assemble spontaneously following purification to give a catalytically active ligase. The improved polymerase also can synthesize the template RNAs that encode each of these three ligase fragments.
This result represents the most complex functional RNA that has been synthesized by a ribozyme from mononucleotide substrates, and it suggests that a similar in vitro evolution strategy could lead to further improvements that enable the polymerase to synthesize itself. However, the fidelity of RNA synthesis is modest, especially on the most challenging templates. Thus, only a very small fraction of the assembled ligase ribozymes has catalytic activity, due to the presence of disabling mutations. Further efforts to improve the polymerase will need to select more strongly for the accurate synthesis of complex RNAs to enable the generalized replication of functional RNAs.
According to the formulation first devised by Manfred Eigen (4) and broadly adopted by others in the field, the fitness of a replicating, evolving entity is determined by three key variables: the rate of amplification (A), the fidelity (quality) of information transfer from parent to progeny (Q), and the rate of decomposition (D). The net production of new copies of a replicating entity is proportional to the current number of copies multiplied by the factor A·Q – D. Studies aiming to develop an RNA replicase ribozyme have generally focused on the production of full-length products (A) rather than on the production of accurate, full-length products (A·Q). Future efforts to improve the activity of the RNA polymerase ribozyme will need to address the latter if the aim is to develop RNA-based life in the laboratory.
Results
Evolution of Polymerase Ribozymes That Synthesize Functional Ribozymes.
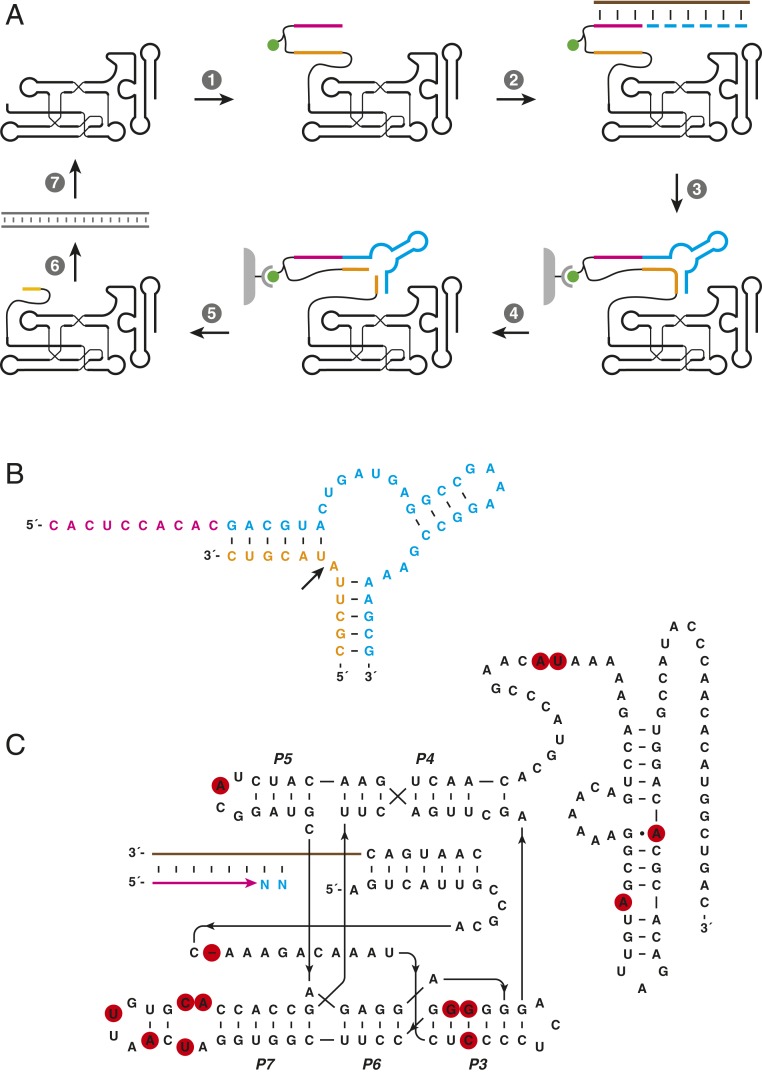
The class I RNA polymerase ribozyme has previously been evolved in vitro for the ability to synthesize functional RNA aptamers (14). Those efforts resulted in the “24-3” variant, which has substantially improved activity compared with its predecessors in copying highly structured RNA templates and in synthesizing a variety of functional RNAs, including simple ribozymes. A more stringent selection procedure was devised to take advantage of this improved activity by requiring the synthesis of a functional hammerhead endonuclease ribozyme. The population of evolved RNAs from which the 24-3 polymerase was isolated were covalently attached to a synthetic oligonucleotide containing an RNA primer, biotin moiety, and the substrate for the hammerhead ribozyme (Fig. 1A). These RNAs were challenged to extend the attached primer on a separate RNA template using NTP substrates, resulting in synthesis of the catalytic motif of the hammerhead ribozyme (Fig. 1B).
Fig. 1.
In vitro evolution of the 38-6 RNA polymerase ribozyme. (A) Scheme for selective amplification of polymerase ribozymes that synthesize a functional hammerhead ribozyme. (1) Attachment to the polymerase of an RNA primer (magenta), biotin (green), and the RNA substrate (orange) to be cleaved by the hammerhead. (2) Hybridization of the primer to an RNA template (brown) that encodes the hammerhead. (3) Extension of the primer by polymerization of NTPs (cyan), followed by biotin capture on streptavidin magnetic beads (gray). (4) Cleavage of the attached RNA substrate by the hammerhead, releasing the polymerase from the beads. (5) Recovery of functional polymerases. (6) Reverse transcription and PCR amplification. (7) Transcription to generate progeny polymerases. (B) Sequence and secondary structure of the hammerhead ribozyme (cyan), together with the primer used to initiate its synthesis and the RNA substrate. The arrow indicates the site of cleavage. (C) Sequence and secondary structure of the 38-6 RNA polymerase. Red circles indicate mutations relative to the 24-3 polymerase (14). Stem elements P3–P7 within the core domain are labeled.
The products of the primer extension reaction were captured via binding of biotin to streptavidin-coated beads. Then the captured products were incubated under conditions that enable a functional hammerhead to cleave a site within the RNA substrate region lying between the primer and the polymerase, thereby releasing the polymerase from the beads. The released polymerase ribozymes then were reverse-transcribed, PCR-amplified, and forward-transcribed to yield a progeny population of ribozymes to begin the next round of evolution.
The in vitro evolution procedure was continued for 14 rounds, with the time allowed for polymerization decreasing progressively from 2 h to 5 min. Selection stringency was further increased during the final eight rounds by adding a gel-shift selection step in which only those polymerases that had extended the primers to full length, as reflected by corresponding mobility in a denaturing polyacrylamide gel, were carried forward to the biotin capture step and subsequent hammerhead cleavage.
After the final round of evolution (round 38 in total), individuals were cloned from the population and sequenced. This analysis revealed three distinct sequence families, the largest of which contained the most efficient polymerases. The most active of these individuals, termed “38-6,” differs from 24-3 by 14 mutations (Fig. 1C). These mutations are distributed throughout both the core ligase domain and the structurally distinct accessory domain of the ribozyme. Most of the mutations occur in the same regions that distinguish 24-3 from earlier versions of the polymerase. The most notable structural change is disruption of the distal portion of the P7 stem, opening the L7 loop that closes the stem. Previous biochemical and structural studies have suggested that this region is not functionally important (10, 25, 26), but that no longer appears to be the case.
The Evolved Polymerase Can Synthesize Complex RNAs.
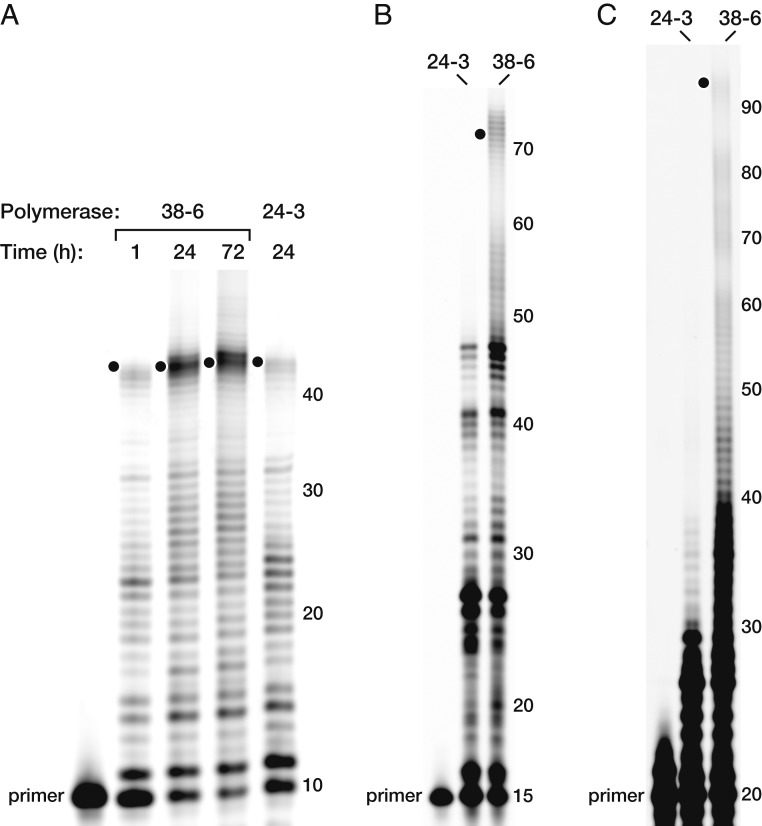
The 38-6 ribozyme has markedly improved RNA-dependent RNA polymerase activity compared with the 24-3 predecessor, especially in the synthesis of complex, structured RNAs. When challenged to synthesize the hammerhead ribozyme on a separate RNA template using a separate RNA primer, 24-3 generates full-length product in 2.4% yield after 24 h, whereas 38-6 achieves a 2.0% yield in 1 h and a 24% yield in 24 h (Fig. 2A). The synthesis of more complex RNAs shows even greater improvement. The RNA-catalyzed synthesis of yeast phenylalanyl tRNA, starting from a primer that contains the first 15 nucleotides and requiring the addition of 61 nucleotides, was barely detectable even after 5 d using the 24-3 polymerase, with a yield of 0.07%. In contrast, the 38-6 polymerase yielded 2.4% full-length product after 5 d (Fig. 2B).
Fig. 2.
Synthesis of functional RNAs molecules by the 24-3 and 38-6 polymerases. (A) Synthesis of the hammerhead ribozyme, as shown in Fig. 1B. (B) Synthesis of yeast phenylalanyl-tRNA after 5 d. (C) Synthesis of the class I ligase after 5 d. Reaction conditions: 100 nM polymerase, 80 nM primer, 100 nM template, 4 mM each NTP, and 200 mM MgCl2 at pH 8.3 and 17 °C. Numbers at the right indicate product lengths, including the primer. Black dots indicate full-length products.
As shown in Fig. 2 and described previously (27), the polymerase generates some products that are several nucleotides longer than full length. This is due to extension beyond the canonical template region and into the adjacent region of the template comprised of a hexadenylate spacer and nucleotides that are complementary to the “processivity tag” (12) of the polymerase. This behavior suggests that the processivity tag, although beneficial for activity, may ultimately be dispensable.
The 38-6 polymerase is a descendant of the class I ligase ribozyme and retains the same catalytic core as the ligase, with 77% sequence identity in this 97-nucleotide region (10). The ligase is the fastest and most structurally complex ribozyme that has been evolved starting from a pool of random-sequence RNAs (25). As a highly demanding test of the 38-6 polymerase, it was challenged to synthesize the b1-207t variant of the class I ligase, using a primer that contains the first 20 nucleotides of the ligase. The yield of full-length products in this reaction was 0.12% after 5 d (Fig. 2C). Notably, the 24-3 predecessor is unable to synthesize the class I ligase in detectable yield, even after long incubation. Nonetheless, the yield of full-length ligase obtained with 38-6 is low, and the 20-nucleotide primer provides a significant portion of the ligase motif, including residues that coordinate the catalytic Mg2+ ion within the ribozyme’s active site (26).
The Polymerase Can Synthesize Its Own Ancestor in Three Fragments.
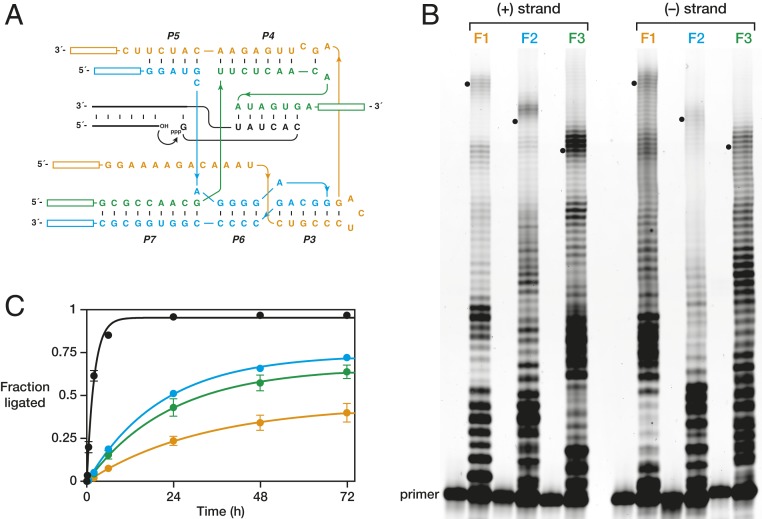
A divide-and-conquer strategy was adopted to enable the 38-6 polymerase to synthesize the class I ligase in its entirety, splitting the ligase into three fragments that can assemble noncovalently to form a functional ribozyme (Fig. 3A). The sites of fragmentation were chosen within stem-loop elements that are known to be tolerant of insertions and are not critical for the ribozyme to adopt its correctly folded structure (8, 26). The sites of fragmentation have been used previously to split the ligase or its descendants to provide a complex that retains catalytic activity (22, 24). One split was at the distal end of the P5 stem between nucleotides C40 and G41, and the other was at the distal end of the P7 stem, replacing the L7 loop with two C·G base pairs to stabilize the interaction between fragments 2 and 3. The resulting three-part class I ligase is only approximately sevenfold less active than the contiguous form of the b1-207t ribozyme (SI Appendix, Fig. S1).
Fig. 3.
Synthesis of the three-fragment form of the class I ligase ribozyme by the 38-6 polymerase. (A) Sequence and secondary structure of the class I ligase, divided into fragments 1 (orange), 2 (cyan), and 3 (green), each with flanking primer regions of 12 nucleotides each (boxes). The substrates for ligation are shown in black, with a curved arrow indicating the site of ligation. (B) Synthesis of each of the three fragments (+strand) and their complements (−strand) by the 38-6 polymerase. Reaction conditions: 100 nM polymerase, 80 nM primer, 100 nM template, 4 mM each NTP, and 200 mM MgCl2 at pH 8.3 and 17 °C for 3 d. Black dots indicate full-length products. (C) Activity of the three-fragment ligase ribozyme, either with all fragments synthesized by T7 RNA polymerase protein (black, kobs = 0.49 ± 0.02 h−1) or with two fragments synthesized by the protein and the third synthesized by the 38-6 polymerase (fragment 1, orange, kobs = 0.031 ± 0.004 h−1; fragment 2, cyan, kobs = 0.049 ± 0.001 h−1; fragment 3, green, kobs = 0.043 ± 0.004 h−1). Values are based on three replicates, with SE. Reaction conditions: 1 µM each fragment, 20 µM 5′-substrate (S2), 80 µM 3′-substrate with attached template (S3), 60 mM MgCl2, 200 mM KCl, and 0.6 mM EDTA at pH 8.3 and 23 °C.
To enable the RNA-catalyzed synthesis of the entire class I ligase and its complement, the primers used to initiate synthesis must not include any nucleotides of the ribozyme. Thus, external primer sites were added to both ends of each fragment (Fig. 3A). Including these primer regions, the lengths of fragments 1, 2, and 3 are 64, 54, and 49 nucleotides, respectively, and similarly for their complements. The class I ligase assembled from the three fragments is approximately eightfold less active compared with an assembled three-fragment ligase that does not include the added primer regions (SI Appendix, Fig. S1).
On dividing the class I ligase into three fragments, each fragment is much more readily synthesized by the 38-6 polymerase compared to the synthesis of the contiguous ligase. Following a 3-d incubation, fragments 1, 2, and 3 were generated from their corresponding RNA templates in yields of 1.1%, 1.0%, and 5.7%, respectively (Fig. 3B). The polymerase was also able to synthesize the complement of each fragment, in yields of 0.53%, 0.91%, and 2.0%, respectively.
Each of the 95 nucleotides that compose the class I ligase were susceptible to mutation during synthesis by the polymerase ribozyme, with the expectation that many of these mutations would be detrimental to the activity of the ligase. If only one of the ligase fragments is synthesized by the ribozyme and the other two are prepared using T7 RNA polymerase protein, then there is ∼10-fold decreased ligase activity compared with the situation in which all three fragments are synthesized by the protein (Fig. 3C). If the ligase is assembled from three fragments that have been synthesized by the 38-6 ribozyme, then there is an ∼8,000-fold reduction of ligase activity, with an observed rate of only 6.1 ± 0.2 × 10−5 h−1 (SI Appendix, Fig. S2). This rate is only sevenfold faster than that of the uncatalyzed, RNA-templated reaction. Thus, while the improved polymerase ribozyme has the ability to synthesize large complex RNAs, including a three-fragment form of its own ancestor, the fidelity of synthesis is insufficient to enable a significant fraction of the products to retain the information necessary for catalytic function.
Fidelity of the Evolved Polymerase.
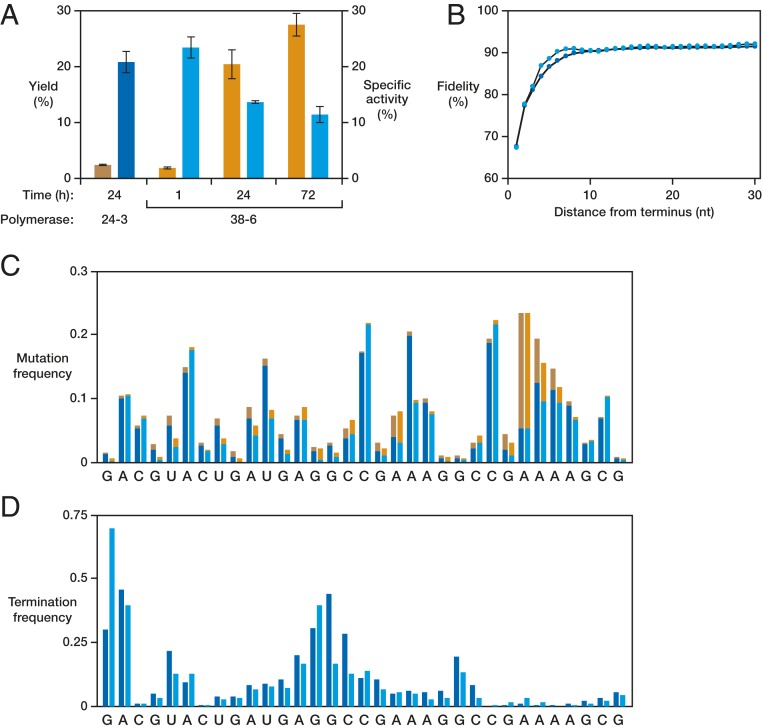
The fidelity of RNA polymerization by the 38-6 ribozyme was investigated more thoroughly to assess the incidence of deleterious mutations that compromise the activity of the assembled class I ligase. Using an unstructured C-rich template that has been the subject of previous studies (12–14), full-length RNA products were generated within minutes using either the 24-3 or 38-6 ribozyme. Sequence analysis of these full-length products revealed an average fidelity of 97% and 96% per nucleotide position for the 24-3 and 38-6 polymerase, respectively (SI Appendix, Table S1). If one instead considers all oligomeric products, irrespective of length, then the fidelity of 24-3 is only 92% (14). This result indicates that partial-length products that contain a mutation are less likely to be extended to full length compared with accurately synthesized intermediates. Similar behavior has been observed with other variants of the class I polymerase (10, 13).
The trade-off between yield and fidelity is even more apparent for the most challenging templates. When the 38-6 polymerase was pushed to its limits to generate the fragments that assemble to form the class I ligase, sequence analysis of full-length fragment 1 revealed a fidelity of only 83% per nucleotide position due to point mutations, as well as frequent insertions with an occurrence of 1.7% per nucleotide position (SI Appendix, Table S2). There are notable hotspots for substitution and deletion mutations, in some cases exceeding a frequency of 40%. The 38-6 polymerase ribozyme can generate far more complex RNA products than could be achieved previously, but only at the expense of reduced fidelity for the most challenging cases.
Deep sequencing analysis was carried out to investigate more thoroughly the relationship between yield and fidelity, both of which are important for the synthesis of functional RNAs. For the RNA-catalyzed synthesis of the hammerhead ribozyme, the fidelity of the 24-3 and 38-6 polymerases is similar for comparable yields of full-length products (Fig. 4). The hammerhead is synthesized in an ∼2% yield by the 24-3 polymerase after 24 h and in the same yield by the 38-6 polymerase after 1 h, with the products of these two reactions having similar specific activity (Fig. 4A). However, if the reaction with the 38-6 polymerase is allowed to continue for 24 h or 72 h, by which time the yield has increased to 20% or 28%, respectively, then the specific activity of the resulting materials decreases significantly. Thus, it appears that at longer incubation times, the 38-6 polymerase is able to continue the extension of partial-length products that contain a mutation, improving the yield of full-length products but resulting in decreased fidelity of those full-length products.
Fig. 4.
Synthesis of the hammerhead ribozyme by the 24-3 and 38-6 polymerases. (A) Yield and specific activity of synthesized products. Yield (orange) is the percent primer extended to give the full-length hammerhead, as shown in Fig. 1B. Specific activity (blue) is the initial rate of reaction, under multiple-turnover conditions, of the ribozyme-synthesized hammerhead relative to that of the protein-synthesized hammerhead (for which kobs = 0.38 min−1). Values are the average of three replicates, with SD. Reaction conditions: 10 to 100 nM hammerhead ribozyme, 4 µM RNA substrate, and 20 mM MgCl2 at pH 8.0 and 23 °C. (B) Fidelity as a function of distance from the 3′-terminal nucleotide for all partial- and full-length products synthesized by either 24-3 in 24 h (dark blue) or 38-6 in 1 h (light blue). Values are the average fidelity per nucleotide within the specified distance upstream from the terminus. (C) Positional mutation frequencies across the entire hammerhead sequence, with substitutions shown in blue and insertions/deletions shown in orange, for 24-3 (darker colors) and 38-6 (lighter colors). (D) Positional termination frequencies across the hammerhead sequence for 24-3 (dark blue) and 38-6 (light blue). Termination frequency is the probability that extension ended immediately before the indicated nucleotide, as determined by analysis of the extension products by high-resolution PAGE (Fig. 2A).
This hypothesis is supported by deep sequencing analysis, which reveals that fidelity is lowest for the last added nucleotide (∼67%) and increases monotonically for nucleotides that are increasingly further upstream from the 3′ terminus (Fig. 4B). Positions that are at least 10 nucleotides upstream from the terminus have an average fidelity of ∼92%. These data are very similar for the 24-3 and 38-6 polymerases. The two polymerases also are very similar with regard to the position-specific frequency of mutation, which ranges from 76% to 99%, depending on the difficulty of copying various template positions (Fig. 4C). The frequency of mutation is greatest at positions that are susceptible to wobble mutation (SI Appendix, Table S3) or that involve a run of consecutive U residues in the template. Where the two polymerases differ is in the propensity for chain termination that results in partial-length products, with 38-6 being substantially less prone to early termination, especially at the most challenging template positions (Fig. 4D). This is seen most clearly at a region of predicted stable secondary structure in the template that corresponds to the central stem-loop of the hammerhead ribozyme. Because the 38-6 polymerase can more effectively traverse such challenging positions, it can generate complex functional RNAs that cannot be synthesized by 24-3 or its evolutionary predecessors.
Discussion
To achieve self-sustained replication, an RNA polymerase ribozyme must be able to synthesize copies of both itself and its complement in sufficient yields to enable exponential growth. Although various ribozymes have been shown to copy themselves either partially or entirely, these reactions have always depended on carefully curated sets of preformed oligonucleotide substrates, thereby restricting the ability of the copied RNA to mutate and evolve (15, 18, 28–30). Another issue that has not been addressed in previous studies, or in the current study, is the need to separate the template and product strands, other than by means such as thermal or chemical denaturation. Strand separation is necessary to liberate the functional strand and to make both strands available to serve as new templates.
The 38-6 polymerase was selected based on its ability to synthesize functional ribozymes from NTP substrates. This polymerase is able to synthesize its own ancestor, the class I ligase, in the form of three fragments that, following purification, can self-assemble to give an active complex. This is the most complex ribozyme ever synthesized by a ribozyme from mononucleotide substrates, and it suggests that further improvements in activity might enable the polymerase to synthesize itself. However, it is also clear that the fidelity of polymerization, especially on the most challenging templates, severely limits the amount of functional information that can be copied. This limitation would prevent the propagation of heritable information across successive generations of RNAs (4).
The high number of mutations seen with the most challenging templates do not appear to result from an increased propensity to incorporate mismatched nucleotides. At similar yields, the error rates of the 24-3 and 38-6 polymerases are similar; however, over the long incubation times needed for the 38-6 polymerase to complete the most difficult syntheses, the number of mutations within the full-length products is substantially higher, causing most of the product molecules to lack function. Prior forms of the class I polymerase generally had difficulty extending past a misincorporated nucleotide, so the full-length products contain fewer mutations compared with the partial-length products (10, 13). The improved activity of 38-6 also appears to enhance its ability to extend mismatched intermediates. In addition, it may have an increased tendency to bypass sites of polymerase stalling by incorporating a mismatched nucleotide.
The selection scheme used to obtain the 38-6 polymerase required the accurate synthesis of a hammerhead ribozyme and thus imposed selection pressure favoring improved fidelity. However, only 11 of the 33 nucleotides that the polymerase was required to synthesize are critical for hammerhead function (31, 32). Furthermore, the short reaction times in the later rounds of evolution placed strong selection pressure on the reaction rate, forcing a trade-off between efficiency and fidelity. Going forward, it will be important to impose greater pressure on fidelity by requiring the polymerase to synthesize more complex RNAs that contain a larger number of nucleotides that are critical for function. The 38-6 polymerase is now able to synthesize such complex RNAs, making it possible to impose highly stringent requirements for fidelity. The greater the efficiency and fidelity of the ribozyme, the more readily it can be evolved toward further improvements in efficiency and fidelity. This bootstrapping process is analogous to what is thought to have driven the evolution of more complex genomes in the RNA world (3, 33).
The dual requirements of efficiency and fidelity for the propagation of heritable information have long been recognized but too often ignored when considering synthetic replicating systems. Eigen’s formulation of the “error threshold” considered the relative advantage of a selectively advantageous individual compared with the population as a whole, demonstrating how that relative advantage must exceed the probability of the individual producing an error copy (4). Note, as described above, that the production of full-length copies (A) and the fraction of those copies that are error-free (Q) are two distinct variables, the product of which reflects the production of accurate, full-length products (A·Q). For RNA replication, the probability of producing an error-free copy is the product of the fidelity of the component nucleotide addition reactions. Thus, there is an inverse relationship between the per-nucleotide fidelity of polymerization and the maximum length of RNA that can be maintained through successive rounds of replication. For any realistic values for selective advantage and per-nucleotide fidelity, the maximum length of RNA that can be maintained is approximately the inverse of the fidelity. The class I ligase contains approximately 100 nucleotides and thus must be copied with an average fidelity of >99%. The 38-6 polymerase contains nearly 200 nucleotides and thus must be copied with an average fidelity of >99.5%.
In the present study, the class I ligase was divided into three fragments that assemble to form an active complex that has only slightly reduced activity compared with the contiguous molecule. Excluding the primer regions, the number of nucleotides that must be synthesized to produce these three fragments are 40 + 30 + 25 = 95. It is likely that the 38-6 polymerase similarly could be divided into fragments that assemble noncovalently to provide a functional molecule. The smaller size of the fragments compared with the contiguous molecule makes them easier to synthesize but does not alleviate the error threshold, which applies to the total number of nucleotides necessary to produce the replicating entity. It is possible that the assembly of components fragments provides a means of “proofreading” to exclude defective fragments, but that does not appear to be the case here. Based on comparison of mixed assemblages of RNA strands produced by either the 38-6 polymerase ribozyme or T7 RNA polymerase protein, the deleterious effects of mutations within each strand appear to be compounded in the assembled complex.
In seeking to surmount the error threshold and to achieve the self-sustained evolution of RNA, it will be necessary either to increase the fidelity of the polymerase ribozyme substantially or to decrease its size substantially. Previous studies have focused on improving the efficiency and sequence generality of RNA-templated RNA polymerization, which are worthy objectives. Going forward, however, these parameters should no longer be regarded as the chief impediment to the construction of synthetic RNA life.
Materials and Methods
Materials.
The sequences of all oligonucleotides used in this study are listed in SI Appendix, Table S4. Synthetic oligonucleotides were either purchased from IDT or prepared by solid-phase synthesis using an Expedite 8909 DNA/RNA synthesizer, with reagents and phosphoramidites from Glen Research. RNA templates were prepared by in vitro transcription of synthetic DNA templates and polymerase ribozymes were prepared by in vitro transcription of double-stranded DNAs that were generated by PCR amplification of the corresponding plasmid DNA (14) (SI Appendix, Materials and Methods). His-tagged T7 polymerase was prepared from Escherichia coli strain BL21 containing plasmid pBH161 (kindly provided by W. McAllister, SUNY Downstate Medical Center, Brooklyn, NY). Hot Start OneTaq, Universal miRNA Cloning Linker, K227Q T4 RNA Ligase 2, terminal transferase, and Q5 Hot Start High-Fidelity DNA Polymerase were obtained from New England BioLabs. MyOne C1 streptavidin magnetic beads, Pierce high-capacity streptavidin agarose beads, SuperScript IV reverse transcriptase, RNase H, Turbo DNase, and TOPO TA Cloning Kit were obtained from Thermo Fisher Scientific. γ-(2-Azidoethyl)-ATP and sulfo-cyanine5-azide were purchased from Jena Bioscience, and NTPs were purchased from Chem-Impex International. All other chemical reagents were obtained from Sigma-Aldrich.
In Vitro Evolution.
Selective amplification of polymerase ribozymes based on their ability to synthesize the hammerhead ribozyme was performed similarly to the procedure used to select polymerases that synthesize RNA aptamers (14). In vitro transcription of DNAs encoding the population of polymerases was performed in the presence of γ-(2-azidoethyl)-ATP, which enabled attachment via click chemistry of a 5′-hexynylated RNA primer (P1a) that was linked to both biotin and the hammerhead substrate (Fig. 1A and SI Appendix, Table S4). Following purification by denaturing polyacrylamide gel electrophoresis (PAGE), 12 nM ribozyme-primer conjugate was mixed with 24 nM RNA template (Tem1) and annealed by first heating to 80 °C for 30 s and then cooling to 17 °C. These materials were added to a reaction mixture containing 4 mM each NTP, 200 mM MgCl2, 50 mM Tris⋅HCl pH 8.3, and 0.05% Tween-20, which was incubated at 17 °C for various times. The reaction was quenched by adding an equal volume of quench buffer (250 mM EDTA pH 8.0, 500 mM NaCl, 5 mM Tris⋅HCl pH 8.0, and 0.025% Tween-20), then mixed with 5 µg of streptavidin magnetic beads per 1 pmol of ribozyme-primer conjugate and allowed to stand at 23 °C for 1 h. The beads had been preblocked by incubating with 1 mg/mL tRNA. The RNA template was removed by three washes with NaOH buffer (25 mM NaOH, 1 mM EDTA, and 0.05% Tween-20), followed by two washes with urea buffer (8 M urea, 1 mM EDTA, and 10 mM Tris⋅HCl pH 8.0).
During the initial rounds of evolution, the beads were washed with binding buffer (300 mM NaCl, 1 mM EDTA, and 10 mM Tris⋅HCl pH 8.0); again blocked by incubating with 1 mg/mL tRNA; washed once more with binding buffer; resuspended in 20 mM MgCl2, 50 mM Tris⋅HCl pH 8.0, and 0.05% Tween-20; and incubated at 23 °C for 30 min to allow hammerhead-catalyzed cleavage of the attached RNA substrate. During the later rounds of evolution, a gel-shift selection step was added after removal of the RNA template and before hammerhead cleavage. For those rounds, following the washes with urea buffer, the reaction products were eluted from the beads by incubating in 95% formamide and 10 mM EDTA at 95 °C for 10 min. Then the polymerase-primer conjugates that had been extended to full length were separated by PAGE, eluted from the gel, and bound to 20 µg of streptavidin magnetic beads that had been preblocked with tRNA. After one wash each with NaOH buffer and urea buffer, the beads were again blocked with tRNA, and hammerhead cleavage was allowed to occur, as described above. For all rounds of evolution, the solution containing the RNAs released from the beads by hammerhead cleavage was passed through a 0.2-µm filter, then the RNAs were reverse-transcribed and PCR-amplified to provide materials to begin the next round of evolution.
After the final round of evolution (round 38 in total), the PCR-amplified DNA was cloned into Escherichia coli using the TOPO-TA cloning kit, and the cells were grown at 37 °C for 16 h on LB agar plates containing 50 µg/mL kanamycin. Individual colonies were picked and grown in 3 mL of LB medium with 50 µg/mL kanamycin at 37 °C for 16 h. Plasmid DNA was harvested using the QIAprep Spin Miniprep Kit (Qiagen) and sequenced by GENEWIZ.
RNA-Catalyzed RNA Polymerization.
Unless noted otherwise, RNA polymerization reactions used 100 nM polymerase, 80 nM primer, and 100 nM template; sequences are provided in SI Appendix, Table S4. The RNAs were annealed by first heating to 80 °C for 30 s then cooling to 17 °C, and were then added to a reaction mixture containing 4 mM each NTP, 200 mM MgCl2, 25 mM Tris⋅HCl pH 8.3, and 0.05% Tween-20, which was incubated at 17 °C for various times. The reactions were quenched with an equal volume of quench buffer, and the products were captured on streptavidin magnetic beads to enable removal of the RNA template, as described above.
For synthesis of either tRNA or the class I ligase, the reaction products were analyzed directly by PAGE. For synthesis of the 11-nucleotide RNA used to measure fidelity, the products were separated by PAGE, and the full-length materials were eluted from the gel and purified by ethanol precipitation. The purified RNAs were ligated to the Universal miRNA Cloning Linker using K227Q T4 RNA Ligase 2 according to the manufacturer’s protocol, after which the RNAs were captured on streptavidin magnetic beads and the ligated products were eluted from the beads, as described above. These RNAs were reverse-transcribed and PCR-amplified using primers Fwd2 and Rev2, then cloned into E. coli, as described above. Individual colonies were picked and sequenced by Eton Bioscience, and sequences were aligned using SnapGene (GSL Biotech).
For synthesis of the hammerhead ribozyme, the extension products were captured, washed, eluted, and purified by ethanol precipitation, as described above. RNA cleavage activity was assayed under multiple-turnover conditions in a reaction mixture containing 10 to 100 nM ribozyme, 4 µM Cy5-labeled RNA substrate (S1), 20 mM MgCl2, 50 mM Tris⋅HCl pH 8.0, and 0.05% Tween-20, which was incubated at 23 °C for 90 min. The reaction was quenched using 95% formamide and 10 mM EDTA, and the products were analyzed by PAGE. Reaction rates were calculated based on a linear fit of the fraction cleaved during the initial phase (<10%) of the reaction. To determine specific activity, these rates were compared with that of a hammerhead ribozyme that had been synthesized by T7 RNA polymerase.
Deep Sequencing of the Hammerhead Ribozyme.
The hammerhead ribozyme was synthesized by either the 24-3 or 38-6 polymerase, as described above, pooling all partial- and full-length products after 24 and 1 h, respectively. The products were ligated to the Universal miRNA Cloning Linker using K227Q T4 RNA Ligase 2 according to the manufacturer’s protocol, then reverse-transcribed using SuperScript IV. The resulting RNA-cDNA complexes were captured on streptavidin magnetic beads and washed to remove unextended DNA primer, and the cDNAs were released by digesting the RNA strand with RNase H. The cDNAs were tailed with poly(C) using terminal transferase according to the manufacturer’s protocol. The enzymes were inactivated by heating at 80 °C for 20 min, then dsDNA was generated by PCR using Q5 Hot Start High-Fidelity DNA Polymerase. Illumina adapter sequences were added to the ends of the cDNA using primers Fwd4 and Rev4, followed by amplification using the Illumina Nextera Index primers. The final dsDNA products, ranging in size from 150 to 250 nucleotides, were purified by agarose gel electrophoresis and then sequenced on an Illumina MiniSeq with a 75-cycle paired-end run, carried out by the Salk Next Generation Sequencing Core.
The sequencing data were processed to categorize all mutations relative to the expected hammerhead ribozyme sequence. The Illumina adapter sequences were trimmed, and unindexed reads were removed using cutadapt v2.4 (34). Then the paired reads were merged using FLASH v1.2.11 (35), with a minimum overlap of four nucleotides and maximum mismatch density of 0.3, and filtered to remove low-quality reads with a Phred score <30 at any position using fastq_quality_filter from FASTX Toolkit v0.0.13 (36). The resulting reads were aligned using bowtie2 (37) in end-to-end mode, with a seed length of five nucleotides and with reference to a sequence consisting of the expected full-length hammerhead and the flanking constant regions, including the complement of the processivity tag: CTACAGGGCACTCCACACGACGTACTGATGAGGCCGAAAGGCCGAAAAGCGTTTTTTGTCATTGTCCTGTAGGCACCATCAAT (constant regions underscored). Samtools v1.9 (38) was then used to prepare a sorted, indexed, and compressed alignment file, and the bamtoaln module of breseq (v 0.27.0) (39) was used to produce a gapped alignment of the reads relative to the reference sequence. Finally, a custom JavaScript (SI Appendix) was used to calculate the numbers of matches, mismatches, deletions, and insertions as a function of template position and read length along the reference sequence. The script was compiled after extracting the Java files using Javac v1.7 (Oracle), and run with Java using the output tables from breseq bamtoaln and a reference length of 83. The resulting data tables were manually processed to generate fidelity tables and position-specific data plots. All raw and processed data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE142114) (40). The average per-nucleotide fidelity was calculated as the geometric mean of the averaged fidelities for each of the four nucleobases.
RNA-Catalyzed Synthesis of the Class I Ligase.
Analytical-scale polymerization reactions used either 0.1 µM RNA template (for fragments 1, 3, and 3′, with prime denoting the complementary strand) or 0.05 µM RNA template (for fragments 2, 1′, and 2′), 0.08 µM primer, and 0.1 µM polymerase ribozyme, which were annealed as described above (sequences listed in SI Appendix, Table S4). These RNAs were added to a reaction mixture containing 4 mM each NTP, 200 mM MgCl2, 25 mM Tris⋅HCl pH 8.3, and 0.05% Tween-20, followed by incubation at 17 °C for 72 h. The reactions were quenched, the RNA template was removed, and the extension products were analyzed by PAGE, as described above.
Preparative-scale reactions were performed under the same conditions as above, except the extension products were captured on streptavidin-coated agarose beads that had been preblocked with 1 mg/mL tRNA. The beads were collected in a 10-mL chromatography column attached to a vacuum manifold and washed twice with NaOH buffer, twice with urea buffer, and twice with binding buffer, then blocked again with 1 mg/mL tRNA. The extension products were eluted from the beads by incubation in 95% formamide and 10 mM EDTA at 95 °C for 10 min, then collected using a centrifugal filter. Free streptavidin monomers were removed using the Monarch RNA Cleanup Kit (New England BioLabs), then the full-length products were separated by PAGE, eluted from the gel, purified by ethanol precipitation, and quantified by comparing their fluorescence intensity to known standards by analytical PAGE.
Purified fragment 1 RNAs were ligated to the Universal miRNA Cloning Linker as described above, then reverse-transcribed using primer Rev3. The RNA/cDNA hybrid was captured on streptavidin magnetic beads and washed once with binding buffer, then the cDNA was eluted with NaOH buffer, which was passed through a 0.2-µm filter, quenched in 100 mM Tris pH 7.5, and ethanol-precipitated. Cytidine residues were added to the 3′ end of the cDNA using terminal transferase according to the manufacturer’s protocol using 1 mM dCTP, followed by incubation at 37 °C for 30 min and then heating at 70 °C for 10 min. The products were PCR-amplified using primers Fwd3 and Rev3, cloned into E. coli, and sequenced by Eton Bioscience, and the sequences were aligned using SnapGene, as described above.
Activity of the Class I Ligase.
Ligation reactions were performed under multiple-turnover conditions (41) in a mixture containing 20 µM 5′-substrate (S2) that had been labeled with Cy5, 80 µM 3′-substrate (S3) that had been chemically triphosphorylated (42), and 1 µM ligase that was provided as either a contiguous strand or three separate fragments. The reaction mixture also contained 60 mM MgCl2, 200 mM KCl, 0.6 mM EDTA, and 50 mM Tris⋅HCl pH 8.3, which was incubated at 23 °C. Aliquots were obtained at various times and quenched by adding four volumes of 80% formamide and 100 mM EDTA, after which the products were analyzed by PAGE.
Supplementary Material
Acknowledgments
We thank Yu-Ting Huang for assistance with preparing RNA primers, templates, and polymerase ribozymes, and to Nasun Ha and the Salk Next Generation Sequencing Core for advice on preparing the hammerhead ribozyme for deep sequencing. This work was supported by grants from the National Aeronautics and Space Administration (NSSC19K0481) and the Simons Foundation (287624). K.F.T. was supported by a predoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada. M.N.S. is Director of the Razavi Newman Integrative Genomics and Bioinformatics Core Facility of the Salk Institute, which is supported by Grants CCSG:P30 014195, GM102491, and AG064049 from the NIH and by the Helmsley Trust.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Sequencing data and corresponding fidelity measurements reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE142114).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914282117/-/DCSupplemental.
References
- 1.Gilbert W., Origin of life: The RNA world. Nature 319, 618 (1986). [Google Scholar]
- 2.Joyce G. F., The antiquity of RNA-based evolution. Nature 418, 214–221 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Joyce G. F., Szostak J. W., Protocells and RNA self-replication. Cold Spring Harb. Perspect. Biol. 10, a034801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eigen M., Self-organization of matter and the evolution of biological macromolecules. Naturwissenschaften 58, 465–523 (1971). [DOI] [PubMed] [Google Scholar]
- 5.Joyce G. F., Bit by bit: The Darwinian basis of life. PLoS Biol. 10, e1001323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attwater J., Holliger P., A synthetic approach to abiogenesis. Nat. Methods 11, 495–498 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Pressman A., Blanco C., Chen I. A., The RNA world as a model system to study the origin of life. Curr. Biol. 25, R953–R963 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Ekland E. H., Szostak J. W., Bartel D. P., Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269, 364–370 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Ekland E. H., Bartel D. P., RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature 382, 373–376 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Johnston W. K., Unrau P. J., Lawrence M. S., Glasner M. E., Bartel D. P., RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 292, 1319–1325 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Zaher H. S., Unrau P. J., Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA 13, 1017–1026 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wochner A., Attwater J., Coulson A., Holliger P., Ribozyme-catalyzed transcription of an active ribozyme. Science 332, 209–212 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Attwater J., Wochner A., Holliger P., In-ice evolution of RNA polymerase ribozyme activity. Nat. Chem. 5, 1011–1018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horning D. P., Joyce G. F., Amplification of RNA by an RNA polymerase ribozyme. Proc. Natl. Acad. Sci. U.S.A. 113, 9786–9791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attwater J., Raguram A., Morgunov A. S., Gianni E., Holliger P., Ribozyme-catalysed RNA synthesis using triplet building blocks. eLife 7, e35255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madhani H. D., Guthrie C., Dynamic RNA-RNA interactions in the spliceosome. Annu. Rev. Genet. 28, 1–26 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Evguenieva-Hackenberg E., Bacterial ribosomal RNA in pieces. Mol. Microbiol. 57, 318–325 (2005).15978067 [Google Scholar]
- 18.Doudna J. A., Couture S., Szostak J. W., A multisubunit ribozyme that is a catalyst of and template for complementary strand RNA synthesis. Science 251, 1605–1608 (1991). [DOI] [PubMed] [Google Scholar]
- 19.Vaidya N., et al. , Spontaneous network formation among cooperative RNA replicators. Nature 491, 72–77 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Rogers T. A., Andrews G. E., Jaeger L., Grabow W. W., Fluorescent monitoring of RNA assembly and processing using the split-spinach aptamer. ACS Synth. Biol. 4, 162–166 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Akoopie A., Müller U. F., Lower temperature optimum of a smaller, fragmented triphosphorylation ribozyme. Phys. Chem. Chem. Phys. 18, 20118–20125 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Vaish N. K., et al. , Zeptomole detection of a viral nucleic acid using a target-activated ribozyme. RNA 9, 1058–1072 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q. S., Cheng L. K. L., Unrau P. J., Characterization of the B6.61 polymerase ribozyme accessory domain. RNA 17, 469–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mutschler H., Wochner A., Holliger P., Freeze-thaw cycles as drivers of complex ribozyme assembly. Nat. Chem. 7, 502–508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ekland E. H., Bartel D. P., The secondary structure and sequence optimization of an RNA ligase ribozyme. Nucleic Acids Res. 23, 3231–3238 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shechner D. M., et al. , Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science 326, 1271–1275 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horning D. P., Bala S., Chaput J. C., Joyce G. F., RNA-catalyzed polymerization of deoxyribose, threose, and arabinose nucleic acids. ACS Synth. Biol. 8, 955–961 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayden E. J., Lehman N., Self-assembly of a group I intron from inactive oligonucleotide fragments. Chem. Biol. 13, 909–918 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Lincoln T. A., Joyce G. F., Self-sustained replication of an RNA enzyme. Science 323, 1229–1232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sczepanski J. T., Joyce G. F., A cross-chiral RNA polymerase ribozyme. Nature 515, 440–442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruffner D. E., Stormo G. D., Uhlenbeck O. C., Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry 29, 10695–10702 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Salehi-Ashtiani K., Szostak J. W., In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414, 82–84 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Wolf Y. I., Koonin E. V., On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol. Direct 2, 14 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011). [Google Scholar]
- 35.Magoč T., Salzberg S. L., FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hannon G. J., FASTX-Toolkit, version 0.0.13. http://hannonlab.cshl.edu/fastx_toolkit/. Accessed 20 February 2010.
- 37.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., et al. ; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deatherage D. E., Barrick J. E., Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151, 165–188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tjhung K. F., Shokhirev M. N., Horning D. P., Joyce G. F., An RNA polymerase ribozyme that synthesizes its own ancestor. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142114. Deposited 16 December 2019. [DOI] [PMC free article] [PubMed]
- 41.Bergman N. H., Johnston W. K., Bartel D. P., Kinetic framework for ligation by an efficient RNA ligase ribozyme. Biochemistry 39, 3115–3123 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Olea C. Jr, Horning D. P., Joyce G. F., Ligand-dependent exponential amplification of a self-replicating L-RNA enzyme. J. Am. Chem. Soc. 134, 8050–8053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.