Significance
Mutations that reduce the function of the de novo DNA methyltransferase, DNMT3A, are very common in patients with clonal hematopoiesis and hematopoietic malignancies. Restoring the function of DNMT3A in mouse hematopoietic cells deficient for this protein corrects the DNA methylation defect in an ordered fashion, and partially restores abnormalities in gene expression and myeloid population skewing. These data may be relevant for therapeutic strategies designed to restore DNMT3A activity in patients with diseases caused by mutations in this gene.
Keywords: DNA methylation, DNA methyltransferase, hematopoiesis, gene expression
Abstract
Mutations in the DNA methyltransferase 3A (DNMT3A) gene are the most common cause of age-related clonal hematopoiesis (ARCH) in older individuals, and are among the most common initiating events for acute myeloid leukemia (AML). The most frequent DNMT3A mutation in AML patients (R882H) encodes a dominant-negative protein that reduces methyltransferase activity by ∼80% in cells with heterozygous mutations, causing a focal, canonical DNA hypomethylation phenotype; this phenotype is partially recapitulated in murine Dnmt3a−/− bone marrow cells. To determine whether the hypomethylation phenotype of Dnmt3a−/− hematopoietic cells is reversible, we developed an inducible transgene to restore expression of DNMT3A in transplanted bone marrow cells from Dnmt3a−/− mice. Partial remethylation was detected within 1 wk, but near-complete remethylation required 6 mo. Remethylation was accurate, dynamic, and highly ordered, suggesting that differentially methylated regions have unique properties that may be relevant for their functions. Importantly, 22 wk of DNMT3A addback partially corrected dysregulated gene expression, and mitigated the expansion of myeloid cells. These data show that restoring DNMT3A expression can alter the epigenetic “state” created by loss of Dnmt3a activity; this genetic proof-of-concept experiment suggests that this approach could be relevant for patients with ARCH or AML caused by loss-of-function DNMT3A mutations.
Mutations in the DNMT3A gene are the most common events associated with age-related clonal hematopoiesis (ARCH) (1–4), and are among the most common initiating mutations in acute myeloid leukemia (AML) (5–7). In patients with AML, heterozygous point mutations that cause missense changes at amino acid R882 (R882H, R882C, R882S, etc.) are by far the most prevalent (5, 8–12). The R882 residue is near the distal dimerization site of the DNA methyltransferase domain; the mutant R882H protein interacts preferentially with wild-type (WT) DNMT3A, which creates a “sink” that traps the WT protein in inactive heterodimers. Since active isoforms of DNMT3B are generally not expressed in AML cells, the de novo methylation capacity of cells with heterozygous DNMT3AR882H mutations is reduced by ∼80%, resulting in thousands of focal differentially methylated regions (DMRs) that occur in the same position in every patient tested (i.e., they are “canonical” in location), and nearly all of which are hypomethylated (13, 14). Although many of these DMRs are located within or near genes, very few closely linked genes have been found to be differentially expressed (13, 14).
In AML cells that are WT for DNMT3A, a large number of CpG islands (CGI) are hypermethylated (14), a finding that is common to many cancer types; however, this phenotype is mitigated in R882 mutant AMLs, suggesting that CGI hypermethylation is, in fact, caused by DNMT3A activity. The genomes of DNMT3AR882 mutant AML samples also have many hypomethylated DMRs that are not associated with CGI, suggesting that there are additional regions in hematopoietic cell genomes where DNA methylation is DNMT3A-dependent (13–18). Likewise, a child with Tatton-Brown-Rahman syndrome caused by an inherited DNMT3AR882H mutation was found to have a focal hypomethylation phenotype in his nonleukemic hematopoietic cells (14), suggesting that this epigenetic state exists before transformation, and may therefore be relevant for the initiation of myeloid malignancies (14, 19–22).
Murine hematopoietic cells that are constitutively deficient for Dnmt3a have thousands of DMRs that possess a focal, canonical hypomethylation phenotype (13, 16). Dnmt3a deficiency is associated with expansion and immortalization of hematopoietic stem cells, a block in hematopoietic differentiation, and the development of myeloid and lymphoid malignancies after a long latent period (17, 23, 24). Young mice with Dnmt3a deficiency have essentially normal blood counts and hematopoietic development, despite the hypomethylation phenotype. While humans with complete DNMT3A deficiency in their bone marrow cells have not been described, many patients with ARCH—and some with AML—have heterozygous loss-of-function mutations in DNMT3A that cause haploinsufficiency (16). In mice, Dnmt3a haploinsufficiency is associated with a very subtle DNA hypomethylation phenotype in hematopoietic cells, myeloid lineage expansion over time, and the development of myeloid malignancies after a very long latent period (∼18 mo), during which cooperating mutations are acquired (16). All of these observations suggest that Dnmt3a haploinsufficiency and deficiency create an epigenetic state that somehow facilitates the acquisition of cooperating mutations and transformation.
In this report, we asked whether restoration of DNMT3A activity in hematopoietic cells with Dnmt3a deficiency could “repair” the hypomethylated regions. To address this issue, we designed a genetic proof-of-principal experiment where we restored DNMT3A expression in the bone marrow cells of adult mice engrafted with Dnmt3a-deficient hematopoietic cells. The restoration of DNMT3A activity resulted in the accurate remethylation of nearly all hypomethylated DMRs over a period of 6 mo. DMRs were remethylated in an orderly fashion; fast vs. slow remethylating DMRs had unique features that suggest that they may be associated with important biological activities. Remethylation of the Dnmt3a-deficient genome partially reversed dysregulated gene expression and myeloid skewing, suggesting that this epigenetic state is at least partially reversible.
Results
Dnmt3a-Deficient Bone Marrow Samples Have a Focal, Canonical Hypomethylation Phenotype.
Mice that lack Dnmt3a at birth are runted and die within ∼3 wk (25), but the hematopoietic cells of these mice can be transplanted into secondary recipients for longitudinal analysis of hematopoietic function. We therefore used a secondary transplantation model for all experimental and control mice in this study, which involved collection of bone marrow cells from 2.5-wk-old littermate-matched Dnmt3a knockout and WT control mice (referred to as Dnmt3a−/− or Dnmt3a+/+ henceforth) and transplantation into lethally irradiated C57BL/6 secondary recipients. The donor mice expressed CD45.2, and the recipients were chosen to express CD45.1, which allowed us to track and sort the donor cell populations for all downstream analyses. We used three pairs of matched 6-wk-posttransplant Dnmt3a−/− and Dnmt3a+/+ mice to define DMRs for all subsequent experiments.
Whole-genome bisulfite sequencing was used for all DNA methylation studies. On average, these datasets covered 20.6 million out of 21.0 million (98%) of the individual CpGs in the mouse genome, with excellent reproducibility between biological replicates (Pearson’s r > 0.8 for each of three samples in either Dnmt3a+/+ or Dnmt3a−/−).
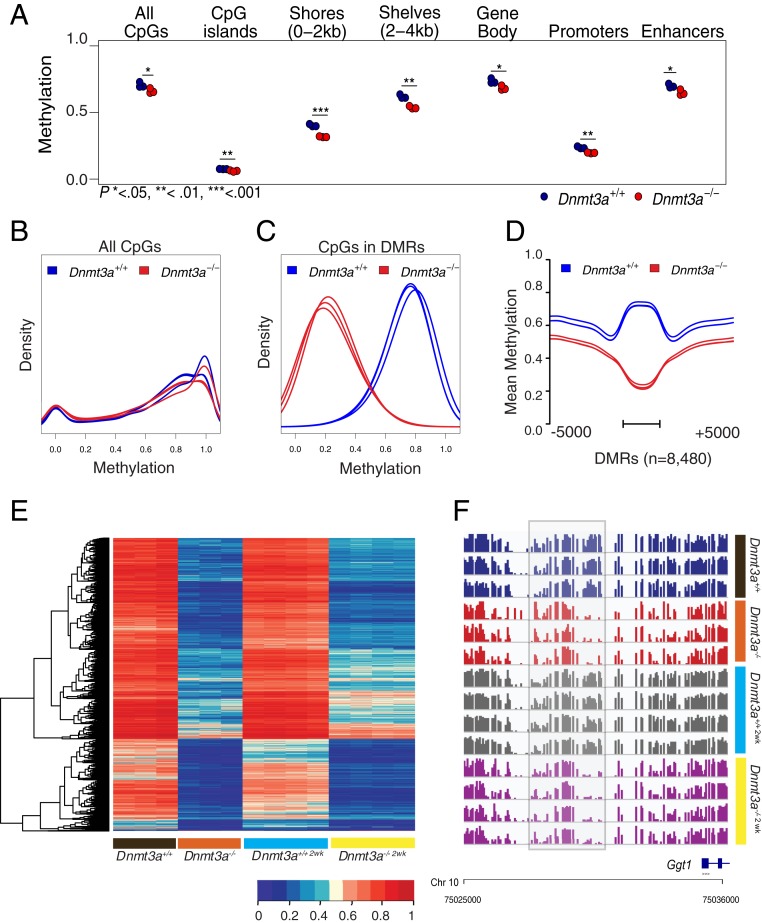
The mean CpG methylation values were significantly lower in all annotated regions of the genome in the Dnmt3a−/− bone marrow samples; CGI, CGI shores, CGI shelves, and promoters had the largest differences (Fig. 1A). The distribution of methylation values across the genome for each sample showed highly similar methylation patterns among individual mice (Fig. 1B). We next identified all DMRs between the Dnmt3a+/+ and Dnmt3a−/− samples, as previously described (14). DMRs were required to have >10 CpGs, a mean methylation difference between the two groups of >0.2, and a false discovery rate (FDR) of <0.05. Contiguous DMRs within 50 base pairs (bp) of each other were merged (the average sizes and CpG content of DMRs are described below). Using these criteria, we identified 8,480 DMRs in the Dnmt3a−/− samples; 8,466 out of 8,480 DMRs (99.83%) were hypomethylated in the Dnmt3a−/− bone marrow samples (Fig. 1 C and D and Dataset S1). In total, these DMRs encompassed ∼7.63 megabases (Mb) of DNA, representing about 2.5% of the genome.
Fig. 1.
DNA methylation phenotypes of Dnmt3a+/+ and Dnmt3a−/− bone marrow cells. (A) Mean CpG methylation levels from whole-genome bisulfite sequencing of bone marrow cells derived from Dnmt3a+/+ (n = 3) and Dnmt3a−/− mice (n = 3), harvested 6 wk after transplantation into lethally irradiated recipients. Mean values for all CpGs and annotated regions of the genome are shown. Hypothesis testing was performed via two-tailed, pairwise t tests, with Bonferroni correction for multiple testing within each genomic region; *P < 0.05; **P < 0.01; ***P < 0.001. Small but statistically significant differences are consistently observed in the methylation status of Dnmt3a+/+ and Dnmt3a−/− bone marrow cells across the genome. (B) Density plot of methylation values from all CpGs for each bone marrow sample shown in A. (C) Density plot of CpG methylation values from 8,480 DMRs defined by comparing the Dnmt3a+/+ (n = 3) and Dnmt3a−/− (n = 3) samples. (D) Aggregate (mean) methylation of 8,480 DMRs for each bone marrow sample. DMRs were scaled to a uniform length and are shown with the adjacent 5 kb of flanking sequence. The bar represents the span of the scaled DMR regions (see Methods). (E) Heatmap showing mean methylation values for the 8,480 DMRs as defined above. Values for the same DMRs were plotted passively for Dnmt3a+/+2wk and Dnmt3a−/−2wk samples (processed directly from 2-wk-old mice, and not transplanted). (F) Primary methylation values for each CpG (shown as a bar from 0 to 100% methylated for each sample) near a Dnmt3a−/− hypomethylated DMR in the 5′ flanking region of the Ggt1 gene. This region was identified as a DMR by comparing methylation values for Dnmt3a+/+ vs. Dnmt3a−/− or Dnmt3a−/−2wk samples (P = 3.20E-08 and 2.10E-15 by Mann−Whitney U test with Bonferroni correction).
The focal and canonical nature of the 8,480 DMRs is revealed in heatmaps that display the average methylation value of each DMR as a unique data point. Methylation patterns of the transplanted Dnmt3a+/+ and Dnmt3a−/− samples are nearly identical to that of primary bone marrow samples harvested from 2-wk-old germline null Dnmt3a mice (Fig. 1E) (16, 25) (referred to as Dnmt3a+/+2wk and Dnmt3a−/−2wk); since the methylation patterns are preserved in Dnmt3a−/− bone marrow cells engrafted into WT mice, the methylation phenotype must represent a cell-autonomous property of bone marrow stem/progenitor cells. An example of a DMR in Dnmt3a−/− vs. Dnmt3a−/−2wk bone marrow samples is shown in Fig. 1F (highlighted by gray box) in the 5′ flanking region of the Ggt1 gene. The DMR pattern is remarkably consistent among all mice with the same genotype (i.e., it is canonical), a finding that was recapitulated for virtually all DMRs examined.
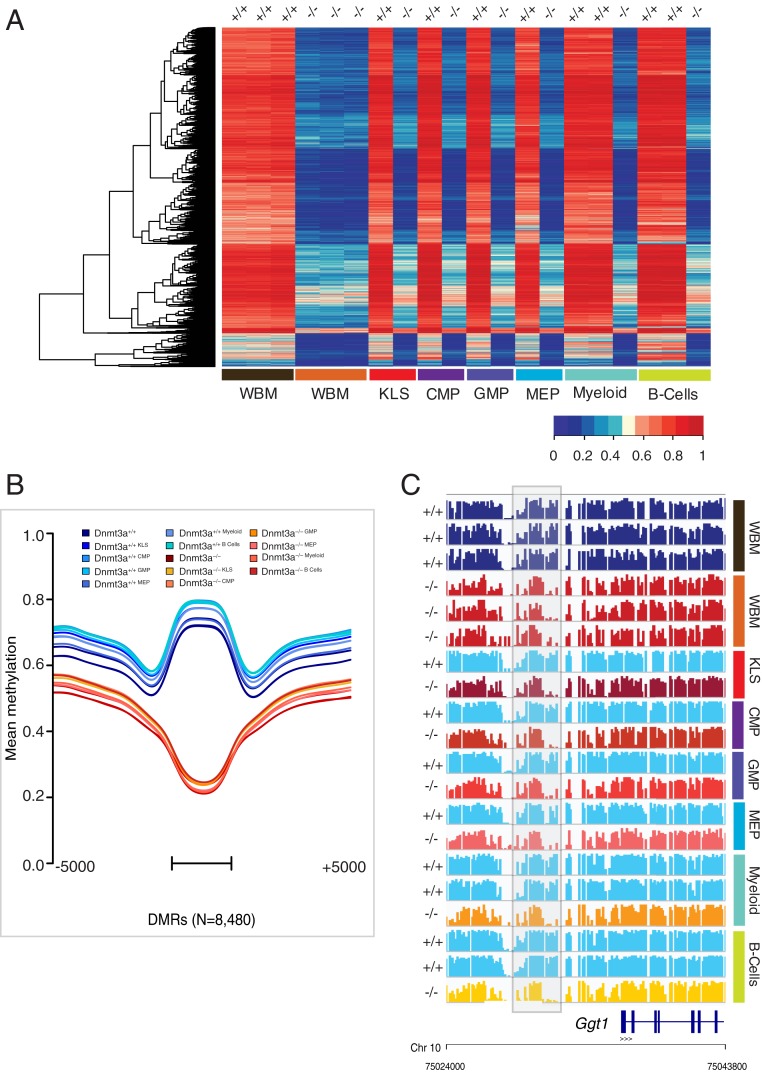
Many CpGs in the DMRs of Dnmt3a−/− samples are almost completely unmethylated (beta values less than 0.05) in total bone marrow samples, which consist of distinct populations of cells representing different hematopoietic lineages (i.e., myeloid, erythroid, lymphoid, etc.). This suggested that the normal Dnmt3a-dependent methylation “marks” may be added in stem/progenitor cells, and then maintained in all lineages (presumably by Dnmt1, and also Dnmt3a) (26). Because of improvements in Whole Genome Bisulfite Sequencing (WGBS) library preparation techniques (that use as little as 50 ng of input DNA), we were able to test this hypothesis with flow cytometry-purified populations of relatively rare progenitor cells. We performed a progenitor-specific WGBS analysis using Dnmt3a+/+ vs. Dnmt3a−/− sorted bone marrow cells from an enriched stem/progenitor cell population (KLS), committed myeloid progenitors (CMP and granulocyte−monocyte progenitor [GMP]), megakaryocyte-erythroid progenitors (MEP), and mature lineage compartments (CD11b+/Gr-1+ myeloid cells, and B220+ B cells). We passively plotted the methylation data from these compartments using the 8,480 DMRs defined above for whole bone marrow (Fig. 2 A and B) and observed a nearly identical methylation pattern in the flow-purified cells from the Dnmt3a+/+ and Dnmt3a−/− cells in all six compartments, which strongly suggests that the normal Dnmt3a-dependent methylation marks were added in stem/progenitor cells, and then maintained in all lineages as they differentiate. Although each of these compartments has a unique methylation signature pertinent to its specialized functions (27), the compartments also have common locations where Dnmt3a acts. Fig. 2C displays an exemplary DMR in the 5′ flanking region of the Ggt1 gene (gray box, the same as that shown in Fig. 1F) with a nearly identical methylation phenotype in all compartments tested.
Fig. 2.
Canonical DMRs identified in whole bone marrow (WBM) cells are conserved in multiple hematopoietic progenitor compartments and lineages. (A) Heatmap showing mean methylation values for the 8,480 DMRs from the unfractionated WBM cells of Dnmt3a+/+ (n = 3) and Dnmt3a−/− mice (n = 3). Values for the same DMRs were plotted passively for the KLS, GMP, CMP, and MEP compartments, and for mature flow-sorted populations consisting of Gr1+ cells (mature myeloid) and B220+ cells (predominantly B cells); “+/+” and “−/−” refer to the Dnmt3a genotype of each population. (B) Aggregate (mean) methylation at 8,480 DMRs as described in A: myeloid progenitor compartments: KLS, GMP, CMP, and MEP and mature populations, neutrophils, and B cells are plotted passively for the same loci. DMRs were scaled to a uniform length and are shown with the adjacent 5 kb of flanking sequence. The bar represents the span of the scaled DMR regions. (C) Primary methylation values for each CpG (shown as a bar from 0 to 100% methylated for each sample) near a Dnmt3a−/− DMR in the 5′ flanking region of the Ggt1 gene (gray box; see Fig. 1F). This region is significantly hypomethylated in total bone marrow samples, and all other purified cellular compartments (P = 3.30E-45 and P < 2.2e-16 by Mann−Whitney U test with Bonferroni correction).
Doxycycline-Regulated Expression of the DNMT3A Transgene in Hematopoietic Cells.
To determine whether expression of DNMT3A could remethylate the DMRs associated with Dnmt3a loss in hematopoietic cells, we developed a transgenic mouse capable of expressing WT human DNMT3A at near-physiologic levels in mouse hematopoietic stem/progenitor cells (HSPCs). A full-length human complementary DNA (cDNA) encoding the WT DNMT3A1 isoform was cloned into a mammalian expression vector directly downstream from a tetracycline responsive element (SI Appendix, Fig. S1A), and a transgenic mouse line was propagated from a founder mouse with a random single-copy insertion on chromosome 1 at position 192,986,250 (based on mm9 coordinates), just downstream from the Fam71a gene. As expected, this transgene is expressed in a doxycycline dose-dependent fashion in mice that also express the rtTA coactivator (ubiquitously expressed via insertion in the Rosa26 locus). The rtTA and DNMT3A transgenic lines were intercrossed with mice heterozygous for the same Dnmt3a mutation described above, a deletion of parts of exons 18 and 19 described originally by Okano et al. (25). With appropriate backcrosses, we created Dnmt3a−/− x rtTA x DNMT3A transgenic mice, which are subsequently referred to as “Dnmt3a null-3A addback mice.” Since these mice die with runting at about 3 wk of age (as expected) (2, 3), we harvested the bone marrow cells of these mice (Ly5.2) at 14 to 18 d of age, and transplanted them into lethally irradiated C57BL/6 recipients (Ly5.1). Engraftment was assessed 4 to 6 wk later by defining the size of the Ly5.2 population in the peripheral blood; engraftment rates of >80% were routinely detected (see below). Within transplanted cohorts, half of the mice were placed on doxycycline chow (10,000 parts per million [ppm], a dose shown to induce near-physiologic levels of DNMT3A in mouse HSPCs; see below), and half received normal chow without doxycycline. Whole bone marrow was harvested from two independent sets of mice at 1, 2, and 4 wk (two each with Dox and two without Dox) and one mouse each at 8, 18, and 24 wk (one with Dox and one without Dox), and these samples were subjected to WGBS, as described below.
To examine the expression of the tetracycline-regulated DNMT3A transgene, we evaluated total RNA-sequencing (RNA-seq) data from the bone marrow cells of mice fed for 2, 8, or 24 wk with or without Dox chow. Dnmt3a gene expression was defined by mapping all reads from the direct location of the Dnmt3a deletion in exons 18 and 19 (in counts per million); human DNMT3A exons 18 and 19 are 95% identical to the mouse (228 of 240 bp are identical), so human DNMT3A messenger RNA reads map to the same sites in the mouse genome. These data defined the loss of reads from these exons in Dnmt3a−/− bone marrow cells, and also defined the numbers of reads found in transplanted Dnmt3a+/+ marrow cells, which were significantly different (P = 0.05). In Dnmt3a null-3A addback mice fed with normal chow, expression levels remain at levels near zero. In mice fed with Dox chow, DNMT3A expression was restored to levels that were similar to that of WT transplanted mice (SI Appendix, Fig. S1B). Immunoblotting of whole bone marrow lysates after feeding Dox chow for 1 wk or 4 wk revealed that transgenic DNMT3A protein levels were similar to that of endogenous Dnmt3a in the bone marrow cells of 2-wk-old WT mice (SI Appendix, Fig. S1C; this polyclonal rabbit antibody cross-reacts with mouse and human DNMT3A). Dnmt3a was undetectable in 6-wk-posttransplant Dnmt3a+/+ marrow, suggesting that Dnmt3a is normally down-regulated in hematopoietic cells after 2 wk of age.
We then defined endogenous and transgenic DNMT3A protein expression in hematopoietic progenitors, and mature, lineage-defined cells, using a quantitative flow-based assay, using fixed, permeabilized cells and a rabbit monoclonal antibody that cross-reacts between human and mouse DNMT3A/Dnmt3a. The mean fluorescent intensity of the DNMT3A-specific signal was used to estimate protein abundance. We detected an induction of DNMT3A protein in all populations analyzed in DNMT3A addback mice, relative to the untreated Dnmt3a null-3A addback mice controls (SI Appendix, Fig. S1D and Dataset S2). After 4 wk of Dox feeding, the levels of induced DNMT3A expression were similar to that of endogenous Dnmt3a in the progenitor and LSK-SLAM populations (SLAM, 1.1-fold; LSK, 0.8-fold; CMP, 0.7-fold; GMP, 1.6-fold; MEP, 1.0-fold). However, mature, lineage-restricted cells (myeloid, erythroid, and B cells) expressed more DNMT3A protein than was detected in WT cells of the same lineage (myeloid, 5.4-fold; erythroid, 4.2-fold; and B cells, 4.8-fold; SI Appendix, Fig. S1D and Dataset S2). This pattern of transgene induction closely parallels expression of the rtTA transactivator in the Rosa26 locus, as previously described (28). These data are also consistent with previous observations showing that the endogenous expression of human DNMT3A is less in the myeloid, erythroid, and lymphoid lineages compared to the LSK, progenitor, and LSK-SLAM populations (SI Appendix, Fig. S1D) (5).
Remethylation of DMRs as a Function of Time and Genomic Context.
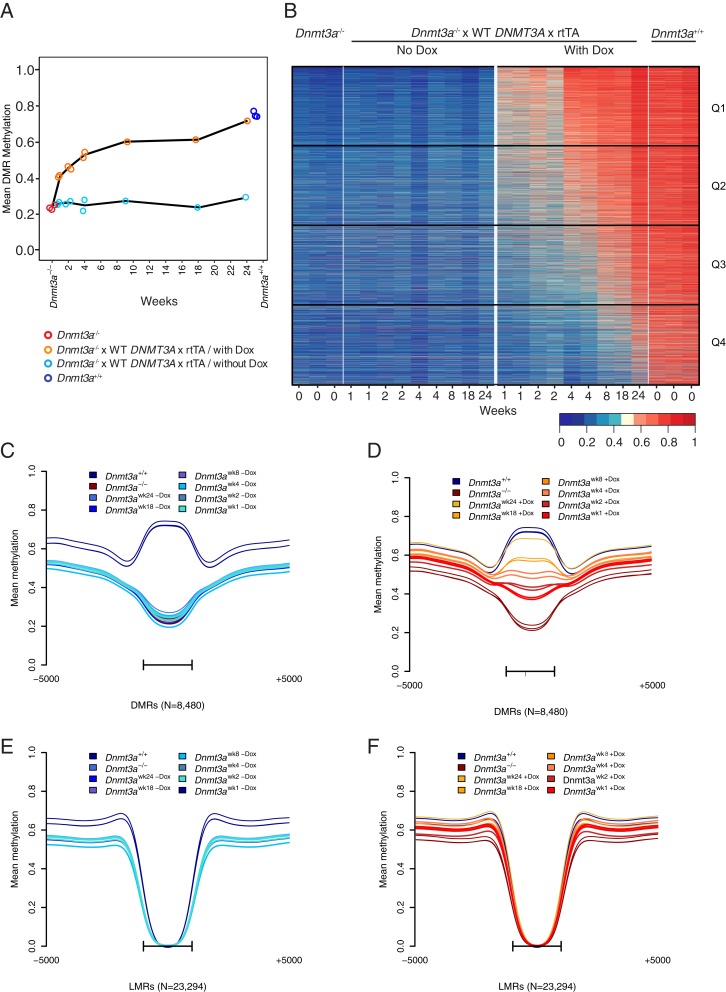
Our WGBS data resulted in coverage of >98% of the CpGs in the mouse reference genome, with a mean of 10× to 12× coverage for each sample. We calculated the mean DMR methylation values for the 8,480 DMRs in the Dnmt3a null-3A addback bone marrow cells derived from mice fed normal vs. Dox chow (10,000 ppm) for 1 to 24 wk. Importantly, none of the Dnmt3a−/− mice used in these studies had any evidence of a hematopoietic malignancy at harvest (e.g., abnormal CBCs or enlarged spleens). The mean methylation values of the DMRs steadily increased as a function of time on Dox chow. The addback of DNMT3A begins to restore methylation at DMRs starting at week 1, with further increases at weeks 2 and 4, and near-complete remethylation after 24 wk of feeding Dox chow (Fig. 3A). Overall, these results suggest that DNMT3A expression restores methylation at some DMRs quickly, and at some DMRs more slowly. Regardless, nearly all DMRs were eventually remethylated (Fig. 3 B–D).
Fig. 3.
Restoration of DNMT3A expression is associated with remethylation of Dnmt3a−/−-dependent DMRs in bone marrow cells. (A) Global mean methylation of the 8,480 DMRs over time (weeks). Red dots represent Dnmt3a−/− as baseline values, followed by values for samples from mice fed with Dox (i.e., DNMT3A reexpression) for the designated time in weeks. There are two independent datasets for weeks 1, 2, and 4, and one dataset each for weeks 8, 18, and 24. By week 24, DNMT3A reexpression causes near-complete remethylation of these DMRs compared to the methylation levels in Dnmt3a+/+ bone marrow (dark blue dots). (B) Heatmap of mean methylation values from the 8,480 DMRs. The columns are grouped by Dnmt3a−/−, Dnmt3a−/− x WT DNMT3A x rtTA without and with Dox, and Dnmt3a+/+. Low methylation levels in the DMRs from marrow derived from the Dnmt3a−/− x WT DNMT3A x rtTA mice without Dox remain unchanged over time. Time-dependent restoration of methylation across all DMRs is observed in Dnmt3a−/− x WT DNMT3A x rtTA/with Dox, where the 24-wk time point shows near-WT levels of methylation. Quartiles of 2,120 DMRs representing fast (Q1), intermediate (Q2 and Q3), and slow (Q4) remethylating regions are shown on the right. (C and D) Aggregate (mean) methylation at 8,480 DMRs from Dnmt3a+/+ and Dnmt3a−/− samples (n = 3, each). DMRs from Dnmt3a−/− x WT DNMT3A x rtTA bone marrow (C) without Dox and (D) with Dox were plotted passively for the same 8,480 DMRs. In both C and D, DMRs have been scaled to a uniform length, and are shown with the adjacent 5 kb of flanking sequence. Bars represent the span of the scaled DMR regions. (E and F) Aggregate (mean) methylation at 23,294 LMRs. Dnmt3a−/− x WT DNMT3A x rtTA (E) without Dox and (F) with Dox were plotted passively for the same LMRs. In both E and F, LMRs have been scaled to a uniform length and are shown with the adjacent 5 kb of flanking sequence. Bars represent the span of the scaled LMR regions.
We evaluated donor cell chimerism in all mice at harvest to determine whether there was selection against the donor Dnmt3a−/− cells (Ly5.2) with Dox feeding, and reciprocal enrichment of Dnmt3a+/+ recipient cells (Ly5.1); if this occurred, the “remethylation response” would simply be due to replacement of the Dnmt3a−/− cells with WT cells. However, the bone marrows of all transplanted mice, with or without Dox chow, remained >80% donor-derived (Dnmt3a−/−, Ly5.2+) at all time points (SI Appendix, Fig. S2). Therefore, the observed remethylation response is not caused by a slow replacement of the Dnmt3a-deficient cells by WT cells.
To determine whether persistent expression of the DNMT3A transgene caused DNA hypermethylation, we evaluated DNA methylation patterns across the entire genome in WT mice containing the rtTA transgene, with or without the WT DNMT3A transgene. These mice were fed Dox chow (10,000 ppm) for 1.5 y, and then bone marrow was harvested for WGBS (SI Appendix, Fig. S3A). No annotated region of the genome was significantly hypermethylated in mice that expressed the DNMT3A transgene for 1.5 y. Further, we evaluated DMRs and low methylation regions (LMRs) for evidence of hypermethylation with long-term DNMT3A expression (SI Appendix, Fig. S3B). We identified 23,294 LMRs (which have very low levels of CpG methylation in both WT and Dnmt3a−/−samples) using the hmr program in the “methpipe” package, using the same parameters described in Spencer et al. (14). LMRs were defined as hypomethylated in all samples (regardless of Dnmt3a genotype) with coverage-weighted mean methylation values of <0.2 encompassing >5 CpGs. Long-term expression of the DNMT3A transgene did not lead to significant hypermethylation of LMRs, where Dnmt3a clearly does not act (SI Appendix, Fig. S3 B, Right). Finally, we evaluated the extent of remethylation in all annotated regions of the genome with Dnmt3a null-3A addback mice (SI Appendix, Fig. S3C). As expected, the methylation values from mice eating normal chow were essentially equivalent to Dnmt3a−/− mice. The mice that ate Dox chow (10,000 ppm) had gradual restoration of methylation at all annotated regions, which were statistically equivalent to that of WT cells at week 24 of Dox feeding. These data suggest that remethylation caused by expression of the DNMT3A transgene in this model system does not result in nonspecific hypermethylation of DNA.
Based on the finding that most remethylation takes place by week 4, we decided to further evaluate the characteristics of fast, intermediate, and slow remethylating DMRs at that time point (Fig. 3B and Dataset S1). We divided the DMRs into quartiles (fastest remethylation in quartile 1, slowest in quartile 4) based on the distribution of differences between Dnmt3a−/− bone marrow and Dnmt3a null-3A addback bone marrow cells from mice fed Dox chow for 4 wk. Mean methylation differences in the DMRs were highest in quartile 1 (Q1) and lowest in quartile 4 (Q4; SI Appendix, Fig. S4 B–E). By definition, the DMRs of Q1 remethylated rapidly, and were virtually all restored to WT values by week 8. In contrast, a small number of Q4 DMRs did not completely remethylate to WT values even by week 24. The specific patterns and rates of remethylation at individual DMRs in individual mice were consistent and reproducible, suggesting that remethylation is an ordered, locus-dependent process that follows a set of specific rules. However, since the bone marrow is a heterogeneous mixture of cells, we also considered the possibility that different rates of remethylation might exist in different subsets of cells (e.g., progenitors, mature myeloid, or mature B cells), due to different rates of cell division, different levels of transgene expression, and/or other factors. We therefore compared WGBS data from purified Kit+Lin− cells, CD11b+/Gr1+ cells, or B220+ cells from a pair of mice engrafted with Dnmt3a null-3a addback bone marrow and fed normal chow vs. Dox chow, for 2 wk (SI Appendix, Fig. S5). As expected, DMRs in the purified populations fed normal chow (i.e., no DNMT3A expression) were essentially identical to that of Dnmt3a−/− total bone marrow DMRs. In bone marrow from the mouse fed with Dox chow for 2 wk, remethylation was apparent in all compartments, but was greatest in the mature myeloid cells, suggesting that this rapidly dividing population (with higher levels of DNMT3A transgene expression than progenitors; SI Appendix, Fig. S1D) is remethylated more quickly than the B cell population. Remethylation of the Kit+/Lin− progenitor population was similar to that of total bone marrow cells at 2 wk. These data suggest that mature myeloid cells do contribute disproportionately to the early remethylation phenotype, but that the general pattern of remethylation is similar in progenitor and lineage-restricted cells.
To determine whether these fast vs. slow remethylating DMRs were associated with specific genomic properties, we performed a series of studies to better define their characteristics. First, we quantified the numbers of DMRs within each quartile that were annotated to specific regions of the genome. CGI, shores, and intergenic regions tended to be enriched for DMRs associated with the slow remethylating quartiles (SI Appendix, Fig. S6A). The faster remethylating DMRs of Q1 were enriched in gene bodies, and had the lowest number of DMRs mapping to CGI, shores, and intergenic regions. The physical width of DMRs was smallest in Q1 and greatest in Q4 (870.1 ± 560.7 bp vs. 925.3 ± 819.3 bp; P < 0.05) (SI Appendix, Fig. S6B). The CpG content of DMRs was the lowest in Q1 and highest in Q4 (18.9 ± 10.4 CpGs/DMR vs. 24.3 ± 18.2 CpGs/DMR; P < 0.001) (SI Appendix, Fig. S6C). Finally, using RNA-seq data from the single-cell sequencing samples described below, we found that the mean levels of gene expression were significantly higher in Q1 > Q2 > Q3 > Q4 in both the Dnmt3a+/+ and Dnmt3a−/− samples, when considering all genes in all cells (SI Appendix, Fig. S7); this suggests that genes associated with fast remethylating DMRs are more transcriptionally active.
We annotated pathways associated with genes within 5 kb of the nearest DMR using Protein Analysis Through Evolutionary Relationships (http://pantherdb.org/) (29–32). We compared the gene lists using Mus musculus as the reference organism, and identified biological processes associated with each quartile (SI Appendix, Fig. S8). Genes near faster remethylating DMRs were significantly enriched for pathways involving hematopoiesis, cell death, development and differentiation, signaling, and transcription, whereas genes associated with more slowly remethylating DMRs were mainly enriched for cell death pathways. Using the HOMER algorithm (33), we searched for sequence motifs that were enriched in the DMRs of Q1 vs. Q4, but none were identified as statistically significant.
Gene Expression Patterns Associated with Dnmt3a-Deficient Hematopoietic Cells.
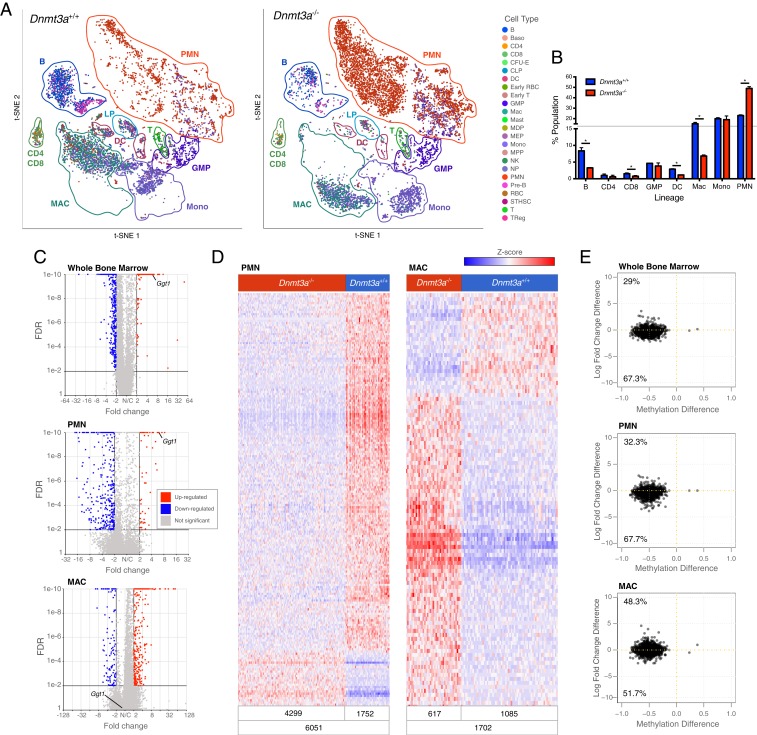
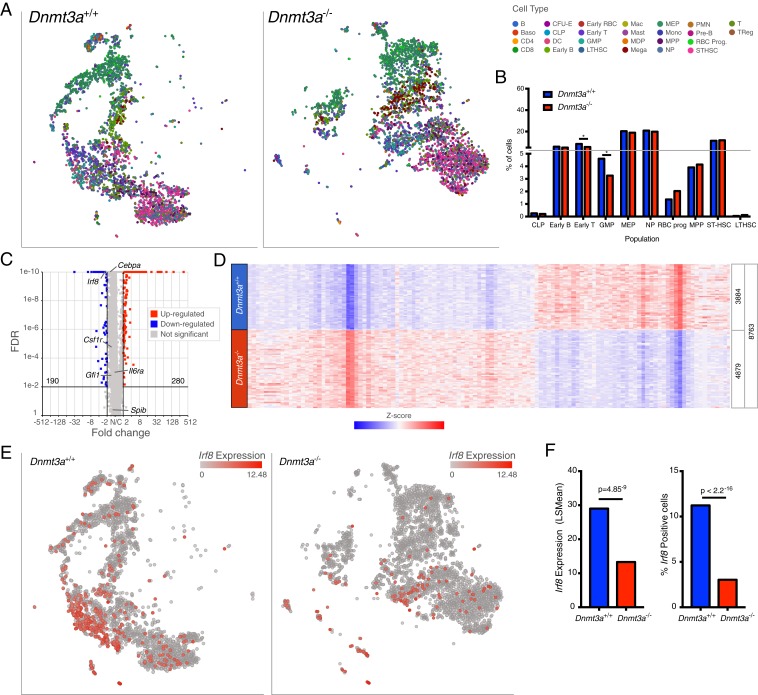
Bulk RNA-seq studies of Dnmt3a+/+ vs. Dnmt3a−/− total bone marrow cells revealed only 106 differentially expressed genes (DEGs), including Dnmt3a itself, as expected (Dataset S3). Some of these genes were on the X and Y chromosomes, since the sexes of the Dnmt3a+/+ vs. Dnmt3a−/− samples were different. Although the small number of changes in gene expression are consistent with previously published studies (14, 16, 18), we suspected that gene expression changes in subpopulations of cells were being masked with RNA-seq from a pool of all bone marrow cells. We therefore performed single-cell RNA-seq (scRNA-seq) on total bone marrow cells from mice that had been transplanted 12 wk earlier with Dnmt3a+/+ vs. Dnmt3a−/− donors, using the 10× Genomics Chromium platform, as previously described (16, 34). We evaluated cryopreserved bone marrow cells with or without sorting for viability from the same donor mice, and the results were essentially equivalent; we therefore pooled the data for the analyses shown in Fig. 4. After applying standard approaches for data filtration using Partek Flow software, we inferred the lineage of each cell using a k-nearest neighbor algorithm trained on the Haemopedia Database (35) (Fig. 4A). Although all lineages were present in both genotypes, the Dnmt3a−/− marrow samples had a significant reduction in the proportions of B cells, CD8+ T cells, dendritic cells, and macrophages, and a significant enrichment in mature myeloid cells (polymorphonuclear cells [PMNs]; Fig. 4B). Identification of DEGs for whole bone marrow, PMNs, and macrophage populations are shown using volcano plot projections in Fig. 4C, and expression data are shown in Datasets S4–S6. In all three populations, the vast majority of genes were not differentially expressed. Using data from all cells (whole bone marrow), 172 genes were significantly up-regulated in Dnmt3a-deficient cells (fold change ≥ 2.0, FDR ≤ 0.01), and 716 genes were significantly down-regulated (fold change ≤ −2.0, FDR ≤ 0.01). In an analysis restricted to PMNs, 59 genes were significantly up-regulated, and 329 genes were down-regulated; in macrophages, 340 genes were significantly up-regulated, and 110 genes were down-regulated. The Ggt1 gene, which has a canonical DMR in its 5′ flanking region (Figs. 1 and 2), is expressed at significantly higher levels in Dnmt3a−/− PMNs, but is not dysregulated in macrophages (Ggt1 labels in Fig. 4C); since PMNs represent the most abundant population in the bone marrow, this change can also be detected in the whole bone marrow data (Dataset S4) and bulk RNA-seq data (Dataset S3). Of the 67 DEGs up-regulated in Dnmt3a−/− bone marrow cells identified by bulk RNA-seq, 8 were concordantly and significantly up-regulated in the scRNA-seq whole bone marrow sample data; of the 39 down-regulated DEGs, 5 were concordantly down-regulated using the scRNA-seq data. However, most of the genes that were differentially expressed in small populations of cells were not detected in the bulk RNA-seq data (Dataset S3), or by using scRNA-seq data pooled from all bone marrow cells (Dataset S4).
Fig. 4.
Dnmt3a−/− bone marrow cells have population-restricted DEGs, and a myeloid lineage bias. (A) The t-SNE projections of scRNA-seq data from whole bone marrow cells derived from mice transplanted with Dnmt3a+/+ (n = 2; Left) or Dnmt3a−/− donors (n = 2; Right), and showing known hematopoietic populations based on Haemopedia gene expression profiling (35). (B) Population fractions associated with the scRNA-seq data shown in A; * indicates FDR < 0.05 via Fisher’s exact test with multiple hypothesis correction. (C) Volcano plots showing DEGs in whole bone marrow, PMNs, and macrophage populations (MACs) from Dnmt3a−/− mice (FDR ≤ 0.01, FC ≤ −2, ≥ 2). (D) Heatmaps of normalized (Z-score) expression values for 388 DEGs identified in PMNs (59 up-regulated and 329 down-regulated) and 450 DEGs identified in macrophages (340 up-regulated and 110 down-regulated). Numbers of cells represented in each grouping are shown at the bottom of each heatmap. (E) Methylation difference of each DMR associated with a gene within 5 kb (x axis), plotted against the difference in expression values between Dnmt3a+/+ and Dnmt3a−/− cells (y axis). Inset numbers indicate the fraction of genes in each quadrant. Nearly all DMRs associated with genes are hypomethylated, and the majority of these genes are expressed at lower levels in Dnmt3a−/− cells.
To evaluate the consistency of differential gene expression among cells assigned to the same lineage, we created heatmaps showing the expression values for all DEGs for lineage-defined cells from each genotype (Fig. 4D). In both PMNs and macrophages, most cells from each lineage have a canonical expression phenotype for each DEG, revealing that these are consistent effects across many similar cells, and not driven by small outlier populations.
Finally, we evaluated the relationship between DMRs and gene expression for each of these populations (Fig. 4E). We determined the average expression values for each of the 2,513 genes located within 5 kb of the 8,480 DMRs defined in Fig. 1. The difference in methylation values between Dnmt3a+/+ vs. Dnmt3a−/− cells is shown on the x axes; nearly all DMRs are hypomethylated in Dnmt3a−/− bone marrow cells. The differences between the average expression values for each of the DMR-associated genes are represented on the y axes. For each population, the majority of DMR-associated genes had slightly lower expression levels in Dnmt3a−/− cells. Overall, differences in expression were not significantly correlated with methylation differences, demonstrating the complex and difficult-to-predict relationships between methylation patterns and gene expression in hematopoietic cells (7, 13, 14, 16, 17, 36).
Differentially Expressed Genes in HSPCs.
The scRNA-seq data described above clearly demonstrate a shift in the proportions of mature myeloid and macrophage-like cells in Dnmt3a-deficient bone marrow, suggesting that an alteration in the expression of one or more lineage-defining factors may occur in Dnmt3a-deficient progenitors. Because progenitor cells are rare, we performed scRNA-seq on progenitor-enriched (Kit+/Lineage depleted) bone marrow cells derived from mice transplanted with Dnmt3a+/+ vs. Dnmt3a−/− bone marrow cells (Fig. 5). Representations of the cellular composition of these samples are shown in t-distributed stochastic neighbor embedding (t-SNE) plots in Fig. 5A, and the fractions of defined populations are quantified in Fig. 5B. Most populations are equivalent in the two samples, but the GMP population was significantly smaller in the Dnmt3a−/− sample. DEGs defined for all of the cells in this dataset are displayed in a volcano plot (Fig. 5C) and an expression heatmap (Fig. 5D and Dataset S7). A total of 280 genes were significantly up-regulated in Dnmt3a−/− progenitors, and 190 genes were down-regulated; most genes exhibited a trend toward down-regulation that was not significant (note the downward shift in nonsignificant genes between a fold change of −2 and +2). Gene ontology pathways enriched in the down-regulated gene set were most highly enriched for processes associated with neutrophil aggregation, inflammatory responses to antigenic stimuli, and leukocyte migration involved in inflammatory responses, among others (SI Appendix, Fig. S9). We specifically evaluated the expression of a set of well-characterized genes known to be important for myeloid vs. monocyte/macrophage lineage determination (i.e., Spi1, Gfi1, IL6ra, Csf1r, Cebpa, and Irf8; expression levels for each are identified on the volcano plot in Fig. 5C) (37–52). Irf8 was expressed at significantly reduced levels in the Dnmt3a-deficient progenitors, but the other genes were not dysregulated. The expression of Irf8 in individual cells (using the same t-SNE projection as used in Fig. 5A) is shown in Fig. 5E. Ifr8 is expressed in fewer cells in the Dnmt3a−/− sample, and the expression level per cell is also significantly reduced (Fig. 5F). Since Irf8 is known to drive progenitor cells toward the monocyte/macrophage lineage at the expense of myeloid cells, its down-regulation in Dnmt3a−/− progenitors would be expected to increase myeloid differentiation at the expense of the monocyte/macrophage lineage.
Fig. 5.
Dnmt3a−/− progenitors express reduced Irf8. (A) The t-SNE projections of scRNA-seq data from Dnmt3a+/+ (Left) and Dnmt3a−/− (Right) lineage-negative (i.e., depleted for cells expressing B220, CD19, CD11b, Ter119, and/or CD71), c-KIT−positive cells with known populations assigned according to Haemopedia gene expression profiling. (B) Population distributions assigned using the scRNA-seq data shown in A; * indicates FDR < 0.05 via Fisher’s exact test and multiple hypothesis correction. (C) Volcano plot showing DEGs in lineage-negative, c-KIT−positive cells from Dnmt3a−/− mice (FDR ≤ 0.01, fold change ≤ −2, ≥ 2). The values for several transcription factors associated with myelomonocytic lineage determination are shown. (D) Heatmap of normalized (Z-score) expression values for 470 DEGs (280 up-regulated, and 190 down-regulated) identified in lineage-negative, c-KIT−positive cells defined by the comparison of Dnmt3a+/+ vs. Dnmt3a−/−cells. Numbers of cells represented in each grouping are shown at the right of the heatmap. (E) Cells colored red according to levels of Irf8 expression in Dnmt3a+/+ (Left) and Dnmt3a−/− (Right) lineage-negative, c-KIT−positive cells. (F) Mean expression levels (Left) and fractions of cells positive for Irf8 relative to total cells (Right).
Addback of DNMT3A Partially Corrects Myeloid Skewing and Gene Expression Changes.
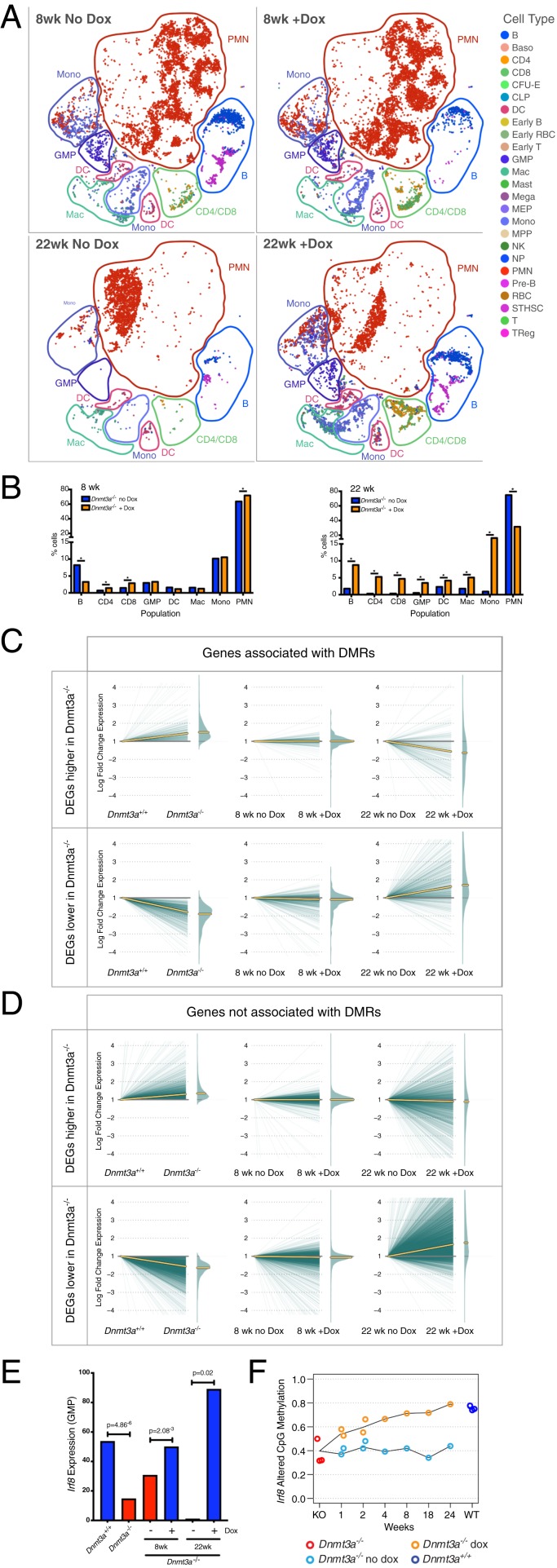
To determine whether reactivation of DNMT3A expression altered hematopoietic cell populations and/or gene expression over time, we transplanted Dnmt3a−/− bone marrow cells into four secondary recipients, documented engraftment 4 wk later, and then placed two mice on Dox chow (10,000 ppm) and two mice on normal chow; mice with and without Dox feeding were harvested 8 and 22 wk later, and bone marrow cells were subjected to scRNA-seq. We identified the transcriptional outputs of all cells, and defined clusters with lineage annotation (Fig. 6A); quantification of all cell types in the 8- and 22-wk addback samples are shown in Fig. 6B. Although skewing toward the myeloid lineage was apparent in the sample pair harvested after 8 wk, there were minimal changes in the proportions of all cell types with Dox feeding. In the sample pair harvested at 22 wk, a dramatic skewing of the sample toward myeloid cells was apparent in the Dnmt3a−/− mouse that was fed normal chow. However, with Dox feeding, a partial restoration of other lineages was apparent, and the proportion of myeloid cells in the bone marrow was significantly reduced.
Fig. 6.
Restoring DNMT3A expression partially corrects myeloid skewing and dysregulated gene expression in Dnmt3a−/− bone marrow cells. (A) The t-SNE projections of scRNA-seq data of whole bone marrow from recipient mice transplanted with Dnmt3a null-3A addback bone marrow and fed normal chow (no Dox), or 10,000 ppm Dox chow (with Dox) for the indicated times. Known populations are annotated according to Haemopedia gene expression profiling. (B) Population percentages identified using the scRNA-seq data from 8 wk (Top) or 22 wk (Bottom) of Dox chow feeding; * indicates FDR < 0.05 via Fisher’s exact test with multiple hypothesis correction. (C) Expression changes in DEGs within 5 kb of a DMR, defined by comparing Dnmt3a+/+ vs. Dnmt3a−/− cells. Plots are split into up-regulated genes (Top) or down-regulated genes (Bottom). Left shows the log fold change difference of each gene in Dnmt3a+/+ vs. Dnmt3a−/− cells. Middle and Right passively plot the expression values for the same sets of DEGs, displaying the effects of feeding Dox for 8 or 22 wk. Yellow lines indicate the median expression value for each set of genes. (D) Identical to C, but displaying DEGs not associated with a DMR. (E) Fraction of GMPs positive for Irf8 expression in Dmnt3a+/+ (blue) vs. Dnmt3a−/− (red) bone marrow cells, identified by scRNA-seq. The expression of Irf8 in GMPs is significantly increased after 8 or 22 wk of feeding Dox chow (red vs. blue bars). (F) Remethylation of differentially methylated CpGs in the Irf8 gene body with Dox feeding (orange line). No remethylation was detected in mice fed with normal chow (blue line).
We evaluated global trends in gene expression associated with these two pairs of addback samples. Fig. 6C displays the global pattern of expression of genes located within 5 kb of a DMR, with genes expressed at higher levels in Dnmt3a−/− bone marrow cells on top and those at lower levels on the bottom. When the expression values of the same genes were “passively” plotted for the 8-wk addback samples (Fig. 6 C, Middle), no trend toward correction of gene expression was apparent. However, with 22 wk of Dox feeding, a reduction in the expression of the up-regulated genes was apparent, and, conversely, there was a tendency toward increased expression of the down-regulated genes. For genes that were not associated with DMRs (Fig. 6D), there was no evidence of correction at 8 wk of Dox feeding, but, at 22 wk, the down-regulated genes displayed a tendency toward increased expression, while expression of regulated genes was not altered.
Because Irf8 was expressed at lower levels in the progenitors of Dnmt3a−/− mice, we evaluated its expression in the 8- and 22-wk addback sample pairs (Fig. 6D). Mean expression levels of Irf8 in GMPs increased significantly after 8 and 22 wk of Dox feeding. We also evaluated DNA methylation within the body of the Irf8 gene (Fig. 6E), and found that CpGs within the gene body were canonically hypomethylated in Dnmt3a−/− bone marrow cells. These gene body CpGs were remethylated as a function of time, with nearly complete correction at week 24. Since gene body methylation is often associated with high levels of gene expression (53–56), the loss of methylation of the Irf8 gene body may contribute to its decreased expression, while its restoration corresponds to increased expression.
We also evaluated the expression and remethylation patterns of cells of Ggt1, which has a canonical DMR in its 5′ flanking region (Figs. 1 and 2), and which is expressed at higher levels in Dnmt3a−/− PMNs (Fig. 6). We evaluated the expression of Ggt1 in individual cells from Dnmt3a+/+ and Dnmt3a−/− bone marrow samples (SI Appendix, Fig. S10A): Its expression is essentially restricted to PMNs; the fraction of PMNs expressing Ggt1 increases from 0.27% (WT) to 3.25% (Dnmt3a-deficient), and the level of Ggt1 RNA per expressing cell increases from 2.91 in WT cells to 17.6 in Dnmt3a−/− cells, (6.05-fold, P < 0.01; SI Appendix, Fig. S10B). Data from the 8-wk addback sample pair revealed a 3.3-fold reduction of Ggt1+ cells with Dox feeding (6.03% vs. 1.81%), and a significant reduction in mean Ggt1 expression per expressing cell (SI Appendix, Fig. S10B). The 22-wk sample from Dnmt3a-deficient mice (“no Dox”) revealed a striking increase in Ggt1-expressing PMNs (12.96%) that was reduced with Dox feeding (0.92%); likewise, the average expression level of Ggt1 in the 22-wk no Dox sample was even more elevated than in earlier samples, and was reduced to near-baseline levels in mice that were fed Dox chow (SI Appendix, Fig. S10B). Finally, the hypomethylated DMR located ∼5 kb upstream from Ggt1 was progressively remethylated over time with Dox chow feeding, exhibiting near-WT levels of methylation at week 24 (SI Appendix, Fig. S10C).
Discussion
In this report, we describe an experimental system that allowed us to determine the accuracy and rate of DNA remethylation in hematopoietic cells that were constitutively deficient for Dnmt3a. The bone marrow cells of these mice had a focal, canonical DNA hypomethylation phenotype that was minimally changed after transplantation into secondary recipients. Using a “tet-on” system to express WT human DNMT3A1 in these bone marrow cells, we found that DNA remethylation was detectable within a week, and nearly complete within 6 mo. The pattern of remethylation was highly ordered and consistent from mouse to mouse, suggesting that DMRs possess unique, locus-dependent features that help to define the rate at which they are remethylated. Long-term, enforced expression of transgenic DNMT3A did not result in hypermethylation of regions where Dnmt3a does not normally act, and did not cause global hypermethylation of the genome. Dnmt3a-deficient hematopoietic cell genomes can therefore be accurately remethylated in a genetically defined system. Using scRNA-seq approaches, we were able to define population-restricted DEGs in Dnmt3a-deficient hematopoietic cells, and show that DNMT3A addback caused a general trend toward correction of gene expression over time. This response was also associated with the gradual correction of the myeloid-enriched phenotype of Dnmt3a-deficient bone marrow cells.
The expression of WT human DNMT3A in Dnmt3a-deficient bone marrow cells resulted in rapid changes in DNA methylation in most DMRs, but some remethylated more quickly than others. The speed of remethylation of individual DMRs was ordered and stereotypical, suggesting that DMRs have unique, context-dependent properties that affect their ability to be recognized by or acted on by DNMT3A. These data strongly suggest that Dnmt3a must interact differently with specific regions of the genome, and these regions are defined by signals that do not require any prior exposure to Dnmt3a during development. The nature of these signals is currently unclear: we were not able to identify any sequence motifs within DMRs that distinguished fast vs. slow remethylating regions, nor were there any geographically restricted regions of the genome where remethylation occurred more rapidly. However, the recent identification of H3K36me2 and H3K36me3 as recruiters of DNMT3A to specific regions of the genome may offer a clue regarding how these regions may be targeted for methylation by DNMT3A (57).
The phenotypes associated with Dnmt3a deficiency—and remethylation—developed over extended periods of time, for unclear reasons. Even though this model system is based on constitutive Dnmt3a deficiency, the myeloid skewing phenotype was only minimally apparent at 2 wk of age, and became more pronounced over time; this suggests that acquired genetic and/or epigenetic events are very important for the shift toward myeloid enrichment. Many of the downstream effects of remethylation (i.e., correction of gene expression and myeloid skewing) occurred long after remethylation was detected. Although the reasons for this are not yet clear, we suggest that remethylation of late precursors and more mature cells—the most prevalent cells in the marrow—may have only small effects on gene expression or cell fate determination, since these cells may be irreversibly committed to a program regardless of their methylation status. If the key changes in methylation must occur in HSPCs, the consequences may occur more slowly, since these cells divide only infrequently, and since it can take weeks to months for phenotypes to appear in their progeny. Clearly, additional studies will be required to better understand the kinetics of the observed phenotypes in this system.
Previous gene expression studies of Dnmt3a haploinsufficient or deficient mouse bone marrow cells, and studies of human AML samples initiated by DNMT3A mutations, have failed to yield highly reproducible expression signatures (13, 14, 16). Indeed, our bulk sequencing studies of Dnmt3a−/− bone marrow cells revealed very few DEGs. However, scRNA-seq of Dnmt3a−/− bone marrow samples revealed hundreds of highly reproducible DEGs that were usually cell lineage- and/or developmental stage-specific. The relationships of these expression changes with DMRs were locus- and DMR-specific, and did not conform to a set of rules that could be applied globally. With DNMT3A addback, a general trend toward a correction of the gene expression patterns was detected, but the relationship of corrected expression and local remethylation was not clear for most genes. These findings demonstrate the continuing difficulty of establishing rules linking patterns of local DNA methylation and gene expression, a decades-old conundrum (58, 59); clearly, many factors besides DNA methylation are important for determining gene expression levels at any given locus.
The scRNA-seq data revealed hundreds of genes that are dysregulated in specific populations of Dnmt3a-deficient cells, but the importance of any individual gene for the observed hematopoietic phenotypes is not yet clear. We evaluated two genes (one up-regulated and one down-regulated) in greater detail to illustrate some of the features associated with potentially relevant candidates. Ggt1 encodes gamma-glutamyltransferase 1, an enzyme that catalyzes reduced glutathione to cysteine and glycine, delivering cysteine to the cell to facilitate the synthesis of glutathione, a major intracellular antioxidant that protects cells from oxidant stresses (60). Although its expression has not previously been described in either normal or malignant myeloid cells, it is up-regulated in the myeloid cells of Dnmt3a-deficient mice; a canonical hypomethylated DMR located several kilobases upstream of Ggt1 is remethylated with DNMT3A addback, and the gene is eventually down-regulated. The location of this DMR suggests that it may act as a proximal regulatory element or enhancer for this gene when unmethylated. However, no functional studies of this element have yet been described, so the importance of this DMR for Ggt1 up-regulation in myeloid cells will require functional validation. In contrast, Irf8 is a transcription factor that is down-regulated in Dnmt3a-deficient progenitor cells, where its gene body is hypomethylated; expression of this gene is known to induce progenitors to commit to the monocyte/macrophage lineage. Down-regulation of Irf8 is associated with the induction of neutrophilic maturation (38, 50–52, 61), which is observed in Dnmt3a haploinsufficient and deficient hematopoietic cells. DNMT3A addback was associated with Irf8 gene body remethylation, restoration of its expression, and the partial correction of time-dependent myeloid skewing; although the mechanisms responsible are not yet clear, the relationship of gene body methylation with gene expression is well described (53–56). Although extensive functional studies will be needed to fully understand the roles of individual genes for the phenotypes of Dnmt3a-deficient hematopoietic cells, these two genes provide examples of how candidate genes can be selected for functional validation.
The experimental system described here has caveats that may have influenced some of our observations. For example, we expressed a human DNMT3A transgene that was regulated indirectly by the Rosa26 locus, not the Dnmt3a locus. Although the transgene was expressed at near-physiologic levels in HSPCs, it was relatively overexpressed in committed progenitors and more mature cells of all lineages; this may explain why remethylation occurred more quickly in mature cells of the myeloid lineage. Further, endogenous Dnmt3a is clearly down-regulated in normal bone marrow cells a few weeks after birth, but the transgene is persistently expressed in Dox-fed animals throughout life. Finally, we did not evaluate addback phenotypes in Dnmt3a haploinsufficient mice (which are more like human patients with heterozygous loss-of-function mutations), because these mice have very subtle phenotypes (16) that would have been very difficult to evaluate with this model system. Although these experimental details may have influenced some phenotypes, we suggest that the fundamental conclusion of the study is still valid: enforced expression of DNMT3A in Dnmt3a-deficient bone marrow cells causes ordered and accurate DNA remethylation as a function of time.
In summary, these data suggest that CpGs that would normally be methylated by Dnmt3a remain “recognizable” in the genomes of cells that have never previously expressed the enzyme, and that some of the effects of Dnmt3a deficiency can be corrected by genetically reexpressing the WT protein. Although this approach would be impractical in humans with loss-of-function or dominant-negative DNMT3A mutations, it may be possible to engineer small molecules that can increase DNMT3A activity by altering its ability to homodimerize or homotetramerize, or that inactivate mutant forms of the enzyme. Although much additional work will be required to determine whether this will be possible, the current study does suggest that the genome retains signals that can direct DNMT3A to its proper sites of action, restore normal DNA methylation at these sites, and partially correct the phenotypes associated with Dnmt3a deficiency. This suggests that patients with ARCH or AML caused by mutations in DNMT3A could derive therapeutic benefit from an approach that restores the full activity of this enzyme.
Methods
Generation and Propagation of Mice.
A construct containing a tetracycline-inducible, WT human DNMT3A1 cDNA (SI Appendix, Fig. S1A) was introduced into the C57Bl6/J genome, and founders were crossed with R26-M2rtTA mice in the B6 background. A complete description can be found in Results and SI Appendix, Supplementary Methods.
Immunoblotting/Flow Cytometry.
Bone marrow cells from mouse femurs were utilized for flow cytometry applications. Antibodies against well-characterized cell surface receptors were used for identification and sorting of defined populations. Details can be found in SI Appendix, Supplementary Methods.
Bisulfite Sequencing.
Bisulfite sequencing was performed, as described in Cole et al. (16), aligned with biscuit, and DMRs were called using metilene (62), and smoothed/combined using the bsseq package (63). Full details and parameters are in SI Appendix, Supplementary Methods.
Total RNA Sequencing, Processing, and Analysis.
The Illumina TruSeq Stranded kit was used to produce bulk RNA-seq data, which used TopHat (64) and Stringtie (65) for alignment and expression quantification. DEGs were called using edgeR (66, 67). Additional details can be found in SI Appendix, Supplementary Methods.
Single-Cell RNA Sequencing, Processing, and Analysis.
Single-cell RNA libraries were created using the 10× Genomics Chromium Single Cell 5′ Kit, aligned with CellRanger, and assigned to lineages using cellMatch. The characteristics of each sample are summarized in Dataset S8. Additional filtering and defining of DEGs were done using Partek Flow software. Full details and parameters used are in SI Appendix, Supplementary Methods.
Supplementary Material
Acknowledgments
This study was supported by NIH Grants CA101937 and CA197561, and the Foundation for Barnes-Jewish Hospital (all to T.J.L.); and NIH Grant CA211782 (to C.A.M.). The Siteman Cancer Center Flow Cytometry Core (NCI Grant P30CA91842) provided expert support for all flow sorting studies.
Footnotes
Competing interest statement: T.J.L. is engaged in a scientific collaboration with Rigel Pharmaceuticals to develop drugs that inhibit the function of the DNMT3A R882H mutation. He has received two honoraria from Rigel Pharmaceuticals to give presentations at their company in South San Francisco, CA. None of this work is described in this paper.
Data deposition: All sequencing data for all studies were deposited to the NCBI, https://www.ncbi.nlm.nih.gov/bioproject (BioProject ID PRJNA483874).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918611117/-/DCSupplemental.
References
- 1.Steensma D. P., et al. , Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaiswal S., et al. , Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie M., et al. , Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 20, 1472–1478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genovese G., et al. , Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley T. J., et al. , DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch J. S., et al. , The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley T. J., et al. ; Cancer Genome Atlas Research Network , Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcucci G., Haferlach T., Döhner H., Molecular genetics of adult acute myeloid leukemia: Prognostic and therapeutic implications. J. Clin. Oncol. 29, 475–486 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Thol F., et al. , Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J. Clin. Oncol. 29, 2889–2896 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Yan X. J., et al. , Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 43, 309–315 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Ribeiro A. F., et al. , Mutant DNMT3A: A marker of poor prognosis in acute myeloid leukemia. Blood 119, 5824–5831 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Shen Y., et al. , Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood 118, 5593–5603 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Russler-Germain D. A., et al. , The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25, 442–454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spencer D. H., et al. , CpG Island hypermethylation mediated by DNMT3A is a consequence of AML progression. Cell 168, 801–816.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Challen G. A., et al. , Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 44, 23–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole C. B., et al. , Haploinsufficiency for DNA methyltransferase 3A predisposes hematopoietic cells to myeloid malignancies. J. Clin. Invest. 127, 3657–3674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong M., et al. , Loss of Dnmt3a immortalizes hematopoietic stem cells in vivo. Cell Rep. 23, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong M., et al. , Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat. Genet. 46, 17–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollink I. H. I. M., et al. , Acute myeloid leukaemia in a case with Tatton-Brown-Rahman syndrome: The peculiar DNMT3A R882 mutation. J. Med. Genet. 54, 805–808 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Kosaki R., Terashima H., Kubota M., Kosaki K., Acute myeloid leukemia-associated DNMT3A p.Arg882His mutation in a patient with Tatton-Brown-Rahman overgrowth syndrome as a constitutional mutation. Am. J. Med. Genet. A. 173, 250–253 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Shen W., et al. , The spectrum of DNMT3A variants in Tatton-Brown-Rahman syndrome overlaps with that in hematologic malignancies. Am. J. Med. Genet. A. 173, 3022–3028 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Tatton-Brown K., et al. ; Clinical Assessment of the Utility of Sequencing and Evaluation as a Service (CAUSES) Research Study; Deciphering Developmental Disorders (DDD) Study , The Tatton-Brown-Rahman syndrome: A clinical study of 55 individuals with de novo constitutive DNMT3A variants. Wellcome Open Res. 3, 46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Challen G. A., et al. , Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell 15, 350–364 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayle A., et al. , Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood 125, 629–638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okano M., Bell D. W., Haber D. A., Li E., DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Jones P. A., Liang G., Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 10, 805–811 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farlik M., et al. , DNA methylation dynamics of human hematopoietic stem cell differentiation. Cell Stem Cell 19, 808–822 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takiguchi M., et al. , Variability of inducible expression across the hematopoietic system of tetracycline transactivator transgenic mice. PLoS One 8, e54009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashburner M., et al. , Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mi H., Muruganujan A., Casagrande J. T., Thomas P. D., Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi H., Poudel S., Muruganujan A., Casagrande J. T., Thomas P. D., PANTHER version 10: Expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44, D336–D342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mi H., Thomas P., PANTHER pathway: An ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. 563, 123–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petti A. A., et al. , A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat. Commun. 10, 3660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Graaf C. A., et al. , Haemopedia: An expression Atlas of murine hematopoietic cells. Stem Cell Reports 7, 571–582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glass J. L., et al. , Epigenetic identity in AML depends on disruption of nonpromoter regulatory elements and is affected by antagonistic effects of mutations in epigenetic modifiers. Cancer Discov. 7, 868–883 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisenbeis C. F., Singh H., Storb U., Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 9, 1377–1387 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Tamura T., Kurotaki D., Koizumi S., Regulation of myelopoiesis by the transcription factor IRF8. Int. J. Hematol. 101, 342–351 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Chen H. M., et al. , Neutrophils and monocytes express high levels of PU.1 (Spi-1) but not Spi-B. Blood 85, 2918–2928 (1995). [PubMed] [Google Scholar]
- 40.Pahl H. L., et al. , The proto-oncogene PU.1 regulates expression of the myeloid-specific CD11b promoter. J. Biol. Chem. 268, 5014–5020 (1993). [PubMed] [Google Scholar]
- 41.Person R. E., et al. , Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat. Genet. 34, 308–312 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hock H., et al. , Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18, 109–120 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Peters M., et al. , Extramedullary expansion of hematopoietic progenitor cells in interleukin (IL)-6-sIL-6R double transgenic mice. J. Exp. Med. 185, 755–766 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schirmacher P., et al. , Hepatocellular hyperplasia, plasmacytoma formation, and extramedullary hematopoiesis in interleukin (IL)-6/soluble IL-6 receptor double-transgenic mice. Am. J. Pathol. 153, 639–648 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai X. M., et al. , Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99, 111–120 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Sasmono R. T., et al. , A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood 101, 1155–1163 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Cecchini M. G., et al. , Role of colony stimulating factor-1 in the establishment and regulation of tissue macrophages during postnatal development of the mouse. Development 120, 1357–1372 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Zhang D. E., et al. , Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 94, 569–574 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura A., et al. , Accelerated apoptosis of peripheral blood monocytes in Cebpb-deficient mice. Biochem. Biophys. Res. Commun. 464, 654–658 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Scheller M., et al. , Altered development and cytokine responses of myeloid progenitors in the absence of transcription factor, interferon consensus sequence binding protein. Blood 94, 3764–3771 (1999). [PubMed] [Google Scholar]
- 51.Tamura T., Nagamura-Inoue T., Shmeltzer Z., Kuwata T., Ozato K., ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity 13, 155–165 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Tsujimura H., Nagamura-Inoue T., Tamura T., Ozato K., IFN consensus sequence binding protein/IFN regulatory factor-8 guides bone marrow progenitor cells toward the macrophage lineage. J. Immunol. 169, 1261–1269 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Kulis M., et al. , Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 44, 1236–1242 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Yang X., et al. , Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26, 577–590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duymich C. E., Charlet J., Yang X., Jones P. A., Liang G., DNMT3B isoforms without catalytic activity stimulate gene body methylation as accessory proteins in somatic cells. Nat. Commun. 7, 11453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hellman A., Chess A., Gene body-specific methylation on the active X chromosome. Science 315, 1141–1143 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Weinberg D. N., et al. , The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baylin S., Bestor T. H., Altered methylation patterns in cancer cell genomes: Cause or consequence? Cancer Cell 1, 299–305 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Singal R., Ginder G. D., DNA methylation. Blood 93, 4059–4070 (1999). [PubMed] [Google Scholar]
- 60.Heisterkamp N., Groffen J., Warburton D., Sneddon T. P., The human gamma-glutamyltransferase gene family. Hum. Genet. 123, 321–332 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Holtschke T., et al. , Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87, 307–317 (1996). [DOI] [PubMed] [Google Scholar]
- 62.Jühling F., et al. , metilene: Fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 26, 256–262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansen K. D., Langmead B., Irizarry R. A., BSmooth: From whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 13, R83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pertea M., et al. , StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson M. D., Oshlack A., A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.