Significance
The G-patch domain is found in eukaryotic and viral proteins involved in protein–protein and protein–nucleic acid interactions. Some G-patch proteins play a vital role in the spliceosome by stimulating the DEAH-box ATPases Prp2 and Prp43; however, their structural characterization and exact binding mode have remained elusive. By studying the Prp2-specific G-patch domain of Spp2 by means of X-ray crystallography and nuclear magnetic resonance spectroscopy, we could show that the G-patch is mostly disordered in solution but adopts a defined fold on binding to Prp2. The disordered nature of Spp2 might be important for the positioning of Prp2 during its recruitment to the spliceosome.
Keywords: G-patch, DEAH-box ATPase, spliceosome, helicase, Prp2
Abstract
The spliceosome consists of five small RNAs and more than 100 proteins. Almost 50% of the human spliceosomal proteins were predicted to be intrinsically disordered or to contain disordered regions, among them the G-patch protein Spp2. The G-patch region of Spp2 binds to the DEAH-box ATPase Prp2, and both proteins together are essential for promoting the transition from the Bact to the catalytically active B* spliceosome. Here we show by circular dichroism and nuclear magnetic resonance (NMR) spectroscopy that Spp2 is intrinsically disordered in solution. Crystal structures of a complex consisting of Prp2-ADP and the G-patch domain of Spp2 demonstrate that the G-patch gains a defined fold when bound to Prp2. While the N-terminal region of the G-patch always folds into an α-helix in five different crystal structures, the C-terminal part is able to adopt two alternative conformations. NMR studies further revealed that the N-terminal part of the Spp2 G-patch, which is the most conserved region in different G-patch proteins, transiently samples helical conformations, possibly facilitating a conformational selection binding mechanism. The structural analysis unveils the role of conserved residues of the G-patch in the dynamic interaction mode of Spp2 with Prp2, which is vital to maintain the binding during the Prp2 domain movements needed for RNA translocation.
Intrinsically disordered proteins (IDPs) or protein regions (IDRs) lack stable secondary and tertiary structures under physiological conditions and in the absence of an interaction partner or ligand. However, IDPs and IDRs might undergo a disorder-to-order transition on binding to their targets, which has been referred to as “coupled folding and binding” or “induced folding.” The disorder of proteins provides functional advantages like the adaptability to different binding partners or the ability to bind with high specificity but relatively low affinity, which enables fast association or dissociation of the IDPs/IDRs to dynamic complexes (1–3).
IDPs and IDRs are widespread throughout the family of proteins involved in pre-mRNA splicing, as it has been predicted that approximately 45% of the human spliceosomal proteins are fully disordered and approximately 80% of the spliceosomal proteins contain disordered regions with a length of at least 30 residues (4). The spliceosome is a highly dynamic multimegadalton ribonucleoprotein (RNP) complex that catalyzes the removal of noncoding introns from precursor messenger RNAs (pre-mRNAs) in the nucleus of eukaryotic cells (5–7). The spliceosomes of all organisms consist of five uracil-rich small nuclear RNAs (UsnRNAs), a defined set of proteins stably associated with these UsnRNAs forming the UsnRNPs, and a large number of additional spliceosomal proteins that might bind to the spliceosome only transiently (8). Notably, the total number of spliceosomal proteins varies significantly with the organism (9).
For each intron to be excised, the spliceosome newly assembles onto the pre-mRNA in a stepwise order. Initially, the U1 snRNP binds to the 5′ splice site of the pre-mRNA and the U2 snRNP to the branch point sequence, resulting in formation of the spliceosomal A complex. Subsequently, the U4/U6.U5 tri-snRNP joins the spliceosome, leading to the catalytically inactive Bact complex. In a subsequent step, the catalytically active B* complex is formed, which catalyzes the first transesterification reaction. Then further remodeling leads to the formation of complex C, and the second transesterification reaction takes place. Finally, the spliceosome is disassembled, resulting in the release of the spliced mRNA and the intron lariat.
During assembly, splicing reaction, and disassembly, the spliceosome undergoes large compositional and conformational changes, including the remodeling of RNA–RNA, RNA–protein, and protein–protein interactions (8). These rearrangements are mainly driven by eight DExD/H-box ATPases, which use the energy from ATP hydrolysis to unwind dsRNA and/or to remodel RNA–protein interactions (10–12). These ATPases belong to the helicase superfamily 2 (SF2) and share a common fold containing two RecA-like domains that form the helicase core (13). Additional domains located C-terminal of the helicase core have been shown to act as a binding platform for interaction partners or to have a regulatory effect on ATPase activity (14, 15). The transition of the spliceosome into the catalytically active B* complex is promoted by the DEAH-box ATPase Prp2 together with its cofactor Spp2 (16–19).
The Prp2-specific cofactor Spp2 belongs to the family of G-patch–containing proteins. The name-giving glycine-rich patch (G-patch) consists of at least six conserved glycines and was first identified in RNA-associated proteins by bioinformatic analysis (20). G-patch proteins are absent in archaea and bacteria but present in some retroviral proteins and widespread among eukaryotes, where the G-patch domain can be found in various RNA- and DNA-binding proteins (21). The G-patch protein Ntr1 strongly enhances the ATPase and helicase activities of the DEAH-box helicase Prp43 (22, 23), in contrast to Spp2, which stimulates only the RNA-dependent ATPase activity of isolated Prp2 and not any helicase activity (24). Actually, no helicase activity could be observed for isolated Prp2 or the Prp2-Spp2 complex within the spliceosome (25, 26). This appears to be consistent with the recently determined cryo-electron microscopy (cryo-EM) structures of the Bact complex, as Prp2 binds just to single-stranded RNA at the periphery of the spliceosome (27, 28). These cryo-EM structures do not provide any information regarding Spp2, however.
In addition to the cryo-EM structures, biophysical studies on spliceosomal DEAH-box ATPases Prp16 and Prp22 also argue in favor of translocation along a single-stranded RNA as the primary function of these proteins rather than RNA duplex unwinding (29). Recent structural studies have demonstrated that RNA translocation is a highly dynamic process that requires DEAH-box ATPases to toggle between open and closed conformations of the helicase core (30, 31).
To gain insight into the structure of Spp2 and to understand how its interaction to Prp2 could be maintained despite the pronounced domain movements needed during RNA translocation, we examined unbound Spp2 in solution by means of circular dichroism (CD) and nuclear magnetic resonance (NMR) spectroscopy and determined five crystal structures of Prp2 in complex with the G-patch domain of Spp2 (Spp2G-patch). Spp2G-patch is unfolded in solution and only transiently samples an α-helical conformation in its N-terminal part. This N-terminal amphipathic helix stably binds Prp2 mainly via hydrophobic interactions with a conserved hydrophobic patch at the winged-helix (WH) domain. The C-terminal part binds to the RecA2 domain and can adopt two alternative conformations. Although the binding of Spp2G-patch does not induce major structural effects on Prp2 when compared with structures of Prp2 alone, the crystallographic snapshots of the interaction demonstrate that the N-terminal part of the G-patch serves as an anchor point, whereas the remaining regions associate in a much more flexible manner that likely guarantees its attachment to the mobile RecA2 domain.
Results
Purified scSpp2 Is Intrinsically Disordered.
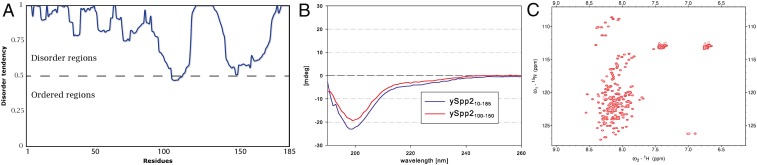
The G-patch protein Spp2 from Saccharomyces cerevisiae (scSpp2) consists of 185 amino acids and is predicted to be intrinsically disordered (Fig. 1A); however, experimental data regarding the folding and oligomerization state of scSpp2 have not been available (32). Since the expression of full-length scSpp2 in Escherichia coli was unsuccessful, truncated scSpp210–185 lacking the N-terminal nine residues was used for further experiments. Size exclusion chromatography of scSpp210–185 resulted in an apparent molecular mass of approximately 65 kDa, roughly three times higher than the calculated molecular mass of scSpp210–185. In contrast, multiangle light scattering identified this truncated Spp2 as a monomer, indicating that the protein behaves like a partially or fully unfolded protein (SI Appendix, Fig. S1). In addition, CD measurements were performed with scSpp210–150 as well as with the further truncated version scSpp2100–150 containing only the G-patch region. The CD spectra clearly show the absence of stable secondary structure elements for both samples (Fig. 1B). To study the properties of scSpp2 in more detail, a heteronuclear singular quantum coherence (HSQC) NMR spectrum of 15N-labeled scSpp210–185 was recorded (Fig. 1C). The NMR spectrum was of high quality with a narrow distribution of the cross peaks around 8 ppm of the 1H frequency, which is typical for the spectra of intrinsically disordered proteins.
Fig. 1.
Disorder and secondary structure analysis of yeast Spp2 in solution. (A) Analysis of the scSpp2 amino acid sequence by the Genesilico MetaDisorder prediction server. The probability to form disordered regions (y-axis) is shown with respect to the residue number (x-axis). (B) CD spectra of scSpp210–185 (blue line) and scSpp2100–150 (red line). The CD in mdeg (y-axis) is shown with respect to the wavelength (x-axis). (C) Two-dimensional 1H-15N HSQC spectrum of scSpp210–185 at 288 K and pH 6.5.
Crystal Structure of the ctPrp2-ctSpp2G-patch Complex.
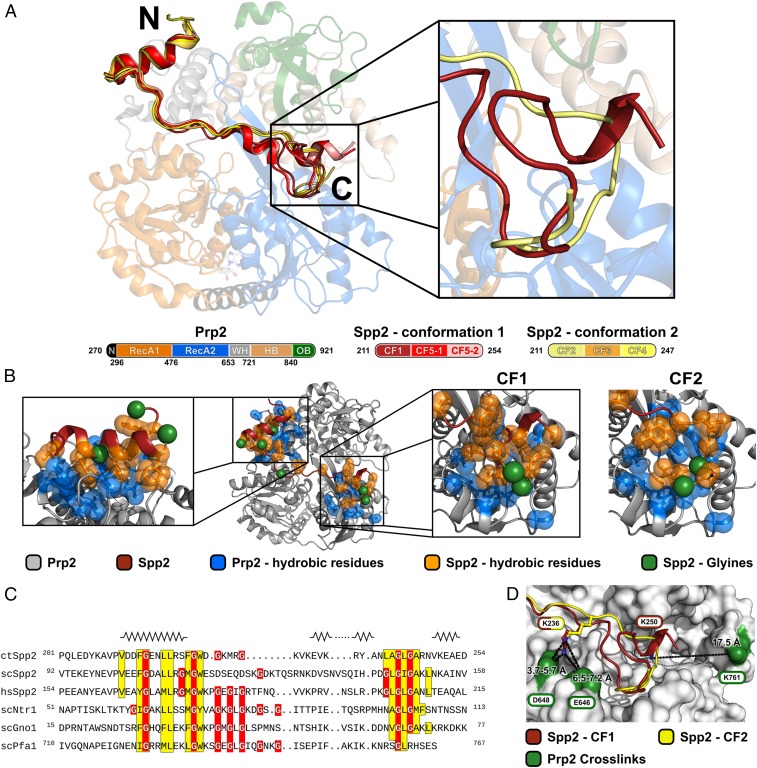
Since all attempts to crystallize the yeast Prp2-Spp2100–150 complex were fruitless, we used the ortholog proteins of Chaetomium thermophilum (ct), as this approach had previously been successful for the crystallization of other spliceosomal proteins, such as Prp2, Prp22, Prp43, Brr2, and Cwc27 (31, 33–38). The ctSpp2 consists of 313 residues, with the G-patch domain located between residues 211 and 254. For crystallization experiments ctSpp2211–254 was generated, and the stable complex of the truncated proteins ctPrp2270–921 and ctSpp2211–254 was prepared. Five different crystal forms (CF1 to CF5) of this ctPrp2-ctSpp2211–254 complex in four unique crystal packing arrangements were obtained, and all crystal structures were solved and refined.
In all crystal forms of the ctPrp2-ctSpp2211–254 complex, the N-terminal residues 212 to 222 of the G-patch domain form an amphipathic α-helix that is bound to the WH domain (Fig. 2A). The α-helix is terminated by a sharp kink, followed by a region exhibiting an extended conformation reaching the β-hairpin of the RecA2 domain. The C-terminal part of the ctSpp2 G-patch adopts two alternative conformations in the five different crystal structures. In conformation 1 (CF1 and CF5), this part interacts with the upper region of the RecA2 domain and adopts a loop-like conformation. In both molecules of CF5, this section is preceded by a short (less than two turns long) α-helix not present in CF1, while in both crystal forms, another short α-helical region is formed by the C-terminal residues of the G-patch. Formation of this short α-helix present only in CF5 is most likely induced by crystal contacts with the G-patch of the symmetry-related molecule (SI Appendix, Fig. S2). While in conformation 1 the polypeptide chain is kinked at position Y238, in conformation 2 it forms a roughly straight extended conformation up to N240, followed by a turn redirecting it to bind to a similar region of the RecA2 domain as observed for the C terminus of conformation 1. The G-patch of conformation 1 structures could be resolved up to D254, and in conformation 2 only residues up to R247 were defined in the electron density map.
Fig. 2.
Structural overview of the ctSpp2 G-patch bound to ctPrp2. (A) The model of the ctPrp2-ctSpp2211–254 complex depicted as a cartoon model, with ctPrp2 displayed semitransparently. N-terminal residues (270 to 296) of ctPrp2 are shown in black, the RecA1 domain (297 to 476) is shown in orange, the RecA2 domain (477 to 653) is shown in blue, the WH domain (654 to 721) is shown in gray, the HB domain (722 to 840) is shown in wheat, and the OB domain (841 to 921) is shown in green. Two alternative conformations of the ctSpp2 G-patch were found in complex structures obtained from five crystal forms (CFs). ctSpp2211–254 molecules exhibiting conformation 1 are depicted in different shades of red, whereas molecules belonging to conformation 2 are displayed in different shades of yellow. (Right) A zoomed-in view of the alternative conformations at the C-terminal end of the G-patch with one representative for each conformation. (B) Overview of hydrophobic interactions between ctSpp2211–254 and ctPrp2. Hydrophobic residues of ctSpp2 are shown in orange, while hydrophobic residues of ctPrp2 within 8 Å of the conserved ctSpp2211–254 hydrophobic residues are displayed in blue. Glycine residues of ctSpp2211–254 are highlighted as green spheres. (C) Sequence alignment of the G-patch domains of Spp2 from C. thermophilum, S. cerevisiae, and H. sapiens together with Ntr1, Gno1, and Pfa1 from S. cerevisiae. Conserved hydrophobic residues are highlighted in yellow, and glycine residues are shown in red. Secondary structure elements present in any of the five crystal forms are displayed on top of the corresponding segment of the sequence. The N-terminal amphipathic helix, as well as a hydrophobic stretch at the C-terminal end, are highly conserved. (D) Residues identified to cross-link to the lysines K236 and K250 of ctSpp2211–254 are shown as sticks and the cross-linked residues on ctPrp2 are shown in green.
The Spp2 G-patch binds Prp2 mainly via hydrophobic interactions (Fig. 2B). The sequence alignment of Spp2 from Chaetomium thermophilum, S. cerevisiae, and Homo sapiens together with the S. cerevisiae G-patch domains of Prp43-binding G-patch proteins Ntr1, Gno1, and Pfa1 highlight the conservation of the hydrophobic residues of the N-terminal amphipathic α-helix and a hydrophobic stretch at the C-terminal end (Fig. 2C). The hydrophobic side of the N-terminal α-helix faces a highly conserved hydrophobic patch on the surface of the WH (Fig. 2C and SI Appendix, Fig. S13A). Both alternative C-terminal conformations bind the same conserved hydrophobic region on the RecA2 domain. In addition to these hydrophobic interactions, the N-terminal α-helix is anchored via π-stacking between F214(ctSpp2) and Y697(ctPrp2) and the two hydrogen bond pairs G215(ctSpp2)-N694(ctPrp2) and R229(ctSpp2)-T686(ctPrp2) (SI Appendix, Fig. S3A). The conformation 1 is stabilized by polar interactions between L241(ctSpp2) and Y592(ctPrp2), as well as between K250(ctSpp2) and Y489(ctPrp2), constituting the C-terminal fragment of ctSpp2211–254 (SI Appendix, Fig. S3B). Conformation 2 is stabilized by a hydrogen bond formed between G243(ctSpp2) and E486(ctPrp2) (SI Appendix, Fig. S3C).
Apart from a set of conserved glycines, the middle section of the G-patch connecting the N-terminal α-helix with the C-terminal hydrophobic stretch shows a very low degree of conservation (Fig. 2C). In all crystal forms except CF5, this linker region does not directly interact with Prp2 and exhibits significantly elevated B-factors, implicating its greater flexibility (SI Appendix, Fig. S4). B-factors of the linker in CF5 are not elevated, as its conformation is stabilized by crystal contacts with the G-patch of a symmetry-related complex (SI Appendix, Fig. S2).
Overall, the G-patch domain can be divided into three parts: the N-terminal helix, the linker region, and the C-terminal interaction site. To test the contribution of each of these parts to the binding of ctSpp2G-patch to ctPrp2, we performed binding affinity experiments with truncated versions of the G-patch domain via isothermal titration calorimetry (ITC) (SI Appendix, Fig. S5). ctSpp2G-patch binds ctPrp2 with an affinity of Kd = 0.72 µM (SI Appendix, Fig. S5A). The construct lacking the C-terminal stretch with the alternative conformations (ctSpp2G-patchΔC) binds with roughly one order of magnitude decreased affinity (Kd = 9 µM) (SI Appendix, Fig. S5B). While the C-terminal part contributes significantly to the interaction, it is not able to bind alone, as the C-terminal part lacking the linker and the N-terminal helix (ctSpp2G-patchΔNL) exhibits no binding at all (SI Appendix, Fig. S5D). The construct consisting exclusively of the N-terminal helix (ctSpp2G-patchΔLC) exhibits a Kd of 40.3 µM (SI Appendix, Fig. S5C). Despite the high flexibility of the linker region and lack of detectable interactions in the crystal structures, it has an impact on the binding of the G-patch motif, as reflected by a fourfold decreased binding affinity of ctSpp2G-patchΔLC compared with ctSpp2G-patchΔC. Hence, the N-terminal helix is sufficient for the binding and indicates that this region functions as an anchor point for the binding of the G-patch motif to Prp2.
The functional impact of the conserved glycine residues, a hallmark of the G-patch, has remained elusive so far. Analysis of the ctPrp2-ctSpp2211–254 complex crystal structure revealed that the observed combination of φ and ψ torsion angles of the Spp2 glycine residues 223, 226, and 230, which are highly conserved throughout all G-patch proteins, are only favored for glycine residues according to the Ramachandran plot (SI Appendix, Fig. S6). These glycines are located at pronounced kinks that redirect the polypeptide chain and would be unfavored for nonglycine residues. In addition, G226 performs a CH/π interaction with W224, which encapsulates the highly conserved tryptophan between this glycine and a hydrophobic patch on Prp2 (SI Appendix, Fig. S7 B and C). A similar glycine–tryptophan interaction is observed in the trp cage miniprotein, in which a glycine within a nearby loop has been found to play a role in stabilizing the trp cage fold (39–41). To verify the importance of these three glycines, they were mutated to serine, and the binding to ctPrp2 was analyzed via a glutathione S-transferase (GST) pulldown assay (SI Appendix, Fig. S7A). Consistent with the results from the Ramachandran plot analysis, the ctSpp2 single point mutations G223S, G226S, and G230S showed significantly reduced binding to GST-ctPrp2 compared to wild-type ctSpp2 (SI Appendix, Fig. S7A, lanes 1 to 3). Double and triple mutations G223/226S and G223/226/230S completely abolished the protein interaction, highlighting the importance of these eponymous conserved glycine residues (SI Appendix, Fig. S7A, lanes 4 and 5).
In all five complex structures, Prp2 contains a bound ADP molecule. A comparison with the previously reported structures of the Prp2-ADP complex reveals that the nucleotide conformation as well as the conformation of the Prp2 catalytic center are not affected by the binding of the G-patch (SI Appendix, Fig. S8A) (35). This is also reflected in a virtually identical Kd of ∼172 nM for the binding of ADP in either the absence or the presence of the G-patch (SI Appendix, Fig. S8B) (35). The binding of ATP in dependence of the G-patch could not be tested using ATP analogs via ITC, as AMPPCP does not bind to Prp2 (35). For the ATP analogs AMPPNP and ATPγS, ATPase activity in the presence of Prp2 was observed, which did not allow successful ITC binding experiments (SI Appendix, Fig. S9). This activity could be caused either by the hydrolysis of the ATP analog or by an ATP contamination. Although the Prp2–Spp2 interaction seems to not have a major influence on the global conformation of Prp2, stabilization of the conformation of the RecA2 β-hairpin can be observed (SI Appendix, Fig. S10).
Cross-Linking Data Confirm the Spp2-Binding Site in Solution.
To exclude the possibility that the binding mode of Spp2 observed in the crystal structure is induced solely by packing of the molecules in the crystal lattice, in-solution chemical cross-linking was performed on the complex of ctPrp2 with full-length ctSpp2, and cross-linked peptides were identified by mass spectrometry. Chemical cross-linking was performed with BS3, a lysine-directed homo-bifunctional cross-linking reagent with an 11.4-Å spacer arm, and also with EDC, a hetero-bifunctional cross-linking reagent, leading to formation of a covalent bond between primary amines (Lys) and carboxyl-containing amino acid residues (Asp, Glu).
Two out of the five lysine residues located within the G-patch and thus present in the crystal structure were cross-linked to ctPrp2, namely K236 and K250, which form cross-links to E646/D648 and K761 of ctPrp2, respectively. The first cross-link corresponds to a zero-length cross-link, while the second cross-link corresponds to lysine residues within the distance of the spacer arm of BS3. These results are in good agreement with the crystal structure of the ctPrp2-ctSpp2211–254 complex (Fig. 2D).
Since the in-solution cross-linking experiments were performed with full-length ctSpp2, five more cross-links outside of the crystallized Spp2 fragment could be identified. These correspond to the lysine residues K17, K26, K72, and K296 in ctSpp2, cross-linked to the ctPrp2 residues K760, K761, K532/K535, and K761, respectively (SI Appendix, Fig. S11 A and B). Taken together, the cross-linking experiments with both reagents show that the mode of binding of ctSpp2211–254 to ctPrp2 in solution is maintained in the reported crystal structures of the complex, and that the interactions between both proteins extend beyond the crystallized ctSpp2 fragment. Cross-linking studies on the spliceosomal Bact complex from yeast have shown that regions other than the G-patch of scSpp2 also interact with scPrp2, as well as with other spliceosomal factors (27).
The G-Patch of Spp2 Populates Transient Helical Conformations in Solution.
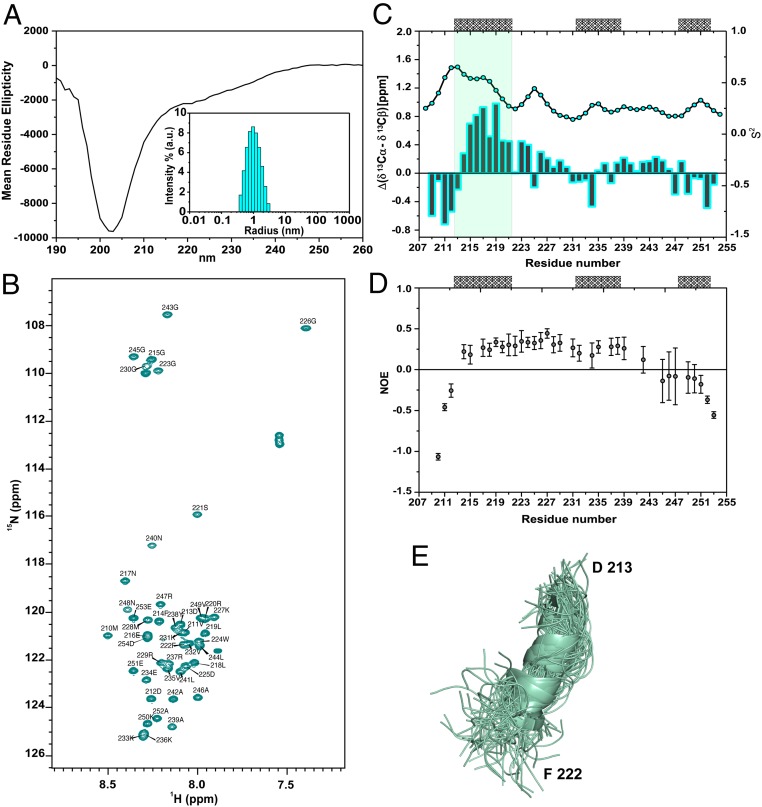
To obtain residue-specific insight into the conformational space sampled by G-patch proteins in solution and before complex formation, a slightly larger ctSpp2 G-patch region (ctSpp2208–254) was generated, as a higher yield of the purified recombinant protein could be obtained. CD on ctSpp2208–254 displayed a minimum around 203 nm, typical of an unstructured protein that does not possess a stable secondary structure (Fig. 3A), in agreement with the results for scSpp2 (Fig. 1). In addition, dynamic light scattering showed that ctSpp2208–254 is predominantly monomeric in solution (Fig. 3A). To perform the sequence-specific assignment of its backbone resonances, the 3D triple-resonance NMR experiments with HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, and HNCO were recorded using 13C/15N-labeled ctSpp2208–254. Analysis of the corresponding spectra enabled assignment of the backbone resonances of all nonproline residues of ctSpp2208–254 (Fig. 3B).
Fig. 3.
Transient helical conformations in the G-patch of ctSpp2 before complex formation. (A) Far-UV CD spectrum (mean residue ellipticity vs. wavelength, in nm) of ctSpp2208–254. (Inset) DLS measurements demonstrating that ctSpp2208–254 is predominantly monomeric in solution. (B) Two-dimensional 1H-15N-HSQC spectrum of ctSpp2208–254. The sequence-specific assignment of the backbone resonances is indicated. (C) Residue-specific ΔCα-ΔCβ secondary chemical shifts of ctSpp2208–254 together with S2 parameters derived by TALOS+. Positive values of ΔCα-ΔCβ indicate a propensity for α-helical conformations, while negative values indicate a propensity to form extended structures. The positions of α-helices observed in ctSpp2208–254 when in complex with Prp2 are shown on top. (D) Heteronuclear steady-state {1H,15N} NOE as a function of residue number. Error bars represent the SDs and were calculated as described in Materials and Methods. (E) Ensemble of α-helical conformations populated by residues D213 to F222 of ctSpp2208–254.
On the basis of the NMR chemical shifts, the secondary structure of ctSpp2208–254 was studied with single-residue resolution. Particularly useful for this analysis are the Cα and Cβ chemical shifts (42). Fig. 3C shows the variation in ΔδCα-ΔδCβ values along the primary sequence of ctSpp2208–254, that is, the deviation between the experimental chemical shifts and those expected for a random coil peptide. Stretches of consecutive residues with positive ΔδCα-ΔδCβ values are characteristic for the population of helical conformations, while regions with negative ΔδCα-ΔδCβ values identify the β-structure. For ctSpp2208–254, the magnitude of ΔδCα-ΔδCβ values was <1.0 (Fig. 3C), in agreement with the absence of a rigid secondary structure (Fig. 3A), which would result in ΔδCα-ΔδCβ values >3.0. At the same time, a pronounced tendency of residues F214 to S221 to populate helical conformations was detected (Fig. 3C). Quantitative analysis of the ΔδCα-ΔδCβ values indicated that approximately 20% of the conformations sampled by F214 to S221 are helical (43).
Next, the dynamic properties of ctSpp2208–254 were investigated. Subjecting the assigned chemical shifts to the software TALOS+ (44) provided estimates for the general order parameter, S2. S2 values describe the extent of motions on a picosecond-nanasecond time scale, providing important information on the level of spatial restriction within the molecular reference frame. Low-order parameters throughout the backbone of ctSpp2208–254 were found (Fig. 3C), indicating a low motional restriction typical for IDPs. The least dynamic residues were F214 to S221, which transiently populate the helical structure. The decreased mobility of this region was further supported by 15N spin relaxation measurements. The N- terminal and C-terminal residues starting at residue L244 had negative steady-state {1H,15N} nuclear Overhauser effect (NOE) values (Fig. 3D), indicating that they are the most dynamic parts of ctSpp2208–254. In between, steady-state {1H,15N} NOE values were ∼0.25. Notably, residues F214 to S221 did not have higher steady-state {1H,15N} NOE values than the subsequent residues up to A239 (Fig. 3D), suggesting that the elevated general order parameters S2 of F214 to S221 are caused by slower motions, which affect the chemical shifts but not the steady-state {1H,15N} NOE. Such slower motions are consistent with transient helix formation. Taken together, the NMR spectroscopy data demonstrate that the conserved N-terminal part of the G-patch of ctSpp2 populates transient helical conformations in solution and a part of the conserved hydrophobic C-terminal stretch shows increased dynamics, in agreement with the alternative conformations of this region found in the crystal structures (Fig. 3E).
Discussion
G-patch proteins play a vital role in the functioning of the spliceosomal DEAH-box ATPases Prp2 and Prp43, and thus their interplay has been the subject of numerous studies. While in both cases, the interaction of the G-patch protein with the corresponding ATPase is essential to fulfill their roles in the spliceosomal context, they modulate the activity of its ATPase differently (18, 19, 23, 45). Spp2 enhances the ATPase activity of Prp2 only in the presence of RNA and is not able to induce any helicase activity (24, 25). In contrast, Ntr1 stimulates the Prp43 ATPase activity in the presence and in absence of RNA, and greatly enhances the helicase activity (22, 23, 37). Although the functional impact of the G-patch proteins on the spliceosomal DEAH-box ATPases and ultimately its role during splicing have been studied extensively, not much about their interaction mode is known. It has been demonstrated that a stretch of 60 amino acids containing the conserved G-patch domain of Ntr1 is sufficient for Prp43 interaction and stimulation (22, 46). With this information, we designed a minimal Spp2 construct to gain molecular insight into the interaction of the G-patch with the DEAH-box ATPase Prp2 by means of X-ray crystallography and to study its properties in solution using CD and NMR spectroscopy.
Our data show that both N-terminally truncated scSpp210–185 and the G-patch domains of scSpp2 and ctSpp2 are intrinsically disordered in solution (Figs. 1 and 3). However, in the ctPrp2-ctSpp2211–254 complex, the N-terminal region of the G-patch adopts a helical conformation, which could be confirmed to be transiently present in only 20% of the molecules in solution (Figs. 2A and 3). This amphipathic α-helix binds primarily via hydrophobic interactions to the ctPrp2 WH domain in all crystal structures (Fig. 2). The C-terminal end of the crystallized G-patch exhibits two alternative conformations in different crystal structures, both of which bind mainly to the RecA2 domain also via hydrophobic interactions (Fig. 2). This C-terminal hydrophobic stretch and the N-terminal α-helix are connected by a linker region that does not directly interact with ctPrp2. Elevated B-factor values in CF1-4 suggest a high flexibility of this region (SI Appendix, Fig. S4).
The G-patch regions of Spp2 and Ntr1 share the property of being intrinsically disordered in solution (22). Interestingly, CD spectra of the Ntr1 G-patch showed no secondary structures, but the formation of α-helices could be induced on the addition of trifluoroethanol. These helices likely correspond to the N-terminal α-helix observed for ctSpp2211–254 upon interaction with ctPrp2. In fact, this part of the G-patch is highly conserved in G-patch proteins interacting with either Prp2 or Prp43, and the presence of hydrophobic residues occurring in a 3–4 pattern (HxxH, HxxxH) strongly argues in favor of the formation of an amphipathic α-helix as well in all G-patch proteins interacting with Prp43 (Fig. 2C). Binding experiments of different truncated versions of ctSpp2G-patch show that this N-terminal helix is sufficient for binding to the DEAH-box ATPase and thus likely displays the major anchor point of the G-patch motif (SI Appendix, Fig. S5). The C-terminal end, which also shows a high degree of conservation of hydrophobic residues and glycines, contributes significantly to the overall binding of the G-patch domain despite its rather flexible binding implied by two alternative conformations (Fig. 2A and SI Appendix, Fig. S5B). A reduction in affinity slightly above one order of magnitude of ctSpp2G-patchΔC highlights the importance of this region in the interaction, although it is not able to bind alone (SI Appendix, Fig. S5D). In contrast, the flexible linker region of Spp2 does not show any sequence conservation. The lack of detectable interactions of this region in all crystal structures, together with the elevated B-factor values, suggest only minor contributions to the binding of the G-patch. Indeed, this is reflected by a milder fourfold reduction in the affinity of ctSpp2G-patchΔLC compared with ctSpp2G-patchΔC (SI Appendix, Fig. S5C). Furthermore, the occurrence of glycines in all G-patch domains might guarantee its intrinsic flexibility and allow unique conformations on complex formation. In particular, glycines 223, 226, and 230 of ctSpp2 exhibit conformations allowed only for glycine residues, and mutation to serine strongly weakens the binding of the ctSpp2G-patch to ctPrp2 (SI Appendix, Fig. S7). In addition, the hydrophobic patches on ctPrp2 involved in the interaction with the N- and C-terminal parts of ctSpp2211–254 are also conserved among Prp2 and Prp43 (SI Appendix, Fig. S13). The level of conservation of the interacting regions on both Prp2/Prp43 and the different G-patch domains suggests that interaction of the G-patch with Prp43 might be comparable to that seen in the ctPrp2-ctSpp2211–254 complex. In fact, a previously published study on the scPrp43–scNtr1 interaction proposes a binding site similar to that seen in the ctPrp2-Spp2211–254 complex structure (22). By superposition-based modeling of ctSpp2211–254 onto the ADP-bound scPrp43 structure, several of the identified cross-links between scPrp43 and scNtr1 were confirmed to be in good agreement with respect to the observed distances in the model, thereby suggesting a similar binding mode as seen for ctSpp2211–254 and ctPrp2 (SI Appendix, Fig. S14 A and B).
Both Spp2 and Ntr1 have been proposed to bind to the C-terminal domain of their respective ATPase, and the OB-fold domain has been credited with a special role in this interaction (19, 22, 47). While we can confirm that the N-terminal α-helix of the G-patch indeed binds to the WH domain belonging to the C-terminal domains, our complex structure does not show any direct interactions between ctSpp2211–254 and the OB-fold domain (Fig. 2A). However, the C-terminal stretch of the ctSpp2 G-patch binds to a hydrophobic patch on the RecA2 domain located at the interface between the OB-fold and RecA2 domains. Conformational perturbations due to mutations introduced into the OB-fold domain (19), or even the complete truncation of this domain (47), could indirectly lead to the observed impacts on the binding and function of the G-patch protein. In particular, the β-hairpin of the RecA2 domain, which is involved in numerous contacts with the OB-fold domain, may be structurally influenced by artificial variations of the OB-fold domain. This could lead to an indirect effect on the binding of the G-patch, since its linker region extends close to this structural feature.
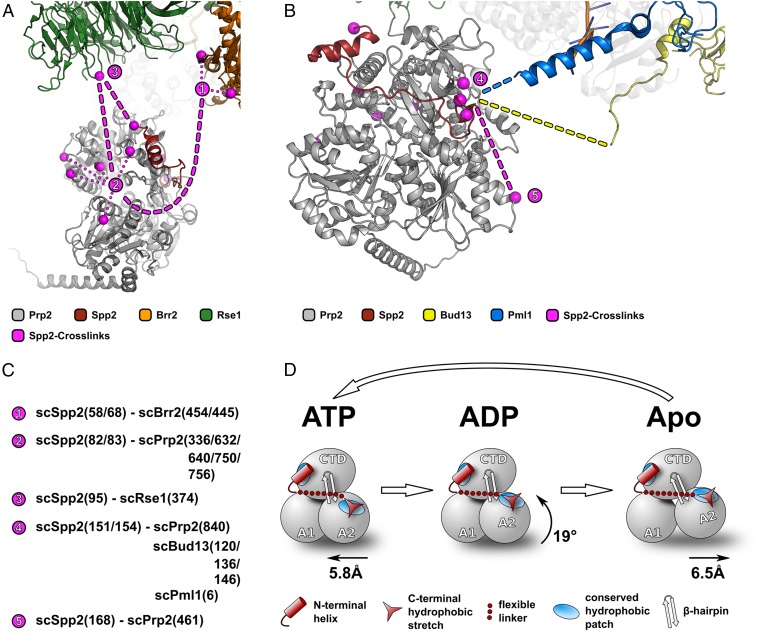
Cryo-EM structures of the Bact complex unveiled the location of Prp2 within the spliceosome, but due to the high flexibility of Spp2 and the limited resolution of cryo-EM 3D reconstructions at the periphery of such dynamic complexes, no structural information on the G-patch protein has been available so far (27, 28, 48, 49). Using the ctPrp2-ctSpp2211–254 complex structure, it is now possible to situate the Spp2 G-patch within the Bact complex and in concert with available cross-link data estimate the interaction network of flanking regions of the G-patch (SI Appendix, Fig. S15) (27). The association of Spp2 with the spliceosome can be divided into three different interaction regions. The N-terminal part contacts spliceosomal factors Brr2 and Rse1, as well as Prp2 (Fig. 4 A and B). The interaction between Spp2 and Prp2 at this site seems to be highly dynamic, as multiple cross-links were found on the ATPase. Cross-links between the C-terminal end of the G-patch and the OB-fold domain located in close proximity are in good agreement with the ctPrp2-ctSpp2211–254 complex structure (Fig. 4 B and C). In addition, this part of the G-patch contacts nearby regions of Bud13 and Pml1, which are not visible in the cryo-EM 3D reconstruction. C-terminally from the G-patch, Spp2 contacts Prp2 at a more distant location of the RecA2 domain. In summary, the cross-link data published by Rauhut et al. (27) are in good agreement with our ctPrp2-ctSpp2211–254 complex structure. In addition, our cross-link data confirm that also parts outside the ctSpp2 G-patch can contact ctPrp2, as seen for S. cerevisiae Spp2 and Prp2 (SI Appendix, Fig. S11).
Fig. 4.
Spp2 in the spliceosome and its conformational adaptability to different DEAH-box ATPase conformations. Using a combination of the ctPrp2-ctSpp2211–254 complex structure together with the cryo-EM and cross-linking data from Rauhut et al. (27), how Spp2 interacts with the spliceosome can be estimated. All spliceosomal factors are depicted as cartoon models, and cross-links of Spp2 are highlighted as pink spheres. The estimated path of Spp2 is displayed as a pink dashed line, and main contact sites are numbered starting from the most N-terminal cross-linked residue. (A) N-terminally from the G-patch, Spp2 cross-links with Brr2 and Rse1, as well as with Prp2. (B) The C-terminal end of the Spp2 G-patch cross-links with the OB-fold domain of Prp2 and parts of Bud13 and Pml1. Cross-linked regions of Bud13 and Pml1 are not part of the cryo-EM model and are symbolically depicted as dashed lines. C-terminally from the G-patch, Spp2 contacts another part of the RecA2 domain. (C) Overview of Spp2 cross-links numbered as in A and B. (D) Schematic representation of the catalytic states of Prp2 during one translocation cycle. The RecA2 domain is the most mobile domain during this process, and due to the versatile conformations of the conserved C-terminal stretch together with the flexible linker region, the Spp2 G-patch is able to adapt to the individual conformations of the RecA2 domain.
The numerous interactions of Spp2 with Prp2 and the spliceosomal factors Brr2, Rse1, Bud13, and Pml1 might play roles in the proper positioning of the ATPase after recruitment to the spliceosome. It has been shown that Prp2 is able to join the spliceosomal complex in the absence of Spp2, but the catalytic activation of the spliceosome can occur only when Spp2 is present (18, 24). This hints at the possibility that Prp2 is recruited first to a site distant to the functional site and has to be repositioned by Spp2. In fact, yeast two-hybrid and pulldown experiments have shown direct interactions of Prp2 and Brr2 independent of Spp2 (50, 51). Interestingly, neither cross-linking studies nor cryo-EM structures of the Bact-complex could verify this interaction, but instead Prp2 is just located in close proximity to Brr2 (27, 28). However, this does not exclude the possibility that Prp2 is directly associated with Brr2 at a prior timepoint, and it could be feasible that Prp2 is initially recruited via interactions with Brr2 close to its target site (SI Appendix, Fig. S12). If Spp2 is already present in the Bact-complex during the recruitment of Prp2 via Brr2, the G-patch protein could recognize the nearby Prp2 and ensure the proper positioning at its target site. Alternatively, Prp2 is preloaded with Spp2 on recruitment to Brr2 and the G-patch protein could detect the functional site via its interactions with Brr2, Rse1, Bud13, and Pml1 and thereby reposition Prp2 to its designated site.
The intrinsic flexibility of Spp2 might provide key advantages for this repositioning role. On one hand, the flexibility allows Spp2 to reach more distant parts of the spliceosome while remaining attached to either Prp2 or the functional site components. This avoids the necessity of larger conformational rearrangements at the target site to correctly place Prp2. On the other hand, Spp2 is able to connect four different spliceosomal factors (Brr2, Rse1, Bud13, and Pml1) to Prp2 while having only a minimal impact on the accessibility of the pre-mRNA. The functional target of Prp2 is a single-stranded stretch of the pre-mRNA protruding out from the interior of the spliceosome, and a structured protein assuming the role of positioning Prp2 for its interaction with the RNA could occlude the RNA, thereby restricting its accessibility. Once Prp2 is correctly positioned, it is able to load the ssRNA, which together with the G-patch stimulates the ATPase/translocation activity of the DEAH-box ATPase to remodel the spliceosome (24, 31, 37).
The intrinsic flexibility of proteins is known to be connected to many advantages in binding to interaction partners, such as increased adaptability and specificity, and in the case of G-patch proteins, this flexibility seems to be crucial for coping with the conformational dynamics of Prp2 and Prp43 (1–3). It has been shown that during the RNA translocation of DEAH-box ATPases, the RecA2 domain is the most mobile domain, rotating 19° on ATP hydrolysis, moving away from the RecA1 domain by 6.5 Å after ADP release, and returning to a closed conformation on ATP binding (Fig. 4D) (31). Since the C-terminal hydrophobic stretch of ctSpp2211–254 is located on the RecA2 domain of Prp2, it must be able to adapt to the movements of the RecA2 domain during translocation to remain constantly bound. This is achieved on one hand by the presence of a flexible linker connecting the anchoring α-helix with the C-terminal interacting fragment of the G-patch and on the other hand by a highly flexible interaction mode of the C-terminal ctSpp2211–254 region with the ctPrp2 RecA2 domain (Fig. 2A and SI Appendix, Fig. S4). The binding at this site relies mainly on hydrophobic interactions, which has the advantage of requiring only a few polar interactions to be broken upon conformational changes (SI Appendix, Fig. S3). This allows this part of the G-patch to adopt different alternative conformations, as seen in the crystal structures (Fig. 2A). In addition, NMR data indicate that in solution, the C-terminal part of the ctSpp2 G-patch exhibits increased flexibility, consistent with the alternative conformations observed in the crystal structures (Fig. 3D).
Although we were able to observe two different interaction modes of this region in the ADP-bound state of Prp2, other conformations might occur in different nucleotide-bound states of Prp2. Conserved glycines at the beginning of the linker and in the C-terminal hydrophobic stretch likely enhance the flexible properties of these two regions and ultimately ensure that the C-terminal end of the G-patch is able to stay in contact with the hydrophobic patch on the RecA2 domain at every nucleotide-specific conformational state of the DEAH-box ATPase (Figs. 2C and 4D). In contrast, the N-terminal amphipathic α-helix of the G-patch shows no variability in binding to the WH domain in all crystal structures (Fig. 2A). In addition, NMR studies indicate that the flexibility of these residues is lower compared with other regions of the G-patch (Fig. 3C). This favors the binding of the ctSpp2 G-patch to ctPrp2 because of the decreased entropic penalty that the G-patch has to pay when it assumes a rigid conformation in the bound state. Thus, crystallographic and NMR data suggest that the N-terminal part of the G-patch acts as the main anchoring point to Prp2.
Although this part of the G-patch transiently samples helical conformations with an average of 20% (ensemble and time average), it does not provide sufficient evidence for a binding based on conformational selection (Fig. 3E). Transient sampling of a folded region does not facilitate a conformational selection mechanism in any case, as is seen for other IDPs, such as NTAIL (52, 53). Assuming this to be the case, Prp2 might be able to select and further stabilize already transiently populated helical conformations of the N-terminal parts, and the C-terminal parts could subsequently bind to Prp2 as well. Instead, a folding on binding mechanism cannot be excluded, as the majority of the Spp2 G-patch is not found in the prefolded state.
Due to the stimulating effect of G-patch proteins on the functions of Prp2 and Prp43, it has also been speculated that the G-patch might directly interact with either the ATP- or RNA-binding sites (22, 24). Our ctPrp2-ctSpp2211–254 complex structures demonstrate that the G-patch does not bind anywhere close to the adenosine nucleotide-binding site and thus is not able to directly alter the catalytic center (Fig. 2A). In contrast, the C-terminal end of the Spp2 G-patch is located in close proximity to the entrance of the RNA-binding tunnel, as is seen in the RNA complex structures of Prp43 and Prp22 (31, 37). However, assuming the same binding of the C-terminal end to the RecA2 domain in the ATP and Apo states, this part of the G-patch is still approximately 12 to 13 Å away from the closest RNA backbone phosphate (SI Appendix, Fig. S16). This distance makes involvement of the G-patch in interactions with the RNA unlikely. Instead of directly interacting and altering binding sites for ATP/ADP and RNA, the G-patch might have an effect on the global dynamics of the DEAH-box ATPase and thereby affect functional properties, such as ligand binding and catalytic efficiency, in an indirect manner. For example, the N-terminal amphipathic α-helix binds to the WH domain, which serves as hinge region for the opening of the RNA-binding tunnel (Fig. 2A) (37). This interaction might have an impact on the binding of the RNA, which in turn could lead to the observed RNA-dependent stimulation of the ATPase activity of Prp2 and Prp43 by its respective G-patch protein (22, 24).
On the other hand, the C-terminal part of the G-patch interacts with the highly mobile RecA2 domain and might affect the mobility of this domain (Fig. 4D) (31). In addition, although the linker region of the G-patch does not interact with the β-hairpin of the RecA2 domain, it folds over this structural feature like a safety belt in all ctPrp2-ctSpp2211–254 complex structures and stabilizes its conformation, which has been found to be rather flexible in the absence of Spp2 (SI Appendix, Fig. S10) (35). Since the mobility of the RecA2 is tightly linked to ATP hydrolysis, an influence of the G-patch on the dynamics of this domain might have a regulatory effect on binding constants of ATP/ADP, as well as on the catalysis rate of ATP.
To gain more in-depth insight into the exact modus operandi of the G-patch proteins, extensive studies using other techniques are needed. Single-molecule experiments, as well as stopped-flow kinetics with Prp2 and Prp43 in the presence and absence of their respective G-patch proteins, could provide more detailed information about the functional implications and complement our structural data.
Materials and Methods
Detailed descriptions of the experimental procedures are provided in SI Appendix, Materials and Methods. All Spp2 and Prp2 constructs were expressed in E. coli Rosetta II (DE3), and the first affinity chromatography purification step was performed according to the tag used (His6-, Strep-, or GST-tag). scSpp2 constructs were then further purified by anion-exchange chromatography. Finally, all Spp2 and Prp2 constructs were subjected to gel filtration chromatography.
The structural properties of the scSpp2 and ctSpp2208–254 were analyzed in solution by CD and NMR spectroscopy. ctSpp2208–254 was also subjected to dynamic light scattering experiments.
Five crystal structures of the ctPrp2-ctSpp2G-patch complex have been solved. The highest resolution limit was assessed using a minimum I/σ(I) value of 1.5 and a minimum CC1/2 value of 60% as cutting criteria. X-ray diffraction data statistics are summarized in SI Appendix, Table S1. The structures was solved via molecular replacement using the ctPrp2-ADP structure (Protein Data Bank ID code 6FA5) as a template. All structures were refined to reasonable R factors (SI Appendix, Table S1).
The biological relevance of the complex crystal structures was verified via in-solution chemical cross-linking. Binding properties of the ctSpp2 G-patch motif were examined via ITC and GST pulldown experiments using either different truncations or glycine mutants of the G-patch motif, respectively.
Data Availability.
The coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID codes 6RM8 [CF1], 6RM9 [CF2], 6RMA [CF3], 6RMB [CF4], and 6RMC [CF5]). Raw and annotated MS data from the search using Mass Matrix software (54) are available via the ProteomeXchange at PRIDE database (PXD015793). Chemical shift data have been submitted to the Biological Magnetic Resonance Bank (BMRB) (accession no. 50014).
Supplementary Material
Acknowledgments
We thank DESY Hamburg, a member of the Helmholtz Association of German Research Centers, for providing the experimental facilities. Parts of this research were carried out at PETRA III, and we are thankful for the assistance with the use of European Molecular Biology Laboratory beamlines P13 and P14. We thank Dr. Stefan Becker for help with the production of ctSpp2208-254. This work was supported by Deutsche Forschungsgemeinschaft Grants SFB860 TP A02, A10, B02, and Z01 (to R.F., H.U., and M.Z.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.S. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6RM8 [CF1], 6RM9 [CF2], 6RMA [CF3], 6RMB [CF4], and 6RMC [CF5]).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907960117/-/DCSupplemental.
References
- 1.Dyson H. J., Wright P. E., Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Stein A., Pache R. A., Bernadó P., Pons M., Aloy P., Dynamic interactions of proteins in complex networks: A more structured view. FEBS J. 276, 5390–5405 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Tompa P., Structure and Function of Intrinsically Disordered Proteins (CRC Press, 2009). [Google Scholar]
- 4.Korneta I., Bujnicki J. M., Intrinsic disorder in the human spliceosomal proteome. PLoS Comput. Biol. 8, e1002641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoskins A. A., Moore M. J., The spliceosome: A flexible, reversible macromolecular machine. Trends Biochem. Sci. 37, 179–188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matera A. G., Wang Z., A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15, 108–121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Will C. L., Lührmann R., Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, a003707 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wahl M. C., Will C. L., Lührmann R., The spliceosome: Design principles of a dynamic RNP machine. Cell 136, 701–718 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Fabrizio P., et al. , The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell 36, 593–608 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Cordin O., Hahn D., Beggs J. D., Structure, function and regulation of spliceosomal RNA helicases. Curr. Opin. Cell Biol. 24, 431–438 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Ding S. C., Pyle A. M., Molecular mechanics of RNA translocases. Methods Enzymol. 511, 131–147 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozgur S., et al. , The conformational plasticity of eukaryotic RNA-dependent ATPases. FEBS J. 282, 850–863 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Fairman-Williams M. E., Guenther U.-P., Jankowsky E., SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 20, 313–324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cordin O., Beggs J. D., RNA helicases in splicing. RNA Biol. 10, 83–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kudlinzki D., Schmitt A., Christian H., Ficner R., Structural analysis of the C-terminal domain of the spliceosomal helicase Prp22. Biol. Chem. 393, 1131–1140 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Kim S. H., Lin R. J., Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 16, 6810–6819 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King D. S., Beggs J. D., Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res. 18, 6559–6564 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy J., Kim K., Maddock J. R., Anthony J. G., Woolford J. L. Jr, The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1, 375–390 (1995). [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman E. J., et al. , Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol. Cell. Biol. 24, 10101–10110 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aravind L., Koonin E. V., G-patch: A new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 24, 342–344 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Robert-Paganin J., Réty S., Leulliot N., Regulation of DEAH/RHA helicases by G-patch proteins. BioMed Res. Int. 2015, 931857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christian H., Hofele R. V., Urlaub H., Ficner R., Insights into the activation of the helicase Prp43 by biochemical studies and structural mass spectrometry. Nucleic Acids Res. 42, 1162–1179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka N., Aronova A., Schwer B., Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 21, 2312–2325 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warkocki Z., et al. , The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev. 29, 94–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bao P., Höbartner C., Hartmuth K., Lührmann R., Yeast Prp2 liberates the 5′ splice site and the branch site adenosine for catalysis of pre-mRNA splicing. RNA 23, 1770–1779 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S. H., Smith J., Claude A., Lin R. J., The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 11, 2319–2326 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rauhut R., et al. , Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353, 1399–1405 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Yan C., Wan R., Bai R., Huang G., Shi Y., Structure of a yeast activated spliceosome at 3.5 Å resolution. Science 353, 904–911 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Semlow D. R., Blanco M. R., Walter N. G., Staley J. P., Spliceosomal DEAH-box ATPases remodel pre-mRNA to activate alternative splice sites. Cell 164, 985–998 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boneberg F. M., et al. , Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis. RNA 25, 685–701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamann F., Enders M., Ficner R., Structural basis for RNA translocation by DEAH-box ATPases. Nucleic Acids Res. 47, 4349–4362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozlowski L. P., Bujnicki J. M., MetaDisorder: A meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinformatics 13, 111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Absmeier E., et al. , A noncanonical PWI domain in the N-terminal helicase-associated region of the spliceosomal Brr2 protein. Acta Crystallogr. D Biol. Crystallogr. 71, 762–771 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Absmeier E., Becke C., Wollenhaupt J., Santos K. F., Wahl M. C., Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2. Cell Cycle 16, 100–112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitt A., Hamann F., Neumann P., Ficner R., Crystal structure of the spliceosomal DEAH-box ATPase Prp2. Acta Crystallogr. D Struct. Biol. 74, 643–654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tauchert M. J., Fourmann J.-B., Christian H., Lührmann R., Ficner R., Structural and functional analysis of the RNA helicase Prp43 from the thermophilic eukaryote Chaetomium thermophilum. Acta Crystallogr. F Struct. Biol. Commun. 72, 112–120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tauchert M. J., Fourmann J.-B., Lührmann R., Ficner R., Structural insights into the mechanism of the DEAH-box RNA helicase Prp43. eLife 6, e21510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulrich A., Wahl M. C., Structure and evolution of the spliceosomal peptidyl-prolyl cis-trans isomerase Cwc27. Acta Crystallogr. D Biol. Crystallogr. 70, 3110–3123 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Neidigh J. W., Fesinmeyer R. M., Andersen N. H., Designing a 20-residue protein. Nat. Struct. Biol. 9, 425–430 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Naduthambi D., Zondlo N. J., Stereoelectronic tuning of the structure and stability of the trp cage miniprotein. J. Am. Chem. Soc. 128, 12430–12431 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Zondlo N. J., Aromatic-proline interactions: Electronically tunable CH/π interactions. Acc. Chem. Res. 46, 1039–1049 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wishart D. S., Sykes B. D., The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4, 171–180 (1994). [DOI] [PubMed] [Google Scholar]
- 43.Marsh J. A., Singh V. K., Jia Z., Forman-Kay J. D., Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Protein Sci. 15, 2795–2804 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu H.-Y., Shen X.-H., Ni R.-S., Qian X.-Y., Gao X., [Expression of E-cadherin and uPA and their prognostic value in carcinoma of human larynx.] Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 44, 1024–1028 (2009), Chinese. [PubMed]
- 45.Tsai R.-T., et al. , Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol. Cell. Biol. 27, 8027–8037 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fourmann J.-B., et al. , The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction. eLife 5, e15564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walbott H., et al. , Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 29, 2194–2204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haselbach D., et al. , Structure and conformational dynamics of the human spliceosomal Bact complex. Cell 172, 454–464.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Zhang X., et al. , Structure of the human activated spliceosome in three conformational states. Cell Res. 28, 307–322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Nues R. W., Beggs J. D., Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157, 1451–1467 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H.-L., Cheng S.-C., The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol. Cell. Biol. 32, 5056–5066 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Longhi S., Structural disorder within paramyxoviral nucleoproteins. FEBS Lett. 589, 2649–2659 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Dosnon M., et al. , Demonstration of a folding after binding mechanism in the recognition between the measles virus NTAIL and X domains. ACS Chem. Biol. 10, 795–802 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Xu H., Freitas M. A., MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics 9, 1548–1555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID codes 6RM8 [CF1], 6RM9 [CF2], 6RMA [CF3], 6RMB [CF4], and 6RMC [CF5]). Raw and annotated MS data from the search using Mass Matrix software (54) are available via the ProteomeXchange at PRIDE database (PXD015793). Chemical shift data have been submitted to the Biological Magnetic Resonance Bank (BMRB) (accession no. 50014).