Significance
Plants use phytochrome photoreceptors to monitor for the presence of competitors for photosynthetically active sunlight. They do so by sensing near-proximity or direct shade (“shade signals”) from neighboring vegetation. This study defines the functional role, and molecular mechanism of action, of Pseudo-Response Regulator proteins (PRRs) (integral components of the plant central circadian oscillator) in regulating growth responses to such signals. These PRRs are shown to bind, intranuclearly, directly to members of the Phytochrome Interacting Factor (PIF) family of transcription factors, repressing their capacity to transcriptionally activate a subset of their Direct Target Genes. These findings show that the plant clock communicates with the genome using direct, physical interaction of multiple central oscillator components with the transcriptional-regulatory machinery of the cell.
Keywords: PIF proteins, PRR proteins, shade avoidance, gene expression regulation, phytochrome
Abstract
Light-environment signals, sensed by plant phytochrome photoreceptors, are transduced to target genes through direct regulation of PHYTOCHROME-INTERACTING FACTOR (PIF) transcription factor abundance and activity. Previous genome-wide DNA-binding and expression analysis has identified a set of genes that are direct targets of PIF transcriptional regulation. However, quantitative analysis of promoter occupancy versus expression level has suggested that unknown “trans factors” modulate the intrinsic transcriptional activation activity of DNA-bound PIF proteins. Here, using computational analysis of published data, we have identified PSEUDO-RESPONSE REGULATORS (PRR5 and PRR7) as displaying a high frequency of colocalization with the PIF proteins at their binding sites in the promoters of PIF Direct Target Genes (DTGs). We show that the PRRs function to suppress PIF-stimulated growth in the light and vegetative shade and that they repress the rapid PIF-induced expression of PIF-DTGs triggered by exposure to shade. The repressive action of the PRRs on both growth and DTG expression requires the PIFs, indicating direct action on PIF activity, rather than a parallel antagonistic pathway. Protein interaction assays indicate that the PRRs exert their repressive activity by binding directly to the PIF proteins in the nucleus. These findings support the conclusion that the PRRs function as direct outputs from the core circadian oscillator to regulate the expression of PIF-DTGs through modulation of PIF transcriptional activation activity, thus expanding the roles of the multifunctional PIF-signaling hub.
Plants utilize a set of sensory photoreceptors to monitor their environment for the presence, absence, directionality, periodicity, intensity, and color of the impinging light and respond by adapting their growth and development to optimize solar energy capture for photosynthesis (1, 2). These receptors decode the information in the incoming light signals and direct these adaptational responses in the recipient plant to match the prevailing environmental conditions. Among these receptors the phytochrome (phy) family (phyA, B, C, D, and E in Arabidopsis) track the red (R) and far-red (FR) wavelengths of the spectrum by switching reversibly between the biologically inactive, R-absorbing, Pr conformer and the biologically active, FR-absorbing, Pfr conformer. Research over the last two decades, focused on defining the molecular mechanism by which the photoactivated receptor transduces its signals to the cellular response network, has identified a short pathway directly from the phy molecule to the transcriptional-regulatory machinery of target genes (3). This pathway involves rapid (1 to 2 min) translocation of the activated Pfr form into the nucleus, where it binds to PHYTOCHROME-INTERACTING transcription FACTOR (PIF) proteins, inducing their phosphorylation, ubiquitination, and degradation (4–8).
In dark-germinated seedlings, phy activation (Pfr formation) occurs for the first time upon initial exposure to light, triggering a rapid decline of the existing high PIF levels down to a new, lower steady state in the light, set by the balance between the continuing rates of synthesis and degradation. This balance is dynamic in normal light-grown plants, resulting in temporally fluctuating PIF abundance in proportion to the level of Pfr in the cell. Three major environmental parameters control this dynamic balance by modulating Pfr levels (3, 9): 1) the nightly darkness of the normal diurnal cycle (10), 2) vegetative shade from neighboring foliage (1, 2, 11), and 3) elevated temperatures (12, 13). Each of these factors decreases Pfr levels, concomitantly allowing increases in PIF levels 1) during the night, 2) in the shade, and 3) at higher ambient temperatures, respectively. The PIF transcription factors in turn regulate expression of target genes, which then dictate growth and developmental responses at the seedling level through a downstream transcriptional network (3, 9).
Parallel chromatin immunoprecipitation sequencing (ChIP-seq) and RNA sequencing (RNA-seq) analysis has identified 338 genes that are direct targets of transcriptional regulation by one or more of the “PIF quartet” (PIF1, PIF3, PIF4, and PIF5) in dark-grown seedlings (14, 15). A subpopulation of these Direct Target Genes (DTGs) display rapid responses in expression upon initial exposure to light (in 1 h or less), consistent with responsiveness to the rapid decline in PIF abundance (16–18). A further subset of the PIF-induced (light-repressed) DTGs also shows rapid reversal of light repression upon exposure to vegetative shade (shade induction) in parallel with increases in PIF levels (4, 17, 19, 20). Similarly, some of these DTGs display a progressive increase in expression during the night under diurnal conditions in parallel with increasing PIF levels, as well as progressively increasing elongation growth rates, up to a maximum just before dawn (3, 10, 21, 22). A notable exception to this pattern is PIF7. Photoactivated phyB binding to PIF7 upon the initial dark-to-light transition does not induce rapid degradation (23). A phosphorylated form of PIF7 does, however, accumulate upon constant light exposure. This form is rapidly dephosphorylated upon exposure to vegetative shade, and this induces nuclear translocation and increased binding of PIF7 to target genes, with increased expression (24, 25).
The above parallel ChIP-seq and RNA-seq analysis revealed varying levels of participation of the individual PIFs in the binding to, and regulation of expression of, the individual DTGs, from individually PIF-specific to robustly shared by all four PIFs (14, 15). Unexpectedly, however, no quantitative correlation was found between the level of promoter occupancy by an individual PIF (determined by ChIP-seq) and the level of transcriptional activation of that DTG (determined by RNA-seq) (15). These findings suggest the existence of some form of “postoccupancy modulation” of the intrinsic transcriptional activation activity of the DNA-bound PIF proteins. One possible mechanistic basis for this phenomenon could be modulation by other proteins, such as coactivators or corepressors (26–28).
We have explored this possibility of modulation by other proteins here. Based on the rationale that colocalization with the PIFs on the promoters of DTGs could reflect modulation of PIF transcriptional activity, we have used a computational approach to identify factors [from an array of published genome-wide ChIP studies (29)] that associate with these promoters in the vicinity of the PIFs. Of the several factors that did show enriched localization near the PIF-binding sites, we examined the functional relevance to PIF-regulated expression of the two strongest candidates, Pseudo-Response Regulators 5 and 7 (PRR5 and PRR7), using a combination of mutant analysis, photobiological analysis, and protein interaction assays.
Results
The PRRs Colocalize with the PIF-Quartet Proteins at the PIF-Binding Sites on the Promoters of PIF-DTGs.
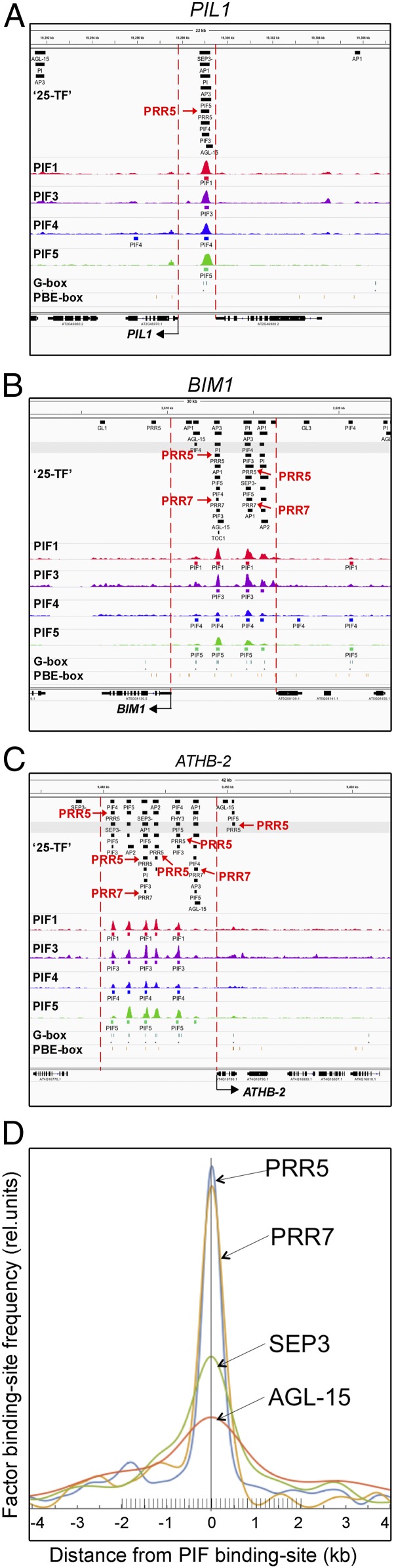
To search for candidates that might function as modulators of PIF transcriptional activity, we compared the distribution of published, ChIP-defined, genome-binding sites for 25 transcription factors (“25 TFs”) (29) with that of the PIF proteins in our 338 PIF-DTGs (14, 15). These 25 TFs represent a diversity of TFs that function in a variety of growth and developmental processes. Our analysis revealed a striking coincidence between the binding sites of a subset of the 25 TFs and those of the PIF quartet (PIF1, PIF3, PIF4, and PIF5) (Fig. 1 A–C and SI Appendix, Fig. S1). Quantitative analysis of the distance of the binding sites for each TF from those of the PIFs in the PIF-DTG promoters showed a range of levels of coincidence, with the strongest being that displayed by PRR5 and PRR7 (Fig. 1D). The robustness of the co-occupancy frequency across the PIF-DTG promoters and the tight proximity of clustering around the PIF-binding sites displayed by PRR5 and PRR7 suggest these factors as potential “transinfluences” (26, 28) capable of modulating PIF transcriptional activity.
Fig. 1.
PRR proteins colocalize with PIF transcription factors on the promoters of PIF- DTGs. (A–C) Browser images of ChIP-seq–determined binding distributions of the PIFs (colored peaks) (15) and the 25-TF transcription factors (upper short, black horizontal bars; illegible factor names are listed for each gene in SI Appendix, Table S1) (29) across the promoter regions (between vertical red dashed lines) of selected PIF-DTGs (gene structures depicted below as black boxes linked by lines). Transcription direction is indicated by arrows. Locations of G-box (CACGTG) and PIF-binding E-box (PBE-box, CACATG/CATGTG) sequence motifs are shown by short, vertical black lines below the ChIP-seq images. PRR5- and PRR7-binding sites are highlighted in red. (D) Proximity of 25-TF–binding sites to PIF-binding sites. Curves depict the frequency of the binding sites for individual transcription factors in the 25-TF set as a function of distance from the PIF-binding sites in DTGs. The four TFs most robustly colocalized with the PIFs (PRR5, PRR7, SEP3, and AGL-15) are shown.
The PRRs and PIFs Conversely Modulate Phenotypic Responses to Light.
It is well-established that the PIF quartet (30) and PIF7 (23) function to promote skotomorphogenesis in dark-grown seedlings. By contrast, examination of the phenotypes of multiple, dark-grown prr mutant combinations [including prr5prr7prr9 triple mutants (prr579)] and PRR-overexpression (PRR-OX) lines has revealed no detectable role for these three PRRs in skotomorphogenic development (31–33) (SI Appendix, Fig. S2).
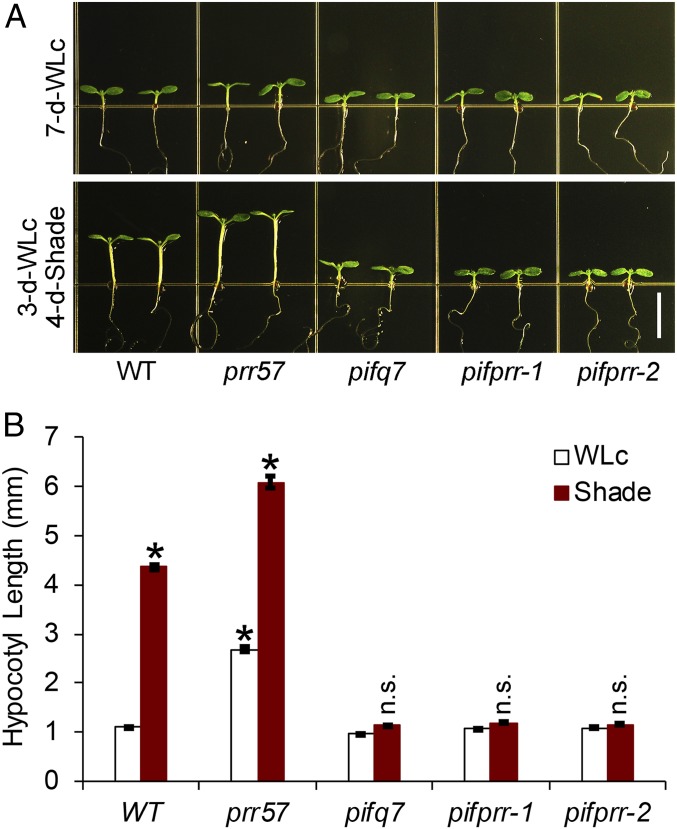
On the other hand, the prr mutants displayed longer hypocotyls than wild-type (WT) when grown in continuous red light (Rc) (31–35) (SI Appendix, Fig. S2), as well as in continuous white light (WLc) (Fig. 2). Conversely, the PRR-OX lines showed marked hypersensitivity to Rc (36) (SI Appendix, Fig. S2), as well as to WLc, although to a lesser extent for the latter (Fig. 2). These phenotypes are the converse of those found for the PIFs, where pif mutants display hypersensitivity to Rc (23, 30, 37) (SI Appendix, Fig. S2) and overexpressors display hyposensitivity (6, 37–39).
Fig. 2.
The PIFs and PRRs conversely modulate seedling responsiveness to vegetative shade. (A) Phenotypes of WT, pif1pif3pif4pif5 (pifq) quadruple mutant, pif7 single mutant, pifqpif7 (pifq7) quintuple mutant, prr57 double mutant, prr579 triple mutant, and PRR5ox and PRR7ox overexpressor seedlings grown at 21 °C either in WLc for 7 d (7-d-WLc) (Upper) or in WLc for 3 d followed by FR-supplemented WLc (shade; R:FR ratio of 0.3) for 4 d (3-d-WLc 4-d-shade) (Lower). (Scale bar, 5 mm.) (B) Quantification of hypocotyl lengths of the genotypes shown in A. Data represent the mean and SE from at least 30 seedlings. (Left) Asterisks indicate that the hypocotyl lengths of shade-treated seedlings are statistically significantly different from the corresponding WLc controls by Student’s t test (P < 1e-10). n.s. indicates “not significantly different” (P > 0.1). (Right) Asterisks indicate that the hypocotyl lengths of shade-treated seedlings are statistically significantly different from the shade-treated WT control by Student’s t test (P < 1e-10).
The PRRs and PIFs Conversely Modulate Phenotypic Responses to Vegetative Shade.
Previously, the PIF quartet (40) and PIF7 (24), respectively, have been shown separately to contribute partially to the shade-induced elongation of hypocotyls (Fig. 2). We have constructed and examined a pif1pif3pif4pif5pif7 quintuple mutant (designated pifqpif7) and found that it is conspicuously unresponsive to shade (Fig. 2 A and B, Left). This finding establishes that the five PIF-quintet members (PIFs1, -3, -4, -5, and -7) are collectively fully responsible for promoting hypocotyl cell elongation in response to shade.
In striking contrast to the PIFs, we found that the prr mutants display enhanced responsiveness to shade compared to WT (Fig. 2 A and B, Right). Conversely, the PRR5ox and PRR7ox overexpressors are significantly less responsive to shade than the WT (Fig. 2 A and B, Right). These phenotypes indicate that the PRRs suppress hypocotyl elongation both in light and in response to vegetative shade.
PIF-Quintet Members Function Differentially in Stimulating Shade-Induced Expression of PIF-DTGs.
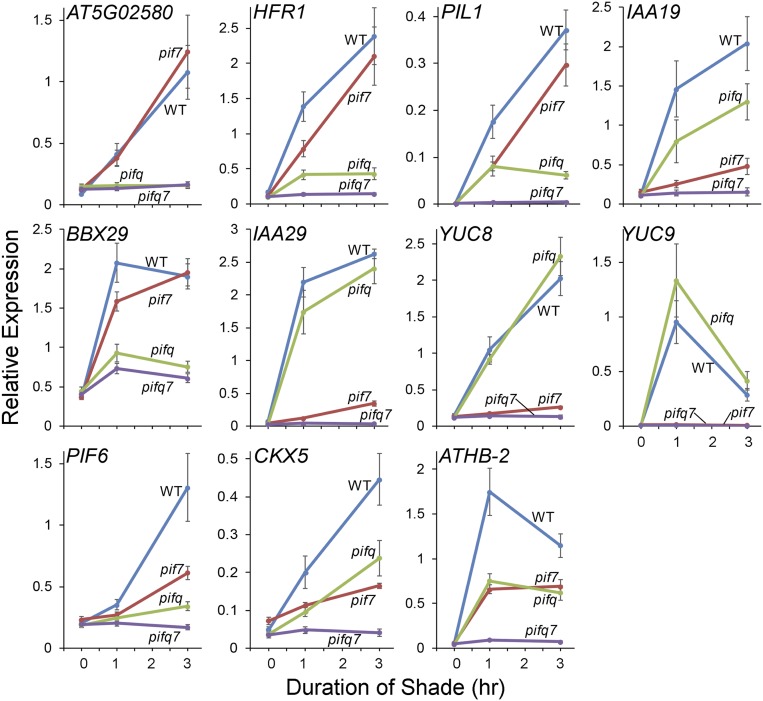
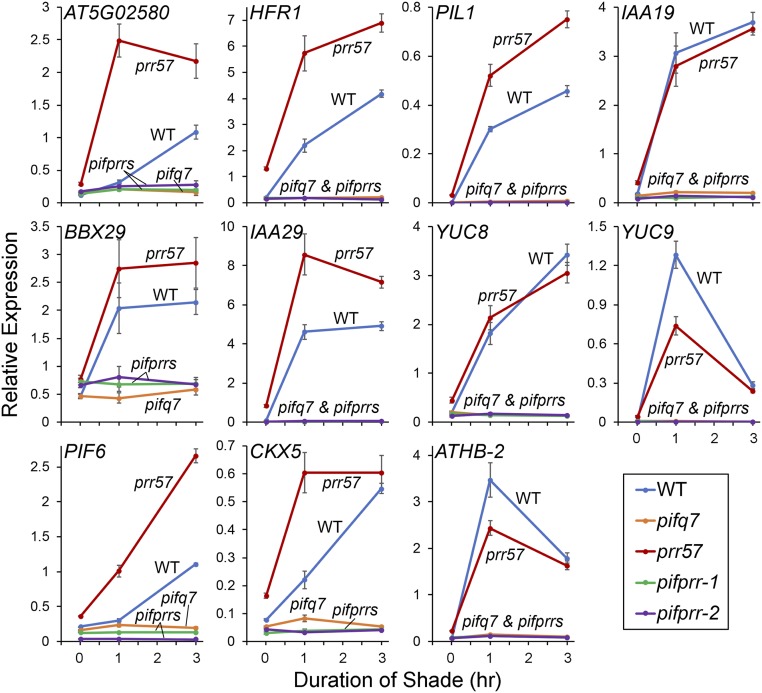
Given the lack of shade responsiveness of pifqpif7 mutant seedlings (Fig. 2 A and B, Left), we examined the shade responsiveness of a set of marker genes (previously established as PIF-DTGs) that are rapidly induced by shade in the WT (19, 24, 40). These genes display remarkable divergence in dependency on the PIF quartet, on the one hand, and on PIF7, on the other hand, for their shade responsiveness (Fig. 3). Notable extreme examples of this converse pattern are the gene AT5G02580, which shows complete dependence on the PIF quartet and no detectable dependence on PIF7, and IAA29, YUC8, and YUC9, which show no apparent dependence on the PIF quartet, but almost complete dependence on PIF7. The remaining genes that were tested displayed varying degrees of intermediate patterns between these two extremes. These findings indicate a spectrum of combinatorial activities of the five quintet members toward these DTGs. One major pattern that emerges is the dominant role played by PIF7 in regulating auxin-signaling–related genes in response to shade (especially IAA29, YUC8, and YUC9), consistent with a previous report (24).
Fig. 3.
The PIF quintet acts collectively but differentially to induce DTG expression in response to vegetative shade. Time-course analysis of expression of the indicated PIF-DTGs in WT, pifq-quadruple, pif7 monogenic, and pifqpif7 quintuple mutant seedlings grown at 21 °C for 5 d in WLc (time 0) followed by 1 to 3 h FR-light–supplemented WLc (shade; R:FR ratio of 0.3). Transcript levels were determined using RT-qPCR at the times indicated. Error bars represent the SEM of three biological replicates.
Regardless of these findings, however, all of the genes tested are, without exception, essentially completely unresponsive to the shade signal in the absence of all five PIFs in the quintuple pifqpif7 mutant (Fig. 3). Together with the phenotypic data (Fig. 2), these findings indicate that the five PIF-quintet members are collectively fully responsible for promoting hypocotyl cell elongation in response to shade and that they do so via regulation of a network of DTGs.
PRRs Enhance Light-Imposed Repression and Repress Shade-Elicited Stimulation of PIF-Induced Gene Expression.
Consistent with the visible phenotypes, we detected no consistent, significant differences between the dark-grown prr mutant, PRR7 overexpressor, and WT seedlings in the expression levels of the set of marker PIF-DTGs (SI Appendix, Fig. S3). Nor did we observe any consistent differences in the rates of light-induced changes in transcript levels across these marker PIF-DTGs between these genotypes upon initial light exposure (SI Appendix, Fig. S3).
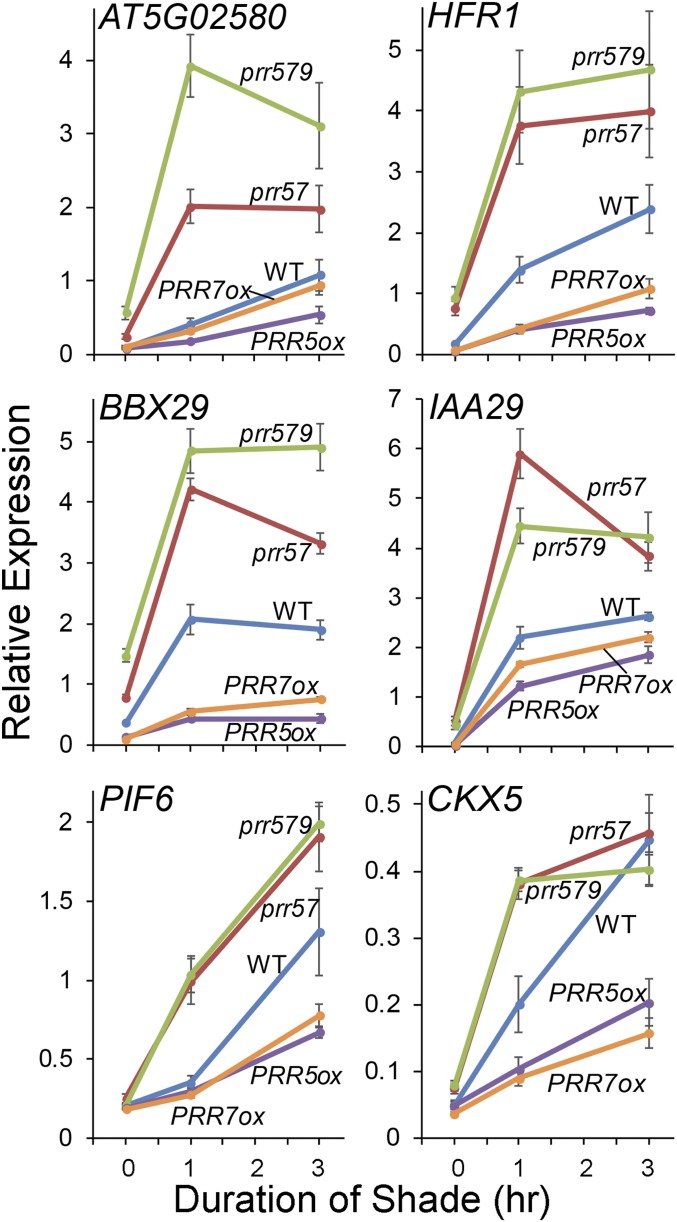
By contrast, the rapid increase in expression of the marker genes induced by shade is enhanced in the prr57 double and prr579 triple mutants relative to the WT, and reduced in the PRR overexpressors, in a subset of these DTGs (Fig. 4 and SI Appendix, Fig. S4). Also notable, although quantitatively less evident at the scale shown in Fig. 4, there is a tendency for the expression levels of these genes to be somewhat higher in the prr mutants and lower in the overexpressors than the WT in long-term white light. Together, these data indicate that the PRRs function to repress expression of PIF-induced DTGs, both under long-term exposure to light and in response to shade in those light-grown plants.
Fig. 4.
The PRRs repress shade-induced expression of PIF-DTGs. Time-course analysis of expression of the indicated PIF-DTGs in WT, prr57 double-mutant, prr579 triple-mutant, and PRR5ox and PRR7ox overexpressor seedlings grown at 21 °C for 5 d in WLc (time 0) followed by 1 to 3 h FR-light–supplemented white light (shade; R:FR ratio of 0.3). Transcript levels were determined using RT-qPCR at the times indicated. Error bars represent the SEM of three biological replicates.
The PRRs Require PIFs in Order to Display Repression of Seedling Shade Responsiveness.
To determine whether the enhanced shade responsiveness of the prr mutants requires the presence of PIFs, we examined the shade responsiveness of a pifqpif7prr57 septuple mutant (designated pifprr-1 and pifprr-2 in two independent lines) compared to the pifqpif7 quintuple mutant, prr57 double mutant, and WT seedlings. The data show that the shade-induced elongation of pifqpif7prr57 seedlings is dramatically reduced relative to the prr57 seedlings, down to the level of the pifqpif7 mutant (Fig. 5). These data establish that the reversal of PRR-imposed repression (derepression) of hypocotyl shade responsiveness in the prr mutants does indeed require the presence of the PIFs.
Fig. 5.
The PRRs require PIFs to modulate seedling responsiveness to vegetative shade. (A) Phenotypes of WT, prr57 double-mutant, pifq7 quintuple-mutant, pifq7prr57-1 (pifprr-1), and pifq7prr57-2 (pifprr-2) septuple-mutant seedlings grown at 21 °C either in WLc for 7 d (7-d-WLc) (Upper) or in WLc for 3 d followed by FR-light–supplemented WLc (shade; R:FR ratio of 0.3) for 4 d (3-d-WLc 4-d-shade) (Lower). (Scale bar, 5 mm.) (B) Quantification of hypocotyl lengths of the genotypes shown in A. Data represent the mean and SE from at least 30 seedlings. Asterisks indicate that the hypocotyl lengths of WT shade, prr57 WLc, and prr57 shade seedlings are statistically significantly different from the WT WLc control by Student’s t test (P < 1e-10). n.s. indicates that the hypocotyl lengths of pifq7 shade, pifprr-1 shade, and pifprr-2 shade seedlings are not significantly different from their respective WLc controls (P > 1e-10).
The PRRs Require PIFs in Order to Display Repression of Shade-Induced Expression of DTGs.
To determine whether the derepression of PRR-imposed suppression of the shade responsiveness of PIF-DTGs, which occurs in the prr mutants (Fig. 4), also requires the presence of the PIFs, we examined the expression of our marker gene set in the pifqpif7prr57 (abbreviated pifprrs), pifqpif7 (pifq7), and prr57 mutant combinations compared to WT. The data show no discernible difference in expression of these genes between the pifqpif7prr57 (pifprrs) septuple and pifqpif7 quintuple mutants (Fig. 6), indicating that the complete loss of detectable shade-induced expression of PIF-DTGs that occurs in the pifqpif7 mutant is unaffected by the additional loss of PRR5 and PRR7 in the pifqpif7prr57 mutant. These data establish that the reversal of PRR-imposed suppression of shade-responsive expression of DTGs in the prr mutants requires the presence of the PIFs. Together with the phenotypic data, these results imply, that, in the WT, the PRRs function normally to suppress shade-induced DTG expression and consequent hypocotyl cell elongation by repressing the promotive activity of the PIFs, rather than repressing another separate independent pathway.
Fig. 6.
The PRRs require PIFs to exert control of shade-induced expression of PIF-DTGs. Time-course analysis of expression of the indicated PIF-DTGs in WT, prr57 double-mutant, pifq7 quintuple-mutant, pifq7prr57-1 (pifprr-1), and pifq7prr57-2 (pifprr-2) septuple-mutant (collectively, pifprrs) seedlings grown at 21 °C for 5 d in WLc (time 0) followed by 1 to 3 h FR-light–supplemented white light (shade; R:FR ratio of 0.3). Transcript levels were determined using RT-qPCR at the times indicated. Error bars represent the SEM of three biological replicates.
The PRRs Interact Directly with the PIFs.
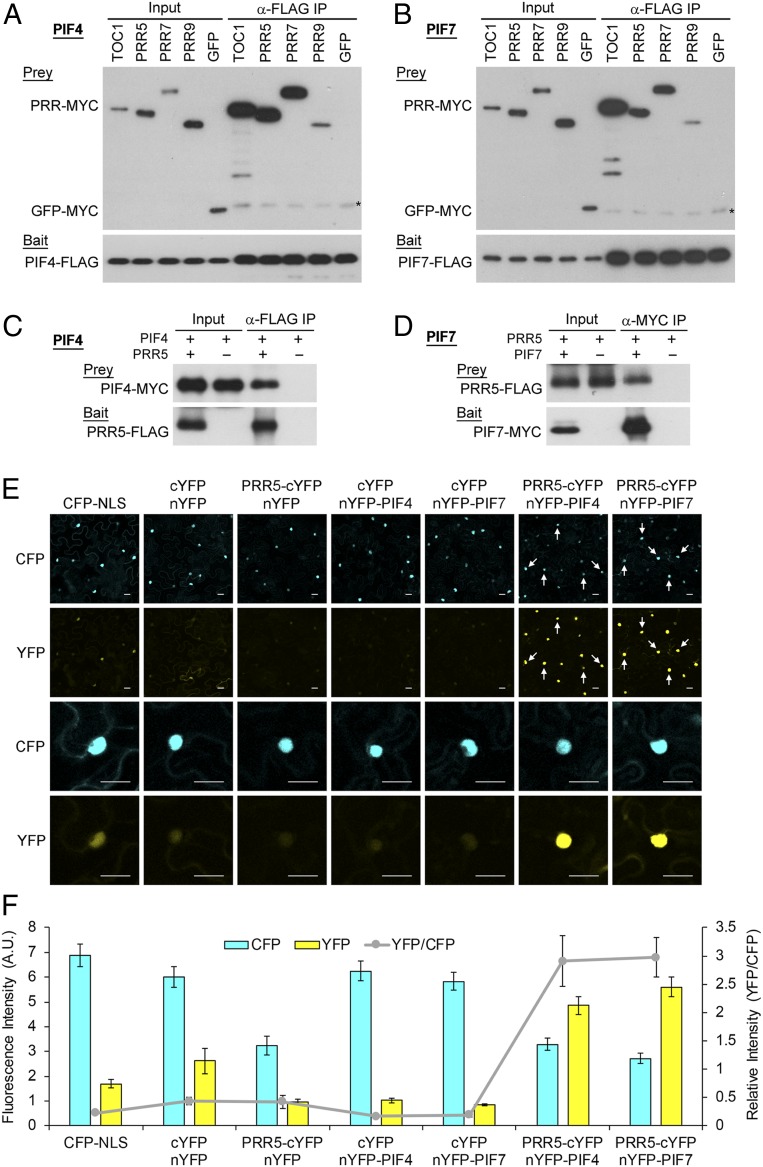
The mechanism of this repression in principle could be direct or indirect. However, given the colocalization of the PRRs with the PIFs on the promoters of PIF-DTGs (Fig. 1), and the established direct intranuclear binding of TOC1 and the PRRs to PIF3 in planta (21, 22), as well as the binding of TOC1 to PIF4 in transfected protoplasts (41) and to other PIFs in yeast two-hybrid assays (42), we investigated the PRRs for potential direct binding to shade-active PIF7, as well as PIFs 3 and 4, using coimmunoprecipitation (co-IP) and Bimolecular Fluorescence Complementation (BiFC) assays.
The co-IP interaction assays using in-vitro–synthesized proteins show that all PRRs tested (TOC1 and PRRs 5, 7, and 9) bind to all PIFs tested (PIFs 3, 4, and 7) under these conditions (Fig. 7 A and B and SI Appendix, Fig. S5). Interestingly, the data suggest that TOC1 has the highest affinity (at least for PIFs 3 and 7), whereas PRR9 has the weakest affinity. Quasi–in-vivo co-IP assays using a transiently expressed subset of these proteins in Nicotiana benthamiana leaf cells indicate that PIF4 and PIF7 (those tested) can likely interact with PRR5 in the living plant cell (Fig. 7 C and D), consistent with the previous results showing TOC1 interaction with PIF3 and PIF4 (22, 43). Moreover, BiFC assays show that these interactions between PRR5 and PIF4 or PIF7 do indeed occur in the living cell and are focused in the nucleus, as indicated by colocalization with the nuclear-marker internal controls (cyan fluorescent protein fused to a nuclear-localization-signal-encoded peptide (CFP-NLS)) (Fig. 7 E and F), and as also shown for PIF3 (21).
Fig. 7.
The PRRs directly interact with PIFs in the nucleus. (A and B) In vitro co-IP assays: Interactions of PIF4 (A) or PIF7 (B) with TOC1 and PRRs following synthesis in vitro. FLAG-tagged PIF4 (A) or PIF7 (B) served as bait. Coexpressed, MYC-tagged TOC1 and PRRs served as prey. GFP-MYC served as the negative control. Asterisks indicate nonspecific cross-reacting bands. (C and D) Quasi–in-vivo co-IP assays: Interactions of PIF4 (C) or PIF7 (D) with PRR5 following expression in Nicotiana leaf cells. PRR5-FLAG (C) or PIF7-MYC (D) served as bait, while coexpressed PIF4-MYC (C) or PRR5-FLAG (D) served as prey. (E and F) BiFC assays: Direct interactions of PIF4 or PIF7 with PRR5 in the nucleus of living cells. (E) Top two rows: Low-magnification confocal microscopic images showing fields of leaf cells, expressing pairwise combinations of the constructs indicated at the top. The N-terminal fragment of YFP (nYFP) was fused to PIF4 (nYFP-PIF4) or PIF7 (nYFP-PIF7) and the C-terminal fragment (cYFP) was fused to PRR5 (PRR5-cYFP). Full-length CFP was fused to a nuclear localization signal motif (CFP-NLS) which served as a coexpressed, internal nuclear-marker control. White arrows highlight examples of PRR5-PIF4 and PRR5-PIF7 interactions, and CFP-NLS localization in the same nucleus. Lower two rows: High-magnification images showing the same single nucleus for CFP and YFP from the top rows. CFP, CFP fluorescence. YFP, YFP fluorescence. (Scale bars, 20 µm.) (F) Quantification of fluorescence intensity of CFP and YFP across fields of multiple coexpressing nuclei and the relative intensities, indicated as the ratio of YFP/CFP. Data represent the mean and SE from at least 30 nuclei from multiple fields.
Collectively, these data indicate that each of the PRR factors, as well as TOC1, can likely directly interact with all PIF members in the nucleus. The intranuclear interactions of PRR5 with PIF4 and PIF7, the two major regulators of the shade avoidance response, imply that the negative regulation of PIF promotive activity by the PRR factors is exerted through these interactions, resulting in attenuated expression of the PIF-DTGs. Preliminary domain analysis of PRR5 using in vitro co-IP assays with PIF4 indicates, intriguingly, that the N-terminal Pseudo-Receiver–containing domain is dominantly responsible for interaction with the PIF molecule (SI Appendix, Fig. S6).
Discussion
It is well recognized that, in general, the effectiveness with which a transcription factor can activate its target genes can be regulated at many levels, which include (but are not limited to) regulation of factor abundance, chromatin-regulated accessibility of DNA-binding sites, DNA-binding-motif sequence variation, and coregulator (coactivator or corepressor) proteins that can cooperatively enhance or repress the intrinsic activity of the transcription factor (26–28). However, what determines the genome-wide binding pattern or regulation of a given transcription factor is frequently unresolved. Here, we have provided evidence that the PRR proteins of Arabidopsis function to directly attenuate the intrinsic transcriptional-activation capacity of members of the PIF subfamily of bHLH factors through physical interaction with these factors in both light-exposed and vegetatively shaded plants.
Because the PRRs are integral components of the central circadian oscillator of plant cells (41, 44–47), their repression of the transcription of PIF-target genes reveals a direct and immediate functional link between the clock and the genome. It is now well-established that the PIF family functions as a transcriptionally centered signaling hub at the convergence of multiple pathways (3, 4, 9). These include the phy, CRY, and UVR8 photosensory/thermosensory pathways; the hormones gibberellin, ethylene, and brassinosteroids; and sucrose (3, 4, 9, 11, 43, 48, 49). Together with previous reports of TOC1 and PRR regulation of PIF-mediated, night-induced, end-of-day FR-induced, and thermo-induced gene expression and hypocotyl elongation (21, 22, 43), our present data simultaneously support the addition of circadian clock output signaling to these converging pathways, show PRR regulation of shade responsiveness, and suggest the mechanism by which these components directly regulate the transcriptional machinery of PIF-DTGs.
Because these PRR family members (including TOC1, also known as PRR1) are expressed in sequential, overlapping waves across the 24-h diurnal cycle from morning until midnight (21, 22, 34, 50, 51), it can be deduced that they sustain a continual envelope of transcriptional repression of PIF-induced DTGs, during both the daylight hours and postdusk darkness. Collectively, this endows the PRR family with the dual functions 1) of dampening excessive responsiveness, both to frequently alternating sun-shade exposure and to elevated temperatures during daylight, and, as shown by Soy et al. (22), 2) of timing the release from constrained growth toward dawn during the prolonged dark periods of short-day photocycles. Although night is not the same as shade, as Casal (2) notes, it is notable that the phy-PIF module uses the same core molecular mechanism to transduce the sensing of these two distinct environmental states directly to the same transcriptional networks. The reason for the apparent absence of PRR repression of PIF activity in dark-grown seedlings (SI Appendix, Fig. S2) is unknown. One possibility is that the levels of the PRRs are insufficient to impact the collective activities of the PIFs, which are present at much higher, saturating levels in dark-grown seedlings than in the light (3, 4, 30). Under this scenario, the reversible antagonistic interplay between the PIFs and PRRs would come into play only when levels of the PIFs have declined enough in the light to allow PRR input.
In establishing a genetic framework for the present study, we have extended understanding of the degree of redundancy among the PIF family members in regulating shade avoidance and, conversely, have uncovered a previously unappreciated level of specificity among the PIFs in regulating the expression of individual DTGs. The complete absence of observable shade responsiveness in the pif1pif3pif4pif5pif7 quintuple mutant (designated pifqpif7) demonstrates that the five PIF-quintet members (PIFs 1, 3, 4, 5, and 7) are collectively entirely responsible for this response at both the phenotypic and gene-expression levels. Strikingly, however, examination of the separate contributions of PIF7 and the PIF quartet to the induction of individual PIF-DTGs reveals a marked divergence in target-gene specificity between the two: from complete PIF7 specificity (IAA29, YUC8, and YUC9), versus complete quartet specificity (AT5G02580), through varying intermediate levels of additive contributions of all five PIFs. The data also show that PIF7 has the dominant role in regulating the auxin pathway that drives hypocotyl cell elongation, consistent with the work of Li et al. (24).
It is notable that, despite this general pattern of combinatorial activities among the PIFs toward their DTGs, the PRRs exert their repressive activity on all of the tested target genes. This result indicates that the PRRs impose this activity on all five PIF-family members, regardless of which dominates in transcriptional induction.
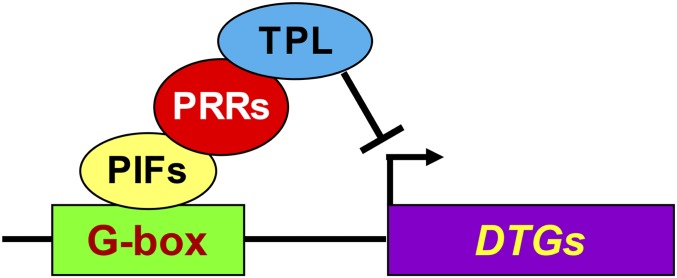
The colocalization of the PRRs and PIFs at the G-box motifs on PIF-DTG promoters (Fig. 1 and SI Appendix, Fig. S1) (29, 52), coupled with the evidence of physical interactions presented above (Fig. 7) and elsewhere (21, 22), indicate that the PRRs either bind indirectly to the promoters through the PIFs or bind directly to the promoter DNA, close to the PIFs, as well as to the PIF proteins themselves. Although, in principle, the PRRs could repress PIF transcriptional activation activity by inhibiting PIF binding to its DNA target or by binding to the G-box motifs with displacement of the PIFs, this would not seem to be compatible with the ChIP-seq–detected colocalization of the PRRs and PIFs on PIF-DTG promoters. Similarly, although the PRRs have been reported to bind to a TGTG DNA motif (44), we did not find this motif enriched or conserved in the PRR DNA-binding regions of PIF-DTGs. In addition, we were unable to detect direct binding of PRR5 to the DNA fragment containing the PIF-binding sites in the PIL1 promoter, using in vitro electrophoretic mobility shift (EMSA) assays (SI Appendix, Fig. S7), despite the presence of two TGTG motifs within that fragment. We also did not observe any direct binding to, or physical displacement of PIF4 from, a G-box motif using this assay (SI Appendix, Fig. S8). Our co-IP data further establish that the PRRs do not require DNA to interact with the PIFs (Fig. 7 C and D). Thus, although it has been reported that the G-box motifs, enriched in PRR-binding promoter locations, are necessary for transcriptional regulation by the PRRs (52), the most likely configuration seems to be indirect binding through the PIF proteins at those locations (Fig. 8).
Fig. 8.
Proposed mechanism of PRR-imposed repression of PIF transcriptional activation activity. PIF proteins bind sequence specifically to G-box CACGTG (or related PBE-box, CACATG) cis-elements in the promoters of PIF-DTGs. PRR proteins bind to the DNA-bound PIF activators and recruit TPL or TPR transcriptional corepressors to the promoter, repressing PIF-driven expression of the DTGs.
Previous studies have provided evidence that the PRRs function as repressors of core circadian-gene expression, specifically CCA1 and LHY (47, 53, 54), as well as of other direct targets of the PRRs, genome-wide (52, 55, 56). Evidence that the PRRs interact with the transcriptional corepressor family TOPLESS/TOPLESS RELATED (TPL/TPR) has led further to the conclusion that the PRRs facilitate repression of CCA1 and LHY1 expression by recruiting TPL/TPR to the promoters of these genes (54). Based on these reports and our present data, we propose the model in Fig. 8 for the mechanism by which the PRRs repress shade-induced PIF-DTGs. This model proposes that the PRR-TPL/TPR corepressor complex is recruited to target genes by promoter-bound PIF transcription factors, rather than binding directly to the DNA of those target genes.
The evidence that the PRR attenuation of the shade-induced phenotypic and gene-expression responses require the presence of one or more of the PIF quintet raises the question of whether the PRRs exert their repressive activity on various cellular processes exclusively through the PIFs or also through repression of other transcription factors. Although the PRRs have been reported to stabilize the floral regulator CONSTANS (57), we found no reports of direct repression of other transcription factors. It will be interesting to see whether the clock employs direct interaction of the PRRs with transcription factors other than the PIFs to repress other pathways.
Taken together, the findings presented here and previously (17, 22–25) provide additional insights into the continuum of overlapping and differential regulatory functions displayed by the members of the multifunctional PIF-signaling hub. Of particular note here is the apparently exclusive control of the rapid induction of auxin biosynthetic and regulatory genes by PIF7. These findings also provide robust evidence that multiple integral components of the central plant circadian oscillator connect immediately to downstream transcriptional networks through direct binding to, and repression of, the members of the PIF-transcription-factor quintet that comprise the signaling hub. In addition to potentially expanding the spectrum of known transcriptional pathways that are clock modulated, the data provide insights into the mechanisms by which the clock coordinately and pleiotropically influences a diversity of cellular activities. Because a significant fraction of the PIF-DTGs genes themselves encode a diversity of transcription factors (14, 15), this configuration provides a conduit through which clock output signals are transduced and immediately amplified to synchronously control multiple cellular pathways involved in growth and development.
Materials and Methods
Detailed information on plant materials and growth conditions, computational analyses, gene expression analysis, in vitro and quasi–in-vivo co-IP assays, BiFC assays, and EMSA are provided in SI Appendix, Materials and Methods.
Data Availability Statement.
All data discussed in the paper will be made available to readers from the corresponding author on request. A metadata sheet with the ChIP-seq datasets and computational analysis procedures have been deposited in the Gene Expression Omnibus database under accession number GSE136843.
Supplementary Material
Acknowledgments
We thank Ken S. Heyndrickx and Klaas Vandepoele for sharing genomic datasets of their 25 TFs and providing analytical support; David Somers, Eva Farre, Norihito Nakamichi, and Takeshi Mizuno for sharing plant materials; and Hongquan Yang and Wenxiu Wang for sharing BiFC constructs. This work was supported by NIH Grant 5R01GM047475-24 and US Department of Agriculture Agricultural Research Service Current Research Information System Grant 2030-21000-051-00D (to P.H.Q.).
Footnotes
The authors declare no competing interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE136843).
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918317117/-/DCSupplemental.
References
- 1.Franklin K. A., Quail P. H., Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casal J. J., Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64, 403–427 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Leivar P., Monte E., PIFs: Systems integrators in plant development. Plant Cell 26, 56–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leivar P., Quail P. H., PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ni W., et al. , PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8, 15236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ni W., et al. , A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344, 1160–1164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X., Paik I., Zhu L., Huq E., Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 20, 641–650 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Pham V. N., Kathare P. K., Huq E., Phytochromes and phytochrome interacting factors. Plant Physiol. 176, 1025–1038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paik I., Kathare P. K., Kim J. I., Huq E., Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 10, 1035–1046 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soy J., et al. , Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 71, 390–401 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galvāo V. C., et al. , PIF transcription factors link a neighbor threat cue to accelerated reproduction in Arabidopsis. Nat. Commun. 10, 4005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung J. H., et al. , Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Legris M., et al. , Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y., et al. , A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9, e1003244 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeiffer A., Shi H., Tepperman J. M., Zhang Y., Quail P. H., Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7, 1598–1618 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monte E., et al. , The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. U.S.A. 101, 16091–16098 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leivar P., Monte E., Cohn M. M., Quail P. H., Phytochrome signaling in green Arabidopsis seedlings: Impact assessment of a mutually negative phyB-PIF feedback loop. Mol. Plant 5, 734–749 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leivar P., et al. , Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21, 3535–3553 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornitschek P., et al. , Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Kohnen M. V., et al. , Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28, 2889–2904 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin G., et al. , Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr. Biol. 28, 311–318.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Soy J., et al. , Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. U.S.A. 113, 4870–4875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leivar P., et al. , The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20, 337–352 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., et al. , Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X., et al. , Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. eLife 7, e31636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biggin M. D., Animal transcription networks as highly connected, quantitative continua. Dev. Cell 21, 611–626 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Lelli K. M., Slattery M., Mann R. S., Disentangling the many layers of eukaryotic transcriptional regulation. Annu. Rev. Genet. 46, 43–68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X., O’Shea E. K., Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Mol. Cell 42, 826–836 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heyndrickx K. S., Van de Velde J., Wang C., Weigel D., Vandepoele K., A functional and evolutionary perspective on transcription factor binding in Arabidopsis thaliana. Plant Cell 26, 3894–3910 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leivar P., et al. , Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto Y., et al. , Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol. 44, 1119–1130 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Nakamichi N., et al. , The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 46, 609–619 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Nakamichi N., Kita M., Ito S., Yamashino T., Mizuno T., PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46, 686–698 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Ito S., et al. , Genetic linkages between circadian clock-associated components and phytochrome-dependent red light signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 48, 971–983 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Kaczorowski K. A., Quail P. H., Arabidopsis PSEUDO-RESPONSE REGULATOR7 is a signaling intermediate in phytochrome-regulated seedling deetiolation and phasing of the circadian clock. Plant Cell 15, 2654–2665 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato E., Nakamichi N., Yamashino T., Mizuno T., Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 43, 1374–1385 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Huq E., Quail P. H., PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21, 2441–2450 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khanna R., et al. , The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19, 3915–3929 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Sady B., Kikis E. A., Monte E., Quail P. H., Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 2232–2237 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leivar P., et al. , Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24, 1398–1419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang W., et al. , Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Yamashino T., et al. , A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44, 619–629 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Zhu J. Y., Oh E., Wang T., Wang Z. Y., TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 7, 13692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gendron J. M., et al. , Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McClung C. R., The plant circadian oscillator. Biology (Basel) 8, E14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez S. E., Kay S. A., The plant circadian clock: From a simple timekeeper to a complex developmental manager. Cold Spring Harb. Perspect. Biol. 8, a027748 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Somers D. E., The Arabidopsis clock: Time for an about-face? Genome Biol. 13, 153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedmale U. V., et al. , Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164, 233–245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma A., et al. , UVR8 disrupts stabilisation of PIF5 by COP1 to inhibit plant stem elongation in sunlight. Nat. Commun. 10, 4417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farré E. M., Liu T., The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr. Opin. Plant Biol. 16, 621–629 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Mizuno T., Nakamichi N., Pseudo-response regulators (PRRs) or true oscillator components (TOCs). Plant Cell Physiol. 46, 677–685 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Liu T. L., Newton L., Liu M. J., Shiu S. H., Farré E. M., A G-box-like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol. 170, 528–539 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakamichi N., et al. , PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L., Kim J., Somers D. E., Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U.S.A. 110, 761–766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakamichi N., et al. , Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. U.S.A. 109, 17123–17128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu T., Carlsson J., Takeuchi T., Newton L., Farré E. M., Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 76, 101–114 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Hayama R., et al. , PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 36, 904–918 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper will be made available to readers from the corresponding author on request. A metadata sheet with the ChIP-seq datasets and computational analysis procedures have been deposited in the Gene Expression Omnibus database under accession number GSE136843.