Significance
Synthetic nanomaterials are being sought to shuttle therapeutic payloads directly into the cell nucleus as a major target for chemo- and gene-based therapies. However, it remains uncertain whether and how synthetic entities are able to bypass the nuclear pore complexes (NPCs) that regulate transport into and out of the nucleus. We have constructed biocompatible polymer vesicles that infiltrate NPCs and resolved their nuclear uptake mechanism in vitro and in vivo. Their ability to deliver payloads directly into cell nuclei is further validated by transmission electron microscopy.
Keywords: drug delivery, polymersome, nanocarrier, nuclear pore complex, nucleocytoplasmic transport
Abstract
Organelle-specific nanocarriers (NCs) are highly sought after for delivering therapeutic agents into the cell nucleus. This necessitates nucleocytoplasmic transport (NCT) to bypass nuclear pore complexes (NPCs). However, little is known as to how comparably large NCs infiltrate this vital intracellular barrier to enter the nuclear interior. Here, we developed nuclear localization signal (NLS)-conjugated polymersome nanocarriers (NLS-NCs) and studied the NCT mechanism underlying their selective nuclear uptake. Detailed chemical, biophysical, and cellular analyses show that karyopherin receptors are required to authenticate, bind, and escort NLS-NCs through NPCs while Ran guanosine triphosphate (RanGTP) promotes their release from NPCs into the nuclear interior. Ultrastructural analysis by regressive staining transmission electron microscopy further resolves the NLS-NCs on transit in NPCs and inside the nucleus. By elucidating their ability to utilize NCT, these findings demonstrate the efficacy of polymersomes to deliver encapsulated payloads directly into cell nuclei.
Nucleocytoplasmic transport (NCT) describes the exchange of molecular cargoes across the nuclear envelope (NE) that encloses the nucleus from the cytoplasm in eukaryotic cells (1, 2). This is mediated by ∼60-nm-diameter channels in the NE known as nuclear pore complexes (NPCs) (3), which form the sole aqueous gateways to the genome (4). NPCs are permeable to small molecules less than 40 kDa (or ∼5 nm), but the entry of large nonspecific entities is impaired (5, 6). This selective barrier functionality is attributed to several highly dynamic, intrinsically disordered proteins known as phenylalanine–glycine nucleoporins (FG Nups) that are located within the NPC central channel (7). Exclusive NPC access is reserved for soluble transport receptors known as karyopherins (or Kaps) (8) that usher biochemically specific cargoes bearing nuclear localization signals (NLSs) into the nucleus. To traverse the NPC barrier, Kaps such as the classical import receptor Kapβ1 (importinß1) exert multivalent binding interactions with the FG Nups (9). Meanwhile, Kapβ1 forms a heterodimer (10) with an adaptor Kapα (Kapα•Kapβ1), which authenticates and binds to NLSs. To complete nuclear import, NLS-cargoes that transit in NPCs have to be released into the nucleus. This is orchestrated by the small GTPase Ran (11), which binds Kapβ1 in its GTP-bound form (RanGTP) to concomitantly release Kapα and NLS-cargoes inside the nucleus.
In housing the genome, the nuclear interior constitutes a principal target for applications such as chemotherapy (12) and gene therapy (13). This has motivated efforts to engineer synthetic nonviral delivery systems (vectors), which are highly sought-after based on their tunable payload capacities, reduced pathogenicity, and low immunogenicity (14). Nevertheless, it remains challenging to engineer organelle-specific, nuclear-targeting nanoparticles that shuttle therapeutic payloads directly into the nucleus (15). Key challenges include biocompatibility, biological stability, degradation, encapsulation efficiency, detection, and targeting accuracy (16).
Successful entry into the nuclear interior requires passage through the intracellular barrier presented by NPCs. This can be achieved by conjugating the simian virus 40 (SV40) large T antigen monopartite NLS (17), the bipartite NLS from nucleoplasmin (18), or the HIV-1 transactivating protein (TAT) peptide (19) to synthetic entities. These include 39-nm-diameter gold nanoparticles (20), 50-nm-diameter silica nanoparticles (21), 60-nm-diameter magnetite nanoparticles (22), 25-nm- and 150-nm-diameter chitosan nanoparticles (23), and 234-nm-diameter polymeric nanoparticles (24). Still, several aspects of nuclear targeting are unresolved (16). As a case in point, it is confounding how nanoparticles (23–25) whose sizes exceed the maximum pore diameter are able to traverse the NPC. Thus, it remains poorly understood how synthetic entities bypass the selective NPC barrier and are taken up into the nucleus.
Here, we have undertaken detailed biophysical, ultrastructural, and cellular studies to resolve the nuclear targeting mechanism of NLS-conjugated polymer vesicles, also called polymersomes (26). These are sought after for diagnostic and therapeutic applications (27–29) given their ability to function as molecular nanocarriers (NCs) that encapsulate diverse hydrophilic and hydrophobic entities within their aqueous lumens and membranes, respectively. Based on their biocompatibility, superior structural stability over liposomes (30), and ease of surface modifications for biological specificity, our results underscore the role of NLS-conjugated polymersome NCs as promising candidates for nuclear targeting applications.
Results
NC Design and Characterization.
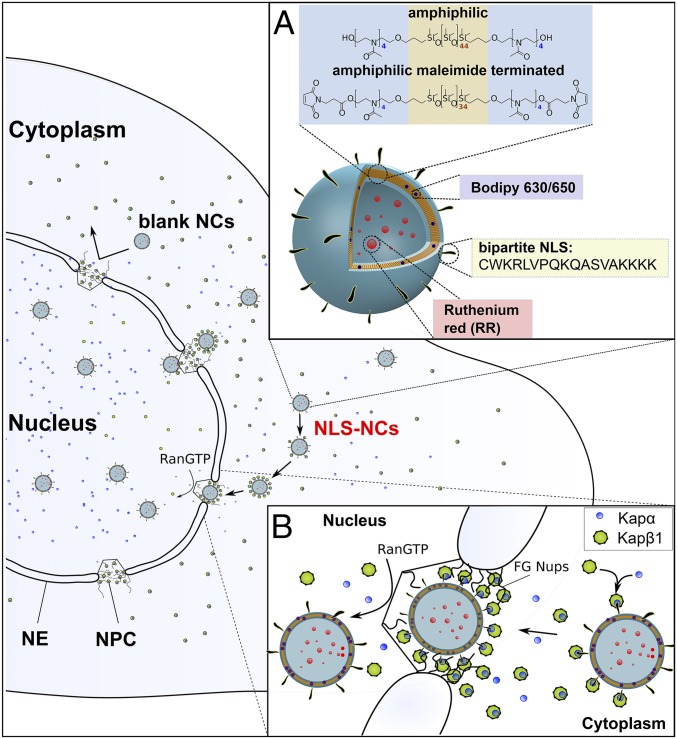
Polymersomes self-assembled from poly(2-methyl-2-oxazoline)-block-poly-(dimethylsiloxane)-block-poly(2-methyl-2-oxazoline) (PMOXA-PDMS-PMOXA) triblock copolymers are known to exhibit low toxicity in vitro and in vivo (31, 32), and did not provoke an innate immune response in mice following intraperitoneal injection (33). Here, we synthesized two variants: PMOXA4-PDMS44-PMOXA4 (Mn = 4,000 Da) and a maleimide-terminated derivative Mal-PMOXA4-PDMS34-PMOXA4-Mal (Mn = 3,800 Da) that were obtained with a molecular weight dispersity of 1.7 and 2.8, respectively (SI Appendix, Fig. S1). In this regard, polydispersity may be advantageous for the formation of polymersomes leading to more uniform size distributions (34). Both polymers are optimized for polymersome self-assembly based on their average amphiphilic block-copolymer ratio (35) (i.e., f, the hydrophilic molecular mass fraction in relation to the total molecular mass) that was 29% and 32%, respectively. Afterward, we conjugated the bipartite nucleoplasmin NLS (18) (CWKRLVPQKQASVAKKKK; M = 2,127 Da) via a catalyst free thiol-ene click reaction (SI Appendix, Fig. S2) to render the maleimide-terminated NCs (henceforth NLS-NCs) viable for NCT (Fig. 1). Additionally, non–NLS-conjugated NCs were assembled from PMOXA4-PDMS44-PMOXA4 exclusively (denoted as blank NCs) and used as nonspecific controls throughout this study. For clarity, both NLS-NCs and blank NCs are collectively referred to as NCs.
Fig. 1.
Organelle-specific targeting of polymersome NCs into the cell nucleus. (A) NLS-NCs self-assemble from amphiphilic PMOXA-PDMS-PMOXA triblock copolymers. Two model compounds are used to test for nuclear delivery: Ruthenium Red (RR) that is encapsulated within the NLS-NC lumen, and Bodipy 630/650 that incorporates into its polymeric membrane. (B) The nuclear transport mechanism involves Kapα•Kapβ1 that 1) authenticates NLS-NCs for selective NPC transport, 2) binds to FG Nups, and 3) releases NLS-NCs into the nucleus upon binding RanGTP.
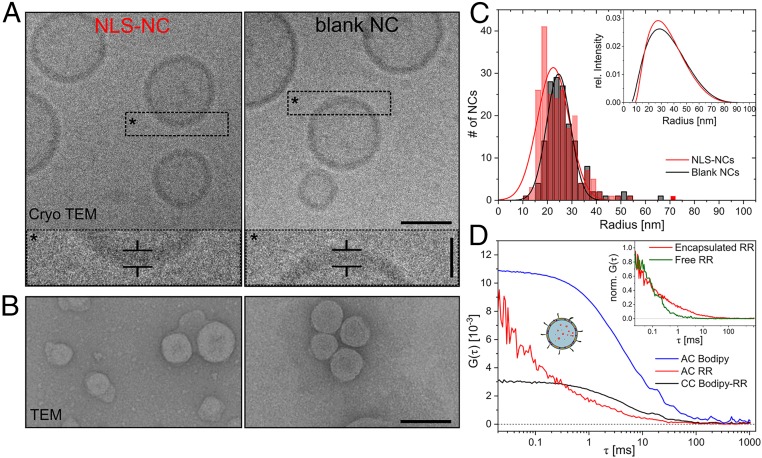
To facilitate nuclear uptake, we extruded NCs that were compatible with the size of the NPC channel. Cryoelectron microscopy (cryo-EM) revealed spherical NLS-NCs and blank NCs comprising hollow lumens enclosed by polymeric membranes that were 8.4 ± 1.1 and 8.2 ± 1.5 nm thick, respectively (Fig. 2A). Meanwhile, transmission electron microscopy (TEM) provided radial distributions of 22 ± 13 and 25 ± 9 nm for NLS-NCs and blank NCs (Fig. 2 B and C), respectively. This was consistent with dynamic light scattering (DLS) analysis, which reported hydrodynamic radii (Rh) of 28 ± 13 and 29 ± 14 nm, and polydispersity indices of 0.22 and 0.23 for NLS-NCs and blank NCs, respectively (Fig. 2 C, Inset). Static light scattering (SLS) was also employed to evaluate NC radius of gyration (Rg), structure, and mass. Knowing both Rg and Rh allowed us to calculate a form factor, r = Rg/Rh, which approached unity, thereby indicating that the NCs exhibited a membrane-enclosed vesicular structure (36) (i.e., hollow spheres) (SI Appendix, Fig. S3 and Table S1). Moreover, the supramolecular NC mass was determined to be 88.3 ± 2.1 MDa, which corresponds to 22,100 polymer chains per NC on average. This equates to an approximate concentration of 23 nM for a 2 mg/mL stock solution for both blank NCs and NLS-NCs, respectively. Because NLSs are not fluorescent, we could not directly measure the number of NLSs per NLS-NC. Instead, we conjugated SAMSA fluorescein probes to maleimide-terminated NCs to act as NLS surrogates. Thereafter, fluorescence correlation spectroscopy gave an estimate of 27 ± 9 NLSs per NC (SI Appendix, Fig. S4 and Table S2) bearing in mind that 1) maleimide end groups hydrolyze over time and/or 2) SAMSA-NC binding might reduce chromophore brightness. Following NLS conjugation, ζ-potential measurements yielded 18.7 ± 1.7 mV for NLS-NCs and 25.5 ± 9.4 mV for blank NCs, respectively.
Fig. 2.
Polymersome NC characterization. (A) Cryo-EM resolves the spherical shape and hollow interior of NLS-NCs (Left) and blank NCs (Right). (Scale bar: 50 nm.) Corresponding zoom-ins show the membrane thickness of individual NCs (*). (Scale bar: 20 nm.) (B) TEM micrographs enable general size analysis of NLS-NCs (Left) and blank NCs (Right). (Scale bar: 100 nm.) (C) Size distribution of NLS-NCs and blank NCs as determined by TEM and verified by DLS (Inset). (D) Dual-color fluorescence lifetime cross-correlation spectroscopy (dcFLCCS) (black) verifies RR encapsulation (red) inside the NLS-NC lumen and Bodipy insertion inside its membrane (blue), respectively. (Inset) Comparison of free and encapsulated RR.
NCs were further evaluated by dual-color fluorescence lifetime cross-correlation spectroscopy (dcFLCCS) upon incorporating Bodipy630/650 dye (hereafter Bodipy) and Ruthenium Red (λEx = 536 nm; hereafter RR) as model cargoes. Specifically, the lipophilic Bodipy incorporates into the polymeric membrane, whereas the hydrophilic RR is encapsulated within the aqueous NC lumen (SI Appendix, Fig. S5 A and B). Indeed, dcFLCCS confirmed their simultaneous incorporation and encapsulation within NCs (Fig. 2D). This is evident from the large cross-correlation (CC) amplitude and the pronounced shift of the autocorrelation (AC) curves toward longer diffusion times in comparison to freely diffusing RR (Fig. 2 D, Inset) or Bodipy (SI Appendix, Fig. S5C). Fitting the RR AC curve to SI Appendix, Eq. S3.1 gave a diffusion coefficient D = 6.5 ± 0.7 µm2/s that corresponds to Rh = 35.5 ± 1.0 nm by invoking the Stokes–Einstein equation. Likewise, we obtained D = 6.7 ± 0.2 µm2/s and Rh = 35.0 ± 0.8 nm from the Bodipy AC curve. The fraction of NCs with coexisting RR and Bodipy obtained from the cross-correlation is 39% for NLS-NCs and 30% for blank NCs (SI Appendix, Fig. S5D).
Kapα•Kapβ1 Binding to NLS-NCs.
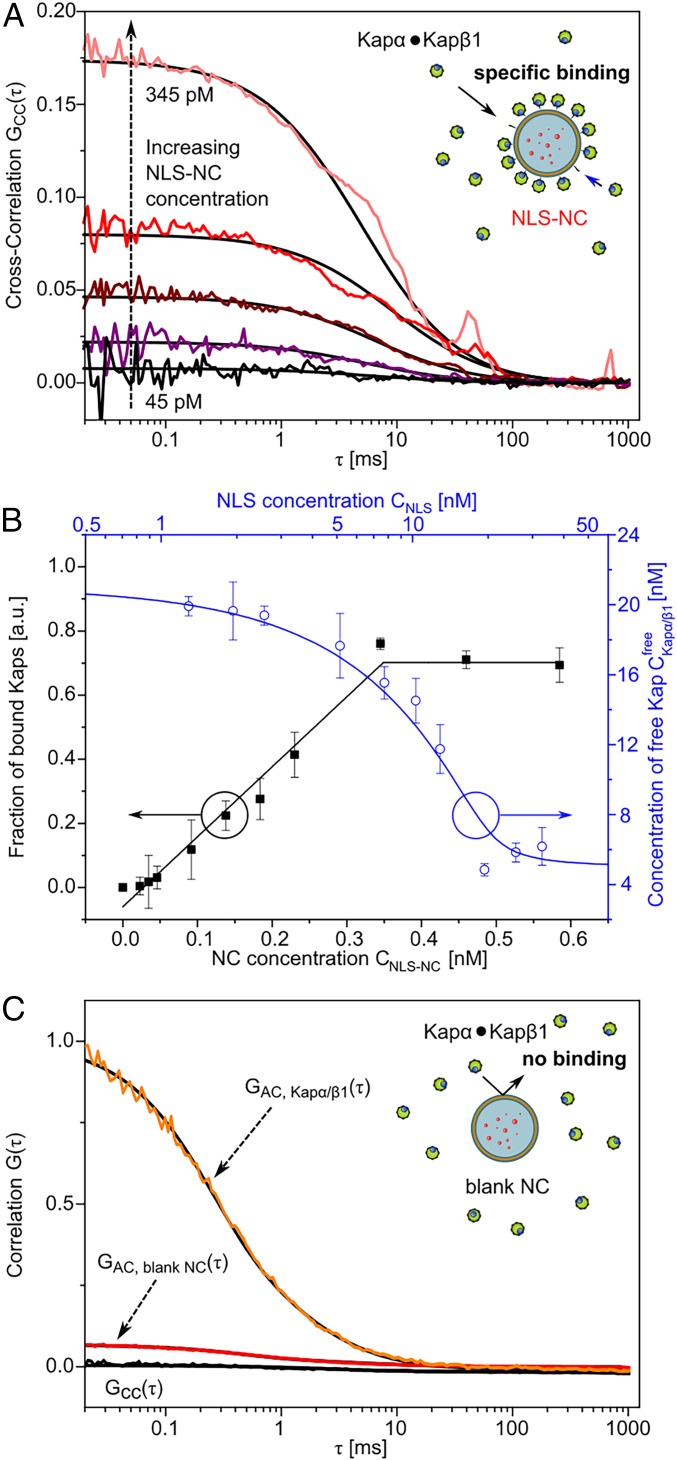
Next, we used dcFLCCS to quantify the equilibrium binding affinity of Kapα•Kapβ1 to the NLS-NCs (37) (SI Appendix). Bodipy-only NLS-NCs (without RR) were titrated in the range of 25 to 590 pM against a constant concentration of 200 nM Kapα, 18 nM unlabeled Kapβ1, and 2 nM Atto-550-labeled Kapβ1. Labeled Kapβ1 was required to facilitate dcFLCCS measurements. Here, the CC amplitude between Kapα•Kapβ1 and NLS-NCs increased with NLS-NC concentration as the increased availability of NLS binding sites shifts the equilibrium toward NLS-NC bound Kapα•Kapβ1 (Kapα•Kapβ1•NLS-NC; Fig. 3A). The corresponding relative CC amplitude (38) (SI Appendix, Eq. S6.1) allows to calculate the binding curve of Kapα•Kapβ1•NLS-NC formation (black squares in Fig. 3B). Fitting the binding curve to a multiple independent binding site model (SI Appendix, Eq. S6.2) yields a maximum of 57 ± 3 Kapα•Kapβ1 copies per NLS-NC at saturation (>345 pM), which is consistent with the estimated number of NLSs per NC. This also gives an apparent binding affinity KD ≤ 0.4 nM for Kapα•Kapβ1-NLS binding (SI Appendix, Eq. S6.2) that is comparable to literature values (39).
Fig. 3.
Kapα•Kapβ1 binding to NLS-NCs. (A) The binding strength of Kapα•Kapβ1 to NLS-NCs was assessed via dcFLCCS. Increasing concentrations of NLS-NCs were titrated against 20 nM Kapβ1 and 200 nM Kapα. Signal cross-correlation GCC(t) increases due to the increasing degree of binding. (B) Binding curve fitting (black) obtained from the fraction of bound Kaps (black squares) via the relative cross-correlation amplitude yields 57 ± 3 Kapα•Kapβ1 complexes per NLS-NC with KD ≤ 0.4 nM. The drop in free Kapα•Kapβ1 concentration (blue circles) is accurately simulated (blue line) based on the assumption that the maximum number of bound Kapα•Kapβ1 complexes per NLS-NC is equal to the number of competent NLS binding sites. See SI Appendix, section 6 for details. (C) Blank NCs are not recognized by Kapα•Kapβ1 complexes and show no signal cross-correlation (black).
Upon binding to NLS-NCs, free Kapα•Kapβ1 complexes in solution are depleted and their concentration decreases with an increase in NLS binding sites (blue circles in Fig. 3B). Based on fitting parameters from SI Appendix, Eq. S6.2, this reduction can be accurately simulated as a function of the total NLS concentration (blue line in Fig. 3B and SI Appendix, Eqs. S6.3–S6.6).
Meanwhile, free Atto-550 did not interact with the NLS-NCs, thereby indicating that Kapα•Kapβ1 bound NLS-NCs specifically (SI Appendix, Fig. S6). Moreover, it is evident from the lack of cross-correlation (Fig. 3C) that Kapα•Kapβ1 did not interact with blank NCs.
Kapα•Kapβ1 Mediates NLS-NC–FG Nup Interactions.
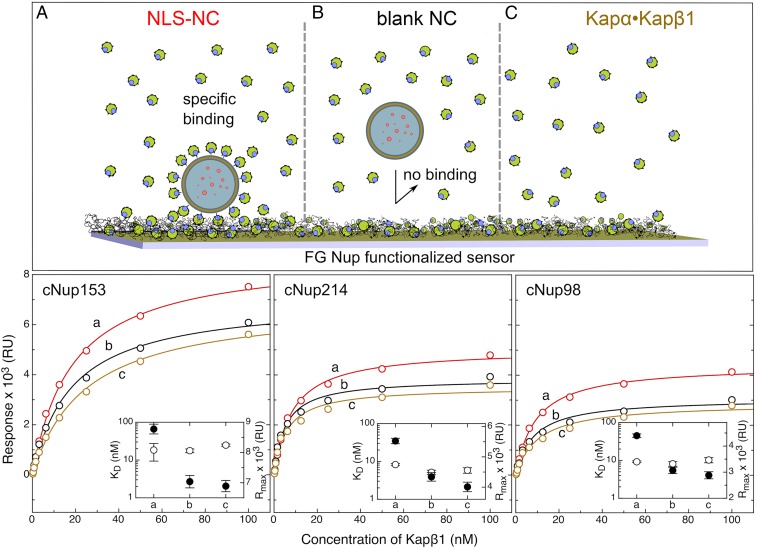
Multivalent interactions between Kaps and FG Nups facilitate selective transport across the NPC (40). We ascertained the binding of Kapα•Kapβ1•NLS-NCs to three FG Nups (cNup98, cNup214, and cNup153) by surface plasmon resonance (SPR) (SI Appendix, Fig. S7) (9). Langmuir isotherm analysis (Fig. 4) indicates that the apparent binding affinity (KD) of Kapα•Kapβ1•NLS-NCs to cNup153 (and also cNup214 and cNup98) does not differ from that of standalone Kapα•Kapβ1, being 18.4 ± 9.0 and 24.8 ± 1.7 nM, respectively. This is likely due to a fraction of free Kapα•Kapβ1 that is generally present with Kapα•Kapβ1•NLS-NCs at equilibrium. Regardless, their binding to the FG Nups provoked a maximal binding response (in resonance units [RU]) that was ∼2 kRU higher than standalone Kapα•Kapβ1. This correlates to an increase of bound mass, which can be calculated from the relation 1,300 RU = 1 ng/mm2 (41) to give 1.5 ng/mm2 or ∼9 NLS-NCs per µm2.
Fig. 4.
Kapα•Kapβ1 mediates NLS-NC binding to the FG Nups. (A) Kapα•Kapβ1•NLS-NCs elicit the highest FG Nup- binding response as measured by SPR. (B) Blank NCs do not bind the FG Nups in the presence of Kapα•Kapβ1. (C) When NCs are absent, the binding response of standalone Kapα•Kapβ1 is similar to B. This indicates that the large binding response of Kapα•Kapβ1•NLS-NCs results from the added mass of each NLS-NC. Langmuir isotherm fits (solid lines where a, b, and c correspond to A, B, and C, respectively) yield the maximal response signal (Rmax; filled circles) and the equilibrium dissociation constants (KD; open circles) shown in the Insets and summarized in SI Appendix, Table S3.
As controls, blank NCs mixed with Kapα•Kapβ1 elicited a similar binding response to standalone Kapα•Kapβ1 in terms of its magnitude, which signified a lack of NC binding to the FG Nups (SI Appendix, Fig. S7 A–C). Also, neither blank NCs nor NLS-NCs showed FG Nup binding in the absence of Kapα•Kapβ1 (SI Appendix, Fig. S7D). This verifies that Kapα•Kapβ1 mediates the selective binding of the NLS-NCs to the FG Nups as a prerequisite to bypass the NPC selective barrier (SI Appendix, Table S3).
RanGTP Regulates NLS-NC Nuclear Uptake in Permeabilized Cells.
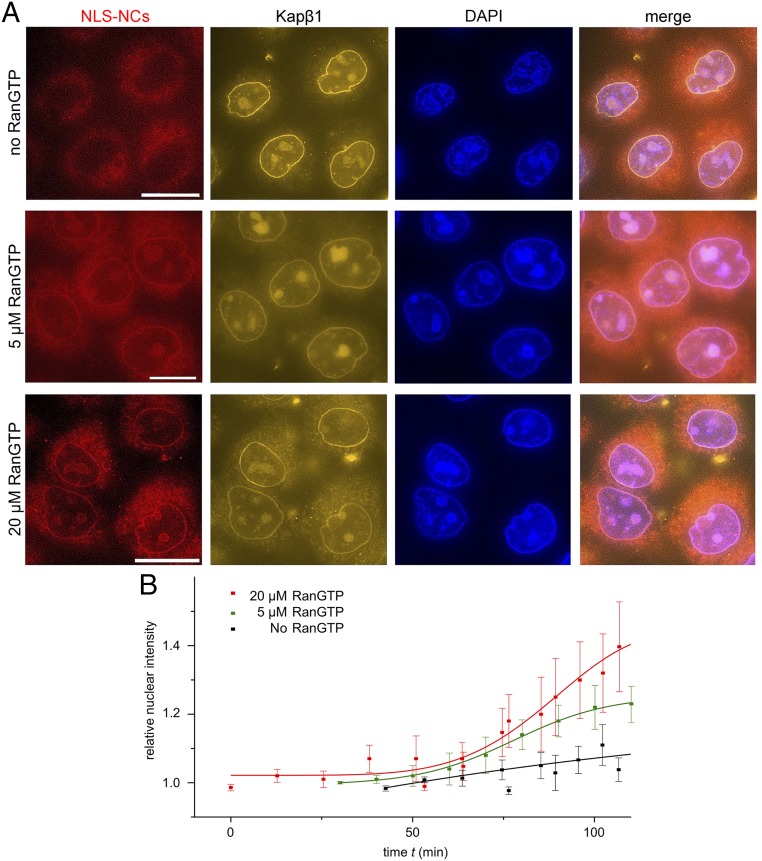
During import, RanGTP binds to Kapβ1 to trigger the release of Kapα and its cargoes in the nucleus (10, 11). To evaluate this, we employed a permeabilized cell assay using a so-called “Ran mix” (40) that includes RanGDP, key transport factors, and an energy-regenerating system that reactivates an enzyme known as Ran guanine nucleotide exchange factor (or RanGEF) that converts RanGDP to RanGTP in the nucleus. In doing so, we sought to ascertain whether RanGTP promoted NLS-NC nuclear uptake. This was carried out by varying the amount of RanGDP in Ran mix from 0 to 5 and 20 µM whilst keeping the NLS-NC concentration constant. Negligible amounts of Bodipy-labeled NLS-NCs were detected in the nucleus after a 2-h incubation when RanGTP was absent (Fig. 5A). However, near physiological concentrations (42), 5 µM RanGTP was sufficient to drive nuclear NLS-NC uptake, whereas 20 µM RanGTP enhanced it (Fig. 5B). Meanwhile, the signal of Atto550-labeled Kapβ1 at the nuclear envelope indicated the presence of Kapα•Kapβ1 or Kapα•Kapβ1•NLS-NCs on transit at the NPCs in all three cases. In other words, Kapα•Kapβ1 is necessary but insufficient for the nuclear uptake of NLS-NCs. In marked contrast, blank NCs did not pass through NPCs in the presence of Ran mix (SI Appendix, Fig. S8). Hence, NLS-NCs require Kaps to enter NPCs, whereas nuclear uptake requires RanGTP to bind Kapβ1 and release Kapα and the NLS-NCs from the NPCs (40).
Fig. 5.
NLS-NCs depend on RanGTP for nuclear uptake in permeabilized cells. (A) NLS-NC nuclear import is highest in 20 µM RanGTP and lowest when RanGTP is absent. (Scale bar: 10 µm.) (B) Time-lapse imaging indicates that the nuclear import rates of NLS-NCs increase with RanGTP concentration.
Resolving NLS-NCs That Infiltrate the Nucleus in Live Cells.
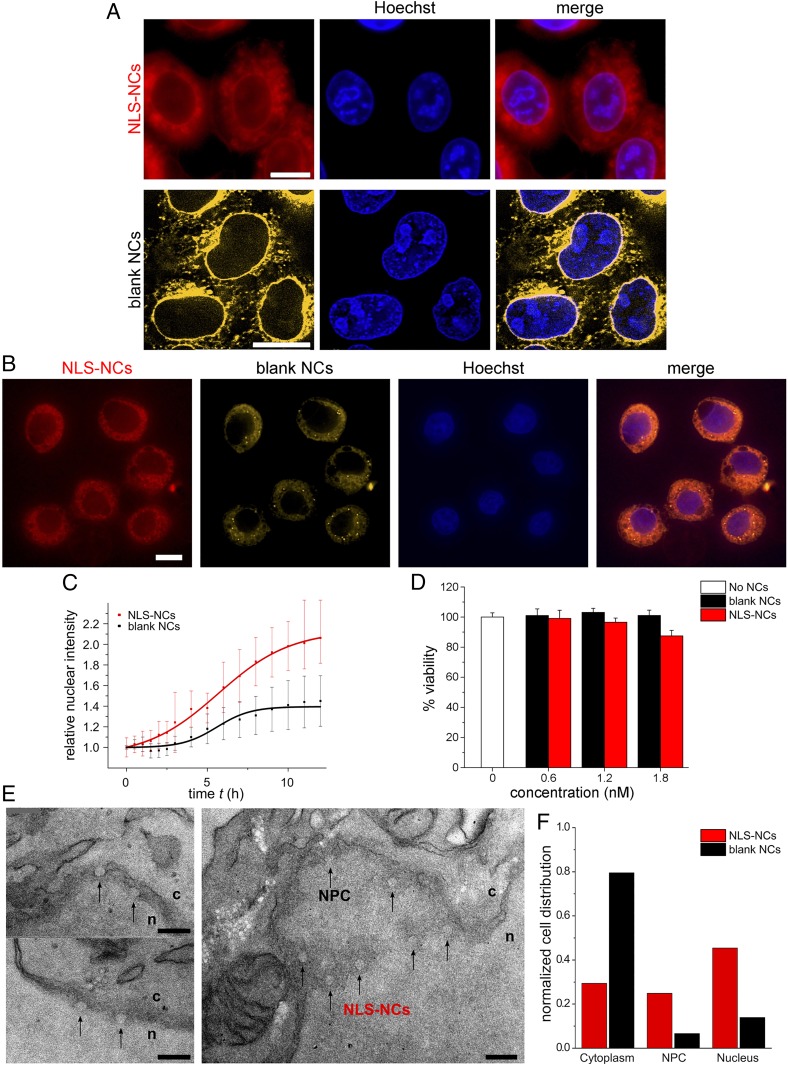
Next, we studied the nuclear uptake of Bodipy-NLS-NCs and Nile Red (NR)-labeled blank NCs into live HeLa cells by time-lapse fluorescence microscopy (Fig. 6 A and B). PMOXA-PDMS-PMOXA NCs enter cells through an endosomal escape pathway (31). Subsequently, NLS-NC uptake into the nucleus doubled after 12 h and was consistently more pronounced than blank NCs (Fig. 6C). In contrast, blank NCs were predominant in the cytoplasm and along the nuclear envelope (Fig. 6A). Such differences are further evident by comparing between their respective nuclear and cytoplasmic signals as time progresses. Whereas blank NCs plateau at similar relative intensities in both compartments within 12 h, NLS-NCs continue to accumulate in the nucleus but not in the cytoplasm (SI Appendix, Fig. S9). Still, neither NLS-NCs nor blank NCs were toxic to the HeLa cells up to 48 h (Fig. 6D). Indeed, differences in the nuclear uptake behavior of NLS-NCs and blank NCs are evident following coincubation in the same cells (Fig. 6B).
Fig. 6.
Nuclear uptake and ultrastructural analysis of NLS-NCs in HeLa cells. (A) Fluorescence imaging shows that NLS-NCs import into the nuclei of live cells, whereas blank NCs are largely rejected. (Scale bar: 10 µm.) (B) This is most striking when both NLS-NCs and blank NCs are coincubated in the same cells. (Scale bar: 10 µm.) (C) Time-lapse imaging over 12 h reveals that nuclear import rate is enhanced for NLS-NCs in comparison to the passive diffusion of blank NCs. (D) NLS-NC and blank NC-treated HeLa cells remain viable after 48 h. (E) TEM ultrastructural analysis resolves NLS-NCs (black arrows) that traverse NPCs to enter the cell nucleus. c, cytoplasm; n, nucleus. (Scale bar: 200 nm.) (F) Statistical distribution of NLS-NCs (n = 292 in 56 cells) in comparison to blank NCs (n = 166 in 21 cells; SI Appendix, Fig. S10).
TEM ultrastructural analysis was then used to verify whether individual NLS-NCs did indeed translocate through NPCs and if they retained their structure following NCT. Unlike inorganic nanoparticles, however, it is formidable to resolve the polymeric NCs in the crowded cellular environment due to their low image contrast. We therefore reasoned that an EDTA regressive staining protocol (43) could chelate and deplete uranyl stained material (e.g., chromatin) so as to enhance the visibility of the NCs (Materials and Methods). This revealed features that bore the distinct circular imprints of NLS-NCs (Fig. 6E) and blank NCs (SI Appendix, Fig. S10), respectively. Overall, we found that 30%, 25%, and 45% NLS-NCs were localized in the cytoplasm, at NPCs, and within the nuclear interior, respectively (Fig. 6F). Their localization was irrespective of NLS-NC size, which is 69 ± 12 nm in diameter (n = 292; SI Appendix, Fig. S10). In comparison, a 77% majority of blank NCs measuring 69 ± 14 nm in diameter (n = 166) were found in the cytoplasm, which indicates that their localization was size independent. Thus, a majority of blank NCs was prevented from entering the nucleus despite being comparable in size to the NLS-NCs, as well as NPCs that were 61 ± 16 nm wide (n = 197; SI Appendix, Fig. S10).
Discussion
In this work, we have developed polymersome NCs and mapped their transport pathway into the cell nucleus at the molecular, ultrastructural, and cellular levels. Our findings show that NLS-NCs emulate authentic cargo specificity to bypass NPCs and enter into the cell nucleus. Moreover, they are biocompatible, have low cytotoxicity (28), and possess a superior structural integrity that is compliant to changes in shape without rupturing (44). Crucially, NLS-NCs harbor aqueous lumens that are amenable to the encapsulation of various molecular payloads as a prerequisite. This is important for delivering nuclear specific drug compounds (12), protein-based therapeutics (16), and plasmids in gene-based therapies (45). Moreover, their ∼8-nm-thick polymer membranes may offer enhanced stability in cellular environments or when binding to proteins (e.g., Kapα•Kapβ1) in comparison to liposomes (46). Not least, the relative ease of conjugating different peptides to these polymersomes by thiol-ene “click” reactions makes them attractive as candidates for diverse applications in nanomedicine.
We have further shown that: 1) Kapα•Kapβ1 authenticates and binds NLS-NCs; 2) the presence of multiple Kapα•Kapβ1 copies per NLS-NC ensures its binding to the FG Nups, but may further facilitate efficient transport through the NPC, as has been suggested for large cargoes (47–49); and 3) RanGTP is required to displace the NLS-NCs from NPCs into the nuclear interior by binding Kapβ1. Indeed, NPCs exclude blank NCs in the absence of the above molecular interactions.
This study includes a methodological advance to resolve individual NCs in cells using a TEM-based ultrastructural analysis. Our TEM results verify that NLS-NCs, but not blank NCs, can successfully transit and traverse NPCs. This is unexpected, as the largest TEM-resolved entities to reside in the NPC are 39-nm-diameter gold nanoparticles (20) and 30- to 40-nm-diameter viral capsids (50, 51). Interestingly, interactions between Kapα•Kapβ1•NLS-NCs and the FG Nups might lead to a reduction of the NPC barrier (52), and/or may alter pore shape (53). This might explain how NPCs accommodate NLS-NCs despite bearing comparable diameters. In addition, the NLS-NCs may deform (44) as they pass through the pore, although the resolution of the current experiments precludes such observations. Still, it is difficult to rationalize how large NLS-conjugated nanoparticles (23–25) (between 143 and 234 nm in size) that significantly exceed the NPC diameter may traverse it. Hence, TEM-based ultrastructural analysis would be essential to verify these reports, such as to ensure that degradation did not occur prior to import (16). Regardless, our analysis shows that the NLS-NCs are appropriately sized for traversing NPCs, which sets a maximal design cutoff for future nuclear targeting systems. Future efforts will reveal how adjusting NLS-NC size, the degree of NLS functionalization, membrane thickness, etc., can optimize nuclear uptake. Other challenges include achieving a controlled release of NLS-NC payloads within the nucleus and studying how cells might respond to degraded NLS-NC material.
Materials and Methods
Synthesis.
See SI Appendix for details.
Polymersome Preparation.
NLS-NCs were prepared via a solvent-free method. Here, a homogeneous amphiphilic polymer film was deposited onto the bottom of a round-necked flask. This consisted of 1.9 mg of PMOXA4-PDMS44-PMOXA4 (95 wt%) and 0.1 mg of PMOXA4-PDMS34-PMOXA4 (5 wt%) where 34% of all polymer end groups had been substituted with active maleimide linking sites, i.e., Mal-PMOXA4-PDMS34-PMOXA4-Mal (SI Appendix). Hence, at least 1.6% of each polymersome is composed of Mal-PMOXA4-PDMS34-PMOXA4-Mal. Polymersome self-assembly followed film rehydration and desorption in 1 mL of 75-µM RR in PBS. The heterogeneous polymersome dispersion was extruded 15 times through a polycarbonate membrane of 50-nm pore size (Whatman Nuclepore Track Etch Membrane). Excess RR was removed via size exclusion chromatography through a Sephadex G-25 column (GE Healthcare Life Science HiTrap Desalting Column). Cysteine-terminated bipartite nuclear localization sequences from nucleoplasmin 2 (CWKRLVPQKQASVAKKKK; M = 2,127 Da; GenScript; Lot No. 91262870001/PE3665) were then conjugated to the polymersome structure by a spontaneous thiolene click reaction. After 12 h, excess cysteine was added to quench unreacted maleimide sites in an overnight reaction. Pure NLS-conjugated polymersomes (NLS-NCs) were obtained by dialyzing out free NLS and free cysteine against PBS using dialysis tubing with a molecular mass cutoff of 3.5 kDa in 2-h triplicates. When required, NLS-NCs were also labeled with 200 nM lipophilic Bodipy 630/650 or 1 µM lipophilic Nile red 552/636, respectively. Negative control blank polymersome NCs (blank NCs) were prepared from 2 mg of PMOXA4-PDMS44-PMOXA4 exclusively using the same preparation and purification procedures. Depending on the experiment, blank NCs were also labeled with 1 µM lipophilic Nile red 552/636.
Protein Expression, Purification, and Labeling.
Cysteine-tagged FG domains of human Nup214, Nup98, and Nup153 were cloned, expressed, and purified as described (9). Kapα, Kapβ1, and RanGDP were also expressed and purified as described (40). Kapβ1 labeling by Atto-550 succinimidyl ester (Atto-550 NHS-ester or simply Atto-550) was carried out in PBS buffer using a standard procedure (Invitrogen). Conjugation efficiency was determined by spectrophotometry (Nanodrop 2000).
Cryo-EM.
Four microliters of 2 mg/mL polymersome dispersions were dropped onto glow discharged carbon-coated lacey copper grids (300 mesh; Electron Microscopy Science). Samples were blotted in a commercial vitrification system (Vitrobot Mark IV; Thermo Fisher) and after plunge freezing the grids were transferred at −178 °C into a Gatan 626 cryoholder (Gatan) and imaged in a Tecnai F20 microscope (Thermo Fisher) operated at 200 kV. Resulting cryo-EM images were recorded with a BM-Ceta camera (4,096 × 4,096 pixels; Thermo Fisher).
TEM.
Samples were imaged on a Philips CM100 microscope operating at 100-kV acceleration voltage and equipped with a charge-coupled device (CCD) camera. Dilute NC solutions (5 µL of 0.2 mg/mL polymersomes) were deposited onto prehydrophilized carbon-coated 400 mesh copper grids and negatively stained with 2% uranyl acetate solution. Size analysis was carried out using ImageJ (54), taking at least 150 individual NC specimens for evaluation. Diameters were calculated by taking the average between the minor and major axes of each individual NC.
Dynamic and Static Light Scattering.
DLS and SLS experiments were performed on a commercial goniometer (LS Instruments) equipped with a 30-mW HeNe laser (wavelength, 633 nm) and two parallel avalanche photomultiplier detectors (APDs). The detected count rate was set to 40 kHz via an automatic laser intensity regulation function. After-pulsing effects were antagonized by pseudo–cross-correlation between the signals detected in the two APDs. The scattering intensity of freshly extruded polymersomes was measured in dust-free 10-mm high-precision quartz cells, which were placed in an optically matching thermostat vat at 298 K.
ζ Potential.
All measurements were performed on a Zetasizer Nano ZSP (Malvern Instruments) at 298 K. NLS-NC and blank NC dispersions of 0.5 mg/mL were diluted 20-fold in Millipore water to 25 µg/mL and <10 mM salt concentration. Each sample was measured in triplicate to determine the average ζ potential.
Dual-Color Fluorescence Lifetime Cross-Correlation Spectroscopy.
Measurements were performed on an Olympus IX73 inverted microscope stand equipped with a 1.2 N.A. water-immersion 60× superapochromat objective (UplanSApo; Olympus) and suitable emission and excitation bandpass filters (Semrock and AHF). Two pulsed diode lasers (LDH-P-FA-530 and LDH-D-C-640; PicoQuant) were operated at 40 MHz for pulse interleaved excitation dcFLCCS (Sepia II; PicoQuant). Emitted photons were detected in two separated channels coupled with two SPAD detectors (SPCM CD3516H; Excelitas) and a time-correlated single-photon counting unit to generate picosecond histograms also called lifetime spectra (16-ps resolution; HydraHarp 400) from the statistical photon arrival times. The laser powers were set to 20 µW for the LDH-P-FA-530 and to 17 µW for the LDH-D-C-640 laser and the intensity fluctuation recorded for 120 s with a correlation integration time taken as 2 s. The confocal volume was calibrated using free dyes of known diffusion constants D (using Rhodamine B in excitation channel 530 with D = 426.4 µm2/s at 298 K and a structural parameter of S = 4, and Atto-655NHS ester in excitation channel 640 with D = 403.6 µm2/s at 298 K and a structural parameter of S = 4). All measurements were performed 20 µm away from the coverslip.
SPR.
SPR binding assays were performed using a BiacoreT200 (GE Healthcare) at 25 °C using four flow cells as described previously (9, 41). See SI Appendix for details.
Cell Culture.
HeLa cells were cultured in Dulbecco’s modified Eagle medium GlutaMAX-I (DMEM) (Gibco Life Sciences) and supplemented with 10% (vol/vol) FBS (BioConcept), 100 units/mL penicillin, and 100 µg/mL streptomycin (Sigma-Aldrich). Cells were maintained at 37 °C and 5% CO2.
Permeabilized Cell Assays.
HeLa cells were cultured in eight-well glass bottom µ-slides (ibidi) up to 80% confluency in DMEM mixed with 10% (vol/vol) FBS. The cells were washed three times with PBS before permeabilization in digitonin solution (40 µg/mL in transport buffer) for 5 min (40). This was followed by a triple wash in PBS buffer, followed by nuclear staining with DAPI (Sigma-Aldrich), and another triple wash with PBS. Excess buffer was wicked off and the permeabilized cells incubated with 300 µL of Ran mix for 30 min (containing 1 µM Kapβ1; 2 µM Kapα; 5 µM, 20 µM, or no RanGDP; 1 mM GTP [Roche]; 1 µM NTF2; 100 μM ATP [Roche]; 4 mM creatine phosphate [Roche]; 20 U/mL creatine kinase [Roche]). Thirty microliters of either 6 nM Bodipy 630/650-labeled NLS-NCs or 6 nM Nile Red 552/636-labeled blank NCs were added to the Ran mix solution in order to obtain a final NC concentration of 0.6 nM. A DeltaVision wide-field fluorescence microscope was used for time-lapse measurements over 120 min with images taken every 10 min. Studies on permeabilized cells were repeated three times at each experimental condition.
Live-Cell Imaging.
Nuclear uptake into HeLa cells was studied via wide-field fluorescence microscopy. Cultured cells were seeded in eight-well glass bottom µ-slides (ibidi) using DMEM with 10% (vol/vol) FBS as nutrition medium and grown until they reached a confluency of 50 to 80%. In all live-cell studies, we stained the cell nuclei with Hoechst (Thermo Fisher) and used DMEM without phenol red (Gibco). To begin with the experimental assay, a concentration of 0.6 nM Bodipy 630/650-labeled NLS-NCs or 0.6 nM Nile Red 552/636-labeled blank NCs were added to the cell medium. The cells were then transferred to an Olympus IX71 stand that was preheated to 37 °C with a 5% CO2 atmosphere. A DeltaVision core wide-field fluorescence microscope was equipped with a Photometrics CoolSNAP HQ2 camera coupled to an interline CCD transfer and was operated via SoftWorx 4.12 software. A 60× oil objective was applied for imaging. Relative nuclear fluorescence intensities were determined via signal colocalization with the chromatin stain Hoechst. NC uptake kinetics was followed over 12 h with time-lapse images taken every 30 min for the first 3 h, followed by images being recorded every 1 h for the next 9 h. Cell studies were repeated three times at each experimental condition.
Fluorescence Image Analysis.
NC uptake in both permeabilized HeLa cells and live HeLa cells was analyzed by three-dimensional (3D) deconvolution fluorescence microscopy. Pixel saturation due to the cellular accumulation of NCs throughout the experimental time course was avoided by determining the optimal exposure prior to image acquisition. Datasets were recorded over a Z-stack range of 15 µm at 0.35-µm z intervals at each time point. These were analyzed using Imaris software with an integrated ImarisCell module based on the fluorescence intensities of Bodipy 630/650-labeled NLS-NCs and Nile Red 552/636-labeled blank NCs, respectively. Each signal was summed over the entire 3D cell volume at each time point. Nuclear uptake was determined from the signals that colocalized with chromatin staining by Hoechst in the same 3D volume. Fluorescence intensities in the cytoplasm were obtained by subtracting the nuclear intensities from the sum intensity over the entire cell volume (SI Appendix, Fig. S9). Relative fluorescence intensities at each time point were calculated by normalizing the average detected photon count per unit volume by its corresponding value at time equals zero (i.e., at the start of the experiment). This analysis was applied to a total of 28 cells incubated with NLS-NCs and 18 cells incubated with blank NCs.
Cell Viability.
Cell viability was determined using a CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS; Invitrogen) based on a standard protocol. HeLa cells were seeded in a 96-well plate (5,000 cells per well). After 24 h, cells were dosed with increasing concentrations of blank NCs or NLS-NCs (0.16, 0.35, and 0.65 mg/mL, final concentrations) and incubated further for 48 h. The MTS reagent (20 µL) was added to each well and after 2-h absorbance at 490 nm was measured using a Spectramax plate reader. Background absorbance was subtracted from each well, and data were normalized to control untreated cells. Experiments were done in quadruplicate (n = 4), and data were plotted using Origin (OriginLab).
Ultrastructural Analysis.
HeLa cells were seeded in 100 × 21-mm cell culture dishes (Thermo Fisher) and grown in DMEM media (Gibco) containing 10% (vol/vol) FBS. The cells were then incubated for 12 h at 37 °C in 5% CO2 with 0.6 nM RR-NLS-NCs or 0.6 nM RR-blank NCs, respectively. Thereafter, the cells were washed with PBS three times and collected into cell pellets that were immediately frozen with Karnovsky fixative and embedded in Epon resin. Ultrathin 50-nm sections were cut and mounted onto nickel grids and treated according to a regressive EDTA staining protocol (43). To do so, the thin sections were floated over a 6%age aqueous uranyl acetate solution for a reaction time of 5 min. Subsequent rinsing with double distilled H2Odd followed a second floating step on a 0.2 M EDTA/water solution. The pH of the solution was raised to 7.0 by adding 1 N sodium hydroxide drop by drop over a 30-min time course. The sections were rinsed again with H2Odd and stained with lead citrate for 5 min before rinsing them another time with H2Odd. The samples were imaged on a Philips CM100 transmission electron microscope operated at 100-kV acceleration voltage and equipped with a CCD camera. Subsequent statistical and size distribution analysis was carried out in ImageJ (54).
Data Availability.
All data are included in the paper and SI Appendix.
Supplementary Material
Acknowledgments
We thank A. Dinu for polymer synthesis support; C. Tiberi from the Center for Cellular Imaging and Nanoanalytics; S. Erpel and M. Dürrenberger from the Nanoimaging Facility; K. Schleicher, W. Heusermann, and N. Ehrenfeuchter from the Imaging Core Facility; T. Sharp from the Biophysics Facility; X. Huang for help with NLS-NC preparation; and M. Ghelfi for microscopy assistance. Furthermore, we are grateful to F. Stellacci, J. Gaitzsch, and W. Meier. C.G.P. and R.Y.H.L. acknowledge the National Centre of Competence in Research Molecular Systems Engineering and the University of Basel for financial support. C.Z. is supported by a Swiss Nanoscience Institute PhD fellowship. Further support to R.Y.H.L. is provided by Swiss National Science Foundation Grant 31003A_170041.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916395117/-/DCSupplemental.
References
- 1.Fahrenkrog B., Aebi U., The nuclear pore complex: Nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 4, 757–766 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Görlich D., Kutay U., Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Beck M., et al. , Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306, 1387–1390 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Strambio-De-Castillia C., Niepel M., Rout M. P., The nuclear pore complex: Bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11, 490–501 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Timney B. L., et al. , Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 215, 57–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popken P., Ghavami A., Onck P. R., Poolman B., Veenhoff L. M., Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol. Biol. Cell 26, 1386–1394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakiyama Y., Mazur A., Kapinos L. E., Lim R. Y. H., Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy. Nat. Nanotechnol. 11, 719–723 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Christie M., et al. , Structural biology and regulation of protein import into the nucleus. J. Mol. Biol. 428, 2060–2090 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Kapinos L. E., Schoch R. L., Wagner R. S., Schleicher K. D., Lim R. Y. H., Karyopherin-centric control of nuclear pores based on molecular occupancy and kinetic analysis of multivalent binding with FG nucleoporins. Biophys. J. 106, 1751–1762 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart M., Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Görlich D., Panté N., Kutay U., Aebi U., Bischoff F. R., Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15, 5584–5594 (1996). [PMC free article] [PubMed] [Google Scholar]
- 12.Wang D., Lippard S. J., Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 4, 307–320 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Verma I. M., Somia N., Gene therapy—promises, problems and prospects. Nature 389, 239–242 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Glover D. J., Lipps H. J., Jans D. A., Towards safe, non-viral therapeutic gene expression in humans. Nat. Rev. Genet. 6, 299–310 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Mintzer M. A., Simanek E. E., Nonviral vectors for gene delivery. Chem. Rev. 109, 259–302 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Tammam S. N., Azzazy H. M. E., Lamprecht A., How successful is nuclear targeting by nanocarriers? J. Control. Release 229, 140–153 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Kalderon D., Richardson W. D., Markham A. F., Smith A. E., Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311, 33–38 (1984). [DOI] [PubMed] [Google Scholar]
- 18.Robbins J., Dilworth S. M., Laskey R. A., Dingwall C., Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell 64, 615–623 (1991). [DOI] [PubMed] [Google Scholar]
- 19.Truant R., Cullen B. R., The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell. Biol. 19, 1210–1217 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panté N., Kann M., Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell 13, 425–434 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan L., et al. , Nuclear-targeted drug delivery of TAT peptide-conjugated monodisperse mesoporous silica nanoparticles. J. Am. Chem. Soc. 134, 5722–5725 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Xu C., et al. , Monodisperse magnetite nanoparticles coupled with nuclear localization signal peptide for cell-nucleus targeting. Chem. Asian J. 3, 548–552 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tammam S. N., Azzazy H. M. E., Breitinger H. G., Lamprecht A., Chitosan nanoparticles for nuclear targeting: The effect of nanoparticle size and nuclear localization sequence density. Mol. Pharm. 12, 4277–4289 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Misra R., Sahoo S. K., Intracellular trafficking of nuclear localization signal conjugated nanoparticles for cancer therapy. Eur. J. Pharm. Sci. 39, 152–163 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Anajafi T., et al. , Nuclear localizing peptide-conjugated, redox-sensitive polymersomes for delivering curcumin and doxorubicin to pancreatic cancer microtumors. Mol. Pharm. 14, 1916–1928 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Discher B. M., et al. , Polymersomes: Tough vesicles made from diblock copolymers. Science 284, 1143–1146 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Leong J., Teo J. Y., Aakalu V. K., Yang Y. Y., Kong H., Engineering polymersomes for diagnostics and therapy. Adv. Healthc. Mater. 7, e1701276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palivan C. G., et al. , Bioinspired polymer vesicles and membranes for biological and medical applications. Chem. Soc. Rev. 45, 377–411 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Tanner P., et al. , Polymeric vesicles: From drug carriers to nanoreactors and artificial organelles. Acc. Chem. Res. 44, 1039–1049 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Bermúdez H., Hammer D. A., Discher D. E., Effect of bilayer thickness on membrane bending rigidity. Langmuir 20, 540–543 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Einfalt T., et al. , Biomimetic artificial organelles with in vitro and in vivo activity triggered by reduction in microenvironment. Nat. Commun. 9, 1127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broz P., et al. , Cell targeting by a generic receptor-targeted polymer nanocontainer platform. J. Control. Release 102, 475–488 (2005). [DOI] [PubMed] [Google Scholar]
- 33.De Vocht C., et al. , Assessment of stability, toxicity and immunogenicity of new polymeric nanoreactors for use in enzyme replacement therapy of MNGIE. J. Control. Release 137, 246–254 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Mantha S., Qi S. H., Barz M., Schmid F., How ill-defined constituents produce well-defined nanoparticles: Effect of polymer dispersity on the uniformity of copolymeric micelles. Phys. Rev. Mater. 3, 026002 (2019). [Google Scholar]
- 35.Hill J. P., Shrestha L. K., Ishihara S., Ji Q., Ariga K., Self-assembly: From amphiphiles to chromophores and beyond. Molecules 19, 8589–8609 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stauch O., Schubert R., Savin G., Burchard W., Structure of artificial cytoskeleton containing liposomes in aqueous solution studied by static and dynamic light scattering. Biomacromolecules 3, 565–578 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Krüger D., Ebenhan J., Werner S., Bacia K., Measuring protein binding to lipid vesicles by fluorescence cross-correlation spectroscopy. Biophys. J. 113, 1311–1320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bacia K., Schwille P., Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat. Protoc. 2, 2842–2856 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Catimel B., et al. , Biophysical characterization of interactions involving importin-alpha during nuclear import. J. Biol. Chem. 276, 34189–34198 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Kapinos L. E., Huang B., Rencurel C., Lim R. Y. H., Karyopherins regulate nuclear pore complex barrier and transport function. J. Cell Biol. 216, 3609–3624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoch R. L., Kapinos L. E., Lim R. Y. H., Nuclear transport receptor binding avidity triggers a self-healing collapse transition in FG-nucleoporin molecular brushes. Proc. Natl. Acad. Sci. U.S.A. 109, 16911–16916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaillan-Huntington C., Braslavsky C. V., Kuhlmann J., Stewart M., Dissecting the interactions between NTF2, RanGDP, and the nucleoporin XFXFG repeats. J. Biol. Chem. 275, 5874–5879 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Bernhard W., A new staining procedure for electron microscopical cytology. J. Ultrastruct. Res. 27, 250–265 (1969). [DOI] [PubMed] [Google Scholar]
- 44.Liu J., et al. , DNA-mediated self-organization of polymeric nanocompartments leads to interconnected artificial organelles. Nano Lett. 16, 7128–7136 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Yin H., et al. , Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15, 541–555 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Allen T. M., Cullis P. R., Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Rev. 65, 36–48 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Schleicher K. D., et al. , Selective transport control on molecular velcro made from intrinsically disordered proteins. Nat. Nanotechnol. 9, 525–530 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Tu L. C., Fu G., Zilman A., Musser S. M., Large cargo transport by nuclear pores: Implications for the spatial organization of FG-nucleoporins. EMBO J. 32, 3220–3230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paci G., Lemke E. A., Molecular determinants of large cargo transport into the nucleus. bioRxiv:10.1101/695080 (6 July 2019). [DOI] [PMC free article] [PubMed]
- 50.Au S., Panté N., Nuclear transport of baculovirus: Revealing the nuclear pore complex passage. J. Struct. Biol. 177, 90–98 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Rabe B., Vlachou A., Panté N., Helenius A., Kann M., Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc. Natl. Acad. Sci. U.S.A. 100, 9849–9854 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim R. Y. H., et al. , Nanomechanical basis of selective gating by the nuclear pore complex. Science 318, 640–643 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Onischenko E., et al. , Natively unfolded FG repeats stabilize the structure of the nuclear pore complex. Cell 171, 904–917.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the paper and SI Appendix.