Abstract
While single-cell sequencing technologies have revealed tissue heterogeneity, resolving mixed cellular libraries into cellular clones is essential for many pooled screens and clonal lineage tracing. Fluorescent proteins are limited in number, while DNA barcodes can only be read after cell lysis. To overcome these limitations, we used influenza virus hemagglutinins to engineer a genetically encoded cell-surface protein barcoding system. Using antibodies paired to hemagglutinins carrying combinations of escape mutations, we developed an exponential protein barcoding system which can label 128 clones using seven antibodies. This study provides a proof of principle for a strategy to create protein-level cell barcodes that can be used in vivo in mice to track clonal populations.
Keywords: virology, cell barcoding, protein engineering
The recent explosion of single-cell analyses has revealed the heterogeneity underlying the development of normal and pathological tissues. The ability to track individual cellular clones in complex mixtures is essential for experimental validation of results from observational single-cell studies. While DNA barcoding allows labeling of extremely complex (>109) mixtures (1), it is limited by the need to lyse cells for barcode readout. Fluorescent labeling allows for nondestructive clonal tracking but is limited by the repertoire of fluorescent proteins (2). Recent work from the Blainey group using optical phenotyping is an impressive expansion of the phenotyping possibilities of pooled screens, but the sequence-based barcode readout still cannot be performed on viable cells (3). We have reported cell barcoding using linear epitopes fused to a cell-surface protein (Pro-Codes) which enables the barcoding and analysis of 364 cell populations with single-cell resolution (4).
To expand the tools available, we investigated the use influenza virus hemagglutinin (HA) as a cell-surface accessible barcode. The selection of influenza virus HA was driven by the ability of the virus to escape the adaptive immune response via antigenic drift. We engineered a library of HA variant cell-surface barcodes which require only seven antibodies for analysis. This approach allows for an exponential increase in barcode diversity, with each additional antibody–escape mutation pair doubling library size.
Results
Selection of the Scaffold Protein, HA, from an Influenza A Virus.
When searching for a protein to serve as a programmable cell-surface barcode we began with several guiding principles: 1) surface expression, 2) antibodies targeting exposed epitopes, 3) loss of antibody binding with point mutations, 4) robustness to mutational burden, and 5) minimal cross-reactivity between mutations. We selected the HA of the A/Shanghai/1/13 (H7N9) virus, a recently characterized potential pandemic strain (5, 6), for its use as a cell-surface barcode.
HA Expression and Characterization of Escape Mutants.
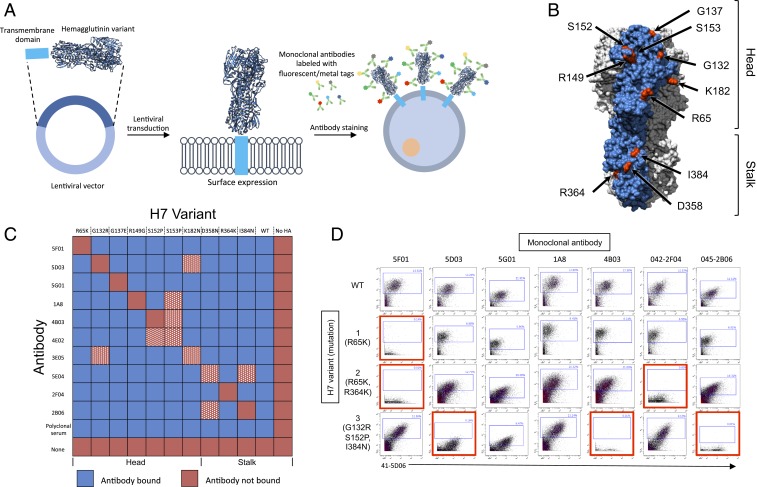
We introduced H7-HA into HEK-293T cells using a lentivirus and measured expression with extracellular antibody staining (Fig. 1A). We selected 10 published antibody–escape mutation pairs (7, 8), with seven mutations in the HA head and three in the stalk (Fig. 1B). The corresponding escape mutations to monoclonal antibodies 07-5F01, 07-5D03, 07-5G01, 1A8, 07-4B03,07-4E02, 07-3E05, 41-5E04, 42-2F04, and 45-2B06 are R65K, G132R, G137E, R149G, S152P, S153P , K182N, D358N, R364K, and I384N, respectively. Mutant cross-reactivity was determined in all 100 escape mutant–antibody pairs (Fig. 1C) and we selected seven pairs that showed no cross-reactivity (Fig. 1C). We then tested the tolerance of H7-HAs to multiple mutations and generated H7 HA variants containing one to three escape mutations. All tested variants showed excellent cell-surface expression and staining with antibodies targeting nonmutated residues (Fig. 1D).
Fig. 1.
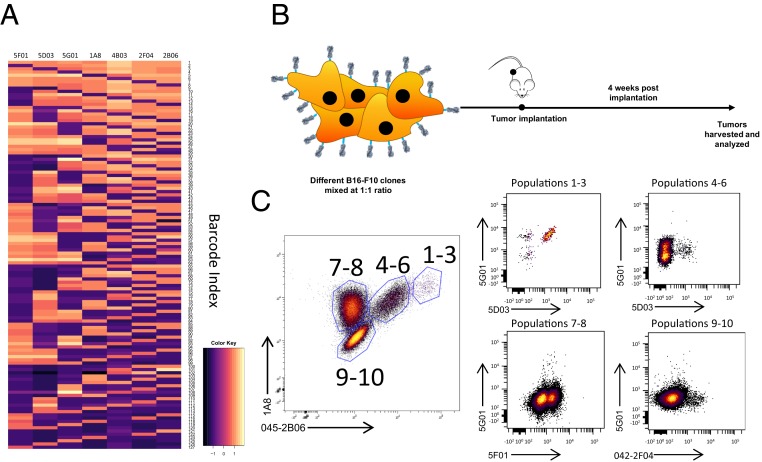
(A) Schematic of barcoding approach. (B) Modeling of A/Shanghai/1/2013 H7 HA was done using PyMOL (Protein Data Bank ID code 4LN3). Escape mutation sites highlighted in red. Single monomer highlighted in blue. (C) Heat map of monoclonal antibody binding to HEK-293T cells transfected with H7 escape mutants. Antibody–escape mutant pairs removed from the library due to cross-reactivity in white and red hatched squares. (D) Wild-type or H7 variants analyzed by mass cytometry (CyTOF). Loss of antibody binding highlighted in red.
HA Barcoding Resolves >100 Populations with Single-Cell Resolution.
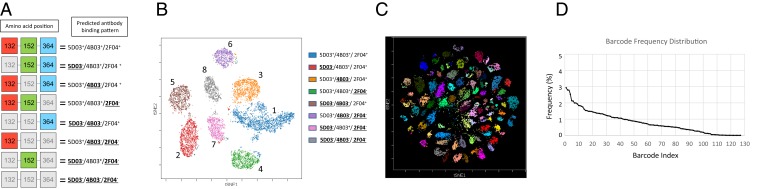
To test our ability to distinguish a mixture of HA-labeled cells we constructed a small-scale barcode library where 3/7 escape mutation positions were combinatorially mutated, leaving 23 (8) HA variants (Fig. 2A). HEK-293T transduced with this pool were easily deconvoluted into eight populations that were distinguishable when plotted in three dimensions (Movie S1). Due to the difficulty in visualizing high-dimensional data using two-dimensional cytometry plots, this minilibrary was also visualized using the viSNE dimensionality reduction algorithm (9, 10), which also showed eight distinct populations corresponding to each HA barcode (Fig. 2B). Next, we generated a library of HA barcodes where all seven escape mutation positions were combinatorially mutated, leading to 27(128) possible HA barcodes. Remarkably, in HEK-293T cells transduced with this library we found that 103 HA variants could be readily identified, with most barcodes showing excellent cluster separation (Fig. 2C), with an even distribution of HA barcodes (Fig. 2D). We also generated a binding heat map for each barcode detected, enabling a granular analysis of the binding of each antibody to each barcode (Fig. 3A).
Fig. 2.
(A) Predicted staining pattern of eight-member H7 library. Mutated positions in gray. (B) viSNE analysis of minilibrary in HEK-293T cells. (C) viSNE analysis of full H7 barcode library in HEK-293T cells. (D) Frequency distribution of detected HA barcodes.
Fig. 3.
(A) Heat map of antibody binding in each identified HA barcode. (B) Schematic of B16 implantation. (C) Fluorescence-activated cell sorter analysis of B16 tumor cells 4 wk postimplantation. Gate names correspond to unique populations identified.
Tumor Cells Expressing HA Barcodes Persist in Immunocompetent Mice.
To test our HA barcodes in vivo, we generated stable cell lines of 10 HA-barcoded B16 melanoma cells, which were mixed and implanted in immunocompetent C57BL/6 mice (Fig. 3B). Notably, tumors were found to maintain HA surface expression after 4 wk of in vivo growth (Fig. 3C)
Discussion
We engineered the influenza virus HA protein to create a library of H7 variants that serve as cell-surface barcodes which can be analyzed with a small number of antibodies. We constructed an exponentially large repertoire of protein variants that can function as barcodes, with N escape mutation–antibody pairs leading to 2N potential barcodes. We used seven escape mutant–antibody pairs (Fig. 1C) that did not exhibit any cross-reactivity with each other and generated a library of >100 HA barcodes. Since influenza viruses can express HAs of different subtypes with minimal antibody cross-reactivity, if a second HA with seven distinct epitopes were added that would allow for 16,384 (214) unique barcodes, showing that each barcode can effectively label a cell of interest and that the library can be used in a highly multiplexed manner with a simple extracellular staining protocol. We also successfully implanted HA barcode-labeled B16 tumor cells into immune-competent mice and were able to detect HA expression after 4 wk of growth, making this system potentially useful for in vivo tracking.
Our HA barcodes are easily able to maintain expression with multiple mutations introduced. This is likely due to HA’s increased tolerance to mutations as a result of strong selective pressure. Moreover, the generalizability of our approach is an added advantage, which utilizes large collections of antibodies and escape mutants available for other HA family members. Each additional antibody–escape mutation pair introduced doubles the number of barcodes available, allowing the construction of extremely complex protein barcode libraries that can be resolved using small numbers of antibodies. An extracellular protein-based cell barcoding system provides a tool for studying complex mixtures of living cells at single-cell resolution. An application for our system is the clonal tracking of tumor growth. The HA barcoding system could allow for a large expansion in the number of individual clones that can be tracked by purely extracellular staining and allows for tracked clones to be recovered alive and further studied, providing a distinct approach to labeling and studying complex cellular mixtures.
Materials and Methods
HA H7 Library Preparation.
All H7 constructs were cloned into a lentiviral vector downstream of a CAG promoter. The HA barcode library was generated via assembly PCR using gBlocks ordered from IDT (Integrated DNA Technologies, Inc.); see Dataset S2 for all sequences. All PCRs were performed using Phusion High-Fidelity PCR Master Mix (Thermo Fisher Scientific) with 5 μM of each primer, according to manufacturer instructions.
Vector Production and Transduction.
Lentiviral vectors were produced and cells were transduced as previously described (11). Twenty-four hours prior to lentiviral transduction, 5 × 104 cells per well were seeded in a six-well plate and transduced the following day.
Cell Culture.
HEK-293T cells were maintained as previously described (11). B16-F10 cells were maintained in DMEM-F12 supplemented with 10% fetal bovine serum (FBS) and 1% penicillin + streptomycin.
Monoclonal Antibodies.
Monoclonal antibodies were generated as previously described (7, 12).
Flow Cytometry and Mass Cytometry.
For antibody staining, cells were blocked in phosphate-buffered saline with 2% FBS for 15 min at 4 °C and incubated with antibody mix for 25 min at 4 °C, washed, and analyzed on either the BD Fortessa or BD LSRII (Becton Dickinson). For mass cytometry, cells were stained for viability with Cell-ID Intercalator-103Rh for 15 min at 37 °C prior to antibody staining and were then analyzed as previously described (4). Analysis was performed using CytoBank or FlowJo Software (FlowJo, LLC).
Melanoma Tumor Model.
Female C57BL/6 mice were purchased from The Jackson Laboratory. All animal experiments were performed in accordance with the Icahn School of Medicine Animal Care and Use Committee. Four- to 6-wk-old female mice were anesthetized by intraperitoneal injections of ketamine/xylazine and implanted intradermally with 100,000 B16-F10 cells. Four weeks postimplantation tumors were harvested, minced, incubated with 1.67 Wunsch U/mL Liberase and 0.2 mg/mlDNase for 30 min at 37 °C, homogenized by pipetting, and filtered through a 70-mm nylon filter. Cells were returned to culture prior to staining and flow cytometry analysis.
Data Availability.
See Datasets S1 and S2 for mass cytometry data and H7 HA sequences.
Supplementary Material
Acknowledgments
Work in the P.P. and F.K. laboratories was supported by National Institute of Allergy and Infectious Diseases grant R01 AI128821 and Centers of Excellence for Influenza Research and Surveillance contract HHSN272201400008C. Work in the B.D.B. laboratory was supported by NIH grant R33CA223947 and a Technology Award from the Cancer Research Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919182117/-/DCSupplemental.
References
- 1.Bhang H. E., et al. , Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 21, 440–448 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Livet J., et al. , Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Feldman D., et al. , Optical pooled screens in human cells. Cell 179, 787–799.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wroblewska A., et al. , Protein barcodes enable high-dimensional single-cell CRISPR screens. Cell 175, 1141–1155.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvier N. M., Palese P., The biology of influenza viruses. Vaccine 26 (suppl. 4), D49–D53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao R., et al. , Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368, 1888–1897 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Henry Dunand C. J., et al. , Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe 19, 800–813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadlbauer D., Amanat F., Strohmeier S., Nachbagauer R., Krammer F., Cross-reactive mouse monoclonal antibodies raised against the hemagglutinin of A/Shanghai/1/2013 (H7N9) protect against novel H7 virus isolates in the mouse model. Emerg. Microbes Infect. 7, 110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Maaten L., Hinton G., Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
- 10.Amir A. D., et al. , viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullokandov G., et al. , High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods 9, 840–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith K., et al. , Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 4, 372–384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
See Datasets S1 and S2 for mass cytometry data and H7 HA sequences.