Significance
Genetic modification of plant cell wall polymers is key to improvement of lignocellulosic biomass for forage, fuel, and renewable chemicals. However, such modifications can often lead to ectopic activation of defense responses and reductions in biomass yield. Here, we show that defense gene induction in transgenic Arabidopsis thaliana with altered lignin content and composition through down-regulation of two different lignin pathway enzymes results from the ectopic expression of a pectin-degrading enzyme in vascular tissue, leading to release of cell wall epitopes that serve as signals for defense gene activation. This knowledge provides a basis for uncoupling lignin modification from ectopic defense gene induction.
Keywords: cell wall remodeling, lignin modification, defense response, elicitor, polygalacturonase
Abstract
There is considerable interest in engineering plant cell wall components, particularly lignin, to improve forage quality and biomass properties for processing to fuels and bioproducts. However, modifying lignin content and/or composition in transgenic plants through down-regulation of lignin biosynthetic enzymes can induce expression of defense response genes in the absence of biotic or abiotic stress. Arabidopsis thaliana lines with altered lignin through down-regulation of hydroxycinnamoyl CoA:shikimate/quinate hydroxycinnamoyl transferase (HCT) or loss of function of cinnamoyl CoA reductase 1 (CCR1) express a suite of pathogenesis-related (PR) protein genes. The plants also exhibit extensive cell wall remodeling associated with induction of multiple cell wall-degrading enzymes, a process which renders the corresponding biomass a substrate for growth of the cellulolytic thermophile Caldicellulosiruptor bescii lacking a functional pectinase gene cluster. The cell wall remodeling also results in the release of size- and charge-heterogeneous pectic oligosaccharide elicitors of PR gene expression. Genetic analysis shows that both in planta PR gene expression and release of elicitors are the result of ectopic expression in xylem of the gene ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1), which is normally expressed during anther and silique dehiscence. These data highlight the importance of pectin in cell wall integrity and the value of lignin modification as a tool to interrogate the informational content of plant cell walls.
Plant cell wall polymers are cross-linked in the wall matrix. The nature of this cross-linking regulates plant growth and serves as a sensor between the cell cytoplasm and the environment. Alterations in cell wall integrity affect cell wall architecture and trigger compensatory changes in cell wall properties (1). Lignin is a major polymer in secondary cell walls, and engineered plants with low lignin levels have reduced biomass recalcitrance, leading to enhanced sugar release for biofuel production and improved forage digestibility (2). However, modification of lignin content and/or composition can result in severe defects in plant growth (2–5) and alterations in plant immunity manifested as either enhanced susceptibility (6) or enhanced resistance through activation of endogenous defense pathways (7, 8). The molecular mechanisms underlying how lignin modifications are perceived in the cell wall and the subsequent signals that are transduced remain unknown. Understanding these is of critical importance for developing improved forages and sources of new bioproducts and fuels.
The “oligosaccharin hypothesis” (9) was first proposed to explain how specific fungal cell wall structures elicit plant defenses (10, 11). It was later expanded (see reviews: refs. 12 and 13) to include plant cell wall-derived oligosaccharides, now referred to as part of a larger group of molecules known as damage-associated molecular patterns (DAMPs), and bacterial lipooligosaccharides, both of which can trigger defense responses and/or impact plant growth and development (14–17). Release of DAMPs triggers the biosynthesis of stress hormones such as salicylic acid (SA) (18), jasmonic acid (19), and ethylene (20), and the generation and accumulation of reactive oxygen species (21). These signals can, in turn, lead to the production of antimicrobial metabolites such as phytoalexins (22), or the synthesis of defense response proteins such as pathogenesis-related (PR) proteins (23), including defensins (24). The defense-inducing plant cell wall-derived DAMPs that have been structurally characterized, to date, are either β-1,3 glucans (25) or α-1,4 oligogalacturonides (OGs, mainly pectic homogalacturonan [HG] fragments) (14, 26–28). A putative OG receptor has also been discovered (28).
Cell walls of alfalfa plants with reduced lignin levels resulting from down-regulation of hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase (HCT) show increased extractability of pectic elicitors of PR protein-encoding transcripts (7). Elicitors of different sets of defense response genes are generated in cell walls of Arabidopsis thaliana with lignin composition altered through up- or down-regulation of the late lignin pathway enzyme ferulate 5-hydroxylase (F5H) (29). These elicitors have yet to be structurally characterized, and whether their release is a direct or indirect result of altered cell wall structure or integrity is unclear. Here, we utilize A. thaliana lines independently modified in expression of HCT or cinnamoyl CoA reductase (CCR), the penultimate enzyme in monolignol biosynthesis, to probe biochemically and genetically the links between lignin content, cell wall integrity, and defense signaling. These lines exhibit extensive but differential transcriptional reprogramming, but share constitutive expression of many PR genes associated with extensive cell wall remodeling. PR gene expression and increased extractability of cell wall-derived elicitors of PR genes are a result of the ectopic expression of the ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1). Our data highlight the importance of pectin for defense signaling, and place active cell wall remodeling as a central process in the release of pectin-derived DAMPs in lignin-modified plants.
Results
Defense Response Genes Are Ectopically Expressed in the HCT-RNA Interference Line and the ccr1 Mutant of Arabidopsis.
Consistent with the position of HCT in the monolignol pathway (SI Appendix, Fig. S1), reduction of this enzyme’s expression leads to a large reduction in levels of both guaiacyl (G, derived from coniferyl alcohol) and syringyl (S, derived from sinapyl alcohol) units in lignin, which is therefore highly enriched in H units derived from p-coumaryl alcohol (3, 30). Loss of function of CCR 1 (CCR1) in Arabidopsis results in large reductions in both G and S units, with a significant but less extreme increase in H units (31). The changes in lignin content and composition in the Arabidopsis lines used in the present work were confirmed by thioacidolysis (see below) and microscopic analysis of inflorescence stems for ultraviolet autofluorescence and Mäule staining (SI Appendix, Fig. S2 A and B). Altered lignin content and composition were seen in both xylem vessels and fibers. The ccr1 mutant plants used here were semidwarf, and the HCT-RNA interference (RNAi) lines also showed the previously reported reductions in plant stature (32).
HCT down-regulation induces expression of PR proteins in A. thaliana (7). To determine whether loss of function of CCR1 induces similar defense responses, we performed Affymetrix microarray analysis on RNA extracted from stems of wild-type, HCT-RNAi, and ccr1 mutant plants (Dataset S1). The ccr1 and HCT-RNAi lines shared similar numbers of differentially up-and down-regulated genes (SI Appendix, Fig. S3), including a number of defense response genes (Table 1 and Dataset S1). There were fewer strongly down-regulated genes (more than 10-fold difference from wild type) in the ccr1 mutant than in the HCT-RNAi line (Dataset S1). Reduction of HCT expression by ∼90% (Dataset S1) led to large-fold increases in levels of transcripts encoding the PR genes PR1, PR10, PR13, PR2 (β-glucanase), PR5 (thaumatin-like), and PR14 (lipid transfer protein), other genes associated with biotic stress and SA-mediated responses, and a phosphate cotransporter responsive to abiotic stress (Table 1 and Dataset S1). Loss of function of CCR1 also led to massive induction of a set of genes associated with seed development and lipid transport/storage (albumin, oleosin, and lipid transfer protein), many of which were not induced in the HCT-RNAi line (Dataset S1). PR1, PR2, PR13, and PR14 were highly overexpressed in both lines, although PR5 was down-regulated in the ccr1 mutant as opposed to its near 50-fold induction in the HCT-RNAi line (Table 1). MapMan analysis (SI Appendix, Fig. S4 A and B) showed that, although both Arabidopsis lines ectopically expressed genes in the same ontology categories, the differentially expressed genes in each category were often different.
Table 1.
The 16 most highly up-regulated defense response genes in A. thaliana HCT-RNAi transgenic lines and ccr-1 mutant plants, as compared to wild type (WT)
| Gene ID | HCT-RNAi/WT ratio | ccr-1/WT ratio | Annotation |
| AT4G23600 | 165.38 | 43.27 | CORI3, CORONATINE INDUCED 1, induced by phytotoxin coronatine, response to wounding, salt, ABA and JA |
| AT5G42800 | 50.44 | 10.46 | Dihydroflavonol reductase, conversion of dihydroquercetin to leucocyanidin; oxidative and abiotic stress response |
| AT1G75040 | 49.26 | 0.25 | PR-5; thaumatin-like protein involved in response to pathogens |
| AT3G28360 | 48.32 | 8.17 | P-glycoprotein 16 (PGP16) ABC transporter, transmembrane domain |
| AT1G80130 | 43.27 | 10.43 | Tetratricopeptide repeat (TPR)-like superfamily protein; response to oxidative stress |
| AT3G04710 | 42.37 | 3.20 | PR-10; tetratricopeptide (TPR) protein with potential to interact with Hsp90/Hsp70 as cochaperone |
| AT2G14610 | 40.26 | 20.56 | PR-1; salicylic-acid responsive; it is a useful molecular marker for the SAR response |
| AT3G57510 | 35.04 | 90.23 | Encodes ADPG1, a PGase protein involved in silique and anther dehiscence |
| AT3G06340 | 35.64 | 2.30 | DNAJ heat shock N-terminal domain-containing protein |
| AT2G38530 | 34.27 | 85.21 | Lipid transfer between membranes; predicted to be a member of PR-14 PR protein family |
| AT3G57260 | 32.86 | 19.37 | PR-2; beta 1,3-glucanase |
| AT1G56650 | 32.61 | 7.06 | Putative MYB domain containing transcription factor involved in anthocyanin metabolism; response to salt stress |
| AT4G24260 | 31.25 | 1.50 | Endo-1,4-β-glucanase; KOR3 is induced by nematodes and is expressed in syncitia induced by Heterodera schachtii |
| AT3G28220 | 28.46 | 4.98 | TRAF-like family protein; response to salt stress |
| AT4G37980 | 27.28 | 18.32 | Elicitor-activated gene 3-1 (ELI3-1); plant-type hypersensitive response |
| AT1G72260 | 23.72 | 89.19 | PR-13; encodes a thionin which is a cysteine-rich protein having antimicrobial properties; Thi2.1 |
Transcriptome analysis was by DNA microarray analysis. The list of genes is extracted from Dataset S1.
Cell Wall Remodeling Genes Are Ectopically Expressed in Reduced Lignin Plants.
Many cell wall metabolism genes, including a number involved in pectin degradation, were differentially up-regulated in the two lignin-modified lines (Table 2 and SI Appendix, Table S1). For example, ADPG1 (gene number AT3G57510), previously ascribed a role in silique and anther dehiscence and expressed during cell separation processes in wild-type plants (ref. 33 and SI Appendix, Fig. S5A), represented one of the most highly induced (by over 90-fold) genes in the ccr1 transcriptome (Table 2 and SI Appendix, Fig. S5B). This same gene was induced 30-fold in the HCT-RNAi line, along with multiple pectate lyases and pectin acetyl/methyl esterases (Table 2 and SI Appendix, Fig. S5B). An unannotated gene, AT1G64405, encoding a 118-amino acid protein with a serine-rich motif known to be strongly expressed in abscission zones (34), was also induced in both ccr1 (3.92-fold) and HCT-RNAi (2.17-fold) lines. Remarkably, however, other than ADPG1 and the peroxidase AT5G51890, none of the cell wall metabolism enzyme genes up-regulated in the HCT-RNAi line was up-regulated in the ccr1 mutant, and vice versa (Table 2 and SI Appendix, Table S1).
Table 2.
Pectin modifying genes differently expressed in HCT-RNAi and ccr-1 mutant plants
| Gene ID | HCT-RNAi/WT ratio | ccr-1/WT ratio | Annotation |
| AT3G57510 | 35.04 | 90.23 | Encodes ADPG1, a PGase protein involved in silique and anther dehiscence |
| AT3G27400 | 3.74 | Pectin lyase-like superfamily protein | |
| AT1G57590 | 3.55 | Pectinacetylesterase family protein | |
| AT5G47500 | 3.15 | Pectin lyase-like superfamily protein | |
| AT1G10640 | 3.12 | Pectin lyase-like superfamily protein | |
| AT5G49180 | 14.18 | Encodes a putative pectin methylesterase; the gene is preferentially expressed in floral buds | |
| AT1G70720 | 10.30 | Plant invertase/pectin methylesterase inhibitor superfamily protein | |
| AT5G07410 | 8.89 | Pectin lyase-like superfamily protein | |
| AT1G56100 | 8.85 | Plant invertase/pectin methylesterase inhibitor superfamily protein | |
| AT2G36710 | 7.75 | Pectin lyase-like superfamily protein | |
| AT4G15750 | 7.72 | Plant invertase/pectin methylesterase inhibitor superfamily protein | |
| AT5G07430 | 4.15 | Pectin lyase-like superfamily protein | |
| AT3G07820 | 3.63 | Pectin lyase-like superfamily protein | |
| AT1G48100 | 3.62 | Pectin lyase-like superfamily protein | |
| AT1G02790 | 3.25 | Encodes a exopolygalacturonase | |
| AT5G19730 | 3.13 | Pectin lyase-like superfamily protein | |
| AT5G48140 | 2.52 | Pectin lyase-like superfamily protein |
Glycome Epitope Profiling Reveals Extensive Cell Wall Remodeling in Reduced Lignin Plants.
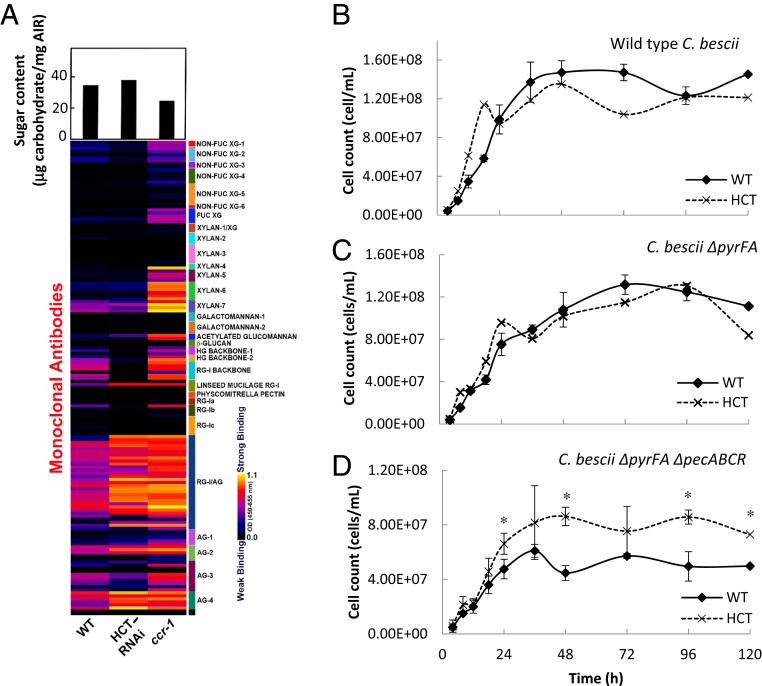
To determine whether the changes in lignin content/composition and/or induction of cell wall-degrading enzymes affected cell wall integrity in the reduced lignin lines, we first examined changes in water-extractable cell wall glycan epitopes by screening the unprocessed water extracts from the HCT-RNAi and ccr1 lines with an array of monoclonal antibodies (35) directed against diverse noncellulosic plant cell wall glycan epitopes. The water extracts of cell walls from the two mutant plant lines were highly enriched in a range of arabinogalactan and rhamnogalacturonan I (RG-I) epitopes when compared to the extracts from the wild-type plants (Fig. 1A). The water extracts of the ccr1 line showed high contents of RG-I backbone, HG, and xylan (Fig. 1A) epitopes. In order to identify overall changes in cell wall structure and glycan composition resulting from the lignin modifications, glycome profiling of inflorescence stem cell walls (alcohol-insoluble residues [AIRs]) was carried out by sequentially extracting the walls with a range of solvent treatments of increasing harshness and screening the neutralized and dialyzed wall extracts with the monoclonal antibody toolkit (SI Appendix, Fig. S6). The resulting glycome profiles showed several noteworthy changes, especially for ccr1. The most distinctive change was a dramatic shift in components containing RG/arabinogalactan epitopes from the harshest extract (postchlorite 4M KOH) to the less harsh treatments (oxalate and carbonate) in both lignin-modified lines compared to the wild type (SI Appendix, Fig. S6, green dotted boxes). In addition, xylan epitopes (both backbone and side-chain epitopes) and pectic backbone epitopes (both HG and RG-I) were more prominent in these early cell wall extracts (SI Appendix, Fig. S6, white dotted boxes). Diverse xyloglucan epitopes also were more easily extractable from the cell walls of the two mutant lines, especially for ccr1, than observed for wild-type walls. Finally, ccr1 cell walls were less tightly integrated, as indicated by the higher recovery of cell wall carbohydrates in wall extracts compared with wild type (SI Appendix, Fig. S6, bar graphs).
Fig. 1.
Cell wall remodeling in reduced lignin Arabidopsis lines. (A) (Lower) Glycome analysis of cell wall polysaccharides from water extracts of cell walls of control, HCT-RNAi, and ccr1 mutant Arabidopsis stems. The nondialyzed water-soluble cell wall extracts were screened against a panel of 155 monoclonal antibodies directed against diverse epitopes in noncellulosic plant glycans. The resulting heat map depicts antibody binding strength based on optical density (OD) depicted as a color gradient ranging from black (no binding) to yellow (strongest binding), as indicated by the key at the lower right. Antibodies are grouped into clades according to the glycans that they predominantly recognize, as indicated by the panel on the right side of the glycome profiles. Upper shows carbohydrate recovery from water-extracted AIRs. Details of monoclonal antibodies are given in ref. 29. (B–D) Lignin modification alleviates the need for pectinase action to enable a cellulolytic bacterium to access Arabidopsis biomass. (B) Growth of wild-type (WT) C. bescii on ground biomass from WT and HCT-down-regulated Arabidopsis plants. (C) Growth of C. bescii JWCB005 (ΔpyrFA). (D) Growth of C. bescii JWCB010 (ΔpyrFA ΔpecABCR). Cells were collected and stained with acridine orange at the times shown and counted using an epifluorescence microscope with a counting chamber lens. Two biological replicates were taken. Asterisks indicate significant differences from WT (P < 0.05) by pairwise multiple comparison Tukey test.
Next, the components of the water extracts from the AIRs were separated by anion exchange chromatography coupled with ELISA-based epitope detection (Epitope Detection Chromatography [EDC]), to examine heterogeneity of the released polysaccharides (SI Appendix, Supplementary Materials and Methods). The fractions were eluted from the anion exchange column by a step gradient of 20%, 30%, and 40% followed by 40 to 100% of 0.6 M sodium chloride, and epitopes were determined by ELISA using a bank of monoclonal antibodies (SI Appendix, Fig. S7). The 20% fraction from both the HCT-RNAi and ccr1 lines contained a large new peak of xylan antibody-reactive material (SI Appendix, Fig. S7C), and smaller peaks were found in the 30% and 40% fractions. Overall, glycome profiling revealed major changes in pectin and xylan extractability from cell walls of the HCT-RNAi and ccr1 mutant lines, confirming extensive cell wall remodeling.
Lignin Modification Removes the Requirement for Pectin Degradation for Access to Cell Wall Polysaccharides.
Targeting pectin modification alone can strongly reduce recalcitrance of lignocellulosic biomass (36), and the cellulolytic thermophilic bacterium Caldicellulosiruptor bescii cannot grow on plant biomass in the absence of a functional pectinase gene cluster (37). Because the Arabidopsis lines studied here exhibit elevated endogenous pectinase gene expression, we employed C. bescii in a bioassay to examine the impacts of lignin modification in HCT-RNAi plants on cell wall remodeling in the context of accessibility of cell walls to microbial deconstruction. In a preliminary experiment, ground biomass from wild-type and HCT-antisense alfalfa plants described previously (7) was compared as the carbon source for growth of wild-type C. bescii, a control auxotrophic strain (ΔpyrFA) used for gene disruption, and the same strain in which the organism’s pectinase gene cluster had been ablated (ΔpecABCR) (SI Appendix, Fig. S8). Cell counts for wild-type C. bescii and the ΔpyrFA mutant increased in a similar manner over the duration of the experiment (SI Appendix, Fig. S8 A and B). However, growth of the pectinase cluster deletion mutant was strongly reduced compared to wild type when the bacteria were grown on wild-type alfalfa biomass, but achieved that of wild-type bacteria when grown on biomass from HCT-antisense alfalfa (SI Appendix, Fig. S8C).
We then examined the growth of the C. bescii strains on HCT-RNAi (Fig. 1 B–D) and ccr1 mutant Arabidopsis biomass (SI Appendix, Fig. S9). Growth of wild-type or ΔpyrFA bacteria was essentially the same on wild-type or HCT down-regulated Arabidopsis biomass (Fig. 1 B and C). However, as observed for alfalfa (SI Appendix, Fig. S8 A–C), down-regulation of HCT allowed the pectinase cluster mutant to achieve higher growth than on cell walls from wild-type plants (Fig. 1D). The same was true for growth of wild-type and mutant C. bescii on ccr1 mutant biomass (SI Appendix, Fig. S9). As a control, we examined biomass from additional Arabidopsis lines with loss of function or overexpression of F5H; these lines, in which lignin composition but not content was affected, do not constitutively express PR genes or ADPG1 (29). Biomass from none of these lines could support wild-type growth of the pectinase deletion mutant (SI Appendix, Fig. S8 D–I). Together, these data suggest that ectopic activation of endogenous pectin degrading enzymes, including pectin lyases and ADPG1, causes cell wall remodeling that overcomes the requirement for an active pectinase gene cluster to enable growth of C. besciii on plant biomass.
Elicitors of PR Proteins Are Released from Cell Walls of Reduced Lignin Plants.
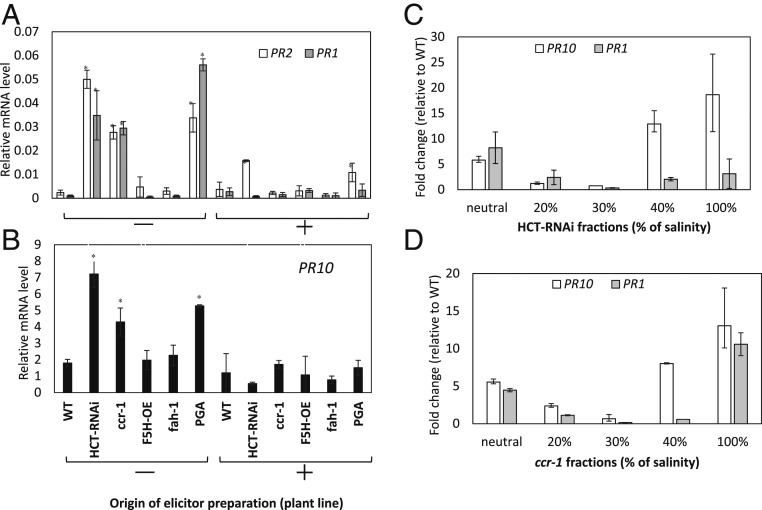
To determine whether the cell wall material that exhibited enhanced extractability in the HCT-RNAi and ccr1 Arabidopsis lines contained signal molecules for induction of PR genes, we prepared AIRs from stems of lignin-modified and control plants and extracted them in water at room temperature. Extracts were then added to Arabidopsis suspension cells that were harvested after 12 h for RNA extraction and determination of PR1, PR2, and PR10 transcript levels. Polygalacturonic acid (PGA) was included as a positive control elicitor, and this and water-soluble extracts from the plant cell walls were also pretreated with commercial polygalacturonase (PGase). PGA induced the three PR genes, and this induction was strongly reduced if the PGA was pretreated with PGase (Fig. 2 A and B). Extracts from cell walls of the HCT-RNAi and ccr1 lines induced PR1, PR2, and PR10 transcripts to well above the levels in cells treated with the water-soluble fraction of cell walls from wild-type plants (Fig. 2 A and B). Pretreatment of the extracts with PGase fully or partially destroyed the ability to induce PR genes (Fig. 2 A and B). The elicitor activity in the extracts exhibited some specificity for PR gene induction. For example, neither the seed storage albumin (SESA2) and 12S globulin genes (CRA2) that are highly induced in the ccr1 mutant nor the glucose 6-phosphate/phosphate cotransporter (GPT2) that is highly induced in the HCT-RNAi line (Dataset S1) was induced by the water-soluble extracts from either the HCT-RNAi or ccr1 mutant line (SI Appendix, Table S2). ADPG1 was also not induced, and CYP81D11, a gene that is strongly induced by elicitors released from F5H overexpressing Arabidopsis (29), was repressed by the water-soluble elicitors (SI Appendix, Table S2).
Fig. 2.
Defense gene expression in Arabidopsis cell cultures in response to water-soluble elicitors from cell walls of wild-type (WT), HCT-RNAi, ccr1, and f5h (fah-1) mutant and F5H overexpressor (OE) Arabidopsis plants. (A) PR1 and PR2 induction in response to crude elicitors from the plant lines shown. (B) PR10 induction in response to crude elicitors from the plant lines shown. (C) PR10 and PR1 induction by ion exchange fractionated elicitors from cell walls of HCT-RNAi plants. (D) PR10 and PR1 induction by ion exchange fractionated elicitors from cell walls of ccr1 plants. The elicitor activity of selected fractions was determined by measuring their ability to induce defense gene transcripts (PR2, PR10, PR1) in cell suspension cultures. Analysis of transcript levels in cell cultures was by qRT-PCR performed with total messenger RNA (mRNA) from suspension cells harvested 12 h postelicitation, and incubated in the dark at 25 °C. Transcript levels are expressed relative to AtPP2A. Results are means ± SD of three biological replicates. Asterisks in A and B indicate significant differences from WT (P < 0.05) by pairwise multiple comparison Tukey test. Elicitor extracts were prepared from the AIR fraction of cell walls. Extracts were added directly to cell cultures (−), or pretreated with PGase (+). PGA was also tested as elicitor for a positive control. Elicitor fractions are as shown in SI Appendix, Fig. S7 A–D.
The fractions from anion exchange chromatography of the released elicitor material (SI Appendix, Fig. S7) were desalted, and tested for their ability to induce defense genes in Arabidopsis suspension cells (Fig. 2 C and D). Ability to induce PR1 and PR10 was found in multiple fractions (neutral, 40%, and 40 to 100% salt) from the water extracts of cell walls from the HCT-RNAi and ccr1 lines. Notably, the 20% and 30% fractions that contained released xylan epitopes (SI Appendix, Fig. S7C) did not exhibit much elicitor activity (Fig. 2 C and D). The water-extracted AIRs from the wild-type and HCT-RNAi lines were then separated on the basis of size by gel permeation chromatography (SI Appendix, Fig. S10A). The two elution profiles were quantitatively and qualitatively different, with the extract from the HCT-RNAi line containing more pectic material and additional peaks of lower molecular weight when compared to the wild type (SI Appendix, Fig. S10A). Some pectic material eluted after the galacturonic acid standard, suggesting that its nature caused it to stick to the gel permeation column. Three peaks with highest uronic acid content plus one late-eluting peak from the HCT-RNAi line were assayed for elicitor activity; all induced PR10 expression (SI Appendix, Fig. S10B). Together with the ion exchange data, these results suggest that the elicitors released from Arabidopsis cell walls as a response to lignin modification are heterogeneous with respect to both size and charge.
Pretreatment of water extracts from cell walls of the lignin-modified plants with PGase gave partial to complete abolition of PR gene induction (Fig. 2 A and B). To explore further the nature of the elicitors, the extracts were pretreated with specific plant cell wall-degrading enzymes (SI Appendix, Table S3). After incubation and destruction of the added enzymatic activity by autoclaving, the extracts were assayed for elicitor activity. Digestion with commercial PGase and arabinanase eliminated the activity of the extracts from HCT-RNAi and ccr1 lines to induce expression of PR1 and PR10, and treatment with fucosidase or xyloglucanase partially reduced this ability.
ADPG1 Is Required for PR Gene Expression in HCT-RNAi and ccr1 Plants.
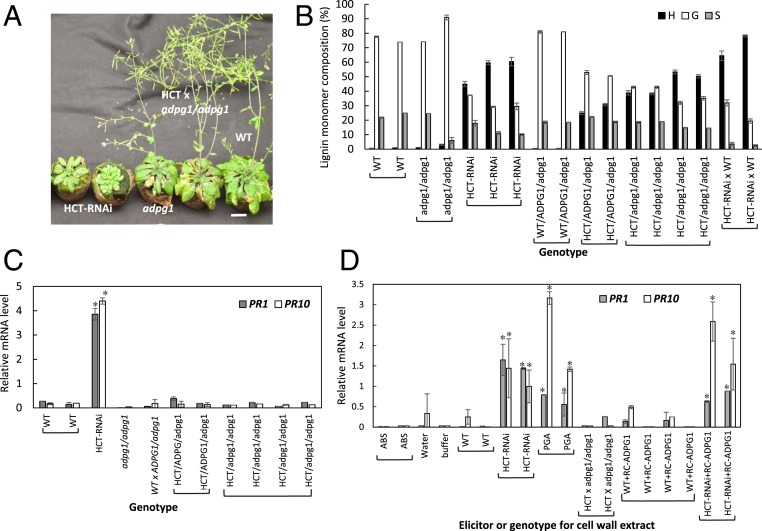
To determine whether ectopic expression of ADPG1 is necessary for induction of PR genes in stems of the lignin-modified plants, we generated crosses to introduce the HCT-RNAi or a ccr1-3 mutant allele into a homozygous adpg1 loss-of-function mutant background. Homozygous lines for both the lignin pathway gene and the adpg1 mutation were selected, as well as lines heterozygous for the functional ADPG1 allele. Because loss of function of ADPG1 imparted a defect/delay in both anther dehiscence and silique opening (SI Appendix, Fig. S11A), mechanical manipulation was necessary to obtain seeds from these crossed plants. The HCT-RNAi line is dwarf (Fig. 3A) with reduced rosette diameter and inflorescence stem length (SI Appendix, Fig. S10B), whereas the homozygous adpg1 mutant exhibits a wild-type growth phenotype as regards plant size (Fig. 3A and SI Appendix, Fig. S11B) and wild-type lignin composition (Fig. 3B). Loss of function of ADPG1 partially restored inflorescence height in the HCT-RNAi background, with a similar effect on rosette diameter (SI Appendix, Fig. S11B). In inflorescence stems expressing the HCT-RNAi construct, loss of function of ADPG1 did not affect the accumulation of high levels of H units as a result of the block in the pathway to G and S units (Fig. 3B), but completely prevented induction of both PR1 and PR10 transcripts (Fig. 3C). Heterozygous lines with loss of function of only one of the two copies of ADPG1 gave the same phenotype as lines with loss of function of both alleles (Fig. 3C), perhaps because ectopically expressed ADPG1 activity is only just sufficient to release elicitor molecules.
Fig. 3.
ADPG1 is required for elicitor release and PR gene induction in HCT-RNAi Arabidopsis. (A) Overall growth phenotypes of wild-type (WT), HCT-RNAi, adpg1, and HCT-RNAi adpg1 lines at 8 wk old. (Scale bar, 2 cm.) (B) Lignin content and composition of the above lines as determined by thioacidolysis. (C) PR1 and PR 10 transcript levels in the above lines. (D) Induction of PR1 and PR10 transcripts in Arabidopsis cell cultures by elicitors derived from WT, HCT-RNAi, and HCT-RNAi adpg1 Arabidopsis. Cell wall extracts from WT and HCT-RNAi plants were also pretreated with RC-ADPG1 prior to testing of elicitor activity. Genotypes are represented as uppercase (WT) and lowercase (mutant) alleles. HCT-RNAi x WT is heterozygous for the RNAi construct. Error bars represent SD of three technical replicates (individual assays). Separate bars are shown for biological replicates. Asterisks indicate significant differences from WT (P < 0.05) by pairwise multiple comparison Tukey test.
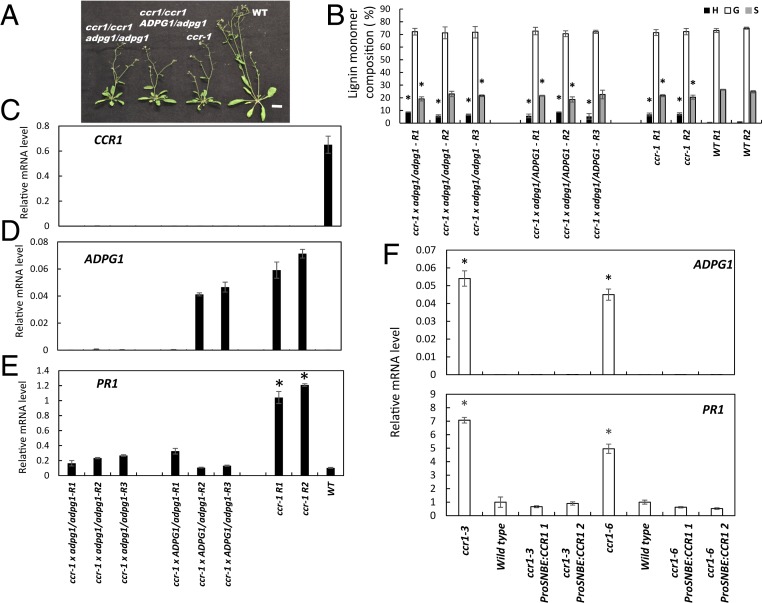
The ccr1-3 mutant displays reduced fertility and a semidwarf growth pattern (38, 39) with smaller rosette and shorter primary inflorescence stem when compared with wild-type plants (Fig. 4A and SI Appendix, Fig. S11 C and D). Loss of function of ADPG1 did not restore fertility, rosette size, or height of primary inflorescence stems in the ccr1 mutant background (Fig. 4A and SI Appendix, Fig. S11 C and D), or the elevated levels of H units (Fig. 4B). However, loss of function of ADPG1 in the ccr1 mutant background did result in strong reduction of PR1 expression in inflorescence stems (Fig. 4 C–E). As was the case in the crosses with the HCT-RNAi line, plants that were heterozygous for the disrupted ADPG1 allele showed a phenotype similar to those that were homozygous. Taken together, the data show that ectopic ADPG1 expression is required for PR induction in stems of both the HCT-RNAi and ccr1 lines.
Fig. 4.
ADPG1 is required for PR1 induction in the ccr1 mutant of Arabidopsis. (A) Phenotype of F2 progeny of the cross ccr1 x adpg1. Wild-type (WT), ccr1, and F2 plants are 7 wk old. (Scale bar, 2 cm.) (B) Lignin content and composition of F2 progeny of the cross ccr1 x adpg1 as determined by thioacidolysis. Results are means ± SD of five technical replicates. Individual biological replicates are indicated by R1, R2, etc. (C) Levels of CCR1 transcripts in WT and ccr1/adpg1 mutant lines. (D) Levels of ADPG1 transcripts in WT and ccr1/adpg1 mutant lines. (E) Levels of PR1 transcripts in WT and ccr1/adpg1 mutant lines. Genotypes are represented as uppercase (WT) and lowercase (mutant) alleles. Transcript analyses were performed on 8-wk-old plants, and expressed relative to ACT2. In C–E, each replicate is a pool of four plants of the same genotype. (F) Restoring CCR expression to xylem prevents ADPG1-mediated induction of PR proteins in the Arabidopsis ccr1 mutant. PR1 transcript levels were determined in inflorescence stems of ccr1 mutant plants (two independent lines, ccr1-3 and ccr1-6), and the ccr1 mutants in which a WT copy of CCR1 was expressed under a xylem-specific promoter (ProSNBE:CCR1) (see ref. 40 for details of the lines). Results are mean ± SD of three biological replicates; WT PR1 transcript value is normalized to 1.0. Asterisks in B, E, and F indicate significant differences from WT (P < 0.05) by pairwise multiple comparison Tukey test.
Restoration of lignin deposition in the xylem tissues of the Arabidopsis ccr1 mutant by expressing the CCR1 open reading frame under the control of a vessel-specific promoter restores growth, likely as a result of improved vascular function (40). To address whether the signals for activation of PR gene expression originate in xylem, we determined the levels of ADPG1 and PR1 transcripts in inflorescence stems of two alleles of the ccr1 mutant and their corresponding complemented lines (ProSNBE:CCR1). The expression of PR1 was reduced to or below the level of the wild-type control on restoration of lignin deposition specifically in the xylem of different independent complemented lines (Fig. 4F), and ADPG1 expression was essentially undetectable in all lines other than the original ccr1 mutants. These data suggest that defense gene activation in stems is a result of expression of ADPG1 primarily in xylem vessels.
ADPG1 Is Required for Release of Cell Wall-Derived Elicitors.
To address whether ADPG1 is responsible for release of pectin-derived signal molecules in stems of low-lignin Arabidopsis, we first examined the preference of this enzyme for various components of the cell wall pectin fraction. In comparison with two other Arabidopsis PGases, ADPG1 was reported to possess low specific activity with PGA as substrate (41). We expressed recombinant ADPG1 (RC-ADPG1) in Escherichia coli, purified the enzyme (SI Appendix, Fig. S12), and compared its activity against that of pure PGA, on apple pectin (consisting largely of methylated galacturonic acid residues), RG-I from tobacco suspension cultures, and RG-II from red wine. RC-ADPG1 exhibited highest activity against apple pectin (comprising a mixture of HG, RG-I, and RG-II), with between approximately fivefold and sevenfold lower specific activity with purified PGA, RG-I, and RG-II (SI Appendix, Table S4). Its relative preference for apple pectin was greater than that of the two commercial PGases.
Water extracts were then prepared from AIR preparations from cell walls of HCT-RNAi adpg1 plants, and the elicitor activity of these fractions was compared with that of the same fractions from wild-type and HCT-RNAi plants using the Arabidopsis cell culture bioassay. Loss of function of ADPG1 resulted in loss of the PR gene-inducing activity seen in water extracts from the HCT-RNAi line with functional ADPG1 (Fig. 3D), indicating that ADPG1 is necessary for elicitor generation. Pretreatment of water extracts from cell walls of wild-type plants with RC-ADPG1 did not result in appearance of elicitor activity in the extract (Fig. 3D), indicating that the elicitor(s) was not present in these extracts in a latent form that could be activated through the action of ADPG1. Furthermore, preincubation of extracts from cell walls of the HCT-RNAi plants with ADPG1 did not reduce their PR induction activity, consistent with the enzyme being able to release a pectic-derived signal(s) from the cell wall without destroying it.
We then examined the effects of RC-ADPG1 on the elicitor activity of water extracts from cell walls of homozygous ccr1 adpg1 mutant plants, using an Arabidopsis leaf injection assay. Injection of water extract from the ccr1 mutant, or PGA, strongly induced PR1 transcripts (SI Appendix, Fig. S13). As in the cell culture assays with extract from HCT-RNAi adpg1 plants, injection of water extract from ccr1 adpg1 plants did not induce PR1 transcripts. However, PR1 transcripts were induced if the water extract was preincubated with RC-ADPG1 (SI Appendix, Fig. S13). Thus, cell wall remodeling releases material harboring latent elicitors, but ADPG1 is required for their conversion to active species.
Discussion
Lignin Modification Is Associated with Extensive Cell Wall Remodeling.
Lignin is an important component of both preexisting and inducible defense responses in plants (6). Various hypotheses have been put forward to explain why reduction in lignin content often appears to enhance rather than reduce plant disease resistance (7, 8); these include antimicrobial activity of lignin pathway intermediates (42) and release of elicitor-active molecules from incorrectly assembled plant cell walls (43). The present results provide a mechanistic explanation for the latter hypothesis.
Impacts of lignin modification on plant gene expression may be initiated through alterations in cell wall integrity, likely, in part, operating through effects on cross-linking to other cell wall components such as pectin and/or hemicellulose. Although pectin is generally considered in the context of primary cell walls, its importance for secondary wall structure is probably greater than previously realized, as lignin modification can affect the expression of pectin-related genes (44, 45), and, conversely, pectin modification affects the expression of lignin pathway genes (46) and lignin content (47). It has been proposed that pectin forms a nucleation site for lignification in alfalfa (48), and partially methylated pectins can interact with lignin polymers composed of coniferyl alcohol to form hydrophobic clusters in vitro, suggesting that activity of pectin methyl esterases might regulate pectin−lignin interactions (49). Moreover, PGase genes have been shown to exhibit high expression during the differentiation of tracheary elements in Zinnia elegans (50) or during secondary wall formation in trees such as aspen and poplar (51, 52), suggesting that pectin modification might function in cell wall remodeling associated with lignin deposition. Although the exact nature of lignin−pectin interactions in the secondary cell wall is not clear, some studies indirectly support the importance of such interactions in orchestrating cell wall integrity (19, 53). This concept is supported further by the ability of C. bescii with a deleted pectinase gene cluster to grow on HCT-RNAi and ccr1 Arabidopsis; clearly, disruption of pectin is critical for opening the cell wall structure to degradation by other enzymes to release the sugars necessary for bacterial growth. It is not possible to determine which of the many induced cell wall-degrading enzymes is responsible for the overall changes in cell wall integrity in the HCT-RNAi and ccr1 lines; although ADPG1 is the most strongly induced pectin-degrading enzyme and the only one induced in common between the two lines, a number of pectin lyase genes are also induced.
In both HCT-RNAi and ccr1 mutant lines, pectins were more easily extractable from cell walls in water, oxalate, or carbonate compared to wild-type cell walls. Increased extractability of pectic backbone epitopes is one of the cell wall remodeling features previously shown in response to abiotic stresses such as low soil moisture availability in stem wood (54). Arabinogalactan and RG-I are the predominant polysaccharide epitopes in the water extracts of HCT-RNAi and ccr1 cell walls, based on glycome and compositional analysis showing increased levels of monosaccharides that constitute these types of molecules (namely fucose, arabinose, galactose, rhamnose, xylose, and galacturonic acid). However, heteroxylans are also preferentially released from ccr1 cell walls.
Lignin Modification Uncovers Latent Cell Wall-Derived Elicitors of Defense Gene Expression.
Molecules or epitopes present on cell wall components with the ability to activate defense pathways have been termed DAMPs (23). To date, they have been shown to be OGs of different sizes originating from pectin, or oligoglucosides (55). The DAMP concept is, in essence, a restatement of the earlier oligosaccharin hypothesis (9, 12) formulated in a series of seminal papers that described plant cell wall structures that elicited plant defenses and/or impacted plant growth and development (14, 15, 27, 56, 57). Subsequent studies on oligosaccharins derived from xyloglucans or pectin (13, 16) led to the hypotheses that plants possess specific receptors for such molecules that may act to transduce signals from the cell wall during attempted penetration catalyzed by pathogen-derived wall-degrading enzymes, and that the effects of oligosaccharins on growth and development may operate through antagonism of auxin action (16, 58). Genetic approaches have been applied to understand oligosaccharin signaling and its potential dual role in defense and development (59, 60), but, in most cases, the elicitor molecules investigated have been limited to synthetic homo-OGs, so the extent of the repertoire of DAMPs/oligosaccharins that function naturally in plant defense has remained unclear.
Analyses of the HCT-RNAi and ccr1 mutant reported here, along with Arabidopsis plants with loss of function or overexpression of the F5H that serves as the entry point to S lignin biosynthesis (29), show that different types of lignin modification lead to release of different elicitors that activate different defense response pathways (PR proteins in the present case; genes involved in response to oomycetes or tritrophic interactions with insects in the case of F5H misregulation) (29). These elicitors, even as crude water-soluble extracts, do not exhibit cross-reactivity for defense gene induction. The pectic framework clearly has the structural complexity to provide such diverse and apparently specific elicitors. Based on the results of ion exchange and size fractionation, the actual elicitor molecules are likely polymorphic, containing epitopes that confer activity along with additional nonactive portions.
The elicitor-active components from both HCT-RNAi and ccr1 lines are destroyed by digestion with PGase and arabinan-1,5-α-l-arabinosidase. This suggests that they are derived from RGs. Classical RG-I contains, among other substitutions, linear five-linked arabinan side chains attached to a central polymer consisting of alternating galacturonic acid and rhamnose residues, whereas RG-II contains highly complex side chains consisting of multiple sugar types attached to a linear chain of α-1,4−linked galacturonic acid residues, with a few arabinose units only attached as end-groups (61). The preference of ADPG1 for apple pectin rather than PGA suggests that the elicitors, or at least their precursors, may contain methylated HG.
Consistent with lignin modification being the primary reason for cell wall remodeling and elicitor release, complementation of the ccr1 mutant with a wild-type copy of CCR1 with expression targeted to xylem prevented the induction of PR1 in stems. This suggests that lignifying xylem cells are the origin of the released elicitors, although some lignification is also restored in fibers of the ProSNBE:CCR1 line (40).
ADPG1 Is Required for Release of Elicitors of PR Genes.
ADPG1 is highly induced in both HCT-RNAi and ccr1 lines, but is not induced in F5H misregulated lines in which lignin composition but not lignin content is altered (29). This PGase is the only pectin-modifying enzyme that is induced in both the HCT-RNAi and ccr1 lines, and loss of function of ADPG1 results in reduction of PR gene expression in HCT-RNAi and ccr1 genetic backgrounds and the loss of elicitor activity in extracts from cell walls of HCT-RNAi/adpg1 plants. However, the observation that water extracts from cell walls of ccr1 adpg1 mutant plants possess elicitor activity only after treatment with RC-ADPG1 suggests that the enzyme has a specific role in elicitor release, and is not itself necessary for the cell wall remodeling that results in solubilization of latent elicitors. ADPG1 is normally expressed in siliques and anthers prior to dehiscence, where it is likely that it degrades pectin to cause cell wall breakage, as its loss of function delays, or, in the case of strong alleles, prevents anther dehiscence (41). Anther dehiscence is also prevented by loss of function of NST1 in Medicago truncatula (62), or NST1 and NST2 in Arabidopsis (63). These NST genes encode NAC family transcription factors that regulate lignin deposition in secondary cell walls (63). The fact that both lignin and pectin modification impact anther dehiscence is consistent with a role for pectin in a structural complex with lignin.
The action of ADPG1 in vivo must be limited, specific, and perhaps localized for it to release elicitor-active molecules without destroying them. Furthermore, induction of ADPG1 does not appear to be a result of the activity of the pectic elicitors released from cell walls of these lines. Thus, it is likely that signaling to induce ADPG1 occurs first, with resulting release of pectic/oligosaccharide elicitors that then activate defense responses. In the model in Fig. 5, an initial stimulus (perhaps a released cell wall component or a physical change in the wall recognized by receptors in the plasma membrane) activates ADPG1 transcription. Several receptors that monitor the “status” of cell wall components have recently been identified (64). The ADPG1 enzyme releases oligogalacturonide elicitors from RG-I and/or RG-II, which, either directly or after processing, may be recognized by the wall-associated kinases which have the ability to bind OGs and PGA (59). This reception results in elevated levels of SA [inferred for Arabidopsis ccr1 mutants and directly demonstrated in previous studies on the ccr1 mutant of M. truncatula (65)] and HCT-down-regulated alfalfa and Arabidopsis lines (7, 66) and consequent induction of PR genes. Assuming that the cell culture system used allows elicitor-mediated induction of all genes irrespective of their tissue specificity, induction of genes such as SESA2 and CRA2 in the ccr1 mutant is likely a secondary effect, as these genes are not induced by the released elicitors.
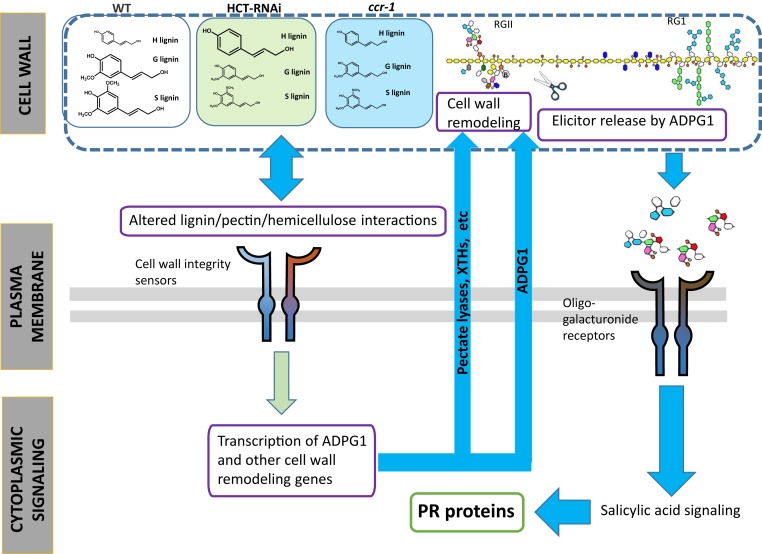
Fig. 5.
Model for the activation of PR genes in HCT-RNAi and ccr1 Arabidopsis plants. In the proposed model, changes in lignin content in xylem cells of HCT-RNAi or ccr1 Arabidopsis are perceived initially by the cell through activation of plasma membrane-localized cell wall integrity receptors. This results in initiation of a signaling cascade that induces the expression of cell wall remodeling genes, including PECTATE LYASES, XYLOGLUCAN ENDO-TRANSGLYCOSYLASES (XTHs), and ADGP1. ADPG1 activity may contribute to solubilization of pectin, but is necessary for release of elicitor fragments, most likely from RG-II. The soluble elicitors activate expression of PR defense response genes through a signaling pathway involving SA (66). Many of the other transcriptomic changes occurring in the lignin-modified plants, such as the activation of seed-specific genes in stems of ccr1, may result from secondary effects. The modification of pectin is also, at least in part, responsible for the reduced recalcitrance of the biomass.
The suite of cell wall disassembly genes that is induced in the transcriptomes of the HCT-RNAi and ccr1 lines is, in many ways, reminiscent of the genes active in plant abscission zones (32, 34, 67). Interestingly, it has been suggested that PR proteins are part of the proteinaceous cell wall components in the protective layer of abscission zones (68), and, extrapolating from the present data, ADPG1 may therefore be a component of the signaling that strengthens defenses in the exposed surfaces postabscission, triggered initially by altered lignin−pectin interactions.
Plants with modified lignin content and/or composition provide an excellent model system for deciphering the complexity of latent signal molecules sequestered within plant cell walls and characterization of their receptors. Improved approaches for the analysis of plant cell wall-released pectic fractions will facilitate these efforts (69). Understanding how plants remodel their cell walls as a result of engineered structural perturbations may allow us to better design improved lignocellulosic energy crops by optimizing bioprocessing quality, yield, and stress resistance.
Experimental Procedures
Detailed descriptions of the experimental methods are provided in SI Appendix, Supplementary Materials and Methods. These include growth of plants, all chemical analytical methods, glycome profiling, generation of and assay of elicitors, and C. bescii bioassays. A. thaliana HCT-RNAi and ccr1 lines (ccr1-3 and ccr1-6 mutants) in ecotype Columbia-0 were obtained from Clint Chapple, Purdue University. The adpg1 mutant of Arabidopsis was obtained from the Arabidopsis Biological Resource Center. The ccr1 ProSNBE:CCR1 line of Arabidopsis in which CCR1 is expressed under a vessel-specific promoter in the ccr1 mutant background has been described previously (40).
All data discussed in the paper will be made available to readers. The microarray datasets supporting the results of this article are available in the National Center for Biotechnology Information Gene Expression Omnibus data repository under the accession number GSE125721, title “Transcriptomic analysis of lignin mutants in Arabidopsis” (70).
Supplementary Material
Acknowledgments
We thank Dr. Nan Lu for critical reading of the manuscript. This work was supported by the University of North Texas, and by the BioEnergy Science Center and the Center for Bioenergy Innovation (Oak Ridge National Laboratory), US Department of Energy (DOE) Bioenergy Research Centers supported by the Office of Biological and Environmental Research in the DOE Office of Science. S.P.-A. was supported by a Marie Curie Intra European Fellowship within the 7th European Community Framework Programme (Grant PIEF-GA-2013-625270). Generation of the Complex Carbohydrate Research Center (CCRC) series of plant cell wall glycan-directed monoclonal antibodies used in this work was supported by the National Science Foundation Plant Genome Program (Grants DBI-0421683 and IOS-0923992). We thank Marie-Christine Ralet and Fabienne Guillon for access to INRA-RU1 EDC antibodies, and Yuhong Tang for assistance with microarray analysis.
Footnotes
The authors declare no competing interest.
Data deposition: All data discussed in the paper will be made available to readers. The microarray datasets supporting the results of this article are available in the NCBI Gene Expression Omnibus (GE) data repository under the accession number GSE125721, title “Transcriptomic analysis of lignin mutants in Arabidopsis.”
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914422117/-/DCSupplemental.
References
- 1.Voxeur A., Höfte H., Cell wall integrity signaling in plants: “To grow or not to grow that’s the question.” Glycobiology 26, 950–960 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Chen F., Dixon R. A., Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25, 759–761 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Shadle G., et al. , Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68, 1521–1529 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Weng J. K., et al. , Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 22, 1033–1045 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J. I., Ciesielski P. N., Donohoe B. S., Chapple C., Li X., Chemically induced conditional rescue of the reduced epidermal fluorescence8 mutant of Arabidopsis reveals rapid restoration of growth and selective turnover of secondary metabolite pools. Plant Physiol. 164, 584–595 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miedes E., Vanholme R., Boerjan W., Molina A., The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 5, 358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallego-Giraldo L., Jikumaru Y., Kamiya Y., Tang Y., Dixon R. A., Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol. 190, 627–639 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Zhao Q., Dixon R. A., Altering the cell wall and its impact on plant disease: From forage to bioenergy. Annu. Rev. Phytopathol. 52, 69–91 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Albersheim P., Darvill A. G., Oligosaccharins. Sci. Am. 253, 58–64 (1985).3906895 [Google Scholar]
- 10.Ayers A. R., Valent B., Ebel J., Albersheim P., Host-pathogen interactions: XI. Composition and structure of wall-released elicitor fractions. Plant Physiol. 57, 766–774 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharp J. K., McNeil M., Albersheim P., The primary structures of one elicitor-active and seven elicitor-inactive hexa(beta-D-glucopyranosyl)-D-glucitols isolated from the mycelial walls of Phytophthora megasperma f. sp. glycinea. J. Biol. Chem. 259, 11321–11336 (1984). [PubMed] [Google Scholar]
- 12.Darvill A., et al. , Oligosaccharins—Oligosaccharides that regulate growth, development and defence responses in plants. Glycobiology 2, 181–198 (1992). [DOI] [PubMed] [Google Scholar]
- 13.Côté F., Hahn M. G., Oligosaccharins: Structures and signal transduction. Plant Mol. Biol. 26, 1379–1411 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Hahn M. G., Darvill A. G., Albersheim P., Host-pathogen interactions: XIX. The endogenous elicitor, a fragment of a plant cell wall polysaccharide that elicits phytoalexin accumulation in soybeans. Plant Physiol. 68, 1161–1169 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.York W. S., Darvill A. G., Albersheim P., Inhibition of 2,4-dichlorophenoxyacetic acid-stimulated elongation of pea stem segments by a xyloglucan oligosaccharide. Plant Physiol. 75, 295–297 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fry S. C., Aldington S., Hetherington P. R., Aitken J., Oligosaccharides as signals and substrates in the plant cell wall. Plant Physiol. 103, 1–5 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche P., et al. , Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell 67, 1131–1143 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Choi H. W., et al. , Activation of plant innate immunity by extracellular high mobility group box 3 and its inhibition by salicylic acid. PLoS Pathog. 12, e1005518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamann T., The plant cell wall integrity maintenance mechanism—Concepts for organization and mode of action. Plant Cell Physiol. 56, 215–223 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Gravino M., Savatin D. V., Macone A., De Lorenzo G., Ethylene production in Botrytis cinerea- and oligogalacturonide-induced immunity requires calcium-dependent protein kinases. Plant J. 84, 1073–1086 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Galletti R., et al. , The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 148, 1695–1706 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darvill A. G., Albersheim P., Phytoalexins and their elicitors—A defense against microbial infection in plants. Annu. Rev. Plant Physiol. 35, 243–275 (1984). [Google Scholar]
- 23.Ferrari S., et al. , Oligogalacturonides: Plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 4, 49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomma B. P., Cammue B. P., Thevissen K., Plant defensins. Planta 216, 193–202 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Klarzynski O., et al. , Linear beta-1,3 glucans are elicitors of defense responses in tobacco. Plant Physiol. 124, 1027–1038 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bishop P. D., Makus D. J., Pearce G., Ryan C. A., Proteinase inhibitor-inducing factor activity in tomato leaves resides in oligosaccharides enzymically released from cell walls. Proc. Natl. Acad. Sci. U.S.A. 78, 3536–3540 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nothnagel E. A., McNeil M., Albersheim P., Dell A., Host-pathogen interactions. XXII. A galacturonic acid oligosaccharide from plant cell walls elicits phytoalexins. Plant Physiol. 71, 916–926 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benedetti M., et al. , Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc. Natl. Acad. Sci. U.S.A. 112, 5533–5538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallego-Giraldo L., et al. , Elicitors and defense gene induction in plants with altered lignin compositions. New Phytol. 219, 1235–1251 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Li X., Bonawitz N. D., Weng J.-K., Chapple C., The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22, 1620–1632 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mir Derikvand M., et al. , Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227, 943–956 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Hoffmann L., et al. , Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16, 1446–1465 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.González-Carranza Z. H., Elliott K. A., Roberts J. A., Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J. Exp. Bot. 58, 3719–3730 (2007). [DOI] [PubMed] [Google Scholar]
- 34.González-Carranza Z. H., et al. , A novel approach to dissect the abscission process in Arabidopsis. Plant Physiol. 160, 1342–1356 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pattathil S., et al. , A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153, 514–525 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M., et al. , Downregulation of pectin biosynthesis gene GAUT4 leads to reduced ferulate and lignin-carbohydrate cross-linking in switchgrass. Commun Biol 2, 22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung D., et al. , Deletion of a gene cluster encoding pectin degrading enzymes in Caldicellulosiruptor bescii reveals an important role for pectin in plant biomass recalcitrance. Biotechnol. Biofuels 7, 147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones L., Ennos A. R., Turner S. R., Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26, 205–216 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Thévenin J., et al. , The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol. Plant 4, 70–82 (2011). [DOI] [PubMed] [Google Scholar]
- 40.De Meester B., et al. , Vessel-specific reintroduction of CINNAMOYL-COA REDUCTASE1 (CCR1) in dwarfed ccr1 mutants restores vessel and xylary fiber integrity and increases biomass. Plant Physiol. 176, 611–633 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogawa M., Kay P., Wilson S., Swain S. M., ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21, 216–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barber M. S., McConnell V. S., DeCaux B. S., Antimicrobial intermediates of the general phenylpropanoid and lignin specific pathways. Phytochemistry 54, 53–56 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Giberti S., Bertea C. M., Narayana R., Maffei M. E., Forlani G., Two phenylalanine ammonia lyase isoforms are involved in the elicitor-induced response of rice to the fungal pathogen Magnaporthe oryzae. J. Plant Physiol. 169, 249–254 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Wróbel-Kwiatkowska M., Starzycki M., Zebrowski J., Oszmiański J., Szopa J., Lignin deficiency in transgenic flax resulted in plants with improved mechanical properties. J. Biotechnol. 128, 919–934 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Goulao L. F., Vieira-Silva S., Jackson P. A., Association of hemicellulose- and pectin-modifying gene expression with Eucalyptus globulus secondary growth. Plant Physiol. Biochem. 49, 873–881 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Xiao C., et al. , Activation tagging of Arabidopsis POLYGALACTURONASE INVOLVED IN EXPANSION2 promotes hypocotyl elongation, leaf expansion, stem lignification, mechanical stiffening, and lodging. Plant J. 89, 1159–1173 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Hao Z., et al. , Loss of Arabidopsis GAUT12/IRX8 causes anther indehiscence and leads to reduced G lignin associated with altered matrix polysaccharide deposition. Front. Plant Sci. 5, 357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wi S. G., Singh A. P., Lee K. H., Kim Y. S., The pattern of distribution of pectin, peroxidase and lignin in the middle lamella of secondary xylem fibres in alfalfa (Medicago sativa). Ann. Bot. 95, 863–868 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lairez D., et al. , Aggregation during coniferyl alcohol polymerization in pectin solution: A biomimetic approach of the first steps of lignification. Biomacromolecules 6, 763–774 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Demura T., et al. , Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc. Natl. Acad. Sci. U.S.A. 99, 15794–15799 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aspeborg H., et al. , Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol. 137, 983–997 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geisler-Lee J., et al. , Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 140, 946–962 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Habrant A., Gaillard C., Ralet M. C., Lairez D., Cathala B., Relation between chemical structure and supramolecular organization of synthetic lignin-pectin particles. Biomacromolecules 10, 3151–3156 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Pattathil S., et al. , Cell wall ultrastructure of stem wood, roots, and needles of a conifer varies in response to moisture availability. Front. Plant Sci. 7, 882 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vorwerk S., Somerville S., Somerville C., The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 9, 203–209 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Tran Thanh Van K., et al. , Manipulation of the morphogenetic pathways of tobacco explants by oligosaccharins. Nature 314, 615–617 (1985). [Google Scholar]
- 57.Eberhard S., et al. , Pectic cell wall fragments regulate tobacco thin-cell-layer explant morphogenesis. Plant Cell 1, 747–755 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aldington S., McDougall G. J., Fry S. C., Structure-activity relationships of biologically active oligosaccharides. Plant Cell Environ. 14, 625–636 (1991). [Google Scholar]
- 59.Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G., A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vallarino J. G., Osorio S., Signaling role of oligogalacturonides derived during cell wall degradation. Plant Signal. Behav. 7, 1447–1449 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ridley B. L., O’Neill M. A., Mohnen D., Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57, 929–967 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Zhao Q., et al. , An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 63, 100–114 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M., The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He Y., Zhou J., Shan L., Meng X., Plant cell surface receptor-mediated signaling—A common theme amid diversity. J. Cell Sci. 131, jcs209353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Man Ha C., et al. , Ectopic defense gene expression is associated with growth defects in Medicago truncatula lignin pathway mutants. Plant Physiol. 181, 63–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gallego-Giraldo L., Escamilla-Trevino L., Jackson L. A., Dixon R. A., Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc. Natl. Acad. Sci. U.S.A. 108, 20814–20819 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J., Chun J.-P., Tucker M. L., Transcriptional regulation of abscission zones. Plants (Basel) 8, 154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim J., et al. , Examination of the abscission-associated transcriptomes for soybean, tomato, and Arabidopsis highlights the conserved biosynthesis of an extensible extracellular matrix and boundary layer. Front. Plant Sci. 6, 1109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voxeur A., et al. , Oligogalacturonide production upon Arabidopsis thaliana-Botrytis cinerea interaction. Proc. Natl. Acad. Sci. U.S.A. 116, 19743–19752 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dixon R. A., Transcriptomic analysis of lignin mutants in Arabidopsis. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125721. Deposited 28 January 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.