Significance
When an animal forms a memory, synapses in specific brain pathways change their strength. Pinpointing which synapses and pathways are modulated in any given learning paradigm, however, is technically challenging and needs to be performed one candidate connection at a time. Here we present SYNPLA, a tool to quickly detect strengthened synapses in genetically or anatomically defined pathways across the brain. To do so, we exploit the temporary translocation of AMPA receptor GluA1 into newly strengthened synapses. Using an assay that can identify proteins less than 40 nm away, we label only synapses that contain both GluA1 and a presynaptic protein exogenously expressed in a specific pathway. SYNPLA thus provides a pathway- and synapse-specific screening tool for memory formation.
Keywords: proximity ligation assay, synaptic potentiation, fear conditioning, defense conditioning, GluA1
Abstract
Which neural circuits undergo synaptic changes when an animal learns? Although it is widely accepted that changes in synaptic strength underlie many forms of learning and memory, it remains challenging to connect changes in synaptic strength at specific neural pathways to specific behaviors and memories. Here we introduce SYNPLA (synaptic proximity ligation assay), a synapse-specific, high-throughput, and potentially brain-wide method capable of detecting circuit-specific learning-induced synaptic plasticity.
Changes in synaptic strength have been hypothesized to underlie learning and memory since before the time of Hebb. The best-studied form of Hebbian synaptic plasticity is long-term potentiation (LTP), which underlies the formation of fear conditioning memories (1). Because of the inherent subjectivity of the term “fear” (2), we will use the term “defense conditioning.” Work over the last two decades has uncovered the molecular mechanisms of LTP in great detail, revealing that the expression of plasticity is mediated by the rapid synaptic insertion of GluA1 subunit–containing AMPA receptors mobilized from an extrasynaptic pool (3). These GluA1-containing receptors are then likely replaced by GluA1-lacking AMPA receptors within 15 to 72 h (4–6), making synaptic GluA1 a marker of recently potentiated synapses. LTP appears to be very general at glutamatergic synapses in the central nervous system, including in the cortex (4), amygdala (7), ventral tegmental area (8), nucleus accumbens (9), striatum (10) and lateral habenula (11). Furthermore, plasticity mediated by trafficking of GluA1 subunit–containing AMPA receptors appears to play an important part in several neuropsychiatric disorders, such as addiction (12), autism (13), and depression (11).
Relating behaviorally induced plasticity to its synaptic substrate remains an important technical challenge. Electrophysiological or optical methods provide mechanistic insight but are low throughput and require specialized equipment (7). Somatic expression of immediate early genes (IEG) such as cfos can be used to screen for brain areas activated during plasticity (14) but lack synaptic resolution. We therefore sought to develop a pathway-specific histological method for detecting behaviorally induced changes.
Here we present a synaptic proximity ligation assay (SYNPLA), a method that uses the proximity ligation assay (PLA) to detect synaptic insertion of GluA1-containing AMPA receptors in defined circuits. PLA is a highly sensitive and specific biochemical method that reliably detects the close (<40 nm) juxtaposition of two proteins in situ (15) (Fig. 1A). To detect nearby molecules A and B, PLA uses a standard primary antibody raised against A and another raised against B. These antibodies are detected by secondary antibodies conjugated to unique oligonucleotides (Ao, Bo). A second set of oligonucleotides, ABo1 and ABo2, that are complementary to parts of both Ao and Bo is added. Only when Ao and Bo are sufficiently close can ABo1 and ABo2 be ligated to form a circle. This circle is then amplified (>1,000-fold) via rolling-circle amplification to form a nanoball of DNA, which can be reliably probed with complementary fluorescent nucleotides and observed as a punctum with light microscopy. A key advantage of PLA over traditional immunostaining is the absolute requirement for a pair of proteins to produce a PLA signal—a logical AND—which confers high specificity and thus reduces the false positive rate. Moreover, the 1000-fold signal amplification renders each positive signal very bright and therefore easily distinguishable from background, allowing results to be imaged quickly and at lower resolution than would otherwise be necessary for resolving synaptic contacts.
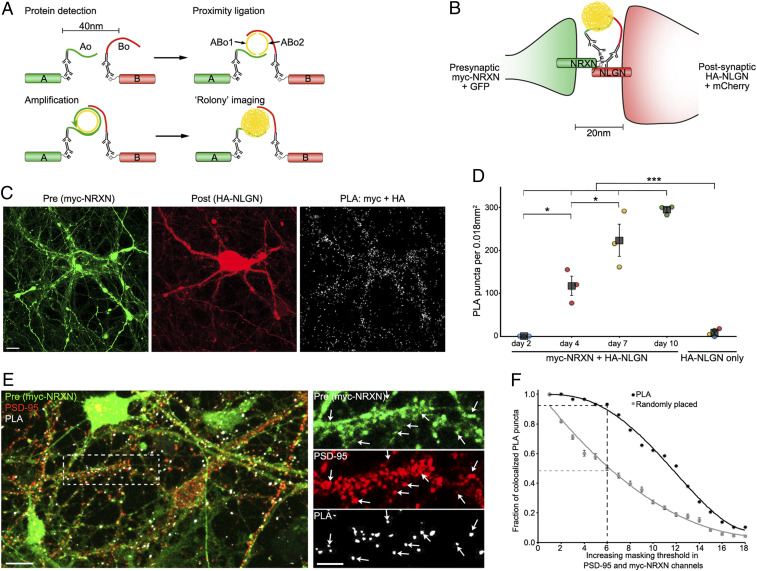
Fig. 1.
SYNPLA detects synapse formation and labels synapses. (A) Diagram of a general PLA reaction. Note that A and B must be sufficiently close to permit complementary binding and ligation of ABo1 and ABo2 and subsequent amplification to make a rolony. (B) Diagram of PLA targeting recombinant presynaptic myc-NRXN and postsynaptic HA-NLGN. (C) PLA rolonies (white dots, Right) formed between postsynaptic cultured mouse hippocampal neurons expressing HA-NLGN + mCherry (red; Middle) and cocultured presynaptic neurons expressing myc-NRXN + GFP (green; Left). (Scale bar, 10 μm.) (D) PLA puncta in each sample (circles) and average (squares) ± SEM for indicated expression and days in vitro (color indicates data acquired on the same day). ***P < 0.001, two-way ANOVA (see Methods); *P < 0.05, one-way ANOVA (with the Tukey–Kramer post hoc test). (E) PLA reaction between endogenous GluA1 and recombinant myc in cultured rat hippocampal neurons (exposed to cLTP; see Fig. 2) expressing myc-NRXN and cytosolic fluorescent protein (green), subsequently immunostained for endogenous PSD-95 (red). (Scale bars, 10 μm, Left; 5 μm, Right.) Arrows represent dendritic spines labeled by presynaptic myc-NRXN, PSD-95, and PLA puncta. (F) PLA puncta localize to synapses. In PSD-95 and myc-NRXN channels (from E, Right), pixels were set to zero (masked) for values below a progressively increasing threshold (x axis). Puncta (PLA or randomly placed) display colocalization if nonzero pixels exist within 0.14 μm in both thresholded channels. For the indicated threshold, ∼95% of PLA but only ∼50% of randomly placed (mean ± SEM of 30 placements) puncta colocalized with pre- and postsynaptic markers. See SI Appendix, Fig. S3 and Materials and Methods for details.
Results
We first confirmed previous experiments (16) suggesting that PLA could be used to detect direct protein–protein interactions across the synaptic cleft in dissociated cultured neurons (Fig. 1 B–D). We exploited the developmental window (days 2 to 10) during which synapses are formed in this preparation (17). In some neurons we expressed the presynaptic protein neurexin 1b with a myc tag (myc-NRXN) along with cytosolic GFP; in other neurons we expressed neuroligin 1, the normal postsynaptic binding partner of NRXN (18), with an HA tag (HA-NLGN) and cytosolic mCherry. Consistent with the normal time course of synapse formation (17), PLA reactions using antibodies to myc and HA produced an increasing number of PLA puncta, whereas cultures expressing only NLGN-HA led to minimal PLA products, highlighting the specificity and high signal to noise ratio of SYNPLA (Fig. 1 C and D and SI Appendix, Figs. S1 and S2). To demonstrate that SYNPLA labels synapses, we measured colocalization between PLA puncta (generated as described below) and both presynaptic signal and postsynaptic PSD-95 (visualized with immunofluorescence; see Fig. 1 E and F and SI Appendix, Fig. S3 for details). We also performed PLA using antibodies to endogenous pre- and postsynaptic proteins (SI Appendix, Fig. S4), showing protein overexpression is not required. These results indicate that SYNPLA can detect synapses expressing recombinant and/or endogenous proteins with a high signal to noise ratio.
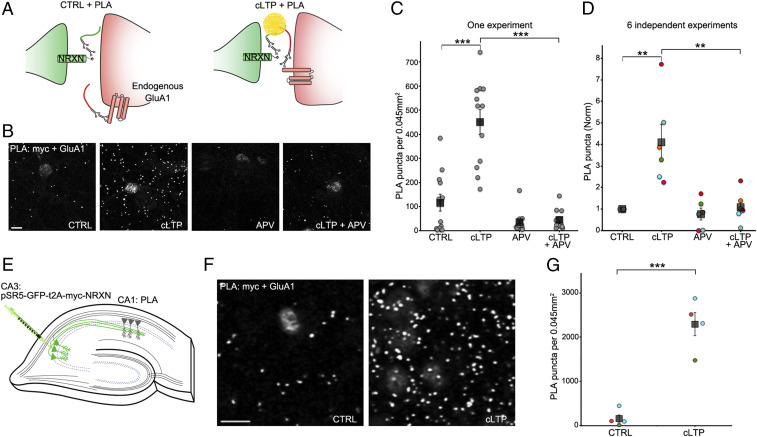
We reasoned that since the 20 nm synaptic cleft is less than the 40 nm PLA capture radius, SYNPLA could detect selectively the apposition of presynaptic NRXN with GluA1 inserted into the postsynapse during LTP. In contrast, no PLA products should be produced between NRXN and the more distant extrasynaptic pool of uninserted GluA1-containing AMPA receptors (Fig. 2A). We expressed myc-NRXN in dissociated cultured neurons (Figs. 1 E and F and 2 B–D) or in region CA3 of cultured (organotypic) hippocampal brain slices (19) (Fig. 2 E–G), and we performed PLA using antibodies to myc and the extracellular domain of GluA1 on the dissociated cultured neurons or region CA1 of organotypic slices. Although we observed few PLA puncta in control conditions, chemically induced LTP (cLTP) (20) greatly increased the number PLA puncta (Fig. 2 B–G; fourfold, P < 0.01, one-way ANOVA followed by Tukey–Kramer post hoc test in dissociated cultures; 12-fold, P < 0.001, paired t test in organotypic slices). The increase in the number of PLA puncta was blocked by addition of APV, a blocker of LTP induction (21), to neurons prior to cLTP (Fig. 2 B–D; P < 0.01, one-way ANOVA followed by Tukey–Kramer post hoc test). These results indicate that SYNPLA can employ recombinantly expressed presynaptic myc-NRXN and endogenous postsynaptic GluA1 to detect synaptic plasticity under cultured conditions.
Fig. 2.
SYNPLA detects synaptic potentiation in hippocampal primary cultures and organotypic slices. (A) Diagram of SYNPLA targeting presynaptic myc-NRXN and endogenous GluA1. cLTP induces mobilization of GluA1 from an extrasynaptic pool and insertion into the postsynaptic density, decreasing the distance between the targeted proteins and permitting PLA. (B) Representative images of SYNPLA reactions performed on myc-NRXN-expressing cultured rat hippocampal neurons at 14 d in vitro for the indicated conditions; PLA is shown in gray. (Scale bar, 10 μm.) (C) Quantification of a single SYNPLA experiment (as in F). The number of SYNPLA puncta detected in each field of view (circles) and the average (squares) ± SEM across all fields of view (12 per condition) are shown. ***P < 0.001, one-way ANOVA followed by the Tukey–Kramer post hoc test. (D) Quantification of six independent experiments (indicated by different colors); the average number of SYNPLA puncta across 10 to 12 fields of view for each experiment (circles, normalized to control [CTRL]); for each experiment, a significant difference was measured between CTRL and cLTP conditions (P values between 0.00004 and 0.02), and the average PLA signal across experiments (squares) ± SEM is shown. **P < 0.01, one-way ANOVA (with the Tukey–Kramer post hoc test). (E) Diagram of SYNPLA in rat organotypic hippocampal slices. Sindbis virus expressing myc-NRXN is injected in the presynaptic CA3 region. SYNPLA between myc-NRXN and endogenous GluA1 is measured in the postsynaptic CA1 region. (F) Representative images of SYNPLA under the indicated conditions in region CA1 of organotypic slice cultures. (Scale bar, 10 μm.) (G) Quantification of SYNPLA puncta in organotypic slices from three independent experiments (indicated with different colors, four slices per condition [one circle per slice]); squares indicate average ± SEM across experiments. ***P < 0.001, paired t test.
To test if SYNPLA can detect synaptic plasticity following memory formation in vivo, we injected the auditory cortex and/or the medial geniculate nucleus of the thalamus of rats with a virus expressing myc-NRXN and cytosolic GFP. We then exposed the rats to a cued defense-paired conditioning protocol, wherein a 10 s tone (conditioned stimulus) is immediately followed by a brief foot shock (Fig. 3 A–C). Such protocols have been shown to produce LTP-like plasticity at synapses onto the lateral amygdala (LA) (7). Control animals received either no viral injection, no conditioning, or unpaired conditioning, wherein the tone and the foot shock are not temporally paired. We perfused control or conditioned animals 30 min after conditioning and then postfixed and sectioned the brains at 50 μm.
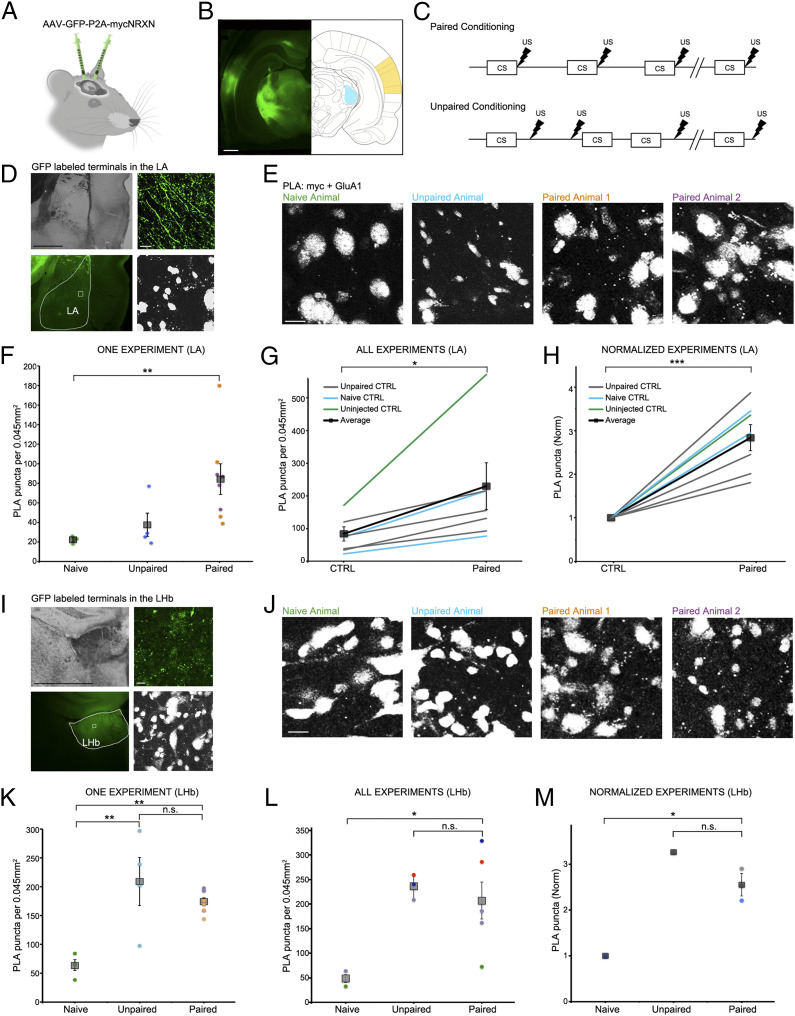
Fig. 3.
SYNPLA detects potentiated synapses between the medial geniculate nucleus or auditory cortex and the LA as well as the LHb following defense conditioning. (A) Injection of AAV9-GFP:P2A:myc-NRXN into the auditory cortex and/or medial geniculate nucleus. (B) GFP expression in the cell bodies at the injection sites (auditory cortex: yellow, medial geniculate nucleus: blue). (Scale bar, 1 mm.) (C) Diagrams of paired (Top) and unpaired (Bottom) defense conditioning paradigms; tone (CS; 10 s) and shock (thunderbolt) were delivered where indicated. (D) Representative images of presynaptic GFP-labeled fibers terminating onto the LA under low (Left; Top: transmitted light, Bottom: GFP signal, with LA traced in white) and high (Right; Top: GFP signal, Bottom: PLA) magnification in an animal that received paired conditioning. (Scale bar, Left, 1 mm; Right, 10 μm.) (E) Representative images of a single SYNPLA experiment in the LA from animals subjected to the indicated conditions; colors correspond to symbols in F. (Scale bar, 10 μm.) Note that nuclei show nonspecific staining after PLA in ex vivo slices; this signal is subtracted during image analysis (see Materials and Methods). (F) Quantification of the experiment shown in E. The number of SYNPLA puncta detected in one field of view (circles) and the average across all fields of view (squares) ± SEM are shown. **P < 0.01; one-way ANOVA with Tukey–Kramer post hoc test. (G) Quantification of all in vivo experiments, showing SYNPLA puncta per animal (average across all fields of view; gray) under the indicated control conditions and the average across animals (squares) ± SEM. *P < 0.05; paired t test. See SI Appendix, Fig. S6 for information regarding specific injection sites used in these experiments. SI Appendix, Fig. S7 shows similar results with data normalized to GFP-labeled fiber intensity. (H) Same data as in G, normalized to control. ***P < 0.001; paired t test. (I) Representative images of presynaptic GFP-labeled fibers terminating onto the LHb under low (Left; Top: transmitted light, Bottom: GFP signal, with LHb traced in white) and high (Right; Top: GFP signal, Bottom: PLA) magnification in an animal that received paired conditioning. (Scale bar, Left, 1 mm; Right, 10 μm.) (J) Representative images of one SYNPLA experiment in the LHb from animals subjected to the indicated conditions; colors correspond to symbols in K. (Scale bar, 10 μm.) (K) Quantification of the experiment shown in J. The number of SYNPLA puncta detected in each field of view (circles; four per animal) and the average across all fields of view (squares) ± SEM are shown. **P < 0.01; one-way ANOVA with Tukey–Kramer post hoc test; n.s., nonsignificant. (L) Quantification of in vivo experiments of all animals displaying green fibers in LHb (two naïve, three unpaired, and five paired animals), showing SYNPLA puncta per animal (average across all fields of view [four per animal]; each color represents distinct experiments) under the indicated conditions and the average across animals (gray squares) ± SEM. *P < 0.05; one-way ANOVA with Tukey–Kramer post hoc test. (M) Same data as in L for experiments with a naïve animal control, normalized. *P < 0.05; one-way ANOVA with Tukey–Kramer post hoc test.
We subjected tissue sections to SYNPLA and imaged the LA region containing GFP-labeled presynaptic fibers (Fig. 3D). We found that uninjected and naïve animals and animals receiving unpaired conditioning displayed few PLA puncta, whereas animals receiving paired conditioning displayed, on average, a threefold increase in PLA puncta (Fig. 3 F–H; in Fig. 3H P < 0.001, paired t test; see also SI Appendix, Figs. S6 and S7). The large fractional increase in GluA1-containing receptors at synapses during learning is consistent with a synaptic plasticity model wherein GluA1-containing receptors are added during plasticity and replaced within about 24 h with GluA1-lacking receptors (4). Different cell populations in the LA have distinct projections and inputs (22), which likely contributes to the observed within-animal variability (Fig. 3F). Variability across different experiments (conducted on different days) was also observed (e.g., Fig. 3G), but tissue from paired animals consistently displayed more PLA signal than control animals processed in the same day.
Subsequently, we examined regions outside the LA that showed GFP staining, indicating these regions received inputs from the injected regions. One such region was the lateral habenula (LHb), which has been coined the brain “disappointment” center and, when stimulated, is aversive (23, 24). While naïve animals (injected with the virus expressing myc-NRXN and cytosolic GFP) displayed few PLA puncta in the LHb, both unpaired and paired animals showed significantly elevated PLA puncta (Fig. 3 I–M). This result suggests that plasticity between the injected regions and LHb occurred if animals received a shock, regardless of the relation to the tone. Moreover, we also examined if there was synaptic plasticity in the auditory cortex and the medial geniculate nucleus, where the myc-NRXN virus was injected. Interestingly, in the medial geniculate nucleus we saw that animals that received paired conditioning had about fivefold more SYNPLA, whereas in the auditory cortex, we observed low signal in both paired and unpaired animals (SI Appendix, Fig. S8). In general, these results indicate that SYNPLA can detect synaptic plasticity induced by learning in brain regions where it is expected and under expected conditions. Furthermore, other circuits, which may not normally be examined for plasticity during a particular learning protocol, may be discovered to display plasticity under particular aspects of a learning protocol.
Discussion
Here we have demonstrated that SYNPLA can reliably detect synaptic potentiation in circuits participating in newly formed memories. While there are both intra-animal variability and interexperiment variability in SYNPLA signals, cLTP and classical defense conditioning reliably resulted in significant differences between control and test conditions in each independent experiment (see Materials and Methods for potential sources and recommendations to reduce experimental variability). SYNPLA can be used to probe plasticity in anatomically defined pathways as in the present study or in genetically defined pathways by limiting the expression of myc-NRXN only to neurons expressing Cre recombinase. Simultaneous screening of multiple pathways in the same animal might also be possible by exploiting orthogonal PLA probes (25). Unlike IEG-based screening approaches, which assay cell-wide changes, SYNPLA is synapse and pathway specific and directly assesses synaptic plasticity. Moreover, it is fast and easy to perform and so could be scaled up as a powerful brain-wide screen for behaviorally induced plasticity.
Materials and Methods
Information about the experimental models used (primary neuronal cultures, organotypic slices, and animals for ex vivo SYNPLA), colocalization analysis, and statistics can be found in the SI Appendix. Animal procedures were approved by the Cold Spring Harbor Laboratory or University of California San Diego Animal Care and Use Committee and were carried out in accordance with NIH standards.
Proximity Ligation Assay.
After fixation in 4% paraformaldehyde, we performed PLA using Duolink PLA reagents (Sigma Aldrich) largely according to the manufacturer’s instructions. For experiments on primary neuronal cultures, samples were incubated in 4 mM glycine for 5 min after fixation. Organotypic or ex vivo slices were permeabilized for 30 min using 0.2% TritonX-100. We then blocked all samples using Duolink blocking buffer (organotypic slices were blocked with BlockOneHisto [Nacalai Tesque]) at 37 °C for 1 h. We then applied primary antibodies (for Fig. 1 C and D, goat anti-myc [ab9132, Abcam] and rabbit anti-HA [ab9110, Abcam], both at 1:10,000 dilution; for Figs. 1 E and F, 2, and 3, goat anti-myc [ab9132, Abcam], 1:2,000 dilution and rabbit anti-GluA1 [AGC-004, Alomone Labs], 1:100 dilution [1:50 for Fig. 3]) in Duolink antibody diluent for 1 h at room temperature or overnight at 4 °C. For SI Appendix, Fig. S2 different primary antibodies were used as negative controls: guinea pig anti-GluA1 (AGP-009, Alomone Labs), rabbit anti-cFos (Cell Signaling Technology, #2250); all were at 1:100 dilution. For SI Appendix, Fig. S4 an antibody targeting endogenous NRXN (goat polyclonal from Abcam, ab77596) was used (1:100 dilution) instead of the anti-myc antibody. After washing 3 × 10 min in PLA wash buffer A, we probed the primary antibodies using Duolink PLA probes goat PLUS and rabbit MINUS at 37 °C for 2 h and then used Duolink In Situ Detection Reagents FarRed (Sigma Aldrich) to detect the proximity between the proteins of interest. Importantly, we found that performing the PLA reaction shortly after fixation greatly improved the SYNPLA signal to noise ratio, especially in ex vivo slices. Reactions performed on different days (with different stock reagents, etc.) could introduce variability; thus, we recommend that control and test conditions be processed in parallel with the same reagents (e.g., Fig. 3G).
Imaging.
To image PLA signals in an unbiased way, fields of view were selected using the myc-NRXN infection channel (i.e., GFP or mCherry channel) for all experiments. For Fig. 1 C and D, we subjected three separate coverslips containing both myc-NRXN- and HA-NLGN-expressing neurons and one coverslip containing only HA-NLGN-expressing neurons to PLA at each time point. Per coverslip, we acquired three z stacks containing a single mCherry-expressing cell using a 63× oil objective on a Zeiss laser scanning 780 confocal. For Fig. 2 B and C, we acquired 12 z stacks per condition (one coverslip per condition). In Fig. 2D, each dot is the average number of PLA puncta in 10 to 12 images of a single experiment, normalized to its control sample for each of six independent experiments. For Fig. 2 F and G, we used one to two slices per condition for each experiment. Three independent repeats are shown, totaling four slices per condition. For Fig. 3, we acquired 2 to 5 z stacks in the LA of each slice (one slice per animal). Fig. 3 F and K shows the number of PLA puncta detected for each z stack for the indicated animals. In Fig. 3 G and L, each dot represents the average number of PLA puncta for all individual animals. Fig. 3H (Fig. 3M) shows the same data as Fig. 3G (Fig. 3L), but the average PLA signal in each paired animal was normalized to its control. All images used for experiments shown in Figs. 1 E and F, 2, and 3 were acquired on an Olympus FV1000 confocal microscope with a 60× oil immersion objective. All images were obtained blind to condition.
PLA Puncta Detection.
For all experiments in primary cultures, we quantified the number of PLA dots across the entire thickness of the cell layer using a custom Fiji macro (consisting primarily of a maxima finding function). For organotypic slices samples, we quantified the number of PLA puncta in the CA1 region using a custom Fiji macro in a single z plane, ∼2 to 3 μm below the slice surface. PLA signal decreases to noise levels deeper into the tissue (SI Appendix, Fig. S5), likely due to limited reagent penetration. For ex vivo experiments, we quantified PLA across the entire thickness of the samples to account for tissue irregularity. Briefly, in ImageJ (Fiji), nuclei were subtracted from each z plane using the particle analysis and the image calculator tool. Next, a fast Fourier transform (FFT) band pass filter was applied to the resulting stack to enhance small particle contrast. Finally, using a threshold of 3 to 6% highlighting the brightest pixels, puncta were detected with the particle analysis tool (4 to 25 pixels in size, corresponding to 0.16 to 1 μm2). All images were analyzed blind to condition.
Data Availability.
All data relevant to this study are included in this paper or in the SI Appendix.
Supplementary Material
Acknowledgments
The following funding sources are acknowledged: NIH (Grants 5RO1NS073129 and 5RO1DA036913 to A.M.Z.; Grant 29R01MH049159 to R.M.; Grant U01MH109113 to A.M.Z. and R.M.), the Brain Research Foundation (Grant BRF-SIA-2014-03 to A.M.Z.), Intelligence Advanced Research Projects Activity (IARPA) (Grant MICrONS D16PC0008 to A.M.Z.), the Simons Foundation (Grant 382793/SIMONS to A.M.Z.), a Paul Allen Distinguished Investigator Award (A.M.Z.), the Shiley Foundation (R.M.), a Boehringer Ingelheim Fonds PhD fellowship (J.M.K.), and a Genentech Foundation PhD fellowship (J.M.K.). Resources from UCSD School of Medicine Microscopy Core were used to gather data (funded by P30 NS047101).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919911117/-/DCSupplemental.
Change History
July 21, 2021: The Acknowledgments have been updated.
References
- 1.LeDoux J. E., Emotion circuits in the brain. Annu. Rev. Neurosci. 23, 155–184 (2000). [DOI] [PubMed] [Google Scholar]
- 2.LeDoux J. E., Coming to terms with fear. Proc. Natl. Acad. Sci. U.S.A. 111, 2871–2878 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huganir R. L., Nicoll R. A., AMPARs and synaptic plasticity: The last 25 years. Neuron 80, 704–717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi T., Svoboda K., Malinow R., Experience strengthening transmission by driving AMPA receptors into synapses. Science 299, 1585–1588 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Matsuo N., Reijmers L., Mayford M., Spine-type-specific recruitment of newly synthesized AMPA receptors with learning. Science 319, 1104–1107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCormack S. G., Stornetta R. L., Zhu J. J., Synaptic AMPA receptor exchange maintains bidirectional plasticity. Neuron 50, 75–88 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Rumpel S., LeDoux J., Zador A., Malinow R., Postsynaptic receptor trafficking underlying a form of associative learning. Science 308, 83–88 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Gao C., Wolf M. E., Dopamine alters AMPA receptor synaptic expression and subunit composition in dopamine neurons of the ventral tegmental area cultured with prefrontal cortex neurons. J. Neurosci. 27, 14275–14285 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf M. E., Tseng K. Y., Calcium-permeable AMPA receptors in the VTA and nucleus accumbens after cocaine exposure: When, how, and why? Front. Mol. Neurosci. 5, 72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong Q., Znamenskiy P., Zador A. M., Selective corticostriatal plasticity during acquisition of an auditory discrimination task. Nature 521, 348–351 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li K., et al., βCaMKII in lateral habenula mediates core symptoms of depression. Science 341, 1016–1020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lüscher C., Malenka R. C., Drug-evoked synaptic plasticity in addiction: From molecular changes to circuit remodeling. Neuron 69, 650–663 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim C.-S., et al., Pharmacological rescue of Ras signaling, GluA1-dependent synaptic plasticity, and learning deficits in a fragile X model. Genes Dev. 28, 273–289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzowski J. F., et al., Mapping behaviorally relevant neural circuits with immediate-early gene expression. Curr. Opin. Neurobiol. 15, 599–606 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Söderberg O., et al., Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Peikon I. D., et al., Using high-throughput barcode sequencing to efficiently map connectomes. Nucleic Acids Res. 45, e115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basarsky T. A., Parpura V., Haydon P. G., Hippocampal synaptogenesis in cell culture: Developmental time course of synapse formation, calcium influx, and synaptic protein distribution. J. Neurosci. 14, 6402–6411 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Südhof T. C., Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoppini L., Buchs P. A., Muller D., A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods 37, 173–182 (1991). [DOI] [PubMed] [Google Scholar]
- 20.Otmakhov N., et al., Forskolin-induced LTP in the CA1 hippocampal region is NMDA receptor dependent. J. Neurophysiol. 91, 1955–1962 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Collingridge G. L., Kehl S. J., McLennan H., Excitatory amino acids in synaptic transmission in the Schaffer collateral-commissural pathway of the rat hippocampus. J. Physiol. 334, 33–46 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeDoux J., The amygdala. Curr. Biol. 17, R868–R874 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Proulx C. D., Hikosaka O., Malinow R., Reward processing by the lateral habenula in normal and depressive behaviors. Nat. Neurosci. 17, 1146–1152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shabel S. J., Wang C., Monk B., Aronson S., Malinow R., Stress transforms lateral habenula reward responses into punishment signals. Proc. Natl. Acad. Sci. U.S.A. 116, 12488–12493 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leuchowius K.-J., et al., Parallel visualization of multiple protein complexes in individual cells in tumor tissue. Mol. Cell. Proteomics 12, 1563–1571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relevant to this study are included in this paper or in the SI Appendix.