Recent genetic evidence has highlighted how the IFNL4 gene acts in a counterintuitive manner, as patients with a nonfunctional IFNL4 gene exhibit increased clearance of hepatitis C virus (HCV) but also increased liver inflammation. This suggests that the IFNL4 gene acts in a proviral and anti-inflammatory manner. These surprising but quite clear genetic data have prompted an extensive examination of the basic characteristics of the IFNL4 gene and its gene product, interferon lambda 4 (IFN-λ4). We have investigated the expression of the IFNL4 gene and found it to be poorly induced by viral infections. A thorough investigation of the IFNL4 promoter revealed a highly conserved and functional promoter, but also one that lacks the defining characteristic of interferons (IFNs), i.e., the ability to be effectively induced by viral infections. We suggest that the unique function of the IFNL4 gene is related to its noncanonical transcriptional regulation.
KEYWORDS: hepatitis C virus, interferons, transcriptional regulation
ABSTRACT
Interferon lambda 4 (IFN-λ4) is a recently identified enigmatic member of the interferon (IFN) lambda family. Genetic data suggest that the IFNL4 gene acts in a proviral and anti-inflammatory manner in patients. However, the protein is indistinguishable in vitro from the other members of the interferon lambda family. We have investigated the gene regulation of IFNL4 in detail and found that it differs radically from that of canonical antiviral interferons. Being induced by viral infection is a defining characteristic of interferons, but viral infection or overexpression of members of the interferon regulatory factor (IRF) family of transcription factors only leads to a minute induction of IFNL4. This behavior is evolutionarily conserved and can be reversed by inserting a functional IRF3 binding site into the IFNL4 promoter. Thus, the regulation of the IFNL4 gene is radically different and might explain some of the atypical phenotypes associated with the IFNL4 gene in humans.
IMPORTANCE Recent genetic evidence has highlighted how the IFNL4 gene acts in a counterintuitive manner, as patients with a nonfunctional IFNL4 gene exhibit increased clearance of hepatitis C virus (HCV) but also increased liver inflammation. This suggests that the IFNL4 gene acts in a proviral and anti-inflammatory manner. These surprising but quite clear genetic data have prompted an extensive examination of the basic characteristics of the IFNL4 gene and its gene product, interferon lambda 4 (IFN-λ4). We have investigated the expression of the IFNL4 gene and found it to be poorly induced by viral infections. A thorough investigation of the IFNL4 promoter revealed a highly conserved and functional promoter, but also one that lacks the defining characteristic of interferons (IFNs), i.e., the ability to be effectively induced by viral infections. We suggest that the unique function of the IFNL4 gene is related to its noncanonical transcriptional regulation.
INTRODUCTION
Interferons (IFNs) are divided into three families according to their distinct receptor utilization, namely, type I (e.g., interferons alpha and beta [IFN-α/β]), type II (interferon gamma [IFN-γ]), and type III (interferon lambda [IFN-λ]). Both type I and type III IFNs are potent antiviral cytokines (1). Type II IFN also possesses some antiviral effect, but its primary function is to link the innate and adaptive parts of the immune system (2). Type I and type III IFNs play a key role in innate immunity to viral infection, and their expression is induced by viral infection in both immune and nonimmune cells (3–5). Humans possess four type III IFN genes that are known as IFNL1, IFNL2, IFNL3, and IFNL4. IFNL1, IFNL2, and IFNL3 share a high degree of similarity and were identified by two independent teams in 2003 (6, 7) as a novel family of genes encoding virally induced IFNs. In the following decade, genome-wide association studies linked clearance of hepatitis C virus (HCV) to genetic variation within the type III IFN loci (8–11), and this subsequently led to the discovery of the IFNL4 gene when genetic data were compared with RNA sequencing analysis (12). The DNA sequence similarity between the IFNL4 gene and the IFNL1 to IFNL3 genes is relatively low, and at the protein level the identity is approximately 28% (13). The IFNL4 gene is well conserved among mammals, except in rodents, where the gene is absent.
Upon identification of the IFNL4 gene, the authors also identified a dinucleotide variant (ΔG/TT, rs368234815) situated in the first exon of the IFNL4 gene (12). The IFNL4-TT frameshift mutation was introduced during early human evolution, before the “out of Africa” scenario (14). There has subsequently been a positive selection for the IFNL4-TT allele that has resulted in this being the major allele in humans, but with major variations in allele frequency between different human populations. The ΔG/TT variation is associated with the rate of spontaneous HCV clearance as well as with the response to treatment (12). IFNL4-ΔG is the ancestral allele and generates the full-length interferon lambda 4 (IFN-λ4) protein, whereas the IFNL4-TT allele leads to a frameshift and therefore to aborted expression of IFN-λ4. Surprisingly, patients harboring the functional IFNL4-ΔG allele have a lower HCV clearance rate than that of patients who have the IFNL4-TT allele (12). Interestingly, the IFNL4-ΔG allele is also associated with lower levels of liver inflammation and fibrosis in HCV-infected patients (15–17), as well as in patients with nonalcoholic fatty liver disease (18, 19). Thus, the genetic evidence suggests that in vivo, the IFNL4 gene acts in a proviral and anti-inflammatory manner, quite in contrast to the proinflammatory and antiviral effect of other type III IFNs. Here, we will use the term “canonical IFNs” to refer to all type I and type III IFN genes except IFNL4.
Biochemically, the IFN-λ4 protein acts in a manner similar to that of the proteins encoded by the canonical type III IFN genes. It signals through the canonical IFN-λ receptor complex (20) and induces a set of genes highly similar to that induced by the canonical members of the IFN-λ family (20, 21). Despite its clear antiviral activity in vitro, the causal role of the IFN-λ4 protein in lower HCV clearance rates is supported by the finding that a single amino acid substitution of a proline to a serine at position 70 (IFN-λ4 P70S) in IFN-λ4 substantially affects the antiviral activity of the protein (22). HCV patients harboring the impaired IFN-λ4 S70 variant display lower IFN-stimulated gene (ISG) expression levels but better treatment response rates and better spontaneous clearance rates than those in patients carrying the fully active IFN-λ4 P70 variant (22). Finally, comprehensive genome-to-genome analysis in chronically infected HCV patients supports a role for the IFNL4 gene as the causative gene. In that study, IFNL4 genotype determined viral load and affected the evolution of HCV quasispecies within patients (23). Thus, the result is a paradox in which the IFN-λ4 protein is antiviral in vitro but appears to be proviral and anti-inflammatory in vivo.
Several attempts have been made to measure induction of the IFNL4 gene during viral infection, both in vivo and in vitro. Expression of IFNL4 mRNA was not detected in peripheral blood mononuclear cells from chronically infected HCV patients (24). Similarly, several other studies measured either no or very low IFNL4 mRNA expression in liver samples from patients with nonviral liver disease, chronic hepatitis B virus infection, or chronic HCV infection (25–27). Finally, both stimulation with pathogen-associated molecular patterns (PAMP) and viral infection of different hepatoma cell lines or primary human hepatocytes resulted in minimal expression of IFNL4 but strong expression of the canonical IFNL3 (28). Together these observations led to speculation that the IFNL4 promoter is nonfunctional and that this is part of a general trend during human evolution to abrogate expression of the IFN-λ4 protein.
It is currently unclear why the IFN-λ4 protein became a liability in humans, whereas it is retained in most other mammalian species. To explain the unique genetic observations linked to the IFNL4 gene, we searched for characteristics of the IFNL4 gene that differentiate it from the canonical IFNs. Here, we demonstrate that the IFNL4 promoter is indeed functional and that the mode of gene regulation differs substantially between the IFNL4 gene and canonical IFNs. Furthermore, we show that the unique features of the IFNL4 promoter are evolutionarily conserved.
RESULTS
Expression of IFNL4 mRNA is negligible upon viral infection in different human cell types.
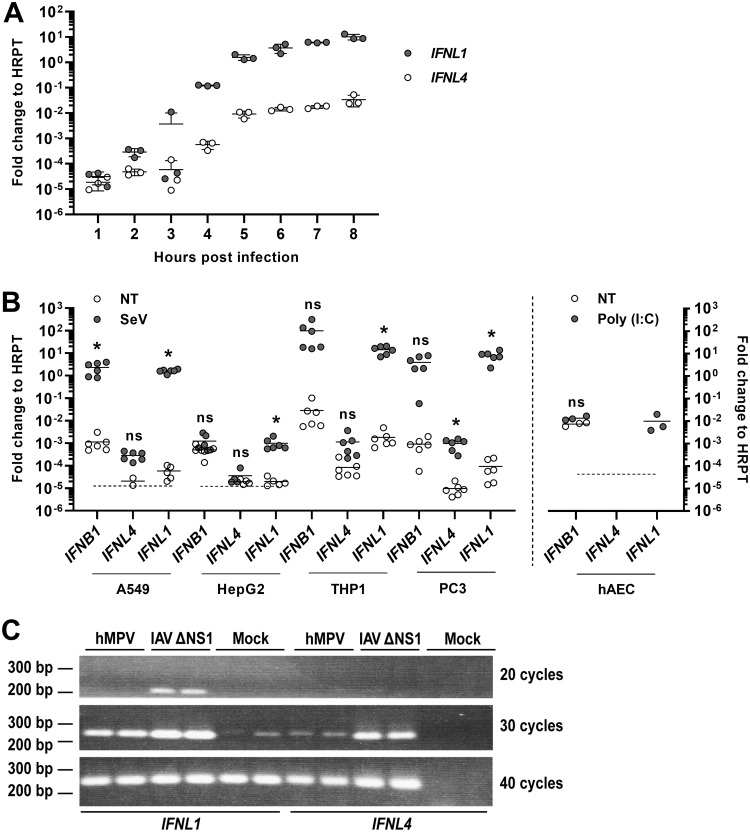
The expression of most IFNs is rapidly induced following a viral infection. Upon infection of human A549 cells with Sendai virus (SeV), expression of IFNL1 mRNA increased more than 100,000-fold within 5 to 6 h before it appeared to reach a plateau (Fig. 1A). The expression of IFNL4 mRNA also increased significantly within the first 5 to 6 h of infection, but far less than that of IFNL1. At 8 h after infection, the expression level of IFNL4 appeared to reach a plateau and was 300-fold lower than that of IFNL1. To further investigate the rather low expression level of IFNL4 mRNA, we compared the expression of this gene, as well as those of IFNL1 and IFNB1, in four different human cell lines with or without viral infection. Based on earlier results, we chose to measure mRNA levels at 6 h after infection or mock infection. As expected, infection with SeV led to a robust expression of IFNB1 and IFNL1 mRNA in A549, THP-1, and PC-3 cells, and a moderate expression of IFNL1 but not IFNB1 in HepG2 cells (Fig. 1B). In contrast, the expression of IFNL4 was much lower than that of the two canonical IFNs and, even after viral infection, expression of IFNL4 rarely exceeded the baseline expression of IFNB1 and IFNL1. With the exception of the HepG2 cells, the absolute expression of IFNB1 and IFNL1 was a 1,000-fold higher than that of IFNL4 following viral infection. Of note, in both A549 and HepG2 cells, the IFNL1 and IFNL4 mRNA levels were close to the detection limit in the absence of viral infection, and for several of the replicates, no product was detected within 40 cycles; those measuring points were thus omitted (Fig. 1B). Cell lines sometimes exhibit minor or major defects in innate immune pathways, and therefore we tested the expression of IFNL4 mRNA in primary human airway epithelial cell (hAEC) cultures stimulated with poly(I·C) and compared it to that of IFNB1 and IFNL1 mRNAs (Fig. 1B). In these cells, poly(I·C) stimulation led to a robust expression of IFNL1 mRNA, whereas IFNB1 mRNA levels were relatively high even in the absence of poly(I·C) and did not increase further after poly(I·C) treatment. In contrast, IFNL4 mRNA levels were below the detection limit both with and without poly(I·C) stimulation.
FIG 1.
Expression of the IFNL4 gene is not virus inducible. (A) A549 (an adenocarcinomic alveolar basal epithelial cell line) cells were infected with SeV, and the expression levels of IFNL4 and IFNL1 genes were quantified by reverse transcription quantitative real-time PCR (RT-qPCR) at the indicated time points. Data were calculated relative to internal expression of HPRT. The experiment was performed in biological triplicates, and data are presented as a scatter plot with mean ± standard deviation (SD) (n = 3). (B) A549, HepG2 (a hepatocellular carcinoma cell line), macrophage-like differentiated THP-1 (a monocyte-like cell line before differentiation), and PC-3 (a prostate cancer cell line) cells were mock infected or infected with SeV for 6 h, whereas human airway epithelial cell (hAEC) cultures were mock treated or treated with poly(I·C) for 18 h. Expression levels of IFNB1, IFNL4, and IFNL1 genes were quantified by RT-qPCR. Data were calculated relative to internal expression of HPRT. The dashed line indicates the detection limit in those cases where mRNA levels could not be determined for some replicates because they were too low. The experiment was performed in biological sextuplicates (left panel) or triplicates (right panel), and data are presented as a scatter plot with mean ± SD (n = 3 or 6). Statistical significance was determined using an unpaired t test. *, 0.01 < P < 0.05; ns, P ≥ 0.05. (C) A549 cells were mock infected or infected with human metapneumovirus (hMPV) or influenza A virus (IAV) ΔNS1 for 18 h. Expression levels of IFNL1 and IFNL4 mRNA were assessed by semiquantitative PCR and vizualised by agarose gel electrophoresis. The experiment was performed in biological duplicates.
The overall low expression of IFNL4 mRNA was surprising to us because a substantial expression of the IFNL4 gene was previously reported to occur in epithelial cells infected by human metapneumovirus (hMPV) (29). Therefore, we repeated this experiment by measuring induction of IFNL transcripts in A549 cells infected by an influenza A virus (IAV) variant lacking its NS1 gene (IAV ΔNS1) or by hMPV. In contrast to previously published data, human metapneumovirus (hMPV) infection did not lead to any substantial expression of IFNL4 mRNA in our experiments, but it did induce the expression of IFNL1 mRNA (Fig. 1C). As expected, infection with IAV ΔNS1 resulted in a strong induction of IFNL1 mRNA but only a minor induction of IFNL4 mRNA. Altogether, IFNL4 expression is negligible compared to that of IFNL1 in A549 cells after hMPV or IAV ΔNS1 infection (Fig. 1C). This leads us to suggest that IFNL4 is a noncanonical IFN, as it lacks the otherwise defining characteristic of IFNs, i.e., their strong induction upon viral infection.
The IFNL1 but not the IFNL4 promoter is activated by viral infection or overexpression of MAVS.
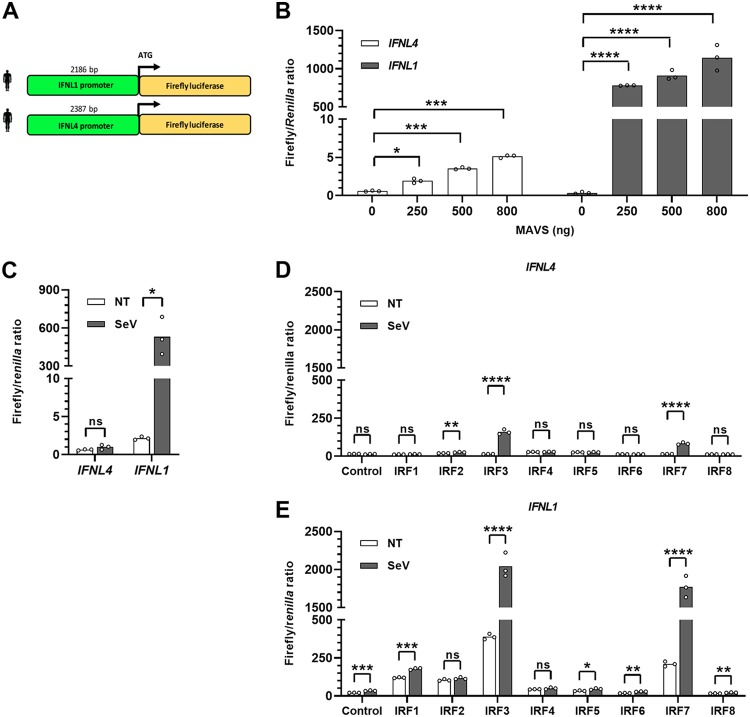
To create a system that allowed us to characterize the IFNL4 promoter in detail, we cloned a 2,186-bp fragment of the IFNL1 promoter and a 2,387-bp fragment of the IFNL4 promoter in front of a firefly luciferase gene (Fig. 2A). Because interferon regulatory factor 3 (IRF3) is a crucial transcription factor governing expression of canonical IFNs, we used two ways of activating IRF3, namely, overexpression of the adaptor protein mitochondrial antiviral-signaling protein (MAVS), which signals upstream of IRF3, or infection with SeV. First, HEK293T cells were transfected with either the IFNL1 or IFNL4 promoter reporter construct along with increasing amounts of a construct encoding MAVS (Fig. 2B). The results show that the IFNL1 promoter is potently induced by MAVS overexpression, whereas this is not the case for the IFNL4 promoter. Even at the largest amount of MAVS-encoding construct used, the IFNL1 promoter gives an approximately 230-fold stronger luciferase activity than that of the IFNL4 promoter. Next, we repeated the experiment using SeV infection instead of overexpression of MAVS, with the similar result that the IFNL1 but not the IFNL4 promoter was potently induced by SeV infection (Fig. 2C). This is consistent with previous results showing a very low IFNL4 mRNA expression level in human cell lines.
FIG 2.
The IFNL4 promoter is not activated by the same stimuli that readily activate the IFNL1 promoter. (A) The promoter regions of the human IFNL1 and IFNL4 genes corresponding to 2,186 and 2,387 bp, respectively, upstream of their translation start site were inserted in front of the firefly luciferase coding sequence in the pGL3.1-Basic plasmid. (B) HEK293T cells were cotransfected with a pGL3.1 plasmid containing the IFNL1 or IFNL4 promoter, a plasmid constitutively expressing the Renilla luciferase gene, and increasing amounts of a plasmid expressing MAVS. At 24 h posttransfection, firefly/Renilla luciferase activity was quantified. The experiment was performed in biological triplicates and the data are presented as a bar chart and scatter plot with mean ± SD (n = 3). (C) HEK293T cells were cotransfected with a pGL3.1 plasmid containing the IFNL1 or IFNL4 promoter and a plasmid constitutively expressing the Renilla luciferase gene. At 24 h posttransfection, the cells were infected with SeV for another 24 h before quantification of firefly/Renilla luciferase activity. One representative out of two independent experiments is shown, each with biological triplicates. The data are presented as a bar chart and scatter plot with mean ± SD (n = 3). (D and E) IRF3-knockout HEK293T cells were cotransfected with a pGL3.1 plasmid containing the IFNL1 (D) or IFNL4 (E) promoter in front of a firefly luciferase gene, a plasmid constitutively expressing the Renilla luciferase gene, and a plasmid constitutively expressing IRF1, IRF2, IRF3, IRF4, IRF5, IRF6, IRF7, or IRF8. At 24 h posttransfection, the cells were infected with SeV for another 24 h before quantification of firefly/Renilla luciferase activity. One representative out of two independent experiments is shown, each with biological triplicates. The data are presented as a bar chart and scatter plot with mean ± SD (n = 3). Statistical significance was determined using an unpaired t test. ****, P < 0.0001; ***, 0.0001 < P < 0.001; **, 0.001 < P < 0.01; *, 0.01 < P < 0.05; ns, P ≥ 0.05.
The IFNL1 but not the IFNL4 promoter is activated by IRF3 and IRF7 following SeV infection.
The transcription factors IRF3 and IRF7 both play an important role in controlling expression of canonical IFNs. However, they are member of a larger family consisting of nine members, IRF1 to IRF9. It is possible that activation of the IFNL4 promoter requires other members of the IRF family than those needed for expression of the canonical IFNs. Therefore, we transfected expression constructs encoding the different IRF members into IRF3-knockout HEK293T cells together with either the IFNL4 or the IFNL1 promoter reporter construct and then infected the cells with SeV before measuring luciferase activity. Overexpression of both IRF3 and IRF7 led to a small, yet significant induction of the IFNL4 promoter upon virus stimulation. However, this induction is negligible compared to that of the IFNL1 promoter (Fig. 2D) and underlines the unique regulatory requirement of the IFNL4 promoter. In contrast, the IFNL1 promoter was induced, as expected, by both IRF3 and IRF7 (Fig. 2E).
Is the behavior of the IFNL4 promoter evolutionarily conserved?
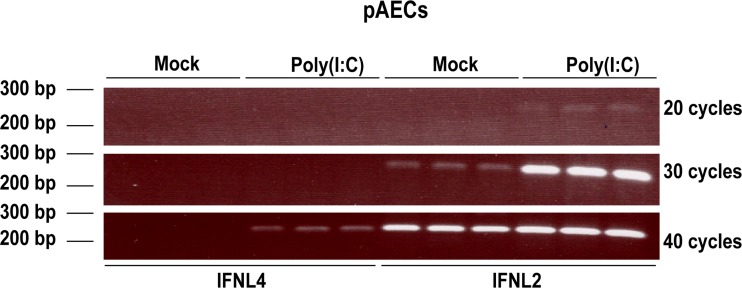
The above-described experiments showed that the human IFNL4 promoter is functionally different from the human IFNL1 promoter. However, since there has been selection against a functional IFNL4 gene in humans (SNP ΔG/TT, rs368234815), it is possible that low IFNL4 promoter activity is a result of an evolutionary pressure to reduce IFNL4 expression. If this is the case, the low activity of the IFNL4 promoter should be specific to humans, as selection against an active form of the IFNL4 gene (ΔG/TT, rs368234815) is a recent event in human evolution (14). To investigate this, we tested the expression of IFNL4 mRNA in pigs, as well as the activity of IFNL4 promoters from several mammalian species. We used primary porcine airway epithelial cell cultures stimulated with poly(I·C) and measured IFNL2 and IFNL4 mRNA levels by semiquantitative PCR (semi-qPCR) (Fig. 3). The expression of the porcine IFNL2 gene is clearly induced by poly(I·C) after 30 cycles, whereas IFNL4-specific primers only yielded a faint band after 40 cycles, indicating a rather low expression of IFNL4 following poly(I·C) stimulation. This suggests that the difference in the expression of IFNL4 compared to that observed for canonical IFNs is not unique to humans and, accordingly, seems not to be the result of the recent selection against a functional IFNL4 gene in humans.
FIG 3.
The IFNL4 gene is poorly expressed in primary porcine epithelial cells. Porcine airway epithelial cells (pAECs) were treated with 10 μg/ml poly(I·C) via the basolateral surface for 18 h. Expression levels of IFNL2 and IFNL4 mRNA were assessed by semiquantitative PCR and visualized by agarose gel electrophoresis. The experiment was performed in biological triplicates.
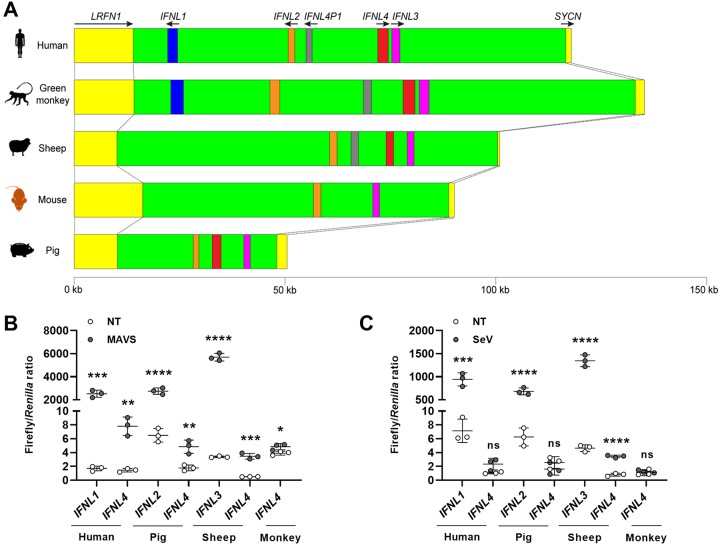
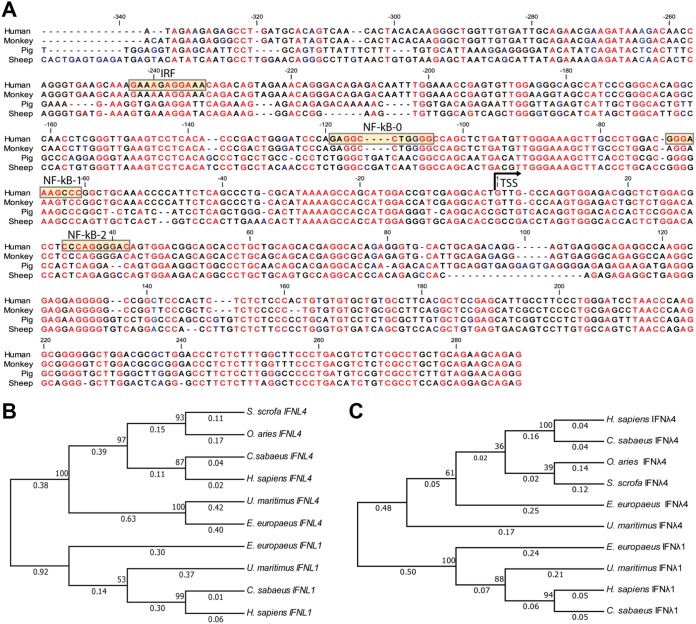
This observation prompted us to investigate the activities of the IFNL4 promoter from a number of mammalian species. The leucine-rich repeat and fibronectin type III domain containing 1 (LRFN1) and syncollin (SYCN) genes, which flank each side of the type III IFN loci, are evolutionarily conserved among mammals (Fig. 4A). We used those genes to mark the borders of the type III IFN loci and then analyzed the genomic loci in a number of mammalian species to identify IFNL4 orthologues. We analyzed the genomic loci from humans, African green monkeys, sheep, pigs, and house mice because in these species the type III IFN loci are fully sequenced. The IFNL1 gene is present in humans and the African green monkey but is absent in sheep, mice, and pigs, whereas the IFNL2 and/or IFNL3 genes can be found in all five species (Fig. 4A). Many mammals seem to have two highly similar IFNL4 genes, but in humans, one of these IFNL4 genes has become a pseudogene, whereas mice have no IFNL4 genes. As for sheep and pigs, sheep, like many other ungulates, possess two IFNL4 genes, while pigs only possess one IFNL4 gene. Thus, humans and pigs have a single IFNL4 gene, while African green monkeys and sheep have two IFNL4 genes. We performed an evolutionary analysis of the IFNL4 genes we identified and concluded that independent duplications of the IFNL4 gene must have occurred several times during mammalian evolution. This makes it difficult to precisely distinguish between true paralogues and orthologous genes, and we therefore chose for our investigation those IFNL4 genes that had the same orientation as the human IFNL4 gene.
FIG 4.
The weak functionality of the IFNL4 promoter is evolutionarily conserved among mammals. (A) Overview of the type III IFN loci in humans, African green monkeys, sheep, mice, and pigs. The type III IFN genes illustrated by different colors share a synteny in mammals and are flanked on each side by the leucine-rich repeat and fibronectin type III domain containing 1 (LRFN1) and syncollin (SYCN) genes. Black arrows indicate the direction of transcription for each gene. (B) HEK293T cells were cotransfected with a pGL3.1 plasmid containing IFNL4 or the IFNL1, IFNL2, or IFNL3 promoter from humans, African green monkeys, pigs, or sheep; a plasmid constitutively expressing the Renilla luciferase gene; and a plasmid constitutively expressing MAVS. At 24 h posttransfection, firefly/Renilla luciferase activity was quantified. One representative out of two independent experiments is shown, each with biological triplicates. The data are presented as a scatter plot with mean ± SD (n = 3). (C) HEK293T cells were cotransfected with a pGL3.1 plasmid containing IFNL4 or the IFNL1, IFNL2, or IFNL3 promoter from humans, African green monkeys, pigs, and sheep and a plasmid constitutively expressing the Renilla luciferase gene. At 24 h posttransfection, the cells were infected with SeV for another 24 h before quantification of firefly/Renilla luciferase activity. One representative out of two independent experiments is shown, each with biological triplicates. The data are presented as a scatter plot with mean ± SD (n = 3). Statistical significance was determined using an unpaired t test. ****, P < 0.0001; ***, 0.0001 < P < 0.001; **, 0.001 < P < 0.01; *, 0.01 < P < 0.05; ns, P ≥ 0.05.
To describe if the functionality of the IFNL4 promoter is evolutionary conserved, we cloned the IFNL4 promoters from African green monkeys, sheep, and pigs and added them to our comparison. As a control, we also cloned the IFNL2 or IFNL3 promoter from these species, except from the African green monkey, where it was not possible due to technical limitations. To compare the different IFNL4 promoters, HEK293T cells were transfected with the different type III IFN promoter reporter constructs along with a construct encoding MAVS (Fig. 4B). Our comparison showed that the IFNL4 promoter is induced by overexpression of MAVS to a much lower degree than the IFNL1, IFNL2, and IFNL3 promoters in all four species that we have tested here. The same results are also seen when we stimulated cells with SeV infection instead of MAVS overexpression (Fig. 4C). One concern in using nonhuman promoters in HEK293T cells is the compatibility of those promoters with the human system. However, we consistently saw strong induction of the IFNL1, IFNL2, and IFNL3 genes and, furthermore, the data from the primary pig airway epithelial cells was in agreement with the data obtained using the pig promoters in the HEK293T cells. Together, the results suggest that the behavior of the IFNL4 promoter appears to be conserved among mammals.
The IFNL4 promoter is highly conserved and forms a separate evolutionary clade.
The evolutionarily conserved behavior of the IFNL4 promoter prompted us to examine the degree of sequence conservation within the IFNL4 promoter region. Figure 5A shows a nucleotide alignment of the first 600 bp of the IFNL4 promoters, counting from the translational start site, from those species examined in the previous promoter activity assay. We chose the translational start site to anchor this alignment for the following two reasons: (i) it is clearly defined for all species, and (ii) regulatory elements can be found both upstream and downstream of the transcriptional start site. The analysis demonstrated a high degree of conservation, suggesting that there has been a significant selective pressure on this region throughout mammalian evolution.
FIG 5.
Conservation of the IFNL4 promoter among mammals. (A) Alignment of the proximal/core promoter and the 5′ UTR of IFNL4 from humans, African green monkeys, pigs, and sheep. The transcriptional start site (TSS), as well as putative IRF and NF-κB binding sites, are indicated in the human sequence. (B) Phylogenetic tree of IFNL1 and IFNL4 from human (Homo sapiens), African green monkey (Chlorocebus sabaeus), sheep (Ovis aries), pig (Sus scrofa), western European hedgehog (Erinaceus europaeus), polar bear (Ursus maritimus) based on the sequence of their proximal/core promoter and 5ʹ UTR. The sequences were aligned using ClustalW, and the tree was generated by the neighbor-joining method. (C) Same tree as in panel B but using the amino acid sequences of the proteins instead. Both trees are drawn to scale, with branch lengths measured in the number of substitutions per site.
Next, we calculated an evolutionary tree from a set of IFNL4 and IFNL1 promoter sequences. The promoter sequences form two clearly separated clades according to which genes they belong to, confirming our previous conclusion that the IFNL4 promoter is both evolutionarily and functionally distinct from the canonical IFN promoters (represented here by the IFNL1 promoter) (Fig. 5B). We also calculated a tree based upon the protein sequences of IFN-λ1 and IFN-λ4 from the same set of species and, as previously observed (3), the IFN-λ4 sequences form a separate and distinct clade (Fig. 5C).
The basic IFNL4 promoter is functional and can be activated by an enhancer element present in the IFNL1 promoter.
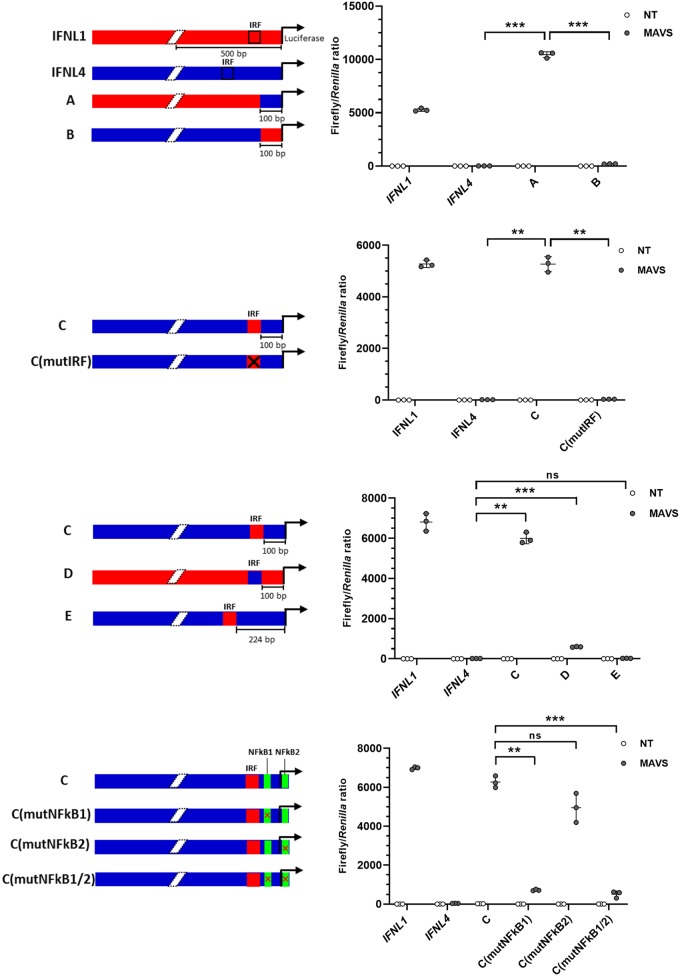
A promoter region can be described, in a somewhat simplified way, as consisting of a core promoter where the RNA polymerase II complex assembles and a series of upstream and/or downstream regulatory elements. To test if the core promoter of IFNL4 is functional, we constructed chimera A by combining 100 bp of proximal IFNL4 promoter and the upstream 1,989 bp of the IFNL1 promoter (Fig. 6A). Chimera A is active upon overexpression of MAVS, whereas the inverse chimera B with the IFNL1 core promoter fused to the upstream regulatory elements of the IFNL4 promoter was largely inactive, indicating that the IFNL4 core promoter can be activated by enhancer elements present in the IFNL1 promoter. To determine if the IRF binding site of the IFNL1 promoter functions as an enhancer in the context of the IFNL4 promoter, we constructed chimera C. In chimera C, the IRF binding motif from the IFNL1 promoter was inserted into the IFNL4 promoter in the same position that it occupies in IFNL1 promoter, which is 100 bp upstream of the transcription start site (TSS) (Fig. 6B). Chimera C is active, whereas chimera C with a mutated IRF site (mutIRF) is inactive (the IRF binding site was mutated by changing nucleotides CAGTTTC of this motif into AAGCAGA). Thus, by inserting a functional IRF binding site into the context of the IFNL4 promoter, the promoter becomes inducible by viral infection. These results confirm that the IFNL4 promoter is a potentially functional core promoter sequence.
FIG 6.
Insertion of a functional IRF binding site in the IFNL4 promoter renders it virus inducible. (A to D) HEK293T cells were cotransfected with a pGL3.1 plasmid containing the IFNL1 or IFNL4 promoter or chimeras thereof, a plasmid constitutively expressing the Renilla luciferase gene, and a plasmid constitutively expressing MAVS. At 24 h posttransfection, firefly/Renilla luciferase activity was quantified. One representative out of two independent experiments is shown, each with biological triplicates. The data are presented as a scatter plot with mean ± SD (n = 3). (A) In chimeras A and B, the first 100 bp upstream of the TSS has been swapped between the IFNL1 and IFNL4 promoters, as indicated by the colors in the diagram. (B) In chimera C, the IRF binding site from IFNL1 was inserted into the IFNL4 promoter at the same relative position it occupied in IFNL1. In chimera C (mutIRF), the IFNL1 IRF binding site in chimera C was mutated to render it inactive. (C) In chimera D, the IRF binding site in IFNL1 was replaced with the putative IRF binding site from IFNL4. In chimera E, the putative IRF binding site in IFNL4 was replaced with the IRF binding site from IFNL1. (D) In chimera C (mutNF-κB1), the NF-κB binding site located upstream of the TSS in IFNL4 was mutated to render it inactive. In chimera C (mutNF-κB2), the NF-κB binding site located downstream of the TSS in IFNL4 was mutated to render it inactive. In chimera C (mutNF-κB1/2) both NF-κB binding sites were simultaneous mutated. Statistical significance was determined using analysis of variance (ANOVA) and Dunnett’s T3 multiple-comparison test. ***, 0.0001 < P < 0.001; **, 0.001 < P < 0.01; ns, P ≥ 0.05.
A nonfunctional and out of place IRF binding site is found within the IFNL4 promoter.
Above, we demonstrated that insertion of the IRF site from the IFNL1 promoter into the IFNL4 promoter resulted in a virus-inducible activation of the IFNL4 promoter. However, one putative IRF site exists within the IFNL4 promoter. Replacing the canonical IRF site found in the IFNL1 promoter with the putative IRF binding site from the IFNL4 promoter resulted in chimera D (Fig. 6C). Chimera D exhibited only minimal activity upon MAVS overexpression, implying that the IRF binding site of IFNL4 is not functional within the context of the IFNL1 promoter. Not only does the sequence of the IRF binding sites differ between the IFNL1 and IFNL4 promoters, but their positions differ as well. While the IRF binding site in the IFNL1 promoter is located relatively close to the TSS (100 bp from the TSS), the IRF binding site in the IFNL4 promoter is located further upstream (224 bp from the TSS). This is more distant from the TSS compared to most canonical IFN promoters. In chimera E, we inserted the IRF motif of the IFNL1 promoter into the position of the putative IRF binding site already found within the IFNL4 promoter. Thus, chimera C and E differ only by the position of the inserted IRF binding site, yet only chimera C is activated by MAVS overexpression, which shows that the position of the IRF binding site does matter (Fig. 6C). In conclusion, although the IFNL4 promoter contains a putative IRF binding site, it seems to be nonfunctional.
The IFNL4 promoter contains functional NF-κB sites.
Efficient induction of IFN normally requires collaboration between the transcription factors IRF3 and NF-κB, and we therefore searched for NF-κB binding sites at the proximal end of the IFNL4 promoter. Two such sites were identified. To test if these sites potentially could be involved in conferring transcriptional activity, we mutated them individually or together in context of the transcriptionally active chimera C (Fig. 6D). Both mutNF-κB1 and mutNF-κB1/2 have reduced transcription, whereas mutNF-κB2 has a minimal effect (Fig. 6D). This indicates that the NF-κB1 site is required for transcriptional activity of chimera C, whereas the NF-κB2 site is less important.
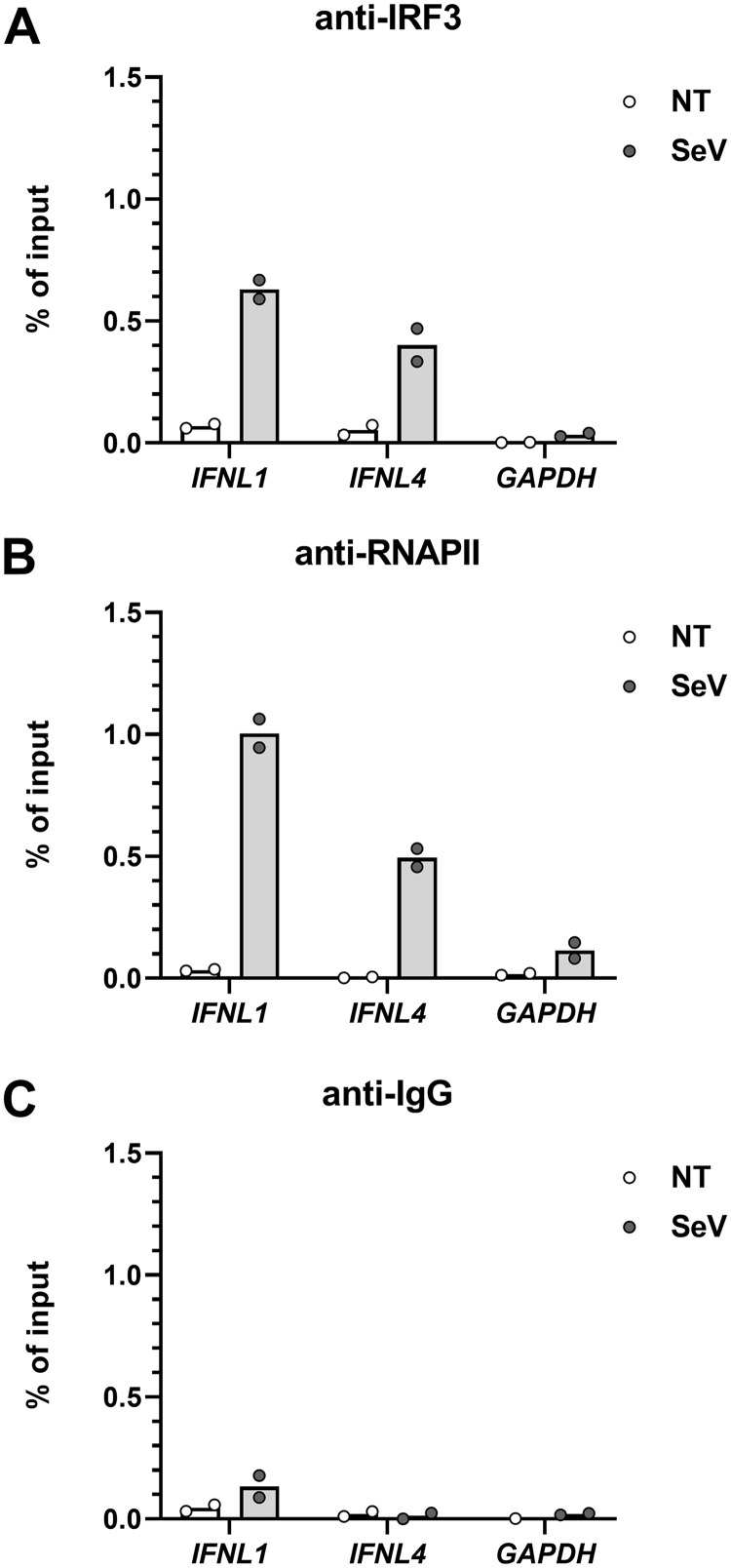
Both IRF3 and RNA polymerase II are recruited to the IFNL4 promoter in response to viral infection.
To investigate if the IRF3 transcription factor and RNA polymerase II are recruited to the promoter of IFNL1 and IFNL4, we performed chromatin immunoprecipitation (ChIP) assays on A549 cells infected by SeV. For this purpose, we used antibodies against IRF3 and RNA polymerase II, as well as an antibody against IgG as a control (Fig. 7A to C). The data show that IRF3 is recruited to the promoter region of both IFNL1 and IFNL4 but with a tendency toward lower enrichment of IRF3 on the IFNL4 promoter region after virus infection compared to that on IFNL1 (Fig. 7A). Moreover, we found that RNA polymerase II occupies both promoters after SeV infection, but more RNA polymerase II was detected at the IFNL1 promoter (Fig. 7B). Thus, decreased IRF3 and RNA polymerase II recruitment can account for at least some of the low response to viral infection exhibited by the IFNL4 promoter compared to the that of IFNL1 promoter.
FIG 7.
Recruitment of IRF3 and RNA polymerase II to the IFNL4 and IFNL1 promoters. A549 cells were mock infected or infected with SeV for 4 h before performing chromatin immunoprecipitation (ChIP) with antibodies against IRF3 (A), RNA polymerase II (B), and IgG as the control (C). One representative out of two independent experiments is shown, each with biological duplicates. The data are presented as a bar chart and scatter plot with mean.
DISCUSSION
Based on genetic data, unique properties have been attributed to the IFNL4 gene. IFNL4 protects against liver fibrosis (17, 19), and hence it is assumed to help dampen or control inflammation in both HCV-infected patients and patients with nonalcoholic steatosis (NASH) (15). Furthermore, its ablation facilitates both spontaneous and treatment-induced HCV clearance (12). These observations are counterintuitive when taking into account that IFNs are normally both antiviral and proinflammatory and suggest that the IFNL4 gene is functionally distinct from those encoding the canonical IFNs. We have conducted a series of experiments to biochemically characterize the IFN-λ4 protein, but found that the biochemical properties of the IFN-λ4 protein are highly similar to those of other members of the IFN-λ family, i.e., the IFN-λ4 protein signals through the same receptor and induces a set of genes highly similar to those induced by the IFN-λ3 protein (20, 21).
A defining characteristic of IFNs is their ability to be induced by viral infection through the transcription factors IRF3 or IRF7 (30). Here, we demonstrate that, while the IFNL4 gene has a putative IRF3/7 binding site, this site is either nonfunctional or functions very poorly, resulting in little or no transcription of the IFNL4 gene in response to viral infections. This agrees well with previous data showing very low levels of IFNL4 mRNA in the liver of HCV-infected patients (25) but contradicts other findings claiming that IFNL4 mRNA expression is induced by viral infection in vitro (29, 31).
A previously published study showed that hMPV infection led to a strong induction (approximately 40,000-fold) of IFNL4 mRNA expression in A549 cells compared to that in noninfected cells (29). However, when we tested IFNL4 gene expression in hMPV-infected A549 cells, we found that the IFNL4 expression was weak and much lower than IFNL1 expression under the same conditions. A different study showed induction of the IFNL4 mRNA, measured as fold induction, in SeV-infected PC-3 cells (31). We measured IFNL4 mRNA levels in PC-3 cells under similar conditions and, although there was an induction of the IFNL4 mRNA in infected versus noninfected cells, the expression level was low and comparable to the level of IFNL1 mRNA found prior to viral infection. During a viral infection, large amounts of canonical IFNs are produced and, in that context, the rather marginal IFNL4 expression upon viral infection is unlikely to have significant role. Hence, we do not believe that the low level of IFNL4 mRNA produced by PC-3 or A549 cells is of physiological relevance.
We performed a series of assays on different IFNL4 and IFNL1 promoter chimeras using a luciferase reporter system in HEK293 cells. Those data clearly show that inserting a functional IRF3 site at the proper position renders the IFNL4 promoter virus inducible. We speculate that the IFNL4 promoter is regulated by yet unknown stimuli, and the evolutionary conservation of both the sequence of the IFNL4 promoter region and its functional characteristics supports this. Our efforts to identify such a stimulus has so far been in vain, but our preliminary data suggest that neither endoplasmic reticulum (ER) stress, nor lipopolysaccharides (LPS), nor activation of the inflammasome leads to activation of the IFNL4 gene.
Other examples of noncanonical regulation of IFN genes exist. The IFNE gene, in contrast to the other type I IFN genes, is not induced by viral infection but is specifically expressed in the female reproductive tract, where it protects against infection. Furthermore, the expression of the IFNE gene is regulated by estrogen (32).
To sum up, we conclude that the regulation of the IFNL4 gene is a conserved feature among mammals and that the IFNL4 gene is a noncanonical member of the type III IFN family, as it is regulated in a unique way that differs from that of the canonical IFNs.
MATERIALS AND METHODS
Cells.
Cell lines used in this study were PC-3 (prostate cancer cell line), A549 (human lung epithelial cells), HepG2 (liver hepatocellular carcinoma), THP-1 (human monocytic cells), and IRF3-knockout HEK293T cells (human embryonic kidney cells). PC-3 cells were cultured in F12K medium supplemented with 2 mM l-glutamine, 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. THP-1 cells were cultured in the RPMI 1640 medium supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. All other cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Well-differentiated human airway epithelial cell cultures from three biological donors were isolated, established, and maintained as previously described (33). Primary porcine tracheobronchial airway epithelial cells were isolated from postmortem material obtained from specific pathogen-free (SPF) pigs. Isolation of cells was performed with protease and DNase digestion, and primary porcine tracheobronchial cells were cultured as previously described (33) but with a few modifications. For cellular differentiation, the human epidermal growth factor concentration was increased 10-fold in the air-liquid interface medium, whereas retinoic acid was 2-fold more concentrated. The epithelial layer was allowed to differentiate for at least 4 weeks prior to stimulation experiments.
Plasmids.
The different promoter sequences used in this study were cloned from genomic DNA using standard cloning techniques. The sources were THP-1 cells for the human IFNL1 and IFNL4 promoters, Vero cells for the African green monkey IFNL4 promoter, Texel sheep peripheral blood mononuclear cells for the sheep IFNL3 and IFNL4, and Göttingen minipig tissue for the pig IFNL2 and IFNL4 promoters. Because the transcriptional start site has not been experimentally verified for most of these genes, we chose to always include the 5′ UTR in the cloned sequence. The sequences were cloned into the multiple cloning site of the pGL3.1-Basic plasmid (Promega), and afterwards the remaining sequence between the inserted promoter sequence and the translational start site of the firefly luciferase gene was deleted by site-directed mutagenesis. The different chimeras and mutants of the human IFNL1 and IFNL4 promoters were constructed using standard cloning techniques and site-directed mutagenesis. In chimera A, nucleotides −1 to −100 of the IFNL1 promoter were replaced by nucleotides −1 to −100 of the IFNL4 promoter. In chimera A, nucleotides −1 to −100 of the IFNL4 promoter were replaced by nucleotides −1 to −98 of the IFNL1 promoter. In chimera C, nucleotides −101 to −132 of the IFNL4 promoter were replaced with nucleotides −101 to −134 of the IFNL1 promoter. In chimera C (mutIRF), nucleotides −129 to −123 from the IFNL1 promoter in chimera were changed from CAGTTTC to AAGCAGA. In chimera D, nucleotides −117 to −132 of the IFNL1 promoter were replaced with nucleotides −225 to −235 of the IFNL4 promoter. In chimera E, nucleotides −225 to −235 in the IFNL4 promoter were replaced with nucleotides −117 to −132 of the IFNL1 promoter. In chimera C (mutNF-κB1), nucleotides −69 to −66 of the IFNL4 promoter in chimera C were changed from ACCC to CCCT. In chimera C (mutNF-κB2), nucleotides 32 to 35 of the IFNL4 promoter in chimera C were changed from CCCA to GGGT. Chimera mutNF-κB1/2 is a combination of the two previous chimeras.
The plasmids used for expressing IRF2, IRF4, and IRF8 (pEF-IRF2, pSV-LS-IRF4, and pICSBP) were kind gifts from Takashi Fujita (Kyoto University, Japan), whereas the plasmids for expression of IRF3 and MAVS (pcDNA3-IRF3 [catalog no. 32713] and pEF-BOS-MAVS [catalog no. 27224]) were from Addgene. The plasmids used for expressing IRF1, IRF5, IRF6, and IRF7 were constructed in our laboratory. The sequences of all plasmids used in this study were verified by Sanger sequencing (GATC Biotech).
Infections and stimulations.
For quantitative PCR (qPCR) assays, cells were seeded in 6-well tissue culture plates at a density of 6 × 105 cells per well. For the THP-1 cells, phorbol 12-myristate 13-acetate (PMA) was added to a final concentration of 100 nM to trigger differentiation into macrophages. After resting for 24 h, the cells were infected with 40 hemagglutinating units (HAU) SeV (Cantell strain) per well for 6 h. Infection of A549 cells with hMPV was performed as previously described (29).
For luciferase assays, cells were infected 24 h posttransfection with 20 HAU SeV (Cantell strain) per well for 24 h.
For ChIP assays, cells were seeded in 150-mm-diameter tissue culture dishes at a density of 1 × 107 cells per dish. After resting for 24 h, the cells were infected with 560 HAU SeV (Cantell strain) per dish for 4 h.
For qPCR and semi-qPCR, human or porcine airway epithelial cells were treated with 10 μg/ml poly(I·C) via the basolateral surface for a duration of 18 h prior to cell lysis and total RNA extraction.
Quantitative real-time and semiquantitative PCR.
Total RNA was extracted with the E.Z.N.A. total RNA kit I (Omega Bio Tek) according to the manufacturer’s instructions. cDNA synthesis was performed with 0.5 μg RNA using RevertAid reverse transcriptase and a random hexamer primer according to the manufacturer’s instructions (Thermo Fisher Scientific). The cDNA was quantified by qPCR using SYBR green I (Roche) and a LightCycler 480 instrument II (Roche). The following primers were used: human IFNB1, forward ACGCCGCATTGACCATCTAT and reverse GTCTCATTCCAGCCAGTGCT; human IFNL1, forward TTCCAAGCCCACCACAACTG and reverse GTGACTCTTCCAAGGCGTCC, human IFNL4, forward CGGCCTGCCTTGAGCTG and reverse GGGTTTGTGACGCCTCTTCT; and human HPRT1, forward CCCTGGCGTCGTGATTAGTG and reverse CACCCTTTCCAAATCCTCAGC. The cycling parameters were 95°C for 10 min followed by 40 cycles of 95°C for 10 s, 60°C for 5 s, and 72°C for 4 s. The crossing points of the amplification curves were determined using the second derivative method on the LightCycler 480 instrument II software 1.5 (Roche). The hypoxanthine-guanine phosphoribosyltransferase (HPRT) gene (HPRT1) was used as a reference gene. The level of mRNA was normalized against internal HPRT mRNA content. Relative mRNA levels of each target gene were calculated using the formula 2−[Cp(target) − Cp(control)].
Semiquantitative PCR.
The following primers were used: human IFNL1, forward GGAAGCAGTTGCGATTTAGCC and reverse GACTCTTCCAAGGCGTCCCT; human IFNL4, forward TTGGCTTCCCTGACGTCTCT and reverse CTCTTCCTCGTAGCGGTCCC; pig IFNL2, forward GTCCCTCTTGGAGGACTGGA and reverse CTGAGCTGGGACACAGGC; and pig IFNL4, forward GATGTCCGTCGCCTCTTGTA and reverse GCGTCTCTTCCTCATAGTGGT. The PCR was performed as described above for qPCR and run for 20, 30, or 40 cycles. The PCR products were run on a 1% agarose Tris-borate-EDTA (TBE) gel with a GeneRuler 100-bp Plus DNA ladder (Thermo Fisher Scientific).
Transfections.
For transfection, cells were seeded in 12-well tissue culture plates at a density of 4 × 105 cells per well. After resting for 24 h, cells were transfected using polyethylenimine (PEI). For all transfections, a total of 2 μg plasmid was gently mixed with 6 μg PEI in a total volume of 200 μl and incubated for 15 min at room temperature before adding it dropwise to the cells. The 2 μg of plasmid always included 50 ng plasmid constitutively expressing Renilla luciferase and 1,000 ng pGL3.1 plasmid containing a type III IFN promoter. Unless otherwise stated, 400 ng plasmid expressing one of the IRFs or MAVS was included in those experiments that required expression of these proteins. To reach the desired total of 2 μg plasmid for each transfection, an empty pcDNA3.1 plasmid was added accordingly.
Luciferase assays.
At 24 h after transfection/infection, cells were lysed with passive lysis buffer (Promega) and firefly and Renilla luciferase activity in the lysate was then measured using the dual-luciferase reporter assay system (Promega) according to the manufacturer’s instructions.
Chromatin immunoprecipitation.
ChIP was performed as previously described (34). The antibodies used were mouse IgG isotype control (Thermo Fisher Scientific), IRF-3 (D83B9) rabbit monoclonal antibody (MAb) (Cell Signaling Technology), mouse anti-RNA polymerase II C-terminal domain (CTD) repeat YSPTSPS antibody (8WG16; abcam) and rabbit anti-histone H3 antibody (abcam). The coimmunoprecipitated DNA was quantified by qPCR as described above, except the cDNA synthesis step was omitted. The following primers were used: human IFNL1, forward TGAGGCCAGTTGGCTGAAAG and reverse GGAGCCTGATGAGGGAACAG; human IFNL4, forward TCAACACTACACAAGGGCTGG and reverse CGGTTTCCAAATTGTCTCTGTCC; and GAPDH, forward GCGTGTAAGGGTCCCCGTCCT and reverse GTTCAACTGGGCACGCACCGA.
Alignments and phylogenetic trees.
All of the sequences used for alignments and phylogenetic trees were extracted from the corresponding genomic reference sequences from NCBI. The accession numbers of these are NC_000019 (human), NC_023647 (African green monkey), NC_010448 (domesticated pig), NC_040265.1 (domesticated sheep), NW_007907093 (polar bear), and NW_006804147 (western European hedgehog). Sequences were aligned using ClustalW, and phylogenetic trees were generated using the neighbor-joining method in MEGA7.
Statistical analysis.
Statistical analysis was performed in GraphPad Prism 8.2.
Data availability.
Detailed information about the plasmids and plasmid sequences used in this study is available upon request.
ACKNOWLEDGMENTS
This study was supported by grants from the Chinese Scholarship Council (H.Z.), the Swiss National Science Foundation (grant number 179260 to R.D.), the Danish Council for Independent Research (grant number 7016-00331B to R.H.), the Riisfort Foundation (R.H.), and the Toyota Foundation (R.H.).
We thank Peter Stäheli, University of Freiburg, for the kind gift of RNA from hMPV- and IAV ΔNS1-infected A549 cells, as well as Martin Kristian Thomsen, University of Aarhus, for the kind gift of Göttingen minipig tissue.
H.Z., M.M., E.T.-D., K.G.W., N.H.H., J.V.-N., L.L., R.D., and H.H.G. designed, performed, and analyzed experiments. R.D., A.L.N., H.H.G., and R.H. supervised the research. H.Z., E.T.-D., H.H.G., and R.H. conceived the project and prepared the manuscript. All authors commented on the manuscript.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Donnelly RP, Kotenko SV. 2010. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res 30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green DS, Young HA, Valencia JC. 2017. Current prospects of type II interferon gamma signaling and autoimmunity. J Biol Chem 292:13925–13933. doi: 10.1074/jbc.R116.774745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wack A, Terczyńska-Dyla E, Hartmann R. 2015. Guarding the frontiers: the biology of type III interferons. Nat Immunol 16:802–809. doi: 10.1038/ni.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazear HM, Schoggins JW, Diamond MS. 2019. Shared and distinct functions of type I and type III interferons. Immunity 50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye L, Schnepf D, Staeheli P. 2019. Interferon-lambda orchestrates innate and adaptive mucosal immune responses. Nat Rev Immunol 19:614. doi: 10.1038/s41577-019-0182-z. [DOI] [PubMed] [Google Scholar]
- 6.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 7.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol 4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 8.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 9.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Muller T, Bahlo M, Stewart GJ, Booth DR, George J. 2009. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. 2009. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 11.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, O’Brien TR. 2013. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruening J, Weigel B, Gerold G. 2017. The role of type III interferons in hepatitis C virus infection and therapy. J Immunol Res 2017:7232361. doi: 10.1155/2017/7232361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Key FM, Peter B, Dennis MY, Huerta-Sánchez E, Tang W, Prokunina-Olsson L, Nielsen R, Andrés AM. 2014. Selection on a variant associated with improved viral clearance drives local, adaptive pseudogenization of interferon lambda 4 (IFNL4). PLoS Genet 10:e1004681. doi: 10.1371/journal.pgen.1004681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohlenberg M, Terczynska-Dyla E, Thomsen KL, George J, Eslam M, Gronbaek H, Hartmann R. 2018. The role of IFN in the development of NAFLD and NASH. Cytokine doi: 10.1016/j.cyto.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Bochud PY, Bibert S, Kutalik Z, Patin E, Guergnon J, Nalpas B, Goossens N, Kuske L, Mullhaupt B, Gerlach T, Heim MH, Moradpour D, Cerny A, Malinverni R, Regenass S, Dollenmaier G, Hirsch H, Martinetti G, Gorgiewski M, Bourliere M, Poynard T, Theodorou I, Abel L, Pol S, Dufour JF, Negro F, Swiss Hepatitis C Cohort Study Group, ANRS HC EP 26 Genoscan Study Group . 2012. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology 55:384–394. doi: 10.1002/hep.24678. [DOI] [PubMed] [Google Scholar]
- 17.Eslam M, McLeod D, Kelaeng KS, Mangia A, Berg T, Thabet K, Irving WL, Dore GJ, Sheridan D, Grønbæk H, Abate ML, Hartmann R, Bugianesi E, Spengler U, Rojas A, Booth DR, Weltman M, Mollison L, Cheng W, Riordan S, Mahajan H, Fischer J, Nattermann J, Douglas MW, Liddle C, Powell E, Romero-Gomez M, George J, International Liver Disease Genetics Consortium . 2017. IFN-lambda3, not IFN-lambda4, likely mediates IFNL3-IFNL4 haplotype-dependent hepatic inflammation and fibrosis. Nat Genet 49:795–800. doi: 10.1038/ng.3836. [DOI] [PubMed] [Google Scholar]
- 18.Petta S, Valenti L, Tuttolomondo A, Dongiovanni P, Pipitone RM, Cammà C, Cabibi D, Di Marco V, Fracanzani AL, Badiali S, Nobili V, Fargion S, Grimaudo S, Craxì A. 2017. Interferon lambda 4 rs368234815 TT>δG variant is associated with liver damage in patients with nonalcoholic fatty liver disease. Hepatology 66:1885–1893. doi: 10.1002/hep.29395. [DOI] [PubMed] [Google Scholar]
- 19.Eslam M, Hashem AM, Leung R, Romero-Gomez M, Berg T, Dore GJ, Chan HL, Irving WL, Sheridan D, Abate ML, Adams LA, Mangia A, Weltman M, Bugianesi E, Spengler U, Shaker O, Fischer J, Mollison L, Cheng W, Powell E, Nattermann J, Riordan S, McLeod D, Armstrong NJ, Douglas MW, Liddle C, Booth DR, George J, Ahlenstiel G, International Hepatitis C Genetics Consortium . 2015. Interferon-lambda rs12979860 genotype and liver fibrosis in viral and non-viral chronic liver disease. Nat Commun 6:6422. doi: 10.1038/ncomms7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamming OJ, Terczyńska-Dyla E, Vieyres G, Dijkman R, Jørgensen SE, Akhtar H, Siupka P, Pietschmann T, Thiel V, Hartmann R. 2013. Interferon lambda 4 signals via the IFNlambda receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J 32:3055–3065. doi: 10.1038/emboj.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauber C, Vieyres G, Terczynska-Dyla E, Anggakusuma, Dijkman R, Gad HH, Akhtar H, Geffers R, Vondran FWR, Thiel V, Kaderali L, Pietschmann T, Hartmann R. 2015. Transcriptome analysis reveals a classical interferon signature induced by IFN lambda 4 in human primary cells. Genes Immun 16:414–421. doi: 10.1038/gene.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terczyńska-Dyla E, Bibert S, Duong FHT, Krol I, Jørgensen S, Collinet E, Kutalik Z, Aubert V, Cerny A, Kaiser L, Malinverni R, Mangia A, Moradpour D, Müllhaupt B, Negro F, Santoro R, Semela D, Semmo N, Heim MH, Bochud P-Y, Hartmann R. 2014. Reduced IFNlambda4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nat Commun 5:5699. doi: 10.1038/ncomms6699. [DOI] [PubMed] [Google Scholar]
- 23.Ansari MA, Pedergnana V, C LCI, Magri A, Von Delft A, Bonsall D, Chaturvedi N, Bartha I, Smith D, Nicholson G, McVean G, Trebes A, Piazza P, Fellay J, Cooke G, Foster GR, Consortium S-H, Hudson E, McLauchlan J, Simmonds P, Bowden R, Klenerman P, Barnes E, Spencer C. 2017. Genome-to-genome analysis highlights the effect of the human innate and adaptive immune systems on the hepatitis C virus. Nat Genet 49:666–673. doi: 10.1038/ng.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freije CA, Caron R, Uhl SA, Chen ST, Rosenberg BR, Eitson JL, Rice CM, Imanaka N, Talal A, Jacobson IM, Zeremski M, Schoggins JW. 2018. Genetic variation at IFNL4 influences extrahepatic interferon-stimulated gene expression in chronic HCV patients. J Infect Dis 217:650–655. doi: 10.1093/infdis/jix593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amanzada A, Kopp W, Spengler U, Ramadori G, Mihm S. 2013. Interferon-lambda4 (IFNL4) transcript expression in human liver tissue samples. PLoS One 8:e84026. doi: 10.1371/journal.pone.0084026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konishi H, Motomura T, Matsumoto Y, Harimoto N, Ikegami T, Yoshizumi T, Soejima Y, Shirabe K, Fukuhara T, Maehara Y. 2014. Interferon-lambda4 genetic polymorphism is associated with the therapy response for hepatitis C virus recurrence after a living donor liver transplant. J Viral Hepat 21:397–404. doi: 10.1111/jvh.12154. [DOI] [PubMed] [Google Scholar]
- 27.Murakawa M, Asahina Y, Kawai-Kitahata F, Nakagawa M, Nitta S, Otani S, Nagata H, Kaneko S, Asano Y, Tsunoda T, Miyoshi M, Itsui Y, Azuma S, Kakinuma S, Tanaka Y, Iijima S, Tsuchiya K, Izumi N, Tohda S, Watanabe M. 2017. Hepatic IFNL4 expression is associated with non-response to interferon-based therapy through the regulation of basal interferon-stimulated gene expression in chronic hepatitis C patients. J Med Virol 89:1241–1247. doi: 10.1002/jmv.24763. [DOI] [PubMed] [Google Scholar]
- 28.Hong M, Schwerk J, Lim C, Kell A, Jarret A, Pangallo J, Loo Y-M, Liu S, Hagedorn CH, Gale M, Savan R. 2016. Interferon lambda 4 expression is suppressed by the host during viral infection. J Exp Med 213:2539–2552. doi: 10.1084/jem.20160437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baños-Lara MDR, Harvey L, Mendoza A, Simms D, Chouljenko VN, Wakamatsu N, Kousoulas KG, Guerrero-Plata A. 2015. Impact and regulation of lambda interferon response in human metapneumovirus infection. J Virol 89:730–742. doi: 10.1128/JVI.02897-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikushima H, Negishi H, Taniguchi T. 2013. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harbor Symp Quant Biol 78:105–116. doi: 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- 31.Minas TZ, Tang W, Smith CJ, Onabajo OO, Obajemu A, Dorsey TH, Jordan SV, Obadi OM, Ryan BM, Prokunina-Olsson L, Loffredo CA, Ambs S. 2018. IFNL4-deltaG is associated with prostate cancer among men at increased risk of sexually transmitted infections. Commun Biol 1:191. doi: 10.1038/s42003-018-0193-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fung KY, Mangan NE, Cumming H, Horvat JC, Mayall JR, Stifter SA, De Weerd N, Roisman LC, Rossjohn J, Robertson SA, Schjenken JE, Parker B, Gargett CE, Nguyen HP, Carr DJ, Hansbro PM, Hertzog PJ. 2013. Interferon-epsilon protects the female reproductive tract from viral and bacterial infection. Science 339:1088–1092. doi: 10.1126/science.1233321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hr J, Dijkman R. 2015. Characterization of human coronaviruses on well-differentiated human airway epithelial cell cultures. Methods Mol Biol 1282:73–87. doi: 10.1007/978-1-4939-2438-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y, Blechingberg J, Fernandes AM, Li S, Fryland T, Borglum AD, Bolund L, Nielsen AL. 2015. EWS and FUS bind a subset of transcribed genes encoding proteins enriched in RNA regulatory functions. BMC Genomics 16:929. doi: 10.1186/s12864-015-2125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Detailed information about the plasmids and plasmid sequences used in this study is available upon request.