The main barrier to HIV cure is the ability of a genetically diverse pool of proviruses, integrated into the genomes of infected CD4+ T cells, to persist despite long-term suppressive combination antiretroviral therapy (cART). CD4+ T cells, however, constitute a heterogeneous population due to their maturation across a developmental continuum, and the genetic “landscapes” of latent proviruses archived within them remains incompletely understood. We applied phylogenetic techniques, largely novel to HIV persistence research, to reconstruct within-host HIV evolutionary history and characterize proviral diversity in CD4+ T cell subsets in five individuals on long-term cART. Participants varied widely in terms of proviral burden, genetic diversity, and age distribution between CD4+ T cell subsets, revealing that proviral landscapes can differ between individuals and between infected cell types within an individual. Our findings expose each within-host latent reservoir as unique in its genetic complexity and support personalized strategies for HIV eradication.
KEYWORDS: HIV, reservoir, persistence, CD4+ T cells, cellular subsets, genetic compartmentalization, proviral age, cell subsets, human immunodeficiency virus
ABSTRACT
The HIV reservoir, which comprises diverse proviruses integrated into the genomes of infected, primarily CD4+ T cells, is the main barrier to developing an effective HIV cure. Our understanding of the genetics and dynamics of proviruses persisting within distinct CD4+ T cell subsets, however, remains incomplete. Using single-genome amplification, we characterized subgenomic proviral sequences (nef region) from naive, central memory, transitional memory, and effector memory CD4+ T cells from five HIV-infected individuals on long-term combination antiretroviral therapy (cART) and compared these to HIV RNA sequences isolated longitudinally from archived plasma collected prior to cART initiation, yielding HIV data sets spanning a median of 19.5 years (range, 10 to 20 years) per participant. We inferred a distribution of within-host phylogenies for each participant, from which we characterized proviral ages, phylogenetic diversity, and genetic compartmentalization between CD4+ T cell subsets. While three of five participants exhibited some degree of proviral compartmentalization between CD4+ T cell subsets, combined analyses revealed no evidence that any particular CD4+ T cell subset harbored the longest persisting, most genetically diverse, and/or most genetically distinctive HIV reservoir. In one participant, diverse proviruses archived within naive T cells were significantly younger than those in memory subsets, while for three other participants we observed no significant differences in proviral ages between subsets. In one participant, “old” proviruses were recovered from all subsets, and included one sequence, estimated to be 21.5 years old, that dominated (>93%) their effector memory subset. HIV eradication strategies will need to overcome within- and between-host genetic complexity of proviral landscapes, possibly via personalized approaches.
IMPORTANCE The main barrier to HIV cure is the ability of a genetically diverse pool of proviruses, integrated into the genomes of infected CD4+ T cells, to persist despite long-term suppressive combination antiretroviral therapy (cART). CD4+ T cells, however, constitute a heterogeneous population due to their maturation across a developmental continuum, and the genetic “landscapes” of latent proviruses archived within them remains incompletely understood. We applied phylogenetic techniques, largely novel to HIV persistence research, to reconstruct within-host HIV evolutionary history and characterize proviral diversity in CD4+ T cell subsets in five individuals on long-term cART. Participants varied widely in terms of proviral burden, genetic diversity, and age distribution between CD4+ T cell subsets, revealing that proviral landscapes can differ between individuals and between infected cell types within an individual. Our findings expose each within-host latent reservoir as unique in its genetic complexity and support personalized strategies for HIV eradication.
INTRODUCTION
Although combination antiretroviral therapy (cART) suppresses human immunodeficiency virus 1 (HIV) replication (1, 2), it is not curative and must be maintained for life (3). This is due to a small pool of long-lived primarily CD4+ T cells that harbor integrated HIV in a transcriptionally reduced state and persist despite cART but that can be reactivated at any time to produce infectious virus (4–8). In general, the HIV reservoirs of individuals who initiated cART during chronic infection are genetically diverse (9–15), which is to be expected, given that seeding of the reservoir begins immediately following infection (16–19) and continues as long as HIV is actively replicating in vivo (20, 21). Recent longitudinal studies confirm this: proviral sequences dating as far back as transmission are present in many individuals’ reservoirs (20–22) though some are enriched for proviruses seeded around the time of cART (20, 22). Populations of cells harboring identical proviruses or identical integration sites also feature prominently in the reservoir, indicating that clonal expansion of latently infected cells also drives HIV persistence (15, 23–29).
HIV eradication will thus require an in-depth understanding of latent HIV genetic composition and persistence in CD4+ T cells, but this is complicated by the fact that CD4+ T cells mature along a program of development and thus constitute a heterogeneous population (30, 31). Upon encountering their cognate antigen, naive T (TN) cells develop into effector and memory cell subsets which include, from least to most differentiated, stem-cell-like memory (TSCM), central memory (TCM), transitional memory (TTM), effector memory (TEM), and finally terminally differentiated (TTD) cells (31). Though HIV DNA is reproducibly detected in all of these subsets during long-term cART (13, 32–38), it has been hypothesized that less differentiated memory T cell subsets may represent the most durable sites for long-term HIV persistence in peripheral blood (32, 34, 35, 37). This is an intuitive notion, given that the longevity of CD4+ T cell subsets (30) and the half-life of proviral DNA in these cells (34, 39) decrease with differentiation, but studies analyzing proviral sequences within CD4+ T cell subsets are limited and have yielded somewhat conflicting observations. Buzon et al. observed that proviruses isolated from less differentiated, longer-lived memory CD4+ T cells, in particular, TSCM and TCM cells, were phylogenetically most closely related to early pre-cART plasma sequences, suggesting that HIV strains circulating in early infection were more likely to persist in these cell subsets (34). Chomont et al. hypothesized that, due to their differential survival and proliferation dynamics, TCM and TTM cells may come to define distinct HIV reservoirs after long-term cART, with increased turnover of shorter-lived TTM cells being associated with reduced within-subset proviral genetic distances compared to that of the longer-lived TCM cells (32). However, counter to the notion that highly differentiated CD4+ T cell subsets represent less stable reservoirs, Imamichi et al. documented a proviral sequence that persisted for 17 years solely within TEM cells, indicating that these subsets can harbor latent HIV for extended periods through proliferation (40). Only one study to our knowledge has applied formal tests for genetic compartmentalization to proviruses archived within CD4+ T cell subsets (29), while none have applied methods to infer latent HIV sequence age distributions within these subsets. Furthermore, TN cells, despite generally harboring lower proviral burdens than memory T cell subsets (32, 33, 36), may nevertheless represent a key HIV reservoir (37, 38), but knowledge of proviral diversity within this subset remains limited (13).
If the longest lived, least differentiated CD4+ T cell subsets constitute the most stable and privileged HIV reservoirs, one could hypothesize that, during long-term cART, the oldest within-host latent HIV sequences would tend to be found therein. Our recent development of a phylogenetic method to infer within-host latent HIV sequence ages allows this hypothesis to be tested (21). One could further hypothesize that, after years of cART, the most evolutionarily distinct within-host HIV lineages and/or the greatest overall proviral diversity will be found within the longest lived, least differentiated CD4+ T cell subsets as more rapid turnover in the shorter lived, more differentiated subsets may lead to more frequent elimination of unique proviral lineages therein. To this end, we investigated proviral diversity, genetic compartmentalization, and latent HIV sequence age distribution in CD4+ T cell subsets during long-term cART and interpreted this information in light of HIV’s pre-cART within-host evolutionary history in five HIV-infected individuals with long-term viremia suppression.
RESULTS
Proviral burden in CD4+ T cell subsets.
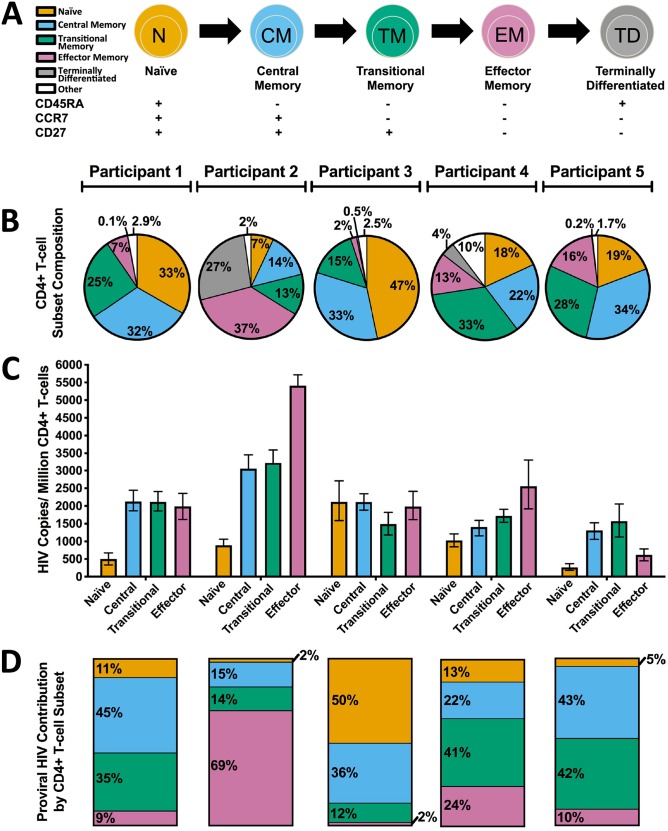
Five HIV-infected adults who initiated cART during chronic infection and for whom longitudinal archived pre-cART plasma samples were available were recruited for study (Table 1). At enrollment, participants had been receiving cART for a median of 9 years and had a median of 8 (interquartile range [IQR], 5 to 14) archived pre-cART plasma samples available for study. Peripheral blood mononuclear cells (PBMCs) were isolated at enrollment and sorted into CD4+ naive (TN), central memory (TCM), transitional memory (TTM), and effector memory (TEM) cell subsets based on their expression of CD45RA, CCR7, and CD27 (Fig. 1 and 2A) and assessed for purity as described previously in Chomont et al. (32) (see also Materials and Methods). CD4+ T cell subset frequencies varied markedly between participants: TCM cells, for example, constituted from 14% to 34% (median, 32%; IQR, 22 to 33%) of total CD4+ T cells (Fig. 2B). Proviral DNA levels in CD4+ T cell subsets also varied. Nevertheless, in 4 of 5 participants, TN cells harbored the lowest absolute proviral burdens (median, 890 HIV copies/million cells; IQR, 383 to 1,573 HIV copies/million cells) (Fig. 2C) and contributed the least to the total proviral burden in peripheral blood CD4+ T cells (median, 11%; IQR, 3 to 32%) (Fig. 2D), while memory subsets (TCM, TTM, and TEM cells together) contributed the most to the total proviral burden in CD4+ T cells (median, 89%; IQR, 68 to 97%). This observation is consistent with studies identifying TN cells as a minor yet potentially important HIV reservoir (37, 38).
TABLE 1.
Participant and HIV sequence data set details
| Participantno. | Time infectedpre-cART (yr) | No. of pre-cARTplasma timepoints | Time oncART (yr) | Total no. of sequences (no. kept) by sourcea |
||||
|---|---|---|---|---|---|---|---|---|
| Pre-cARTplasma | TN cells | TCM cells | TTM cells | TEM cells | ||||
| 1 | 10 | 14 | 9 | 102 (93) | 31 (29) | 26 (24) | 26 (24) | 27 (16) |
| 2 | 11 | 17 | 9 | 162 (130) | 20 (18) | 13 (13) | 18 (15) | 15 (2) |
| 3 | 5 | 8 | 8 | 122 (113) | 16 (13) | 19 (15) | 38 (35) | 18 (15) |
| 4 | 2 | 5 | 9 | 72 (60) | 16 (16) | 20 (16) | 26 (16) | 18 (8) |
| 5 | 14 | 2 | 16 | 41 (38) | 20 (20) | 16 (16) | 24 (11) | 17 (10) |
Kept sequences refer to distinct sequences that were free from hypermutation, other defects, and within-host recombination.
FIG 1.
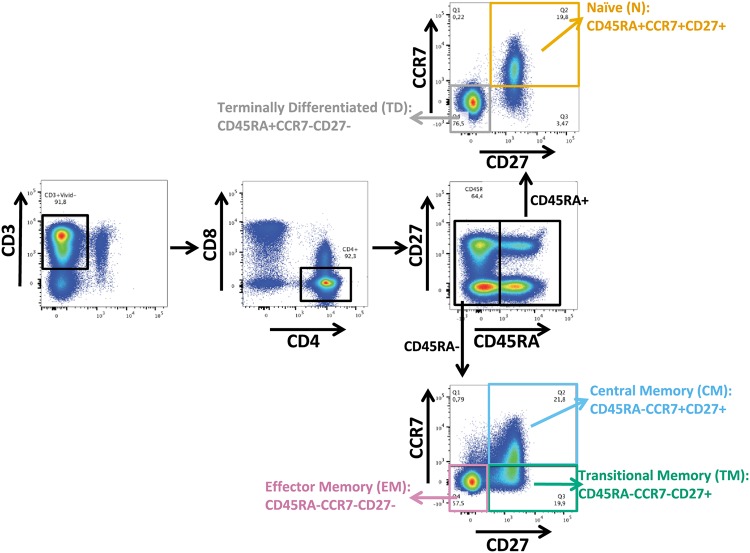
Gating strategy for sorting of CD4+ T cell subsets. Representative flow cytometry plot illustrating how participant CD4+ T cells were isolated after staining with the indicated antibodies. CD4 T cell subsets were defined as follows: TN, CD45RA+ CCR7+ CD27+; TCM, CD45RA− CCR7+ CD27+; TTM, CD45RA− CCR7− CD27+; TEM, CD45RA− CCR7− CD27−.
FIG 2.
Frequency and latent HIV reservoir burden in CD4+ T cell subsets. (A) Schematic of CD4+ T cell differentiation, with corresponding cell surface markers below. All subsequent figures use this color scheme to denote specific CD4+ T cell subsets. (B) Frequency of CD4+ T cells within each subset. The “other” category denotes CD4+ T cells that express other combinations of the markers listed in panel A that are rare and have no reported function (80). (C) Proviral DNA burden, measured as the number of HIV Gag copies/million cells, in each CD4+ T cell subset. Error bars represent 95% Poisson confidence intervals calculated from merged replicate wells. (D) Proportion of the total proviral burden harbored by each CD4+ T cell subset.
Within-host HIV sequence characterization.
We next characterized HIV RNA sequences from longitudinal archived plasma samples collected prior to cART initiation, as well as proviral sequences from TN, TCM, TTM, and TEM cells isolated during long-term cART, using subgenomic single-genome amplification and sequencing. The nef gene was selected based on its relatively high within-host diversity, its richness in phylogenetic signal, and its representativeness of within-host HIV evolution and diversity relative to the rest of the viral genome (21); nef also represents the gene most likely to be intact within proviruses sampled during long-term cART (15, 41). All further references to “sequences” in this text refer to nef sequences. The numbers of HIV RNA and proviral DNA sequences collected from each participant (total and intact/distinct) are reported in Table 1. Of note, we observed identical HIV sequences more frequently among the more differentiated CD4+ T cell subsets though this did not achieve statistical significance (Fig. 3). Participant 2 was remarkable in that this participant’s TEM cells harbored the highest proviral burden of all CD4+ T cell subsets, but 93% (14/15) of the intact proviral sequences recovered from TEM cells were identical, such that only two distinct sequences were isolated from this subset (Table 1). Together, these observations are consistent with reports that the more differentiated latently infected CD4+ T cell subsets tend to be more clonally expanded (13, 29).
FIG 3.
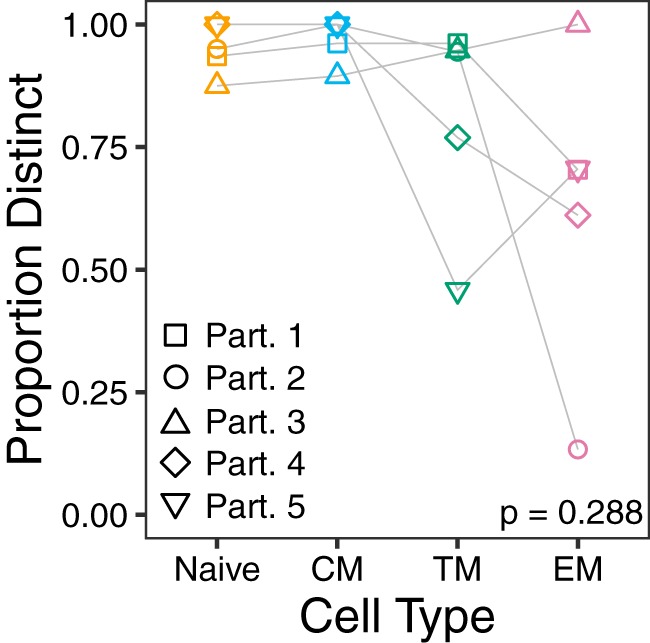
Sequence distinctness in CD4+ T cell subsets. Data points denote the frequency of distinct (unique) proviral sequences per subset per participant. Frequencies across subsets were compared using the Friedman rank sum test. CM, central memory; TM, transitional memory; EM, effector memory.
Proviral ages and genetic compartmentalization in CD4+ T cell subsets.
Our main goal was to characterize within-host proviral landscapes within CD4+ T cell subsets during long-term cART and to interpret this information in the context of HIV’s within-host pre-cART evolutionary history (Fig. 4; see also Fig. 8). For each participant, we used Bayesian methods via MrBayes (42) to infer a random sample of between 15,000 and 30,000 within-host phylogenetic trees from alignments of pre-cART plasma HIV RNA and post-cART proviral sequences and used these trees to investigate the diversity, genetic compartmentalization, and age distribution of proviruses persisting in CD4+ T cell subsets. Conditioning analyses across a distribution of trees mitigates the inherent uncertainty in reconstructing within-host HIV evolutionary relationships and is particularly useful for phylogenetic tests (e.g., for genetic compartmentalization [43] and evolutionary distinctiveness [44]) that are sensitive to tree topology.
FIG 4.
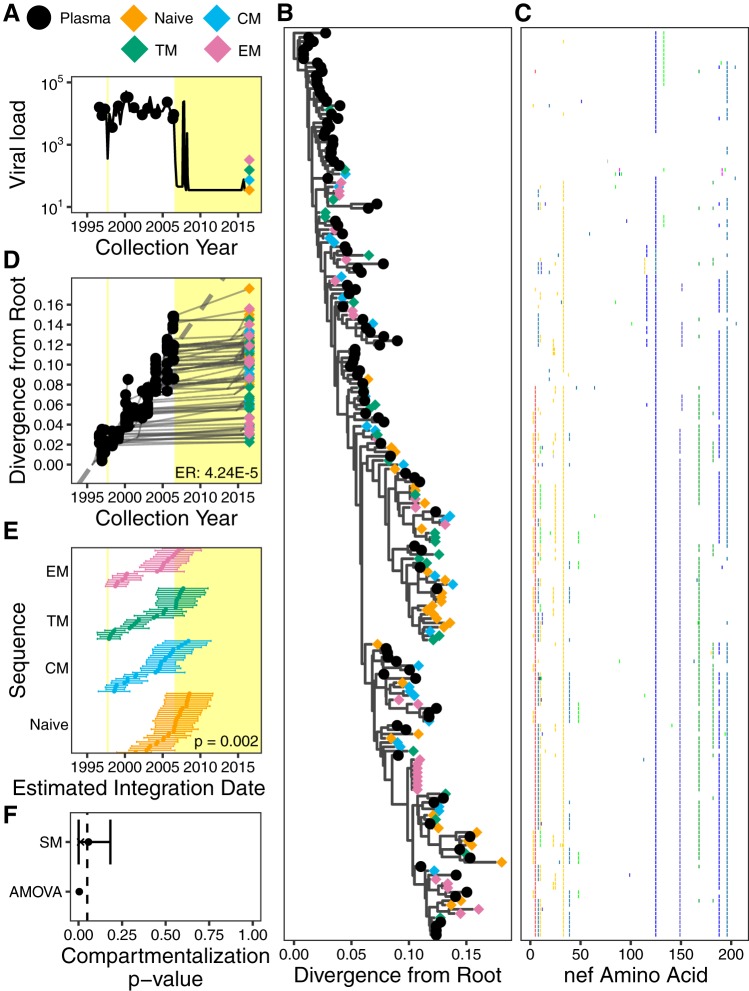
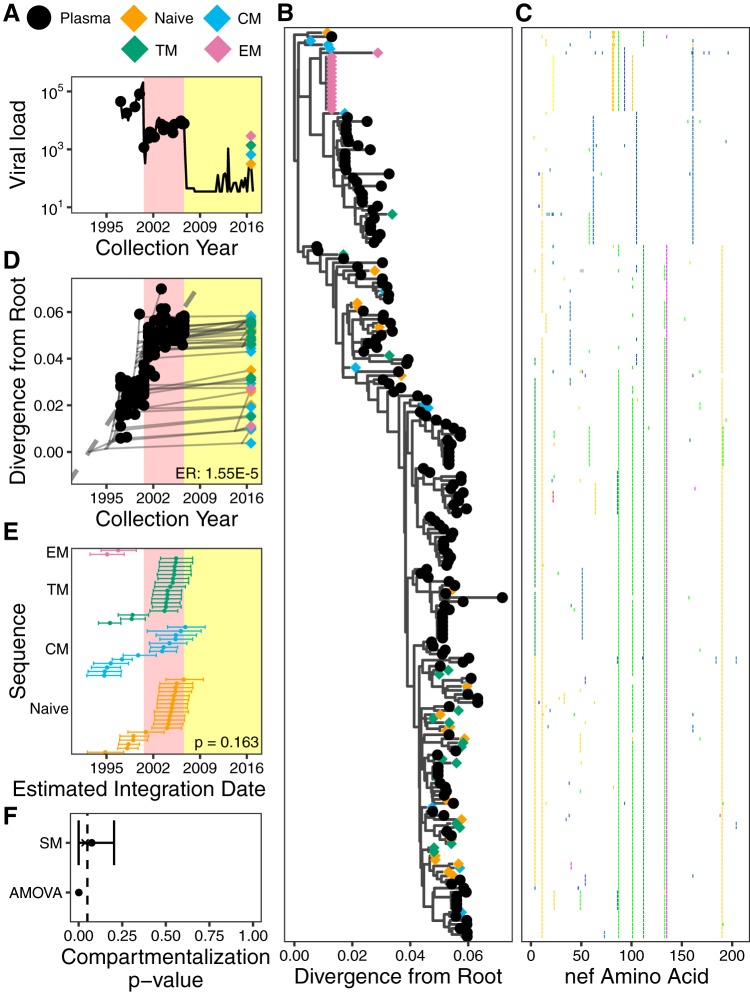
Participant 1. (A) Plasma viral load and sampling history. Yellow shading denotes periods on cART. (B) The phylogeny with the highest likelihood derived from Bayesian inference, rooted using root-to-tip regression. (C) Highlighter plot showing amino acid differences with respect to the consensus of the sequences collected at the earliest plasma HIV RNA sampling time point. (D) Linear model (dashed gray diagonal) relating plasma HIV RNA collection dates to their respective distances from the root of the phylogeny shown in panel B. Thin gray lines show the phylogenetic relationships between the sequences. HIV RNA nef evolutionary rate (ER), expressed in estimated nucleotide substitutions per site per day, is shown in the bottom right corner. (E) The 95% highest posterior density (HPD) intervals of the integration date estimates of the proviral sequences, derived from all phylogenies generated by the Bayesian analysis. The points represent mean values. The distribution of mean values across the CD4+ T cell subsets is compared using a Kruskal-Wallis test (P value at bottom right). (F) P values of proviral genetic compartmentalization across the CD4+ T cell subsets, derived from AMOVA and Slatkin-Maddison (where the latter values are derived from all phylogenies generated by the Bayesian analysis). Here, the circle represents the overall mean P value, the x represents the mean weighted by the likelihood of the phylogeny tested, and the bars represent the 95% HPD interval.
FIG 8.
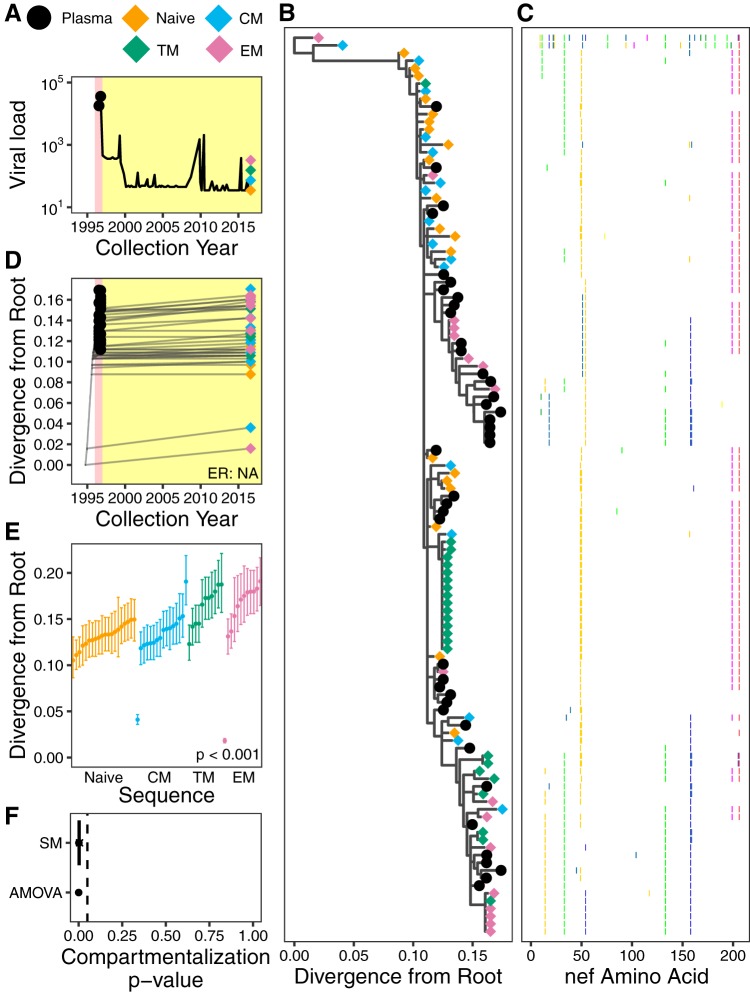
Participant 5. (A) Plasma viral load and sampling history. Yellow shading indicates cART, and pink shading denotes nonsuppressive dual therapy (also in panel D). (B) The phylogeny with the highest likelihood derived from Bayesian inference, rooted using the HXB2 reference strain as an outgroup. (C) Highlighter plot showing amino acid differences with respect to the consensus of the sequences collected at the earliest plasma HIV RNA sampling time point. (D) Linear model (dashed gray diagonal) relating plasma HIV RNA collection dates to their respective distances from the root of the phylogeny shown in panel B. Gray lines show the phylogenetic relationships of the sequences. HIV RNA nef evolutionary rate (ER) could not be determined for this participant due to insufficient molecular clock signal. (E) The 95% highest posterior density (HPD) intervals of root-to-tip patristic distances, expressed as the number of estimated nucleotide substitutions/site, derived from all phylogenies generated by the Bayesian analysis. The points represent mean values. The distribution of mean values across the CD4+ T cell subsets is compared using the Kruskal-Wallis test (P value at bottom right). (F) P values of proviral genetic compartmentalization across the CD4+ T cell subsets, as described in the legend of Fig. 4F.
Participant 1.
Participant 1, for whom a limited number of proviral sequences were previously characterized from PBMCs (21), was diagnosed with HIV in August 1996 and did not initiate suppressive cART until August 2006 (Fig. 4A). Using single-genome amplification, we isolated 102 plasma HIV RNA sequences over an ∼10-year period pre-cART and 110 proviral sequences from CD4+ T cell subsets collected after ∼10 years of cART (Table 1 and Fig. 4A). Optimal rooting of phylogenies inferred from these sequences, where the root represents the most recent common ancestor of the within-host data set, revealed strong molecular clock signal, as demonstrated by the robust linear relationship between the divergence from the root and the collection date of longitudinally sampled pre-cART plasma sequences (representative phylogeny, sequence diversity plot, and linear model are shown in Fig. 4B to D). Averaged over all within-host phylogenies, the mean pre-cART plasma HIV RNA nef evolutionary rate was estimated as 4.24 × 10−5 (95% highest posterior density [HPD], 2.64 × 10−5 to 5.94 × 10−5) substitutions per nucleotide site per day for this participant. Proviral sequences sampled from CD4+ T cell subsets after ∼10 years on cART were interspersed throughout the tree (Fig. 4B), consistent with the notion that HIV sequences are continually seeded into the reservoir during uncontrolled infection and can persist for a decade or longer (20, 21, 45). Identical proviral sequences were also observed, including seven in TEM cells (Fig. 4B), consistent with clonal expansion of latently HIV-infected cells (15, 23–29).
Our ability to estimate a within-host rate of HIV evolution from longitudinal pre-cART plasma HIV RNA sequences allows us to estimate the integration date of a given proviral sequence from its root-to-tip genetic distance (21). Moreover, our reconstruction of thousands of trees per participant allows us to compute a 95% highest posterior density (HPD) around this date estimate (21; see also Materials and Methods) (Fig. 4E). Distinct proviral sequences within a given CD4+ T cell subset differed widely in terms of their estimated integration dates. The oldest provirus isolated from participant 1’s TEM cells, for example, was estimated to have integrated in 1998 (i.e., it was almost 18 years old at time of sampling) while the youngest was estimated to have integrated in 2007. Furthermore, the estimated integration date distributions of distinct proviruses differed significantly between CD4+ T cell subsets in this participant (Kruskal-Wallis test, P = 0.002) (Fig. 4E). However, in contrast to our hypothesis that the longer lived, less differentiated memory T cell subsets would harbor the oldest proviruses, the three memory subsets did not differ in terms of their proviral age distributions (P > 0.2 for all pairwise comparisons), and TN cells harbored the youngest proviruses on average (P < 0.05 in all pairwise comparisons). While some proviral integration date point estimates were after cART initiation, in all cases the 95% HPD estimates extended into the pre-cART period (Fig. 4E), and it is also notable that this participant experienced a major viremic episode at 1 year post-cART, which could have led to reservoir reseeding (Fig. 4A). These observations therefore do not constitute conclusive evidence of ongoing HIV replication during cART.
We next investigated whether distinct proviral sequences within CD4+ T cell subsets displayed population structure or genetic compartmentalization. In actively replicating viral populations, compartmentalization often arises due to restricted gene flow between compartments, but in the context of the HIV reservoir, it may arise due to differential seeding of within-host HIV strains into, or differential subsequent persistence within, distinct CD4+ T cell subsets. Differential clonal expansion dynamics between latently infected CD4+ T cell subsets could also lead to the detection of population structure if identical sequences are retained in the analysis (46, 47); to avoid this, we analyzed distinct sequences per subset only. As recommended (48), we applied two tests capable of detecting population structure across the four subsets (TN, TCM, TTM, and TEM cells): analysis of molecular variance (AMOVA) (49), a genetic-distance-based test, and the Slatkin-Maddison (SM) test, a tree-based test (43). Both of these tests have been applied to within-host HIV data sets (47, 48) including the latent reservoir (46, 48, 50). AMOVA yielded a P value of 0.004, while application of the SM test to all 30,000 within-host phylogenies yielded an overall mean P value of 0.057, a weighted mean P value of 0.011 (where the latter takes the tree likelihoods into account), and a 95% HPD of 0.000 to 0.182 (Fig. 4F). These results indicate that proviral sequences archived within participant 1’s CD4+ T cell subsets exhibit modest yet statistically significant compartmentalization, which is consistent with our detection of significant differences in proviral age distributions between subsets.
Participant 2.
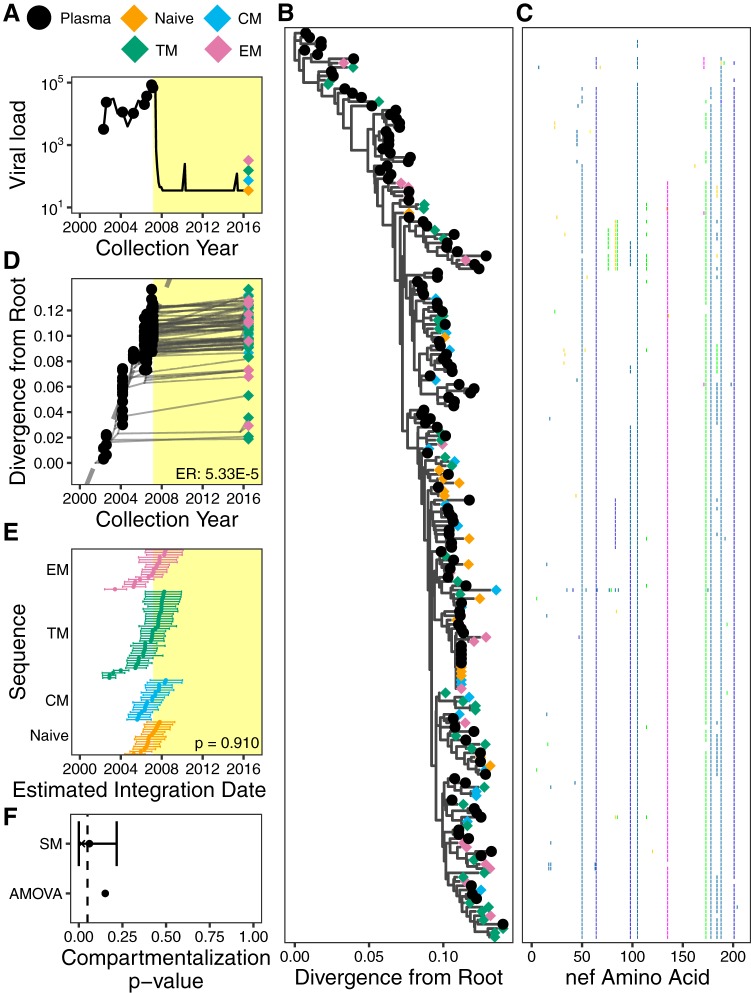
Participant 2, for whom a small number of proviruses isolated from PBMCs were also previously characterized (21), was diagnosed with HIV in April 1995, initiated incompletely suppressive dual ART in July 2000, and initiated suppressive cART in August 2006 (Fig. 5A). We isolated 162 plasma HIV RNA sequences over an ∼9.5-year period pre-cART and 66 proviral sequences from different CD4+ T cell subsets collected ∼10 years post-cART (Table 1). Optimal rooting of participant 2’s phylogenies (Fig. 5B, representative tree, and Fig. 5C, diversity plot) yielded a mean pre-cART plasma HIV RNA nef evolutionary rate estimate of 1.55 × 10−5 (95% HPD, 8.83 × 10−6 to 2.29 × 10−5) substitutions per nucleotide site per day (Fig. 5D shows representative data). Proviruses isolated from CD4+ T cell subsets were interspersed throughout the phylogeny and included sequences from TN, TCM, and TEM cells that branched close to the root, consistent with the notion that, even after years of cART, multiple CD4+ T cell subsets still harbor proviruses archived near the time of HIV transmission (16). Notably, only two distinct proviral sequences were isolated from this participant’s TEM cells, both of which fell within a clade close to the tree root, yielding proviral integration date estimates of 1995 and 1996, respectively (Fig. 5E). The isolation of the former sequence 14 times supports clonal expansion as a major driver of HIV persistence (23–26, 51, 52) where clones may arise very early in infection (45). In contrast, HIV sequences isolated from the other T cell subsets exhibited broader age distributions (e.g., proviruses isolated from TN cells dated to between 1994 and 2006). Overall, TEM cells harbored the oldest proviruses in this participant, but this was not statistically significant (Kruskal-Wallis test, P = 0.163) (Fig. 5E), in part because we recovered only two distinct sequences from TEM cells. Despite this, both genetic distance and tree-based tests indicated that distinct proviruses isolated from participant 2’s CD4+ T cell subsets exhibited modest genetic compartmentalization: AMOVA yielded a P value of <0.001, while the weighted average P value from SM tests was 0.037 (95% HPD, 0.00 to 0.202) (Fig. 5F).
FIG 5.
Participant 2. The panels are as described in the legend of Fig. 4. Pink shading in panels A, D, and E denotes a period of nonsuppressive dual therapy.
Participant 3.
Participant 3 was diagnosed with HIV in 2002 and initiated suppressive cART in May 2007 (Fig. 6A). We isolated 122 plasma HIV RNA sequences over an ∼5-year period pre-cART and 91 proviral sequences from CD4+ T cell subsets collected ∼9 years post-cART (Table 1). Optimal rooting of participant 3’s phylogenies (Fig. 6B, representative tree, and Fig. 6C, diversity plot) yielded a mean pre-cART plasma HIV RNA nef evolutionary rate estimate of 5.33 × 10−5 (95% HPD, 3.18 × 10−5 to 7.85 × 10−5) substitutions per nucleotide site per day (representative data are shown in Fig. 6D). Proviral sequences were again interspersed throughout the tree, where the four proviruses closest to the root were isolated from TTM and TEM cells and dated to various times in late 2002 and 2003. This participant harbored an identical HIV sequence that was isolated from TEM cells and repeatedly from TN and TCM cells, suggesting that latently infected cells can both differentiate and clonally expand to sustain the HIV reservoir. Overall, the proviruses isolated from participant 3’s CD4+ T cell subsets did not significantly differ in terms of their estimated integration dates (Kruskal-Wallis test, P = 0.910) (Fig. 6E), and support for genetic compartmentalization was not consistent between tests: whereas the weighted average P value for SM was 0.015 (HPD, 0.000 to 0.217), AMOVA yielded a P value of 0.151 (Fig. 6F). Taken together, our findings indicated that while participant 3 harbored diverse proviruses, there was no strong evidence that these differed in genetic composition or age between CD4+ T cell subsets.
FIG 6.
Participant 3. The panels are as described in the legend of Fig. 4.
Participant 4.
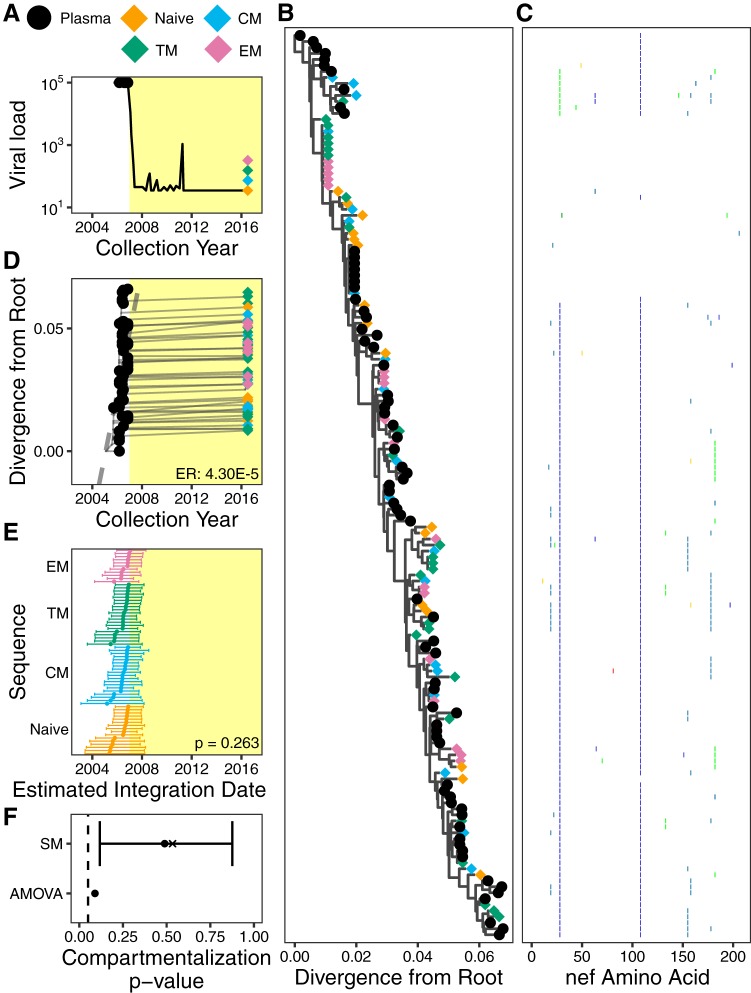
Participant 4 was diagnosed with HIV in 2005 and initiated suppressive cART in November 2006 (Fig. 7A). We isolated 72 plasma HIV RNA sequences at 5 time points between September 2005 and November 2006, prior to cART initiation, and 80 proviral sequences from CD4+ T cell subsets collected ∼9.5 years post-cART (Table 1). Consistent with cART initiation in relatively early infection, within-host HIV diversity was relatively limited (Fig. 7B, representative tree, and Fig. 7C, diversity plot). Despite this, there was sufficient molecular clock signal in the data to infer a mean pre-cART plasma HIV RNA nef evolutionary rate of 4.30 × 10−5 (95% HPD, 1.44 × 10−5 to 7.67 × 10−5) substitutions per nucleotide site per day (representative data are shown in Fig. 7D). Proviral sequences were again interspersed throughout the tree. Notably, one HIV sequence, which branched close to the root, was repeatedly isolated from all memory CD4+ T cell subsets (Fig. 7B). Overall, proviruses isolated from the different CD4+ subsets did not differ from one another in terms of their estimated integration dates (Kruskal-Wallis test, P = 0.263) (Fig. 7E), and no evidence of genetic compartmentalization was detected by either test (Fig. 7F).
FIG 7.
Participant 4. The panels are as described in the legend of Fig. 4.
Participant 5.
Participant 5 was diagnosed with HIV in 1985, was subsequently exposed to nonsuppressive ART (though full treatment history is unavailable), and initiated cART in January 1997. Only two pre-cART plasma samples were available, from July and October of 1996, drawn while the participant was receiving nonsuppressive dual ART (Fig. 8A). A total of 41 plasma HIV RNA sequences were isolated from these two samples, and 77 proviral sequences were isolated from CD4+ T cell subsets collected nearly 20 years after cART initiation (Table 1). However, due to the limited time span of plasma sampling, participant 5’s data set lacked sufficient molecular clock signal to root the tree using standard approaches (see Materials and Methods). We therefore outgroup-rooted the tree using the HIV subtype B consensus sequence HXB2 (Fig. 8B, representative tree, and Fig. 8C, diversity plot); however, the molecular clock signal in the data still did not meet our predefined significance criterion (Fig. 8D). We thus cannot reliably estimate the within-host HIV evolutionary rate or proviral integration dates for participant 5.
Nevertheless, the data still yield useful insights. First, the identification of two highly divergent proviral sequences that branched close to the root, one from TCM cells and the other from TEM cells, suggests that these sequences were seeded into the reservoir well before the first plasma sampling in 1996; however, it should be noted that within-host HIV sequences can sometimes evolve toward the subtype consensus (53, 54), so this observation alone does not prove that these proviruses are old. The remainder of proviruses were interspersed with the plasma HIV RNA sequences, suggesting that they were seeded into the reservoir around the time of cART initiation. Moreover, the root-to-tip distances of HIV sequences isolated from the CD4+ T cell subsets were significantly different; consistent with our original hypothesis, proviral sequences from TN and TCM cells were, on average, closer to the root (suggesting they were older) whereas those from the more differentiated T cell subsets were more distant from the root (suggesting that they were younger) (Kruskal-Wallis test, P < 0.001) (Fig. 8E). Both ANOVA and SM detected highly significant genetic compartmentalization between proviruses archived in different CD4+ T cell subsets (AMOVA, P < 0.001; SM weighted mean, 0.005; HPD, 0.000 to 0.0057) (Fig. 8F). Identical proviral sequences were also observed, including one sequence isolated 13 times from TTM cells and one sequence isolated from both TTM and TEM cells (Fig. 8B).
No consistent trend in the proviral ages of unique versus duplicated sequences.
Clonal expansion of CD4+ T cells is a major driver of HIV persistence (23–26, 51, 52), but the relationship between clonal expansion and proviral age remains unknown. To investigate this, we calculated the average proviral ages of unique versus duplicated sequences identified in participants 1 to 4 (proviral dating could not be performed for participant 5). We found no consistent evidence, however, to support duplicated (presumably clonally expanded) proviral sequences as being on average younger or older than unique ones (paired t test, P = 0.384) (Fig. 9).
FIG 9.
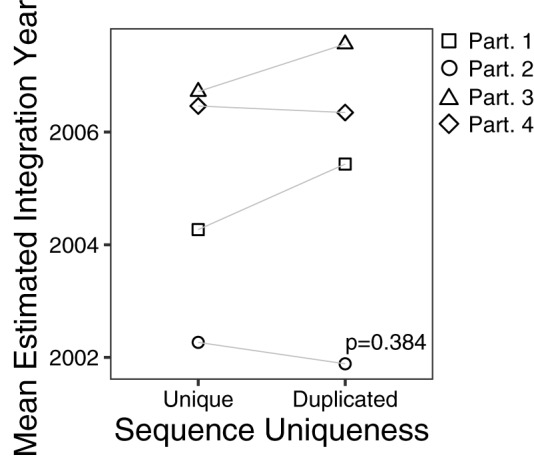
Mean estimated proviral integration year of unique versus duplicated sequences. Data points correspond to the mean proviral integration year of distinct sequences, stratified by whether they are unique (i.e., observed only once) or duplicated (i.e., observed more than once). The P value was calculated using a paired t test between the mean proviral integration year of unique versus duplicated sequences.
No consistent patterns in proviral diversity or distinctiveness among CD4+ T cell subsets.
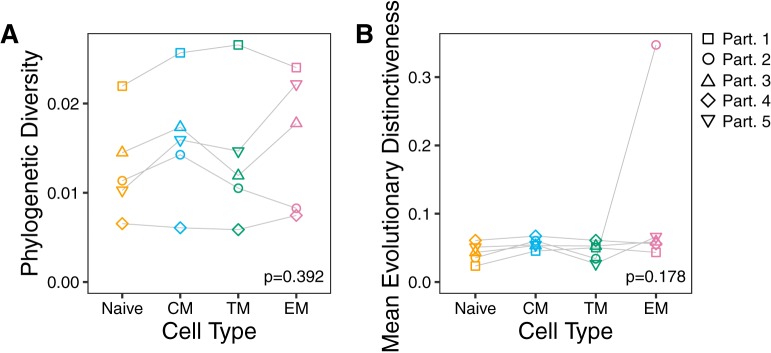
Up until this point, each participant’s data set was examined separately for evidence that their CD4+ T cell subsets harbored genetically distinct proviral populations. In closing, we attempted to glean more general inferences about proviral genetic composition within CD4+ T cell subsets during cART by analyzing data across individuals. In particular, we sought to test the hypothesis that less differentiated memory T cell subsets, by virtue of their longer life-spans, tend to harbor the most genetically diverse and/or distinctive proviral populations. First, we quantified the phylogenetic diversity of distinct proviral sequences isolated from each participant’s CD4+ T cell subsets (55). This provides a measure of the overall genetic variation within each subset. Second, we quantified the average evolutionary distinctiveness of these same sequences (44). This provides a measure of the overall level of topological isolation of sequences within each subset (e.g., subsets rich in sequences that branch deeply within the tree would yield a high distinctiveness score). However, comparisons across participants yielded no significant differences in within-host proviral phylogenetic diversity (Friedman test, P = 0.392) or mean evolutionary distinctiveness metrics across subsets (Friedman test, P = 0.178) (Fig. 10A and B). Overall, no consistent trends were apparent that would allow us to generalize with respect to CD4+ T cell differentiation stage and the genetic properties of proviral sequences archived therein.
FIG 10.
Cross-participant analysis of proviral diversity and distinctiveness within CD4+ T cell subsets. (A) Normalized phylogenetic diversity in the number of nucleotide substitutions per site per tip of distinct sequences in each participant, stratified by specific CD4+ T cell subset. (B) Mean evolutionary distinctiveness of distinct sequences in each participant, stratified by specific CD4+ T cell subset. Evolutionary distinctiveness is a unitless measurement. P values were calculated using a Friedman rank sum test.
DISCUSSION
We observed marked interindividual differences in the extent to which proviruses persisting within distinct CD4+ T cell subsets during long-term cART differed in their genetic composition and estimated ages. Two participants (3 and 4) displayed no evidence of proviral genetic compartmentalization across CD4+ T cell subsets, while three participants (1, 2, and 5) did. For participants 1 and 2, however, the extent of compartmentalization was modest; only participant 5 demonstrated strong compartmentalization (this is evident from the phylogenies shown in Fig. 8B, where proviruses from the less differentiated TN and TCM cell subsets are largely concentrated in subclades closer to the root, while those from the more differentiated TTM and TEM cell subsets, the latter in particular, are largely concentrated in subclades more divergent from the root). However, it is relevant that participant 5 experienced frequent viremic episodes during cART, which may have influenced reservoir dynamics compared to those with a scenario of sustained long-term viremia suppression.
While some level of proviral genetic compartmentalization was detected in 3/5 (60%) of participants, a frequency that is broadly consistent with the recent report by De Scheerder et al. (29), its manifestation was inconsistent between individuals. Notably, we found no consistent evidence to support our original hypothesis that, of the memory CD4+ T cell subsets, TCM cells would tend to harbor the oldest, most genetically diverse and/or distinctive proviral populations by virtue of their increased stability as a reservoir. In all but participant 5, TCM cell-derived proviral sequences were distributed throughout the phylogenies and intermixed with sequences from other subsets. Moreover, in the four participants for whom proviral ages could be estimated (participants 1 to 4), TCM cell proviral ages were not significantly different from those of other memory subsets (all pairwise comparisons, P > 0.8) (data not shown). Other reasonable hypotheses exist: for example, as HIV preferentially infects HIV-specific T cells (56) one could posit that, over time, repeated antigen-driven stimulation of latently infected T cells would lead to their differentiation, such that the oldest proviruses would eventually accumulate in the most differentiated CD4+ T cell subsets. Alternatively, given that HIV is capable of infecting resting CD4+ T cells including TN cells (36, 57), one might hypothesize that, if most of these fail to subsequently encounter their cognate antigen, TN cells may come to harbor the oldest proviruses over time. However, we observed no overall patterns that could support any such generalizations. In fact, other than our observation that identical proviruses tended to be enriched in more differentiated T cell subsets (Fig. 3), which is consistent with the findings of others (13, 29), we did not observe any consistent overall relationship between T cell differentiation stage and the age, diversity, and/or distinctiveness of proviral sequences archived therein.
These results extend our understanding of proviral composition in peripheral blood CD4+ T cells during long-term cART. The observation that proviral sequences were interspersed throughout phylogenetic reconstructions of pre-cART HIV evolutionary history, where the oldest sequences were estimated to have integrated more than 20 years prior to sampling, confirms that the reservoir is continually seeded during uncontrolled infection and that these lineages can persist for decades (32, 45). Our frequent isolation of identical proviral sequences, sometimes representing very old viral lineages (e.g., a 21.5-year-old sequence was repeatedly recovered from participant 2’s TEM cells) and in other cases isolated from multiple CD4+ T cell subsets (e.g., a 10-year-old sequence was repeatedly recovered from participant 4’s TCM, TTM, and TEM cells), further supports CD4+ T cell differentiation and clonal expansion as mechanisms to sustain the HIV reservoir (13, 15, 23–29).
Some caveats and limitations of our study should be noted. First, as we performed subgenomic sequencing, we do not know whether our sequences derive from intact, replication-competent proviruses; in fact, it is likely that most do not, given that the vast majority of proviruses persisting during long-term cART are defective (40, 58–60). It is further conceivable that, during uncontrolled HIV infection, cells harboring replication-competent proviruses may not persist as long as those harboring defective ones, as proposed in a recent study that revealed that a major portion of the replication-competent reservoir dates to around the time of cART initiation (22). Indeed, participant 5’s proviral landscape differed from the others in that the majority of recovered proviruses were interspersed with plasma HIV RNA sequences isolated prior to cART initiation (Fig. 8B), suggesting that this individual's reservoir is skewed toward sequences seeded around the time of cART initiation (22, 34) (though proviral seeding order should be interpreted with caution as the trees of this participant were outgroup rooted). This participant also experienced frequent episodes of uncontrolled viremia during cART, which could have led to gradual clearance of cells harboring older proviruses through repeated restimulation. A second caveat is that because we performed subgenomic proviral sequencing without integration site determination (61), we cannot conclusively state that identical proviral sequences derive from cellular differentiation and/or clonal expansion events; in fact, there is no optimal subgenomic HIV region that can be used to identify unique full-genome HIV sequences 100% of the time (62). A third caveat is that stem cell memory CD4+ T (TSCM) cells, which are a rare yet important HIV reservoir (34), would have been sorted into the TN cell subset as they also express CD45, CCR7, and CD27. Due to the rarity of TSCM cells, however, we do not anticipate that these will have substantially skewed our genetic characterization of TN cells, which is supported by Zerbato et al. who reported that depletion of TSCM from TN cells via CD95 had no effect on total proviral levels measured in TN cells (36). A final caveat is that only peripheral blood CD4+ T cells were studied, while the majority of memory CD4+ T cells reside in tissues (31).
Nevertheless, a strength of our study is the availability of archived plasma dating back to the mid-1990s, which allows us to interpret proviral diversity in context of HIV’s within-host evolutionary history and make inferences regarding reservoir longevity and dynamics. Another strength is our use of Bayesian methods (42) to generate a distribution of phylogenies instead of inferring a single tree via maximum likelihood (20–22) or neighbor-joining (25, 32) methods, allowing us to account for the uncertainty in reconstructing the evolutionary relationships between sampled viral sequences.
In conclusion, our study underscores the genetic complexity of proviruses persisting in CD4+ T cell subsets during long-term cART and the unique individuality of each participant studied. Other than our observation that identical proviral sequences tended to be observed more often in more differentiated memory CD4+ T cell subsets, we observed no consistent relationship between T cell differentiation stage and the age, diversity, and/or distinctiveness of proviral sequences archived therein. Our findings underscore the importance of developing cure strategies that will take intra- and interindividual reservoir genetic diversity into account, including personalized strategies capable of eradicating HIV from a variety of cell types.
MATERIALS AND METHODS
Study participants and ethics statement.
Five participants who initiated cART a median of 10 years following HIV diagnosis (range, 2 to 14 years) and who subsequently received cART for a median 9 years (range, 8 to 16 years) were recruited through the British Columbia Centre for Excellence in HIV/AIDS (Table 1). Participants provided a single large blood donation from which peripheral blood mononuclear cells (PBMCs) were isolated by standard density gradient separation (Histopaque-1077; Sigma), counted (TC20 automated cell counter; Bio-Rad), and cryopreserved at –150°C in 90% fetal bovine serum and 10% dimethyl sulfoxide (DMSO). A median 8 (range, 2 to 17) longitudinal pre-cART plasma samples per participant, dating back to 1996, were also available for analysis. The study was approved by the Providence Health Care/University of British Columbia and Simon Fraser University research ethics boards. All participants provided written informed consent.
Isolation of CD4+ T cell subsets.
CD4+ T cells were purified from a minimum of 350 million cryopreserved PBMCs by negative magnetic selection (StemCell Technologies), after which naive (TN), central memory (TCM), transitional memory (TTM), and effector memory (TEM) CD4+ T cells were isolated as described in Chomont et al. (32). Briefly, purified CD4+ T cells were labeled with a live/dead dye (Invitrogen) and Alexa Fluor 700 (AF700)-labeled mouse anti-human CD3 antibody, pacific blue (PB)-labeled mouse anti-human CD8, allophycocyanin (APC)-labeled mouse anti-human CD4 antibody, APC-Cy7-labeled mouse anti-human CD45RA antibody, phycoerythrin (PE)-Cy7-labeled rat anti-human CCR7 antibody, and PE-labeled mouse anti-human CD27 antibody (all from BD Biosciences) and separated using a FACSAria cell sorter (BD Biosciences) (Fig. 1). Purity of all isolated subsets was verified by flow cytometry after staining for CD3, CD4, CD8, CD45RA, CCR7, CD27, and viability, and percent purity was confirmed as follows: TN median, 95.9% (IQR, 95.6% to 98.2%); TCM median, 98.3% (IQR, 97.3% to 99.5%); TTM median, 98.9% (IQR, 98.9% to 99.6%); and TEM median, 98.5% (IQR, 98.5% to 99.4%).
HIV reservoir quantification.
For each participant, genomic DNA was extracted from a median of 3.1 ×106 (IQR, 2.1 × 106 to 4 × 106) purified CD4+ T cells per subset using a Purelink Genomic DNA Mini kit (Invitrogen). The number of HIV Gag copies/million CD4+ T cells was determined using droplet digital PCR (ddPCR) as previously described (63), where HIV Gag and human RPP30 reactions were conducted in parallel, and copy numbers were normalized to the quantity of input DNA to determine the number of HIV copies/million CD4+ T cells. Briefly, in each ddPCR reaction mixture, between 9 and 13 ng (RPP30) or between 131 and 1,152 ng (HIV Gag) of genomic DNA was combined with 10 units of XhoI restriction enzyme (New England Biolabs), ddPCR Supermix for Probes (no dUTPs) (Bio-Rad), primers (final concentration, 900 nM; Integrated DNA Technologies), probe (final concentration, 250 nM; Integrated DNA Technologies), and nuclease-free water. Primer and probe sequences were as follows: RPP30 forward primer, GATTTGGACCTGCGAGCG; RPP30 probe, VIC-CTGACCTGA-ZEN-AGGCTCT-3IABKFQ; RPP30 reverse primer, GCGGCTGTCTCCACAAGT; HIV Gag forward primer, TCTCGACGCAGGACTCG; HIV Gag probe, FAM-CTCTCTCCT-ZEN-TCTAGCCTC-3IABKFQ; HIV Gag reverse primer, TACTGACGCTCTCGCACC. Droplets were prepared using an Automated Droplet Generator (Bio-Rad) and cycled as follows: 95°C for 10 min; 40 cycles of 94°C for 30 s and 53°C for 1 min; and 98°C for 10 min. Droplets were analyzed on a QX200 Droplet Reader (Bio-Rad) using QuantaSoft software (version 1.7.4; Bio-Rad), and data from a median of 4 replicate wells (IQR, 4 to 6 wells) were merged prior to analysis.
Single-genome HIV amplification and sequencing.
HIV RNA was extracted from 0.5-ml pre-cART plasma aliquots using a NucliSENS EasyMag system (bioMérieux), while genomic DNA was extracted from purified CD4+ T cells using an Invitrogen genomic DNA isolation kit. Single-genome amplification of a subgenomic HIV fragment (nef) was performed by limiting dilution using high-fidelity enzymes and sequence-specific primers optimized for amplification of multiple HIV group M subtypes. For HIV RNA extracts, cDNA was generated using Expand reverse transcriptase (Roche). Next, cDNA as well as genomic DNA extracts was endpoint diluted such that ∼25 to 30% of the resulting nested PCRs, performed using an Expand High Fidelity PCR system (Roche), would yield an amplicon. Negative PCR controls were included in every run. Primers used for cDNA generation/1st round PCR were Nef8683F_pan (forward; 5′-TAGCAGTAGCTGRGKGRACAGATAG-3′) and Nef9536R_pan (reverse; 5′-TACAGGCAAAAAGCAGCTGCTTATATGYAG-3′). Primers used for the 2nd round of PCRs were Nef8746F_pan (forward; 5′-TCCACATACCTASAAGAATMAGACARG-3′) and Nef9474R (reverse; 5′-CAGGCCACRCCTCCCTGGAAASKCCC-3′). Amplicons were sequenced on an ABI 3130xl or 3730xl automated DNA sequencer using BigDye, version 3.1 chemistry (Applied Biosystems). Chromatograms were base called using Sequencher, version 5.0 (Gene Codes).
Alignment and phylogenetic inference.
Sequences were aligned using MAFFT, version 7.313 (64), and manually inspected and edited using AliView, version 1.23 (65). Sequences exhibiting gross defects (e.g., large deletions), mixed bases, hypermutated sequences (identified using Hypermut, version 2.0 [66], with the consensus of a participant’s earliest sequences used as the reference), or within-host recombinants (identified using RDP4 Beta 95 [67]) were discarded. Duplicate sequences (identified using seqinr, version 3.4-5, of the R package [68]) were discarded for the purpose of inferring phylogenies. We used jModeltest, version 2.1.10 (69), to select the substitution model with the lowest Bayesian information criterion (BIC) for each participant except participant 4, for whom we used the best substitution model lacking gamma rate heterogeneity (as models featuring the latter yielded trees that overestimated within-host branch lengths). For each participant, Markov chain Monte Carlo (MCMC) methods were used to build a random sample of phylogenies. Two parallel runs for each participant with MCMC chains of 10 or 20 million generations sampling every 1,000 generations were performed in MrBayes, version 3.2.7 (42), using the substitution model selected as described above and default priors. Convergence of chains was assessed by ensuring the deviation of split frequencies was <0.01 (using MrBayes [42]) and that the effective sampling size of all parameters was >200 (using Tracer, version 1.7.1 [70]). We discarded 25% burn-in from each chain and parallel chains combined in R, version 3.6.0 (71), resulting in 30,000 phylogenies per participant for participants 1, 3, and 4 and 15,000 phylogenies per participant for participants 2 and 5. Phylogenies were plotted using the R package ggtree, version 1.17.4 (72, 73).
Age reconstruction, compartmentalization, and phylogenetic statistics.
We used a published within-host phylogenetic approach to infer proviral sequence ages (21), with the following modifications. Instead of inferring a single maximum likelihood phylogeny per participant, we analyzed all of the phylogenies inferred by the Bayesian method described above. Briefly, each phylogeny was rooted using root-to-tip regression implemented in the R package ape, version 5.3 (74). Then, a linear regression was fitted to each phylogeny that related the collection date of each plasma HIV RNA sequence to its divergence from the root using R (71). We assessed molecular clock and model fit by using a delta Akaike information criterion (ΔAIC) (75), computed as the difference between the AIC of the null model (a zero slope) and the AIC of the linear regression. We determined a within-host data set to have a sufficient molecular clock signal if at least 50% of the trees produced a linear regression with a ΔAIC of at least 5 (a threshold that, in our experiments, corresponds to a P value of 0.0081 when a log-likelihood ratio test is used) and with a root date estimate whose lower limit for the 95% confidence interval was before the first sampled time point. Each linear regression was then used to estimate the integration date of each proviral sequence from its divergence from the root.
Sequence compartmentalization was assessed using two methods: analysis of molecular variance (AMOVA), a genetic distance-based test (49), and Slatkin-Maddison (SM), a tree-based test (43). For the SM test and all other subsequent tests that specifically assessed proviral composition within CD4+ T cell subsets, the within-host phylogenies were pruned to retain only proviral sequences, using the R package ape (74). AMOVA was implemented in the R package pegas, version 0.11 (76), while SM was implemented in the R package slatkin.maddison, version 0.1.0 (77). Phylogenetic diversity (55) and evolutionary distinctiveness (44) were computed using custom scripts implemented in R. Phylogenetic diversity was calculated by summing the edge lengths of a pruned phylogeny retaining only sequences from a particular subset and then dividing by the number of sequences in the subset. Prior to assessing phylogenetic distinctiveness, all within-host phylogenies were first outgroup rooted (OGR) with the evolutionary placement algorithm (EPA) implemented in RAxML, version 8.2.12 (78) with the HIV-1 subtype B reference strain HXB2 (GenBank accession number K03455) as the outgroup. Phylogenetic distinctiveness was then computed as described in May (44) with the R package ape (74). Paired t tests, Kruskal-Wallis rank sum tests, and Friedman rank sum tests were computed using R (71). Computer parallelization was performed using GNU Parallel (79).
Data availability.
Previously deposited sequences in this study were obtained from GenBank and have the following accession numbers: MG822918, MG822919, MG822923 to MG822933, MG822935 to MG822997, MG822999 to MG823016, MG823018 to MG823039, MG823041 to MG823050, MG823052 to MG823101, MG823106 to MG823112, MG823114 to MG823123, MG823125 to MG823143. GenBank accession numbers for new sequences used in this study are MN600002 to MN600712.
ACKNOWLEDGMENTS
This research was funded in part by team grant HB1-164063 (to Z.L.B. and M.A.B.) and project grant PJT-159625 (to Z.L.B. and J.B.J.) from the Canadian Institutes of Health Research (CIHR), by the National Institutes of Health under award number NIHR21A127029 (to Z.L.B., M.A.B., and A.F.Y.P.), and by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health with cofunding from the National Institute on Drug Abuse, the National Institute of Mental Health, and the National Institute of Neurological Disorders and Stroke under award number UM1AI126617 (to Z.L.B. and M.A.B.). N.N.K. was supported by a CIHR Frederick Banting and Charles Best M.Sc. award and now holds a CIHR Vanier Doctoral Award. M.A.B. holds a Canada Research Chair, Tier 2, in Viral Pathogenesis and Immunity. N.C. is supported by a Research Career Award from the Quebec Health Research Fund (FRQS). Z.L.B. is supported by a scholar award from the Michael Smith Foundation for Health Research.
We thank Bemuluyigza Baraki for technical assistance, Conan Woods for database assistance, and Don Kirkby and Walter Scott for assistance with high-performance computing. We thank Julio Montaner, Rolando Barrios, Mark Hull, Silvia Guillemi, and the BC Centre for Excellence in HIV/AIDS for support. We gratefully thank the study participants, without whom research would not be possible.
REFERENCES
- 1.Hogg RS, Heath KV, Yip B, Craib KJ, O'Shaughnessy MV, Schechter MT, Montaner JS. 1998. Improved survival among HIV-infected individuals following initiation of antiretroviral therapy. JAMA 279:450–454. doi: 10.1001/jama.279.6.450. [DOI] [PubMed] [Google Scholar]
- 2.Palella FJ Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med 338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 3.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 4.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 6.Davey RT Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, Kovacs JA, Polis MA, Walker RE, Falloon J, Masur H, Gee D, Baseler M, Dimitrov DS, Fauci AS, Lane HC. 1999. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A 96:15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 8.Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, Khoruts A, Estes JD, Anderson J, Callisto SP, Schmidt TE, Thorkelson A, Reilly C, Perkey K, Reimann TG, Utay NS, Nganou Makamdop K, Stevenson M, Douek DC, Haase AT, Schacker TW. 2015. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A 112:E1126–1134. doi: 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kearney M, Spindler J, Shao W, Maldarelli F, Palmer S, Hu SL, Lifson JD, KewalRamani VN, Mellors JW, Coffin JM, Ambrose Z. 2011. Genetic diversity of simian immunodeficiency virus encoding HIV-1 reverse transcriptase persists in macaques despite antiretroviral therapy. J Virol 85:1067–1076. doi: 10.1128/JVI.01701-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evering TH, Mehandru S, Racz P, Tenner-Racz K, Poles MA, Figueroa A, Mohri H, Markowitz M. 2012. Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog 8:e1002506. doi: 10.1371/journal.ppat.1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Josefsson L, von Stockenstrom S, Faria NR, Sinclair E, Bacchetti P, Killian M, Epling L, Tan A, Ho T, Lemey P, Shao W, Hunt PW, Somsouk M, Wylie W, Douek DC, Loeb L, Custer J, Hoh R, Poole L, Deeks SG, Hecht F, Palmer S. 2013. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci U S A 110:E4987–E4996. doi: 10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Stockenstrom S, Odevall L, Lee E, Sinclair E, Bacchetti P, Killian M, Epling L, Shao W, Hoh R, Ho T, Faria NR, Lemey P, Albert J, Hunt P, Loeb L, Pilcher C, Poole L, Hatano H, Somsouk M, Douek D, Boritz E, Deeks SG, Hecht FM, Palmer S. 2015. Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV Type 1 DNA pool during effective HIV therapy. J Infect Dis 212:596–607. doi: 10.1093/infdis/jiv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. 2017. Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep 21:813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salantes DB, Zheng Y, Mampe F, Srivastava T, Beg S, Lai J, Li JZ, Tressler RL, Koup RA, Hoxie J, Abdel-Mohsen M, Sherrill-Mix S, McCormick K, Overton ET, Bushman FD, Learn GH, Siliciano RF, Siliciano JM, Tebas P, Bar KJ. 2018. HIV-1 latent reservoir size and diversity are stable following brief treatment interruption. J Clin Invest 128:3102–3115. doi: 10.1172/JCI120194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinzone MR, VanBelzen DJ, Weissman S, Bertuccio MP, Cannon L, Venanzi-Rullo E, Migueles S, Jones RB, Mota T, Joseph SB, Groen K, Pasternak AO, Hwang W-T, Sherman B, Vourekas A, Nunnari G, O'Doherty U. 2019. Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nat Commun 10:728. doi: 10.1038/s41467-019-08431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. 1998. Early establishment of a pool of latently infected, resting CD4+ T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A 95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schacker T, Little S, Connick E, Gebhard-Mitchell K, Zhang ZQ, Krieger J, Pryor J, Havlir D, Wong JK, Richman D, Corey L, Haase AT. 2000. Rapid accumulation of human immunodeficiency virus (HIV) in lymphatic tissue reservoirs during acute and early HIV infection: implications for timing of antiretroviral therapy. J Infect Dis 181:354–357. doi: 10.1086/315178. [DOI] [PubMed] [Google Scholar]
- 18.Archin NM, Vaidya NK, Kuruc JD, Liberty AL, Wiegand A, Kearney MF, Cohen MS, Coffin JM, Bosch RJ, Gay CL, Eron JJ, Margolis DM, Perelson AS. 2012. Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. Proc Natl Acad Sci U S A 109:9523–9528. doi: 10.1073/pnas.1120248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, Smith JY, Brinkman AL, Peter LE, Mathew SI, Smith KM, Borducchi EN, Rosenbloom DI, Lewis MG, Hattersley J, Li B, Hesselgesser J, Geleziunas R, Robb ML, Kim JH, Michael NL, Barouch DH. 2014. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512:74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodin J, Zanini F, Thebo L, Lanz C, Bratt G, Neher RA, Albert J. 2016. Establishment and stability of the latent HIV-1 DNA reservoir. Elife 5:e18889. doi: 10.7554/eLife.18889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones BR, Kinloch NN, Horacsek J, Ganase B, Harris M, Harrigan PR, Jones RB, Brockman MA, Joy JB, Poon AFY, Brumme ZL. 2018. Phylogenetic approach to recover integration dates of latent HIV sequences within-host. Proc Natl Acad Sci U S A 115:E8958–E8967. doi: 10.1073/pnas.1802028115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrahams M-R, Joseph SB, Garrett N, Tyers L, Moeser M, Archin N, Council OD, Matten D, Zhou S, Doolabh D, Anthony C, Goonetilleke N, Karim SA, Margolis DM, Pond SK, Williamson C, Swanstrom R. 2019. The replication-competent HIV-1 latent reservoir is primarily established near the time of therapy initiation. Sci Transl Med 11:eaaw5589. doi: 10.1126/scitranslmed.aaw5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. 2014. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. 2014. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bui JK, Sobolewski MD, Keele BF, Spindler J, Musick A, Wiegand A, Luke BT, Shao W, Hughes SH, Coffin JM, Kearney MF, Mellors JW. 2017. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog 13:e1006283. doi: 10.1371/journal.ppat.1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo H-H, Hua S, Chen H-R, Ouyang Z, Reddy K, Dong K, Ndung'u T, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. 2017. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest 127:2689–2696. doi: 10.1172/JCI93289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullins JI, Frenkel LM. 2017. Clonal expansion of human immunodeficiency virus-infected cells and human immunodeficiency virus persistence during antiretroviral therapy. J Infect Dis 215:S119–S127. doi: 10.1093/infdis/jiw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Gurule EE, Brennan TP, Gerold JM, Kwon KJ, Hosmane NN, Kumar MR, Beg SA, Capoferri AA, Ray SC, Ho YC, Hill AL, Siliciano JD, Siliciano RF. 2018. Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc Natl Acad Sci U S A 115:E2575–E2584. doi: 10.1073/pnas.1720665115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Scheerder MA, Vrancken B, Dellicour S, Schlub T, Lee E, Shao W, Rutsaert S, Verhofstede C, Kerre T, Malfait T, Hemelsoet D, Coppens M, Dhondt A, De Looze D, Vermassen F, Lemey P, Palmer S, Vandekerckhove L. 2019. HIV rebound is predominantly fueled by genetically identical viral expansions from diverse reservoirs. Cell Host Microbe 26:347–358.e7. doi: 10.1016/j.chom.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Restifo NP, Gattinoni L. 2013. Lineage relationship of effector and memory T cells. Curr Opin Immunol 25:556–563. doi: 10.1016/j.coi.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farber DL, Yudanin NA, Restifo NP. 2014. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol 14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel M-R, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy J-P, Haddad EK, Sékaly R-P. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bacchus C, Cheret A, Avettand-Fenoel V, Nembot G, Melard A, Blanc C, Lascoux-Combe C, Slama L, Allegre T, Allavena C, Yazdanpanah Y, Duvivier C, Katlama C, Goujard C, Seksik BC, Leplatois A, Molina JM, Meyer L, Autran B, Rouzioux C, Group O. 2013. A single HIV-1 cluster and a skewed immune homeostasis drive the early spread of HIV among resting CD4+ cell subsets within one month post-infection. PLoS One 8:e64219. doi: 10.1371/journal.pone.0064219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, Pereyra F, Zurakowski R, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. 2014. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med 20:139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, Archin NM. 2014. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol 88:14070–14077. doi: 10.1128/JVI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zerbato JM, Serrao E, Lenzi G, Kim B, Ambrose Z, Watkins SC, Engelman AN, Sluis-Cremer N. 2016. Establishment and reversal of HIV-1 latency in naive and central memory CD4+ T cells in vitro. J Virol 90:8059–8073. doi: 10.1128/JVI.00553-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zerbato JM, McMahon DK, Sobolewski MD, Mellors JW, Sluis-Cremer N. 2019. Naive CD4+ T cells harbor a large inducible reservoir of latent, replication-competent HIV-1. Clin Infect Dis 69:1919–1925. doi: 10.1093/cid/ciz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venanzi Rullo E, Cannon L, Pinzone MR, Ceccarelli M, Nunnari G, O’Doherty U. 2019. Genetic evidence that Naive T cells can contribute significantly to the HIV intact reservoir: time to re-evaluate their role. Clin Infect Dis 69:2236–2237. doi: 10.1093/cid/ciz378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaafoura S, de Goer de Herve MG, Hernandez-Vargas EA, Hendel-Chavez H, Abdoh M, Mateo MC, Krzysiek R, Merad M, Seng R, Tardieu M, Delfraissy JF, Goujard C, Taoufik Y. 2014. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4+ memory T Cells. Nat Commun 5:5407. doi: 10.1038/ncomms6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imamichi H, Natarajan V, Adelsberger JW, Rehm CA, Lempicki RA, Das B, Hazen A, Imamichi T, Lane HC. 2014. Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus. AIDS 28:1091–1099. doi: 10.1097/QAD.0000000000000223. [DOI] [PubMed] [Google Scholar]
- 41.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, Bertagnolli LN, Capoferri AA, Kufera JT, Timmons A, Nobles C, Gregg J, Wada N, Ho YC, Zhang H, Margolick JB, Blankson JN, Deeks SG, Bushman FD, Siliciano JD, Laird GM, Siliciano RF. 2019. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566:120–125. doi: 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slatkin M, Maddison WP. 1989. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 123:603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.May RM. 1990. Taxonomy as destiny. Nature 347:129–130. doi: 10.1038/347129a0. [DOI] [Google Scholar]
- 45.Coffin JM, Wells DW, Zerbato JM, Kuruc JD, Guo S, Luke BT, Eron JJ, Bale M, Spindler J, Simonetti FR, Hill S, Kearney MF, Maldarelli F, Wu X, Mellors JW, Hughes SH. 2019. Clones of infected cells arise early in HIV-infected individuals. JCI Insight 4:128432. doi: 10.1172/jci.insight.128432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller RL, Ponte R, Jones BR, Kinloch NN, Omondi FH, Jenabian MA, Dupuy FP, Fromentin R, Brassard P, Mehraj V, Chomont N, Poon AFY, Joy JB, Brumme ZL, Routy JP, Orchid Study Group . 2019. HIV diversity and genetic compartmentalization in blood and testes during suppressive antiretroviral therapy. J Virol 93:e00755-19. doi: 10.1128/JVI.00755-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bull M, Learn G, Genowati I, McKernan J, Hitti J, Lockhart D, Tapia K, Holte S, Dragavon J, Coombs R, Mullins J, Frenkel L. 2009. Compartmentalization of HIV-1 within the female genital tract is due to monotypic and low-diversity variants not distinct viral populations. PLoS One 4:e7122. doi: 10.1371/journal.pone.0007122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zarate S, Pond SL, Shapshak P, Frost SD. 2007. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J Virol 81:6643–6651. doi: 10.1128/JVI.02268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics 131:479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buzon MJ, Codoner FM, Frost SD, Pou C, Puertas MC, Massanella M, Dalmau J, Llibre JM, Stevenson M, Blanco J, Clotet B, Paredes R, Martinez-Picado J. 2011. Deep molecular characterization of HIV-1 dynamics under suppressive HAART. PLoS Pathog 7:e1002314. doi: 10.1371/journal.ppat.1002314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohn LB, Silva IT, Oliveira TY, Rosales RA, Parrish EH, Learn GH, Hahn BH, Czartoski JL, McElrath MJ, Lehmann C, Klein F, Caskey M, Walker BD, Siliciano JD, Siliciano RF, Jankovic M, Nussenzweig MC. 2015. HIV-1 integration landscape during latent and active infection. Cell 160:420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, Wells D, Su L, Luke BT, Halvas EK, Besson G, Penrose KJ, Yang Z, Kwan RW, Van Waes C, Uldrick T, Citrin DE, Kovacs J, Polis MA, Rehm CA, Gorelick R, Piatak M, Keele BF, Kearney MF, Coffin JM, Hughes SH, Mellors JW, Maldarelli F. 2016. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A 113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herbeck JT, Nickle DC, Learn GH, Gottlieb GS, Curlin ME, Heath L, Mullins JI. 2006. Human immunodeficiency virus type 1 env evolves toward ancestral states upon transmission to a new host. J Virol 80:1637–1644. doi: 10.1128/JVI.80.4.1637-1644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dapp MJ, Kober KM, Chen L, Westfall DH, Wong K, Zhao H, Hall BM, Deng W, Sibley T, Ghorai S, Kim K, Chen N, McHugh S, Au L, Cohen M, Anastos K, Mullins JI. 2017. Patterns and rates of viral evolution in HIV-1 subtype B infected females and males. PLoS One 12:e0182443. doi: 10.1371/journal.pone.0182443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10. doi: 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- 56.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 57.Dai J, Agosto LM, Baytop C, Yu JJ, Pace MJ, Liszewski MK, O'Doherty U. 2009. Human immunodeficiency virus integrates directly into naive resting CD4+ T cells but enters naive cells less efficiently than memory cells. J Virol 83:4528–4537. doi: 10.1128/JVI.01910-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez G, Xu X, Chermann JC, Hirsch I. 1997. Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J Virol 71:2233–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho YC, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. 2016. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 22:1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Einkauf KB, Lee GQ, Gao C, Sharaf R, Sun X, Hua S, Chen SM, Jiang C, Lian X, Chowdhury FZ, Rosenberg ES, Chun TW, Li JZ, Yu XG, Lichterfeld M. 2019. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J Clin Invest 129:988–998. doi: 10.1172/JCI124291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laskey SB, Pohlmeyer CW, Bruner KM, Siliciano RF. 2016. Evaluating clonal expansion of HIV-infected cells: optimization of PCR strategies to predict clonality. PLoS Pathog 12:e1005689. doi: 10.1371/journal.ppat.1005689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang SH, Ren Y, Thomas AS, Chan D, Mueller S, Ward AR, Patel S, Bollard CM, Cruz CR, Karandish S, Truong R, Macedo AB, Bosque A, Kovacs C, Benko E, Piechocka-Trocha A, Wong H, Jeng E, Nixon DF, Ho YC, Siliciano RF, Walker BD, Jones RB. 2018. Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J Clin Invest 128:876–889. doi: 10.1172/JCI97555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rose PP, Korber BT. 2000. Detecting hypermutations in viral sequences with an emphasis on G → A hypermutation. Bioinformatics 16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- 67.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Charif D, Lobry JR. 2007. SeqinR 1.0–2: a contributed package to the R Project for Statistical Computing devoted to biological sequences retrieval and analysis, p 207–232. In Bastolla U, Porto M, Roman HE, Vendruscolo M (ed), Structural approaches to sequence evolution. Springer, Berlin, Germany. [Google Scholar]
- 69.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org. [Google Scholar]
- 72.Yu GC, Smith DK, Zhu HC, Guan Y, Lam T. 2017. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 73.Yu G, Lam TT, Zhu H, Guan Y. 2018. Two methods for mapping and visualizing associated data on phylogeny using Ggtree. Mol Biol Evol 35:3041–3043. doi: 10.1093/molbev/msy194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paradis E, Schliep K. 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 75.Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Automat Contr 19:716. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 76.Paradis E. 2010. pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 77.McAdam P. 2019. slatkin.maddison: perform Slatkin Maddison test on for trait transition across phylogeny. https://github.com/prmac/slatkin.maddison.
- 78.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tange O. 2011. GNU Parallel—the command-line power tool. login 36:42–47. [Google Scholar]
- 80.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Previously deposited sequences in this study were obtained from GenBank and have the following accession numbers: MG822918, MG822919, MG822923 to MG822933, MG822935 to MG822997, MG822999 to MG823016, MG823018 to MG823039, MG823041 to MG823050, MG823052 to MG823101, MG823106 to MG823112, MG823114 to MG823123, MG823125 to MG823143. GenBank accession numbers for new sequences used in this study are MN600002 to MN600712.