HSV-1 causes lifelong infection in the human population and can be fatal in neonatal and immunocompromised individuals. There is no vaccine or cure, in part due to the ability of HSV to escape the host immune response by various mechanisms. The HSV particle contains at least 15 envelope proteins, four of which are required for entry and replication. This work suggests a novel role for gC in shielding the HSV entry glycoproteins. gC may function to help HSV escape neutralization by antibodies.
KEYWORDS: glycoproteins, herpes simplex virus, herpesviruses, neutralization, virus entry
ABSTRACT
Viruses have evolved strategies to avoid neutralization by the host antibody response. Herpes simplex virus (HSV) glycoprotein C (gC) functions in viral entry and binds to complement component C3b, inhibiting complement-mediated immunity. We investigated whether gC protects HSV from antibody neutralization. HSV-1 that lacks gC was more sensitive to complement-independent neutralization by a panel of gB monoclonal antibodies than a wild-type gC rescuant virus. The presence of gC decreased neutralization by 2- to 16-fold. The gB in the native envelope of HSV-1 had reduced reactivity with antibodies in comparison to gB from the gC-null virus, suggesting that gC hampers the recognition of gB epitopes in the viral particle. The protein composition of the gC-null virus, including the surface glycoproteins essential for entry, was equivalent to that of the wild type, suggesting that gC is directly responsible for the reduced antibody recognition and neutralization. The neutralizing activity of antibodies to gD and gH antibodies was also increased in HSV lacking gC. Together, the data suggest that HSV-1 gC protects the viral envelope glycoproteins essential for entry, including gB, by shielding them from neutralization as a potential mechanism of immune evasion.
IMPORTANCE HSV-1 causes lifelong infection in the human population and can be fatal in neonatal and immunocompromised individuals. There is no vaccine or cure, in part due to the ability of HSV to escape the host immune response by various mechanisms. The HSV particle contains at least 15 envelope proteins, four of which are required for entry and replication. This work suggests a novel role for gC in shielding the HSV entry glycoproteins. gC may function to help HSV escape neutralization by antibodies.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is an important pathogen that persists in about 90% of the adult population in the developed world. Immune evasion is one mechanism by which HSV remains persistent. Mature HSV glycoprotein C (gC) is a 511-amino-acid protein (1, 2) that plays an important role in HSV infectivity as the primary attachment protein (3). gC is the principal viral ligand for binding to cell surface heparan sulfate proteoglycans (4). gC is one of the main targets for CD4 T-lymphocyte cytotoxicity in human epidermal keratinocytes (5). Complement component C3b binds directly to HSV-1 gC and inhibits complement activation (6, 7). Viral envelope gC and gE have been proposed to protect HSV-1 from neutralization (8). Hook et al. demonstrated that HSV-1 bearing a deletion of both gC and gE genes is neutralized by antibodies to gB, gD, or gH/gL in a manner similar to that of the wild type. Interestingly, the gC-gE-null virus was more sensitive to neutralization by two or more of these antibodies in combination (8). Sera from human natural infections elicit reactions to gB, gD, and gC (9), suggesting that gC is a target of antibody neutralization alongside the essential glycoproteins B and D. Vaccines that contain gB and gC plasmids were shown to be effective in reducing recurrent herpes infection (10) and induce neutralizing antibody (11). Anti-gC antibodies inhibit HSV infection (12). Here, we provide evidence that gC specifically protects gB from antibody recognition of neutralizing epitopes. This function of gC can serve as an immune evasion mechanism that may contribute to HSV persistence in its host.
RESULTS AND DISCUSSION
The absence of gC renders HSV more readily neutralized by antibodies to gB.
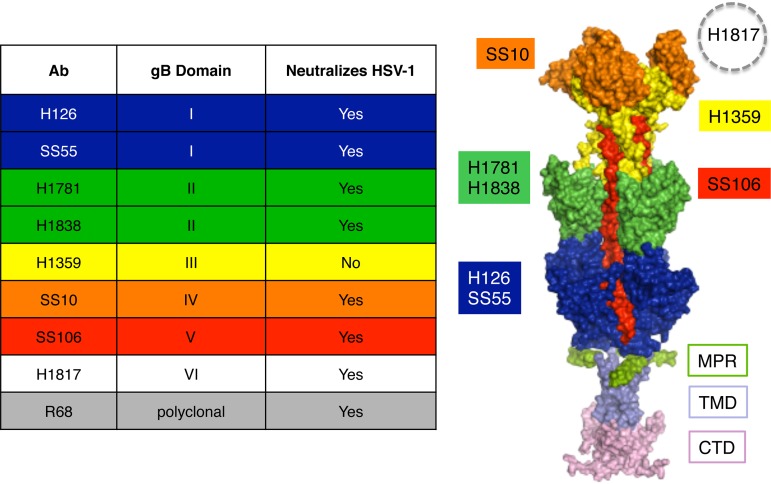
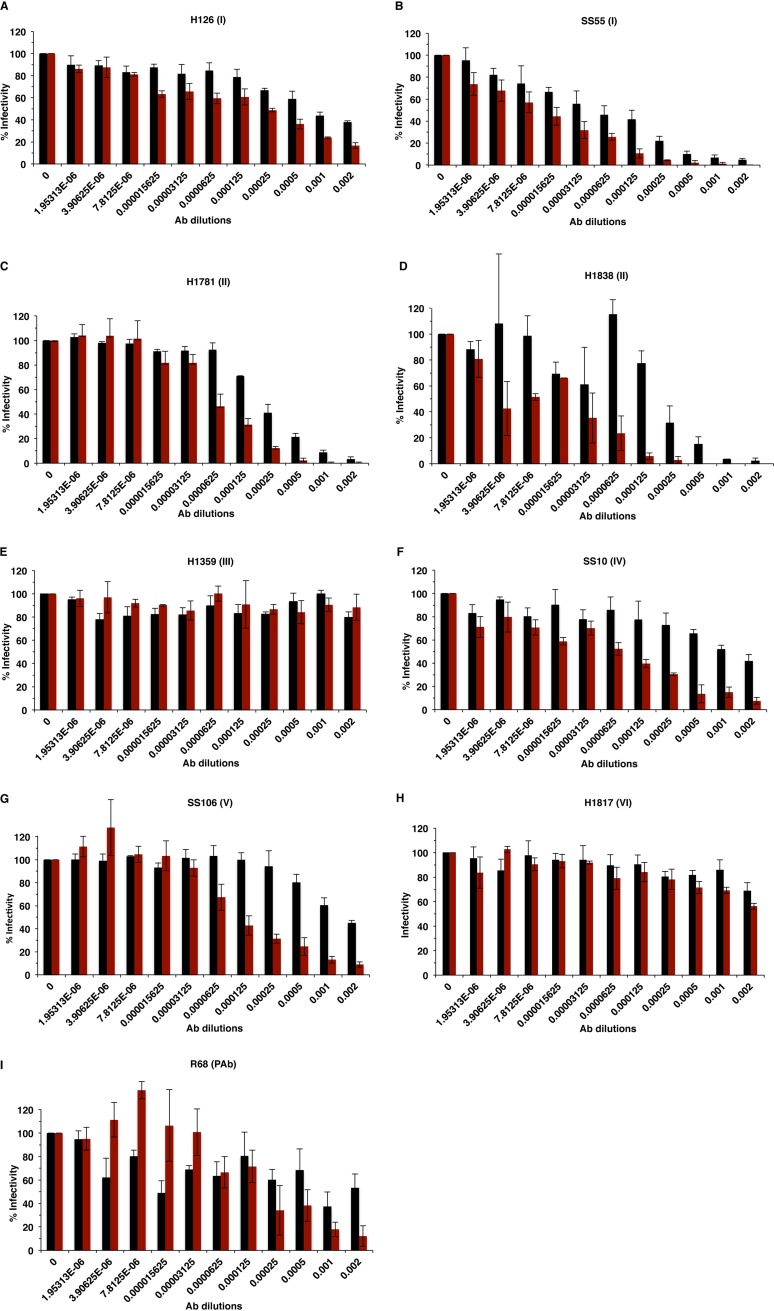
A plaque inhibition assay was used to evaluate the ability of gB antibodies to neutralize HSV-1 infectivity in the presence or absence of gC. A panel of antibodies that recognize distinct domains across the gB ectodomain was used (Fig. 1). Antibody dilutions tested ranged from 1:500 to 1:512,000. Neutralization activity was defined as the ability of an antibody to block >50% of virus infectivity at a dilution of 1:500 or greater. gB antibodies neutralized HSV-1ΔgC2-3 (HSV-1 strain KOS fully deleted of the gC gene) infectivity at a dilution 2 to 16 times higher than that of its rescuant, HSV-1gC2-3R. For example, infectivity of gC-null virus was neutralized by monoclonal antibody (Mab) SS55, which recognizes gB domain I, at a dilution of 1:64,000. This is 4 times higher than the dilution required to neutralize the infectivity of HSV-1gC2-3R (1:16,000) (Fig. 2B). The enhanced ability of gB antibodies to neutralize HSV-1ΔgC2-3 infectivity was also observed with H126 (domain I), H1838 and H1781 (domain II), SS10 (domain IV), SS106 (domain V), and gB-monospecific polyclonal antibody (PAb) R68 (Fig. 2). Notably, 16 times more Mab SS10 or SS106 was necessary to neutralize HSV-1gC2-3R than HSV-1ΔgC2-3 (Fig. 2F and G). The absence of gC did not alter H1817 (domain VI) or H1359 (domain III) neutralization activity (Fig. 2H and E). In our hands, H1817 did not neutralize >50% of infection at the highest concentration tested, in contrast to initial reports in the literature. The gB-specific polyclonal antibody R68 neutralized HSV-1ΔgC2-3 four times better than the rescuant HSV-1gC2-3R (Fig. 2I), suggesting that the profiles of the individual neutralizing monoclonal antibodies (Fig. 2A to D, F, and G) reflect an overall increase in gB sensitivity to neutralization when gC is not present. Together, the results suggest that gC reduces the neutralization activity of antibodies against gB.
FIG 1.
Antibodies to HSV-1 gB and their epitopes. (Left) List of gB antibodies used in this study, the gB domains to which they bind, and HSV neutralization activities. The ectodomain, membrane-proximal region (MPR), transmembrane domain (TMD), and cytoplasmic tail domain (CTD) of the postfusion form of the gB trimer are shown (37). Monoclonal antibody-resistant mutations in domain I, which contains bipartite hydrophobic fusion loops, map to amino acid residue 303 for H126 and residues 203, 335, and 199 for SS55 (38, 39). MAbs H1781 and H1838 bind to domain II and map to residues 454 to 473 and 391 to 410, respectively (13). MAb H1359 binds to domain III and maps approximately to residues 487 to 505 (30). MAb SS10 binds to domain IV and maps to residues 640 to 670 (13). MAb SS106 binds to domain V and maps to a peptide spanning residues 697 to 725 (29). MAb H1817 binds to domain VI (not resolved in the structure) and maps to residues 31 to 43 (13).
FIG 2.
Neutralization of gC-null HSV-1 by antibodies to gB. HSV-1gCR (black bars) or HSV-1ΔgC2-3 (red bars) were incubated with monoclonal antibodies H126 (A), SS55 (B), H1781 (C), H1838 (D), H1359 (E), SS10 (F), SS106 (G), and H1817 (H) or monospecific polyclonal antibody R68 (I) at 37°C for 1 h. Plaque formation was determined on Vero cells.
Several of the neutralizing epitopes on gB that are protected by gC (Fig. 2) are important for entry of HSV into host cells (13). MAbs H126, H1781, H1838, SS55, and SS106 have reduced reactivity with HSV particles that have been exposed to mildly acidic pH, suggesting that these gB epitopes undergo conformational changes associated with membrane fusion (14–17).
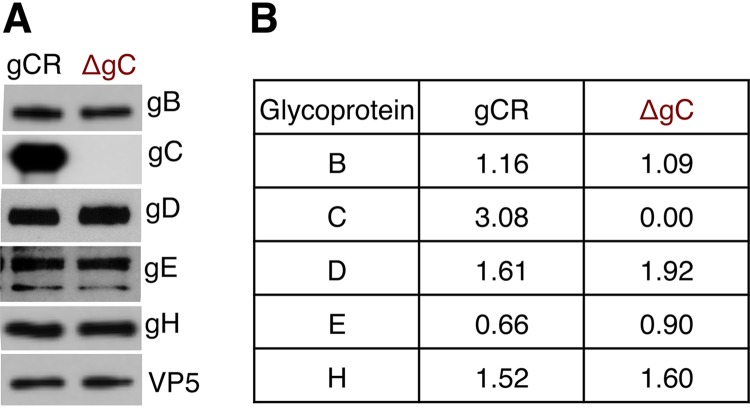
The protein levels of gB, gD, gE, and gH in HSV-1ΔgC2-3 are similar to those in HSV-1gC2-3R.
To assess the possibility that the deletion of gC from HSV-1ΔgC2-3 altered its virion levels of gB, HSV-1ΔgC2-3 was evaluated by Western blotting and compared to HSV-1gC2-3R. Equivalent virion protein 5 (VP5) units of both viruses were analyzed by SDS-PAGE. Virion gB, gD, gE, and gH were detected in HSV-1ΔgC2-3 (Fig. 3A). The mutant and rescuant viruses contained similar levels of the glycoproteins, as evidenced by densitometric analysis (Fig. 3B). As expected, gC was not detected in HSV-1ΔgC2-3 particles (Fig. 3A). Thus, the enhanced ability of gB antibodies to neutralize HSV-1ΔgC2-3 infection is not due to decreased levels of gB in virions but rather more likely because gC shields gB from antibody recognition.
FIG 3.
Entry glycoprotein levels in gC-null HSV-1. (A) HSV-1gCR or HSV-1ΔgC2-3 virions with the same VP5 content were analyzed by SDS-PAGE, followed by Western blotting with antibodies against the indicated proteins. (B) Following densitometric analysis by ImageJ, the ratio of glycoprotein to VP5 density was calculated.
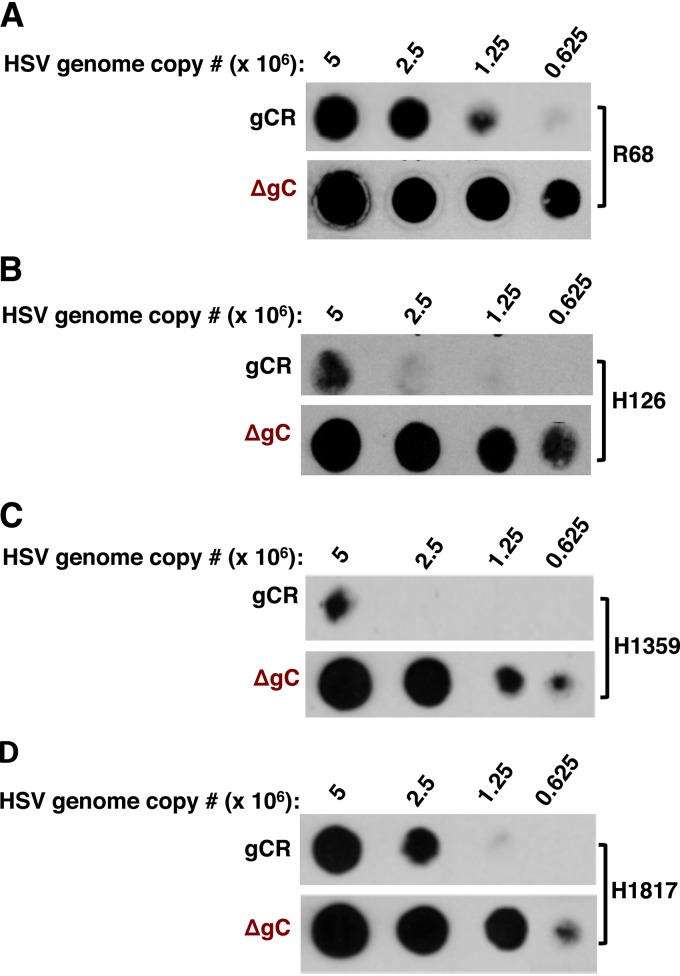
Enhanced reactivity of gB antibodies with HSV-1 that lacks gC.
We evaluated the accessibility of gB epitopes in gC-null HSV-1 by a dot blot immunoassay, which allows analysis of virions in a native state. Virus preparations of equivalent genome copy numbers were blotted onto nitrocellulose membranes and probed with one of several selected antibodies to gB: monospecific polyclonal antibody R68, MAb H126 (I), H1359 (III), or H1817 (VI). For each antibody tested, gB present in HSV-1ΔgC2-3 exhibited greater reactivity than gB in HSV-1gC2-3R (Fig. 4A to D). It would be of interest to confirm the dot blot results with fluid phase analysis of antibody binding. All together, the results suggest that gB epitopes are more accessible when gC is absent, resulting in a higher sensitivity to inhibition by antibody.
FIG 4.
Enhanced reactivity of gB antibodies with gC-null HSV-1. Equivalent genome copy numbers of HSV-1gCR or HSV-1ΔgC2-3 virions were blotted directly onto a nitrocellulose membrane and probed with gB polyclonal antibody R68 (A) or with gB MAb H126 (B), H1359 (C), or H1817 (D).
The absence of gE has little to no effect on HSV-1 neutralization by antibodies to gB.
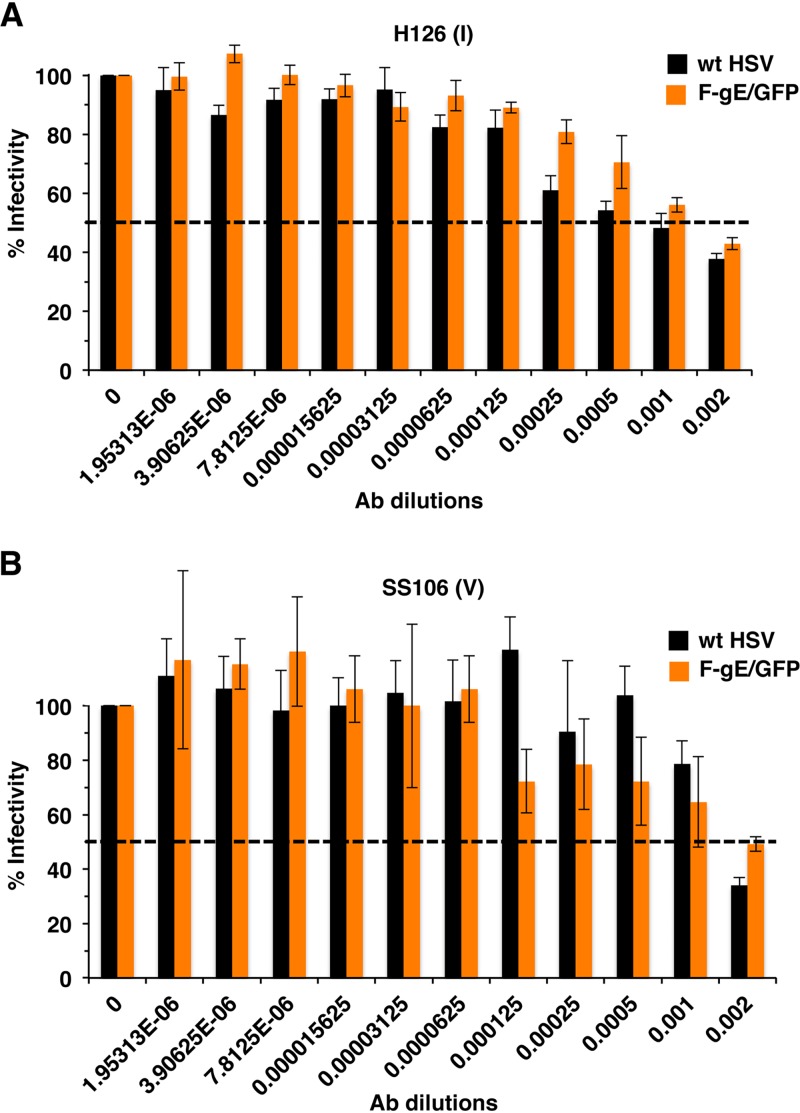
Does deletion of gC from HSV specifically render gB more susceptible to neutralizing antibodies? To address this, we tested another HSV mutant with a deletion of a single envelope glycoprotein gene. In HSV-1gE/GFP, the gE gene is replaced by green fluorescent protein (GFP). HSV-1gE/GFP and its wild-type parent, HSV-1 strain F, were neutralized to the same extent by MAb H126 (domain I) or SS106 (domain V) to gB (Fig. 5A and B). Having tested only two representative neutralizing antibodies, we cannot rule out the possibility that the absence of gE may have a more robust effect on the neutralization activity of antibodies to other gB epitopes or to gD or gH. Overall, these data support the notion that gC specifically protects gB from antibody neutralization.
FIG 5.
Neutralization of gE-null HSV-1 by antibodies to gB. Wild type HSV-1 strain F (black bars) or HSV-1 F-gE/GFP (orange bars) were incubated with monoclonal antibody H126 (A) or SS106 (B) at 37°C for 1 h. Plaque formation was determined on Vero cells. Data are the mean of triplicate samples ± standard deviation. Results are representative of at least two independent experiments.
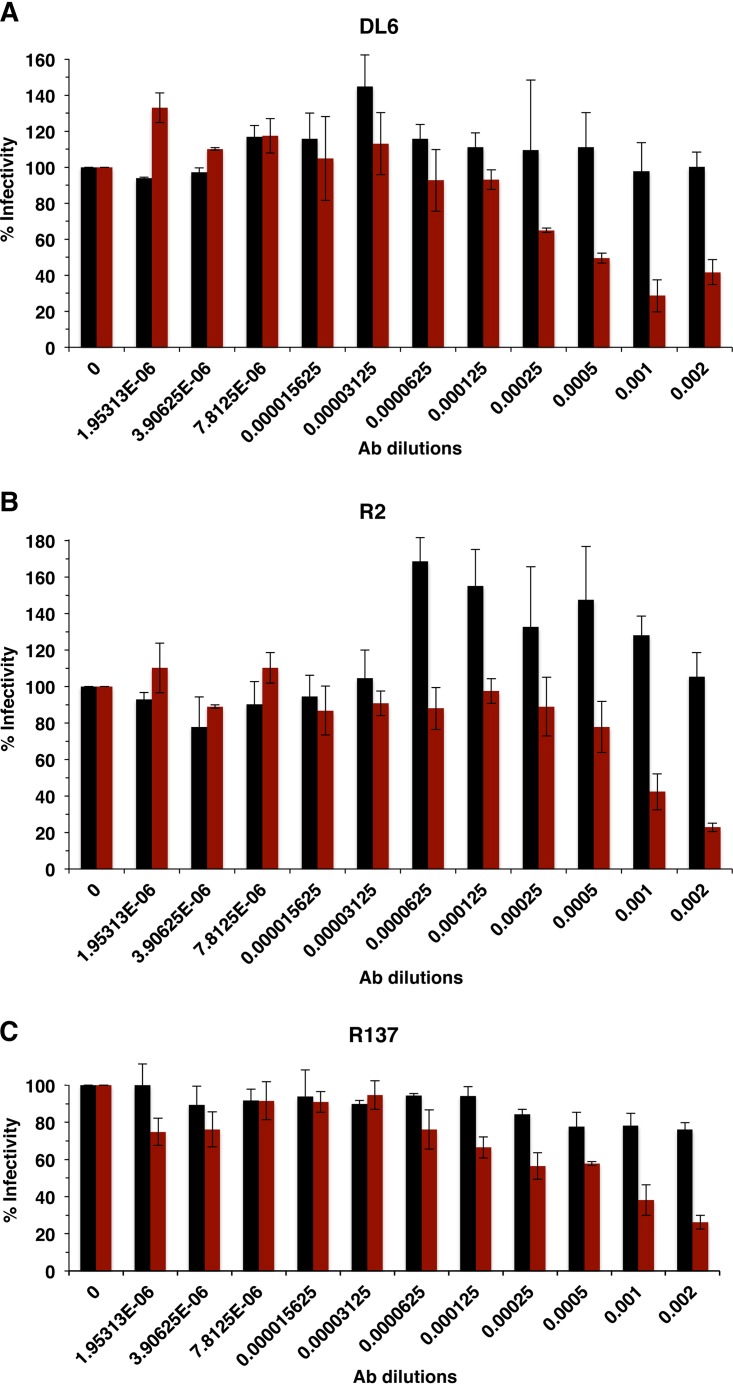
HSV that lacks gC is also more sensitive to neutralization by antibodies to gH and gD.
HSV envelope proteins gD and gH are critical players in HSV entry and are targets of virus-neutralizing antibodies (18, 19). Under the conditions tested, antibodies to gD, DL6 (monoclonal), and R2 (polyclonal) did not neutralize >50% of HSV-1gC2-3R infectivity (Fig. 6A and B). However, the same preparations of DL6 and R2 neutralized HSV-1ΔgC2-3 infectivity at dilutions of 1:2,000 and 1:1,000, respectively (Fig. 6A and B). R137 polyclonal antibody to gH did not neutralize HSV-1gC2-3R at the highest concentration tested. In contrast, >50% of HSV-1ΔgC2-3 infectivity was neutralized at a dilution of 1:500 (Fig. 6C). Thus, gD and gH entry functions are more easily blocked when gC is absent from HSV-1, supporting the idea that in wild-type HSV-1, gC shields the entry glycoproteins from neutralizing antibodies.
FIG 6.
Neutralization of gC-null HSV-1 by antibodies to gD or gH. HSV-1gCR (black bars) or HSV-1ΔgC2-3 (red bars) were incubated with gD MAb DL6 (A), gD PAb R2 (B), or gH PAb R137 (C) at 37°C for 1 h. Plaque formation was determined on Vero cells.
Results presented here support the notion that viral envelope protein gC helps HSV-1 escape neutralization by antibodies in a complement-independent manner. A panel of monospecific antibodies to gB neutralized an HSV-1 gC deletion virus more readily than the wild type. The results suggest that gC shields gB, resulting in poor antibody recognition of gB and reduced neutralizing activity of gB antibodies. Hook et al. (8) demonstrated that deletion of both gC and gE rendered HSV more sensitive to neutralization by pooled IgG from human immune sera or a combination of two or more polyclonal antibodies to gB, gD, or gH/gL. Here, we used a virus with a single deletion for gC and monospecific antibodies. The results suggest a direct role for gC in specifically shielding neutralizing epitopes on gB, which does not conflict with the conclusions of the report by Hook et al. Our data suggest that single deletion of gE from HSV does not change the neutralizing phenotype of gB MAbs H126 and SS106. Thus, the phenotype of the double-deletion HSV may be explained by the absence of gC. Nonetheless, HSV-1 gE is a well-established immune evasion molecule. It binds to the Fc portion of IgG molecules and may prevent neutralization by this mechanism (20, 21). We also observed an increased sensitivity of HSVΔgC to neutralization by gH and gD antibodies.
HSV-1 gC is highly glycosylated. Several viruses use N-glycans on envelope glycoproteins to escape from antibody neutralization. This has been documented for influenza virus, HIV, Nipah virus, and porcine reproductive and respiratory syndrome virus (22–25). The N-glycans of a particular viral glycoprotein typically shield its own epitopes. We propose that HSV-1 gC shields neighboring viral glycoproteins. Direct physical association between gC and gB or other envelope proteins has yet to be conclusively determined. The N-linked oligosaccharides of gC, including the highly glycosylated, mucin-like N-terminal domain, warrant further investigation. An alternative interpretation of the results presented here is that gC may alter the conformation of gB, gD, or gH and render them less susceptible to neutralizing antibody. Further, it is not known whether interactions among gB, gD, and gH/gL are altered in the absence of gC. The stoichiometry of shielding is also curious. How gC manages to shield at least three other viral antigens from neutralization remains to be determined.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (American Type Culture Collection, Manassas, VA) were propagated in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Atlanta, GA) and penicillin, streptomycin, and glutamine (Thermo Fischer Scientific). HSV-1 strain KOS fully deleted for the gC gene, HSV-1ΔgC2-3, and its rescuant, HSV-1gC2-3R (26), were gifts from Curtis R. Brandt (University of Wisconsin, Madison). HSV-1 strain F fully deleted for the gE gene, HSV-1 F-gE/GFP (27), was provided by David C. Johnson, Oregon Health and Sciences University.
Antibodies.
Anti-HSV-1 gB mouse monoclonal antibodies H126 (domain I) (28, 29), H1359 (domain III) (30), and H1817 (domain VI) (13, 30) were purchased from Virusys, Taneytown, MD. Anti-gB mouse monoclonal antibodies SS10 (domain IV), SS55 (domain I) (31), and SS106 (domain V) (13) were gifts from Gary Cohen and Roselyn Eisenberg, University of Pennsylvania. Anti-gB monoclonal antibodies H1781 and H1838 (domain II) were provided by Lenore Pereira, University of California, San Francisco (13, 30). HSV polyclonal antibodies R68 (anti-gB) (6), R2 (anti-gD) (32), R137 (anti-gH), and R47 (anti-gC) (6) and anti-gD monoclonal antibody DL6 (33) were gifts from Gary Cohen and Roselyn Eisenberg. HSV-1 gE monoclonal antibody H1A054-100 was purchased from Virusys. Monoclonal antibody H1A021 to VP5 was purchased from Santa Cruz Biotechnology, Dallas, TX.
Plaque inhibition (neutralization) assay.
Antibodies against HSV gB, gD, or gH were prepared in dilutions ranging from 1:500 to 1:512,000 in culture medium containing FBS that had been inactivated for complement. Antibodies were in different formats: IgG (DL6, H126, H1359, H1817, SS10, SS55, and SS106), ascites fluid (H1781 and H1838), or rabbit serum (R2, R68, and R137). For consistency, the antibody concentration is reported as a dilution. HSV (100 PFU) was added to the antibody and incubated at 37°C for 1 h. The antibody-virus mixture was added to subconfluent Vero cells grown in 24-well plates. At 18 to 24 h postinfection, cells were fixed with methanol-acetone solution. Fixed cells were processed for an immunoperoxidase plaque assay using anti-HSV polyclonal antibody HR50, Pro A-horseradish peroxidase (HRP) secondary antibody, and 4-chloro-1-naphthol substrate for detection of plaques. An antibody is considered neutralizing when there is >50% reduction in plaque number (infectivity) at an antibody dilution of 1:500 or greater.
Western blotting.
HSV in Laemmli buffer was heated to 85°C for 5 min. Proteins were separated on 4-to-20% Tris-glycine gels (Invitrogen, Carlsbad, CA). Gels were transferred to a nitrocellulose membrane and then blocked with 5% milk in 0.2% PBS-Tween for 20 min. The membrane was probed with primary antibody to HSV-1 protein gB, gD, gH, gC, gE, or VP5 overnight. HRP-conjugated secondary antibody was added. Finally, enhanced chemiluminescence substrate (Thermo Fischer Scientific, Waltham, WA) was added, and the membrane was exposed to X-ray film (Genesee, San Diego, CA).
Determination of HSV genome copy number by quantitative real-time PCR.
HSV preparations were first treated with 2 μg/ml DNase (Bio-Rad, Hercules, CA) to remove any viral DNA that was not associated with viral particles. Viral genomic DNA was isolated using a QIAamp blood DNA kit (Qiagen, Valencia, CA). Quantitative PCR was performed using the CFX96 real-time PCR detection system (Bio-Rad). Primers were based on the HSV-1 KOS ICP22 sequence (forward, 5′-GAG TTT GGG GAG TTT G-3′; reverse, 5′-GGC AGG CGG TGG AGA A-3′) (34, 35). A standard curve for quantitative PCR (qPCR) was obtained from known quantities of a plasmid containing the ICP22 coding region.
Dot blot assay.
Serial dilutions of cell-free HSV virions were prepared in serum-free DMEM with 0.2% bovine serum albumin (BSA). Samples were blotted onto a nitrocellulose membrane using a Minifold dot blot system (Whatman) (36). Five percent milk in 0.2% PBS-Tween 20 blocking buffer was added, and the membrane was gently rocked for 20 min. Primary antibody prepared in blocking buffer was added to the membrane overnight. Secondary goat anti-mouse monoclonal HRP-conjugated secondary antibody was added, followed by enhanced chemiluminescent substrate (Thermo Fisher Scientific, Waltham, MA).
ACKNOWLEDGMENTS
We are grateful to Curtis Brandt, Gary Cohen, Roselyn Eisenberg, David Johnson, and Lenore Pereira for providing reagents.
This study was supported by National Institutes of Health (NIH) grant R01 AI119159 (A.V.N) and NIH training grant T32 GM008336 (T.K.S. and K.A.G.).
REFERENCES
- 1.Frink RJ, Eisenberg R, Cohen G, Wagner EK. 1983. Detailed analysis of the portion of the herpes simplex virus type 1 genome encoding glycoprotein C. J Virol 45:634–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu CT, Levine M, Homa F, Highlander SL, Glorioso JC. 1990. Characterization of the antigenic structure of herpes simplex virus type 1 glycoprotein C through DNA sequence analysis of monoclonal antibody-resistant mutants. J Virol 64:856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herold BC, WuDunn D, Soltys N, Spear PG. 1991. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol 65:1090–1098. doi: 10.1006/viro.1994.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WuDunn D, Spear PG. 1989. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol 63:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikloska Z, Cunningham AL. 1998. Herpes simplex virus type 1 glycoproteins gB, gC and gD are major targets for CD4 T-lymphocyte cytotoxicity in HLA-DR expressing human epidermal keratinocytes. J Gen Virol 79:353–361. doi: 10.1099/0022-1317-79-2-353. [DOI] [PubMed] [Google Scholar]
- 6.Eisenberg RJ, Ponce de Leon M, Friedman HM, Fries LF, Frank MM, Hastings JC, Cohen GH. 1987. Complement component C3b binds directly to purified glycoprotein C of herpes simplex virus types 1 and 2. Microb Pathog 3:423–435. doi: 10.1016/0882-4010(87)90012-x. [DOI] [PubMed] [Google Scholar]
- 7.Friedman HM, Cohen GH, Eisenberg RJ, Seidel CA, Cines DB. 1984. Glycoprotein C of herpes simplex virus 1 acts as a receptor for the C3b complement component on infected cells. Nature 309:633–635. doi: 10.1038/309633a0. [DOI] [PubMed] [Google Scholar]
- 8.Hook LM, Huang J, Jiang M, Hodinka R, Friedman HM. 2008. Blocking antibody access to neutralizing domains on glycoproteins involved in entry as a novel mechanism of immune evasion by herpes simplex virus type 1 glycoproteins C and E. J Virol 82:6935–6941. doi: 10.1128/JVI.02599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns TM, Huang ZY, Whitbeck JC, Ponce de Leon M, Lou H, Wald A, Krummenacher C, Eisenberg RJ, Cohen GH. 2014. Dissection of the antibody response against herpes simplex virus glycoproteins in naturally infected humans. J Virol 88:12612–12622. doi: 10.1128/JVI.01930-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mester JC, Twomey TA, Tepe ET, Bernstein DI. 1999. Immunity induced by DNA immunization with herpes simplex virus type 2 glycoproteins B and C. Vaccine 18:875–883. doi: 10.1016/s0264-410x(99)00325-4. [DOI] [PubMed] [Google Scholar]
- 11.Gupta PK, Saini M, Gupta LK, Rao VD, Bandyopadhyay SK, Butchaiah G, Garg GK, Garg SK. 2001. Induction of immune responses in cattle with a DNA vaccine encoding glycoprotein C of bovine herpesvirus-1. Vet Microbiol 78:293–305. doi: 10.1016/s0378-1135(00)00304-7. [DOI] [PubMed] [Google Scholar]
- 12.Adamiak B, Trybala E, Mardberg K, Johansson M, Liljeqvist JA, Olofsson S, Grabowska A, Bienkowska-Szewczyk K, Szewczyk B, Bergstrom T. 2010. Human antibodies to herpes simplex virus type 1 glycoprotein C are neutralizing and target the heparan sulfate-binding domain. Virology 400:197–206. doi: 10.1016/j.virol.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 13.Bender FC, Samanta M, Heldwein EE, de Leon MP, Bilman E, Lou H, Whitbeck JC, Eisenberg RJ, Cohen GH. 2007. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J Virol 81:3827–3841. doi: 10.1128/JVI.02710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dollery SJ, Delboy MG, Nicola AV. 2010. Low pH-induced conformational change in herpes simplex virus glycoprotein B. J Virol 84:3759–3766. doi: 10.1128/JVI.02573-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siekavizza-Robles CR, Dollery SJ, Nicola AV. 2010. Reversible conformational change in herpes simplex virus glycoprotein B with fusion-from-without activity is triggered by mildly acidic pH. Virol J 7:352. doi: 10.1186/1743-422X-7-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dollery SJ, Wright CC, Johnson DC, Nicola AV. 2011. Low-pH-dependent changes in the conformation and oligomeric state of the prefusion form of herpes simplex virus glycoprotein B are separable from fusion activity. J Virol 85:9964–9973. doi: 10.1128/JVI.05291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicola AV. 2016. Herpesvirus entry into host cells mediated by endosomal low pH. Traffic 17:965–975. doi: 10.1111/tra.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krummenacher C, Carfi A, Eisenberg RJ, Cohen GH. 2013. Entry of herpesviruses into cells: the enigma variations. Adv Exp Med Biol 790:178–195. doi: 10.1007/978-1-4614-7651-1_10. [DOI] [PubMed] [Google Scholar]
- 19.Weed DJ, Nicola AV. 2017. Herpes simplex virus membrane fusion. Adv Anat Embryol Cell Biol 223:29–47. doi: 10.1007/978-3-319-53168-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson DC, Frame MC, Ligas MW, Cross AM, Stow ND. 1988. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol 62:1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagashunmugam T, Lubinski J, Wang L, Goldstein LT, Weeks BS, Sundaresan P, Kang EH, Dubin G, Friedman HM. 1998. In vivo immune evasion mediated by the herpes simplex virus type 1 immunoglobulin G Fc receptor. J Virol 72:5351–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vu HL, Kwon B, Yoon KJ, Laegreid WW, Pattnaik AK, Osorio FA. 2011. Immune evasion of porcine reproductive and respiratory syndrome virus through glycan shielding involves both glycoprotein 5 as well as glycoprotein 3. J Virol 85:5555–5564. doi: 10.1128/JVI.00189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skehel JJ, Stevens DJ, Daniels RS, Douglas AR, Knossow M, Wilson IA, Wiley DC. 1984. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci U S A 81:1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 25.Aguilar HC, Matreyek KA, Filone CM, Hashimi ST, Levroney EL, Negrete OA, Bertolotti-Ciarlet A, Choi DY, McHardy I, Fulcher JA, Su SV, Wolf MC, Kohatsu L, Baum LG, Lee B. 2006. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol 80:4878–4889. doi: 10.1128/JVI.80.10.4878-4889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herold BC, Visalli RJ, Susmarski N, Brandt CR, Spear PG. 1994. Glycoprotein C-independent binding of herpes simplex virus to cells requires cell surface heparan sulphate and glycoprotein B. J Gen Virol 75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 27.Farnsworth A, Goldsmith K, Johnson DC. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J Virol 77:8481–8494. doi: 10.1128/jvi.77.15.8481-8494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kousoulas KG, Pellett PE, Pereira L, Roizman B. 1984. Mutations affecting conformation or sequence of neutralizing epitopes identified by reactivity of viable plaques segregate from syn and ts domains of HSV-1(F) gB gene. Virology 135:379–394. doi: 10.1016/0042-6822(84)90194-6. [DOI] [PubMed] [Google Scholar]
- 29.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 30.Pereira L, Ali M, Kousoulas K, Huo B, Banks T. 1989. Domain structure of herpes simplex virus 1 glycoprotein B: neutralizing epitopes map in regions of continuous and discontinuous residues. Virology 172:11–24. doi: 10.1016/0042-6822(89)90102-5. [DOI] [PubMed] [Google Scholar]
- 31.Bender FC, Whitbeck JC, Lou H, Cohen GH, Eisenberg RJ. 2005. Herpes simplex virus glycoprotein B binds to cell surfaces independently of heparan sulfate and blocks virus entry. J Virol 79:11588–11597. doi: 10.1128/JVI.79.18.11588-11597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isola VJ, Eisenberg RJ, Siebert GR, Heilman CJ, Wilcox WC, Cohen GH. 1989. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J Virol 63:2325–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisenberg RJ, Long D, Ponce de Leon M, Matthews JT, Spear PG, Gibson MG, Lasky LA, Berman P, Golub E, Cohen GH. 1985. Localization of epitopes of herpes simplex virus type 1 glycoprotein D. J Virol 53:634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker EB, Pritchard SM, Cunha CW, Aguilar HC, Nicola AV. 2015. Polyethylene glycol-mediated fusion of herpes simplex type 1 virions with the plasma membrane of cells that support endocytic entry. Virol J 12:190. doi: 10.1186/s12985-015-0423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komala Sari T, Pritchard SM, Cunha CW, Wudiri GA, Laws EI, Aguilar HC, Taus NS, Nicola AV. 2013. Contributions of herpes simplex virus 1 envelope proteins to entry by endocytosis. J Virol 87:13922–13926. doi: 10.1128/JVI.02500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sari TK, Gianopulos KA, Nicola AV. 2020. Conformational change in herpes simplex virus entry glycoproteins detected by dot blot. Methods Mol Biol 2060:319–326. doi: 10.1007/978-1-4939-9814-2_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper RS, Georgieva ER, Borbat PP, Freed JH, Heldwein EE. 2018. Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB. Nat Struct Mol Biol 25:416–424. doi: 10.1038/s41594-018-0060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kousoulas KG, Huo B, Pereira L. 1988. Antibody-resistant mutations in cross-reactive and type-specific epitopes of herpes simplex virus 1 glycoprotein B map in separate domains. Virology 166:423–431. doi: 10.1016/0042-6822(88)90513-2. [DOI] [PubMed] [Google Scholar]
- 39.Cairns TM, Fontana J, Huang ZY, Whitbeck JC, Atanasiu D, Rao S, Shelly SS, Lou H, Ponce de Leon M, Steven AC, Eisenberg RJ, Cohen GH. 2014. Mechanism of neutralization of herpes simplex virus by antibodies directed at the fusion domain of glycoprotein B. J Virol 88:2677–2689. doi: 10.1128/JVI.03200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]