In this study, we found that bat CD26s (bCD26s) from different species exhibit large diversities, especially in the region responsible for binding to the receptor binding domain (RBD) of Middle East respiratory syndrome coronavirus (MERS-CoV). However, they maintain the interaction with MERS-RBD at varied affinities and support the entry of pseudotyped MERS-CoV. These bat receptors polymorphisms seem to confer evolutionary pressure for the adaptation of CD26-binding virus, such as the ancestor of MERS-CoV, and led to the generation of diversified CD26-engaging CoV strains. Thus, our data add more evidence to support that bats are the reservoir of MERS-CoV and similar viruses, as well as further emphasize the necessity to survey MERS-CoV and other CoVs among bats.
KEYWORDS: interspecies transmission, MERS-RBD, evolution, bat CD26, receptor, MERS-CoV
ABSTRACT
Continued reports of Middle East respiratory syndrome coronavirus (MERS-CoV) infecting humans have occurred since the identification of this virus in 2012. MERS-CoV is prone to cause endemic disease in the Middle East, with several dozen spillover infections to other continents. It is hypothesized that MERS-CoV originated from bat coronaviruses and that dromedary camels are its natural reservoir. Although gene segments identical to MERS-CoV were sequenced from certain species of bats and one species experimentally shed the virus, it is still unknown whether other bats can transmit the virus. Here, at the molecular level, we found that all purified bat CD26s (bCD26s) from a diverse range of species interact with the receptor binding domain (RBD) of MERS-CoV, with equilibrium dissociation constant values ranging from several to hundreds at the micromolar level. Moreover, all bCD26s expressed in this study mediated the entry of pseudotyped MERS-CoV to receptor-expressing cells, indicating the broad potential engagement of bCD26s as MERS-CoV receptors. Further structural analysis indicated that in the bat receptor, compared to the human receptor, substitutions of key residues and their adjacent amino acids leads to decreased binding affinity to the MERS-RBD. These results add more evidence to the existing belief that bats are the original source of MERS-CoV and suggest that bCD26s in many species can mediate the entry of the virus, which has significant implications for the surveillance and control of MERS-CoV infection.
IMPORTANCE In this study, we found that bat CD26s (bCD26s) from different species exhibit large diversities, especially in the region responsible for binding to the receptor binding domain (RBD) of Middle East respiratory syndrome coronavirus (MERS-CoV). However, they maintain the interaction with MERS-RBD at varied affinities and support the entry of pseudotyped MERS-CoV. These bat receptors polymorphisms seem to confer evolutionary pressure for the adaptation of CD26-binding virus, such as the ancestor of MERS-CoV, and led to the generation of diversified CD26-engaging CoV strains. Thus, our data add more evidence to support that bats are the reservoir of MERS-CoV and similar viruses, as well as further emphasize the necessity to survey MERS-CoV and other CoVs among bats.
INTRODUCTION
Middle East respiratory syndrome coronavirus (MERS-CoV), the causative agent of Middle East respiratory syndrome (MERS), was first identified in 2012 (1). Infection by this virus leads to various symptoms, ranging from asymptomatic cases to severe pneumonia with acute respiratory distress syndrome (ARDS) and multiorgan failure, similar to the diseases caused by severe acute respiratory syndrome coronavirus (SARS-CoV) (2–4). In addition, MERS cases are still occurring and are prone to be an endemic disease in the Middle East. As of February 26, 2019, there were 2345 confirmed MERS cases, including 817 related deaths, with a fatality rate of approximately 35% (5).
MERS-CoV belongs to the family Coronaviridae, which are enveloped viruses with single-stranded positive-sense RNA genomes (6). Coronaviruses (CoVs) can be classified into four genera: alpha-, beta-, gamma-, and delta-CoVs (7). Beta-CoVs are further divided into four lineages (A, B, C, and D) (6). Both MERS-CoV and SARS-CoV are beta-CoVs but belong to different lineages. MERS-CoV is a lineage C virus, while SARS-CoV is a member of lineage B (8). SARS-CoV is hypothesized to have originated in bats and then to have infected humans either indirectly through an intermediate host (e.g., palm civets or raccoon dogs) (9) or without an intermediate host (10). Accumulating data suggest that dromedary camels are the primary reservoir host for MERS-CoV, and multiple independent transmissions seem to have occurred from dromedary camels to humans (11, 12). Recent studies also indicate the susceptibility of alpacas for natural MERS-CoV infection and suggest that alpacas may be another MERS-CoV animal reservoir (13, 14). In addition, gene fragments that are identical to MERS-CoV have also been identified in bats from both Saudi Arabia and Africa (15, 16).
Experimentally, bat CD26s (bCD26s) from a diverse range of bat species can mediate the entry of MERS-CoV (17). Cell lines from certain species of bats also support the replication of MERS-CoV (18). More importantly, Jamaican fruit bats (Artibeus jamaicensis) support the replication of MERS-CoV, with detectable shedding of the virus in the respiratory and intestinal tracts for up to 9 days (19). In contrast to domestic animals such as dromedary camels and alpacas, certain species of bats migrate (20). The characteristics of migration among bats emphasize their roles in transmitting infectious diseases, including CoV-related diseases. Therefore, studying the potential transmission of MERS-CoV among varied species of bats, which are widely distributed around the world, has significant implications for the surveillance and control of MERS-CoV infection.
Viral attachment to susceptible cells and related receptor engagement is the initial step of MERS-CoV infection. Spike (S) protein, integrated into the CoV envelope, can be cleaved into S1 and S2 subunits (21). The S1 subunit is responsible for receptor engagement. The S2 subunit possesses a typical structure of a type I fusion protein and mediates membrane fusion between the viral envelope and host cell membranes (4, 22, 23). For MERS-CoV, the C-terminal domain in S1, which is called the MERS receptor binding domain (MERS-RBD), interacts with the human CD26 (hCD26) receptor (also called DPP IV) (24). The ability of bCD26s from varied species to interact with MERS-RBD would be an indicator of the susceptibility of bats to MERS-CoV infection and, to some extent, suggests the potential of bats for transmitting MERS-CoV.
In this study, we found that bCD26s from many species display varied binding affinity to MERS-RBD. Their interactions supported the entry of pseudotyped MERS-CoV into receptor-expressing cells. Further, structural analysis of a MERS-RBD and Myotis davidii bCD26 complex indicated plasticity at the interaction interface between MERS-RBD and bCD26s, which will shed light on the coevolution between MERS-CoV and bat CD26s.
RESULTS
Interaction between MERS-RBD and bCD26s.
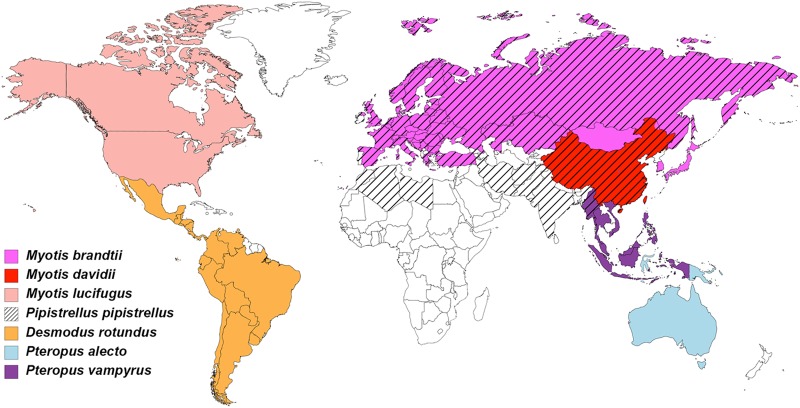
MERS-RBD and bCD26s were expressed in insect cells. bCD26s from seven species in three families were used in this study, namely, Pipistrellus pipistrellus (in Vespertilionidae), Myotis brandtii (in Vespertilionidae), Myotis davidii (in Vespertilionidae), Myotis lucifugus (in Vespertilionidae), Desmodus rotundus (in Phyllostomidae), Pteropus alecto (in Pteropodidae), and Pteropus vampyrus (in Pteropodidae). These bats live in habitats all over the world, except for Greenland (Fig. 1 and see Fig. S1 in the supplemental material).
FIG 1.
Geographic distribution of the bat species discussed in this study. Countries of occurrence for each of the seven bat species in this study were obtained from The International Union for Conservation of Nature and mapped using MapChart.
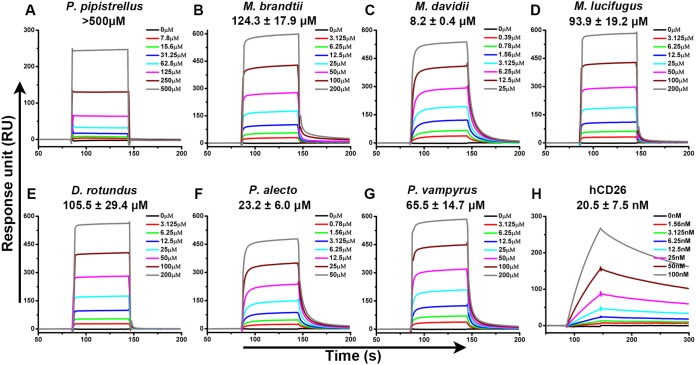
We first evaluated the binding between MERS-RBD and bCD26s by surface plasmon resonance (SPR). Purified MERS-RBD was immobilized on a CM5 chip, and bCD26s were flowed through the chip. As previously reported, MERS-RBD exhibited slow association/dissociation kinetics to interact with hCD26, with an equilibrium dissociation constant (KD) of 20.5 ± 7.5 nM (25). In contrast, the BIAcore binding profiles between MERS-RBD and all of the tested bCD26s displayed fast on and off rates. The calculated KD values varied, with micromolar concentrations ranging from several to hundreds (Fig. 2). In particular, M. davidii bCD26 displayed the highest binding affinity to MERS-RBD, with a KD of 8.2 ± 0.4 μM. Although P. pipistrellus bCD26 is reported to support MERS-CoV entry, the association between the MERS-RBD and P. pipistrellus bCD26 yielded the weakest KD (>500 μM). The interaction between MERS-RBD and the M. brandtii, M. lucifugus, D. rotundus, P. alecto, and P. vampyrus bCD26s were in the tens of micromolar concentration range (Fig. 2 and S2A).
FIG 2.
Specific interaction between MERS-RBD and different bCD26s characterized by SPR. MERS-RBD was immobilized on the chip and tested for binding to various concentrations of the indicated bCD26s or hCD26. The binding profiles are shown. (A) P. pipistrellus bCD26 binding to MERS-RBD. (B) M. brandtii bCD26 binding to MERS-RBD. (C) M. davidii bCD26 binding to MERS-RBD. (D) M. lucifugus bCD26 binding to MERS-RBD. (E) D. rotundus bCD26 binding to MERS-RBD. (F) P. alecto bCD26 binding to MERS-RBD. (G) P. vampyrus bCD26 binding to MERS-RBD. (H) hCD26 binding to MERS-RBD. In each subplot, the concentration of the indicated CD26 used for binding evaluation is listed in the inserted box. The KD were calculated using BIAevaluation software 4.1 and the values are displayed in each subplot. KD values are shown as means ± the standard errors of the mean (SEM) of three independent experiments. The curves are representative of three independent experiments and were generated using Origin8 software.
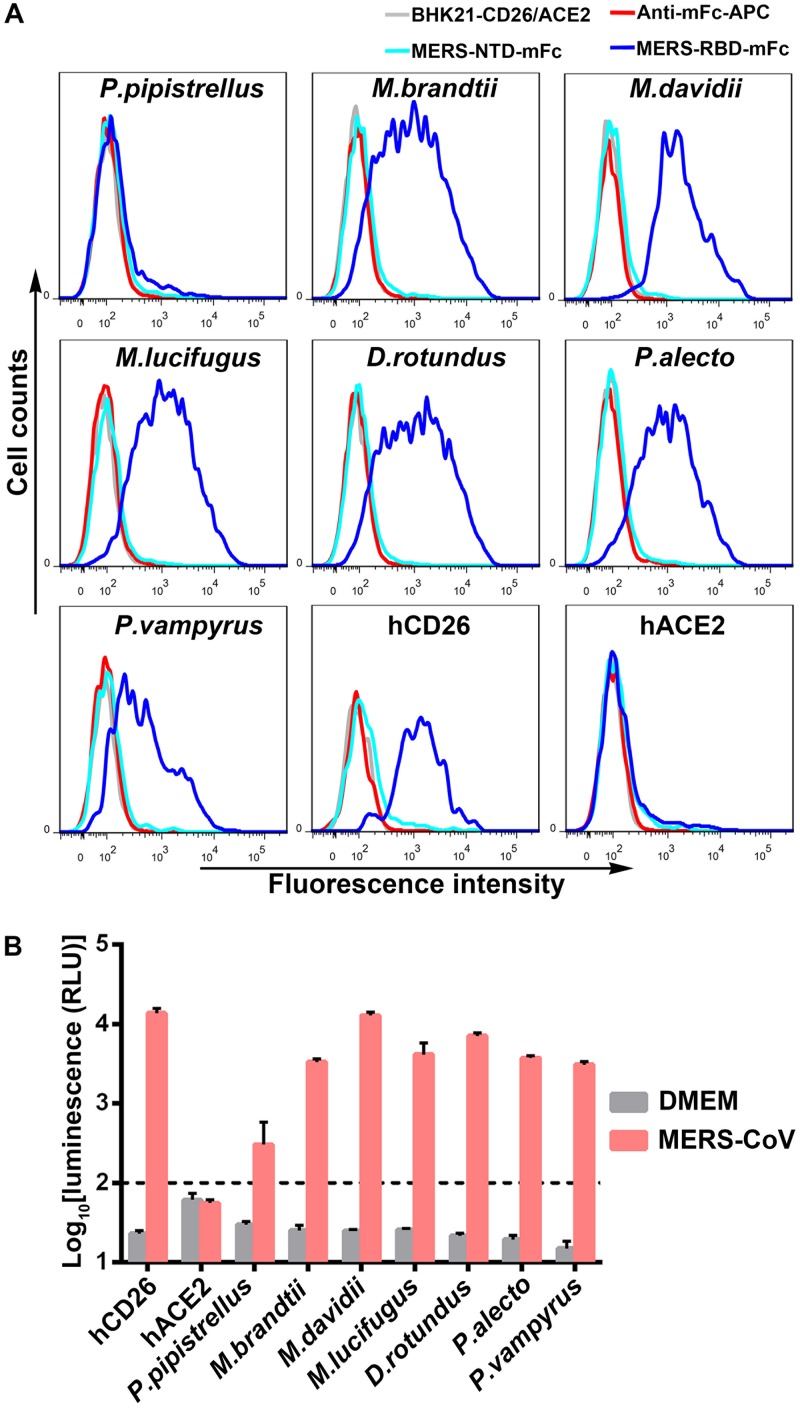
We also applied fluorescence-activated cell sorting (FACS) to test the interaction between MERS-RBD and bCD26s. Each CD26, fused to enhanced green fluorescent protein (eGFP), was transiently expressed in BHK21 cells. MERS-RBD tagged with mouse Fc (MERS-RBD-mFc) was utilized to stain the cells (26). As shown in Fig. 3A, MERS-RBD led to a fluorescence shift in hCD26-expressing cells but not in ACE2-expressing cells, which is the receptor of SARS-CoV and was used as a negative control. Consistently, cells expressing bCD26s displayed shifts to various degrees, except for P. pipistrellus bCD26. No obvious interaction between P. pipistrellus bCD26 and MERS-RBD was detected by FACS. Similarly, no bCD26s interacted with the N-terminal domain of S1 (MERS-NTD) (Fig. 3A).
FIG 3.
Evaluation of MERS-RBD binding to bCD26s by flow cytometry and the ability of bCD26s to support the entry of pseudotyped MERS-CoV. (A) BHK21 cells transiently expressing the indicated protein, which are marked above the boxes, were stained with MERS-RBD-mFc (cyan line) or MERS-NTD-mFc (blue line). In each subplot, the gray line indicates cells without staining. The red line represents cells incubated with the secondary antibody (anti-mFc/APC). The data were collected using a BD FACSCanto and analyzed by FlowJo 7.6. The data are representative of two independent experiments. (B) Infection by lentiviral particles pseudotyped with MERS-CoV S protein. BHK21 cells, which transiently expressed the indicated proteins, were sorted based on the eGFP expression and seeded overnight. The cells were then infected with the pseudotyped MERS-CoV for 5 h. After an additional 48 h of culture, the luciferase activity was determined using a GloMax 96 microplate luminometer (Promega). DMEM without pseudotyped MERS-CoV was used as a negative control. The luminescence value for BHK21 cells with the indicated gene expression was analyzed and transformed into log scale using Prism 6. The values in each column represent the means ± the standard deviations of three replicates. The data displayed are representative of two independent experiments.
The interaction between MERS-RBD and bCD26s initiates virus entry.
After confirmation of the interaction between MERS-RBD and bCD26s, we evaluated whether this interaction supported viral entry by using pseudotyped MERS-CoV. BHK21 cells were transiently transfected with plasmids encoding bCD26s that were fused to eGFP. To enrich transfected cells, eGFP-positive cells were first sorted and then infected with the pseudotyped MERS-CoV. As indicated in Fig. 3B, all tested bCD26s supported the entry of pseudotyped MERS-CoV. Notably, although P. pipistrellus bCD26 displayed too weak an interaction with MERS-RBD to be detected by FACS, P. pipistrellus bCD26 still supported MERS-CoV entry (Fig. 3B), consistent with previous results (17, 24). Therefore, all bCD26s tested bound to MERS-RBD and mediated MERS-CoV entry, despite their different binding affinities.
Molecular basis for the interaction between MERS-RBD and bCD26s.
To elucidate the molecular basis of the interaction between bCD26 and MERS-CoV, dimeric M. davidii bCD26, which exhibited the highest binding affinity among the tested species to MERS-RBD, was purified and mixed with MERS-RBD at a molar ratio of 1:2 to allow for the formation of heterocomplexes. The X-ray diffraction data for the complex crystals were collected, and the complex structure was determined at a resolution of 3.1 Å (Table 1).
TABLE 1.
Crystallographic data collection and refinement statistics for M. davidii bCD26 and MERS-RBD
| Parameter | Value(s) for M. davidii bCD26 and MERS-RBDa |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 114.66, 273.67, 115.23 |
| α, β, γ (°) | 90.00, 119.68, 90.00 |
| Resolution range (Å) | 50–3.10 (3.21–3.10) |
| No. of unique reflections | 110,996 (11,097) |
| Rmergeb | 0.193 (1.015) |
| Rpimc | 0.087 (0.461) |
| CC1/2 | 0.9832 (0.741) |
| I/σI | 8.463 (1.519) |
| Completeness (%) | 99.80 (100.00) |
| Redundancy | 5.7 (5.7) |
| Refinement statistics | |
| Resolution (Å) | 49.81–3.1 |
| No. of reflections | 95,269 |
| Rwork/Rfreed | 0.1994/0.2416 |
| No. of atoms | |
| Protein | 30,730 |
| Water | 0 |
| B-factors (Å2) | |
| Protein | 63.1 |
| Water | |
| RMSD | |
| Bond length (Å) | 0.003 |
| Bond angle (°) | 0.651 |
| Ramachandran plot (%)e | |
| Favored | 94.94 |
| Allowed | 5.04 |
| Outliers | 0.03 |
Where applicable, values for the outmost-resolution shell are given in parentheses.
Rmerge = ΣiΣhkl|Ii|/ΣiΣhklIi, where Ii is the observed intensity and is the average intensity from multiple measurements.
Rpim = Σhkl[1/(N − 1)]1/2 Σi|Ii−<I>|/ΣhklΣiIi, where Ii is the observed intensity and <I> is the average intensity from multiple measurements.
Rwork = ΣǁFoǁFcǁ/Σ|Fo|, where Fo and Fc are the structure-factor amplitudes from the data and the model, respectively. Rfree is the R factor for a subset (5%) of reflections that was selected prior to refinement calculations and was not included in the refinement.
Ramachandran plots were generated by using the program MolProbity.
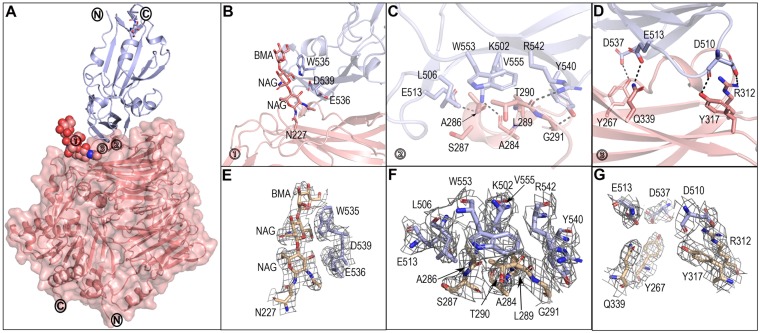
Four pairs of MERS-RBD and monomeric M. davidii bCD26 with a 1:1 binding mode were observed in one crystallographic asymmetric unit. Among the four complexes, the pair of chains A and B displays the best electron density map, which was used for further analysis. The other three complexes exhibit the same structures as the pair of chains A and B, with a root mean square deviation (RMSD) of 0.104 (for 877 Cα atoms) for the pair of chains C and D, 0.205 (for 844 Cα atoms) for the pair of chains E and F, and 0.107 (for 859 Cα atoms) for the pair of chains G and H. In the complex structure, M. davidii bCD26 contains 724 consecutive density-traceable residues, spanning from R38 to P761. In addition, glycosylation modifications were observed at N83, N90, N217, N227, N316, and N491. Homologous to hCD26, M. davidii bCD26 also folded into two structural domains: an α/β hydrolase domain and an eight-bladed β-propeller domain (Fig. 4A). For MERS-RBD, clear electron densities could be traced for 208 residues from V381 to L588. As observed in previous studies, MERS-RBD comprises a core subdomain and an external subdomain that interacts with blades IV and V in M. davidii bCD26 (Fig. 4A).
FIG 4.
Complex structure of MERS-RBD bound to M. davidii bCD26. (A) Overall structure. One 1:1 complex of MERS-RBD and M. davidii bCD26 is shown as a cartoon representation. bCD26 is also displayed in surface representation with 50% transparency. MERS-RBD is labeled in light blue. M. davidii bCD26 is labeled in salmon. The protein N and C termini are marked. The important contact sites are marked with boxed numbers 1 to 3 and are further delineated in panels B to D for interaction details. (B) Interactions contributed by a carbohydrate moiety linked to M. davidii bCD26 N227. The residues involved and the carbohydrates referred to are shown and labeled. (C) Hydrophobic interactions and hydrophilic interactions conferred by the M. davidii bCD26 α3 helix are displayed. (D) H-bonds between M. davidii bCD26 and MERS-RBD. (E to G) Representative electron density maps at the key regions for structures, related to panels B, C, and D, respectively. The density maps are drawn in gray mesh contoured at 1 sigma. The secondary structure elements are specified by ESPript and labeled. See also Table S1 in the supplemental material. All structures were analyzed and depicted by PyMOL.
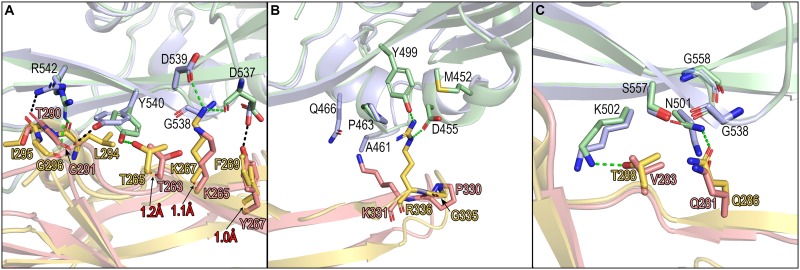
Scrutiny of the buried surface indicated that the glycosylated N227 residue of M. davidii bCD26 binds to MERS-RBD residues W535, E536, and D539 (Fig. 4B and E and Table S1). Consecutive residues 283VAPASVLTG291 in M. davidii bCD26, which fold into the α3 helix and the preceding and extending loops around the α3 helix, are located in the center of the interaction interface. MERS-RBD primarily uses L506, W553, and V555 to interact with M. davidii bCD26 at this region, forming a hydrophobic patch (Fig. 4C and F). Hydrophilic interactions between the two molecules surround this hydrophobic patch. Notably, the potential H-bonds (3.3-Å resolution cutoff) are exclusively mediated by main-chain oxygen or nitrogen in M. davidii bCD26 with the side chains of MERS-RBD residues (including M. davidii bCD26 A284 with MERS-RBD K502, A286 with E513, T290 with R542, and G291 with Y540) (Fig. 4C and F and Table S1). Adjacent to the central region, other potential H-bonds are formed between M. davidii bCD26 Y317 and R312 with MERS-RBD D510, Q339 with E513, and Y267 with D537 (Fig. 4D and G and Table S1).
Variant amino acid interaction details with MERS-RBD between hCD26 and M. davidii bCD26.
Overall, the complex structure of M. davidii bCD26 and MERS-RBD solved in this study resembles the previously reported complexed structure of hCD26 and MERS-RBD (25), suggesting an overall similar recognition mode for MERS-RBD between M. davidii bCD26 and hCD26. However, MERS-RBD exhibits more adaption to interact with hCD26 than with M. davidii bCD26, as indicated by the significantly different binding affinities (20.5 ± 7.5 nM versus 8.2 ± 0.4 μM) (Fig. 2). Consistently, the surface area that is buried in the complex of MERS-RBD/M. davidii bCD26 is 977.7 Å2 for MERS-RBD and 1,040 Å2 for M. davidii bCD26. However, the counterparts in the complex of MERS-RBD/hCD26 are 1,113.4 and 1,203.4 Å2. Moreover, the potential H-bonds between MERS-RBD and M. davidii bCD26 (eight pairs in total) are fewer than in the complex of MERS-RBD and hCD26 (14 pairs in total), as are the van der Waals (vdw) contacts (4.5-Å resolution cutoff) (Table S1).
Further comparison of the interaction details between the two complexes indicated that most residues in M. davidii bCD26 contribute similar contacts as their hCD26 counterparts (Table S1). However, T290 in M. davidii bCD26 substitutes for hCD26 I295, which forms a hydrophobic patch with MERS-RBD L506, W553, and V555 (25). Likely due to the rejection conferred by the hydrophobic MERS-RBD residues, the interaction panel of T290 and its adjacent residues with MERS-RBD shifts, leading to decreased potential H-bonds compared to the human receptor (Fig. 5A and Table S1). In agreement, mimicking the interaction mode between the human receptor and viral ligand by introducing the T290I mutation to M. davidii bCD26 led to a stronger binding affinity for MERS-RBD, with a decrease of >50-fold in the KD compared to the wild type (0.12 ± 0.02 μM versus 8.2 ± 0.4 μM) (Fig. S2B).
FIG 5.
Comparison of the MERS-RBD/hCD26 and MERS-RBD/M. davidii bCD26 pairs for their interaction details. The variations in the MERS-RBD binding interface of M. davidii bCD26 and hCD26 are delineated in panels A to C. MERS-RBD complexed with M. davidii bCD26 is colored in light blue. MERS-RBD complexed with hCD26 is colored in pale green. M. davidii bCD26 is marked in salmon, and hCD26 is marked in yellow-orange. The potential H-bonds between M. davidii bCD26 and MERS-RBD are indicated by black dashed lines. The potential H-bonds between hCD26 and MERS-RBD are indicated by green dashed lines. The red numbers represent the shifted distances of the indicated elements in M. davidii bCD26 compared to hCD26. The structures were analyzed and depicted by PyMOL.
Another evident difference between the two complexes is located in the region distal from CD26 α3. Fewer vdw contacts with MERS-RBD are provided by K331 in M. davidii bCD26 compared to its human equivalent residue of R336 (six in M. davidii bCD26 versus 28 in hCD26) (Fig. 5B). However, the mutation of K331R in M. davidii bCD26 did not strengthen its binding (Fig. S2C), which is likely attributable to the substitution of the most flexible residue G335 in hCD26 with the most rigid residue P330 in M. davidii bCD26, thereby limiting the conformational flexibility of the extending K331. Consistently, although making no contacts with MERS-RBD, the P330G mutation slightly increased the M. davidii bCD26 binding affinity (3.84 ± 0.26 μM for P330G versus 8.2 ± 0.4 μM for the wild type) (Fig. S2D). Furthermore, the combination of K331R with P330G in M. davidii bCD26 resulted in a KD almost 40-fold lower than for the wild type (0.21 ± 0.03 μM for P330G-K331R versus 8.2 ± 0.4 μM for the wild type) (Fig. S2E).
Myotis davidii bCD26 also possess residues that are slightly better adapted for the viral ligand than that in hCD26. For example, mutating V283 to its human counterpart of T288 in M. davidii bCD26 weakened its interaction with MERS-RBD (58.5 ± 6.09 μM for V283T versus 8.2 ± 0.4 μM for the wild type) (Fig. 5C and S2F). Similarly, substituting hCD26 T288 with V288 enhanced its binding to the viral ligand (0.87 ± 0.32 nM for hCD26 T288V versus 20.5 ± 7.5 nM for hCD26) (Fig. S2G).
Effects of glycosylation on the interaction between bCD26s and MERS-RBD.
In addition to the amino acids, glycosylation around the residue equivalent to hCD26 R336 also affects the interactions with MERS-RBD (27, 28). hCD26 possesses no N-linked glycan around R336. In contrast, bCD26s exhibit large diversities (Fig. S3). Consistent to the structural information, M. davidii bCD26 mutant containing N326A (marked MD-gly) exhibited a shift similar to that of M. davidii bCD26, as visualized with Western blotting, suggesting no glycosylation modification at this residue. Although mutants without the indicated potential N-linked glycosylation of P. vampyrus, P. pipistrellus, M. brandtii, and M. lucifugus bCD26s (Materials and Methods) displayed slightly faster SDS-PAGE migration compared to their wild types (Fig. 6A), indicating that these sites are modified by glycans. Two of P. alecto bCD26 mutants (N330A or S332R) seem to migrate similarly to P. alecto bCD26 (Fig. 6A).
FIG 6.
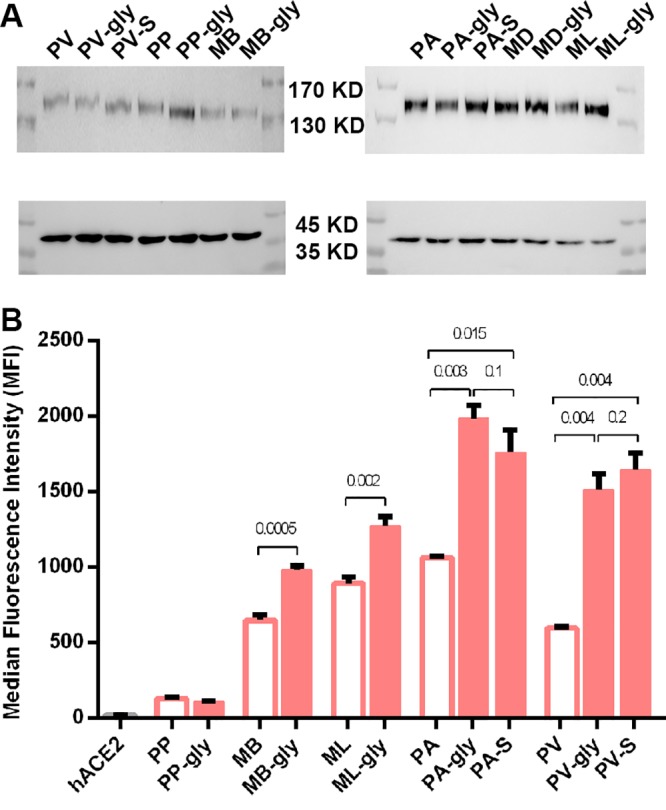
Evaluation of the interaction between MERS-RBD and bCD26s mutants by flow cytometry. (A) Evaluation of the glycosylation at indicated residues by Western blotting. bCD26s containing the indicated mutations were fused to eGFP and transiently expressed on the surfaces of 239T cells. The cells were then collected and subjected to Western blotting with anti-GFP antibody or anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase), which is used as a control. The upper blots represent CD26 and mutants, and the lower blots indicate GAPDH. (B) Transiently transfected cells were collected and sequentially stained with MERS-RBD-mFc and anti-mIgG/APC. The value for each column indicates the average of the MFI of triplicates, and the bar represents the SEM. The data are representative of two independent experiments. The empty column represents the wild type, and the filled one indicates the mutants deleting specific glycosylation (see Materials and Methods). hACE2 is a negative control. Numbers above the column are the P values determined using a Student t test.
We then used MERS-RBD tagged with mFc to stain the cells expressing different bCD26s or their mutants. The median fluorescence intensity (MFI) was collected and used as an indicator of binding affinity between MERS-RBD and different bat receptors. PP-gly did not exhibit obvious interaction with MERS-RBD-mFc, similar to what was observed in its wild type. However, other mutants without potential N-linked glycans around the human R336 equivalent displayed increased binding to MERS-RBD (Fig. 6B and S3), implying the glycans at the indicated sites, if there are any, likely provide steric clashes for the interaction with the viral ligand (Fig. 7).
FIG 7.
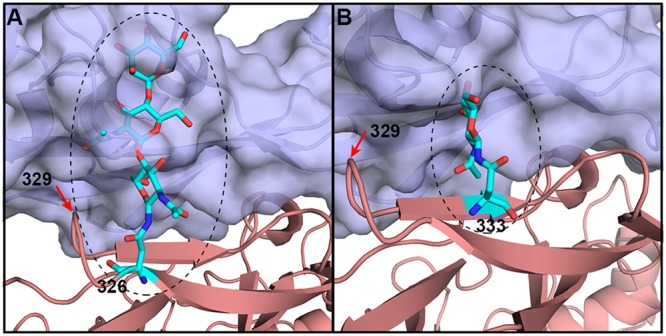
Probable conformations of glycans linked to the indicated residues. bCD26s exhibit multiple glycosylation patterns at the receptor-ligand interface. Based on the structure of M. davidii bCD26 in complex with MERS-RBD, the possible positions and conformations of residue 326- and 333-linked glycans (numbering in M. davidii bCD26) are displayed. (A) Possible glycans on residue 326; (B) possible glycans on residue 333. Certain bCD26s (e.g., P. alecto ) and P. vampyrus) contain glycans linked to residue 329, which is highlighted with the red arrow in A and B.
Collectively, the substitutions of the key residues in the interacting interface of M. davidii bCD26, such as T290 (I295 in hCD26) and K331 (R336 in hCD26), together with the surrounding residues, such as P330 (G335 in hCD26), account for the different binding affinities between MERS-RBD with M. davidii bCD26 and hCD26.
DISCUSSION
Bats have been implicated as the largest reservoir of alpha- and beta-CoVs (7). The bat CoVs HKU4 and HKU5 display the closest phylogenetic relationship with MERS-CoV. Indeed, HKU4 can use the same receptor as MERS-CoV, hCD26, to enter cells (26), indicating the origin of MERS-CoV from bat CoVs. Although no live MERS-CoVs have been isolated from bats to date, gene segments identical to this virus have been sequenced from bats distributed in both Saudi Arabia (Taphozous perforatus) and South Africa (Neoromicia zuluensis) (15, 16). In addition, Jamaican fruit bats support experimental MERS-CoV infection (19). The data in the present study, together with previously published data, demonstrate that bCD26s from a diverse range of species support the entry of both pseudotyped and wild-typed MERS-CoV into cells (17). Therefore, bats may still be infected and transmit the virus even if they are not a direct natural host of MERS-CoV, such as dromedary camels.
In our previous work, we delineated the binding mechanism between MERS-RBD and hCD26, and the key residues responsible for the interaction were determined. Interestingly, among the purified bCD26s, M. davidii bCD26 displayed the least homology to hCD26 in the blade IV and V regions, which are the regions for binding to MERS-RBD. However, M. davidii bCD26 displayed the highest binding affinity to MERS-RBD among the tested bat CD26s. Further structural analysis indicated a similar binding mode of MERS-RBD with hCD26 and M. davidii bCD26, emphasizing the possible origin of MERS-CoV from bats. Similar to hCD26, the glycosylation of the residue equivalent to N229 in hCD26 and the hydrophobic core conferred by the α3 helix establish the interaction with MERS-RBD. In terms of the 13 key residues in the human receptor responsible for the ligand interaction, there are three substitutions in M. davidii bCD26, namely, V283 versus T288, T290 versus I295, and K331 versus R336. Notably, only the substitution of T290 with its counterpart I295 in hCD26 increased the binding affinity to the ligand. The side chain of T290 in bCD26 is avoided by the hydrophobic interface and led to a shift in H-bond pairs. In contrast, neither V283T nor K331R increased the bat receptor binding to MERS-RBD. Further scrutiny of the binding interface uncovered that the rigid P330 N-terminal to K331 in bat receptor, instead of the flexible G335 in the human receptor, limits the orientation of the extended lysine residue and decreases the interaction. The P330G and K331R substitutions would essentially increase the interaction of M. davidii bCD26 with the viral ligand. This structurally detailed information indicates the varied adaptation of the viral ligand to the bat and human receptors.
Compared to the human receptor, bCD26s blades IV and V, which are responsible for binding to MERS-RBD, exhibit less homology to human equivalents than the full proteins (76.64 to 83.94% for blades IV and V versus 82.37 to 88.85% for full proteins) (Fig. S1). The diversity of CD26s among bat species is even higher, with homologies ranging from 73.72 to 99.27% for blades IV and V and 82.26 to 99.87% for full-length CD26s. In addition, around the hCD26 R336 equivalent, bCD26s possess diversified glycosylation patterns, which are present in multiple permissive and nonpermissive CD26 orthologues.
The glycosylation of the receptors also affects their binding to CoVs. It has been reported that N328 of mCD26 is glycosylated and contributes to the inability of mCD26 to interact with MERS-CoV to some extent (28). Both P. vampyrus and P. alecto bCD26s possess a potential N-linked glycosylation at the residue equivalent to mCD26 N328. P. vampyrus bCD26 mutants containing either N331A or S333R exhibited smaller bands than the wild type when tested by Western blotting. In addition, both P. vampyrus bCD26 mutants significantly enhanced the interaction with MERS-RBD, implying that P. vampyrus bCD26 contains N331-linked glycans, which likely hinder the interaction with MERS-RBD. Similarly, both P. alecto bCD26 mutants (N330A or S332R) displayed significantly higher interaction with the viral ligand than the wild type, as determined by FACS. However, the difference in migration between P. alecto bCD26 and the two mutants was not obvious, which is probably due to the short glycans modified at this site, if there are any. Carbohydrates at residue N327/N326 of M. lucifugus and M. brandtii bCD26s likely decreased the association with MERS-RBD, too. Notably, substitutions in the protein residues might also contribute to the change of binding affinity to MERS-RBD, through direct or indirect impact on the overall structures.
A recent report indicates that MERS-CoV can adapt to a semipermissive bCD26 (Desmodus rotundus), acquiring adaptive mutations in five passages (17). Thus, the diversity in bCD26s preserves the interaction with the viral ligand but conferred evolutionary pressure on the ancestor of MERS-CoV or other CD26-binding CoVs, such as HKU4 (26). This pressure likely drives the generation of CoVs with diversified RBDs, thereby forming the reservoir of CoVs and facilitating cross-species transmission.
MATERIALS AND METHODS
Gene construction.
The plasmids used for protein expression and purification were separately constructed by insertion of the coding sequences of MERS-RBD from MERS-CoV strain EMC/2012 (residues E367 to Y606, accession number JX869059), hCD26 (residues S39 to P766, accession number NP_001926), P. pipistrellus CD26 (residues T36 to P760, accession numbers AGF80256.1/KC249974), M. brandtii CD26 (residues G37 to P761, accession number XP_005859434.1), M. davidii CD26 (residues D37 to P761, accession number XP_006766553.1), M. lucifugus CD26 (residues G37 to P761, accession number XP_014301472.1), D. rotundus CD26 (residues S38 to P765, accession number JAA48979.1), P. alecto CD26 (residues S38 to P762, accession number XP_006921185.1), or P. vampyrus CD26 (residues S39 to P763, accession number XP_011356851.1) into the baculovirus transfer vector pFastbac1 (Invitrogen) at the EcoRI and XhoI restriction sites. Mutants were also prepared for M. davidii bCD26, including T295I, K331R, P330G, P330G-K331R, and V283T. hCD26 containing T288V was also constructed. All proteins contained an N-terminal gp67 signal peptide and a C-terminal 6×His tag.
The pEGFP-C1-CD26 plasmids were constructed by cloning the coding region of specific bCD26s or hCD26 into pEGFP-C1 using restriction enzymes XhoI and SmaI. Thus, the proteins were fused to eGFP and expressed on the cell membrane. Similarly, the human ACE2 protein was fused to eGFP by cloning the coding region into pEGFP-N1. bCD26 mutants used to delete specific N-linked glycans were prepared based on the pEGFP-C1-CD26 plasmids. These included N332A in P. pipistrellus bCD26 (PP-gly), N326A in M. brandti bCD26 (ML-gly), N326A in M. davidii bCD26 (MD-gly), N327A in M. lucifugus bCD26 (ML-gly), N331A or S333R in Pteropus vampyrus bCD26 (PV-gly or PV-S), and N330A or S332R in P. alecto bCD26 (PA-gly or PA-S).
Recombinant proteins MERS-RBD-mFc and MERS-NTD-mFc were used in FACS assays. The coding sequences of MERS-RBD (E367-Y606) and MERS-NTD (M1-S353) tagged with the Fc domain of mouse IgG were individually cloned into the pCAGGS expression vector using the EcoRI and XhoI restriction sites. The signal peptide of MERS-CoV S protein (M1-S17) was used for the secretion of both MERS-NTD-mFc and MERS-RBD-mFc.
The full-length coding region of MERS-CoV S protein with a C-terminal FLAG tag was cloned into the pCAGGS vector using the EcoRI and SmaI restriction sites (pCAGGS-MERS-S-flag). The expression of MERS-CoV S protein was tested by Western blotting, and this plasmid was used to package pseudotyped MERS-CoV.
Protein expression and purification.
The Bac-to-Bac baculovirus expression system (Invitrogen) was used to express proteins for crystallization and SPR analysis. The constructed pFastbac1 vectors were transformed into DH10Bac competent cells to generate recombinant bacmids. Transfection of bacmids and virus amplification were conducted in Sf9 cells, while Hi5 cells were used to express proteins. The supernatants of Hi5 cells were collected at 48 h postinfection, and soluble proteins were purified by metal affinity chromatography using a HisTrap HP 5-ml column (GE Healthcare). The samples were then pooled and further purified via gel filtration chromatography with a Superdex 200 column (GE Healthcare) in a buffer composed of 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl.
The mFc recombinant proteins were expressed in HEK293T cells. Plasmids were transiently transfected into cells. After 4 days of expression, supernatants were collected, centrifuged, and mixed with the same volume of binding buffer containing 20 mM Na3PO4 (pH 7.0). The mixtures were then filtered through 0.22-μm-pore-size membranes and flowed through a HiTrap rProtein A FF (GE Healthcare) affinity chromatography column at a maximum flow rate of 1 ml/min. The bound protein was eluted with 0.1 M glycine-HCl (pH 3.0) and collected into tubes containing 200 μl of 1 M Tris-HCl (pH 9.0). mFc fusion proteins were further purified by gel filtration in phosphate-buffered saline (PBS), concentrated, and stored at –80°C.
Sequences used in the alignments.
The GenBank accession numbers of the sequences used for analyzing the conservation of epitopes among bCD26s were as follows: Homo sapiens, NP_001926.2; camel, XP_006176870.1; mouse, NP_034204.1; ferret, ABC72084.1; hamster, NP_001297500.1; guinea pig, XP_012997849.1; Rousettus aegyptiacus, XP_016001925.1; Epomops buettikoferi, AXB27023.1; Pteropus vampyrus, XP_011356851.1; Pteropus alecto, XP_006921185.1; Rhinolophus ferrumequinum, AXB27025.1; Rhinolophus sinicus, XP_019582852.1; Hipposideros armiger, XP_019492386.1; Saccopteryx bilineata, AXB27027.1; Artibeus planirostris, AXB27021.1; Artibeus jamaicensis, AIG55258.1; Carollia perspicillata, AXB27022.1; Desmodus rotundus, JAA48979.1; Miniopterus natalensis, XP_016052462.1; Eptesicus fuscus, XP_008136991.1; Pipistrellus pipistrellus, AGF80256.1; Myotis lucifugus, XP_014301472.1; Myotis davidii, XP_006766553.1; and Myotis brandtii, XP_005859434.1.
Crystallization, data collection, and structure determination.
For protein crystallization, monomeric MERS-RBD was mixed with M. davidii bCD26 at a 1:1 stoichiometry and crystallized by the sitting-drop vapor diffusion method at 18°C with 1 μl of protein solution mixed with 1 μl of reservoir buffer. Diffractable crystals were obtained in 0.2 M imidazole malate (pH 8.5) and 7.5% (wt/vol) PEG 10000. Crystals were cryoprotected in 20% (vol/vol) glycerol in reservoir solution. The diffraction data were collected at the Shanghai Synchrotron Radiation Facility beamline BL19U1. All data sets were processed with HKL2000 software. The complex structure was solved with Phaser (29) by the molecular replacement method using the structure of MERS-RBD/hCD26 as the search model. Restrained rigid-body refinement with REFMAC5 (CPP4 suite) (30) and manual model building with COOT (31) were then performed. Further rounds of refinement were done with Phenix.refine (32), and then the program PROCHECK was used to monitor the stereochemistry of the final structure. Data collection, processing, and refinement statistics are summarized in Table 1. The native data set was collected at 0.979 Å. All structural images were made using PyMOL.
SPR analysis.
Protein interaction was tested by SPR analysis at room temperature (25°C) using a BIAcore 3000. All proteins used for this assay were in buffer containing 10 mM HEPES (pH 7.4), 150 mM NaCl, and 0.005% (vol/vol) Tween 20. MERS-RBD was immobilized onto CM5 chips at approximately 1,000 response units and analyzed for real-time binding by flowing through various concentrations of CD26s. The concentrations of hCD26 were 0, 1.56, 3.125, 6.25, 12.5, 25, 50, 100, and 200 nM. The concentrations of P. pipistrellus bCD26 were 0, 7.8, 15.6, 31.2, 62.5, 125, 250, and 500 μM. The M. davidii bCD26 concentrations were 0, 0.39, 0.78, 1.56, 3.125, 6.25, 12.5, 25, 50, 100, and 200 μM. The other bCD26s were used at 0, 0.78, 1.56, 3.125, 6.25, 12.5, 25, 50, 100, and 200 μM. M. davidii bCD26 V283T was analyzed at 0, 1.56, 3.125, 6.25, 12.5, 25, 50, 100, and 200 μM. M. davidii bCD26 P330G and K331R were used at 0, 0.078, 0.156, 0.3125, 0.625, 1.25, 2.5, 5, 10, 20, and 40 μM. M. davidii bCD26 T290I and P330G-K331R were analyzed at 0, 0.00975, 0.0195, 0.039, 0.078, 0.156, 0.3125, 0.625, 1.25, 2.5, and 5 μM. hCD26 T288V was 0, 0.39, 0.78, 1.56, 3.125, 6.25, 12.5, 25, 50, 100, and 200 nM. After each reaction, the CM5 chip was regenerated using 7 μl of 10 mM NaOH. The binding kinetics were analyzed with BIAevaluation v4.1 using Langmuir binding and steady-state affinity models.
FACS.
For binding tests, the pEGFP-C1 vectors containing the indicated CD26s were transfected into nonsusceptible BHK21 cells using PEI (Sigma) according to the manufacturer’s instructions. The cells were collected 48 h after transfection, suspended in PBS (with 0.5% fetal bovine serum [FBS]), and incubated with individual mouse Fc-fusion proteins at 37°C for 30 min, followed by washing with PBS twice and further incubation with an anti-mouse IgG/APC antibodies. After washing, the cells were analyzed using a BD FACSCalibur. The cells incubated with only the secondary antibody were used as negative controls.
Preparation and infection of pseudotyped MERS-CoV.
The plasmid pNL4-3.luc.R-E- was used to package the pseudotyped MERS-CoV. The pCAGGS-MERS-S-flag and pNL4-3.luc.R-E- plasmids were cotransfected into HEK293T cells at a molar ratio of 2:1. After 48 h, supernatants containing the pseudotyped MERS-CoV were harvested, divided, and stored at –80°C.
For pseudovirus infection assays, the pEGFP-C1-CD26 plasmid was transfected into BHK21 cells. Positive cells, expressing eGFP, were sorted by FACS at 48 h posttransfection and then reseeded into plates overnight. After two washes with PBS, the cells were first incubated with pseudotyped MERS-CoV for 5 h and then cultured with Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS and 0.1% PS (penicillin and streptomycin) for another 48 h. The luciferase activity was determined using a GloMax 96 microplate luminometer (Promega).
Western blotting.
Cells were transfected with CD26s or mutants as described above. At 48 h posttransfection, the medium was removed from each well, and the cells were washed once with PBS. Next, 200 μl of ice-cold lysis buffer containing 20 mM Tris-HCl, 150 mM NaCl, 0.5% NP-40, 0.1% SDS, and proteinase inhibitor was added to each well. The cells were incubated on ice for 10 min with the lysis buffer. The lysates were then transferred to tubes and centrifuged for 5 min at 13,000 rpm. Supernatants were mixed with the loading buffer and separated using a 8% PAGE gel. The samples were then transferred to the NC membrane. After a blocking step with 5% fat-free milk in PBS plus Tween (PBST), the membranes were incubated with either rabbit anti-GFP (1:3,000; Boster Biological Technology) or mouse anti-GAPDH (1:3,000; Abcam) at room temperature for 2 h. The membranes were washed three times with PBST (5 min per wash). Goat anti-IgG/HRP (EasyBio) was then used at a 1:3,000 dilution. After a washing step, the membranes were developed using Amersham ECL Western blotting detection reagents (GE Healthcare) and imaged using Amersham Hyperfilm ECL (GE Healthcare).
Data availability.
The coordinates and structure factor of the structure reported here have been deposited in the Protein Data Bank under PDB ID 6L8Q. All the other relevant data are available from the corresponding authors upon reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of beamline BL19U1 at the Shanghai Synchrotron Radiation Facility for assistance during data collection. We are grateful to Zheng Fan (Institute of Microbiology, Chinese Academy of Sciences [IMCAS]) for technical help with BIAcore experiments and Tong Zhao (IMCAS) for technical support during FACS assays. We are grateful to Hongtao Wu and Chunli Song for performing the Western blot analyses to detect the glycosylation of bCD26s.
This study was supported by the National Key Plan for Scientific Research and Development of China (grant 2016YFD0500305). G.L. is supported by NSFC (grant 31570157). Q.W. is supported by the Youth Innovation Promotion Association CAS (grant 2018119) and NSFC (grants 81922044 and 81973228). J.Y and G.F.G. are supported by the Foundation of the NSFC Innovative Research Group (grant 81621091).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We declare that we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.Chan JFW, Lau SKP, To KKW, Cheng VCC, Woo PCY, Yuen KY. 2015. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Y, Feng Y, Liu D, Gao GF. 2009. Avian influenza virus, Streptococcus suis serotype 2, severe acute respiratory syndrome-coronavirus and beyond: molecular epidemiology, ecology, and the situation in China. Philos Trans R Soc Lond B Biol Sci 364:2725–2737. doi: 10.1098/rstb.2009.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, Xiao G, Xu Y, Yuan F, Zheng C, Liu Y, Yan H, Cole DK, Bell JI, Rao Z, Tien P, Gao GF. 2004. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem Biophys Res Commun 319:283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. 2019. Middle East respiratory syndrome coronavirus (MERS-CoV). World Health Organization, Geneva, Switzerland: https://www.who.int/csr/don/26-february-2019-mers-saudi-arabia/en/. [Google Scholar]
- 6.Knipe DM, Howley PM. 2013. Fields virology, 6th ed Wolters Kluwer/Lippincott/Williams & Wilkins Health, Philadelphia, PA. [Google Scholar]
- 7.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. 2012. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu G, Wang Q, Gao GF. 2015. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol 23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, Poon LL. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 10.Ge X-Y, Li J-L, Yang X-L, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang Y-J, Luo C-M, Tan B, Wang N, Zhu Y, Crameri G, Zhang S-Y, Wang L-F, Daszak P, Shi Z-L. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotten M, Watson SJ, Kellam P, Al-Rabeeah AA, Makhdoom HQ, Assiri A, Al-Tawfiq JA, Alhakeem RF, Madani H, AlRabiah FA, Al Hajjar S, Al-Nassir WN, Albarrak A, Flemban H, Balkhy HH, Alsubaie S, Palser AL, Gall A, Bashford-Rogers R, Rambaut A, Zumla AI, Memish ZA. 2013. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet 382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotten M, Watson SJ, Zumla AI, Makhdoom HQ, Palser AL, Ong SH, Al Rabeeah AA, Alhakeem RF, Assiri A, Al-Tawfiq JA, Albarrak A, Barry M, Shibl A, Alrabiah FA, Hajjar S, Balkhy HH, Flemban H, Rambaut A, Kellam P, Memish ZA. 2014. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio 5:e01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reusken CB, Schilp C, Raj VS, De Bruin E, Kohl RH, Farag EA, Haagmans BL, Al-Romaihi H, Le Grange F, Bosch BJ, Koopmans MP. 2016. MERS-CoV infection of alpaca in a region where MERS-CoV is endemic. Emerg Infect Dis 22:1129–1131. doi: 10.3201/eid2206.152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adney DR, Bielefeldt-Ohmann H, Hartwig AE, Bowen RA. 2016. Infection, replication, and transmission of Middle East respiratory syndrome coronavirus in alpacas. Emerg Infect Dis 22:1031–1037. doi: 10.3201/2206.160192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, Alhakeem R, Durosinloun A, Al Asmari M, Islam A, Kapoor A, Briese T, Daszak P, Al Rabeeah AA, Lipkin WI. 2013. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis 19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ithete NL, Stoffberg S, Corman VM, Cottontail VM, Richards LR, Schoeman MC, Drosten C, Drexler JF, Preiser W. 2013. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis 19:1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letko M, Miazgowicz K, McMinn R, Seifert SN, Sola I, Enjuanes L, Carmody A, van Doremalen N, Munster V. 2018. Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep 24:1730–1737. doi: 10.1016/j.celrep.2018.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caì Y, Yú SQ, Postnikova EN, Mazur S, Bernbaum JG, Burk R, Zhāng T, Radoshitzky SR, Müller MA, Jordan I, Bollinger L, Hensley LE, Jahrling PB, Kuhn JH. 2014. CD26/DPP4 cell-surface expression in bat cells correlates with bat cell susceptibility to Middle East respiratory syndrome coronavirus (MERS-CoV) infection and evolution of persistent infection. PLoS One 9:e112060. doi: 10.1371/journal.pone.0112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munster VJ, Adney DR, van Doremalen N, Brown VR, Miazgowicz KL, Milne-Price S, Bushmaker T, Rosenke R, Scott D, Hawkinson A, de Wit E, Schountz T, Bowen RA. 2016. Replication and shedding of MERS-CoV in Jamaican fruit bats (Artibeus jamaicensis). Sci Rep 6:21878. doi: 10.1038/srep21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dingle H. 1996. Migration: the biology of life on the move. Oxford University Press, New York, NY. [Google Scholar]
- 21.Wang Q, Wong G, Lu G, Yan J, Gao GF. 2016. MERS-CoV spike protein: targets for vaccines and therapeutics. Antiviral Res 133:165–177. doi: 10.1016/j.antiviral.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Lou Z, Liu Y, Pang H, Tien P, Gao GF, Rao Z. 2004. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J Biol Chem 279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, Zhang X, Gao GF. 2017. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raj VS, Mou H, Smits SL, Dekkers DHW, Muller MA, Dijkman R, Muth D, Demmers JAA, Zaki A, Fouchier RAM, Thiel V, Drosten C, Rottier PJM, Osterhaus A, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J, Zhang B, Shi Y, Yan J, Gao GF. 2013. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q, Qi J, Yuan Y, Xuan Y, Han P, Wan Y, Ji W, Li Y, Wu Y, Wang J, Iwamoto A, Woo PC, Yuen KY, Yan J, Lu G, Gao GF. 2014. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe 16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peck KM, Scobey T, Swanstrom J, Jensen KL, Burch CL, Baric RS, Heise MT. 2017. Permissivity of dipeptidyl deptidase 4 orthologs to Middle East respiratory syndrome coronavirus is governed by glycosylation and other complex determinants. J Virol 91:e00534-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peck KM, Cockrell AS, Yount BL, Scobey T, Baric RS, Heise MT. 2015. Glycosylation of mouse DPP4 plays a role in inhibiting Middle East respiratory syndrome coronavirus infection. J Virol 89:4696–4699. doi: 10.1128/JVI.03445-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Read RJ. 2001. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr 57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project Network. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factor of the structure reported here have been deposited in the Protein Data Bank under PDB ID 6L8Q. All the other relevant data are available from the corresponding authors upon reasonable request.