Abstract
Simple Summary
The Ningqiang pony (NQ) is a famous breed with an average adult height at the withers (the ridge between horse’s shoulder blades) of less than 106 cm, but the genetic mechanism responsible for its small stature remains unclear. Research on Chinese pony breeds is a topic of theoretical and practical importance. Long non-coding RNA (lncRNA) has been shown to regulate many biological functions, including growth. Our study, for the first time, reports the lncRNAs transcribed in horse placentas. By comparing 3011 transcripts coding 1464 lncRNAs from NQ ponies and Yili horses (YL) with distinct body sizes, six (TBX3 (T-box3), CACNA1F (L-type voltage-dependent calcium channel a1F), EDN3 (endothelin-3), KAT5 (histone acetyltransferase KAT5), ZNF281 (zinc finger protein 281), TMED2 (transmembrane emp24 domain), and TGFB1 (transforming growth factor beta 1)) out of the 233 genes targeted by differentially expressed lncRNAs were identified as being involved in limb development, skeletal myoblast differentiation, and embryo development. These results provide new insights into the genetic architecture of horse body size.
Abstract
As a nutrient sensor, the placenta plays a key role in regulating fetus growth and development. Long non-coding RNAs (lncRNAs) have been shown to regulate growth-related traits. However, the biological function of lncRNAs in horse placentas remains unclear. To compare the expression patterns of lncRNAs in the placentas of the Chinese Ningqiang (NQ) and Yili (YL) breeds, we performed a transcriptome analysis using RNA sequencing (RNA-seq) technology. NQ is a pony breed with an average adult height at the withers of less than 106 cm, whereas that of YL is around 148 cm. Based on 813 million high-quality reads and stringent quality control procedures, 3011 transcripts coding for 1464 placental lncRNAs were identified and mapped to the horse reference genome. We found 107 differentially expressed lncRNAs (DELs) between NQ and YL, including 68 up-regulated and 39 down-regulated DELs in YL. Six (TBX3, CACNA1F, EDN3, KAT5, ZNF281, TMED2, and TGFB1) out of the 233 genes targeted by DELs were identified as being involved in limb development, skeletal myoblast differentiation, and embryo development. Two DELs were predicted to target the TBX3 gene, which was found to be under strong selection and associated with small body size in the Chinese Debao pony breed. This finding suggests the potential functional significance of placental lncRNAs in regulating horse body size.
Keywords: pony, horse, long non-coding RNA, placenta, TBX3
1. Introduction
Non-coding RNAs (ncRNAs) are a heterogeneous class of RNA molecules transcribed from non(-protein)-coding regions in a genome because they lack an open reading frame and consequently have no protein-coding ability. The ncRNAs are classified into long ncRNAs (lncRNAs, >200 bp) and small ncRNAs (sncRNAs, 18–200 bp) according to their lengths [1,2]. lncRNAs play critical roles in many important biological processes, including cell proliferation and differentiation, signal transduction, stem cell maintenance and metabolism, genome imprinting, chromatin remodeling, regulation of cell cycle, and splicing regulation [3,4]. Many lncRNAs have been identified in a wide range of organisms [5,6], but have been poorly characterized in horses [7]. Only one single study has reported on the identification of horse lncRNAs [7]. It was based on eight types of tissues from 59 horses, producing over 20 million reads of RNA-sequencing (RNA-seq) data, building a database of 20,800 horse lncRNAs [7,8] and supporting the intensive identification of functional lncRNAs in horses.
As one of the five Chinese indigenous pony breeds (Debao, Jianchang, Guizhou, Yunnan, and Ningqiang), the Ningqiang pony (NQ) has an average adult height at withers of less than 106 cm [9]. It is distributed in the northeast of the Shaanxi province but is now close to being endangered. To adapt to the local mountainous environment, NQ has experienced a long history of both natural and human selection for its small stature and an extremely tough body conformation. The Yili horse (YL), a famous racing horse breed with an adult height at the withers of around 148 cm [9], was derived from the Kazakh horse through crossbreeding with Orlov Trotter, Don, Budyonny, and Akhal-Teke horse breeds in the 20th century [10]. The large variation in heights at the withers between NQ and YL provides an opportunity to study their underlying molecular mechanisms.
The placenta is essential for fetus growth and development as it acts as a nutrient sensor to regulate the transfer of nutrients from mother to fetus [11]. Studies have reported a strong association between neonatal body size and placental structure [12]. During pregnancy, the placenta is an important endocrine organ that produces numerous hormones, including estrogens and progesterone, growth hormone, insulin, and insulin-like growth factor 1, most of them crucial in the regulation of fetal growth [13]. A genomic scan showed that the TBX3 gene is under strong positive selection in the Chinese Debao pony [14]; however, the functional validation of TBX3 in horse tissues has not yet been performed due to lack of appropriate samples, i.e., placental tissue.
The development of whole-transcriptome sequencing technology provides an opportunity to efficiently identify and annotate horse lncRNAs. In this study, we conducted RNA-seq analysis of the placental tissues from NQ and YL. After stringent quality control and annotation, the differentially expressed placental lncRNAs between NQ and YL and their target genes were identified. Two lncRNAs were found to regulate TBX3, which may, therefore, play an important role in the development of horse body size. Our study facilitates the further exploration of the fundamental function of lncRNAs in the regulation of horse body size.
2. Materials and Methods
2.1. Ethics Statement
All procedures involving the handling of ponies and horses were approved by the Animal Care and Use Committee of the Chinese Academy of Agricultural Sciences and the Ministry of Agriculture of the People’s Republic of China (IASCAAS-AE-03).
2.2. Animals and Samples
The placental samples of NQ and YL were taken from the Ningqiang national conservation farm in Ningqiang county in Shaanxi province (log: 105°582′ and lat: 32°825′) and Zhaosu horse farm in Zhaosu county in Xinjiang province (log: 81.131 and lat: 43.155). Three YL mares with an average height of 141.3 cm and three NQ mares with an average height of 105 cm were taken. To ensure consistency between biological replications, adult mares about six years old were chosen for placental tissue sampling. To avoid relatedness between the collected samples, we consulted the local technicians and farmers regarding the pedigrees of the mares. These pregnant mares (multiparous) were randomly selected, ensuring no genetic kinship within three generations. After giving birth, the placental tissues from the mares were immediately collected by experienced veterinarians and stored in the RNAlater (Thermo Fisher Scientific, Waltham, MA, USA) at −80 °C. All samples were transported to our laboratory for RNA extraction.
2.3. RNA Extraction, Library Preparation, and Sequencing
Total RNA from all six placental samples was extracted by using the RNeasy Mini Kit (Qiagen, Hilden, Germany). The RNA was quantified according to the RNA integrity number (RIN) value >8 using an Agilent 2100 Bioanalyzer System (Agilent Technologies, Santa Clara, CA, USA). The Illumina TruSeq RNA Library Prep Kit v2 (Illumina, San Diego, CA, USA) was used to construct the cDNA library for RNA-seq. Briefly, after end-repair and adding poly (A) to the 3′ end of the RNA fragments, the sequencing linker was ligated and the products were purified for PCR amplification. The PCR products were separated by 2% agarose gel electrophoresis and fragments varying from 400 to 500 bp were selected to construct sequencing libraries. Finally, the six libraries were sequenced on an Illumina HiSeq 2500 System (Illumina, San Diego, CA, USA) using a 150 × 2 bp paired-end sequencing strategy by Beijing Compass Biotechnology Co. Ltd. (Beijing, China).
2.4. Trimming and Mapping of Reads
To remove low quality reads and adapters, raw sequencing reads were first filtered using Trimmomatic (version 0.33) software (http://www.usadellab.org/cms/index.php?page=trimmomatic) [15]. The reads were filtered using the following steps: first, reads with adapters were removed; second, reads with more than 10% ploy-N were discarded; and third, low-quality reads with more than 50% of bases and Phred scores less than 5 were removed. The Phred scores (Q20, Q30) and GC content of the clean data was calculated. All subsequent analyses were conducted based on the high-quality clean data. The quality of RNA-seq reads was ascertained using the FASTQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). After indexing the reference assembly of horse genome EquCab3.0 with Bowtie-build software (https://sourceforge.net/projects/bowtie-bio/files/bowtie2) [16], all clean reads were mapped with the TopHat2 alignment tool (http://ccb.jhu.edu/software/tophat/index.shtml) [17] using default parameters. The mapped reads of each sample were assembled using Cufflinks v2.1.1 (http://cole-trapnell-lab.github.io/cufflinks/releases/v2.1.1) [18]. To identify the similarity between samples, the read counts were calculated using HTSeq (https://htseq.readthedocs.io/en/release_0.11.1) [19] and normalized using R package DESeq (https://bioconductor.org/packages/release/bioc/html/DESeq.html) [20].
2.5. Step-Wise Filtering for Transcripts
Cufflinks [18] (with the command of ‘min-frags-per-transfrag = 0 and -library-type’) was used to de novo assemble the transcripts from aligned RNA-seq reads and to obtain fragments per kilobase of transcript per million mapped fragments (FKPM) of each gene. Then, all the transcripts were merged using Cuffmerge software (http://cole-trapnell-lab.github.io/cufflinks/cuffmerge/index.html) [18].
2.6. Identification of lncRNAs from the Assembled Transcripts
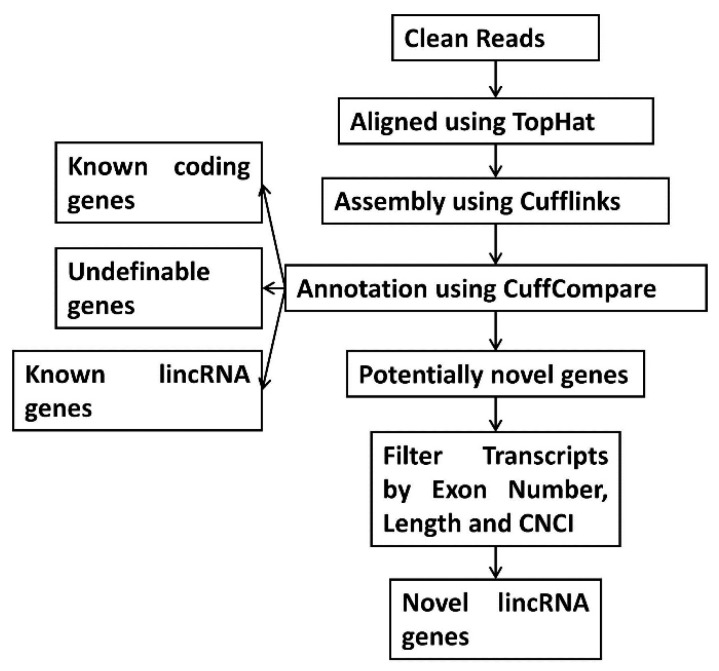
lncRNAs were identified from the assembled transcripts following four steps: (1) Removal of lowly expressed transcripts with FPKM < 0.5, (2) removal of short transcripts < 200 bp and < 2 exons, (3) removal of the transcripts with protein-coding capability using the coding-non-coding index (CNCI) software (https://github.com/www-bioinfo-org/CNCI) with default parameters for a CNCI score ≥ 0 [21], and (4) removal of the transcripts mapped within the 1 kb flanking regions of an annotated gene. For incomplete UTR annotation of protein-coding genes in horses, the transcriptome data used in this study were based on the published refined transcriptome with the candidate lncRNA post-filter 3 removed [22]. The rest of the transcripts with more than 90% located to the “NM_” sequences were also removed. The pipeline used to identify putative lncRNAs from the RNA-seq data is presented in Appendix A Figure A1.
2.7. Quantification of lncRNA Expression
Cuffdiff software (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/index.html) [18] was used to calculate differentially expressed lncRNAs (DELs) between YL and NQ breeds, with a corrected p ≤ 0.05 and fold change (FC) ≥ 2. Volcano plots of the DELs were employed to demonstrate the differences.
2.8. Prediction of Target Genes
Transcripts without coding potential composed our candidate set of lncRNAs. To explore the function of these potential lncRNAs, genes targeted by the placental lncRNAs were first predicted in cis action. The cis role refers to lncRNA action on neighboring target genes [23]. Coding genes from 100 kb upstream and downstream of the lncRNAs were searched. The trans role refers to the influence of lncRNA on the expressions of other genes. Pearson’s correlation coefficients (r) were calculated between the expression levels of the lncRNAs and mRNAs with customized scripts (r ≥ 0.95 or ≤−0.95). For the function of target genes, we referred to the relevant literature.
2.9. Gene Ontology (GO) Analysis
The functional annotation and pathway enrichment of the gene targeted by the DELs were performed using the online analysis tool Gene Ontology Consortium (http://www.geneontology.org/). This is a program combining the enriched GO terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The target genes were mapped to the GO database, and the hypergeometric test was used to determine which GO terms were significantly enriched among the target genes against the background of the reference horse genome EquCab3.0. GO terms were considered to be significantly enriched when they had corrected p-values less than 0.05. A similar method was used to determine which KEGG pathways and relevant complex biological terms were enriched among the target genes (threshold: corrected p-value < 0.05).
2.10. Quantitative Real-Time PCR (qRT-PCR) Validation
To further confirm the reliability of these identified lncRNAs, qRT-PCR assays were performed. First, the total RNA extracted from individual placentas of YL and NQ mares were used for validation. Then, the RNAs were inverse transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (TAKARA, RR047A, Beijing, China). The lncRNAs against the house-keeping gene β-ACTN (Beta-actin) as an endogenous control was quantified using the SYBR Green Master Mix (TAKARA, RR820A, Beijing, China). We randomly selected 10 lncRNAs for the quantification. The primers used in this study are listed in Table 1. qRT-PCR was performed using the following conditions: 95 °C for 60 s, then 40 cycles at 95 °C for 10 s, and the optimized annealing temperatures for 30 s in the Applied Biosystems® 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Each reaction was performed in triplicate for each sample. Gene expression was quantified relative to β-ACTN expression using the comparative cycle threshold (ΔΔCT) method. Then, the log2Fold change (FC) of each gene expression level from qRT-PCR data was calculated for comparison with the expression patterns of RNA-seq data. Log2FC values were calculated for the comparison of YL with NQ.
Table 1.
Primers for quantitative real-time (qRT)-PCR validation.
| lncRNA Name | Length (bp) | Primers | |
|---|---|---|---|
| XLOC_022706 | 204 | F | TTACGAGCATCGCTGGGTTT |
| R | GGTCTTACAGCGACACCACA | ||
| XLOC_021555 | 280 | F | GTTCAATGGAGGAGGGCGAA |
| R | TGGAGCATCTCGCAAAGTGT | ||
| XLOC_048194 | 215 | F | AATGCACTTTTCCGGACCCT |
| R | CCCCAGTTTGATCGTGTGGA | ||
| XLOC_006815 | 217 | F | AACGTCGGTCTCTTTTCCCC |
| R | AGCAAGTTGTTTCGGGTGGA | ||
| XLOC_049268 | 296 | F | TGTCGCCCTTCATCTCTGTG |
| R | TCAGCGGTGGAAGAAAGCAA | ||
| XLOC_038367 | 238 | F | TTGCTGGCTGTTGCTTGAAC |
| R | TCTCCATTGCCAGTTCGGTG | ||
| XLOC_040258 | 178 | F | AAACTAACCAGCTCCCCGTG |
| R | TTGCCAGCCAAAATGCCTTC | ||
| XLOC_045486 | 133 | F | CGGAACCTGTTACACTGCCT |
| R | CTCACACTTCTGCCCCACAT | ||
| XLOC_054035 | 156 | F | CGGACAATTACGCAGCCATG |
| R | CCTCTGCCCTCCTTAGTCCT | ||
| XLOC_061022 | 137 | F | CCGTCTTCCTGTTCTGCACT |
| R | ACATACACCCTGGCCTGTTG | ||
| β-ACTN (Beta-actin) | 182 | F | CAGCCTTCCTTCTTGGGTAT |
| R | TGGCATAGAGGTCTTTACGG | ||
Note: F refers to forward primers, R refers to reverse primers.
3. Results
3.1. Overview of RNA-Seq Data
To identify the unique lncRNAs expressed in horse placenta, a prerequisite was integrating the high-quality and high-depth RNA-seq data. A total of 39.34–47.95 million raw reads were generated across the six samples. After discarding adaptor sequences, the median per-base quality was more than 30 for all the samples and the Q30 rate was more than 83.32% (Supplementary Materials Table S1). Next, we mapped all the clean reads of every sample to the latest reference horse genome EquCab3.0 using TopHat2 software (http://ccb.jhu.edu/software/tophat/index.shtml) [17], achieving alignment rates of approximately 65.50–81.00%.
3.2. Identification of lncRNAs
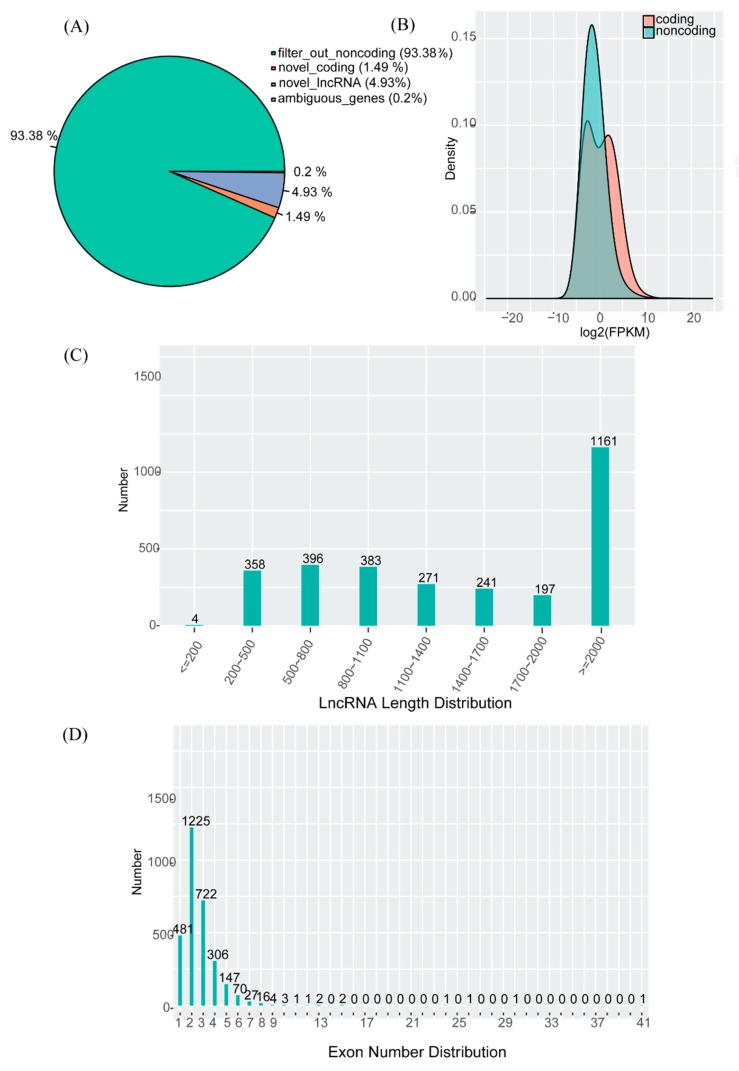
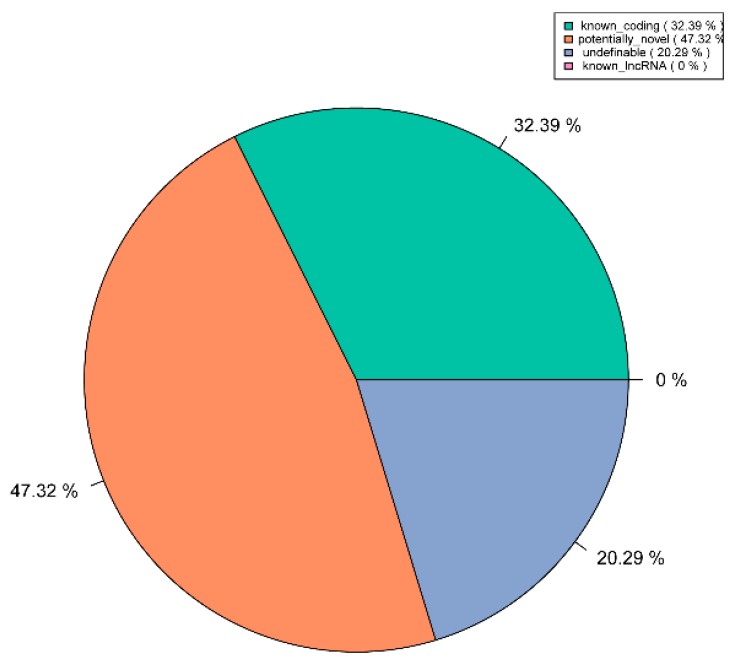
Overall, 20,961 de novo transcripts were captured for analyses. To evaluate the quality of these transcripts, we used Cuffcompare software (http://cole-trapnell-lab.github.io/cufflinks/cuffcompare/index.html) [18] to compare transcripts and coding genes with those in the refGene library. After predicting the coding functions of the lncRNAs that were identified in this study, we classified the novel lncRNAs into three categories with the most common cluster being “potentially_novel” at 47.32%, followed by “known_coding” at 32.39%, and “undefinable” at 20.29% (Appendix A Figure A2). After filtering the potentially_novel transcripts, coding potential analysis was performed using CNCI software (https://github.com/www-bioinfo-org/CNCI) [21]. Finally, we obtained 3011 transcripts and 1464 potential lncRNAs, with 93.38% being non-coding, indicating their reliability (Figure 1A).
Figure 1.
Statistics of horse placental lncRNAs identified in this study. (A) Percentage of potentially novel transcripts. (B) Expression levels of mRNAs and lncRNAs. (C) Length distribution of lncRNAs. (D) Exon number of lncRNAs.
3.3. Features of mRNAs and lncRNAs
To summarize the lncRNAs in our study, we found that 38.56% of the lncRNAs were more than 2000 bp (Figure 1C), whereas 84.02% expanded two or more exons (Figure 1D). All but one of the 1464 lncRNAs were mapped to intronic regions. To ensure high confidence in the lncRNAs identified, we compared them to mRNAs. The preliminary analysis revealed some major differences in gene architecture and expression level among the mRNAs and lncRNAs. For example, the expression levels were higher in mRNAs than lncRNAs (Figure 1B).
3.4. Differentially Expressed lncRNAs and mRNAs
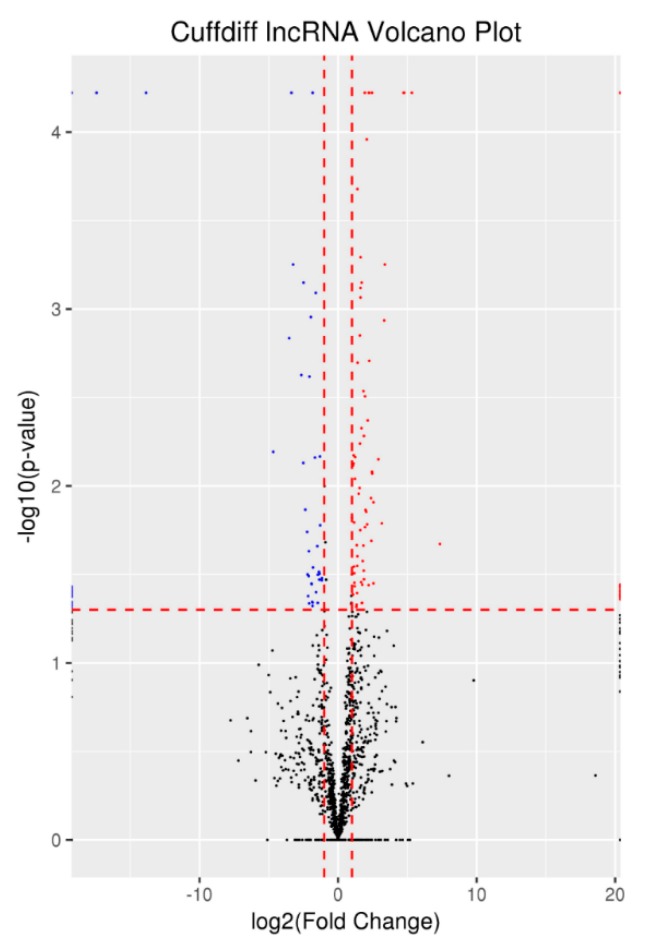
The expression levels were analyzed using Cuffdiff software (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/index.html) [18]. edgeR software (https://bioconductor.org/packages/release/bioc/html/edgeR.html) [24] with parameters of FC ≥ 2.0 and p-value ≤ 0.05 was used to identify DELs between the YL and NQ breeds, resulting in 107 DELs, among which 68 DELs were up-regulated and 39 were down-regulated in YL compared to NQ (Figure 2).
Figure 2.
Differentially expressed lncRNAs. Note: The horizonal dashed red line shows criteria of P values equal to 0.05 while vertical dashed lines show log2FC equal to -1 (left) and 1 (right). The points colored with red and blue show the genes if they are upregulated and downregulated, respectively. Black points mean genes does not meet the significant criteria.
3.5. Predication of the Targeted Genes
To investigate the function of lncRNAs, we predicted the potential target genes of lncRNAs in cis and trans actions. For the cis action of lncRNAs, we searched for protein-coding genes both 10 and 100 kb upstream and downstream of the lncRNAs. We detected 233 targeted genes, among which TBX3, CACNA1F, EDN3, KAT5, ZNF281, and TGFB1 genes are related to body size, which were located near XLOC_021555, XLOC_058541, XLOC_048194, XLOC_032009, XLOC_057255, and XLOC_025357, respectively, thereby suggesting that horse body formation could be regulated by the cis actions of several instead of one lncRNA on neighboring protein-coding genes (Table 2). GO analysis of the lncRNA targeted genes demonstrated that most of these genes were clustered into transcription regulations, developmental protein (p = 0.01) and positive regulation of mitogen-activated protein kinase (MAPK) activity (p = 0.03) pathways. Some genes in the MAPK pathway were also differentially expressed between the inner cell mass and trophectoderm [25]. TBX3 was clustered into the significantly overrepresented term, developmental protein. These findings demonstrate one of the roles of lncRNAs: the regulation of development (Table 3) through their cis actions on their neighboring protein-coding genes, e.g., to regulate the TBX3 protein during body development [26].
Table 2.
Significant differentially expressed lncRNAs and their targeted genes.
| lncRNAs | Log2FC | p | 10 kb Upstream | 10 kb Downstream | 100 kb Upstream | 100 kb Downstream |
|---|---|---|---|---|---|---|
| XLOC_025357 | 1.84 | 5 × 10−5 | ERICH4, BCKDHA, TMEM91, DMAC2, B3GNT8, EXOSC5, B9D2, TGFB1 | |||
| XLOC_038367 | −2.24 | 5 × 10−5 | IQSEC1 | |||
| XLOC_048194 | 1.82 | 5 × 10−5 | EDN3 | |||
| XLOC_051797 | 1.61 | 8 × 10−4 | STX17 | |||
| XLOC_032225 | −3.33 | 1.15 × 10−3 | PHRF1 | LMNTD2, RASSF7 | IRF7, CDHR5, SCT, DEAF1, PHRF1, DRD4 | LMNTD2, HRAS, LOC155356, RASSF7, LRRC56, LOC121596 |
| XLOC_000736 | 3.53 | 1.45 × 10−3 | PLAU, C1H1orf55, CAMK2G | VCL | ||
| XLOC_050107 | 2.64 | 2.35 × 10−3 | IFNE | |||
| XLOC_055900 | −2.14 | 4.25 × 10−3 | IL17REL | IL17REL, TTLL8 | ALG12, PIM3, LOC11177138, CRELD2, ZBED4 | |
| XLOC_021555 | 1.66 | 6.9 × 10−3 | TBX3 | |||
| XLOC_043830 | −2.91 | 7.05 × 10−3 | MFSD1 | MFSD1, LOC15252 | ||
| XLOC_017294 | −2.38 | 0.01 | KIAA1551 | AMN1, ETFBKMT | ||
| XLOC_015361 | −2.56 | 0.01 | TEX44, NMUR1, LOC1214915 | PTMA, PDE6D | ||
| XLOC_056998 | 2.36 | 0.01 | ELF3, RNPEP, TIMM17A | GPR37L1, LOC1146338, LGR6, ARL8A, PTPN7 | ||
| XLOC_016995 | 2.23 | 0.01 | KCNA5 | |||
| XLOC_021660 | −7.36 | 0.02 | RILPL1 | TMED2 | SNRNP35, KMT5A, RILPL1, RILPL2 | TMED2, DDX55, GTF2H3, TCTN2, ATP6VA2, EIF2B1 |
| XLOC_042377 | 2.12 | 0.03 | TSN, NIFK | |||
| XLOC_059454 | 1.91 | 0.03 | CD99, XG | ZBED1, DHRSX | ||
| XLOC_008338 | −2.20 | 0.03 | COQ2, HPSE | LOC163244, PLAC8 | ||
| XLOC_022706 | 2.09 | 0.04 | TBX3 | |||
| XLOC_027025 | −1.31 | 0.04 | LOC17567 | IGSF23 | LOC17567, CEACAM19, CEACAM16 | ZNF18, IGSF23 |
Note: log2FC = log2 of fold change. Genes having possible biological functions contributing to growth are indicated in bold font.
Table 3.
Gene Ontology (GO) results of lncRNA targeted genes.
| Terms | Counts | p | Gene |
|---|---|---|---|
| GO:1903445, protein transport from ciliary membrane to plasma membrane | 2 | 0.018 | RILPL1, RILPL2 |
| GO:0030335, positive regulation of cell migration | 5 | 0.022 | HRAS, FOXF1, LGR6, PLAU, TGFB1 |
| GO:2000630, positive regulation of miRNA metabolic process | 2 | 0.028 | HRAS, RELA |
| GO:0002513, tolerance induction to self-antigen | 2 | 0.028 | FOXP3, TGFB1 |
| GO:0043406, positive regulation of MAP kinase activity | 3 | 0.034 | EDN3, HRAS, TGFB1 |
| Developmental protein | 17 | 0.011 | DEAF1, USP7, CCM2, RNF17, ELF3, TBX3, CAMK2G, EN1, ACKR3, RTL1, CITED2, CXCL17, LBH, HAND1, HES5, FOXC2, OLFM1 |
| Activator | 13 | 0.015 | ATF7IP, ELF3, RELA, ZXDB, FOXP3, MED13L, KAT5, ATF7IP2, CITED2, HAND1, IRF7, FOXF1, FOXC2 |
| Transcription regulation | 31 | 0.025 | DEAF1, ZFP64, ZNF18, ELF3, ASCC1, ZXDB, ZKSCAN5, CITED2, LBH, HAND1, FOXF1, OVOL1, ZNF394, BRD9, KMT5A, ATF7IP, ZNF281, FOXL1, TBX3, RELA, GTF2H3, FOXP3, MED13L, KAT5, ATF7IP2, ZNF789, HES5, DMRTC2, IRF7, FOXC2, NR5A2 |
| ecb05205:Proteoglycans in cancer | 6 | 0.029 | HRAS, HPSE, CAMK2G, SDC4, PLAU, TGFB1 |
Note: Clusters associated with growth are indicated in bold font.
3.6. Validation of lncRNAs by qRT-PCR
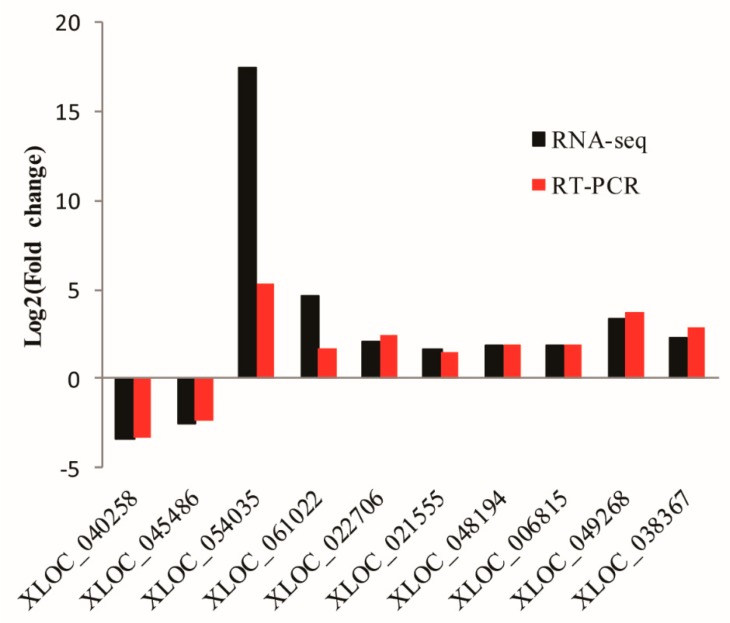
To confirm the expression patterns of the lncRNAs, we performed qRT-PCR analyses on 10 randomly selected lncRNAs identified. Though the theory of RNA-seq and qRT-PCR varies, in RT-PCR, we assumed that the expression of the reference genes is constant for all the samples, whereas in RNA-seq, we assumed that each sample has the same total expressed mRNA. This can be expressed as read count per million mapped reads or RPKM (Reads Per Kilobase per Million mapped reads). When the expression difference exists with sufficient significance, both qRT-PCR and RNA-seq can detect the difference. In this study, qRT-PCR results were aligned with the RNA-seq data, suggesting that the expression patterns based on RNA-seq data were reliable (Figure 3).
Figure 3.
Expression levels of 10 randomly selected lncRNAs based on RNA-seq data and qRT-PCR validation. log2Fold change (FC) > 0 or < 0 indicate the upregulated or down-regulated transcripts in Yili horses compared to Ningqiang ponies, respectively.
4. Discussion
In this study, we generated 1.3 billion 150-bp-long paired-end reads of cDNA from six horse placentas. Using the TopHat (http://ccb.jhu.edu/software/tophat/index.shtml) and Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/releases/v2.1.1) programs, 86.3–89.5% of all the reads were successfully mapped against the current reference horse genome EquCab3.0. After developing the bioinformatics pipeline for processing the large amounts of transcriptome sequences, 1464 multiple-exon lncRNAs were identified in the placentas of YL and NQ breeds, and 361 of them overlapped with a previous study [7], which did not contain the placenta tissue, supporting the tissue-specific expression of lncRNAs [27]. We identified 107 lncRNAs as being differentially expressed between the YL and NQ breeds. The most significant phenotypic variation between these two breeds is height. To the best of our knowledge, unique expression profiles of genes associated with specific variations have already been detected in different breeds of chickens [28], pigs [29], and sheep [30].
Among the 233 lncRNA targeted genes, six were related to body development and formation. For instance, the TBX3 (T-box 3) protein is a downstream target of the transforming growth factor-β1 (TGF-β1) signaling pathway and plays an important role in embryonic development [31]. TBX3 has already been identified as under selection in the Chinese Debao pony [14], suggesting more breeds and samples should be examined to deepen our understanding of the regulation models of horse body size development. TGF-β1 [25] and TMED2 (transmembrane emp24 domain) were also differentially expressed in embryonic cells and placenta [32], proven by the morphogenesis of the embryo and placenta, as well as skeletal muscle physiology [33]. CACNA1F (L-type voltage-dependent calcium channel a1F), involved in the differentiation of adipocytes and related to the etiology of obesity [34], was found to be under selection in the Chinese Debao pony [35]. EDN3 (endothelin-3) was reported to be associated with enteric neuron development and blood circulation, thereby influencing the racing performance of Swedish-Norwegian coldblooded trotter horses [36]. KAT5 (histone acetyltransferase KAT5) chromodomain was found to facilitate SOX4 recruitment to the CALD1 (caldesmon 1) promoter to regulate skeletal myoblast differentiation [37]. As a core transcription factor in embryonic stem cells, ZNF281 (zinc finger protein 281) was found to be related to spontaneous osteochondrogenic differentiation [38]. Other studies showed the strong association of LCORL/NCAPG, HMGA2, PROP1, LASP [39], ZFAT, DIAPH3 [40], ACTN2, ADAMTS17, GH1, ANKRD1 [40], and ACAN [41,42] with miniature size and dwarfism in a variety of pony breeds, including Shetland [41,43], miniature [44], Welsh ponies [45], German warmblood horses [46], American miniature horses, Brazilian ponies [47], and Jeju ponies [48], as well as B4GALT7 [49] and PROP1 [50] in Friesian horses. These genes were not found in this study, which may be due to the expression specificity of lncRNAs in different horse breeds [27] and different omics levels, since TBX3, under strong selection in Chinese ponies, was also identified in this study. This indicated that more breeds and samples are needed to analyze growth regulation during horse development.
We predicted the potential functions of lncRNAs in horse placenta and found that protein-coding genes can interact with lncRNAs through their cis actions. In particular, TBX3, which has been identified as being associated with horse body size [51], was regulated by two lncRNAs in cis actions. We observed that one of these two lncRNAs is located upstream of the enhancer region of the mouse TBX3 (https://enhancer.lbl.gov/cgi-bin/imagedb3.pl?form=presentation&show=1&experiment_id=483&organism_id=1). As a member of the T-box gene family, TBX3 plays an important role in embryonic development and some mutations in this gene cause ulnar-mammary syndrome [26]. In the mouse, TBX3 is recognized as one of the earliest limb initiation factors and loss of functional TBX3 protein in early and later development may disrupt limb initiation and cause digit loss [52]. The higher expression levels of the two TBX3-regulating lncRNAs in YL compared to NQ imply their possible associations with horse height. A certain cluster of lncRNAs in cis actions often target protein-coding genes that are specifically expressed in the growth stage (CACNA1F, EDN3, KAT5, ZNF281, TMED2, and TGFB1). These findings suggest the important role of some lncRNAs in horse body development.
Besides the genes mentioned above, more genes accounting for different characteristics were found. STX17 is a causative gene for melanoma tissue in grey horses [53]. LGR6 (luteinizing hormone receptor 6) is expressed in embryonic hair follicles, and acts as a marker in the most primitive epidermal stem cells [54]. A mutation in IQSEC1 can cause intellectual disability, developmental delay, and short stature [55]. KCNA5 (potassium voltage-gated channel subfamily A member 5) genes, associated with pulmonary arterial hypertension (PAH) [56], may affect the motion of the horse. COQ2 (coenzyme Q2) and PLAC8 (placenta-specific 8) are candidate genes for the onset of type 2 diabetes associated with obesity in rats [57].
This study has limitations due to the use of placenta as the main tissue for one Chinese native pony breed versus the Yili horse. Though placenta is not the best tissue for studying the genetic mechanism of animal growth, it is a hotbed for fetal development and using placental tissue is less invasive for the animals, and so it can be used as an alternative tissue for analysis. To reduce interference in breed-specific tissue, we chose the most different characteristic, body size, as the target. In the future, the molecular components underlying the variation in body size in more southwestern Chinese pony breeds should be investigated.
5. Conclusions
In this study, we identified the first set of 1464 lncRNAs from horse placentas with 107 of them differentially expressed between the Yili horse and Ningqiang pony breeds. The identification of two specific TBX3-regulating lncRNAs suggests their important roles in horse body size. These genetic markers will contribute to the marker-assisted selection of the height of horses.
Acknowledgments
X.L. was supported by the International Postdoctoral Exchange Fellowship Program (2019).
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/10/1/119/s1, Table S1: Quality control of clean reads.
Appendix A
Figure A1.
The pipeline used to identify putative lncRNAs.
Figure A2.
Classification of the novel lncRNAs.
Author Contributions
Conceptualization, X.L. and Y.M.; methodology, X.L. and J.H.; software, J.H.; validation, Y.Z., T.Z. and Y.P.; formal analysis, J.H.; investigation, Y.P.; resources, Y.P.; data curation, Y.Z.; writing—original draft preparation, X.L. and J.H.; writing—review and editing, J.H.; visualization, J.H.; supervision, X.L. and Y.M.; project administration, Y.M., X.L. and Y.P.; funding acquisition, Y.M., X.L. and Y.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31972530, 31772553), the Chinese Academy of Agricultural Sciences (Y2017JC03, Y2018PT68), the Agricultural Science and Technology Innovation Program of China (ASTIP-IAS01), and the Research and Demonstration of Green Sheep Development Technology Integration Model (CAAS-XTCX2016011-02).
Conflicts of Interest
The authors declare no conflict of interest.
Data Availability
All the Illumina sequencing reads have been deposited in the National Genomics Data Center (https://bigd.big.ac.cn) with the accession code (BioProject ID: PRJCA001875).
References
- 1.Diamantopoulos M.A., Tsiakanikas P., Scorilas A. Non-coding RNAs: The riddle of the transcriptome and their perspectives in cancer. Ann. Transl. Med. 2018;6:241. doi: 10.21037/atm.2018.06.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H., Xu Z., Liu D. Small non-coding RNA and colorectal cancer. J. Cell. Mol. Med. 2019;23:3050–3057. doi: 10.1111/jcmm.14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dey B.K., Mueller A.C., Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5:e944014. doi: 10.4161/21541272.2014.944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.St. Laurent G., Wahlestedt C., Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst C., Morton C. Identification and function of long non-coding RNA. Front. Cell. Neurosci. 2013;7:168. doi: 10.3389/fncel.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott E.Y., Mansour T., Bellone R.R., Brown C.T., Mienaltowski M.J., Penedo M.C., Ross P.J., Valberg S.J., Murray J.D., Finno C.J. Identification of long non-coding RNA in the horse transcriptome. BMC Genom. 2017;18:511. doi: 10.1186/s12864-017-3884-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mansour T.A., Scott E.Y., Finno C.J., Bellone R.R., Mienaltowski M.J., Penedo M.C., Ross P.J., Valberg S.J., Murray J.D., Brown C.T. Tissue resolved, gene structure refined equine transcriptome. BMC Genom. 2017;18:103. doi: 10.1186/s12864-016-3451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L., Wang A., Wang L., Li K., Yang G., He R., Qian L., Xu N., Huang R., Peng Z., et al. China National Commission of Animal Genetic Resources. China Agriculture Press; Beijing, China: 2011. Animal genetic resources in China: Pigs. [Google Scholar]
- 10.Liu L.L., Fang C., Liu W.J. Identification on novel locus of dairy traits of Kazakh horse in Xinjiang. Gene. 2018;677:105–110. doi: 10.1016/j.gene.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Jansson T., Powell T.L. Role of the placenta in fetal programming: Underlying mechanisms and potential interventional approaches. Clin. Sci. 2007;113:1–13. doi: 10.1042/CS20060339. [DOI] [PubMed] [Google Scholar]
- 12.Alwasel S.H., Abotalib Z., Aljarallah J.S., Osmond C., Al Omar S.Y., Harrath A., Thornburg K., Barker D.J.P. The breadth of the placental surface but not the length is associated with body size at birth. Placenta. 2012;33:619–622. doi: 10.1016/j.placenta.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 13.Murphy V.E., Smith R., Giles W.B., Clifton V.L. Endocrine regulation of human fetal growth: The role of the mother, placenta, and fetus. Endocr. Rev. 2006;27:141–169. doi: 10.1210/er.2005-0011. [DOI] [PubMed] [Google Scholar]
- 14.Kader A., Liu X., Dong K., Song S., Pan J., Yang M., Chen X., He X., Jiang L., Ma Y. Identification of copy number variations in three Chinese horse breeds using 70K single nucleotide polymorphism BeadChip array. Anim. Genet. 2016;47:560–569. doi: 10.1111/age.12451. [DOI] [PubMed] [Google Scholar]
- 15.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S., Chan C.K.K. Analysis of RNA-Seq Data Using TopHat and Cufflinks. In: Edwards D., editor. Plant Bioinformatics: Methods and Protocols. Springer; New York, NY, USA: 2016. pp. 339–361. [DOI] [PubMed] [Google Scholar]
- 19.Anders S., Pyl P.T., Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S. Analysing RNA-Seq data with the DESeq package. [(accessed on 11 January 2020)]; Available online: https://master.bioconductor.org/help/course-materials/2011/CSAMA/Thursday/Afternoon%20Labs/DESeq.pdf.
- 21.Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., Liu Y., Chen R., Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41:e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hezroni H., Koppstein D., Schwartz M.G., Avrutin A., Bartel D.P., Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11:1110–1122. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang G.Y., Duan A.G., Zhang J.G., He C.Y. Genome-wide analysis of long non-coding RNAs at the mature stage of sea buckthorn (Hippophae rhamnoides Linn) fruit. Gene. 2017;596:130–136. doi: 10.1016/j.gene.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 24.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iqbal K., Chitwood J.L., Meyers-Brown G.A., Roser J.F., Ross P.J. RNA-seq transcriptome profiling of equine inner cell mass and trophectoderm. Biol. Reprod. 2014;90:61. doi: 10.1095/biolreprod.113.113928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bamshad M., Le T., Watkins W.S., Dixon M.E., Kramer B.E., Roeder A.D., Carey J.C., Root S., Schinzel A., Van Maldergem L., et al. The spectrum of mutations in TBX3: Genotype/phenotype relationship in ulnar-mammary syndrome. Am. J. Hum. Genet. 1999;64:1550–1562. doi: 10.1086/302417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kern C., Wang Y., Chitwood J., Korf I., Delany M., Cheng H., Medrano J.F., Van Eenennaam A.L., Ernst C., Ross P., et al. Genome-wide identification of tissue-specific long non-coding RNA in three farm animal species. BMC Genom. 2018;19:684. doi: 10.1186/s12864-018-5037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piórkowska K., Żukowski K., Nowak J., Połtowicz K., Ropka-Molik K., Gurgul A. Genome-wide RNA-Seq analysis of breast muscles of two broiler chicken groups differing in shear force. Anim. Genet. 2016;47:68–80. doi: 10.1111/age.12388. [DOI] [PubMed] [Google Scholar]
- 29.Sodhi S.S., Park W.C., Ghosh M., Kim J.N., Sharma N., Shin K.Y., Cho I.C., Ryu Y.C., Oh S.J., Kim S.H., et al. Comparative transcriptomic analysis to identify differentially expressed genes in fat tissue of adult Berkshire and Jeju native pig using RNA-seq. Mol. Biol. Rep. 2014;41:6305–6315. doi: 10.1007/s11033-014-3513-y. [DOI] [PubMed] [Google Scholar]
- 30.Suárez-Vega A., Gutiérrez-Gil B., Klopp C., Robert-Granie C., Tosser-Klopp G., Arranz J.J. Characterization and comparative analysis of the milk transcriptome in two dairy sheep breeds using RNA sequencing. Sci. Rep. 2015;5:18399. doi: 10.1038/srep18399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J., Weinberg M.S., Zerbini L., Prince S., Bronner M. The oncogenic TBX3 is a downstream target and mediator of the TGF-β1 signaling pathway. Mol. Biol. Cell. 2013;24:3569–3576. doi: 10.1091/mbc.e13-05-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jerome-Majewska L.A., Achkar T., Luo L., Lupu F., Lacy E. The trafficking protein Tmed2/p24β1 is required for morphogenesis of the mouse embryo and placenta. Dev. Biol. 2010;341:154–166. doi: 10.1016/j.ydbio.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giudice J., Loehr J.A., Rodney G.G., Cooper T.A. Alternative splicing of four trafficking genes regulates myofiber structure and skeletal muscle physiology. Cell Rep. 2016;17:1923–1933. doi: 10.1016/j.celrep.2016.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Badou A., Jha M.K., Matza D., Mehal W.Z., Freichel M., Flockerzi V., Flavell R.A. Critical role for the β regulatory subunits of Cav channels in T lymphocyte function. Proc. Natl. Acad. Sci. USA. 2006;103:15529–15534. doi: 10.1073/pnas.0607262103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X.X., Pan J.F., Zhao Q.J., He X.H., Pu Y.B., Han J.L., Ma Y.H., Jiang L. Detecting selection signatures on the X chromosome of the Chinese Debao pony. J. Anim. Breed. Genet. 2018;135:84–92. doi: 10.1111/jbg.12314. [DOI] [PubMed] [Google Scholar]
- 36.Jäderkvist Fegraeus K., Velie Brandon D., Axelsson J., Ang R., Hamilton Natasha A., Andersson L., Meadows Jennifer R.S., Lindgren G. A potential regulatory region near the EDN3 gene may control both harness racing performance and coat color variation in horses. Physiol. Rep. 2018;6:e13700. doi: 10.14814/phy2.13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang S.M., Kim J.W., Kim C.H., An J.H., Johnson A., Song P.I., Rhee S., Choi K.H. KAT5-mediated SOX4 acetylation orchestrates chromatin remodeling during myoblast differentiation. Cell Death Dis. 2015;6:e1857. doi: 10.1038/cddis.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo K.W., Roh K.H., Bhandari D.R., Park S.B., Lee S.K., Kang K.S. ZNF281 knockdown induced osteogenic differentiation of human multipotent stem cells in vivo and in vitro. Cell Transplant. 2013;22:29–40. doi: 10.3727/096368912X654948. [DOI] [PubMed] [Google Scholar]
- 39.Makvandi-Nejad S., Hoffman G.E., Allen J.J., Chu E., Gu E., Chandler A.M., Loredo A.I., Bellone R.R., Mezey J.G., Brooks S.A. Four loci explain 83% of size variation in the horse. PLoS ONE. 2012;7:e39929. doi: 10.1371/journal.pone.0039929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metzger J., Rau J., Naccache F., Bas Conn L., Lindgren G., Distl O. Genome data uncover four synergistic key regulators for extremely small body size in horses. BMC Genom. 2018;19:492. doi: 10.1186/s12864-018-4877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metzger J., Gast A.C., Schrimpf R., Rau J., Eikelberg D., Beineke A., Hellige M., Distl O. Whole-genome sequencing reveals a potential causal mutation for dwarfism in the Miniature Shetland pony. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2017;28:143–151. doi: 10.1007/s00335-016-9673-4. [DOI] [PubMed] [Google Scholar]
- 42.Eberth J.E., Graves K.T., MacLeod J.N., Bailey E. Multiple alleles of ACAN associated with chondrodysplastic dwarfism in Miniature horses. Anim. Genet. 2018;49:413–420. doi: 10.1111/age.12682. [DOI] [PubMed] [Google Scholar]
- 43.Frischknecht M., Jagannathan V., Plattet P., Neuditschko M., Signer-Hasler H., Bachmann I., Pacholewska A., Drögemüller C., Dietschi E., Flury C., et al. A non-synonymous HMGA2 variant decreases height in Shetland ponies and other small horses. PLoS ONE. 2015;10:e0140749. doi: 10.1371/journal.pone.0140749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metzger J., Schrimpf R., Philipp U., Distl O. Expression levels of LCORL are associated with body size in horses. PLoS ONE. 2013;8:e56497. doi: 10.1371/journal.pone.0056497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norton E.M., Avila F., Schultz N.E., Mickelson J.R., Geor R.J., McCue M.E. Evaluation of an HMGA2 variant for pleiotropic effects on height and metabolic traits in ponies. J. Vet. Intern. Med. 2019;33:942–952. doi: 10.1111/jvim.15403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tetens J., Widmann P., Kühn C., Thaller G. A genome-wide association study indicates LCORL/NCAPG as a candidate locus for withers height in German Warmblood horses. Anim. Genet. 2013;44:467–471. doi: 10.1111/age.12031. [DOI] [PubMed] [Google Scholar]
- 47.Bartholazzi Junior A., Quirino C.R., Vega W.H.O., Rua M.A.S., David C.M.G., Jardim J.G. Polymorphisms in the LASP1 gene allow selection for smaller stature in ponies. Livest. Sci. 2018;216:160–164. doi: 10.1016/j.livsci.2018.07.015. [DOI] [Google Scholar]
- 48.Srikanth K., Kim N.Y., Park W., Kim J.M., Kim K.D., Lee K.T., Son J.H., Chai H.H., Choi J.W., Jang G.W., et al. Comprehensive genome and transcriptome analyses reveal genetic relationship, selection signature, and transcriptome landscape of small-sized Korean native Jeju horse. Sci. Rep. 2019;9:16672. doi: 10.1038/s41598-019-53102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leegwater P.A., Vos-Loohuis M., Ducro B.J., Boegheim I.J., van Steenbeek F.G., Nijman I.J., Monroe G.R., Bastiaansen J.W.M., Dibbits B.W., van de Goor L.H., et al. Dwarfism with joint laxity in Friesian horses is associated with a splice site mutation in B4GALT7. BMC Genom. 2016;17:839. doi: 10.1186/s12864-016-3186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orr N., Back W., Gu J., Leegwater P., Govindarajan P., Conroy J., Ducro B., Van Arendonk J.A.M., MacHugh D.E., Ennis S., et al. Genome-wide SNP association–based localization of a dwarfism gene in Friesian dwarf horses. Anim. Genet. 2010;41:2–7. doi: 10.1111/j.1365-2052.2010.02091.x. [DOI] [PubMed] [Google Scholar]
- 51.Kader A., Li Y., Dong K., Irwin D.M., Zhao Q., He X., Liu J., Pu Y., Gorkhali N.A., Liu X., et al. Population variation reveals independent selection toward small body size in Chinese Debao pony. Genome Biol. Evol. 2016;8:42–50. doi: 10.1093/gbe/evv245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emechebe U., Rozenberg J.M., Moore B., Firment A., Mirshahi T., Moon A.M. T-box3 is a ciliary protein and regulates stability of the Gli3 transcription factor to control digit number. eLife. 2016;5:e07897. doi: 10.7554/eLife.07897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sundstrom E., Imsland F., Mikko S., Wade C., Sigurdsson S., Pielberg G.R., Golovko A., Curik I., Seltenhammer M.H., Solkner J., et al. Copy number expansion of the STX17 duplication in melanoma tissue from Grey horses. BMC Genom. 2012;13:365. doi: 10.1186/1471-2164-13-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Snippert H.J., Haegebarth A., Kasper M., Jaks V., van Es J.H., Barker N., van de Wetering M., van den Born M., Begthel H., Vries R.G., et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 55.Ansar M., Chung H.-l., Al-Otaibi A., Elagabani M.N., Ravenscroft T.A., Paracha S.A., Scholz R., Abdel Magid T., Sarwar M.T., Shah S.F., et al. Bi-allelic variants in IQSEC1 cause intellectual disability, developmental delay, and short stature. Am. J. Hum. Genet. 2019;105:907–920. doi: 10.1016/j.ajhg.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pousada G., Baloira A., Vilariño C., Cifrian J.M., Valverde D. Novel mutations in BMPR2, ACVRL1 and KCNA5 genes and hemodynamic parameters in patients with pulmonary arterial hypertension. PLoS ONE. 2014;9:e100261. doi: 10.1371/journal.pone.0100261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sasaki D., Kotoh J., Watadani R., Matsumoto K. New animal models reveal that coenzyme Q2 (Coq2) and placenta-specific 8 (Plac8) are candidate genes for the onset of type 2 diabetes associated with obesity in rats. Mamm. Genome. 2015;26:619–629. doi: 10.1007/s00335-015-9597-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the Illumina sequencing reads have been deposited in the National Genomics Data Center (https://bigd.big.ac.cn) with the accession code (BioProject ID: PRJCA001875).