Abstract
Simple Summary
Identification of mutations in the myostatin gene, affecting the occurrence of the double muscling phenotype in some breeds of beef cattle, was an impetus for further analysis and identification of mutations within this gene in other animal breeds, characterized by increased meat performance parameters. The number of geese in poultry livestock production in Poland is small. The native geese breeds can be successfully used to produce high-quality poultry meat and can be a very good source of goose meat production for regional and organic products. The aim of the study was to identify a mutation in the MSTN (Myostatin) gene and investigate whether this polymorphism can affect body weight in different periods of life in Landes and Kielecka breeds. Measurements of the examined trait were taken with time intervals to demonstrate the putative effect of the identified SNP (Single Nucleotide Polymorphism) on body weight over the course of bird growth. In conclusion, the identified c.1231C>T polymorphism suggests a possible link between the polymorphism and the BW (body weight) of Kielecka geese in the 12th week of life. The most significant factors affecting the BW values in geese are breed and sex.
Abstract
Myostatin, also known as growth differentiation factor 8 (GDF8), belongs to the TGF- superfamily of proteins. MSTN is a highly conserved protein that acts as a negative regulator of skeletal muscle growth. Loss of myostatin functionality causes the phenotype to appear in the form of ‘double musculature’, among others in cattle, sheep, and house mice. The presented results of the research were carried out on two geese breeds—Landes and Kielecka. The aim of the study was to identify mutations in the MSTN gene and study their impact on body weight in both geese breeds in different periods of life. Analysis of the obtained results showed the existence of polymorphism in exon 3 (c.1231C>T) and suggested a possible association (p < 0.05) between BW and genotype in 12 weeks of life in male Kielecka geese breed. The identified polymorphism may be one of the factors important for improving body weight in the studied Kielecka breed, therefore, it is necessary to conduct further research on a larger population of geese breeds in order to more accurately estimate the effect of the identified SNP c.1231C>T on BW in geese.
Keywords: polymorphism, geese, kielecka, landes, MSTN, myostatin
1. Introduction
Proteins belonging to the transforming growth factors beta superfamily (TGF-β) include three isoforms (TGF-β1, -β 2 and -β3), activin and inhibin, growth factors (GDF), bone morphogenetic proteins (BMPs), and anti-Mullerian hormone (AMH), which consists of more than 35 proteins [1]. These proteins play a significant role in embryogenesis in mammals, amphibians, and insects, as well as in bone development, wound healing, hematopoiesis, and in the body’s response to inflammation [2,3,4,5]. Together with growth factors—GDF9 and GDF9b (BMP15)—they also play a significant role in mammal reproductive processes [6,7,8,9]. Myostatin (MSTN), also known as growth and differentiation factor 8 (GDF8), belongs to the TGF-β superfamily of proteins. In pig, buffalo, cow, fish (zebrafish), birds, and domestic mouse, the myostatin gene consists of 3 exons separated from each other by 2 introns and encodes 376 amino acid precursor protein [10]. Myostatin is expressed not only in skeletal muscle but also in the heart and adipose cells of adult animals [11]. Myostatin acts as a negative regulator of muscle growth and is associated with the growth and differentiation of the skeletal muscles of animals [12,13,14,15,16]. The loss of functionality of the MSTN gene caused by mutation occurring in the STOP codon resulted in the double-muscled phenotype in some breeds of beef cattle [12,17] and sheep [13,14,15]. The mutations identified in the MSTN gene were also associated with race outcomes of dogs [16] and horses [18]. Xu et al. [19] identified three SNP polymorphisms in the MSTN gene but only two were associated with breast muscle traits in Pekin duck. The T129C SNP had a significant association with breast muscle thickness, and the second one T952C had a significant association with the fossilia ossis mastoid length. Zhao et al. [20] investigated the effect of Myostatin and Myogenin on Zi and Rhine geese skeletal muscle growth. The authors found a negative association between MSTN gene expression in breast muscle and body weight, breast muscle weight and breast muscle percentage. Recently, there has been a noticeably increased consumer interest in unprocessed, high-quality food. Recent studies proved the high value of lipids and amino acids composition of local geese breeds [21]. It is well accepted that the body weight, as well as qualitative traits in geese, are strongly dependent on genotype [22,23,24]. However, the molecular background of meat production remains to be elucidated. For this study, we choose two local goose breeds significantly different in their muscularity and body weight, as well as the high breeds diversity of these traits. Landes geese, present in Poland since 1986, constitute a phenotypically unified population distinguished by good health, good reproductive traits, and meat efficiency [25]. The second breed, Kielecka goose, is native to southern Poland, characterized by very good muscularity, low fatness but poor meat production [26]. Both breeds have been included in the genetic resources program [27].
The aim of the study was to find and investigate whether the mutation in the MSTN gene affects body weight in different periods of life in Landes and Kielecka breeds.
2. Materials and Methods
2.1. Animals
For the study, 2 geese breeds (246 geese: Kielecka (109:82 Female, 27 Male) and Landes (137:100 Female, 37 Male), significantly different in their muscularity and body weight (BW) parameters as well as the high breeds diversity of this trait were selected for the study. The geese were kept in the same environmental conditions. The birds were fed according to the breeding goose nutrition program (Table S1).
2.2. Measurements of Body Weight
The geese were weighed in WGRS NRIAP (Waterfowl Genetic Resources Station-National Research Institute of Animal Production). Measurements of body weight were made in the 8th, 12th and 95th weeks of life. Birds were weighed on a suspended Axis weight (max. 15 kg, min, 100 g). Birds were put into the scale (with their upper legs) thus that the movement of the bird was prevented—the wings were immobilized. Such a position during the moment of weighing caused relatively fast stabilization of the weight.
2.3. Molecular Analysis
DNA isolation was performed using the Sherlock AX reagent kit (A&A Biotechnology, Gdynia, Poland), according to the protocols provided by the manufacturer. The DNA material was isolated from the feathers, thus there was no need for the consent of the bioethics committee.
2.4. Polymerase Chain Reaction (PCR)
Amplification and sequencing of the 3 myostatin gene exons were performed using the primers shown in Table 1. The PCR reaction was performed in 25 μL volume containing: 12 µL of PCR-grade water, 2.5 µL of PCR buffer with 15 mM MgCl2 (QIAGEN, Hilden, Germany), 5 µL of Q-Solution (5x; QIAGEN), 3 µL of 10 mM dNTPs (APPLIED BIOSYSTEMS, Foster City, CA, USA), 0.25 µL of primer mix (each 100 pmol/µL) (Table 1), 0.25 µL of HotStarTaq DNA polymerase (5 U/µL QIAGEN) and 2 µL of DNA isolate (50ng/ µL). The PCR thermal program was as follows: 15 min of initial activation step at 95 °C (Polymerase is activated by this heating step), 35 cycles of denaturation at 95 °C for 30 s, annealing at 61 °C for 59 s and primer extension at 72 °C for 120 s. The final extension was conducted at 72 °C for 10 min.
Table 1.
Primers used for the amplification and sequencing of the myostatin gene of the Landes and Kielecka geese.
| Primer Name | Primer Sequence | Tm * (°C) | Product Length (bp) | |
|---|---|---|---|---|
| MSTNex3 F | Exon 3 | 5′ TTCCGGTTCCTTTTCCTCTT 3′ | 61 | 577 |
| MSTNex3 R | 5′ TCTGCAGCTTGTGTTGCTCT 3′ | |||
| MSTNex1 F | Exon 1 | 5′ TCAGATTGCATTTGCTTTCA 3′ | 61 | 497 |
| MSTNex1 R | 5′ AGACGAAAGCAGCAGGGTTA 3′ | |||
| MSTNex2 F | Exon 2 | 5′ TTTTTGTTCCCTGTTCAGTAATC 3′ | 61 | 473 |
| MSTNex2 R | 5′ TGCTTTCCAATAAAATGCAAGA 3′ | |||
* PCR annealing temperature.
2.5. Sequencing Method
The PCR product was purified using the EXOSAP—it enzyme (Affymetrix, Santa Clara, CA, USA) and sequenced from both complementary strands using the BigDye Terminator v3.1 sequencing kit (Thermo Fisher Scientific, Waltham, MA, USA) and primers used for PCR reaction separately for forward and reverse primers. The reading of the sequencing reactions was carried out in the 3130xl Genetic Analyzer capillary sequencer (Applied Biosystems, Foster City, CA, USA).
2.6. Alignment
The obtained sequences between individuals of the same breed as well as between breeds were aligned with the goose reference sequence (GenBank No. XM_013178647). We also did the alignment of the MSTN 3 exon for a consensus sequence of the Kielecka and Landes, as well as the goose reference sequence. The BioEdit Sequence Alignment Editor program was used [28].
2.7. Statistical Analyses
Associations between the analyzed features and the examined SNP polymorphism were estimated using the GLM model, using the Duncan multiple range tests. The differences within, between breeds, sexes, and genotypes were estimated according to the statistical model:
| Yijkl = ai + bj + ck | (1) |
where ai = breed (i = Kielecka, Landes), bj = sex (j = male, female), ck = genotype (k = CC, CT, TT).
The Hardy–Weinberg equilibrium was tested using the Court Lab-HW calculator (Michael H. Court (2005–2008)).
2.8. Phylogenetic Analyses
To assess genetic distance of analyzed breeds and other bird species, sequences of MSTN gene of Kielecka and Landes breed as well sequences from GeneBank database for duck (Anas platyrhynchos, GeneBank Accession No XM_005011412.4), chicken (Gallus gallus, GeneBank Accession No NM_001001461.1), and pigeon (Columbia Livia, GeneBank Accession No NM_001282809.1) were used. The MEGA Molecular Evolutionary Genetics A software (MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets) was used to calculate genetic distance based on Tamura–Nei analysis model and construct maximum likelihood phylogenetic tree [29].
3. Results
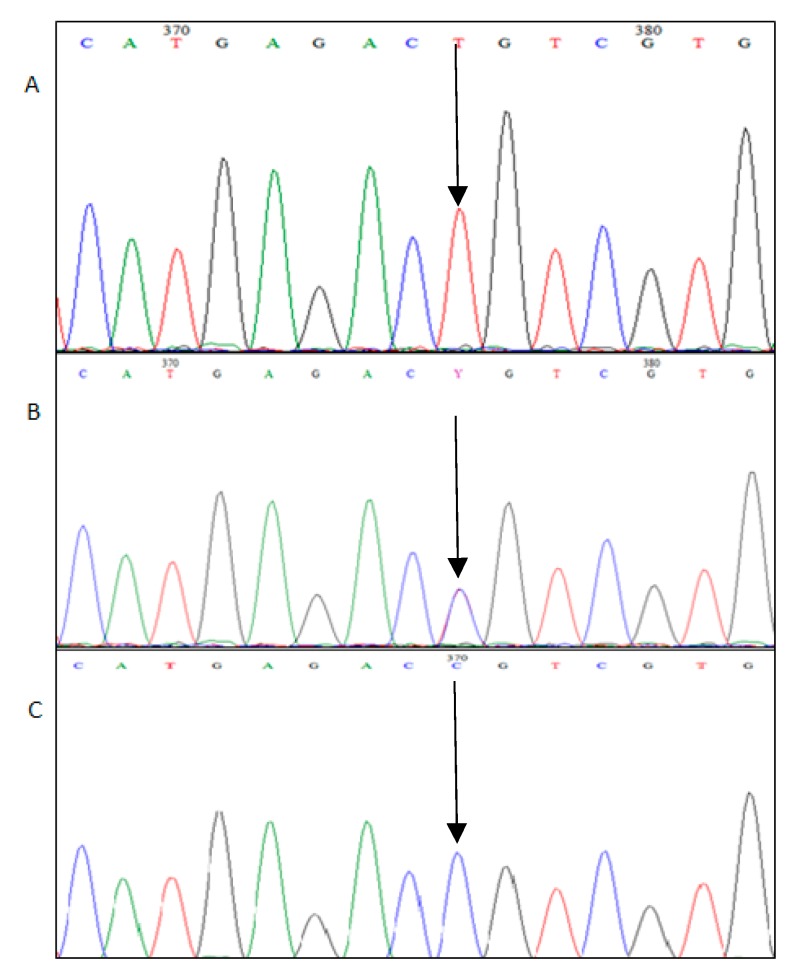
Analysis of the obtained sequences allowed us to identify one mutation in both geese breeds. The SNP c.1231C>T (Figure 1) is a synonymous mutation in 3′ UTR (untranslated region) of the MSTN gene. The mutation depicted in Figure 2 presents the alignment of the MSTN 3 exon fragment of the consensus Kielecka/Landes sequence with the reference sequence (GenBank No. XM_013178647).
Figure 1.
Chromatogram fragment of the Myostatin (MSTN) exon3 containing-c.1231C → T polymorphism. (A) Homozygote TT, (B) heterozygote C/T, (C) homozygote CC.
Figure 2.
Alignment of the fragment of the MSTN gene with SNP c.1231 C>T marked in red.
In Table 2, allele frequencies and genotypes are shown. Both populations are in Hardy–Weinberg equilibrium (HWE) (p < 0.05).
Table 2.
Frequency of c.1231 C>T alleles and genotypes in the MSTN gene in the Landes and Kielecka breeds and p-values for deviation from the Hardy–Weinberg equilibrium.
| Breed | Genotypes | Alleles | HWE p-Value | |||
|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | ||
| Landes | 0.292 | 0.496 | 0.212 | 0.54 | 0.46 | 0.39 |
| Kielecka | 0.248 | 0.458 | 0.294 | 0.477 | 0.523 | 0.99 |
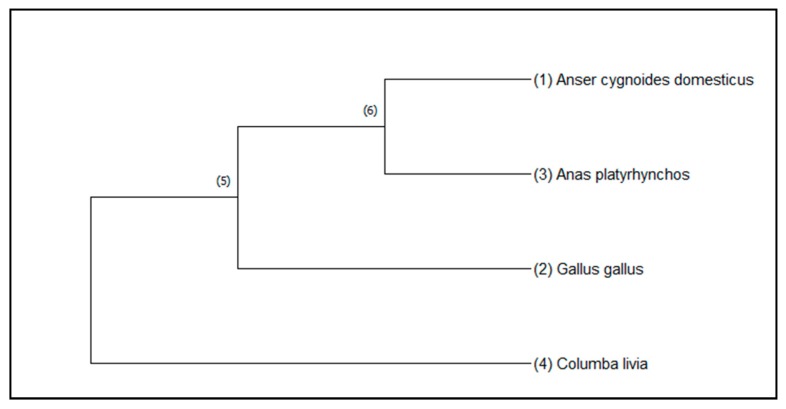
The phylogenetic analysis of a fragment of the MSTN exon3 of two goose breeds studied, and the sequences of duck, chicken, and pigeon are depicted in Figure 3. The closest evolutionary relationship was revealed for goose and duck and for these both species common ancestors are chicken and pigeon.
Figure 3.
Unrooted maximum likelihood tree constructed from the Tamura–Nei model of the MSTN sequences of Kielecka and Landes breed (Anser cygnoides domesticus), Anas platyrhynchos (duck), Gallus gallus (chicken), Columbia Livia (pigeon).; bootstraps of 1000 replicates.
Association Analysis of SNP with Body Weight
The results for BW measurement for each breed, sex, and genotype are shown in Table 3 for the age of 8 weeks, in Table 4 for the age of 12 weeks, and in Table 5 for the age of 95 weeks. The association analyses between the MSTN gene polymorphism and body weight trait for the Landes and Kielecka geese are shown in Figure 4, Figure 5 and Figure 6. All studied groups were analyzed by age, breed, sex, and genotype.
Table 3.
Average measurement results at 8 weeks of life split by genotype, sex, and breed.
| Genotype | Sex | Breed | BW 8 Week [g] | std | N | Min [g] | Max [g] |
|---|---|---|---|---|---|---|---|
| CC | F | KIELECKA | 3051 | 352 | 24 | 2240 | 4030 |
| CC | F | LANDES | 3678 | 264 | 29 | 3340 | 4610 |
| CC | M | KIELECKA | 3623 | 248 | 3 | 3340 | 3800 |
| CC | M | LANDES | 4032 | 318 | 11 | 3660 | 4630 |
| CT | F | KIELECKA | 3043 | 253 | 33 | 2470 | 3700 |
| CT | F | LANDES | 3732 | 237 | 49 | 3250 | 4240 |
| CT | M | KIELECKA | 3479 | 270 | 17 | 2960 | 4000 |
| CT | M | LANDES | 4245 | 289 | 19 | 3720 | 4780 |
| TT | F | KIELECKA | 3079 | 256 | 24 | 2610 | 3750 |
| TT | F | LANDES | 3665 | 298 | 22 | 3060 | 4260 |
| TT | M | KIELECKA | 3440 | 342 | 7 | 2960 | 4040 |
| TT | M | LANDES | 4246 | 495 | 7 | 3760 | 5070 |
Table 4.
Average measurement results at 12 weeks of life split by genotype, sex, and breed.
| Genotype | Sex | Breed | BW 12 Week [g] | std | N | Min [g] | Max [g] |
|---|---|---|---|---|---|---|---|
| CC | F | KIELECKA | 3540 | 285 | 24 | 3090 | 4400 |
| CC | F | LANDES | 4363 | 305 | 29 | 3930 | 5510 |
| CC | M | KIELECKA | 4477 | 536 | 3 | 3960 | 5030 |
| CC | M | LANDES | 4989 | 381 | 11 | 4170 | 5400 |
| CT | F | KIELECKA | 3463 | 272 | 33 | 2900 | 4070 |
| CT | F | LANDES | 4287 | 717 | 49 | 4195 | 5070 |
| CT | M | KIELECKA | 4053 | 357 | 17 | 3470 | 4770 |
| CT | M | LANDES | 5233 | 326 | 19 | 4760 | 6010 |
| TT | F | KIELECKA | 3471 | 308 | 24 | 2480 | 4000 |
| TT | F | LANDES | 4405 | 434 | 22 | 3400 | 5100 |
| TT | M | KIELECKA | 3843 | 315 | 7 | 3320 | 4370 |
| TT | M | LANDES | 5207 | 747 | 7 | 4350 | 6460 |
Table 5.
Average measurement results at 95 weeks of life split by genotype, sex, and breed.
| Genotype | Sex | Breed | BW 95 Week [g] | std | N | Min [g] | Max [g] |
|---|---|---|---|---|---|---|---|
| CC | F | KIELECKA | 4446 | 289 | 24 | 3935 | 5070 |
| CC | F | LANDES | 6152 | 697 | 29 | 4260 | 7945 |
| CC | M | KIELECKA | 4905 | 748 | 3 | 4160 | 5655 |
| CC | M | LANDES | 5776 | 451 | 11 | 5145 | 6425 |
| CT | F | KIELECKA | 4581 | 519 | 33 | 3555 | 5695 |
| CT | F | LANDES | 6345 | 625 | 49 | 5340 | 8260 |
| CT | M | KIELECKA | 4673 | 405 | 17 | 3940 | 5400 |
| CT | M | LANDES | 5968 | 469 | 19 | 5275 | 6870 |
| TT | F | KIELECKA | 4489 | 383 | 25 | 3890 | 5265 |
| TT | F | LANDES | 6191 | 973 | 22 | 4445 | 9085 |
| TT | M | KIELECKA | 4659 | 467 | 7 | 4250 | 5625 |
| TT | M | LANDES | 6236 | 722 | 7 | 5520 | 7430 |
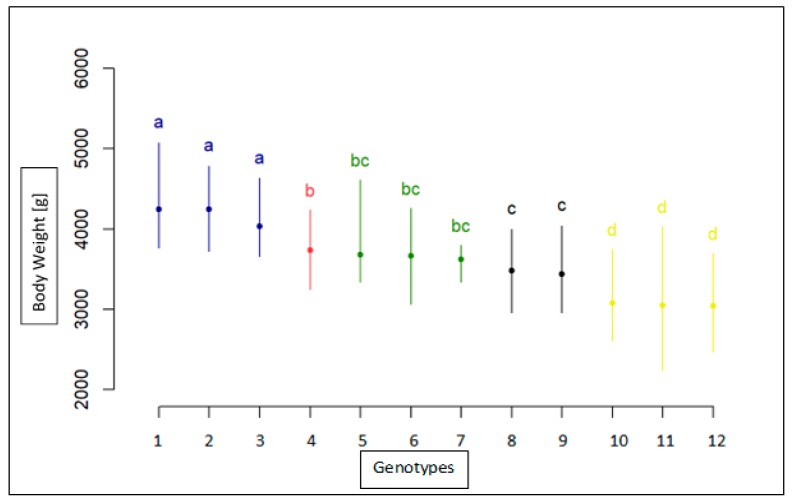
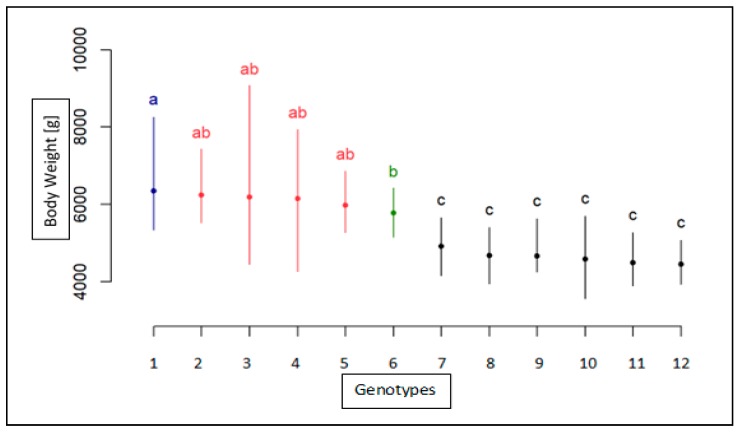
Figure 4.
Statistical characteristics of association of c.1231C>T with body weight in 8-week old geese both breeds(1-TT(Male) Landes, 2-CT(Male)Landes, 3-CC(Male)Landes, 4-CT(Female)Landes, 5-CC(Female)Landes, 6-TT(Female)Landes, 7-CC(Male)Kielecka, 8-CT(Male)Kielecka, 9-TT(Male)Kielecka, 10-TT(Female)Kielecka, 11-CC(Female)Kielecka, 12-CT(Female)Kielecka). Genotypes with different letters show significant differences between groups (a, b, c, d: p ≤0.05).
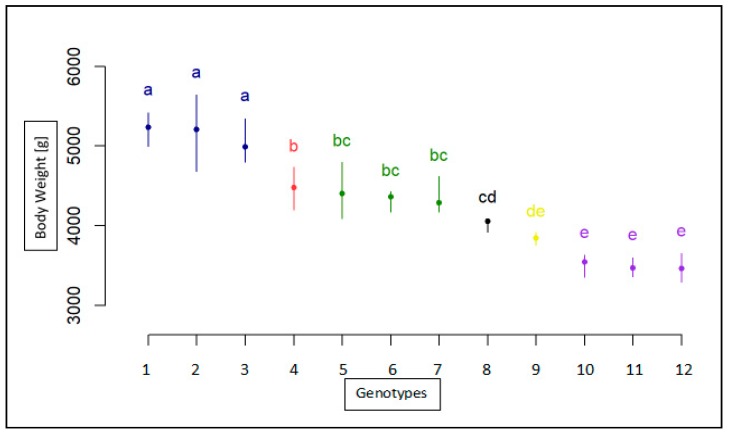
Figure 5.
Statistical characteristics of association of c.1231C>T with body weight in 12-week old geese both breeds(1-CT(Male) Landes, 2-TT(Male)Landes, 3-CC(Male)Landes, 4-CC(Male)Kielecka, 5-TT(Female)Landes, 6-CC(Female)Landes, 7-CT(Female)Landes, 8-CT(Male)Kielecka, 9-TT(Male)Kielecka, 10-CC(Female)Kielecka, 11-TT(Female)Kielecka, 12-CT(Female)Kielecka). Genotypes with different letters show significant differences between groups (a, b, c, d, e: p ≤0.05).
Figure 6.
Statistical characteristics of association of c.1231C>T with body weight in 95-week old geese both breeds (1-CC(Female) Kielecka, 2-CC(Female)Landes, 3-CC(Male)Kielecka, 4-CC(Male)Landes, 5-CT(Female)Kielecka, 6-CT(Female)Landes, 7-CT(Male)Kielecka, 8-CT(Male)Landes, 9-TT(Female)Kielecka, 10-TT(Female)Landes, 11-TT(Male)Kielecka, 12-TT(Male)Landes). Genotypes with different letters show significant differences between groups (a, b, c: p ≤0.05).
In the 8 weeks of life, the male Landes and Kielecka geese were heavier than females of each breed (p < 0.05) and females of Landes were of similar weight as males of Kielecka (p < 0.05) (Table 3, Figure 4). However, there was no association between genotypes and each BW measurement.
At 12 weeks of age, the highest BW measurements were found in Landes males whereas the lowest BW values were noted for Kielecka females. These two groups were significantly different (p < 0.05). For both groups, there was no association between the BW results and the MSTN genotype. The CC homozygotes of Kielecka males and Landes females of all genotypes were of similar weight. The Kielecka male geese with CC genotypes were significantly heavier than individuals with CT and TT genotypes (p < 0.05) (Figure 5). In Table 4, the results of weight measurements with division into breeds and genotypes are shown.
As was depicted in Figure 6, in the 95th week, statistically significant differences in body weight between the breeds were observed (p < 0.05). In the Landes breed, the greatest differences were found between females with CT genotype and males with CC genotype (p < 0.05). Moreover, in the Kielecka breed, there was no statistically significant difference between males and females. In Table 5, the results of body weight in both geese breeds were shown.
4. Discussion
The participation of geese in poultry livestock production in Poland is small. This structure is similar to that observed in the European Union and is mainly caused by the stabilization of meat consumption of particular bird species. In Poland, goose meat production is mainly carried out with the use of W-31 hybrids, derived from the breeding set ♂ W-33 × ♀ W-11, i.e., based on the highly efficient Kołudzka white goose [30]. Recently, there has been a noticeably increased consumer interest in unprocessed, high-quality food. This trend is reflected in food products from small farms, the production of which is often based on the use of breeds with lower yield but characterized by increased resistance on diseases and environmental conditions and good use of feed.
The identification of mutations in the myostatin gene affecting the occurrence of the double-muscled phenotype in some breeds of beef cattle was the impetus for further analysis and identification of mutations within this gene in other animal breeds characterized by increased parameters of meat performance. In the case of cattle, the presence of deletion of 11 nucleotides in the 3 exons of the MSTN gene in homozygous form resulted in muscle hypertrophy presence in Belgian Blue cattle [12]. In sheep of different breeds, the c.1232G>A polymorphism in the 3 ‘UTR region was identified causing a change in the amino acid sequence to the stop codon, resulting in decreased myostatin production and, consequently, increased muscularity in sheep [13,14,15]. In dogs, the 939-940delTG deletion results in a change of cysteine to the stop codon. Dogs with this mutation are characterized by better results in races [16]. In addition, in the case of racehorses, the effect of g.66493737C>T mutation in MSTN on the results of races at different distances was demonstrated [18]. In pigs, 15 polymorphic sites have been identified, and these mutations are associated with the weight of piglets at birth and daily BW gains during the 60–100th day of fattening [31].
In the case of poultry, mutations in the MSTN gene were identified in chickens selected for meatiness (3556T>C; 3581T>A) and egg-laying performance (3360T>C; 3412A>G; 3533A>G; 3624A>T; 3656A>G) [32]. Zhang et al. [33] suggested that the mutation G2283A, detected in MSTN exon 1, has the potential as a genetic marker for body weight traits in the Bian chicken. Five polymorphisms at the 5′ and 3′ promoter sites of the MSTN gene were shown to influence bone growth and fat metabolism [34]. In ducks of Pekin breed, the 129T>C polymorphism was associated with the thickness of the pectoral muscle [19]. In studies carried out by Zhong [20], 6 polymorphisms were identified in the 3 exons of the MSTN gene in Sansui ducks, of which g.106>A was associated with slaughter traits.
In our research, no polymorphism was identified in exon 1 or 2 of the MSTN gene. We found, however, a polymorphism c.1231C>T in the exon 3 of the MSTN gene. The SNP polymorphism was localized 3 bp below CDS (coding DNA sequence) in the 3′ UTR region, which is responsible for translation control [35].
Statistical analyses showed significant differences (p ≤ 0.05) between breeds and between sexes within breeds for the studied trait. The obtained results demonstrate the high impact of breed and sex on body weight in the studied geese breeds, which is consistent with studies conducted elsewhere [22,23]. Based on the analysis of genotypes and the occurrence of significant statistical relationship for the characteristics of BW, the identified polymorphism cannot be clearly linked to the body weight of the studied geese breeds. However, it is believed that conducting further research on a larger population of different geese breeds could estimate more accurately the effect of the identified SNP c.1231C>T on body weight in geese.
5. Conclusions
In conclusion, the identified c.1231C>T polymorphism suggests a possible link between the body weight of Kielecka geese in the 12th week of life. The most significant factors affecting the BW values in geese are breed and sex.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/10/1/10/s1, Table S1: Nutrient contents of feed mixtures.
Author Contributions
Conceptualization, G.S., A.K.-S., A.K.; methodology, G.S., A.K.; software, K.Ż.; validation, K.Ż.; formal analysis, G.S., A.K., A.K.-S., K.Ż.; investigation, G.S., A.K.; resources, G.S., A.K.-S.; data Curation, G.S., K.Ż., M.L., B.G.; writing—original draft Preparation, G.S.; writing—review and editing, G.S., A.K., A.K.-S.; Supervision, A.K.-S. and G.S.; project administration, G.S., A.K.-S.; funding acquisition, A.K.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research Institute grant 04-18-05-11.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Walton K.L., Johnson K.E., Harrison C.A. Targeting TGF-β Mediated SMAD Signaling for the Prevention of Fibrosis. Front. Pharmacol. 2017;8 doi: 10.3389/fphar.2017.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winnier G., Blessing M., Labosky P.A., Hogan B.L. Bone Morphogenetic Protein-4 is Required for Mesoderm Formation and Patterning in the Mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 3.Hogan B.L. Bone Morphogenetic Proteins: Multifunctional Regulators of Vertebrate Development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 4.Letterio J.J., Roberts A.B. Regulation of Immune Responses by TGF-β. Annu. Rev. Immunol. 1998;16:137–161. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- 5.Massagué J. TGF-β Signal Transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 6.Laitinen M., Vuojolainen K., Jaatinen R., Ketola I., Aaltonen J., Lehtonen E., Heikinheimo M., Ritvos O. A Novel Growth Differentiation Factor-9 (GDF-9) Related Factor Is Co-Expressed with GDF-9 in Mouse Oocytes during Folliculogenesis. Mech. Dev. 1998;78:135–140. doi: 10.1016/S0925-4773(98)00161-0. [DOI] [PubMed] [Google Scholar]
- 7.Aaltonen J., Laitinen M.P., Vuojolainen K., Jaatinen R., Horelli-Kuitunen N., Seppä L., Louhio H., Tuuri T., Sjöberg J., Bützow R., et al. Human Growth Differentiation Factor 9 (GDF-9) and Its Novel Homolog GDF-9B Are Expressed in Oocytes during Early Folliculogenesis. J. Clin. Endocrinol. Metab. 1999;84:2744–2750. doi: 10.1210/jc.84.8.2744. [DOI] [PubMed] [Google Scholar]
- 8.Elvin J.A., Yan C., Matzuk M.M. Oocyte-Expressed TGF-β Superfamily Members in Female Fertility. Mol. Cell. Endocrinol. 2000;159:1–5. doi: 10.1016/S0303-7207(99)00185-9. [DOI] [PubMed] [Google Scholar]
- 9.Smołucha G., Piestrzy A. Genetyczny aspekt wysokiej plenności u owiec. Cz. II. Wiadomości Zootechniczne. 2012;2:5–10. [Google Scholar]
- 10.Bellinge R.H.S., Liberles D.A., Iaschi S.P.A., O’Brien P.A., Tay G.K. Myostatin and Its Implications on Animal Breeding: A Review. Wiley Online Library. [(accessed on 26 September 2019)]; doi: 10.1111/j.1365-2052.2004.01229.x. Available online: https://onlinelibrary.wiley.com/doi/full/10.1111/j.1365-2052.2004.01229.x. [DOI] [PubMed]
- 11.Ahad W.A., Andrabi M., Beigh S.A., Bhat R.A., Shah R.A. Applications of Myostatin (MSTN) Gene in the Livestock Animals and Humans: A Review. Int. J. Curr. Microbiol. App. Sci. 2017 doi: 10.20546/ijcmas.2017.609.222. [DOI] [Google Scholar]
- 12.Kambadur R., Sharma M., Smith T.P., Bass J.J. Mutations in Myostatin (GDF8) in Double-Muscled Belgian Blue and Piedmontese Cattle. Genome Res. 1997;7:910–916. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- 13.Clop A., Marcq F., Takeda H., Pirottin D., Tordoir X., Bibé B., Bouix J., Caiment F., Elsen J.-M., Eychenne F., et al. A Mutation Creating a Potential Illegitimate MicroRNA Target Site in the Myostatin Gene Affects Muscularity in Sheep. Nat. Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 14.Hadjipavlou G., Matika O., Clop A., Bishop S.C. Two Single Nucleotide Polymorphisms in the Myostatin (GDF8) Gene Have Significant Association with Muscle Depth of Commercial Charollais Sheep. Anim. Genet. 2008;39:346–353. doi: 10.1111/j.1365-2052.2008.01734.x. [DOI] [PubMed] [Google Scholar]
- 15.Han J., Forrest R.H., Hickford J.G.H. Genetic Variations in the Myostatin Gene (MSTN) in New Zealand Sheep Breeds. Mol. Biol. Rep. 2013;40:6379–6384. doi: 10.1007/s11033-013-2752-7. [DOI] [PubMed] [Google Scholar]
- 16.Mosher D.S., Quignon P., Bustamante C.D., Sutter N.B., Mellersh C.S., Parker H.G., Ostrander E.A. A Mutation in the Myostatin Gene Increases Muscle Mass and Enhances Racing Performance in Heterozygote Dogs. PLoS Genet. 2007;3:e79. doi: 10.1371/journal.pgen.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McPherron A.C., Lee S.-J. Double Muscling in Cattle Due to Mutations in the Myostatin Gene. Proc. Natl. Acad. Sci. USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill E.W., Gu J., Eivers S.S., Fonseca R.G., McGivney B.A., Govindarajan P., Orr N., Katz L.M., MacHugh D. A Sequence Polymorphism in MSTN Predicts Sprinting Ability and Racing Stamina in Thoroughbred Horses. PLoS ONE. 2010;5:e8645. doi: 10.1371/annotation/de9e11b9-eb92-4ee5-a56a-908e06d1ed6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu T.S., Gu L.H., Zhang X.H., Ye B.G., Liu X.L., Hou S.S. Characterization of Myostatin Gene (MSTN) of Pekin Duck and the Association of Its Polymorphism with Breast Muscle Traits. Genet. Mol. Res. 2013;12 doi: 10.4238/2013.February.28.18. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Z.-H., Li H., Yi H.-J., Peng B.-X. The Correlation Between Polymorphisms of the MSTN Gene and Slaughter Traits in Sansui Ducks. Pak. J. Zool. 2016;48:1283–1290. [Google Scholar]
- 21.Haraf G., Wołoszyn J., Okruszek A., Orkusz A., Wereńska M. Nutritional Value of Proteins and Lipids in Breast Muscle of Geese from Four Different Polish Genotypes. Eur. Poult. Sci. 2018;82 doi: 10.1399/eps.2018.224. [DOI] [Google Scholar]
- 22.Kapkowska E., Gumulka M., Rabsztyn A., Poltowicz K., Andres K. Comparative Study on Fattening Results of Zatorska and White Koluda (R) Geese. Ann. Anim. Sci. 2011;11:207–217. [Google Scholar]
- 23.Okruszek A., Wołoszyn J., Haraf G., Orkusz A., Wereńska M. Chemical Composition and Amino Acid Profiles of Goose Muscles from Native Polish Breeds. Poult. Sci. 2013;92:1127–1133. doi: 10.3382/ps.2012-02486. [DOI] [PubMed] [Google Scholar]
- 24.Smolucha G., Kozubska-Sobocinska A., Koseniuk A., Lisowski M., Grajewski B., Zubrzycki I. Comparative Cytogenetics. Volume 12. Pensoft Publishers; Sofia, Bulgaria: 2018. Identification of SNP Polymorphism in the Growth Hormone and Myostatin Genes of the Two Polish Geese Breeds; p. 349. [Google Scholar]
- 25.National Research Institute of Animal Production (NRIAP): Geese Landes Biodiversity, Genetic Resources Programme. [(accessed on 26 September 2019)]; Available online: http://www.bioroznorodnosc.izoo.krakow.pl/drob/gesi/charakterystyka_ras/landes.
- 26.National Research Institute of Animal Production (NRIAP): Geese Kielecka Biodiversity, Genetic Resources Programme. [(accessed on 26 September 2019)]; Available online: http://www.bioroznorodnosc.izoo.krakow.pl/drob/gesi/charakterystyka_ras/kielecka.
- 27.National Research Institute of Animal Production (NRIAP): Geese Biodiversity, Genetic Resources Programme. [(accessed on 26 September 2019)]; Available online: http://www.bioroznorodnosc.izoo.krakow.pl/drob/gesi.
- 28.Hall T.A. BioEdit A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98—References—Scientific Research Publishing. [(accessed on 26 September 2019)]; Available online: https://www.scirp.org/(S(lz5mqp453edsnp55rrgjct55))/reference/ReferencesPapers.aspx?ReferenceID=1383440.
- 29.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gornowicz E., Węglarzy K., Pietrzak M., Bereza M. Slaughter and Meat Traits of Domestic Geese of the Southern Breeds. Wiad. Zootech. 2012;4:5–16. [Google Scholar]
- 31.Stinckens A., Luyten T., Bijttebier J., Van den Maagdenberg K., Dieltiens D., Janssens S., De Smet S., Georges M., Buys N. Characterization of the Complete Porcine MSTN Gene and Expression Levels in Pig Breeds Differing in Muscularity. Anim. Genet. 2008;39:586–596. doi: 10.1111/j.1365-2052.2008.01774.x. [DOI] [PubMed] [Google Scholar]
- 32.Stefaniuk M., Kaczor U., Kulisa M. MSTN gene polymorphism in livestock animals. Postepy Hig. Med. Doswiadczalnej Online. 2014;68:633–639. doi: 10.5604/17322693.1103271. [DOI] [PubMed] [Google Scholar]
- 33.Zhang G., Ding F., Wang J., Dai G., Xie K., Zhang L., Wang W., Zhou S. Polymorphism in Exons of the Myostatin Gene and its Relationship with Body Weight Traits in the Bian Chicken. Biochem. Genet. 2011;49:9–19. doi: 10.1007/s10528-010-9380-x. [DOI] [PubMed] [Google Scholar]
- 34.Wu W., Guo X., Zhang L., Hu D. Association between Single Nucleotide Polymorphisms of Fatty Acid Synthase and Fat Deposition in the Liver of the Overfed Goose. Asian Australas. J. Anim. Sci. 2014;27:1244–1249. doi: 10.5713/ajas.2013.13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matoulkova E., Michalova E., Vojtesek B., Hrstka R. The Role of the 3′ Untranslated Region in Post-Transcriptional Regulation of Protein Expression in Mammalian Cells. RNA Biol. 2012;9:563–576. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.