Abstract
Isolation of cancer cells, bacteria, and viruses from peripheral blood has important applications in cancer diagnosis, therapy monitoring, and drug development. Magnetic particles functionalized with antibodies that target receptors of cancer cells have been shown to isolate such entities using magnetic field gradients. Here, we report enhancement in capture efficiency and specificity by engineering magnetic nanoparticles and integrating them with microfluidics for the enumeration of tumor cells. Nanoparticles were made from iron oxide, coated with poly(ethylene glycol), and conjugated through avidin-biotin chemistry with antibody specifically against epithelial cell adhesion molecule (EpCAM). On exposure of targeted nanoparticles to tumor cells, specific uptake by EpCAM-expressing tumor cells (e.g., BxPC3, a pancreatic cancer cell) was observed, whereas there was negligible uptake by cells with low EpCAM expression (e.g., CCRF-CEM, a leukemia cell). Using an arrangement of magnets called a Halbach array, capture efficiency and specificity towards BxPC3 cells tagged with magnetic nanoparticles were enhanced, compared to conditions without the magnetic field gradient and/or without magnetic particles, either in buffer or in whole blood. These results illustrate that engineered magnetic nanoparticles and their integration with microfluidics have great potential for tumor cell enumeration and cancer prognosis.
Keywords: Magnetophoresis, Targeted streptavidin magnetic nanoparticles, Microfluidic device, Tumor cells, Capture
Graphical Abstract
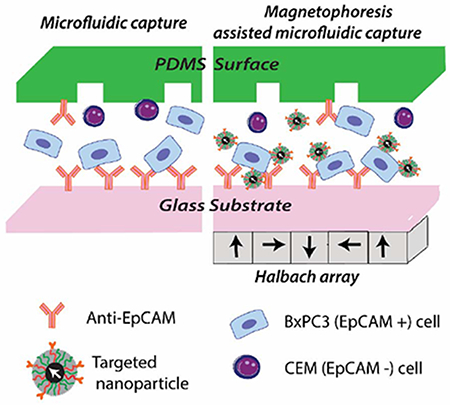
1. Introduction
Magnetophoresis, a nondestructive method for collecting or separating magnetic particles, involves the motion of magnetic particles in a viscous medium under the influence of a magnetic field gradient.1 The choice of magnetic particle, its surface functionalization, and the external field under which capture is performed are some of the critical factors in magnetophoresis.2 Magnetic beads functionalized with targeting moieties are used in blood purification3, removal of bacteria4, 5 from body fluids, and in separation of cancer cells in batch6–8 and continuous flow processes.9–11
At the micro- (<1 μm) and nano-scale (<100 nm), various particle platforms have been explored to isolate and enrich biomarkers and cells.12, 13,14 Capture using particles at the micron scale15 works efficiently in simple cell solutions as they rapidly separate due to the high magnetic moment of the microparticles, resulting in greater forces available for separation.16 However magnetic microparticles are found to be less efficient in capture of cells under flow conditions,9 which has been attributed to poor binding capacity of microparticles for receptors on cells.17 Furthermore, microparticles are often found to aggregate in biological fluids,18, 19 contributing to inefficient capture and recovery in those media. Commercial particles used for capture have also shown significant nonspecific binding,20 thereby affecting selectivity and capture efficiency.
In the ideal case of magnetophoretic capture of tumor cells under flow, one would use particles that are highly selective towards the tumor cells, with minimal interactions (surface binding or uptake) with other cells in the sample. Past studies of magnetophoretic capture of tumor cells have relied on commercial particles7, 10 or particles that are coated with mono- and polysaccharides, all of which suffer from significant non-specific binding to cells,6, 8 potentially limiting specificity. To minimize non-specific interactions with non-targeted cells, here we use magnetic nanoparticles coated with a dense brush of poly(ethylene glycol) (PEG). PEG is a so-called “stealth” polymer that reduces protein binding to the nanoparticles and improves their colloidal stability even in whole blood.21–23 To target the epithelial cell adhesion molecule (EpCAM), a commonly used diagnostic marker for cancer,24 we developed PEG coated magnetic nanoparticles that were functionalized with streptavidin, and then bound to biotinylated anti-EpCAM.
The selectivity of these targeted particles to tumor cells was tested in a microfluidic capture system. Microfluidic devices are often used to isolate and enumerate tumor cells from body fluids.25, 26 They are designed to promote collisions between cells and antibody-functionalized walls (Fig. 1 a) and/or features (e.g. pillars, nanoparticles) resulting in improved capture rates with minimal damage to cells.27–30 To improve throughput, sensitivity, and purity in capture of rare tumor cell populations from body fluids, various magnetophoresis assisted microfluidic capture platforms have been developed.31 When combining microfluidics and magnetophoresis with targeted nanoparticles, the aim is to improve cross-stream migration of cells towards the antibody functionalized surfaces in the microfluidic device, improving contact between surface bound antibodies and their target epitopes on the cell surface. Here, we explore this approach by combining an antibody functionalized herringbone microfluidic capture device with a planar Halbach array and anti-EpCAM-targeted magnetic nanoparticles to capture EpCAM expressing cells from cell mixtures (Fig. 1 b). With the magnetic field gradient generated by the Halbach array under the device, targeted magnetic nanoparticle-bound tumor cells can be forced onto the antibody-coated inner surfaces and captured. At high flow rates, the combined forces also allow for selective capture of tumor cells tagged with the particles, while the non-targeted cells are washed out due to the high flow rate.
Fig. 1.
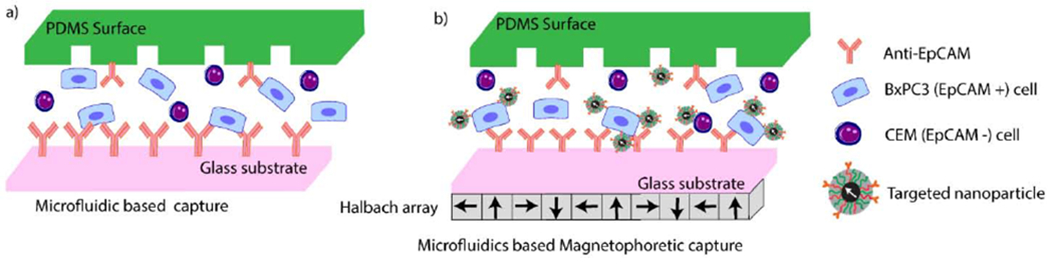
Schematic of tumor cell capture platforms. a) Antibody functionalized microfluidic chip. b) Magnetophoresis assisted microfluidic capture. The drawing is not to scale.
2. Materials and Methods
2.1. Synthesis of magnetic nanoparticles via thermal decomposition synthesis
Magnetic nanoparticles were synthesized by the semi-batch thermal decomposition of iron oleate in the presence of molecular oxygen. The precursor iron oleate was synthesized by reacting iron acetylacetonate (>98% pure, Tokyo Chemical Industry ,TCI America) and oleic acid (90% technical grade, Sigma-Aldrich) at 320°C under Argon atmosphere.32 Particle synthesis was as detailed in Unni et.al.33 A mixture of iron oleate and octadecene (90% technical grade, Sigma-Aldrich) was added in a controlled manner into docosane (90% pure, Sigma-Aldrich) at 350°C for eight hours at 9 mL/h. The synthesis was performed with oxygen feed of 20% oxygen and 80% Ar (Airgas) at a rate of 9.47 sccm, controlled using a mass flow controller (Bronkhorst USA). Additional Argon gas was introduced into the reactor head space to maintain the overall oxygen concentration below 5% and avoid flashing of the organic vapor.
2.2. Surface modification of magnetic nanoparticles
Surface modification to obtain particles with low non-specific cell binding and an antibody targeting agent was achieved by first coating the particles with a silanated poly(ethylene glycol) (PEG), then conjugating a di-carboxy PEG, binding streptavidin, and finally binding the biotinylated antibody. Characterization of the nanoparticles was conducted at each intermediate step. Section 1 of the Supporting Information file contains schematics for all the steps, which are further described below.
2.2.1. PEG coated nanoparticles
2.2.1.1. Preparation of silanated PEG
Silanated PEG was prepared by initially oxidizing commercially procured Poly(ethylene glycol) methyl ether of MW 5000 g/mol (Sigma Aldrich) using Jones reagent as described in Barrera et.al with slight modifications.21 In brief, 50 g of methoxy PEG (m-PEG) was dissolved in 400 mL acetone by heating at 50 °C till the solution turned clear. 16 mL of Jones reagent was added such that for every 1 mol of hydroxy group in PEG, Jones reagent with 2 mol of chromium trioxide (Fisher) was added for effective oxidation.34 The reaction was let to proceed for 24 hours and quenched by addition of 10 mL of isopropyl alcohol. Activated charcoal was added to remove chromium salts from reaction mixtures by adsorption of these salts. The oxidized PEG solution was vacuum filtered, and solvents were removed using a rotary evaporator (Rotovap, Buchi Corporation, USA). The reduced solution was mixed with 1 M hydrochloric acid (at 40 w/v %) and washed with ethyl ether in a separatory funnel. The aqueous layer with oxidized m-PEG was collected and extracted using methylene chloride. The organic solution was dried in the rotovap, washed with cold diethyl ether, and dried in a vacuum oven at room temperature.
Once the number of -COOH groups in the oxidized m-PEG was confirmed by Nuclear Magnetic Resonance Spectroscopy (Fig. S6 in the SI), amidation reaction between carboxyl groups in m-PEG and amine groups in aminopropyltriethoxy silane, APS (TCI, America) was performed. 50 g of mPEG–COOH was melted at 120°C and reacted with 2.33 mL APS (1:1 molar ratio) at 500 mbar in a rotovap for 2 hours to yield silanated PEG.
2.2.1.2. Ligand exchange to obtain PEG coated nanoparticles
To coat the nanoparticles with the PEG silane, 100 mg of oleic acid coated iron oxide nanoparticles in 60 ml of dry toluene (at 2.3 mg iron oxide/mL) were dissolved in 190 ml of dry toluene to obtain a concentration of 0.4 mg/mL. Then, 10.15 g (1.01 mmol) of silanated PEG dissolved in 600 mL of dry toluene. This corresponds to 10x excess of silanated PEG relative to a monolayer at 9.5*1014 ligands/cm2 based on values reported by De Palma et al.35 To catalyze the hydrolysis and condensation of silane groups onto the surface of the nanoparticles, 116.2 μL (2.03 mmol) of acetic acid was added and the solution placed in a shaker for 72 h at room temperature. The particles after ligand exchange were precipitated with diethyl ether and dried before suspending in aqueous media. The particles were purified of unbound excess polymer using a MidiMACS Separator (From Miltenyi) and filtered using a 0.22 um Milex-GP Millipore filter to obtain PEG coated nanoparticles.
2.2.2. Carboxylated PEG coated nanoparticles
2.2.2.1. Preparation of di-carboxylated PEG
Hydroxyl groups in polyethylene glycol (Sigma, MW 10000 g/mol) were converted to carboxy groups to obtain di-carboxylated polyethylene glycol by ring opening polymerization of succinic anhydride (99 % Acros organics) in the presence of DMAP (4-Dimethylaminopyridine, 99%, Fisher). In brief, 8 g of PEG were dissolved in 100 mL of 1,4-dioxane (Certified ACS, Fisher). Then, 0.39 g of 4-DMAP and 0.43 mL of triethyl amine (Fisher, 99% pure) were added to catalyze the ring opening reaction of 0.32 g of succinic acid (4 times molar excess relative to PEG) with PEG. The solvents from di-carboxylated PEG solution were removed in a rotary evaporator. The reduced solution was mixed with 1 M hydrochloric acid (40 w/v %) and washed with ethyl ether in a separatory funnel to remove the DMAP. The aqueous layer containing oxidized PEG was collected and extracted using methylene chloride. The organic solution was then dried in a rotovap, washed with cold diethyl ether and dried in a vacuum oven at room temperature.
2.2.2.2. Estimation of amine groups available for conjugation
After ligand exchange, there were amine groups available on the surface of the PEG nanoparticles arising from unmodified APS remaining during the ligand exchange reaction. The number of amine groups per particle was quantified using the 3-(4-carboxybenzoyl) quinoline-2-carboxaldehyde (CBQCA) assay, as described by the manufacturer (Thermo scientific). For this purpose, 0.25 mg of PEG coated nanoparticles suspended in sodium borate were incubated with 5 uL potassium cyanide (20 mM) and 10 uL ATTO-TAG CBQCA derivatization reagent (4mM) in a 96 well plate and allowed to react for 2 h.. Fluorescent measurements at 465/550 nm were performed in a Spectramax M5 plate reader (Molecular Devices) and calibrated against ethanolamine standards. The number of amine groups per nanoparticle before and after modifying with di-carboxylated PEG was quantified.
2.2.2.3. Conjugation of di-carboxylated PEG to PEG coated nanoparticles
For conjugation of di-carboxylated PEG, 20 mg of PEG coated nanoparticles in water were deprotonated at pH 9 (above the pKa of ~8) and the primary amine groups were reacted with a 10X molar excess of di-carboxylated PEG. First, 2.5 g (0.25 mmol, as amines were estimated at 1250 nanomoles/mg Fe) of di-carboxylated PEG were dissolved in deionized water for 30 minutes and the pH was raised to 4.5 for the deprotonation of carboxylic groups (above pKa of 2.8). For activating dicarboxylic groups, 0.047 g of 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide), EDC (0.25 mmol), and 0.054 g of sulfo- N-hydroxysuccinimide, Sulfo-NHS (0.25 mmol) were added at equimolar ratios to –COOH groups and let to react for 15 minutes. Then, PEG coated nanoparticles were added to the di-carboxylated PEG and amidation reaction was left to proceed for 3 h. The particles conjugated with dicarboxylated PEG were then separated using a MidiMACS separator (from Miltenyi) and any unreacted polymer was washed thoroughly before recovering the di-carboxylated PEG coated nanoparticles.
2.2.3. Anti-EpCAM Nanoparticles
2.2.3.1. Streptavidin coated magnetic nanoparticles
Carboxylic acid groups in 2 mg of carboxylated nanoparticles suspended in 1 mL of water were deprotonated at pH 6. EDC and Sulfo-NHS were added at 1:1 molar ratio of EDC to -COOH on particles and left to activate for 15 minutes. Then, the pH of the nanoparticles was adjusted to 9 and 135 ug (2.4 nanomoles) of streptavidin (MW 55000 g/mol, Sigma-Aldrich) was added. We aimed for 25 molecules per 52 nm carboxylated nanoparticle, assuming an overall 10% conjugation efficiency. The reaction was left for 3 h. Streptavidin coated nanoparticles were purified using a Miltenyi column.
2.2.3.2. Biotin-4-fluorescein assay
Biotin-4-fluorescein (B4F) based quenching assay was performed in order to estimate the number of streptavidin groups available for conjugating the antibody and also to confirm the conjugation of antibody to the particle.36 We took 0.125 mg (in 100 uL) of streptavidin functionalized iron oxide particles in costar black plate and 0.25 uL of 16 uM Biotin-4-Fluorescein (Fisher) in 0.1 M phosphate buffer solution was added in increments of 0.05 to 0.1 uL to the well. Fluorescent measurements were performed at excitation/emission wavelength of 494/525 nm every 2 minutes after addition of Biotin Fluorescein.
2.2.3.3. Antibody conjugation to the nanoparticles
Based on the B4F assay, the number of streptavidin (SaV) molecules per particle was estimated to be ~39. According to the manufacturer, the streptavidin used has 2.26 out of 4 functional units that can bind to a biotin molecule. Thus, we calculated that ~88 molecules could be conjugated to one particle. For every 2 mg of particles (which is 2.14x1012 particles, accounting for 35 nm nanoparticles), 320 pmol of biotinylated antibody was estimated to be used for conjugation. Mass of biotinylated Anti-EpCAM (with molecular weight 150 kDa, from eBioscience, SanDiego) corresponding to 320 pmol that would be used for the conjugations was determined as 0.048 ug. We performed the antibody conjugations by adding 10 times excess of this calculated mass. For every 2 mg of SaV nanoparticles, 0.48 ug of the antibody was added. After 2 h of conjugation, the nanoparticles were purified using a Miltenyi column to remove any unbound antibody.
2.3. Nanoparticle characterization
Physical Size Distribution:
Physical diameters (Dp) of all the nanoparticles were obtained by imaging using a Hitachi H7000 transmission electron microscope at the Interdisciplinary Center for Biotechnology Research (ICBR) of the University of Florida. Images of particles sampled on a formvar/carbon ultrathin B nickel 400 mesh grids (Electron microscopy sciences) were acquired using a Veleta charged-coupled device side-mount camera and were analyzed using ImageJ.
2.3.1. Magnetic Size Distribution:
A Quantum Design magnetic property measurement system (MPMS 3) superconducting quantum inference device (SQUID) magnetometer was used to analyze the magnetic properties of the particles. Magnetization curves at room temperature were obtained for liquid samples in a polytetrafluoroethylene sample holder with 100 μL of the iron oxide nanoparticle suspension in hexane/water. The volume-weighted median magnetic diameter (Dmv) and geometric deviation of the magnetic nanoparticle samples were determined by fitting the superparamagnetic equilibrium magnetization curve to the Langevin function, weighted using a log-normal size distribution.
2.3.2. Hydrodynamic Size Distribution:
Dynamic Light Scattering (DLS) measurements were performed using a Brookhaven Instruments BI90Plus operated at room temperature. The volume weighted lognormal distributions were plotted and the arithmetic mean and standard deviation from lognormal fits were reported.
2.4. Cell lines and blood samples
BxPC-3 cells (CRL-1687, human pancreatic adenocarcinoma) and CCRF-CEM cells (CCL-119, T-cell human acute lymphoblastic leukemia cells) were purchased from American Type Culture Collection (ATCC). Cells were cultured in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% fetal bovine serum (heat inactivated; GIBCO) and 100 units/mL penicillin–streptomycin (PS, Cellgro, Manassas, VA) and incubated at 37°C under 5% CO2 atmosphere. BxPC3 cells were grown as adherent monolayers to 90% confluency, subsequently detached with 0.05% trypsin–0.53 mM ethylenediaminetetraacetic acid (0.05%, Cellgro) and reseeded at a lower concentration for the experiments. For all experiments except visualization using fluorescence microscopy, BxPC3 cells were trypsinized and the tube was let to rotate for 30 min before treatment. Blood was donated in-house by healthy volunteers from the infirmary at student health care center at University Florida under an approved institutional review board protocol. Donations were collected in a vacutainer buffered with sodium citrate.
2.5. Flow cytometry to evaluate affinity of antibody to the cells
In order to evaluate the affinity of the antibody, 105 cells (at n = 3) were trypsinized and let to rotate at 37°C under 5% CO2 atmosphere. They were incubated with 5 uL of Allophycocyanin (ApC) tagged AntiEpCAM (eBioscience, SanDiego, CA) either immediately, 30 min, or 1 hour after trypsinization. Cells were washed after every time point twice with phosphate-buffered saline (PBS) and stored in 4°C in 1% bovine serum albumin in PBS. Flow cytometry was performed using the Canto II fluorescence-activated cell sorter (Becton Dickinson) at ICBR and data acquired was analyzed using FlowJo.
2.6. Quantification of cellular uptake of nanoparticles in BxPC3 and CCRF-CEM cells
For internalization measurements, Anti-EpCAM nanoparticles (targeted particles) and carboxylated nanoparticles (non-targeted particles) were incubated with approximately 2.5 million BxPC3 and CCRF-CEM cells at a concentration of 0.2 mg iron oxide nanoparticles per mL of cell media. Triplicate of each cell/particle mixture was let to rotate at 37 °C under 5% CO2 in the incubator for 1 h under rotation. Nanoparticle assisted enumeration of tumor cells are often performed under similar cell-particle incubation conditions.37, 38 The unbound particles were then washed from cells with PBS by centrifuging at 1250 rpm (Eppendorf Centrifuge 5430R). The cells were further stained with trypan blue and counted using an automated cell counter, Countess II FL. The pellet obtained after the wash was digested in 4 mL of concentrated nitric acid (optima grade, Fisher Scientific) for 24 hours or until all nitric acid evaporated at 101°C. The o-phenanthroline assay was performed to quantify the amount of iron taken up by each cell. In each vial, 96 μL water was added and iron was further reduced with 60 μL of hydroxylamine hydrochloride (8.06 M) for 1 hour. Next, 150 μL of 1,10-phenanthroline monohydrate (13 mM) was added to complex with Fe2+ and promoted using 98 μL of sodium acetate (1.22 M). Absorbance of the 200 μL samples was measured at 508 nm in a SpectraMax M5 microplate reader. The concentration of each sample run in triplicate was determined by relating to a calibration curve prepared using iron standard solution prepared from a Fluka iron standard for inductively coupled plasma. The limit of detection (LOD) and limit of quantification (LOQ) of the assay were 0.04 pgFe/cell and 0.13 pgFe/cell, respectively.
2.7. Visualization of nanoparticle internalization using fluorescence microscopy
For localization measurements, 20,000 BxPC3 cells were incubated with Anti-EpCAM nanoparticles (targeted particles) and carboxylated nanoparticles (non-targeted particles) tagged with AlexaFluor 568 carboxylic acid tris(triethylammonium salt (Molecular probes).The particles at 0.2 mg /mL of media was added for 1 h in Nunc Lab-TeK II 8 well chamber slide. Unbound particles were removed by washing twice with PBS and then cell nuclei were stained using 50 uL of Hoechst 33342 trichloride trihydrate (at 20 mM from life technologies) in 200 uL PBS for 10 min at 37 °C. Cells were washed twice with PBS before imaging using a Keyence BZ-X700.
2.8. Transmission Electron Microscopy (TEM) sample preparation and imaging
For electron microscopy, 2 million BxPC3 or CCRF-CEM cells were incubated with Anti-EpCAM nanoparticles (targeted particles) and carboxylated nanoparticles (non-targeted) particles for 1 hour at 0.2 mg iron oxide nanoparticle per mL of cell media let under rotation at 37°C and 5% CO2. Unbound nanoparticles were washed with 0.1 M Tyrode’s buffer and then cells were fixed with 1% Electron Microscopy grade glutaraldehyde (Ladd Research Industries) in Tyrode’s buffer overnight in the fridge. Pellets were washed with Tyrode’s buffer (2X) then mixed with 3% agar in PBS in a 1.5 mL centrifuge and let to harden in embedding capsules. The cell/agar pellet was trimmed into chunks and washed with Tyrode’s buffer (3X). They were then fixed and stained with 1% osmium tetroxide (Ladd Research Industries) and 1.5% potassium ferricyanide (Sigma Aldrich) for 1 hour. Pellets were washed with Tyrode’s buffer (2X) and then gradually dehydrated using an ethanol gradient (30%, 50%, 70%, 80%, 90%, 100% (2x) and 100 % opened fresh). Cell pellets then infiltrated with epoxy by epoxy/ethanol mixtures (ration, time of 1:1, 1 h; 2:1, overnight; 1:0, 2 h) in a desiccator. After a final infiltration in 100% epoxy with 2,4,6-tri(dimethylaminomethyl)-phenol (DMP from Electron Microscopy Sciences), the pellets were embedded in freshly prepared epoxy with DMP and let to polymerize for 48 h at 65 °C. Resin blocks were trimmed with a razor blade and then sectioned in 70-90 nm sections using a Leica Ultracut UCT ultramicrotome. The thin sections were stained with uranyl acetate and lead citrate for 1 hour and imaged in a Hitachi H7600 at 80 keV at the College of Medicine Electron Microscopy core facility.
2.9. Microfluidic Device Design and Fabrication
Geometrically enhanced mixing (GEM) chips used in this study were developed and reported previously.39 The microfluidic GEM chip consisting of the polydimethylsiloxane (PDMS) structure was bonded to a 3″ × 1″ glass microscope slide. The PDMS structure was fabricated using two-layer soft lithography.40 Silicon wafers were first spin-coated with 50 μm thick SU-8 2035 photoresist (MicroChem, Newton, MA) as the main channel layer. After soft baking, ultraviolet light exposure, and post exposure baking, another layer of SU-8 was added to form the herringbone mixer layer. With precise alignment between the main channel and the mixer, a second exposure was performed to create the herringbone mixer pattern. After development, a silicon master patterned with the complementary structures was obtained. PDMS structures were fabricated by casting a liquid PDMS precursor against the master using Sylgard 184 reagents (Dow Corning, Midland, MI), according to the manufacturer’s instructions. Inlet and outlet wells were created at the channel ends by punching holes in the PDMS sheet. The channel depth, which was controlled by the spin speed of the SU-8, was measured using a Dektak 150 profilometer.
Magnetic field gradients were generated using a planar Halbach array. A Halbach array is a special arrangement of permanent magnets that augments the magnetic field gradients on one side of the array while cancelling the field to near zero on the other side. This is achieved by having a spatially rotating pattern of magnetization. The Halbach array was assembled using B888-2PE-N52 and B888-2PE-N52 magnets as per instructions obtained from K&J Magnetics.
2.10. Cell Capture in Microfluidic Device
2.10.1. Magnetic nanoparticle assisted capture in cell mixtures
BxPC3 cells were detached from the flask using trypsin and pelleted out in media after centrifuging at 200 ref at 25 °C. The pellet was suspended in 1 mL of RPMI media and kept rotating for 30 min at 37°C, 5% CO2 in Eppendorf tubes to make sure EpCAM receptor regeneration occurred. BxPC3 cells were pre-stained with CellMask Deep Red Plasma membrane stain (at 1 μL stain /ml of media (as recommended by life technologies), for 15 min at 37°C in PBS. CCRF-CEM cells was stained with Hoechst 333422 cell-labeling solutions at 1 μL dye/ml of media (20 mM), for 10 min at 37°C in PBS. The two cell lines were mixed at 106/mL CCRF-CEM cells and 104/mL BxPC-3 cells. Non-targeted magnetic nanoparticles and magnetic nanoparticles targeted with antibody were added at 200 ug iron/mL to cell solution and incubated for 1 hour at 37 °C under rotation. The cell mixture was washed in PBS twice at 200 ref for 5 min. To initiate cell capture experiments, one channel volume (~100 μL) of 1 mg/mL avidin (Invitrogen) in PBS was first introduced into the device, followed by incubation for 15 min and then rinsed three times with PBS. Next, one channel volume of biotinylated anti-EpCAM (20 μg/mL) was introduced into the device and incubated for 15 min, rinsed with PBS containing BSA and Tween-20. Finally, 200 uL of cell mixture was pumped into the device by pumping using a syringe pump (KD Legato 111, KD Scientific, Holliston, MA) with a BD syringe connected to the inlet of the device via polymer tubing and a female luer-to-barb adapter (IDEX Health & Science, Oak Harbor, WA) at a flow rate of 2 μL/ s (or other flow rates specified in the text, at n = 3 devices for each experiment). After the cell capture experiment, the microchannel was washed three times with 100 uL PBS. Fluorescent images were taken with DAPI channel (CCRF-CEM) and cy5 channel (BxPC-3) in an Olympus IX71 fluorescence microscope (Olympus America, Melville, NY) with an automated ProScan stage (Prior Scientific, Rockland, MA) and segmentation technique was used to image and count the number of cells captured. The images were analyzed with the cellSens (a software from Olympus) and the cell count was gathered afterwards.
2.10.2. Magnetic nanoparticle assisted capture of BxPC3 cells spiked in blood
For spiking experiments, the BxPC-3 cells were labelled with cell mask deep red solution at (1 μL/ml in 1 mL PBS, 15 min, 37 °C). Nuclei for all cells were stained with Hoechst 333422 cell-labeling solutions at 1 μL/ml of PBS. They were spiked in blood sample to get the concentration of BxPC-3 at 3.5x104/mL and incubated with 200 ug iron/mL for 1 hour at 37 °C under rotation. The samples were not washed due to the nature of blood. The devices were functionalized with biotinylated antibody for capture experiments. Blood samples spiked with cells were introduced into the devices at 2 uL/s at 200 uL/device and they were washed with PBS after cell capture. Device was then washed with 300 uL PBS at 2 uL/s. Images were taken with (4′,6-diamidino-2-phenylindole) DAPI channel (for nuclei of White blood cells and BxPC-3 cells) and cy5 channel (for membrane of BxPC-3 cells).
3. Results and Discussion
3.1. Preparation and characterization of Anti-EpCAM conjugated magnetic nanoparticles
Magnetic nanoparticles, illustrated in Fig. 2a, were prepared in order to test their ability to enhance microfluidic device capture efficiency and specificity in cell mixtures under strong magnetic field gradients. The nanoparticles were synthesized via thermal decomposition33, coated with polyethylene glycol21 , and further modified with streptavidin,41 to which biotinylated Anti-EpCAM antibody was attached as detailed in the methods section and in Section 1 of the Supporting Information (SI). The nanoparticles were synthesized in the presence of oxygen to enhance their magnetic properties. The resulting nanoparticles had a physical diameter (Dp) of 20.3 ± 1.5 nm (Fig. 2d) and magnetic diameter (Dm) of 18.1 ± 0.7 nm (Fig. 3a). Oleic acid on the particles was exchanged with silanated polyethylene glycol (MW 5000 g/mol) to obtain particles with a mean hydrodynamic diameter of 76 nm and distribution standard deviation of 25 nm (Fig. 3b). The number of amine groups from unmodified amino propyl silane covalently attached to the particles during the ligand exchange was estimated to be ~1250 nanomoles of amine/mg particle using the 3-(4carboxybenzoyl)quinoline-2-carboxaldehyde) (CBQCA) assay. To enable streptavidin conjugation, carboxylated PEG nanoparticles were obtained by the amidation reaction between amine groups and carboxylated PEG (MW 10,000 g/mol), prepared by DMAP induced ring opening of succinic acid by hydroxyl groups of PEG and using EDC-sulfo NHS chemistry. The reduction in the nanomoles of amines per mg particle from ~1250 to < 30 nanomoles amines/mg particle (below the limit of detection for the assay, Fig. S1 in the Supplementary Information) was used as confirmation of attachment of the carboxylated PEG to the particles.
Fig. 2.

Characterization and confirmation of conjugation on the particle. a) Schematic of the Anti-EpCAM conjugated nanoparticles. b) Transmission electron microscopy image of the antibody coated nanoparticles. c) B4F assay results confirming streptavidin conjugation. d) Physical, magnetic and hydrodynamic diameter distributions of the antibody conjugated nanoparticles.
Fig. 3.
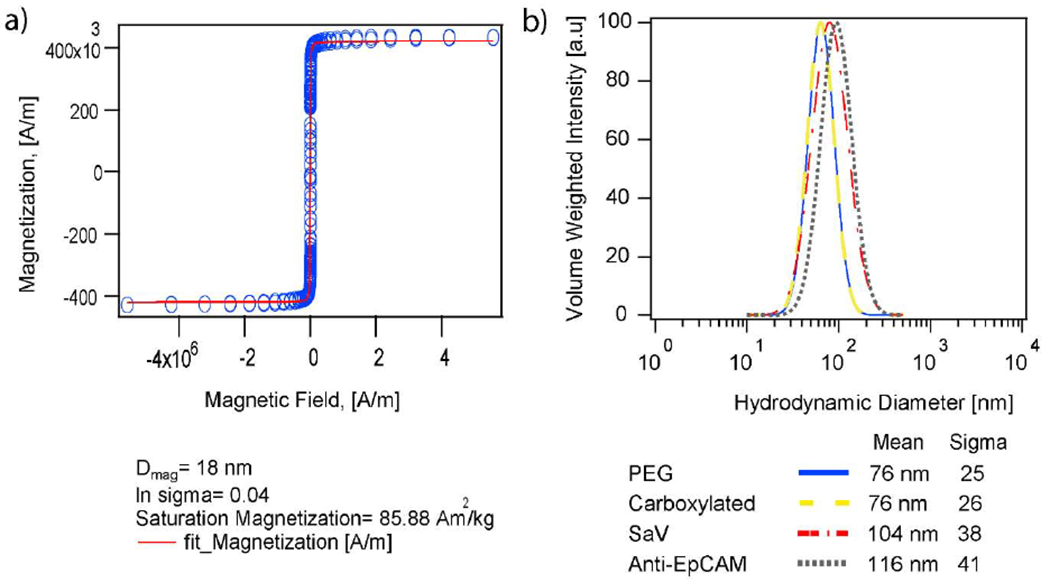
a) Equilibrium magnetization curve for the oleic acid coated particles and Langevin fit performed to extract magnetic diameter of the particles. b) Hydrodynamic size distributions for the nanoparticles after each stage of conjugation.
Primary amines from streptavidin were covalently attached to the carboxylic groups on the nanoparticles using EDC and sulfo-NHS. Conjugation was confirmed using the biotin-4-fluorescein (B4F) assay, as shown in Fig. 2c. The fluorescence signal remained quenched (red open circles in Fig. 2c) until addition of 20 pmol of Biotin-4-fluorescein to 0.125 mg Fe of streptavidin coated iron oxide nanoparticles. This suggests that there were 40 molecules of streptavidin per nanoparticle (assuming 2.26 out of 4 active units were present in streptavidin, as indicated by the manufacturer). Biotinylated anti-EpCAM was further conjugated to the streptavidin to yield magnetic nanoparticles with a mean hydrodynamic diameter of 116 nm and distribution standard deviation of 41 nm (Fig. 2d). After conjugation it was found that with addition of 4 pmol of biotin-4-fluorescein all binding sites of 0.125 mg streptavidin coated nanoparticles were occupied (green closed circles in Fig. 2c) and with further addition of B4F there was an increase in fluorescence intensity. This suggests that approximately 8 molecules of streptavidin per particle remained unconjugated and further suggested that we had attached ~70 molecules of Anti-EpCAM per particle.
The hydrodynamic diameter of the particles was measured after each successive functionalization step and is shown in Fig. 3b. This, combined with TEM images of the particles in Fig. S2 in the SI after each step of surface modification suggests that the nanoparticles do not aggregate and that each nanoparticle is composed of a single core coated with a PEG, streptavidin, and antibody layer.
3.2. Quantification of nanoparticle affinity towards EpCAM positive and EpCAM negative cells
To explore the possibility of enhancing tumor cell capture efficiency and specificity by using magnetic nanoparticles and magnetic field gradients in an antibody-functionalized microfluidic device, we chose BxPC3 cells as a model EpCAM positive cell line42 and CCRF-CEM cells as a model EpCAM negative cell line.43, 44 Flow cytometry studies performed to evaluate the affinity of the Anti-EpCAM antibody to the chosen cell lines showed high specificity of the antibody to BxPC3 cells, as shown in Fig. S3 in the SI.
To assess the interaction of the antibody functionalized particles with the EpCAM positive BxPC3 cells, cells were incubated for 1 hour with 200 ug of both Anti-EpCAM functionalized (targeted) nanoparticles and control carboxylated (non-targeted) nanoparticles. Nanoparticle binding and uptake was quantified using the o-phenanthroline assay.45, 46 Fig. 4 shows that BxPC3 cells had 1.60 pg iron /cell when antibody was attached to the particles, as opposed to 0.52 pg iron/cell when particles without antibody were used. This was a statistically significant enhancement in uptake (p-value < 0.05), confirming affinity and selectivity of targeted nanoparticles to the EpCAM positive cells. To study the specificity of the targeted particles, similar studies were performed with EpCAM negative CCRF-CEM cells. Uptake of the targeted particles by the CCRF-CEM cells was 0.68 pg iron/cell. This was a lower (p-value < 0.05) level of uptake than with BxPC3 cells, suggesting that the antibody conjugated particles had negligible affinity to the EpCAM negative cells. We note in Fig. 4, that cell controls (no nanoparticles) were determined to have ~0.39 pg iron/cell according to the o-phenanthroline assay. This assay is not specific to iron oxide nanoparticles and instead quantifies all forms of iron in the sample. Thus, this value for the cells alone could correspond to endogenous iron content under our culture conditions. Importantly, the amount of iron in the cell-only controls did not differ statistically from that determined in cells exposed to non-targeted particles (p-value > 0.05), suggesting low non-specific uptake of the nanoparticles. In analyzing the data in Fig. 4, one could subtract values for the cell-only controls from those of cells incubated with nanoparticles. In doing so, one must consider the variability of both groups and propagation of error. By doing so, we estimate that the exogenous Fe (due to nanoparticle uptake) for BxPC3 cells is 0.13 ± 0.49 pg iron/cell for the non-targeted particles and 1.14 ± 0.19 pg iron/cell for the targeted particles, and for the CEM cells it is 0.20 ± 0.48 pg iron/cell for the non-targeted particles and 0.29 ± 0.30 pg iron/cell for the targeted particles. These calculations further suggest that uptake of the non-targeted particles is negligible and that the targeted particles are selective towards the BxPC3 cells. We also evaluate the ratio of targeted to non-targeted nanoparticles (from the exogenous Fe calculations) taken up by BxPC3 cell line and see that uptake of the targeted particles was approximately 9-fold higher than that of non-targeted particles. For the CEM cell lines, this ratio is about 1.5, suggesting uptake of the targeted particles to be approximately same as for the non-targeted particles. Further, comparing uptake of the anti-EpCAM targeted particle across the two cell lines, a 4-fold higher uptake is seen by the EpCAM expressing cell line than by the CEM cells. These observations are consistent with the higher level of expression of EpCAM in the BxPC3 cells, relative to the CEM cells, and with the observed affinity of the anti-EpCAM antibody to the BxPC3 cells as seen by flow cytometry. The antibody conjugated particles were used to enhance microfluidic capture of BxPC3 cells from cell mixtures.
Fig. 4.
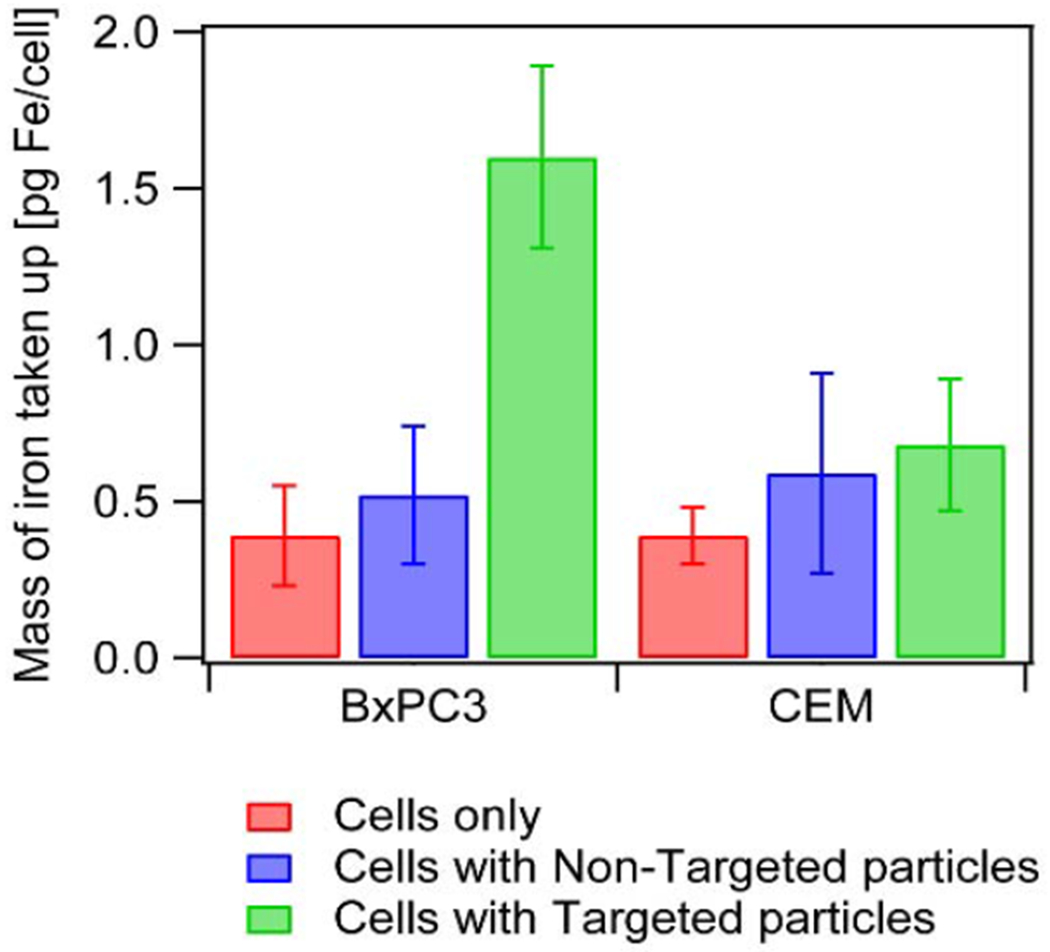
Uptake of iron oxide nanoparticles in BxPC3 and CCRF-CEM cells for cells with no particles (in red), cells incubated with nontargeted particles (in blue), and cells incubated with targeted particles (in green). All experiments were repeated 3 times.
3.3. Visualization of nanoparticle-cell interaction using fluorescence microscopy
We visualized the interaction of targeted and non-targeted nanoparticles with the BxPC3 cells using fluorescence microscopy. As seen in Fig. 5, nanoparticles conjugated with Alexa fluor 568 and tagged with anti-EpCAM (targeted particles) were bound to the surface of the BxPC3 cells. When control carboxylated nanoparticles (non-targeted particles) were tagged with Alexa fluor 568, little evidence of nanoparticle attachment to cells was found.
Fig. 5.
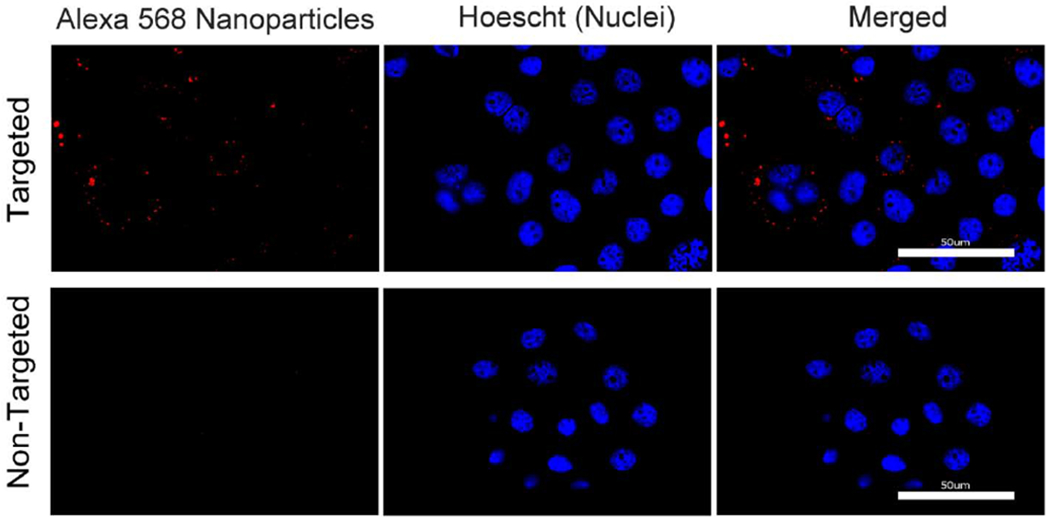
Interaction of targeted (top panel) and non-targeted nanoparticles (bottom panel) with BxPC3 cells.
3.4. Visualization of nanoparticle internalization using transmission electron microscopy
Transmission electron micrographs were taken to visualize internalization of Anti-EpCAM nanoparticles (targeted nanoparticles) and control carboxylated nanoparticles (non-targeted nanoparticles) with BxPC3 and CCRF-CEM cells. As seen in Fig. 6 anti-EpCAM nanoparticles were found predominantly in the lysosomal compartment and some on the surface of the EpCAM positive BxPC3 cells. There was minimal evidence of internalization of the anti-EpCAM nanoparticles in the EpCAM-negative CCRF-CEM cells, as seen in Fig. S4 in the SI. For the non-targeted nanoparticles, we did find evidence of internalization and accumulation in lysosomes of BxPC3 cells (see Fig. S5 in the SI). However, we note that drawing quantitative conclusions regarding nanoparticle uptake by cells using TEM is difficult.47 Thus, based on the combination of quantitative assessment of nanoparticle uptake using the o-phenanthroline assay and qualitative assessment based on confocal microscopy and TEM we believe that the anti-EpCAM nanoparticles have greater uptake in the EpCAM positive BxPC3 cells, where they associate with the cell membrane and internal compartments, such as lysosomes, whereas the same particles have relatively low affinity towards the EpCAM negative CCRF-CEM cells.
Fig. 6.
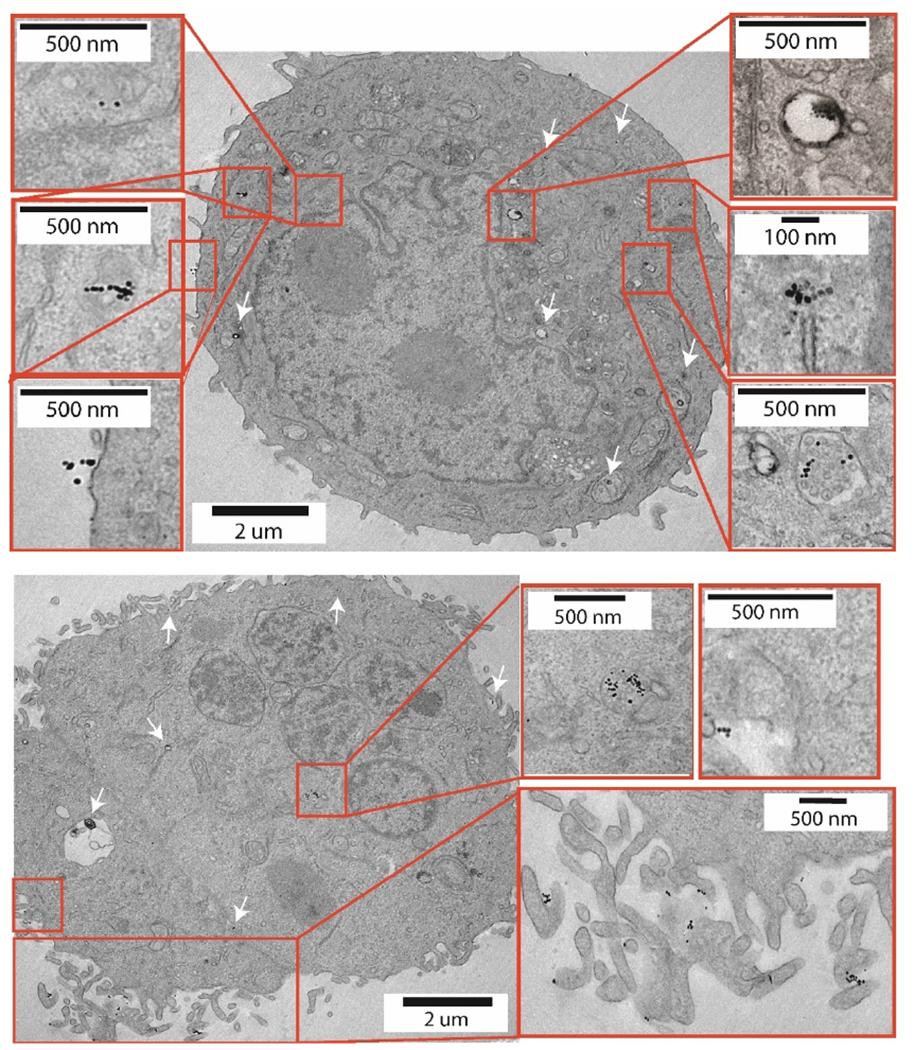
BxPC3 cells with targeted nanoparticles indicated by white arrows.
3.5. Microfluidic capture using magnetic nanoparticles and antibody functionalized microfluidic devices
BxPC3 and CCRF-CEM cells were first stained and then mixed at a ratio of 1:100. For every 200 uL of cell mixture, with 200,000 CCRF-CEM cells and 2,000 BxPC3 cells, 200 ug of Anti-EpCAM nanoparticles were incubated for 1 hour at 37°C, washed, and then introduced into the antibody functionalized microfluidic device. As seen in Fig. 7, at a flow rate of 2 μL/s, without the presence of the Halbach array (HA) ~40 % of the BxPC3 cells were captured. Adding the Halbach array resulted in a 1.6-fold increase in capture efficiency of the BxPC3 cells to ~75% at 2 μL/s. In contrast, the EpCAM negative CCRF-CEM cell line had a capture efficiency of less than 10% with and without the Halbach array. This suggests that even if there is some non-specific uptake of the anti-EpCAM nanoparticles by the EpCAM-negative CCRF-CEM cells, it is not enough to influence the capture efficiency in the antibody-functionalized microfluidic device.
Fig. 7.
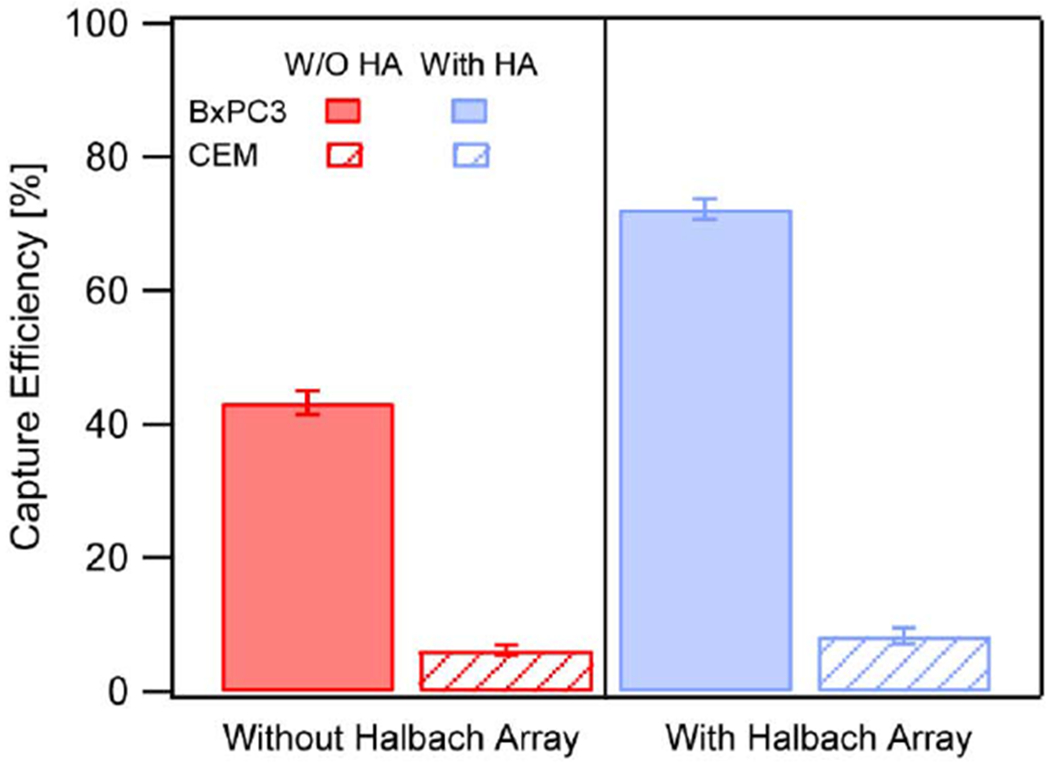
Capture efficiencies of BxPC3 (Red) and CCRF-CEM (blue) cells with (solid bar) and without (dashed bar) application of the Halbach array. All experiments were conducted in 3 different microfluidic devices.
We also evaluated capture efficiency as a function of flow rate, with and without incubation of cells with Anti-EpCAM nanoparticles. Fig. 8 shows that while capture efficiency decreased with increasing flow rate, capture efficiency was enhanced when Anti-EpCAM nanoparticles were used in antibody functionalized microfluidic devices combined with the planar Halbach array. The results indicate that the combination of magnetophoresis and microfluidic device surface affinity results in higher cell capture across a range of flow rates.
Fig. 8.
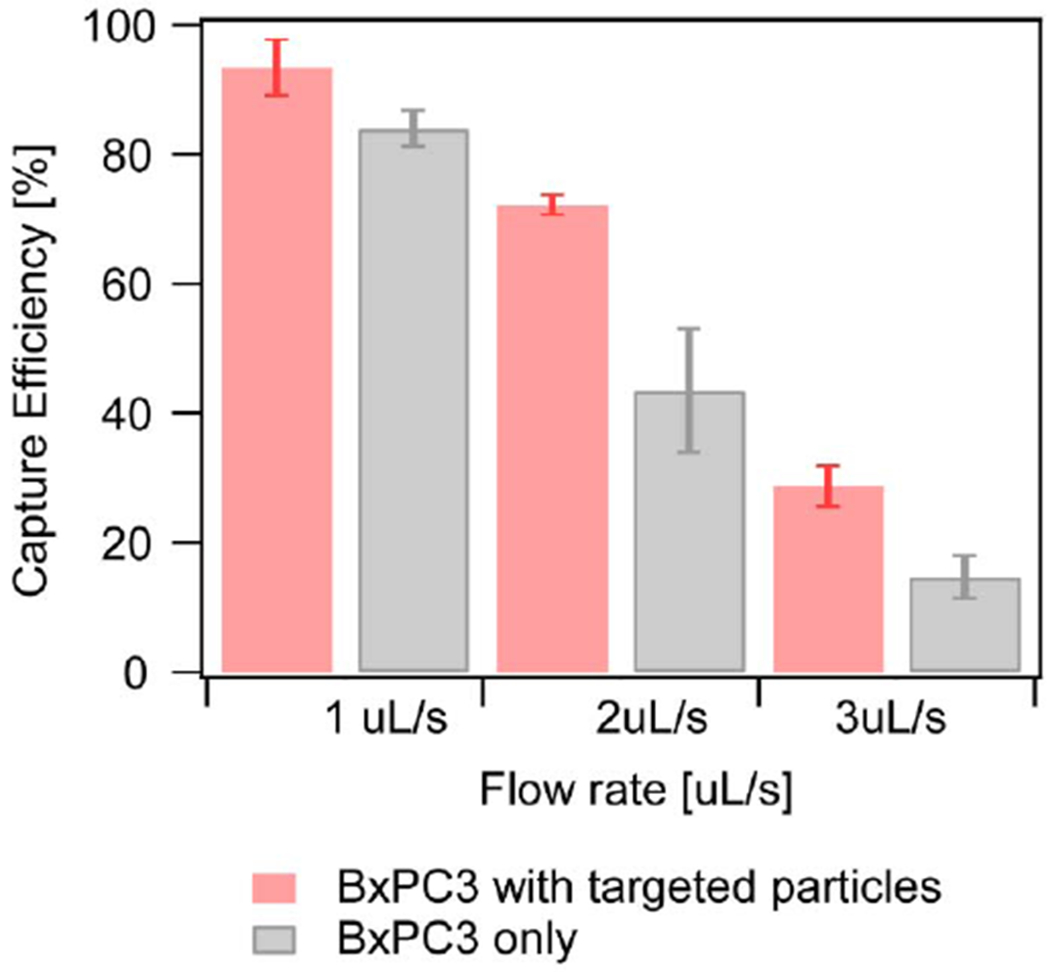
Capture efficiency comparison between cells with targeted particles (in red) and cells only (in grey) All experiments were conducted in 3 different microfluidic devices.
3.6. Capture of BxPC3 cells spiked in whole blood
We evaluated capture efficiency of BxPC3 cells spiked in whole blood. The BxPC3 cells were pre-stained and introduced into heparinized whole blood at 35,000 cells/ml. The antibody conjugated particles were incubated in the blood mixture and then introduced in the antibody functionalized microfluidic device. Capture efficiency of BxPC3 against WBCs was evaluated as discussed in the methods section. As seen in Fig 9a, about 60 % of BxPC3 were captured from the blood mixture, which is comparable to capture efficiency of the BxPC3 in PBS at 2 μL/s. Capture purity, defined as the ratio of the number of BxPC3 captured to the total number of nucleated cells (BxPC3 and WBCs), was calculated to be about 34%. For comparison, we evaluated capture efficiency and purity for BxPC3 cells mixed with CEM cells in PBS. The capture efficiency for this case (Fig. 9a) was similar to that seen in blood, suggesting that the presence of blood plasma and other cells does not significantly impact nanoparticle binding to the BxPC3 cells, nor does it affect magnetic capture of the BxPC3 cells. In contrast, the purity of captured cells was close to 100% for the BxPC3 cells mixed with CEM cells in PBS, while it was about 34% for the BxPC3 cells in whole blood. We attribute this difference to the greater affinity of WBCs for the microfluidic device surface.
Fig. 9.
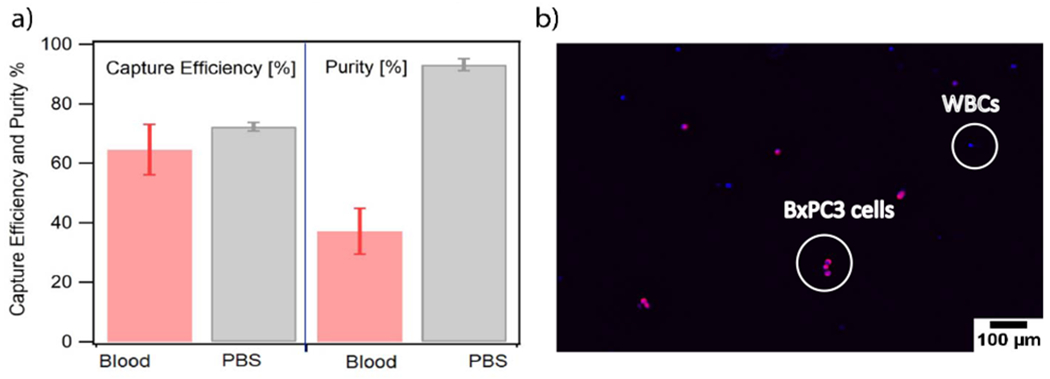
a. Capture efficiency of BxPC3 when spiked in blood mixture in red and the purity of capture in grey; b. Representative image of the BxPC3 cells (membrane stained with cell mask deep red, nuclei stained with Hoescht 333422) captured against white blood cells (Nuclei stained with Hoescht 333422) in whole blood. All experiments were conducted in 3 different microfluidic devices.
Integrating magnetic nanoparticle assisted magnetophoresis with microfluidics allowed for capture of tumor cells in buffer and from whole blood with higher efficiency and specificity. We attribute this enhancement firstly to the PEG coated magnetic nanoparticles modified with an antibody. These nanoparticles had minimal non-specific interactions with non-targeted cells. The improved surface to volume ratio of the nanoparticles allowed for high binding capacity to the targeted EpCAM receptor. Secondly, the magnetic force during capture and removal of magnetic particles in magnetophoresis is influenced not only by the magnetic properties of the particles but also the magnetic field gradient. Here, we used a Halbach array, which results in a stronger magnetic field gradient over a larger fraction of the microfluidic device capture surface, compared with single magnets.48, 49 Separation efficiency during magnetic capture also depends on the geometry of the capture device and on separation time, in the case of batch capture processes, or residence time in the case of flow capture processes. Use of the herringbone structure to distort streamlines and induce chaotic mixing and micro-vortexing effects, maximized collisions and interactions between cells and device surfaces, leading to increased cell capture efficiency.39 Together with engineered targeted nanoparticles and the microfluidic capture system under high field gradients, enhancement in capture of tumor cells in a flow system where the magnetic forces dominate drag forces was demonstrated.
4. Conclusions
We developed targeted magnetic nanoparticles coated with polyethylene glycol and functionalized with biotinylated Anti-EpCAM. These particles exhibited high affinity towards EpCAM expressing tumor cells and negligible non-specific uptake by EpCAM negative cells. It was hypothesized these targeted magnetic nanoparticles would selectively bind EpCAM expressing pancreatic tumor cells from cell mixtures and, that in the presence of large magnetic field gradient they would facilitate enhanced tumor cell capture in devices functionalized with EpCAM antibodies. In this paper, we show that combination of magnetic forces with microfluidic surface capture via antibodies resulted in enhanced capture efficiency and specificity at flow rates as high as 7.2 mL/h from cell mixtures and blood samples spiked with tumor cells. Most studies in the past on magnetophoretic capture of tumor cells have relied on commercial particles7, 10 or particles that are coated with mono- and poly-saccharides that show limiting specificity due to non-specific cell binding and uptake. Further, prior studies using targeted particles for cell capture have considered single cell populations,37 and non-specific uptake of particles by cells other than the tumor cell of interest has not been assessed.38 Here we systematically magnetically enhanced capture of targeted cell populations using antibody targeted PEG coated particles in single cell suspensions, cell mixtures, and whole blood spiked with the targeted cells. We find that the dense PEG coating helps reduce non-specific uptake by CCRF-CEM cells and confirmed this result using fluorescent imaging, transmission electron micrographs and iron quantification assay. We anticipate that our thorough description of consecutive treatments of magnetic nanoparticle surface to provide the functional groups for anti-EpCAM attachment would benefit the development of nanoparticle formulations with improved specific cell binding and limited nonspecific interactions. We believe integration of engineered nanoparticles and microfluidics can be used for efficient capture and characterization of rare populations of tumor cells and can be extended to circulating tumor cells in the future.
Supplementary Material
Acknowledgements
We thank the University of Florida for providing financial support and infrastructure during the course of this research. This work was also partially supported by the National Institutes of Health, through awards K25CA149080 and RO1 AR068324. We thank Dr. Doty Andria for help performing flow cytometry at the ICBR. We are grateful to Dr. Sharon Matthews and Chao Chen, of the University of Florida College of Medicine Electron Microscopy Core Facility, for advice and technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
There are no conflicts to declare.
References
- 1.Zborowski M; Chalmers JJ, Magnetophoresis: Fundamentals and Applications. Wiley Encyclopedia of Electrical and Electronics Engineering: 2015; pp 1–23. [Google Scholar]
- 2.Plouffe BD; Murthy SK; Lewis LH, Fundamentals and application of magnetic particles in cell isolation and enrichment: a review. Reports on Progress in Physics 2015, 78 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herrmann IK; Urner M; Koehler FM; Hasler M; Roth-Z’Graggen B; Grass RN; Ziegler U; Beck-Schimmer B; Stark WJ, Blood Purification Using Functionalized Core/Shell Nanomagnets. Small 2010, 6 (13), 1388–1392. [DOI] [PubMed] [Google Scholar]
- 4.Shen HJ; Wang J; Liu HY; Li ZH; Jiang FL; Wang FB; Yuan Q, Rapid and Selective Detection of Pathogenic Bacteria in Bloodstream Infections with Aptamer-Based Recognition. Acs Applied Materials & Interfaces 2016, 8 (30), 19371–19378. [DOI] [PubMed] [Google Scholar]
- 5.Lattuada M; Ren Q; Zuber F; Galli M; Bohmer N; Matter MT; Wichser A; Bertazzo S; Pier GB; Herrmann IK, Theranostic body fluid cleansing: rationally designed magnetic particles enable capturing and detection of bacterial pathogens. Journal of Materials Chemistry B 2016, 4 (44), 7080–7086. [DOI] [PubMed] [Google Scholar]
- 6.Scarberry KE; Mezencev R; McDonald JF, Targeted removal of migratory tumor cells by functionalized magnetic nanoparticles impedes metastasis and tumor progression. Nanomedicine 2011, 6 (1), 69–78. [DOI] [PubMed] [Google Scholar]
- 7.Bai LL; Du YM; Peng JX; Liu Y; Wang YM; Yang YL; Wang C, Peptide-based isolation of circulating tumor cells by magnetic nanoparticles. Journal of Materials Chemistry B 2014, 2 (26), 4080–4088. [DOI] [PubMed] [Google Scholar]
- 8.Issadore D; Chung J; Shao HL; Liong M; Ghazani AA; Castro CM; Weissleder R; Lee H, Ultrasensitive Clinical Enumeration of Rare Cells ex Vivo Using a Micro-Hall Detector. Science Translational Medicine 2012, 4 (141). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohmer N; Demarmels N; Tsolaki E; Gerken L; Keevend K; Bertazzo S; Lattuada M; Herrmann IK, Removal of Cells from Body Fluids by Magnetic Separation in Batch and Continuous Mode: Influence of Bead Size, Concentration, and Contact Time. Acs Applied Materials & Interfaces 2017, 9 (35), 29571–29579. [DOI] [PubMed] [Google Scholar]
- 10.Shi WT; Wang SQ; Maarouf A; Uhl CG; He R; Yunus D; Liu YL, Magnetic particles assisted capture and release of rare circulating tumor cells using wavy-herringbone structured microfluidic devices. Lab on a Chip 2017, 17 (19), 3291–3299. [DOI] [PubMed] [Google Scholar]
- 11.Zhao WJ; Liu Y; Jenkins BD; Cheng R; Harris BN; Zhang WZ; Xie J; Murrow JR; Hodgson J; Egan M; Bankey A; Nikolinakos PG; Ali HY; Meichner K; Newman LA; Davis MB; Mao LD, Tumor antigen-independent and cell size variation-inclusive enrichment of viable circulating tumor cells. Lab on a Chip 2019, 19 (10), 1860–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Safarik I; Safarikova M, Use of magnetic techniques for the isolation of cells. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences 1999, 722 (1-2), 33–53. [PubMed] [Google Scholar]
- 13.Bucak S; Jones DA; Laibinis PE; Hatton TA, Protein separations using colloidal magnetic nanoparticles. Biotechnology Progress 2003, 19 (2), 477–484. [DOI] [PubMed] [Google Scholar]
- 14.Radisic M; Iyer RK; Murthy SK, Micro- and nanotechnology in cell separation. International Journal of Nanomedicine 2006, 1 (1), 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molday RS; Yen SPS; Rembaum A, APPLICATION OF MAGNETIC MICROSPHERES IN LABELING AND SEPARATION OF CELLS. Nature 1977, 268 (5619), 437–438. [DOI] [PubMed] [Google Scholar]
- 16.Miltenyi S; Muller W; Weichel W; Radbruch A, HIGH-GRADIENT MAGNETIC CELL-SEPARATION WITH MACS. Cytometry 1990, 11 (2), 231–238. [DOI] [PubMed] [Google Scholar]
- 17.Xu HY; Aguilar ZP; Yang L; Kuang M; Duan HW; Xiong YH; Wei H; Wang A, Antibody conjugated magnetic iron oxide nanoparticles for cancer cell separation in fresh whole blood. Biomaterials 2011, 32 (36), 9758–9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sista RS; Eckhardt AE; Srinivasan V; Pollack MG; Palanki S; Pamula VK, Heterogeneous immunoassays using magnetic beads on a digital microfluidic platform. Lab on a Chip 2008, 8 (12), 2188–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith JE; Sapsford KE; Tan WH; Ligler FS, Optimization of antibody-conjugated magnetic nanoparticles for target preconcentration and immunoassays. Analytical Biochemistry 2011, 410 (1), 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chalmers JJ; Xiong Y; Jin X; Shao M; Tong X; Farag S; Zborowski M, Quantification of Non-Specific Binding of Magnetic Micro- and Nanoparticles Using Cell Tracking Velocimetry: Implication for Magnetic Cell Separation and Detection. Biotechnology and Bioengineering 2010, 105 (6), 1078–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrera C; Herrera AP; Rinaldi C, Colloidal dispersions of monodisperse magnetite nanoparticles modified with poly(ethylene glycol). Journal of Colloid and Interface Science 2009, 329 (1), 107–113. [DOI] [PubMed] [Google Scholar]
- 22.Bohorquez AC; Unni M; Belsare S; Chiu-Lam A; Rice L; Pampo C; Siemann D; Rinaldi C, Stability and Mobility of Magnetic Nanoparticles in Biological Environments Determined from Dynamic Magnetic Susceptibility Measurements. Bioconjugate Chemistry 2018, 29 (8), 2793–2805. [DOI] [PubMed] [Google Scholar]
- 23.Jokerst JV; Lobovkina T; Zare RN; Gambhir SS, Nanoparticle PEGylation for imaging and therapy. Nanomedicine 2011, 6 (4), 715–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Went PT; Lugli A; Meier S; Bundi M; Mirlacher M; Sauter G; Dirnhofer S, Frequent EpCam protein expression in human carcinomas. Human Pathology 2004, 35 (1), 122–128. [DOI] [PubMed] [Google Scholar]
- 25.D S; Z C; M W; Y Z, Nanomaterial-based Microfluidic Chips for the Capture and Detection of Circulating Tumor Cells . 4 ed.; Nanotheranostics . , 2017. August 20 . ; Vol. 1, pp 389–402. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khamenehfar A; Li PCH, Microfluidic Devices for Circulating Tumor Cells Isolation and Subsequent Analysis. Current Pharmaceutical Biotechnology 2016, 17 (9), 810–821. [DOI] [PubMed] [Google Scholar]
- 27.Stott SL; Hsu CH; Tsukrov DI; Yu M; Miyamoto DT; Waltman BA; Rothenberg SM; Shah AM; Smas ME; Korir GK; Floyd FP; Gilman AJ; Lord JB; Winokur D; Springer S; Irimia D; Nagrath S; Sequist LV; Lee RJ; Isselbacher KJ; Maheswaran S; Haber DA; Toner M, Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proceedings of the National Academy of Sciences of the United States of America 2010, 107 (43), 18392–18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagrath S; Sequist LV; Maheswaran S; Bell DW; Irimia D; Ulkus L; Smith MR; Kwak EL; Digumarthy S; Muzikansky A; Ryan P; Balis UJ; Tompkins RG; Haber DA; Toner M, Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450 (7173), 1235–U10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang M; Wen CY; Wu LL; Hong SL; Hu J; Xu CM; Pang DW; Zhang ZL, A chip assisted immunomagnetic separation system for the efficient capture and in situ identification of circulating tumor cells. Lab on a Chip 2016, 16 (7), 1214–1223. [DOI] [PubMed] [Google Scholar]
- 30.Kang JH; Krause S; Tobin H; Mammoto A; Kanapathipillai M; Ingber DE, A combined micromagnetic-microfluidic device for rapid capture and culture of rare circulating tumor cells. Lab on a Chip 2012, 12 (12), 2175–2181. [DOI] [PubMed] [Google Scholar]
- 31.Alnaimat F; Dagher S; Mathew B; Hilal-Alnqbi A; Khashan S, Microfluidics Based Magnetophoresis: A Review. Chemical Record 2018, 18 (11), 1596–1612. [DOI] [PubMed] [Google Scholar]
- 32.Vreeland EC; Watt J; Schober GB; Hance BG; Austin MJ; Price AD; Fellows BD; Monson TC; Hudak NS; Maldonado-Camargo L; Bohorquez AC; Rinaldi C; Huber DL, Enhanced Nanoparticle Size Control by Extending LaMer’s Mechanism. Chemistry of Materials 2015, 27 (17), 6059–6066. [Google Scholar]
- 33.Unni M; Uhl AM; Savliwala S; Savitzky BH; Dhavalikar R; Garraud N; Arnold DP; Kourkoutis LF; Andrew JS; Rinaldi C, Thermal Decomposition Synthesis of Iron Oxide Nanoparticles with Diminished Magnetic Dead Layer by Controlled Addition of Oxygen. ACS Nano 2017, 11 (2), 2284–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lele BS; Kulkarni MG, Single step room temperature oxidation of poly(ethylene glycol) to poly(oxyethylene)-dicarboxylic acid. Journal of Applied Polymer Science 1998, 70 (5), 883–890. [Google Scholar]
- 35.De Palma R; Peeters S; Van Bael MJ; Van den Rul H; Bonroy K; Laureyn W; Mullens J; Borghs G; Maes G, Silane ligand exchange to make hydrophobic superparamagnetic nanoparticles water-dispersible. Chemistry of Materials 2007, 19 (7), 1821–1831. [Google Scholar]
- 36.Mittal R; Bruchez MP, Biotin-4-Fluorescein Based Fluorescence Quenching Assay for Determination of Biotin Binding Capacity of Streptavidin Conjugated Quantum Dots. Bioconjugate Chemistry 2011, 22 (3), 362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HJ; Cho HY; Oh JH; Namkoong K; Lee JG; Park JM; Lee SS; Huh N; Choi JW, Simultaneous capture and in situ analysis of circulating tumor cells using multiple hybrid nanoparticles. Biosensors & Bioelectronics 2013, 47, 508–514. [DOI] [PubMed] [Google Scholar]
- 38.Galanzha EI; Shashkov E; Sarimollaoglu M; Beenken KE; Basnakian AG; Shirtliff ME; Kim JW; Smeltzer MS; Zharov VP, In Vivo Magnetic Enrichment, Photoacoustic Diagnosis, and Photothermal Purging of Infected Blood Using Multifunctional Gold and Magnetic Nanoparticles. Plos One 2012, 7 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheng WA; Chen T; Tan WH; Fan ZH, Multivalent DNA Nanospheres for Enhanced Capture of Cancer Cells in Microfluidic Devices. Acs Nano 2013, 7 (8), 7067–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sia SK; Whitesides GM, Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis 2003, 24 (21), 3563–3576. [DOI] [PubMed] [Google Scholar]
- 41.Quevedo PD; Behnke T; Resch-Genger U, Streptavidin conjugation and quantification-a method evaluation for nanoparticles. Analytical and Bioanalytical Chemistry 2016, 408 (15), 4133–4149. [DOI] [PubMed] [Google Scholar]
- 42.Thege FI; Lannin TB; Saha TN; Tsai S; Kochman ML; Hollingsworth MA; Rhim AD; Kirby BJ, Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: characterization, optimization and downstream analysis. Lab on a Chip 2014, 14 (10), 1775–1784. [DOI] [PubMed] [Google Scholar]
- 43.Zhang JL; Fan ZH, A universal tumor cell isolation method enabled by fibrin-coated microchannels. Analyst 2016, 141 (2), 563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu XL; He RX; Li SS; Cai B; Zhao LB; Liao L; Liu W; Zeng Q; Wang H; Guo SS; Zhao XZ, Magneto-Controllable Capture and Release of Cancer Cells by Using a Micropillar Device Decorated with Graphite Oxide-Coated Magnetic Nanoparticles. Small 2013, 9 (22), 3895–3901. [DOI] [PubMed] [Google Scholar]
- 45.Santiago-Rodriguez L; Lafontaine MM; Castro C; Mendez-Vega J; Latorre-Esteves M; Juan EJ; Mora E; Torres-Lugo M; Rinaldi C, Synthesis, stability, cellular uptake, and blood circulation time of carboxymethyl-inulin coated magnetic nanoparticles. Journal of Materials Chemistry B 2013, 1 (22), 2807–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.E394-15, A., Standard Test Method for Iron in Trace Quantities Using the 1,10-Phenanthroline Method. ASTM International, West Conshohocken, PA, 2015. [Google Scholar]
- 47.Schrand AM; Schlager JJ; Dai LM; Hussain SM, Preparation of cells for assessing ultrastructural localization of nanoparticles with transmission electron microscopy. Nature Protocols 2010, 5 (4), 744–757. [DOI] [PubMed] [Google Scholar]
- 48.Halbach K, DESIGN OF PERMANENT MULTIPOLE MAGNETS WITH ORIENTED RARE-EARTH COBALT MATERIAL. Nuclear Instruments & Methods 1980, 169 (1), 1–10. [Google Scholar]
- 49.Hayden ME; Hafeli UO, ‘Magnetic bandages’ for targeted delivery of therapeutic agents. Journal of Physics-Condensed Matter 2006, 18 (38), S2877–S2891. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


