Abstract
Acidithiobacillus ferrooxidans YNTRS-40 (A. ferrooxidans) is a chemolithoautotrophic aerobic bacterium isolated from Tengchong hot springs, Yunnan Province, China, with a broad growth pH range of 1.0–4.5. This study reports the genome sequence of this strain and the information of genes related to the adaptation of diverse stresses and the oxidation of ferrous iron and sulfur. Results showed that YNTRS-40 possesses chromosomal DNA (3,209,933-bp) and plasmid DNA (47,104-bp). The complete genome of 3,257,037-bp consists of 3,349 CDS genes comprising 6 rRNAs, 52 tRNAs, and 6 ncRNAs. There are many encoded genes associated with diverse stresses adaptation and ferrous iron and sulfur oxidation such as rus operon, res operon, petI, petII, sqr, doxDA, cydAB, and cyoABCD. This work will provide essential information for further application of A. ferrooxidans YNTRS-40 in industry.
Keywords: Acidithiobacillus ferrooxidans, complete genome, stress resistance, ferrous iron oxidation, sulfur oxidation
1. Introduction
Acidithiobacillus usually found in acidic environments with heavy metal and oligotrophic conditions. The members of this genus contribute to the formation of acidic habitats, the acidification of waters [1,2], and the biogeochemical cycle of iron and/or sulfur [3,4]. The Acidithiobacillus genus contains eight validated species, including A. ferrooxidans [5], A. ferriphilus [6], A. ferrivorans [7], A. ferridurans [8], A. albertensis [9], A. thiooxidans [10], A. caldus [11], and A. sulfuriphilus [12]. The former four dominant species can obtain energy for growth by using ferrous iron, elemental sulfur, reduced sulfur compounds, hydrogen, or tetrathionate as electron donors [2,6,8,13,14].
A. ferrooxidans has iron- and sulfur-oxidizing abilities and can grow in the environments with high concentrations of metal ions such as pyritic ore bodies, coal deposits and their acidified drainages [15,16,17,18,19,20]. Therefore, it has potential utilization in microbial electrosynthesis systems, eco-friendly bioleaching technology, biological desulfurization and machining of workpieces [21]. Until now, nine A. ferrooxidans strain genomes (ATCC 23270, ATCC 53993, Hel18, BY0502, CCM 4253, IO-2C, YQH-1, DLC-5, and RVS1) have been available in the public databases. The genomes of these strains only reported chromosome genomes, while the chromosome genomes and plasmid genomes were not detected at the same time. Additionally, only strains ATCC 23270 and ATCC 53993 were complete genome sequences [20,22,23]. The genome of strain YNTRS-40 reported in this paper is complete and contains a plasmid, which is the first report on the simultaneous acquisition of chromosome genome and plasmid genome. The plasmid genome could be helpful in studying the metabolic characteristics of these microbes.
To further investigate the genetic characteristics and application potential of A. ferrooxidans YNTRS-40, its genome was entirely sequenced and annotated. Our findings would be useful for understanding the roles and potential of A. ferrooxidans in the field of geobiology, biomedicine, and technology.
2. Materials and Methods
2.1. Growth Conditions, Genomic DNA Isolation and Morphological Detection
A. ferrooxidans YNTRS-40 was isolated from the soil soaked in drainage water in Tengchong hot springs, Yunnan Province, China. Field sampling and geochemical measurements were reported in 2018 [24]. General features of A. ferrooxidans YNTRS-40 are shown in additional files in Table S1. This strain was grown in modified 9K medium containing 2.4 g (NH4)2SO4, 0.1 g KCl, 0.5 g K2HPO4, 0.5 g MgSO4·7H2O and 0.01 g Ca(NO3)2, 40 g FeSO4·7H2O per liter water with pH 1.75, and incubated at 28 °C for 48 h with agitation at 120 rpm on a shaker. Genomic DNA was obtained using sodium dodecyl sulfate (SDS) method and the Blood & Cell Culture DNA midi kit (QIAGEN, Hilden, Germany) following the manufacturer’s standard protocol. Cell morphology of A. ferrooxidans YNTRS-40 was detected using scanning electron microscopy (Hitachi S-4800, Ibraraki, Japan).
2.2. Genome Sequencing, Assembly and Annotation
The sequencing of the complete genome was performed on Nanopore GridION X5 (Oxford Nanopore Technologies, Oxford, United Kingdom) [25] by constructing a 1D genomic DNA library using the ligation sequencing kit 1D (Oxford Nanopore Technologies, Oxford, United Kingdom). After quality filtering, the high-quality reads were assembled into contigs using the Canu v1.7.11 (Maryland Bioinformatics Labs, Park, MD, USA) [26], and the assembled data was optimized by using Pilon v1.22 (Free Software Foundation, Inc., Boston, MA, USA) [27]. All sequencing project information is in Table S2.
Coding sequences (CDS) in the genome were predicted through Prodigal v2.6.3 (Free Software Foundation, Inc., Boston, Massachusetts) [28]. The tRNAs and rRNAs of the genome were predicted by tRNAscan-SE v2.0 (Lowe Lab, Santa Cruz, CA, USA) [29] and RNAmmer v1.2 (DTU Health Tech, Lyngby, Denmark) [30], respectively. The genome sequence was annotated by the Rapid Annotation Subsystem Technology (RAST) (University of Tennessee, Memphis, TN, USA) [31], the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa Laboratories, Kyoto, Japan) [32], the Clusters of Orthologous Groups (COG) (Bethesda Softworks LLC, Rockville, Maryland) [33] and the Gene Ontology (GO) (Stanford University School of Medicine, Stanford, CA, USA) [34]. The circular map of the genome were obtained using Circos v1.7.11(Canada’s Michael Smith Genome Sciences Centre, Vancouver, Canada) [35].
Based on 16S rRNA gene sequence of strain YNTRS-40 (Accession number: MK811409) and other members of Acidithiobacillus obtained from GenBank, phylogenetic analysis was carried out by MEGA7 software(Temple University, Philadelphia, PA, USA) [36] using CLUSTAL W (EMBL, Heidelberg, Germany) to perform a multiple alignments [37] and the phylogenetic tree was constructed by the neighbor-joining method [38]. Sequence alignment and ANI analysis were performed using MEGA7 software (Temple University, Philadelphia, PA, USA) [36] and JSpeciesWS (Ribocon GmbH, Bremen, Germany) [39] with default settings.
3. Results and Discussion
A. ferrooxidans YNTRS-40 is a Gram-negative non-endospore-forming chemolithoautotrophic aerobic bacteria in the order Acidithiobacillales of the class Acidithiobacillia (Table S1). It possesses resistance against heavy metal and oligotrophic conditions. Although several Acidithiobacillus species with the capacity of iron and sulfur oxidization were identified from acidic environments, their genetic features associated with the resistance to the extreme environment were ambiguous [4]. In this study, A. ferrooxidans YNTRS-40 was isolated and its genome was sequenced to analyze the stress resistance. Microscopically, YNTRS-40 cells displayed rod-shaped and were 0.28–0.40 μm in width and 1.00–1.68 μm in length (Figure 1). This strain grew to a logarithmic stage fastly after 48 h under aerobic conditions at pH 1.75, 28 °C and 120 rpm in a shaker with a modified 9K medium. The complete genome sequences have been submitted to GenBank under the accession number CP040511 (chromosome; Figure 2) and CP040512 (plasmid).
Figure 1.
Scanning electron micrograph of Acidithiobacillus ferrooxidans YNTRS-40.
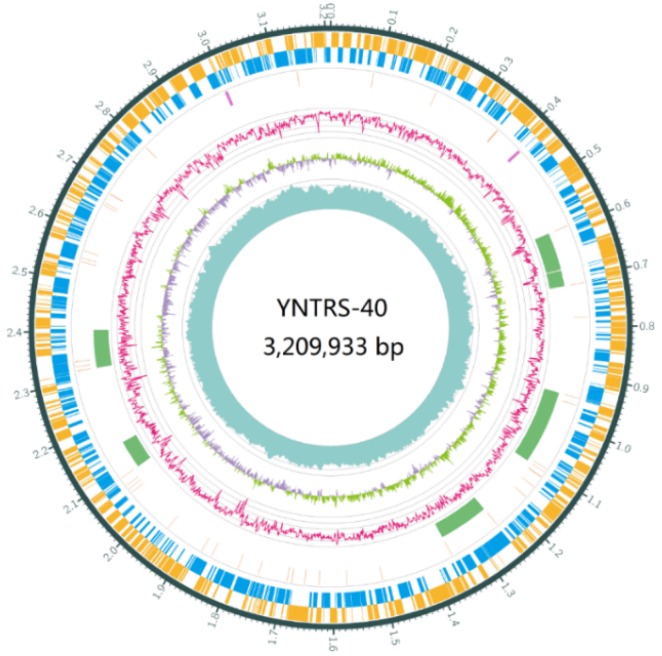
Figure 2.
Circular chromosome genome map of Acidithiobacillus ferrooxidans YNTRS-40. (From the outside to the center, genes on direct strand, genes on complementary strand, tRNAs (orange), rRNAs (purple), CRISPR (blue), and genomic island (green), GC-skew, sequencing depth are displayed).
The genome size of A. ferrooxidans YNTRS-40 is 3,257,037-bp, and the genome contains one circular chromosome of 3,209,933-bp with 58.54% GC content and one circular plasmid (47,104-bp with 56.43% GC content). The circular chromosome comprised 3349 predicted CDS genes, 6 rRNAs, 52 tRNAs and 6 ncRNAs (Table 1), and the circular plasmid contained 70 predicted CDS genes. The statistics and properties of the genome are summarized in Table 1. Total 2015 genes identified from the chromosome were classified into 26 functional categories based on the Cluster of Orthologous Groups (COG; Table 2) [40]. Among all categories, the inorganic ion transport and metabolism category (P, 6.70%), the energy production and conversion category (C, 6.40%) and the defense mechanisms category (V, 4.47%) indicate that the strain YNTRS-40 can grow in the environment with high concentrations of metal ion.
Table 1.
Genome statistics of Acidithiobacillus ferrooxidans YNTRS-40.
| Attribute | Value | % of Total 1 |
|---|---|---|
| Genome size (bp) | 3,257,037 | 100.00 |
| DNA coding (bp) | 2,940,490 | 90.28 |
| DNA G + C (bp) | 1,905,651 | 58.51 |
| DNA scaffolds | 2 | 100.00 |
| Total genes | 3419 | 100.00 |
| Protein coding genes | 3349 | 97.95 |
| RNA genes | 70 | 2.05 |
| Pseudo genes | NA 2 | NA 2 |
| Genes in internal clusters | 8 | 16.21 |
| Genes with function prediction | 1692 | 50.52 |
| Genes assigned to COGs | 1793 | 53.54 |
| Genes with Pfam domains | 2539 | 75.81 |
| Genes with signal peptides | NA 2 | NA 2 |
| Genes with transmembrane helices | NA 2 | NA 2 |
| CRISPR repeats | 0 | 0 |
1 The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome; 2 NA, not applicable.
Table 2.
Number of genes associated with general COGs functional categories.
| Code | Value | % Age 1 | Description |
|---|---|---|---|
| J | 172 | 5.14 | Translation, ribosomal structure, and biogenesis |
| A | 0 | 0 | RNA processing and modification |
| K | 118 | 3.52 | Transcription |
| L | 126 | 3.76 | Replication, recombination, and repair |
| B | 1 | 0.03 | Chromatin structure and dynamics |
| D | 29 | 0.87 | Cell cycle control, Cell division, chromosome partitioning |
| Y | 0 | 0 | Nuclear structure |
| V | 90 | 2.69 | Defense mechanisms |
| T | 87 | 2.60 | Signal transduction mechanisms |
| M | 162 | 4.84 | Cell wall/membrane/envelope biogenesis |
| N | 20 | 0.60 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 14 | 0.42 | Extracellular structures |
| U | 62 | 1.85 | Intracellular trafficking, secretion, and vesicular transport |
| O | 96 | 2.87 | Posttranslational modification, protein turnover, chaperones |
| C | 129 | 3.85 | Energy production and conversion |
| G | 93 | 2.78 | Carbohydrate transport and metabolism |
| E | 137 | 4.09 | Amino acid transport and metabolism |
| F | 53 | 1.58 | Nucleotide transport and metabolism |
| H | 108 | 3.22 | Coenzyme transport and metabolism |
| I | 72 | 2.15 | Lipid transport and metabolism |
| P | 135 | 4.03 | Inorganic ion transport and metabolism |
| Q | 34 | 1.02 | Secondary metabolites biosynthesis, transport, and catabolism |
| R | 138 | 4.12 | General function prediction only |
| S | 51 | 1.52 | Function unknown |
| X | 88 | 2.63 | Mobilome: prophage, transposons |
| - | 1334 | 39.83 | Not in COGs |
1 The total is based on the total number of protein coding genes (3349) in the genome.
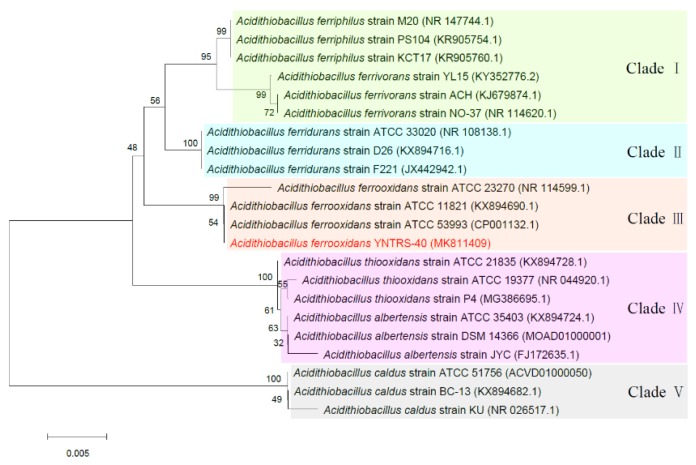
Based on 16S rRNA gene sequence analysis, it can be seen that all strains clustered separately into different clades, such as A. ferriphilus (Clade I), A. ferrivorans (Clade I), A. ferridurans (Clade II), A. ferrooxidans (Clade III), A. thiooxidans (Clade IV), A. albertensis (Clade IV), and A. caldus (Clade V). This finding was similar to a study by Zhang et al. [41] and slightly different from literature, in which A. ferridurans, A. thiooxidans, and A. albertensis clustered into the common clade [2]. The strain YNTRS-40 appeared to represent a coherent group with Acidithiobacillus ferrooxidans ATCC 11821 and Acidithiobacillus ferrooxidans ATCC 53993 (Figure 3). The 16S rRNA gene sequence similarities between the strain YNTRS-40 and the closest relative and the result of ANI analysis among A. ferrooxidans strains, including the strain YNTRS-40, are shown in Tables S3 and S4.
Figure 3.
Phylogenetic tree based on the 16S rRNA gene sequence of Acidithiobacillus ferrooxidans YNTRS-40 and its relatives. Bootstrap values were calculated by MEGA7 using the neighbor-joining method from 1000 replications. Bar, 0.005 nucleotide substitutions per nucleotide position.
3.1. Genomic Features Related to Adaptation to Diverse Stresses
Acidithiobacillus spp. possess extreme environmental resistance, and they can adjust their survival, colonization, growth, and development to extremely acidic conditions (grow optimally at pH 2.0 and survive in pH 1.0–4.5) [42,43,44]. To balance the extracellular and intracellular environment heterogeneity in extremely acidic habitats containing heavy metal ions, these acidophilic microorganisms diverge and evolve to possess the acid and metal resistance [40].
Genome analysis using the Rpsblast and the Interproscan v5.30–69.0 [45] revealed many functional genes involved in the adaptation of strain YNTRS-40 to extreme environments (Table 2 and Tables S5 and S6). Among them, the Cus systems are critical in copper resistance [46]. The expression of proteins such as oxidoreductase and transferase in the cell should be regulated under the environment containing sulfur and metal ions. It has been documented that the oxidoreductase and transferase not only participate in energy generation but also enhance tolerance to environmental stress [47,48,49]. Based on the category of biological process in Table S5, the gene function of strain YNTRS-40, such as response to extracellular stimulus, cellular response to stress, and response to oxidative stress, indicated that it could cope with extreme environmental stress.
Based on the COG analysis (Table 2), the functional genes related to defense mechanisms (V), cell wall/membrane/envelope biogenesis (M), amino acid transport and metabolism (E), inorganic ion transport and metabolism (P) and general function prediction only (R) were slightly more than the other genes. These revealed that this strain exhibited excellent environmental adaptability and has potential applications in the ecological industry, such as sulfur removal from gases, metal extraction from electronic waste [21]. Additionally, the genes associated with function unknown (S) indicated that strain YNTRS-40 might possess some new genes [50].
The plasmid usually contains the genes related to secondary metabolism according to the characteristics of microorganisms [51]. There were 70 CDSs in the circular plasmid of strain YNTRS-40, and 39 CDSs of them were predicted as hypothetical proteins, and the rest were found to be involved in metabolism and defense (Table S7). The RAST annotation results showed that the plasmid comprised all kinds of secondary metabolism-related genes, transcriptional regulatory genes, transposase-related genes, mobile element protein-related genes, and stress-tolerance genes. These indicated that the primary metabolism-related genes were not present in this plasmid, and the presence of plasmid might favor the adaptation of this strain to environmental stress.
3.2. Genomic Features Related to the Oxidation of Ferrous Iron and Sulfur
A. ferrooxidans can gain energy from the oxidation of Fe2+ for growth and survival [5,20]. During the oxidation of Fe2+, most of the electrons are transferred to O2 along the potential gradient, which called downhill potential gradient, while a small part of electrons is transmitted conversely along the potential gradient, which named uphill potential gradient [52]. In the latter process, the NAD(P)H is generated and involved in CO2 fixation and aerobic metabolism [20,52,53]. These two electron transfer pathways, namely the downhill and the uphill potential gradients, are interrelated [52].
The KEGG analysis showed that several coding genes related to the downhill and the uphill electron transfer pathway existed in the strain YNTRS-40 (Table S8). Among them, the rus operon, which consists of cyc2, cyc1, cup, coxB, coxA, coxC, coxD, and rus genes, were found to be involved in electron transfer in the downhill electron pathway [54]. The previous study suggested that the expression and regulation of the rus operon are associated with the substrate electron donor in the environment [54]. The operon might be activated persistently when bacteria use Fe2+ as an electron donor, but only expressed transitorily during the period of early logarithmic growth when cells take S as an electron donor [55,56]. Additionally, the pet I, pet II and res operon were found in the genome of the strain YNTRS-40. Among them, pet I encodes a bc1 complex that participates in the inverse electron transfer when the strain uses Fe2+ as a substrate, and pet II encodes another bc1 complex, which is responsible for forwarding electron transfer when S is used as substrate [57]. The res operon near the pet operon encodes ResB and ResC protein, which might serve as a molecular chaperone in the maturation process of the c1 cytochrome of the bc1 complex [58].
A. ferrooxidans also can obtain the energy required for growth by oxidizing reduced sulfur. Sulfur was found to be a more favorable energy source than Fe2+ because it can provide more ATP than Fe2+ at the same molar level [59,60]. The KEGG results indicated that some genes involved in sulfur oxidation, including sqr, doxDA, cydAB, and cyoABCD genes, which code sulfide quinone reductase, thiosulfate quinone oxidoreductase and thiosulfate dehydrogenase, respectively (Table S8). These genes might be upregulated in strain YNTRS-40 when sulfur is used as a substrate. Additionally, the enzymes encoded by these genes were coupled to a respiratory chain and occurred at different nodes in the respiratory chain [61]. These results suggested that the strain YNTRS-40 has the potential for industrial application through iron and sulfur-oxidizing such as metal bioleaching, gas desulfurization, and bioremediation.
4. Conclusions
The genome of A. ferrooxidans YNTRS-40 revealed that the strain could grow well under extremely acidic conditions containing heavy metal ions and has the ability to remove sulfur from gases and extract metal from solids since it contained various genes participating in the adaptation to environmental stress and the oxidation of ferrous iron and sulfur. This paper is a simultaneous first in reporting the chromosome genome and the plasmid genome of A. ferrooxidans. It could be helpful to research the metabolic characteristics and commercial application potential of A. ferrooxidans YNTRS-40 in the future.
Acknowledgments
Nextomics Biosciences Co., Ltd. is acknowledged for help with DNA sequencing.
Supplementary Materials
https://www.mdpi.com/2076-2607/8/1/2/s1. Table S1: Classification and general features of Acidithiobacillus ferrooxidans YNTRS-40 according to the MIGS recommendations, Table S2: Genome sequencing project information for Acidithiobacillus ferrooxidans YNTRS-40, Table S3: The 16S rRNA gene sequence similarities between the strain YNTRS-40 and the closest relative, Table S4: The result of ANI analysis among Acidithiobacillus ferrooxidans strains including the strain YNTRS-40, Table S5: Partial GO annotation of coding proteins in Acidithiobacillus ferrooxidans YNTRS-40 chromosome genome, Table S6: Identified metal resistance genes in Acidithiobacillus ferrooxidans YNTRS-40, Table S7: RAST categories of CDSs in Acidithiobacillus ferrooxidans YNTRS-40 plasmid genome, Table S8: Identified genes associated with ferrous iron and sulfur oxidation in Acidithiobacillus ferrooxidans YNTRS-40.
Author Contributions
Conceptualization, Y.Z. and L.Y.; methodology, Y.Z.; software, Y.Z. and D.Z.; validation, L.Y., W.W. and S.Z.; formal analysis, L.Y.; investigation, Y.N. and L.Y.; resources, L.Y. and Y.N.; data curation, L.Y.; writing—original draft preparation, Y.Z.; writing—review and editing, L.Y.; visualization, S.Z.; supervision, W.W.; project administration, L.Y.; funding acquisition, L.Y. and W.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Longjiang Scholar Program of Heilongjiang Province, grant number Q201815; National Natural Science Foundation of China, grant number 41471201 and 31100006; Natural Science Foundation of Heilongjiang Province of China, grant number QC2014C023, C201442 and ZD2018005; Support Program of Scientific Research Team and Platform of HBAU, grant number TDJH201809; National Key Research and Development Program of China, grant number 2018YFD0800906; Technology Program of Land Reclamation General Bureau of Heilongjiang, grant number HNK135-04-08 and HKKY190406; Program of Science and Technology Innovation Team of the Heilongjiang Higher Education Institutions, grant number 2012TD006.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Colman D.R., Poudel S., Hamilton T.L., Havig J.R., Selensky M.J., Shock E.L., Boyd E.S. Geobiological feedbacks and the evolution of thermoacidophiles. ISME J. 2018;12:225–236. doi: 10.1038/ismej.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nuñez H., Moya-Beltrán A., Covarrubias P.C., Issotta F., Cárdenas J.P., González M., Atavales J., Acuña L.G., Johnson D.B., Quatrini R. Molecular systematics of the genus Acidithiobacillus: Insights into the phylogenetic structure and diversification of the taxon. Front. Microbiol. 2017;8:30. doi: 10.3389/fmicb.2017.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hongbo Z., Yisheng Z., Xian Z., Lu Q., Menglin S., Yu Y., Yansheng Z., Jun W., Hyunjung K., Guanzhou Q. The dissolution and passivation mechanism of chalcopyrite in bioleaching: An overview. Miner. Eng. 2019;136:140–154. [Google Scholar]
- 4.Xian Z., Xueduan L., Yili L., Fenliang F., Xiaoxia Z., Huaqun Y. Metabolic diversity and adaptive mechanisms of iron- and/or sulfur-oxidizing autotrophic acidophiles in extremely acidic environments. Env. Microbiol. Rep. 2016;8:738–751. doi: 10.1111/1758-2229.12435. [DOI] [PubMed] [Google Scholar]
- 5.Temple K.L., Colmer A.R. The autotrophic oxidation of iron by a new bacterium: Thiobacillus ferrooxidans. J. Bacteriol. 1951;62:605–611. doi: 10.1128/jb.62.5.605-611.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falagán C., Johnson D.B. Acidithiobacillus ferriphilus sp. nov., a facultatively anaerobic iron- and sulfur-metabolizing extreme acidophile. Int. J. Syst. Evol. Microbiol. 2016;66:206–211. doi: 10.1099/ijsem.0.000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallberg K.B., González-Toril E., Johnson D.B. Acidithiobacillus ferrivorans, sp. nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles. 2010;14:9–19. doi: 10.1007/s00792-009-0282-y. [DOI] [PubMed] [Google Scholar]
- 8.Hedrich S., Johnson D.B. Acidithiobacillus ferridurans sp. nov., an acidophilic iron-, sulfur- and hydrogen-metabolizing chemolithotrophic gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 2013;63:4018–4025. doi: 10.1099/ijs.0.049759-0. [DOI] [PubMed] [Google Scholar]
- 9.Bryant R.D., McGroarty K.M., Costerton J.W., Laishley E.J. Isolation and characterization of a new acidophilic Thiobacillus species (T. albertis) Can. J. Microbiol. 1983;29:1159–1170. doi: 10.1139/m83-178. [DOI] [Google Scholar]
- 10.Waksman S.A., Joffe J.S. Microorganisms concerned in the oxidation of sulfur in the soil: II. Thiobacillus Thiooxidans, a new sulfur-oxidizing organism isolated from the soil. J. Bacteriol. 1922;7:239–256. doi: 10.1128/jb.7.2.239-256.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallberg K.B., Lindström E.B. Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology. 1994;140:3451–3456. doi: 10.1099/13500872-140-12-3451. [DOI] [PubMed] [Google Scholar]
- 12.Falagán C., Moya-Beltrán A., Castro M., Quatrini R., Johnson D.B. Acidithiobacillus sulfuriphilus sp. nov.: An extremely acidophilic sulfur-oxidizing chemolithotroph isolated from a neutral pH environment. Int. J. Syst. Evol. Microbiol. 2019;69:2907–2913. doi: 10.1099/ijsem.0.003576. [DOI] [PubMed] [Google Scholar]
- 13.Kelly D.P., Wood A.P. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 2000;50:511–516. doi: 10.1099/00207713-50-2-511. [DOI] [PubMed] [Google Scholar]
- 14.Nitschke W., Bonnefoy V. Energy acquisition in low pH environments. In: Quatrini R., Johnson D.B., editors. Acidophiles: Life in Extremely Acidic Environments. Caister Academic Press; Norfolk, UK: 2016. pp. 19–48. [Google Scholar]
- 15.Davis R.A., Jr., Welty A.T., Borrego J., Morales J.A., Pendon J.G., Ryan J.G. Rio Tinto estuary (Spain): 5000 years of pollution. Environ. Geol. 2000;39:1107–1116. doi: 10.1007/s002549900096. [DOI] [Google Scholar]
- 16.González-Toril E., Llobet-Brossa E., Casamayor E.O., Amann R., Amils R. Microbial ecology of an extreme acidic environment, the Tinto River. Appl. Environ. Microbiol. 2003;69:4853–4865. doi: 10.1128/AEM.69.8.4853-4865.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rawlings D.E. Heavy metal mining using microbes. Annu. Rev. Microbiol. 2002;56:65–91. doi: 10.1146/annurev.micro.56.012302.161052. [DOI] [PubMed] [Google Scholar]
- 18.Rawlings D.E. Characteristics and adaptability of iron- and sulfur-oxidizing microorganisms used for the recovery of metals from minerals and their concentrates. Microb. Cell Fact. 2005;4:13. doi: 10.1186/1475-2859-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sydow A., Krieg T., Mayer F., Schrader J., Holtmann D. Electroactive bacteria—Molecular mechanisms and genetic tools. Appl. Microbiol. Biotechnol. 2014;98:8481–8495. doi: 10.1007/s00253-014-6005-z. [DOI] [PubMed] [Google Scholar]
- 20.Valdés J., Pedroso I., Quatrini R., Dodson R.J., Tettelin H., Blake R., Eisen J.A., Holmes D.S. Acidithiobacillus ferrooxidans metabolism: From genome sequence to industrial applications. BMC Genom. 2008;9:597. doi: 10.1186/1471-2164-9-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shuang Z., Lei Y., Weijia X., Peng C., Yu Z., Weidong W. Acidithiobacillus ferrooxidans and its potential application. Extremophiles. 2018;22:563–579. doi: 10.1007/s00792-018-1024-9. [DOI] [PubMed] [Google Scholar]
- 22.Lei Y., Shuang Z., Weidong W., Huixin H., Yanjie W., Gaobo Y., Peng C. Draft genome sequence of Acidithiobacillus ferrooxidans YQH-1. Genom. Data. 2015;6:269–270. doi: 10.1016/j.gdata.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng C., Lei Y., Zhengrong W., Ruixiang X., Suyue L., Ningbo W., Ning L., Hongyu L. Draft genome sequence of extremely acidophilic bacterium Acidithiobacillus ferrooxidans DLC-5 isolated from acid mine drainage in Northeast China. Genom. Data. 2015;6:267–268. doi: 10.1016/j.gdata.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava S., Briggs B.R., Dong H. Abundance and taxonomic affiliation of molybdenum transport and utilization genes in Tengchong hot springs, China. Environ. Microbiol. 2018;20:2397–2409. doi: 10.1111/1462-2920.14250. [DOI] [PubMed] [Google Scholar]
- 25.Senol Cali D., Kim J.S., Ghose S., Alkan C., Mutlu O. Nanopore sequencing technology and tools for genome assembly: Computational analysis of the current state, bottlenecks and future directions. Brief. Bioinform. 2018;20:1542–1559. doi: 10.1093/bib/bby017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koren S., Walenz B.P., Berlin K., Miller J.R., Phillippy A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyatt D., Gwo-Liang C., LoCascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe T.M., Chan P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:54–57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagesen K., Hallin P., Rødland E.A., Stærfeldt H.-H., Rognes T., Ussery D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2013;42:199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2014;43:261–269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 38.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Michael R., Ramon R.M., Frank O.G., Jörg P. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xian Z., Xueduan L., Liangzhi L., Guanyun W., Danli Z., Yili L., Bo M. Phylogeny, divergent evolution, and speciation of sulfur-oxidizing Acidithiobacillus populations. BMC Genom. 2019;20:438. doi: 10.1186/s12864-019-5827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen A.B., Webb C. Ferrous sulphate oxidation using thiobacillus ferrooxidans: A review. Process. Biochem. 1995;30:225–236. doi: 10.1016/0032-9592(95)85003-1. [DOI] [Google Scholar]
- 43.Wenbin N., Dejuan Z., Feifan L., Lei Y., Peng C., Xiaoxuan Y., Hongyu L. Quorum-sensing system in Acidithiobacillus ferrooxidans involved in its resistance to Cu2+ Lett. Appl. Microbiol. 2011;53:84–91. doi: 10.1111/j.1472-765X.2011.03066.x. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y., Yang M., Yao T., Xiong H. Isolation, identification and arsenic-resistance of Acidithiobacillus ferrooxidans HX3 producing schwertmannite. J. Environ. Sci. 2014;26:1463–1470. doi: 10.1016/j.jes.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Jones P., Binns D., Hsin-Yu C., Fraser M., Weizhong L., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Almárcegui R.J., Navarro C.A., Paradela A., Albar J.P., von Bernath D., Jerez C.A. Response to copper of Acidithiobacillus ferrooxidans ATCC 23270 grown in elemental sulfur. Res. Microbiol. 2014;165:761–772. doi: 10.1016/j.resmic.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Sugio T., Iwahori K., Takeuchi F., Negishi A., Maeda T., Kamimura K. Cytochrome c oxidase purified from a mercury-resistant strain of Acidithiobacillus ferrooxidans volatilizes mercury. J. Biosci. Bioeng. 2001;92:44–49. doi: 10.1016/S1389-1723(01)80197-3. [DOI] [PubMed] [Google Scholar]
- 48.Chengying J., Liu L., Xu G., Xiaoyan Y., Shuangjiang L., Poetsch A. Resolution of carbon metabolism and sulfur-oxidation pathways of Metallosphaera cuprina Ar-4 via comparative proteomics. J. Proteom. 2014;109:276–289. doi: 10.1016/j.jprot.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 49.Yue Z., Mengran Y., Shuang Z., Dan Z., Jiangong D., Weidong W., Lei Y. Iron and sulfur oxidation pathways of Acidithiobacillus ferrooxidans. World J. Microbiol. Biotechnol. 2019;35:60. doi: 10.1007/s11274-019-2632-y. [DOI] [PubMed] [Google Scholar]
- 50.Ruiqi Y., Guangxiu L., Tuo C., Wei Z., Gaosen Z., Sijing C. The complete genomic sequence of a novel cold-adapted bacterium, Planococcus maritimus Y42, isolated from crude oil-contaminated soil. Stand. Genom. Sci. 2018;13:23. doi: 10.1186/s40793-018-0328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mochizuki S., Hiratsu K., Suwa M., Ishii T., Sugino F., Yamada K., Kinashi H. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol. Microbiol. 2003;48:1501–1510. doi: 10.1046/j.1365-2958.2003.03523.x. [DOI] [PubMed] [Google Scholar]
- 52.Barrie Johnson D., Hallberg K.B. Carbon, iron and sulfur metabolism in acidophilic micro-organisms. Adv. Microbiol. Physiol. 2008;54:201–255. doi: 10.1016/S0065-2911(08)00003-9. [DOI] [PubMed] [Google Scholar]
- 53.Ilbert M., Bonnefoy V. Insight into the evolution of the iron oxidation pathways. Biochim. Biophys. Acta. 2013;1827:161–175. doi: 10.1016/j.bbabio.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Yarzabal A., Appia-Ayme C., Ratouchniak J., Bonnefoy V. Regulation of the expression of the Acidithiobacillus ferrooxidans rus operon encoding two cytochromes c, a cytochrome oxidase and rusticyanin. Microbiology. 2004;150:2113–2123. doi: 10.1099/mic.0.26966-0. [DOI] [PubMed] [Google Scholar]
- 55.Quatrini R., Jedlicki E., Holmes D.S. Genomic insights into the iron uptake mechanisms of the biomining microorganism Acidithiobacillus ferrooxidans. J. Ind. Microbiol. Biotechnol. 2005;32:606–614. doi: 10.1007/s10295-005-0233-2. [DOI] [PubMed] [Google Scholar]
- 56.Yarzábal A., Duquesne K., Bonnefoy V. Rusticyanin gene expression of Acidithiobacillus ferrooxidans ATCC 33020 in sulfur- and in ferrous iron media. Hydrometallurgy. 2003;71:107–114. doi: 10.1016/S0304-386X(03)00146-4. [DOI] [Google Scholar]
- 57.Brasseur G., Bruscella P., Bonnefoy V., Lemesle-Meunier D. The bc1 complex of the iron-grown acidophilic chemolithotrophic bacterium Acidithiobacillus ferrooxidans functions in the reverse but not in the forward direction: Is there a second bc1 complex? Biochim. Biophys. Acta. 2002;1555:37–43. doi: 10.1016/S0005-2728(02)00251-7. [DOI] [PubMed] [Google Scholar]
- 58.Quatrini R., Appia-Ayme C., Denis Y., Jedlicki E., Holmes D.S., Bonnefoy V. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genom. 2009;10:394. doi: 10.1186/1471-2164-10-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Findlay A.J., Kamyshny A. Turnover rates of intermediate sulfur species (Sx2-, S0, S2O32-, S4O62-, SO32-) in anoxic freshwater and sediments. Front. Microbiol. 2017;8:2551. doi: 10.3389/fmicb.2017.02551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mangold S., Valdés J., Holmes D., Dopson M. Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front. Microbiol. 2011;2:17. doi: 10.3389/fmicb.2011.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quatrini R., Appia-Ayme C., Denis Y., Ratouchniak J., Veloso F., Valdes J., Lefimil C., Silver S., Roberto F., Orellana O., et al. Insights into the iron and sulfur energetic metabolism of Acidithiobacillus ferrooxidans by microarray transcriptome profiling. Hydrometallurgy. 2006;83:263–272. doi: 10.1016/j.hydromet.2006.03.030. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.