Graphical Abstract
Acetylated lysine binding pocket of BRDT Select BET inhibitors

Importance of and need for male contraceptives
The first oral female contraceptive (oral contraceptive pills, OCP) was introduced in 1960, opening the market to today’s plethora of options for women seeking birth control. Hormonal pills and implants, topical patches, vaginal rings, depo injectables, and intrauterine devices are available to provide safe, reliable, effective and reversible contraception, along with other non-contraceptive benefits.1
However, many women are unable to use hormonal contraceptives due to preexisting medical or pathological conditions.2 Additionally, some women may choose to discontinue hormonal contraceptive use due to associated side effects.3–5 Recently, it was discovered that some women may experience contraceptive failures due to a difference in steroid metabolism that depletes the blood concentration of hormonal contraceptives; even with perfect use of their hormonal contraceptive, these women are at risk of unintended pregnancy.6 Furthermore, some women can find it difficult to obtain contraceptives due to cost of effective prescription contraception and the associated doctors’ visits.7
Contraceptive options for men are limited. Condoms are 98% effective at preventing pregnancy with perfect use, but with average use are only 83–85% effective.7–9 Vasectomies are another major form of male contraception and while reliable are not easily reversible, making the procedure exclusively a long-term contraceptive method.8, 9 Withdrawal has an unintended pregnancy rate of 22%.9 The disproportionate number of contraceptive methods available to men has put the onus of family planning largely on women.
Despite the myriad contraceptive options available, in 2011 the unintended pregnancy rate in the United states was still 45%, down from 51% in 2008, as a percentage of reported pregnancies (Figure 1).10 Most of these unintended pregnancies can be attributed to lack of contraceptive use, but 43% of unintended pregnancies reported were caused by inconsistent or incorrect use of contraceptives.7 Globally, nearly half of pregnancies are unplanned.11
Figure 1.
Reported percentage of pregnancies in the United States in all women.10
Contraceptives are only effective at preventing unintended pregnancies with continued and near-perfect use, and the use of multiple methods at one time (i.e., condoms and OCP) is recommended.9 Clearly, there is a need for a safe, effective, reversible male contraceptive.
Since as early as the 1970s, researchers have been investigating the possibility of a male hormonal contraceptive (MHC), seeking to suppress spermatogenesis by interfering with the normal release of gonadotropin-releasing hormone, and thus the downstream luteinizing hormone and follicle stimulating hormone through negative feedback of exogenous testosterone. Both androgen-only and androgen-progestin combination MHC regimens have been studied and are reviewed elsewhere.11–16
The general consensus from MHC studies is both a positive proof of principle and that androgen-progestin combination regimens are more effective than androgen-only conditions. Common side effects include acne, changes in mood, night sweats, a reversible decrease in testicular volume, and changes in cholesterol profile (decreased HDL, LDL, and total cholesterol). The short durations of treatment (typically no longer than one year) have precluded adequate assessment of cardiovascular or thromboembolic events related to use, or other unknown long-term medical problems. Attempts to create a hormonal contraceptive option for men have proven unsuccessful, due to high prevalence of side effects and lack of universal or uniform efficacy, thus no hormonal regimen has progressed to the approval phase.11, 15
Another area being explored for potential male contraceptive methods is that of physical occlusion of the vas deferens. Several surgical and non-surgical methods are currently under investigation and are reviewed in greater detail elsewhere.11, 12, 14 These options generally show good contraceptive properties in clinical and preclinical trials, establishing proof of principle, but studies demonstrating the reversibility of these methods are still required.
Several non-hormonal contraceptive agents have been studied, though none are currently in clinical trials.17 One of the most well studied potential therapies is gossypol, a naturally occurring phenol originally extracted from the cotton plant. While early studies showed it to be well-tolerated with a strong contraceptive effects, further research indicated inconsistent results as a contraceptive, poor recovery of fertility, and toxicity with prolonged exposure, prohibiting gossypol from use as a modern contraceptive.12, 14 Another interesting and validated target for non-hormonal male contraception is the retinoic acid receptor (RAR), which is discussed in more depth elsewhere.11, 12, 14, 15 Development of RAR antagonists is currently underway, but as yet no RAR antagonists have been shown to inhibit spermatogenesis in humans.12, 15 Other therapies for non-hormonal male contraception are currently being explored, including adrenergic receptors, phenoxybenzamine, prazosin, tamsulosin, adjudin, H2-gamendazole, and others reviewed elsewhere, though none are currently in clinical testing.11, 12, 14–16, 18
Role of bromodomains and BRDT
The concept of epigenetics was first introduced in 1939 and later refined to describe heritable changes in gene expression that are not due to alterations in DNA sequence.19 Practically, epigenetics is the study of differential gene expression. Expression can be modified many ways, including post-translational modification (PTM) of the chromatin structure. Many expression changes are caused by marks written, read, or erased from histone proteins, with specific classes of proteins responsible for each modification. Writers, such as acetylases, methylases, and phosphorylases, add marks to histone proteins. Readers, such as bromodomains, chromodomains, and PHD fingers, recognize the modifications. Erasers, such as deacetylases, demethylases, and phosphatases, remove the post-translational marks. These marks lead to changes in the chromatin structure, either uncoiling the DNA structure and encouraging higher levels of transcription, or further coiling the DNA around histone proteins, leading to decreased gene expression. Many different PTMs are used, including serine/threonine/tyrosine phosphorylation, lysine/arginine methylation, citrullination, ubiquitination, and others, that make up the histone code. Further exploration of PTMs has been reviewed elsewhere.20–24
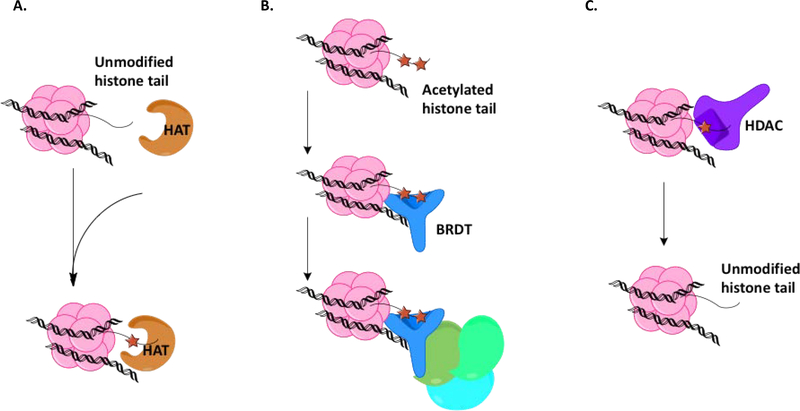
One of the most frequently occurring PTMs is ε-N-acetylation of lysine residues (Figure 2).25 Acetylation levels are strictly controlled by two enzyme families. Lysine acetyltransferases (KATs, or HATs for histone specific acetyltransferases) are responsible for appending acetyl groups onto proteins, acting as the “writers” of this PTM. When the N-ε-lysine sidechains of histone protein tails are acetylated, an unpacking of the chromatin structure is often seen, due to the decreased affinity of the histones for DNA; however, when the positive charge of the lysine tail is masked by the acetyl group, the histone protein loses affinity for the negatively charged phosphate backbone of DNA as the ionic charge-charge interaction is no longer possible.26 Histone deacetylases (HDACs) “erase” acetyl groups from modified proteins, which can lead to condensing of the chromatin structure. The class of proteins responsible for recognizing these acetylated lysines (Kac) and recruiting the appropriate binding partners necessary for chromatin-dependent signal transduction are the bromodomains (BRDs).
Figure 2.
Lysine acetylation post-translational modification. A. Histone acetyl transferase (HAT; orange enzyme) adds acetyl groups (orange stars) to lysine residue of histone tail at two points (H4K5 and H4K8). B. Testis-specific bromodomain BRDT (blue protein) recognizes acetylated lysine residues and recruits binding partners. C. Histone deacetylase (HDAC; purple enzyme) removes acetyl group from lysine residue, leaving an unmodified histone tail.
BRDs are ~110 amino acid interaction modules first discovered in 1992 and named for the brahma gene in Drosophila melanogaster by Tamkun et al.27 Subsequent research has revealed much about BRDs and to date, 61 human BRDs have been identified in 46 different proteins.28 BRD-containing proteins are often large, multidomain proteins associated with chromatin remodeling, transcriptional control, or DNA repair, with the BRD motif surrounded by other epigenetic reader domains. These flanking domains may be tightly linked together to form a single interaction domain, or connected loosely by long, flexible linker regions allowing the motifs to be accommodating of diverse substrates.
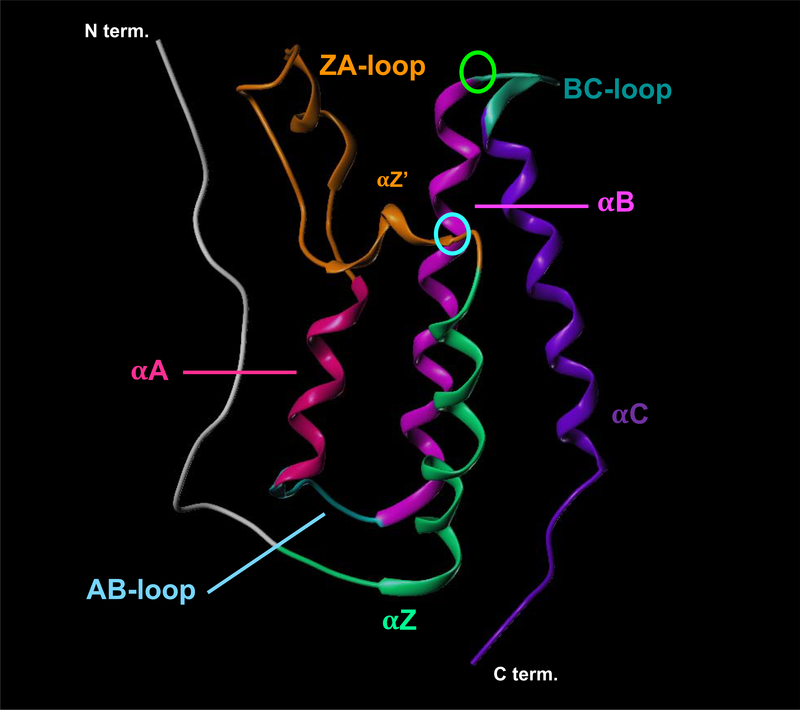
A pivotal study of the human BRDs by Filippakopoulos et al. has shown that despite large sequence variations, all BRDs share the same conserved fold of a left-handed bundle of four alpha helices (αZ, αA, αB, and αC) linked by two highly variable loop regions (ZA and BC loops) (see Figure 3).28 These loops come together to form a long hydrophobic groove (ZA channel) leading to an asparagine residue (Asn109 in the first BRD of BRDT and Asn140 in the first BRD of BRD4) conserved on the BC loop, seen in most BRDs, responsible for Kac binding (Figure 4). The Kac of histone tails are bound by BRDs through a hydrogen bond from the acetyl carbonyl of the Kac to the amide nitrogen of this conserved Asn. A tyrosine residue (Tyr66 in BRDT(1); Tyr97 in BRD4(1)) within the binding site (ZA loop) has also been shown to be essential for Kac binding through a water-mediated hydrogen bond and is found in most BRDs.28, 29
Figure 3.
Ribbon structure of bromodomain containing protein BRD4(1) showing canonical structure of bromodomains. N and C termini are labeled as are canonical secondary structures. Critical Kac-binding Asn residue (Asn109 of BRDT(1)) circled in green, critical Tyr residue (Tyr66 of BRDT(1)) circled in cyan.
Figure 4.
The Kac binding pocket architecture of BRDT(1) shown as semi-transparent surface with residues labeled and depicted in sticks. Landmarks of Kac binding site highlighted in color: KL flank (teal), WPF shelf (orange), Asn rim (magenta), ZA channel and conserved water molecules (red). Critical Asn109 residue circled in green.
By manually aligning and visually inspecting the structures of all 61 human BRDs, Filippakopoulos et al. were able to identify several features conserved throughout the folded protein domain. One of these features is a conserved phenylalanine found in the C terminus of the Z’ helix within the ZA loop. This Phe is part of a structure termed the WPF shelf, named for the tryptophan, proline, and phenylalanine residues (Trp81, Pro82, and Phe83) in BRD4(1) (Figure 4). This general motif is present in most BRDs as a hydrophobic ridge along the rim of the Kac binding pocket, though the exact residues involved are variable. The WPF shelf is often involved in van der Waals interactions with BRD binding partners, potentially contributing to ligand selectivity.
Also conserved within the Kac binding site of BRDs are five structural water molecules forming a network along a groove formed by the ZA loop (the ZA channel). These waters contribute to many water-mediated hydrogen bonds that help stabilize the Kac binding pocket and contribute to ligand binding.29 The importance of these structural waters has been demonstrated by several groups, highlighting the ability of these water molecules to tune selectivity and affinity of BRD-ligands, contributing to the druggability of these proteins.30–34 This suggests that researchers should investigate the conserved waters as targetable groups, rather than displaceable obstacles in drug design. To date, only one potent BRD ligand is known to displace these conserved waters.34
The ZA and BC loops surrounding the Kac binding site are highly variable between BRDs, as are the C and N termini of BRD-containing proteins. This low overall sequence homology between BRD-containing proteins is reflected in the highly diverse surface properties seen, especially in the electrostatic potentials, which range from highly positive to strongly negative in charge, suggesting that BRDs recognize a wide array of binding partners. The variability of the ZA and BC loops at the rim of the Kac binding pocket contributes to ligand specificity.28, 35 In their study of BRD structure, Filippakopoulos et al. also explored BRD interactions with histone Kac sites and discovered that some BRDs also bind to non-histone Kac sites.28 They confirmed that BRDs generally have a low affinity for Kac binding, suggesting that additional interactions may be required for higher affinity target-binding. Supporting this is a previous discovery by Morinière et al., showing that diacetylated motifs were preferentially recognized by murine BRDT over monoacetylated peptides.36 This tendency to bind double-acetylated histone tails is conserved across a number of BRDs, where both Kacs insert into the BRD pocket, but only the N-terminal modification interacts with the conserved Asn residue, while the second Kac hydrogen binds with residues of the Kac-binding pocket and the N-terminal Kac through a water-mediated hydrogen bond.
To further explore the idea that multiple PTMs might affect BRD-binding, an experiment systematically interrogating combinations of acetylated and trimethylated lysines (Kme3) along with phosphoserine (pS) and phospho-threonine (pT) PTMs around each Kac site was developed for a peptide array.28 The results showed that most BRDs tested were highly sensitive to PTMs surrounding the Kac of histone proteins. For example, the second BRD of BRD4 (BRD4(2)) interacted strongly with a diacetylated H3 (H3K4acK9ac) truncated peptide, while it did not interact with peptides acetylated on K4 alone. Moreover, BRD4(2) also showed no interaction with H3K4acK9me3, indicating that BRDs are sensitive not only to the number of PTMs on a given protein but also to the type of PTM displayed. Preferences for a specific number and identity of PTMs flanking the Kac site contribute to BRD specificity, but more studies are required before the role of each human BRD can be definitively established.
BET family bromodomains
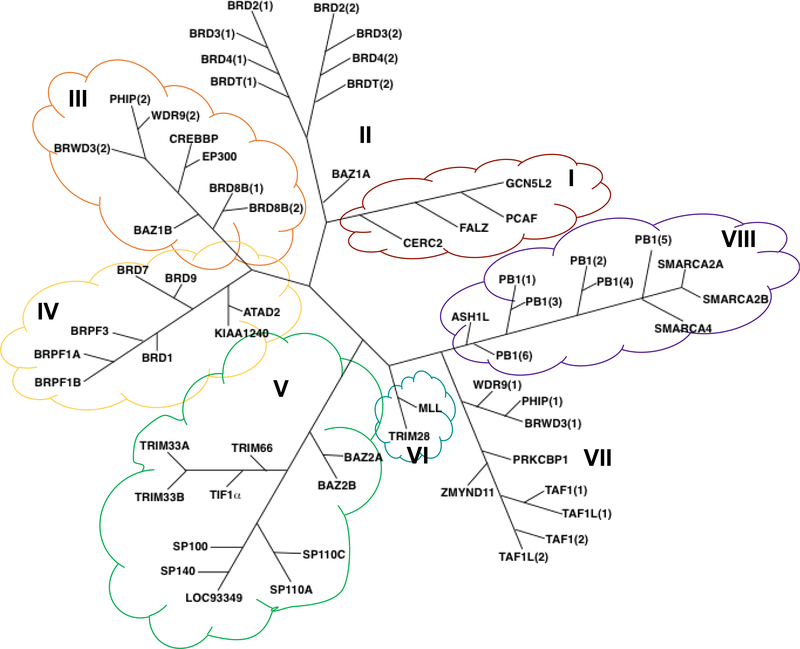
Based on three-dimensional structural alignments, a phylogram was derived clustering human BRDs into eight major families (I-VIII) (Figure 5).28 The most well-studied BRDs come from family II, the bromodomain and extra-terminal (BET) proteins: bromodomain-containing proteins 2, 3, 4 (BRD2, BRD3, BRD4), and testis-specific bromodomain-containing protein (BRDT). The four BET proteins are distinct from other BRD families due to their unique C-terminal extra terminal domain and N-terminal tandem BRDs. The extra terminal domain is responsible for protein-protein interactions involved in recruiting different effector proteins to the chromatin.37, 38 The extra terminal domain is highly conserved across the BET family and also exhibits a helical architecture.39
Figure 5.
Phylogenetic tree of bromodomain containing proteins. The different families are labeled in Roman numerals.
Several conserved motifs are also conserved in the modular architecture of the BET family (Figure 6). A SEED motif at the very C-terminus of the extra terminal domain is composed of serine, glutamate, and aspartate residues. The A motif is located between the tandem BRDs of the BET proteins and contains a conserved stretch of 12 amino acids (KGVKRKADTTTP) that is responsible for the nuclear localization of the BETs; deletion of this signature results in mislocalization of the mutant proteins.40 The BET proteins also contain a conserved ‘motif B’, thought to be responsible for mediating BET protein dimerization, as has been shown in BRD2.41 BRD4 and BRDT also contain a C-terminal domain motif.
Figure 6.
BET bromodomain proteins. A. Electrostatic surface potentials for the first bromodomain of the BET family proteins (blue is negative, red is positive); BRD names and PDB accession codes are shown in the figure. Pink circle highlights critical Asn residue, orange circle shows WPF shelf. B. Structural comparison of the BET family members in mice. The percentages under each bromodomain and the ET domain indicate the percentage of identical amino acid residues in the domain as compared to the corresponding domain in murine BRD2, the founding member of the mouse BET family. The percentages under the CTD are compared to murine BRD4.
The tandem BRDs (BD1 and BD2) of the BET family are conserved in eukaryotes, while plant BET proteins contain only one BRD, supporting the hypothesis that the tandem BRDs arose from a duplication within the ancestral BET gene. Since that time, the first and second have diverged significantly, creating greater similarity between BD1 or BD2 across the mammalian BET family than there is between BD1 and BD2 for a given BET protein. For example, BD1 of BRDT (BRDT(1)) is 73% identical to BRD2(1) and BRDT(2) is 76% identical to BRD2(2), while BRDT(1) is only 40% identical to BRDT(2), though the domains still share high homology and most of the sequence differences lie outside the Kac binding pocket.33, 39 This difference between the tandem BRDs has allowed for probe development, showing proof of principle for selective targeting of BD1 or BD2.42 Chromatin binding is predominantly controlled by BD1, but genetic and chemical-genetic studies have shown roles for both BRDs in transcriptional activation.
The BET proteins are expressed throughout the human body and are responsible for transcriptional regulation in cellular proliferation, mitosis, cell cycle progression, apoptosis, and other cellular functions. BRD2 helps control neuronal differentiation, cell-cycle progression, and cell-cycle exit in neuroepithelial cells in central nervous system development.43 BRD3 is involved in regulating the expression of erythroid and megakaryocyte-specific genes.44 BRD4 is a central regulator of transcriptional elongation by recruiting the positive transcription elongation factor b (P-TEFb) complex to chromatin, leading to the activation of ribonucleic acid polymerase II (RNA Pol II), a protein complex that catalyzes DNA transcription.44 Additional evidence shows BRD4 interacts with a large portion of the genome, allowing the protein to stimulate elongation of both protein-coding and non-coding enhancer RNA transcripts.45 BRD4 also binds many M/G1 genes programmed to be expressed at the end of mitosis, and may even play a role in “bookmarking” certain genes for post-mitotic transcriptional reactivation by enhancing transcription kinetics and helping to decompact chromatin in daughter cells.46, 47 BRDT is required for spermatogenesis, playing an important role in reorganizing chromatin and facilitating histone eviction and replacement by transition proteins during germ cell differentiation, and it also acts as a central regulator of transcriptional elongation through P-TEFb.38, 48
With the central role that BET proteins play in many important cellular functions, misregulation of BET activity is associated with many diverse disease states, as is discussed in more detail elsewhere.44, 49 BRD2 has been shown to play a role in obesity, acute inflammatory response, neuronal development, hematopoiesis, and hematologic malignancies.44, 49–51 BRD3 has been implicated in many aggressive leukemias.44, 49 BRD3 and BRD4 have been involved in chromosomal translocations leading to tumor-causing fusions with nuclear protein in testes (NUT), leading to rare and aggressive carcinomas that have no specific tissue or organ of origin, known as NUT midline carcinomas (NMCs).44 BRD4, in particular, has been implicated in basal-like breast cancer, as a coactivator protein for oncoprotein MYC, and NMCs, as well as many other cancers.44, 49, 52, 53 While BRDT is exclusively expressed in the testes, it has also been seen in lung cancer.54 BETs have also been shown to have a role in tethering viral genomes to mitotic chromosomes.
BET inhibitors
Due to the established role of BET proteins in many disease states, many small molecule inhibitors have been developed. The first potent and selective BET inhibitor was discovered through a phenotypic screen for up-regulators of apolipoprotein A1 (ApoA1) to inhibit inflammation.55, 56 Though the triazolobenzodiazepine I-BET/I-BET-762 was identified without a known target protein, structure-activity relationship (SAR) studies revealed that the benzodiazepine core and the 5-position aryl group were required for activity (Figure 7). Additionally, the C3 stereochemistry strongly affects the potency of the molecule, with the (S)-enantiomer showing activity and the (R)-enantiomer showing no activity. An affinity matrix was used to identify the BET proteins as the ultimate protein target of I-BET, with no other BRDs showing affinity for the molecule; this selectivity was confirmed by thermal stability and other biochemical assays.56 The binding of I-BET to tandem BET proteins was demonstrated by in vitro ITC and SPR experiments, with a Kd of 50.5–61.3 nM, and similar affinity found in a competitive FRET assay where I-BET displaced a tetra-acetylated histone peptide H41–14K5acK8acK12acK16ac (IC50 of 32.5–42.4 nM for tandem BETs).55, 56
Figure 7.
Structures of select BET inhibitors.
Crystal structures with select BET proteins have revealed nearly identical binding modes within the Kac binding pocket: N3 of I-BET binds the conserved Asn140 of BRD4 and makes an additional hydrogen bond to the amide nitrogen while N2 binds the conserved Tyr97 through a structural water molecule. Shape complementarity between ligand and binding site is very good, with van der Waals interactions between the 4-chlorophenyl moiety and the conserved WPF shelf.
The first clinical trial of I-BET was launched in 2012 investigating the effects of the drug on NMC and many other cancers.57 Since then, four other phase I/II clinical trials have begun, though no results have yet been released.58–61
Simultaneously, another triazolodiazepine compound was discovered that selectively inhibited BET proteins, (+)-JQ1, also from an anti-inflammatory phenotypic study.62 Like I-BET, only the (+)-JQ1 enantiomer is active against the BET family, with (−)-JQ1 enantiomer showing no activity against any of the BRDs tested.62 With good selectivity for the BET family proteins, (+)-JQ1 has a Kd of 49 nM (ITC) and an IC50 of 77 nM (AlphaScreen) against BRD4(1). (+)-JQ1 has also been shown to inhibit the interaction of BRD4 with nuclear chromatin in human cells.62 As with I-BET, (+)-JQ1 shows excellent shape complementarity to the Kac binding site. The methylated-1,2,4-triazole moiety of (+)-JQ1 forms hydrogen bonds to the conserved Asn and, through a water molecule, to the Tyr within the Kac binding pocket and the 4-chlorophenyl group interacting with the WPF shelf.62
A widely used chemical probe, (+)-JQ1 has been proven an effective inhibitor of many cancers, including NMC, acute myeloid leukemia, multiple myeloma, and triple-negative breast cancer, among others.62–66 (+)-JQ1 (licensed by Oncoethix and slightly modified to OTX-015b) has been used in five clinical trials treating various cancers and hematologic malignancies.67–71 Though most of these trials have been phase I, determining appropriate doses of OTX-015b, one phase II study treating recurrent glioblastoma multiforme was terminated due to a lack of clinical activity.68
Other triazolodiazepine BET inhibitors include GSK525762, MS417, and CPI-203. Additional chemotypes of BET inhibitors include 3,5-dimethylisoxazoles like I-BET-151, benzimidazoles like BIC1, quinazolinones like PFI-1, and many others reviewed elsewhere.72, 73 As with other drug targets, not all BET inhibitors are drug candidates but instead find use as probe compounds.
The discovery and design of numerous BET inhibitors is ongoing to refine specificity within the BET family. Part of the difficulty of this goal is the nature of the Kac binding site. The binding of a BRD to an acetylated histone tail is a protein-protein interaction. These interfaces are notoriously difficult to inhibit with small molecules due to the broad, shallow, and featureless surfaces involved, making intra-BET family selectivity challenging. While most current BET inhibitors are active against all of the BET proteins, a few have been developed to selectively inhibit the first or second of the BET family’s tandem BDs. Hexahydropyridoindolone Olinone (Figure 7) is a BET-specific inhibitor that prefers BD1 over BD2.74 This selectivity is achieved through a hydrogen bond interaction of the indanone carbonyl with Asp144 of BRD4(1) (and corresponding Asp residues of BRD2(1), BRD3(1), and BRDT(1)). The corresponding residue in BRD4(2) is His435 (and corresponding His residues of BRD2(1), BRD3(1), and BRDT(1)), which crowds out the indolone from the Kac binding pocket.74
RVX-208 and RVX-297 are related quinazolinone BET inhibitors that preferentially bind BD2.42, 75 While both compounds form hydrogen bonds with Asn140 and a water-mediated hydrogen bond to Tyr97 of BRD4(1), the remaining interactions with the Kac binding site are largely hydrophobic and non-specific. RVX-208 and RVX-297 bind to the canonical Asn437 of BRD4(1) but are also able to occupy the entire ZA channel, which is narrowed in BRD2 by the presence of His438 (as compared to Asp144 in BRD4(1)). This tighter pocket of BD2 shows much better shape complementarity with RVX-208 and RVX-297 than BD1, providing some selectivity.42, 75
While intra-BET selectivity continues to prove challenging, the examples of Olinone, RVX-208 and RVX-209 show proof of principle for further selectivity within the family. Continued exploration and exploitation of the protein sequence differences between the BET proteins should allow for tuned specificity.
A newly emerging class of pharmaceuticals is the proteolysis-targeting chimeras (PROTACs), compounds that induce protein knockdown by recruiting the cell’s natural protein degradation processes. PROTACs are chimeric molecules composed of three parts: a target protein ligand, a degradation machinery recruiting unit, and a chemical linker to join the functionalities together. When both ligand moieties have bound their respective partners, the proximity of the proteins leads to the polyubiquitination of the target protein by the E3 ubiquitin ligase complex, and the target protein is subsequently degraded by the proteasome. While first described as large, polar, peptide-based molecules by Sakamoto et al. in 2001, PROTACs have since also been developed into cell-permeable, small-molecule-like entities.76 First generation small-molecule PROTACs matched the peptide-based entities in potency, but exploration of degradation machinery recruiting units (often an E3 ligand component) and linker moieties have greatly increased both potency and selectivity profiles of PROTACs, allowing for some tunability of their effects.77, 78 Additionally, PROTACs have been shown to have catalytic, superstoichiometric target degradation.78 PROTAC development and applications have been reviewed extensively elsewhere.78–81
The BET proteins have been targeted by several PROTAC efforts, most often utilizing (+)-JQ1 or OTX-015 as the BET-targeting ligand. PROTAC ARV-825 combines the BET ligand OTX-015 with pomalidomide, a ligand of the (Cul4-Rbx1-DDB1-)cereblon E3 ubiquitin ligase complex, leading to BET protein degradation (Figure 8). In Burkitt’s lymphoma cell lines, ARV-825 creates a prolonged degradation of BRD4 and a suppression of c-MYC levels and other downstream signalling.82 ARV-825 was found to have similar degradation effects on BRD2 and BRD3 but appears not to have been tested in BRDT. ARV-771 also uses OTX-015 as a BET ligand but contains a Von Hippel-Lindau (VHL)-binding ligand as its E3 recruiting element (Figure 1.2.8.). Like ARV-825, this PROTAC degrades BRD2, BRD3, and BRD4, but it has not been tested against BRDT. ARV-771 dosing leads to BET degradation and depletion of the c-MYC transcript and protein in cells.83 Additionally, while ARV-771 has a similar Kd value to that of (+)-JQ1, the PROTAC demonstrates a more than 10-fold higher efficacy in reducing c-MYC levels. These favorable preclinical assessments of ARV-825 and ARV-771 have created high expectations for clinical testing.
Figure 8.
Structures of select BET-targeting PROTACs.
dBET1 is another PROTAC developed using OTX-015 and pomalidomide, differing from ARV-825 in the linker used to combine these moieties (Figure 8). dBET1 induces a BET-selective cereblon-dependent protein degradation in vitro and in vivo, and additionally was able to delay leukemia progression in mice.84 Quantitative expression proteomics analysis showed a near equal knockdown of BRD2, BRD3, and BRD4 in leukemia cells. However, unlike ARV-825 and ARV-771, dBET1 was also tested against BRDT in a BROMOscan assay (DiscoveRx) and showed equal potency against this testis-specific BET (Kd values ranging from 6 to 19 nM for the BET proteins).84
PROTAC MZ1 is similar in structure to ARV-771, differing in linker length and a slight modification to the VHL-ligand (Figure 8). Interestingly, MZ1 is somewhat selective for BRD4 degradation over BRD2 and BRD3 but was not tested against BRDT.85, 86 Further investigation of this BRD4 selectivity lead to crystallization of the ternary BRD4(2)-MZ1-VHL complex.86 The crystal structure solution revealed that MZI forms specific intermolecular interactions with both BRD4(2) and VHL, but intermolecular interactions are also seen between BRD4(2) and VHL. These extensive, newly identified protein-protein interactions drive the BRD4 selectivity and also lead to increased stabilization of the ternary complex. The ternary complex crystal structure allowed for structure-based drug design efforts resulting in AT1, a PROTAC compound with even greater BRD4 selectivity over BRD2 and BRD3 (again, BRDT was not tested).86 Further exploration of such de novo protein-protein interactions may allow for the selective degradation of other BET proteins, creating intra-family specificity.
BRDT(1) target validation
As mentioned above, BRDT is required for spermatogenesis, encouraging chromosome compaction for the first meiotic division in sperm production. This chromatin reorganization protects the genetic information, and the location and modification state of the remaining histones prepare the genome for reactivation after fertilization. BRDT(1) may act to facilitate post-meiotic chromatin compaction by binding to acetylated histone tails, facilitating the replacement of hyperacetylated histones with sperm-specific, highly basic proteins known as protamines.38, 87 Knock-out experiments have shown that BRDT(1) is essential for spermatogenesis, with male mice maturing to healthy but sterile adults.39, 62, 88, 89
During mouse studies of (+)-JQ1, the researchers noted that the testes of the test animals were unusually small. As a pan-BET inhibitor, (+)-JQ1 also inhibits BRDT, and the small size of the (+)-JQ1-treated testes was due to a lack of mature spermatids.88 As a BET inhibitor, (+)-JQ1 competitively binds BRDT(1), preventing the chromatin remodeling required for spermatogenesis, as shown in Figure 9.A, adapted from the in vitro studies performed by Matzuk and coworkers using both murine (circles) and human (squares) BRDT-(1).88 Breeding studies (Figure 9.B) demonstrated the reversibility of JQ1-induced infertility, where adult males were pretreated with vehicle control or (+)-JQ1 for 6 weeks and then caged continuously with two adult females with continued (+)-JQ1 dosing. Throughout this period, no litters were birthed. The (+)-JQ1 treatment was discontinued and male fertility was restored, with litters born by month 4 of the experiment (one month after treatment cessation). Not only was full fertility restored within one month of treatment cessation, but litter sizes and pups born following (+)-JQ1 treatment proved normal.87
Figure 9.
Inhibition of BRDT(1) by (+)-JQ1. Modified from Matzuk et al.88 A. Competitive inhibition of human (squares) and mouse (circles) BRDT(1) binding to synthetic biotinylated H4Kac4 by JQ1 using proximity detection assays (hBRDT(1) IC50 = 11 nM; mBRDT(1) IC50 = 10 nM). B. Adult males were pretreated for 6 weeks with vehicle control (n = 7) or (+)-JQ1 (50 mg/kg QD; n = 3) and then caged continuously with two females each while continuing 50 mg/kg QD for month 1 and escalating to 75 mg/kg QD for month 2. (+)-JQ1 treatment was stopped at the end of month 2 of mating. Graphical representation of pups born in each month to the females reveals a contraceptive effect evident in months 1–3 (data represent mean ± SEM; *p < 0.001) and durable restoration of fertility at month 4.
This demonstrates the reversibility of BRDT(1) inhibition by (+)-JQ1, and acts as a proof of principle for BRDT(1)-targeted male contraception. As BRDT is expressed only in mid- to later-term spermatocytes and not in mitotically dividing spermatogonia, a BRDT-specific inhibitor would not affect the spermatogonial stem cell population, making any small molecule-BRDT inhibition reversible and leading to no ill effects on subsequent mature spermatids.39, 48 Altogether, this makes BRDT(1) a promising target for a reversible, non-hormonal male contraceptive.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Committee opinion no. 615: Access to contraception. Obstet. Gynecol 2015, 125, 250–255. [DOI] [PubMed] [Google Scholar]
- 2.Curtis KM; Tepper NK; Jataoui TC; Berry-Bibee E; Horton LG; Zapata LB; Simmons KB; Pagano HP; Jamieson DJ; Whiteman MKUS Medical eligibility criteria for contraceptive use, 2016. MMWR Recommendations and Reports 2016, 65, 1–104. [DOI] [PubMed] [Google Scholar]
- 3.Burrows LJ; Basha M; Goldstein AT The effects of hormonal contraceptives on female sexuality: A review. JSM 2012, 9, 2213–2223. [DOI] [PubMed] [Google Scholar]
- 4.Sabatini R; Cagiano R Comparison profiles of cycle control, side effects and sexual satisfaction of three hormonal contraceptives. Contraception 2006, 74, 220–223. [DOI] [PubMed] [Google Scholar]
- 5.Wallwiener CW; Wallwiener LM; Seeger H; Mück AO; Bitzer J; Wallwiener M Prevalence of sexual dysfunction and impact of contraception in female German medical students. JSM 2010, 7, 2139–2148. [DOI] [PubMed] [Google Scholar]
- 6.Lazorwitz A; Aquilante CL; Oreschak K; Sheeder J; Guiahi M; Teal S Influence of genetic variants on steady-state etonogestrel concentrations among contraceptive implant users. Obstet. Gynecol 2019, 133, 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Improving contraceptive use in the United States; Guttmacher Institute: New York, 2008. [PubMed] [Google Scholar]
- 8.Contraception: How effective are birth control methods. https://www.cdc.gov/reproductivehealth/contraception/index.htm (04/20/2018).
- 9.Trussell J Contraceptive failure in the United States. Contraception 2011, 83, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finer LB; Zolna MR Declines in unintended pregnancy in the United States, 2008–2011. New Engl. J. Med 2016, 374, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amory JK Development of novel male contraceptives. Clin. Transl. Sci 2019, XX, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khourdaji I; Zillioux J; Eisenfrats K; Foley D; Smith R The future of male contraception: A fertile ground. Transl. Androl. Urol 2018, 7, S220–S235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murdoch FE; Goldberg E Male contraception: Another holy grail. Bioorg. Med. Chem. Lett 2014, 24, 419–424. [DOI] [PubMed] [Google Scholar]
- 14.Kogan P; Wald M Male contraception: History and development. Urol. Clin. North Am 2014, 41, 145–161. [DOI] [PubMed] [Google Scholar]
- 15.Long JE; Lee MS; Blithe DL Male contraceptive development: Update on novel hormonal and nonhormonal methods. Clin. Chem 2019, 65, 153–160. [DOI] [PubMed] [Google Scholar]
- 16.Thirumalai A; Page ST Recent developments in male contraception. Drugs 2019, 79, 11–20. [DOI] [PubMed] [Google Scholar]
- 17.Male contraceptives on ClinicalTrials.gov. In https://clinicaltrials.gov/ct2/results?cond=male+contraceptive&term=&cntry=&state=&city=&dist=: 2019.
- 18.Amobi NI; Smith IC Differential inhibition in the human vas deferens by phenoxybenzamine: a possible mechanism for its contraceptive action. J. Reprod. Fertil 1995, 103, 215–221. [DOI] [PubMed] [Google Scholar]
- 19.Waddington CH Preliminary notes on the development of the wings in normal and mutant strains of Drosophila. Proc. Natl. Acad. Sci. U. S. A 1939, 25, 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pande V Understanding the complexity of epigenetic target space. J. Med. Chem 2016, 59, 1299–1307. [DOI] [PubMed] [Google Scholar]
- 21.Stricker SH; Köferle A; Beck S From profiles to function in epigenomics. Nat. Rev. Genet 2016, 18, 51–66. [DOI] [PubMed] [Google Scholar]
- 22.Baralle FE; Giudice J Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol 2017, 18, 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aebersold R; Mann M Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [DOI] [PubMed] [Google Scholar]
- 24.Beltrao P; Bork P; Krogan NJ; Noort V Evolution and functional cross-talk of protein post-translational modifications. Mol. Syst. Biol 2013, 9, 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choudhary C; Kumar C; Gnad F; Nielsen ML; Rehman M; Walther TC; Olsen JV; Mann M Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [DOI] [PubMed] [Google Scholar]
- 26.Allfrey VG; Faulkner R; Mirsky AE Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U. S. A 1964, 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamkun JW; Deuring R; Scott MP; Kissinger M; Pattatucci AM; Kaufman TC; Kennison JA brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2SWI2. Cell 1992, 68, 561–572. [DOI] [PubMed] [Google Scholar]
- 28.Filippakopoulos P; Picaud S; Mangos M; Keates T; Lambert J-P; Barsyte-Lovejoy D; Felletar I; Volkmer R; Müller S; Pawson T; Gingras A-C; Arrowsmith, Cheryl H; Knapp S Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen DJ; Ornaghi P; Yang JC; Lowe N; Evans PR; Ballario P; Neuhaus D; Filetici P; Travers AA The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 2000, 19, 6141–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crawford TD; Tsui V; Flynn EM; Wang S; Taylor AM; Côté A; Audia JE; Beresini MH; Burdick DJ; Cummings R; Dakin LA; Duplessis M; Good AC; Hewitt MC; Huang H-R; Jayaram H; Kiefer JR; Jiang Y; Murray J; Nasveschuk CG; Pardo E; Poy F; Romero FA; Tang Y; Wang J; Xu Z; Zawadzke LE; Zhu X; Albrecht BK; Magnuson SR; Bellon S; Cochran AG Diving into the water: Inducible binding conformations for BRD4, TAF1(2), BRD9, and CECR2 bromodomains. J. Med. Chem 2016, 59, 5391–5402. [DOI] [PubMed] [Google Scholar]
- 31.Vidler LR; Brown N; Knapp S; Hoelder S Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J. Med. Chem 2012, 55, 7346–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hewings DS; Rooney TPC; Jennings LE; Hay DA; Schofield CJ; Brennan PE; Knapp S; Conway SJ Progress in the development and application of small molecule inhibitors of bromodomain–acetyl-lysine interactions. J. Med. Chem 2012, 55, 9393–9413. [DOI] [PubMed] [Google Scholar]
- 33.Shadrick WR; Slavish PJ; Chai SC; Waddell B; Connelly M; Low JA; Tallant C; Young BM; Bharatham N; Knapp S; Boyd VA; Morfouace M; Roussel MF; Chen T; Lee RE; Kiplin Guy R; Shelat AA; Potter PM Exploiting a water network to achieve enthalpy-driven, bromodomain-selective BET inhibitors. Biorg. Med. Chem 2018, 26, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Divakaran A; Talluri SK; Ayoub AM; Mishra NK; Cui H; Widen JC; Berndt N; Zhu J-Y; Carlson AS; Topczewski JJ; Schonbrunn EK; Harki DA; Pomerantz WCK Molecular basis for the n-terminal bromodomain-and-extra-terminal-family selectivity of a dual kinase–bromodomain inhibitor. J. Med. Chem 2018, 61, 9316–9334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhalluin C; Carlson JE; Zeng L; He C; Aggarwal AK; Zhou M-M; Zhou M-M Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- 36.Morinière J; Rousseaux S; Steuerwald U; Soler-López M; Curtet S; Vitte A-L; Govin J; Gaucher J; Sadoul K; Hart DJ; Krijgsveld J; Khochbin S; Müller CW; Petosa C Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 2009, 461, 664–668. [DOI] [PubMed] [Google Scholar]
- 37.Florence B; Faller DV You bet-cha: a novel family of transcriptional regulators. Front. Biosci 2001, 6, d1008–1018. [DOI] [PubMed] [Google Scholar]
- 38.Gaucher J; Boussouar F; Montellier E; Curtet S; Buchou T; Bertrand S; Hery P; Jounier S; Depaux A; Vitte AL; Guardiola P; Pernet K; Debernardi A; Lopez F; Holota H; Imbert J; Wolgemuth DJ; Gérard M; Rousseaux S; Khochbin S Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J. 2012, 31, 3809–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berkovits BD; Wolgemuth DJ The role of the double bromodomain-containing BET genes during mammalian spermatogenesis. Curr. Top. Dev. Biol 2013, 102, 293–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukazawa H; Masumi A The conserved 12-amino acid stretch in the inter-bromodomain region of BET family proteins functions as a nuclear localization signal. Biol. Pharm. Bull 2012, 35, 2064–2068. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Gutierrez P; Mundi M; Garcia-Dominguez M Association of bromodomain BET proteins with chromatin requires dimerization through the conserved motif B. J. Cell Sci 2012, 125, 3671–3680. [DOI] [PubMed] [Google Scholar]
- 42.Picaud S; Wells C; Felletar I; Brotherton D; Martin S; Savitsky P; Diez-Dacal B; Philpott M; Bountra C; Lingard H; Fedorov O; Müller S; Brennan PE; Knapp S; Filippakopoulos P RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 19754–19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsume M; Kimura-Yoshida C; Mochida K; Shibukawa Y; Amazaki S; Wada Y; Hiramatsu R; Shimokawa K; Matsuo I Brd2 is required for cell cycle exit and neuronal differentiation through the E2F1 pathway in mouse neuroepithelial cells. Biochem. Biophys. Res. Commun 2012, 425, 762–768. [DOI] [PubMed] [Google Scholar]
- 44.Wang C-Y; Filippakopoulos P Beating the odds: BETs in disease. Trends Biochem. Sci 2015, 40, 468–479. [DOI] [PubMed] [Google Scholar]
- 45.Kanno T; Kanno Y; LeRoy G; Campos E; Sun H-W; Brooks SR; Vahedi G; Heightman TD; Garcia BA; Reinberg D; Siebenlist U; O’Shea JJ; Ozato K BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat. Struct. Mol. Biol 2014, 21, 1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dey A; Nishiyama A; Karpova T; McNally J; Ozato K; Zheng Y Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol. Biol. Cell 2009, 20, 4899–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao R; Nakamura T; Fu Y; Lazar Z; Spector DL Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol 2011, 13, 1295–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berkovits BD; Wolgemuth DJ The first bromodomain of the testis-specific double bromodomain protein Brdt is required for chromocenter organization that is modulated by genetic background. Dev. Biol 2011, 360, 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belkina AC; Denis GV BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer 2012, 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belkina AC; Nikolajczyk BS; Denis GV BET protein function is required for inflammation: Brd2 genetic disruption and BET Inhibitor JQ1 impair mouse macrophage inflammatory responses. J. Immunol 2013, 190, 3670–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S; Walker SR; Nelson EA; Cerulli R; Xiang M; Toniolo PA; Qi J; Stone RM; Wadleigh M; Bradner JE; Frank DA Targeting STAT5 in hematologic malignancies through inhibition of the bromodomain and extra-terminal (BET) bromodomain protein BRD2. Mol. Cancer Ther 2014, 13, 1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi J; Cao J; Zhou BP TWIST-BRD4 complex: potential drug target for basal-like breast cancer. Curr. Pharm. Des. 2015, 21, 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A study to investigate the safety, pharmacokinetics, pharmacodynamics, and clinical activity of GSK525762 in subjects with NUT midline carcinoma (NMC) and other cancers. clinicaltrials.gov (2/9/17).
- 54.Scanlan MJ; Altorki NK; Gure AO; Williamson B; Jungbluth A; Chen Y-T; Old LJ Expression of cancer-testis antigens in lung cancer: definition of bromodomain testis-specific gene (BRDT) as a new CT gene, CT9. Cancer Lett. 2000, 150, 155–164. [DOI] [PubMed] [Google Scholar]
- 55.Nicodeme E; Jeffrey KL; Schaefer U; Beinke S; Dewell S; Chung C-w.; Chandwani R; Marazzi I; Wilson P; Coste H; White J; Kirilovsky J; Rice CM; Lora JM; Prinjha RK; Lee K; Tarakhovsky A Suppression of inflammation by a synthetic histone mimic. Nature 2010, 468, 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung C.-w.; Coste H; White JH; Mirguet O; Wilde J; Gosmini RL; Delves C; Magny SM; Woodward R; Hughes SA; Boursier EV; Flynn H; Bouillot AM; Bamborough P; Brusq J-MG; Gellibert FJ; Jones EJ; Riou AM; Homes P; Martin SL; Uings IJ; Toum J; Clément CA; Boullay A-B; Grimley RL; Blandel FM; Prinjha RK; Lee K; Kirilovsky J; Nicodeme E Discovery and characterization of small molecule inhibitors of the BET family bromodomains. J. Med. Chem 2011, 54, 3827–3838. [DOI] [PubMed] [Google Scholar]
- 57.A study to investigate the safety, pharmacokinetics, pharmacodynamics, and clinical activity of GSK525762 in subjects with NUT midline carcinoma (NMC) and other cancers. In https://ClinicalTrials.gov/show/NCT01587703.
- 58.A dose escalation study to investigate the safety, pharmacokinetics (PK), pharmacodynamics (PD) and clinical activity of GSK525762 in subjects with relapsed, refractory hematologic malignancies. In https://ClinicalTrials.gov/show/NCT01943851.
- 59.Dose escalation and expansion study of GSK525762 in combination with fulvestrant in subjects with estrogen receptor positive (ER+) breast cancer. In https://ClinicalTrials.gov/show/NCT02964507.
- 60.Dose escalation and dose expansion study of GSK525762 in combination with androgen deprivation therapy and other agents in subjects with castrate-resistant prostate cancer. In https://ClinicalTrials.gov/show/NCT03150056.
- 61.A cross-over study to evaluate the effect of itraconazole and rifampicin on the pharmacokinetics (PK) of GSK525762 in healthy female subjects of non child bearing potential. In https://ClinicalTrials.gov/show/NCT02706535.
- 62.Filippakopoulos P; Qi J; Picaud S; Shen Y; Smith WB; Fedorov O; Morse EM; Keates T; Hickman TT; Felletar I; Philpott M; Munro S; McKeown MR; Wang Y; Christie AL; West N; Cameron MJ; Schwartz B; Heightman TD; La Thangue N; French CA; Wiest O; Kung AL; Knapp S; Bradner JE Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zuber J; Shi J; Wang E; Rappaport AR; Herrmann H; Sison EA; Magoon D; Qi J; Blatt K; Wunderlich M; Taylor MJ; Johns C; Chicas A; Mulloy JC; Kogan SC; Brown P; Valent P; Bradner JE; Lowe SW; Vakoc CR RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011, 478, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delmore JE; Issa GC; Lemieux ME; Rahl PB; Shi J; Jacobs HM; Kastritis E; Gilpatrick T; Paranal RM; Qi J; Chesi M; Schinzel A; McKeown MR; Heffernan TP; Vakoc CR; Bergsagel PL; Ghobrial IM; Richardson PG; Young RA; Hahn WC; Anderson KC; Kung AL; Bradner JE; Mitsiades CS BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mertz JA; Conery AR; Bryant BM; Sandy P; Balasubramanian S; Mele DA; Bergeron L; Sims RJ Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vazquez R; Riveiro ME; Astorgues-Xerri L; Odore E; Rezai K; Erba E; Panini N; Rinaldi A; Kwee I; Beltrame L; Bekradda M; Cvitkovic E; Bertoni F; Frapolli R; D’Incalci M The bromodomain inhibitor OTX015 (MK-8628) exerts antitumor activity in triple-negative breast cancer models as single agent and in combination with everolimus. Oncotarget 2017, 8, 7598–7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.A phase 1, dose-finding study of the bromodomain (Brd) inhibitor OTX015/MK-8628 in hematologic malignancies (MK-8628–001). In https://ClinicalTrials.gov/show/NCT01713582.
- 68.A dose-finding study of MK-8628 in participants with recurrent glioblastoma multiforme (MK-8628–002). In https://ClinicalTrials.gov/show/NCT02296476.
- 69.A dose-finding study of OTX105/MK-8628, a small molecule inhibitor of the bromodomain and extra-terminal (BET) proteins, in adults with selected advanced solid tumors (MK-8628–003). In https://ClinicalTrials.gov/show/NCT02259114.
- 70.A dose exploration study with MK-8628 in Participants with selected advanced solid tumors (MK-8628–006). In https://ClinicalTrials.gov/show/NCT02698176.
- 71.A dose exploration study with MK-8628 in participants with selected hematologic malignancies (MK-8628–005). In https://ClinicalTrials.gov/show/NCT02698189.
- 72.Meslamani J; Smith SG; Sanchez R; Zhou M-M Structural features and inhibitors of bromodomains. Drug Discov. Today Technol 2016, 19, 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Romero FA; Taylor AM; Crawford TD; Tsui V; Côté A; Magnuson S Disrupting acetyl-lysine recognition: Progress in the development of bromodomain inhibitors. J. Med. Chem 2016, 59, 1271–1298. [DOI] [PubMed] [Google Scholar]
- 74.Gacias M; Gerona-Navarro G; Plotnikov AN; Zhang G; Zeng L; Kaur J; Moy G; Rusinova E; Rodriguez Y; Matikainen B; Vincek A; Joshua J; Casaccia P; Zhou MM Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression. Chem. Biol 2014, 21, 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kharenko OA; Gesner EM; Patel RG; Norek K; White A; Fontano E; Suto RK; Young PR; McLure KG; Hansen HC RVX-297- a novel BD2 selective inhibitor of BET bromodomains. Biochem. Biophys. Res. Commun 2016, 477, 62–67. [DOI] [PubMed] [Google Scholar]
- 76.Sakamoto KM; Kim KB; Kumagai A; Mercurio F; Crews CM; Deshaies RJ Protacs: Chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 8554–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schneekloth AR; Pucheault M; Tae HS; Crews CM Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg. Med. Chem. Lett 2008, 18, 5904–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ottis P; Crews CM Proteolysis-targeting chimeras: Induced protein degradation as a therapeutic strategy. ACS Chem. Biol 2017, 12, 892–898. [DOI] [PubMed] [Google Scholar]
- 79.Churcher I Protac-induced protein degradation in drug discovery: Breaking the rules or just making new ones? J. Med. Chem 2018, 61, 444–452. [DOI] [PubMed] [Google Scholar]
- 80.Pingyuan W; Jia Z Proteolysis targeting chimera (PROTAC): A paradigm-shifting approach in small molecule drug discovery. Curr. Top. Med. Chem 2018, 18, 1354–1356. [DOI] [PubMed] [Google Scholar]
- 81.Cromm PM; Crews CM Targeted protein degradation: From chemical biology to drug discovery. Cell. Chem. Biol 2017, 24, 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu J; Qian Y; Altieri M; Dong H; Wang J; Raina K; Hines J; Winkler James D.; Crew Andrew P.; Coleman K; Crews Craig M. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol 2015, 22, 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raina K; Lu J; Qian Y; Altieri M; Gordon D; Rossi AM; Wang J; Chen X; Dong H; Siu K; Winkler JD; Crew AP; Crews CM; Coleman KG PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl. Acad. Sci. U. S. A 2016, 113, 7124–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Winter GE; Buckley DL; Paulk J; Roberts JM; Souza A; Dhe-Paganon S; Bradner JE Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015, 348, 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zengerle M; Chan K-H; Ciulli A Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol 2015, 10, 1770–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gadd MS; Testa A; Lucas X; Chan K-H; Chen W; Lamont DJ; Zengerle M; Ciulli A Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol 2017, 13, 514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bryant JM; Berger SL Low-hanging fruit: targeting Brdt in the testes. EMBO J. 2012, 31, 3788–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matzuk Martin M.; McKeown Michael R.; Filippakopoulos P; Li Q; Ma L; Agno, Julio E; Lemieux Madeleine E.; Picaud S; Yu Richard N.; Qi J; Knapp S; Bradner James E. Small-molecule inhibition of brdt for male contraception. Cell 2012, 150, 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shang E; Nickerson HD; Wen D; Wang X; Wolgemuth DJ The first bromodomain of Brdt, a testis-specific member of the BET sub-family of double-bromodomain-containing proteins, is essential for male germ cell differentiation. Development 2007, 134, 3507–3515. [DOI] [PubMed] [Google Scholar]