Abstract
Brain metastasis (BM) predominantly occurs in triple-negative (TN) and epidermal growth factor 2 (HER2)-positive breast cancer (BC) patients, and currently, there is an unmet need for the treatment of these patients. BM is a complex process that is regulated by the formation of a metastatic niche. A better understanding of the brain metastatic processes and the crosstalk between cancer cells and brain microenvironment is essential for designing a novel therapeutic approach. In this context, the aberrant expression of miRNA has been shown to be associated with BM. These non-coding RNAs/miRNAs regulate metastasis through modulating the formation of a metastatic niche and metabolic reprogramming via regulation of their target genes. However, the role of miRNA in breast cancer brain metastasis (BCBM) is poorly explored. Thus, identification and understanding of miRNAs in the pathobiology of BCBM may identify a novel candidate miRNA for the early diagnosis and prevention of this devastating process. In this review, we focus on understanding the role of candidate miRNAs in the regulation of BC brain metastatic processes as well as designing novel miRNA-based therapeutic strategies for BCBM.
Keywords: Breast cancer brain metastasis, miRNA, Brain tumor microenvironment, Blood-brain barrier, CNS metastasis
Introduction
Distant organ metastasis in breast cancer (BC) patients accounts for 90% of deaths [1]. In the central nervous system (CNS), the incidence of brain metastasis (BM) is ten times higher than primary brain lesions [2]. BC is the second most common cancer associated with BM with an incidence of BM, approximately 15-30% of total breast cancer cases [3]. Among different BC subtypes, triple-negative (TN) and HER2-positive BCs are more prone to BM [4]. Around 25% of BC cases show HER2 amplification, and out of these, 30-55% of patients develop BM with a median survival of only 4-14 months [5, 6]. Furthermore, TNBC patients with ER-/PR-/ HER2- status are at high risk of BM recurrence [7].
Despite improvements in BC therapy, the treatment of patients with BM is still disappointingly challenging. BMs are commonly associated with poor prognosis and affect both cognitive and sensory functions of patients and limit the quality of life (QOL) [8]. Several markers, such as age, histology, ER/PR/HER2 status, and the number of non-CNS metastatic sites, have been used to predict BM from non-BM BC patients [9]. Owing to a high level of variability, these predictive markers have limitations.
In order to make progress in this field, there is an urgent need to improve the understanding of the pathobiology of BM, perhaps via first modeling the intricate process of metastasis in the brain microenvironment. The development of a BM is a multistep process, and the metastatic cellular niche is highly dynamic and heterogeneous [10, 11]. Moreover, the brain metastatic cell population harbors a unique genetic and epigenetic profile that distinguishes those cells from similar metastases in other organs [12]. Previous reports suggested an early onset of BM (22 months) after primary diagnosis with TNBC patients as compared to HER2+(30 months) and ER+/HER2-(63.5 months) BC patients [13]. Interestingly, the BBB protects the normal brain, looses its permeability partially or hetrogenously, and transforms into a blood-tumor barrier (BTB) which enhances the accessibily of therapeutic drugs to some extent, but not completly [14]. However the therapeutic role of BTB permeability is not well defined. Smith et al., demonstrated that BTB limits the uptake of chemotherapeutic drugs for BC, such as doxorubicin and paclitaxel into the brain relative to other organs [14, 15], suggesting limitations of BTB for the complete response of these drugs.
Growing evidence has demonstrated the role of miRNA in different steps of the metastatic process, including in BCBM, such as the epithelial to mesenchymal transition (EMT) [16–19], local invasion [20–23], intravasation [24–30], survival in circulation [31–33], extravasation [34], the integrity of the BBB [35–40], niche formation [41, 42], and colonization in the brain parenchyma [43–45]. MiRNAs are 20-22 short nucleotide sequences that often negatively regulate gene expression through the imperfect binding of their seed sequences to the 3’UTR region of target genes [46]. They can cleave or degrade target mRNA when binding with complete complementarity, and thereby inhibit translation of the target [47]. Recent investigations have revealed unique miR expression profiles in different cancer types at different stages, with compelling evidence supportive of miR-based staging and typing [48, 49]. In addition, miRNAs can regulate multiple genes and hence multiple processes simultaneously [50]. Given their ability to modulate the expression of multiple genes at a time, miRs are viewed as attractive therapeutic targets for cancer metastasis, a process mediated by multiple deregulated genes. This review discusses the functional role of miRNAs at different steps of BCBM in hopes of identifying novel miRNA-based therapeutic candidates for the treatment of this devastating process.
Molecular events leading to BM: the role of miRNA
BM is a complex, multi-step, selective process. BM initiates by the dissemination of tumor cells from the primary site to the circulation and known as circulating tumor cells (CTCs). Prior to which these cells undergo EMT transition to invade the extracellular matrix (ECM) at the primary site. Then, to survive anoikis and immunosurveillance, primary tumor cells and CTCs secrete RNA and miRNA encapsulated in exosomes, which further facilitate the survival of metastatic cancer cells at the metastatic site. These miRNAs also transform brain stroma and breach the BBB for BM. Given the coordinated multi-step process that culminates in BM, miRs are perfectly poised to play a cardinal role in BM establishment, given their inherent ability to regulate multiple genes at a given time (Table 1). We have discussed below the role of miRNA at different stages of BM, starting from the primary site of dissemination to brain colonization.
Table 1.
miRNAs mediated regulation of BCBM
| miRNA | Targets | Regulation | References |
|---|---|---|---|
| EMT | |||
| miR-8084 | ING2, p53-BAX | upregulated | [16] |
| miR-484 | PAX-5 | upregulated | [19] |
| miR-708-3p | ZEB1, CDH2 and vimentin | downregulated | [17] |
| miR-210 | E-cadherin (ORF), PAX-5 | upregulated | [19] |
| miR-142-3p | Bach-1, CXCR4, MMP9, and VEGFR | downregulated | [18] |
| miR-199a/214 | Slug | downregulated | [51] |
| miR-3178 | Notch1 | downregulated | [52] |
| miR-212-5p | Prrx2 | downregulated | [53] |
|
miR-29,miR-30 miR-200 family |
ADAM12-L | downregulated | [54] |
| Intravasation | |||
| miR-126 | VEGF/PI3K/AKT axis, MAPK | downregulated | [30] |
| miR-520/373 | ANGPTL4, PTHrP, PAI-1 | downregulated | [25] |
| miR-204 | ANGPT1 and TGFβR2 | downregulated | [24] |
| miR-200 family | IL-8 and CXCL1 | downregulated | [27] |
| miR-105 | ZO-1 | downregulated | [29] |
| Intravascular Microenvironment | |||
| miR-141 | Protection in circulation | upregulated | [31] |
| miR-183 | DAP12/NK cells | downregulated | [32] |
| Extravasation in Brain Microenvironment | |||
| miR-7, let-7c, miR-21 | FasL, SERPIN1 | upregulated | [55] |
| miR-200c | FAP-1 | downregulated | [56] |
| miR-206 | Cx43 | downregulated | [57] |
| miR-19a, miR-32,miR-124a, miR-130b, miR-148a, and miR-583 | PCTH7 | downregulated | [58] |
| miR-125a/b-5p | ET-1 | downregulated | [59] |
| miR-1266, miR-185 and miR-30c | BCL2L1 | downregulated | [60] |
| miR-151-3p | TWIST1 | downregulated | [61] |
| miR-17 | ICAM-1and E-Selectin | downregulated | [62] |
| miR-126 and miR-1185 | VCAM1 | downregulated | [63] |
| miR-483-5p | ALCAM | downregulated | [64] |
| miR-21-3p | L1CAM | upregulated | [34] |
| miR-212 | HBEGF | downregulated | [65] |
| miR-655 | COX2 | downregulated | [66] |
| miR-200b, 200c | ST6GALNAC5 | downregulated | [67, 68] |
| BBB Regulation | |||
| miR-181c | PDPK1 | upregulated | [69] |
| miR-143 | PUMA | upregulated | [35] |
| miR-125a-5p | ICAM-1 | downregulated | [38] |
| miR-1258 | HPSE | downregulated | [40] |
| miR-210 | Occludin, β-catenin | upregulated | [37] |
| Cross Talk and Niche Formation | |||
| miR-26a |
PTEN ATM |
upregulated | [70, 71] |
| miR-19a | PTEN | upregulated | [42] |
| miR-345 | KISS1 | upregulated | [72] |
| miR-124, miR-155, miR-689 | Associated with M1 phenotype of microglia | upregulated | [73] |
| miR-711 and miR-145 | Associated with M2 phenotype of microglia | upregulated | [73] |
| miR-503 |
L1CAM trigger M1–M2 polarization of microglia |
upregulated | [41] |
| Metabolic Reprogramming | |||
| miR-122 | PKM2 , GLUT-1 | upregulated | [74] |
| miR-155 | PIK3R1-PDK/AKT-FOXO3a-cMYC axis | downregulated | [75] |
| miR-7 | RelA | upregulated | [76] |
| Colonization | |||
| miR-200 family (miR-200a,200b, 200c, miR-141, and miR-429) | ZEB1 and ZEB2 | upregulated | [43, 44] |
| miR-147 | ZEB1 | upregulated | |
| miR-126 | IGFBP2, PITPNC1 and MERTK | downregulated | [77] |
miRNA-mediated activation of EMT
Although the EMT is both highly conserved and vital for normal developmental processes [78], it serves an essential role in metastasizing cancer cells [79]. In cancer pathogenesis, EMT promotes the dissemination of the primary tumor [80]. EMT transcription factors (TFs), such as TWIST1, SNAIL1, and SLUG, are contributory to BC metastatic potential and associated with poor prognosis [81]. ADAM12, a long splice variant with a transmembrane domain and member of the disintegrin and metalloproteinase family [82], can be induced by Twist1, thereby promoting tumor invasion via regulation of invadopodia formation and focal adhesions [83]. MiR-34a suppresses BC metastasis by downregulating EMT-TFs (SLUG, TWIST1, and ZEB1/2) and NOTCH1 signaling [81]. Further, ADAM12 is a direct target of the miR-29 and miR-200 families, both involved in BC progression [54]. Aside from regulating EMT-TFs, miRNAs can also regulate cytoskeletal rearrangement in cancer cells by targeting the expression of key molecules and cell signaling pathways involved in cell adhesion [84]. MiR-8084, miR-708-3p, miR-96-182-183 cluster, miR-484, miR-210, and miR-142-3p modulate the invasive potential of BC cells by modulating EMT [16–19]. Recently, it has been shown that miR-124, miR-199a/214, miR-3178, miR-30a, miR-508-3p and miR-212-5p can modulate the level of EMT markers and TFs regulating the expression of E-cadherin in TNBC, a subtype that commonly metastasizes to the brain [51–53, 85].
miRNA-mediated intravasation
Once breast tumor cells change their phenotype through EMT-driven mechanisms, metastasizing tumor cells start the process of metastasis by intravasation into nearby capillaries to facilitate neovascularization for survival [86]. To metastasize at distal sites, cancer cells begin contacting endothelial cells via adhesion molecules and protein receptors [86]. They then follow an amoeboid motility pattern and squeeze themselves between endothelial cells [87]. Some secretory miRNA can regulate the integrity of the endothelium, and thereby the process of intravasation. For instance, miR-105 that is secreted by BC cells disrupts the endothelium by targeting Zonula occludens protein-1(ZO-1), a tight junction protein1 (TJP-1) [29], thus promoting BM. Deryugina et al. discovered an alternative intravasation model suggestive of intravasation within the interior core of a primary tumor in parallel to stromal invasion [88].
Angiogenic factors and growth factors either released by tumor cells or stromal cells individually or during their mutual crosstalk contribute to intravasation [89]. These factors allow tumor cells to invade through the basement membrane, adhere to the endothelial membrane, and pass through endothelial gap junctions to disseminate into the circulation [86]. Although no miRNA has been reported to influence intravasation directly, they can regulate angiogenic signals by targeting angiogenic factors and protein kinases. A recent study discovered a novel role of TGF-β by tumor-associated fibroblasts (TAFs) in the organization of tumor blood capillaries. TAFs enhanced vessel coverage by pericytes, which are vascular cells that support capillaries [90]. In this regard, mRNA profiling of miR-520/373 overexpressing metastatic MDA-MB-231 cells elicited a strong downregulation of TGF-β signaling. It has also been reported that miR-520/373 are instrumental in reducing metastasis through downregulating TGF-β dependent potent angiogenic factors such as plasminogen activator inhibitor-1 (PAI-1), parathyroid hormone-related protein (PTHrP), and angiopoietin-like 4 (ANGPTL4) [25]. In a separate study, miR-204 was shown to suppress vascularization and angiogenesis in vitro and in vivo through targeting pro-angiogenic ANGPT1 and TGFβR2 in BC [24]. The miR-200 family could also play a role in regulating angiogenesis by directly targeting the pro-angiogenic cytokines IL-8 and CXCL1 in endothelial cells [27].
miRNA-mediated survival in the intravascular microenvironment
When a primary tumor grows, circulating tumor cells (CTCs) are shed and enter the circulation. Most CTCs are phagocytosed or undergo apoptosis, leaving behind only a few surviving CTCs to arrive at the targeted organ. Metastatic tumors, as well as CTCs from the primary tumor, may exhibit characteristics different from those of their cell of origin. In order to survive, CTCs must overcome anoikis and immune surveillance once they detach from the primary tumor. One of the tools exploited by CTCs after entering the circulation is platelet activation; by inducing platelet aggregation, tumor cells are protected from immune surveillance, undergo cell arrest within the vasculature, and experience enhanced survival [91, 92]. The CSCs phenotype of BC cells is associated with brain tropism in TNBC patients [93–95]. Debeb et al. have shown that overexpression of miR-141 in the MDA-MB-231 cell line enhances its brain tropism in a tail vein injection mouse model. Further, knockdown of miR-141 inhibited the metastatic ability of inflammatory BC to the brain, suggesting that miR-141 protects cells in the circulation and helps with colonization in the brain [31].
Platelets also contribute to immune evasion by CTCs from scavenging natural killer (NK) cells by enshrouding CTCs and releasing TGFβ and platelet-derived growth factor (PDGF) that directly inhibit the activity of NK cells [96]. Platelet-derived microparticles (PMPs) are major repositories for miRs, and platelets can transfer miRNA contents and modulate gene expression in CTCs [33]. PMP encapsulated miR-183 can suppress NK cell activation, possibly via the silencing of DAP12 a key accessory protein critical for surface NK receptor stabilization and downstream signal transduction [32]. Platelets also contribute to attenuate the early formulation of a metastatic niche [97]. Thus, platelet-derived miRNA also helps in the survival of CTCs after intravasation. The role of miRNA released by CTCs and the intravascular microenvironment in establishing a brain pre-metastatic niche formation warrants further investigation.
Extravasation
Once CTCs are able to survive in circulation, BC cells arrest in blood capillaries and start the process of extravasation, a process coordinated by many oncogenes [98]. Many pairs of ligand-receptor molecules contribute to the process of extravasation, including selectins, integrins, cadherins, CD44, and immunoglobulin superfamily receptors [99, 100].
Extravasation is a rate-limiting step for BCBM, as cancer cells must overcome the initial defenses imposed by astrocytes and other protective factors in the brain microenvironment [101]. Astrocytes that are mobilized to the metastatic brain lesion at a very early stage of colonization induce apoptosis through the FasL-mediated pathway [102]. In recent studies, several miRNAs have been described to target various members of the Fas-mediated apoptotic pathway. For example, miR-7, let-7c, and miR-21 regulate the expression of FasL [55], while miR-200c regulates the induction of apoptosis through CD95 by targeting FAP-1 [56]. Cancer cells release protease inhibitors known as serpins to combat the apoptotic effects exerted by astrocytes. MiR-21 has been shown to inhibit Serpin1, a gene with novel tumor-suppressive effects in gastric cancer [103]. However, its role in BM is unknown. Eventually, astrocytes support CTCs survival in brain parenchyma via establishing connexins (Cx) gap junctions and promote BM [104]. The expression of miR-206 is inversely correlated with Cx43 levels and is associated with decreased proliferation and migration [57]. PCDH7 in brain tropic BCs contributes to establishing Cx43 gap junctions with astrocytes and forms Ca++ channels [104]. A high PCDH7 level in the brain tropic CSC population has been reported and contributes to CSC extravasation, adaptation, and colonization in the new niche formation through the PCDH7-PLCb-Ca2þ-CaMKII/S100A4 pathway involving PCDH7-mediated tumor–astrocyte interaction [95]. In addition, miR-19a, miR-32, miR-124a, miR-130b, miR-148a, and miR-583 have been reported as potential regulators of PCDH7 [58]. However, the role of these miRNAs in PCDH7 regulated BM has yet to be studied [95].
The production of IL6 and IL-8 by cancer cells requires the establishment of gap junctions with astrocytes [105]. These cytokines influence both cell types by inducing the expression of endothelin ligand (ET-1) on astrocytes and endothelin receptors (ETAR and ETBR) on cancer cells [101, 105]. ET-1 is regulated through miR-125a/b-5p in endothelial cells [59]. In addition, the expression of a few genes was found to be dependent on such interaction [106]. Some of them were validated in BM, such as TWIST1, GSTA5, and BCL2L1 [106]. Interestingly, BCL2L1 is regulated by miR-1266, miR-185, and miR-30c [60] in prostate cancer. TWIST1 is regulated by miR-151-3p in BC [61]. These miRNAs are involved in negative regulation of the apoptotic pathway and upregulation of invasion or migration respectively, but the role in BM is not clear yet.
Emerging evidence shows that cell adhesion molecules (CAMs) play an essential role in extravasation through a cell-cell adhesion receptor. In an in vivo model of BM, a subset of adhesion molecules, including E-selectin, VCAM-1, ALCAM, ICAM-1, VLA-4, and a4 were found to be upregulated in the cerebral endothelium when injected intracardially. Conversely, the expression of their ligands (PSGL-1, VLA-4, ALCAM, LFA-1, and VCAM-1) was upregulated in brain tropic cancer cells [107], revealing a crucial role for these CAMs during the initial steps of extravasation. MiRNAs post-transcriptionally regulate CAMs. For instance, TGF-β induced ICAM-1, and E-selectin expression is regulated by miR-17 [62]. MiR-126 and miR-1185 regulate endothelial expression of VCAM1 [63, 108]. ALCAM is reported as a target gene of miR-483-5p [64].
Moreover, cancer cells can invade through the endothelium by projecting invadopodia [109]. Invadopodia are chemosensing protrusions that guide cancer cell extravasation to promote brain tropism in metastasis [110]. PAK1 (P21 (RAC1) Activated Kinase 1) is responsible for guiding cancer cell extravasation in BCBM [110]. PAK1 reduces the expression of miR-132 through the PAK1/ATF2/miR-132 axis. L1CAM, an adhesion molecule, mediates the spread of metastatic cells on the vasculature and additionally mediates interactions between cancer cells and endothelial cells in BM. The depletion of L1CAM in cancer cells fails to co-opt brain capillaries and hence is unsuccessful in promoting metastatic outgrowth. Interestingly, miR-21-3p was reported to be a positive regulator of L1CAM expression [34]. These studies strongly suggest that miRNAs can modulate the expression of various CAMs in cancer, as well as endothelial cells, and thereby play a decisive role in the establishment of metastasis at the distant metastatic site via extravasation.
Reactive astrocytes have been shown to contribute to the formation of a protumorigenic niche via a number of mechanisms involving secreted molecules. In the BCBM mouse model, Massague’s group has identified 17 genes that are specifically correlated with BC-metastasis associated genes. Among these 17 genes, four genes, COX2, EGFR ligand HBEGF, ANGPTL4, and the a2,6-sialyltransferase ST6GALNAC5 were identified as signature molecules of BC metastasis to the brain parenchyma [67]. COX2 is actively involved in BM by regulating the expression of MMP-1 in BC patients, and high expression is reported in BC patients [111]. Interestingly, COX2 expression is associated with BBB permeability . COX2 induces a stem-like cell phenotype by upregulating miR-655 and miR-526b in BC, thereby rendering cells more metastatic [66, 112]. MiR-212 directly targets HBEGF and suppresses cell growth, migration, and invasion [65]. ST6GALNAC5, a direct target of miR-200c, is a specific mediator of BCBM [67]. Conversely, the upregulation of ST6GALNAC5 in brain-tropic BC cells showed a decrease in adhesive properties of the endothelial component of a well-characterized human BBB in vitro model [113]. ST6GALNAC5 can also regulate the EMT process in BM and is a target of miR-200b [68]. Several target genes actively participate in extravasation within the brain parenchyma, although their regulation in context of miRNA is not studied in BCBM.
Blood-brain barrier
The blood-brain barrier (BBB) is a semipermeable barrier comprised of endothelial cells, astrocytes, and pericytes, forming the neurovascular unit [114]. It remains important to study the role of miRNAs in enhancing the permeability of the BBB. Endothelial cells are interconnected with each other via tight junctions, a functionally important component of the BBB, controlling the free flow of substances into the brain parenchyma. Most of the solutes that are allowed to permeate the BBB, such as glucose, macronutrients, and electrolytes, enter via transporters present on the surface of endothelial cells.
Endothelial tight junctions facilitate the transmigration of tumor cells through the BBB [115]. CD44, VEGF, and CXCR4 contribute to the transendothelial migration process by disturbing endothelial integrity [116]. Astrocytes are indispensable for the development and maintenance of the BBB [106]. The intracellular junctions of brain endothelial cells form with tight junction proteins, such as occludin, claudins, and ZO-1 proteins [117]. Disruption of intercellular junctions causes the breakdown of the BBB and transform it into BTB [118, 119].
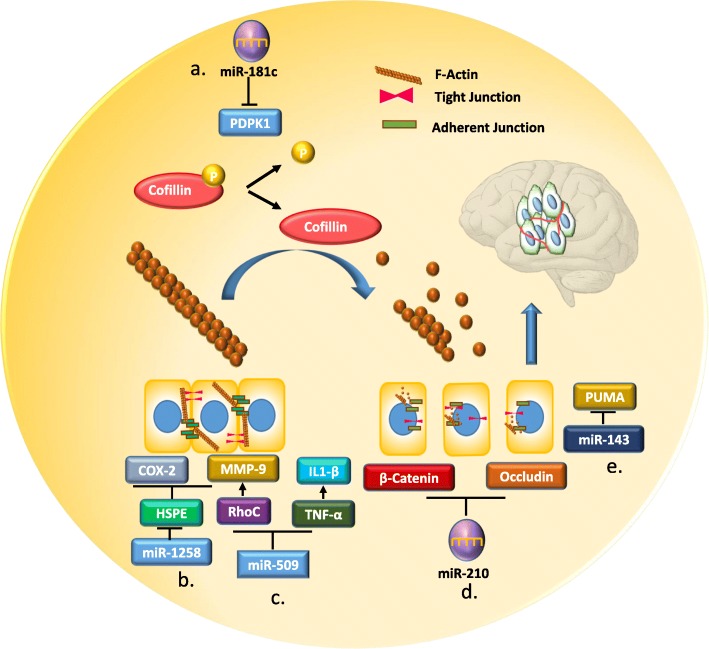
The priming of the pre-metastatic niche, or organotropism, starts before cancer cells reach the metastatic site from the primary tumors via paracrine routes. In this context, miRNAs containing exosomes or extracellular vehicles (EVs) have the ability to modify the brain microenvironment, which leads to enhanced BM despite the barrier function of the BBB [42, 120]. Recently, miRNAs emerged as regulators of tight junction adhesion proteins and their upstream and downstream signaling pathways, playing an important role in maintaining the integrity of the BBB. For instance, miR-181c promotes the destabilization of the BBB through the delocalization of actin fibers via the downregulation of 3-phosphoinositide-dependent protein kinase-1 (PDPK1). PDPK1 degradation by miR-181c leads to the downregulation of phosphorylated cofilin and a resultant activated cofilin-induced modulation of actin dynamics [69]. MiR-143 enhances the permeability of endothelial cells through targeting p53 upregulated modulator of apoptosis (PUMA), and consequently shows a reduction of tight junction proteins (TJPs) [35]. Additionally, miR-125a-5p has been shown to be an important player in the maintenance of the integrity of the BBB. This miRNA can directly regulate barrier function in an in vitro BBB model and can reduce monocyte migration through a BBB cell layer in vitro [38] (Fig. 1).
Fig. 1.
Schematic of miRNA regulatory blood-brain-barrier (BBB) tight junction (TJs) protein. a miR-181c promotes the destruction of the BBB through the delocalization of actin fibers via the downregulation of 3 phosphoinositide-dependent protein kinase-1 (PDPK1). PDPK1 degradation by miR-181c leads to the downregulation of phosphorylated cofilin and the resultant activated cofilin-induced modulation of actin dynamics [69]. b miR-1258 downregulates MMP-9 and COX-2 protein by directly targeting HPSE, hence protecting the BBB from destruction [40]. c miR-509 negatively regulates the expression of two essential genes for brain metastasis, RhoC and TNF-α, which enhance the permeability of the BBB [121]. d miR-210 directly targets β- Catenin and Occudin to disrupt the integrity of the BBB [37]. e MiR-143 decreases the expression of TJs by directly targeting p53 upregulated modulator of apoptosis (PUMA) and increases the permeability of human brain endothelial cells [35]
In BC, miR-1258 expression was directly associated with heparanase expression. Heparanase is a prometastatic enzyme present in BCBM cells that degrades heparan sulfate chains to affect the cytoskeleton and render cells more capable of crossing the BBB [39, 40]. Researchers demonstrated miR-1258 downregulates the phosphorylation of Akt and EGFR signaling along with the repression of MMP-9 and COX-2 protein expression by direct targeting of HPSE [39, 40]. Watabe K et al. observed high expression of miR-509 in primary tumors whereas level was significantly downregulated in BM lesions. Consequently, the reduction of miR-509 in BM lesions induces the expression of two essential genes for BM, RhoC and TNF-α, followed by upregulation of the MMP9 level, which altogether augments the permeability of BBB and penetration of tumor cells in the brain [121]. MiR-210 suppresses the junction proteins and disrupts the BBB in hypoxic-ischemic brain injury [37]. In addition, high expression of miR-210 is associated with poor survival in BC patients [36]. Exosomal profiling done by Dario et al. showed significant upregulation of miR-210 (2 to 6-fold increase) in three brain metastatic BC cell-derived exosomes [122]. Therefore, it is plausible that miR-210-containing exosomes released by the brain may help BC cells breach the BBB.
Crosstalk of cancer cells with brain microenvironment
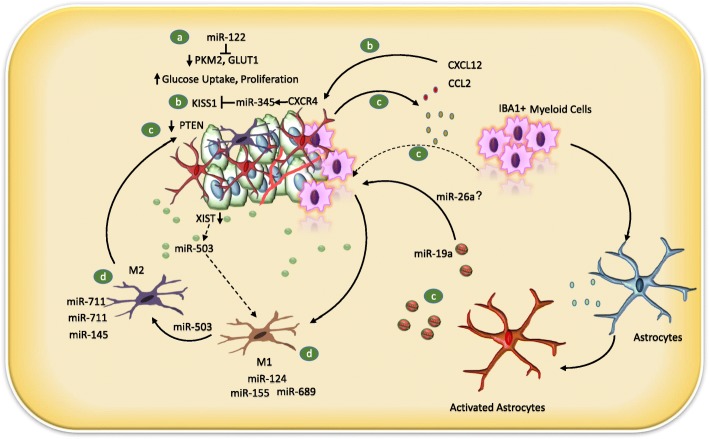
Once infiltrated into the brain tissue, cancer cells encounter a number of host cell types, including pericytes, reactive glia, neural progenitor cells, neurons, and oligodendrocytes [123]. Astrocytes and endothelial cells are the first to encounter incoming metastatic cells. Once normal astrocytes encounter cancer cells, they become reactive astrocytes (RAs) due to a perceived disruption to brain homeostasis. At the initial stages of BCBM, RAs act as a primary host defense system by proficiently limiting the survival of arriving metastatic cells [102], whereas, at later stages, RAs have been actively involved in promoting metastatic outgrowth via secretion of miRNA containing exosomes [42]. Exosomes can successfully form the pre-metastatic niche in the brain by modulating tumor-stroma communications [74]. MiRNAs with gene regulatory functions have emerged as key regulators of the tumor microenvironment [124]. For instance, miR-26a is present in astrocytes and released through exosomes or by endothelial cells. MiR-26 can regulate the growth of brain tumors and radiosensitize tumor cells by targeting PTEN and ATM, respectively [70, 71]. Thus, miR-26a may play a key role in regulating the brain tumor microenvironment.
Zhang et al. (2015) discovered a complicated reciprocal mechanism between brain metastatic BC cells and stromal cells (astrocytes and myeloid cells). In BCBM patients, PTEN loss was observed when compared to primary breast tumors. In addition, miR-19a in the miR-17-92 cluster was identified as a candidate responsible for mediating PTEN suppression from astrocytes to tumor cells via exosomes. Reactive astrocytes secrete interleukins and chemokines, such as CCL2 and CXCL12/SDF1, respectively, playing a mitogenic role. Moreover, human BCBM has higher levels of CCL2 than primary tumors. Interestingly, PTEN has been shown to be instrumental in the regulation of BCBM, as a reduced expression of PTEN leads to enhanced CCL2-mediated recruitment of IBA1+ microglial cells, and thereby establishment of the BM [42].
Astrocytes have also been shown to enhance metastatic growth through enhancing the CXCL12/CXCR4-MIR345-KISS1/KISS1R axis. A significant reduction in KISS1 expression in BCBM patient’s primary tumors has been noticed. Ilya V. Ulasov et al. identified that CXCL12 secreted by astrocytes can induce miRNAs that can directly target KISS1 mRNA in metastatic BC cells and negatively regulate KISS1 expression. In this regard, miR-345 was the only identified miR that directly targets KISS1, which is induced via the treatment of CXCL12 or CCL2 proteins [125–127]. They experimentally confirmed the binding of miR-345 in stably transfected KISS1 3’UTR in MDA231Br cells with astrocyte conditioned media treatment or in the presence of individual recombinant CCl2 or CXCL12 proteins [72]. Interestingly, the downregulation of KISS1 has a stimulating effect on ATG5 expression associated with autophagosome maturation. Finally, they revealed a paracrine loop between KISS1 and the CXCL12-miR-345 that can promote BC cell invasion and survival in the brain.
Microglia are also a crucial component of the brain parenchyma; they constitute about 5-20% of the total CNS population and they are the only brain resident myeloid cells that play an important role in brain homeostasis and immunosurveillance [128–130]. Microglia release cytokines and interleukins that support cancer invasion and colonization of the parenchyma. Microglia can differentiate from the proinflammatory M1 phenotype to the immunosuppressive M2 phenotype based on environmental factors [131]. Interestingly, miRNA can modulate microglial polarization. For instance, miR-124, miR-155, and miR-689 are associated with the M1 phenotype, whereas miR-711 and miR-145 are strongly associated with M2 polarization [73]. MiR-124 is a brain enriched miRNA highly present in resting microglia, and its expression declines with microglial activation [132]. However, the role of miR-124 in BCBM is yet to be studied. Loss of XIST, a long noncoding RNA in tumor cells, causes local immune suppression by converting the microglia to the M2 phenotype through the transport of exosomal miR-503 from the tumor cells [41]. These studies strongly suggest that miRNAs have the ability to modulate microglia activation, and thereby modulate the brain microenvironment and subsequently metastasis partly via immune invasion (Fig. 2).
Fig. 2.
Cross talk of the brain tumor microenvironment with BC cells. a Autocrine and paracrine role of miR-122 in the development of the pre-metastatic niche via regulating glucose metabolism in cancer cells. MiR-122 downregulates the expression of pyruvate kinase isozymes, PKM2, and glucose transporter 1 (GLUT1), and decreases ATP levels in BC cells. MiR-122 reduces glucose consumption in stromal cells and allows more glucose to be accessible to cancer cells, hence facilitating the formation of the metastatic niche and cancer cell growth [74]. b CXCL12 or CCL2 secreted by astrocytes increases the level of miR-345 via CXCR4, which negatively regulates the expression of KISS1 and promotes invasion and survival in the brain [72]. c MiR-19a mediates the suppression of PTEN in cancer cells secreted by activated astrocytes. Reactive astrocytes secrete interleukins and chemokines, such as CCL2 and CXCL12/SDF1. Reduced expression of PTEN leads to enhanced CCL2-mediated recruitment of IBA1+ myeloid cells, and thereby establishment of the brain metastasis (BM) [42]. miR-26a is present in astrocytes and released by astrocytes through exosomes, or it can be secreted by HUVEC cells, but its role in brain niche formation is not clear [70, 71]. d Microglia release cytokines and interleukins that support cancer cells to invade and colonize the parenchyma. In cancer microglia, it can transform from the immunogenic phenotype (M1) to immunosuppressive phenotype and miRNA can modulate microglial polarization. MiR-124, miR-155, and miR-689 are associated with the M1 phenotype, whereas MiR-711 and miR-145 are strongly associated with M2 polarization [128, 129]. Loss of XIST, a long noncoding RNA in tumor cells, causes local immune suppression by converting the microglia to the M2 phenotype through the transport of exosomal miR-503 from the tumor cells [41]
miRNAs and metabolic reprogramming in the brain microenvironment
Adaptation in the pre-metastatic niche is of great importance and starts before the arrival of CTCs to distant sites of metastasis to sustain their survival and growth [133]. Modulation of the tumor microenvironment by metabolic factors is a different aspect of cancer cells and tumor microenvironment crosstalk. Metabolic reprogramming is associated with the deregulation of several pathways controlled by hypoxia-inducible factor 1 alpha, MYC, p53, and miRNAs. MiRNAs target metabolic enzymes, oncogenes, and tumor suppressors involved in metabolic reprogramming, becoming crucial elements in the crosstalk of molecular pathways that promote extravasation and metastasis. In BC, cancer-associated stromal cells rely on glycolysis to provide energy metabolites to cancer cells through monocarboxylate transporters during disease progression [134]. Endothelial cells also rely on glycolytic metabolism to support vessel sprouting for angiogenesis [134]. Emerging evidences in the metabolic reprogramming of the microenvironment identified a prerequisite metabolic condition required to sustain cancer cells in the brain [135]. For example, brain metastatic cells switch to metabolic reprogramming by upregulating the fructose-1, 6-bisphosphatases (FBP2) based gluconeogenesis pathway and amino acid oxidation to survive and grow in the low glucose environment of the brain parenchyma [135].
Interestingly, Emily Wang’s group has studied the autocrine and paracrine role of miR-122 in glucose metabolism in primary BC and pre-metastatic niche development and metastasis [74]. MiR-122 downregulates the expression of pyruvate kinase isozymes, PKM2, and glucose transporter 1 (GLUT1) and decreases ATP levels in BC cells. They demonstrated that cancer cells secreted miR-122, which downregulates glucose uptake in astrocytes as well as lung fibroblasts. Orthotopic xenograft mice with stably overexpressed DCISMCF/miR-122 form smaller tumors than empty vectors. Collectively, they showed that cancer cells could induce glucose reallocation in the pre-metastatic niche by repressing glucose consumption in stromal cells and allowing more glucose to be accessible to cancer cells, hence facilitating metastatic cancer growth. MiR-122 partially exhibits this effect and helps in metabolic reprogramming of the tumor microenvironment by downregulating its metabolic target genes PKM1/2 and GLUT1 in stromal cells in vitro and in vivo. MiR-122 has potential as a predictive marker and therapeutic target for BC metastasis [74].
Furthermore, Chang et al. recently demonstrated the role of miR-155 in glucose metabolism in the TNBC subtype. Utilizing a BC mouse model with miR-155-/- or miR-155+/- backgrounds, they unraveled the miR-155-PIK3R1-PDK/AKT-FOXO3a-cMYC axis that mediates energy metabolism in BC [75]. However, the metastatic potential of miR-155 has not been studied in the context of BM. High glucose uptake is a salient feature of cancer cells [136]. MiR-7 is highly expressed in the brain and promotes glycolysis, as evinced by an increased intracellular ATP/ADP ratio, glucose consumption, and lactic acid production. MiR-7 directly targets the expression of RelA, which regulates the expression of the cell surface glucose transporter, Glut3, hence promoting glycolysis [76, 137]. In human BC cells, miR-7 suppresses the homing and migration potential of human endothelial cells; however, there is the possibility that the opposite may be true and the brain tumor microenvironment may deliver exosomes containing miR-7 to increase the glucose uptake and survival of breast tumor cells in the brain parenchyma. Therefore, its role in the metabolic reprogramming of the brain microenvironment in BCBM is obscure and needs to be studied in detail.
Metastatic colonization
The major problem with the EMT concept is that the appearance of the majority of human metastatic histology samples resembles the epithelial phenotype and usually looks like the primary tumor [138]. Evidence from previous studies suggests that for successful colonization and growth after extravasation to a secondary site to occur, cancer cells have to go through the mesenchymal to epithelial transition (MET) [138]. Interestingly, BCBM is dependent on cellular reprogramming through the EMT to the MET. Yang and colleagues (2012) have demonstrated that Twist1 reversibly regulates the EMT during metastasis. They have also shown that early metastatic colonies elicited strong positive Ki67 expression with low Twist1 expression under reversible EMT conditions, while irreversible EMT resulted in colonies with high Twist1 expression and low Ki67 [139], suggesting that metastatic cancer cells must revert to the epithelial phenotype by a MET in order to grow at a secondary site.
In another study, epithelial markers, such as E-cadherin, β-catenin, connexin 26, and connexin43, were found to be upregulated in BC patients. In contrast, mesenchymal markers FSP1 and vimentin were variably altered in BC, suggesting a partial MET [140]. Shreds of evidence show that miRNA participates in the process of EMT to MET [141]. A well-documented example is the miR-200 family. MiR-200s are associated with poor prognosis of BC [142]. Recently, members of the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429, containing similar consensus seed sequence) have been recognized as new epithelial markers and negative regulators of EMT. The miR-200 family members inhibit the EMT and promote MET transformation in BC cells by directly targeting ZEB1 and ZEB2. The miR-200 family regulates the MET and metastatic colonization in BC, suggesting that flexible transitions between EMT and MET, or epithelial-mesenchymal plasticity, may be crucial at different stages of metastasis [43–45].
Moreover, human BC metastases often show a higher level of E-cadherin than their corresponding primary tumor [140]. Korpal et al. suggested that miR-200s promote metastatic colonization of BC not only by influencing cell-intrinsic epithelial traits through targeting of the Zeb–E-cadherin axis but also by altering the tumor cell-derived secretome through targeting of the Sec23 homolog A, Sec23a-mediated transport pathway. It ultimately targets two metastatic suppressors, insulin-like growth factor binding protein 4 (IGFBP4) and tubule interstitial nephritis antigen-like 1 [142].
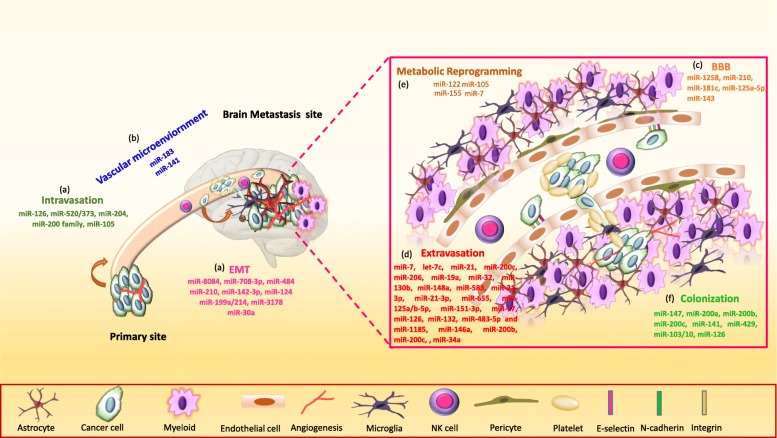
In addition, CTCs increase the level of miR-200s in BC patient serum and cerebrospinal fluid (CSF) with BCBM [143, 144]. Although these studies suggest that extracellular miR-200s are associated with BC metastasis, they did not show that circulating miR-200 miRNAs are functional [145]. Bisrat G. Debeb et al. generated a preclinical mouse model via tail vein injection of epithelial-like inflammatory TNBC and HER2 positive cells and mesenchymal-like lung metastatic cells. The knockdown of miR-141 ceases the BM; however, ectopic expression of miR-141 enhances the brain colonization of inflammatory metastatic cells in vivo. Alternatively, ectopic expression of miR-141 in lung metastatic cells was not sufficient for the onset of BM, suggesting an epithelial phenotype is important at the final step of BM [31]. High expression of ZEB1 and ZEB2 at a tumor invasion front in brain metastatic tissues suggests a role of these EMT regulators in facilitating BM [146]. MiR-126 is reported as a tumor suppressor in various cancers [147–149]. It regulates the migration of endothelial cells towards the metastatic BC cells in vitro and in vivo [77]. Endogenous expression of miR-126 suppresses metastatic colonization by targeting IGFBP2, PITPNC1, and MERTK- novel pro-angiogenic genes and biomarkers of human metastasis [77]. Silencing of miR-126 in poorly metastatic CN34 BC cells results in increased endothelial recruitment and metastatic brain colonization [77]. Overall, the miRNAs are crucial at multiple steps of breast cancer brain metastasis (BCBM) (Fig. 3).
Fig. 3.
miRNAs function at multiple steps of breast cancer brain metastasis (BCBM). MiRNAs regulate key steps of BCBM, (a) breast cancer cell intravasation and dissemination via EMT from the primary site, (b) survival in the circulation/vascular microenvironment, (c) breaching of blood-brain barrier (BBB) integrity, (d) extravasation into brain parenchyma, (e) metabolic reprogramming into the brain microenvironment, and (f) colonization and growth of cancer cells into brain
miRNA and BCBM therapeutics
Despite advances in therapy for BCBM, the exact molecular mechanism, and biomarkers for the diagnosis and prognosis of patients are lacking [150]. Available treatment options include local therapies, such as whole-brain radiation therapy (WBRT), stereotactic radiosurgery (SRS), surgery, chemotherapy, and tyrosine kinase inhibitors (TKIs) [151]. TKIs are promising anticancer agents for HER2-positive BCBM, such as lapatinib, which is a dual TKI that targets both HER2/ErbB2 and EGFR. However, inhibition of kinases is not specific for a single tyrosine kinase, results in reduced specificity with high toxicity [152]. Due to specificity and toxicity issues, monoclonal antibodies emerged as strong therapeutic tools and proved as a potent therapy for cancer treatment. The current focus for TNBC patients is to manipulate the anti-tumor immune response by blocking the activity of immune checkpoint inhibitors. Atezolizumab, a PD-L1 blocking antibody in combination with nab-paclitaxel improved the progression-free survival in PD-L1 positive subgroup in an Impassion130 trial (NCT02425891) [153]. In phase II, ongoing trial (NCT03483012) TNBC patients with BM are treated with SRS alone or in combination with Atezolizumab. There is hope that SRS in combination with atezolizumab, may enhance the immune response to BM patients.
The enduring challenges in BCBM are to develop early detection markers and novel targeted therapies that can cross the BTB and improve the survival rate of BCBM patients. Therefore, miRNAs are emerging as noninvasive, diagnostic, and prognostic markers in BM. The miRNAs present in the blood plasma and CSF are attractive biomarkers for BM, which provide the disease severity, but also impart the prognostic value of the treatment response [154]. Although a considerable number of miRNAs are found inside the cell, many miRNAs are secretory, and their expression elevates or drops in the brain lesions or BM. In this context, miRNA levels can be used to monitor the disease burden, tumor response, and differentiation between brain lesions and metastatic brain tumors [155]. MiR-10 and miR-21 are highly expressed in the cases of GBM and BCBM; however, the miR-200 family can be used to discriminate between GBM and BM [144]. Additionally, miR-223, miR-711, miR-125, and miR-935 signatures were shown to discriminate among medulloblastoma, GBM, breast, and lung cancer BM [156].
Tumor suppressor miRNA with oncogenic targets may enhance the efficacy of treatment in combination with conventional chemotherapy, radiotherapy and immunotherapy in BCBM patients. For instance, miR-770-5p decreases the migration and invasive potential of HER+ breast cancer cells through inhibiting the translation of downstream signaling of PI3K and MAPK, i.e., AKT and ERK, pathways that mediate resistance to anti-HER2 therapies. Additionally, miR-770-5p can increase the responsiveness of trastuzumab and reverse drug resistance [157]. miR-770 also suppresses the doxorubicin-resistance and metastasis of TNBC cells [158, 159]. miR-326, a suppressor of Hedgehog pathway, is inversely correlated with multi drug resistance protein (MRP-1) expression in BC patients and sensitize the response in doxorubicin and etoposide (VP16) in resistant BC cells. miR-21 has been shown to sensitize BC cells to topotecan and taxol [160]. miRNA-143-3p increase the sensitivity of TNBC to paclitaxel by inferenig with CIAPIN1 expression, a cytokine-induced apoptosis inhibitor 1 protein [161]. miR-449 can induce doxorubicin respone in TNBC by downregulating cell cycle related genes [162]. Such combinations, which are already tested in BC preclinical models have the potential to be tested for BCBM. In addition, miRNA with known function in BM such as miR-181c, miR-1258, miR-509, miR-143, miR-122 and miR-19a could be utilized in combination with radiotherapy, anti-HER2 therapies (lapatinib or trastuzumab), chemotherapy or immunotherapies for BM.
Since miRNA can target multiple sets of genes, it is an excellent clinical choice for the heterogeneous population of BM. In this context, miR-7 has been shown to attenuate BC growth by downregulating both EGFR and PKB signaling pathways [156]. MiR-7 also inhibits BCBM by inhibiting the self-renewal capacity of BC stem-like cells by regulating the expression of KLF4 [163]. An additional example is let-7, which targets several oncogenic pathways, including Ras, HMGA2, cyclin d1/2/3, cyclin A, CDK4/6, c-Myc, DICER1, and Lin28, which are responsible for stem cell self-renewal and chemoresistance [164]. The current challenges with the delivery of miRNA into the brain are poor penetration of miRNAs into tumor tissues due to the presence of BBBs, instability of miRNA mimics or inhibitors in the blood circulation, and neurotoxicity and immunotoxicity due to an off-target effect. Therefore, miRNA can be conjugated to drug carrier systems or nanoparticles (NPs) for targeting cancer cells. These miRNA delivery systems have shown minimal toxicities and have the ability to cross the BBB and successfully release the miRNA to promote clinical advancement. Recently, numerous delivery systems have been developed to cross the BBB, such as Cationic lipid nanoparticles (LNP) [165], Cationic Dendrimers PAMAM [166], Poly (lactic acid-co-glycolic) acid (PLGA) nanoparticles [167], Magnetic Nanoparticles [168], and Viral Vector Systems [169, 170]. Although water-soluble polymers, cationic lipids, or liposome nanocarriers are less toxic than a viral vector, the delivery efficiency remains lower [171]. Since leukocytes (including monocytes/macrophages, neutrophils, dendritic cells, and lymphocytes) or MSCs target tumors and can migrate across physiological barriers like the BBB, these cell types are increasingly utilized as carriers to transfer NPs to tumors [172]. As leukocytes/MSCs follow the same pattern of migration as tumor cells to cross the BBB, these cellular mechanisms can be utilized effectively to deliver miRNA conjugated NPs to the BMs. These NPs can be attached to the monocytes/macrophages/MSCs for the delivery of miRNA through various nanotechnology strategies [173].
Additionally, BBB-permeable NPs can be used to deliver miRNAs into the metastatic sites. Recently, Galstyan et al. have used BBB-permeable nano-immunoconjugates for the successful inhibition of GBM growth using mouse models [174]. They have used poly (β-L-malic acid) NPs covalently attached to immune checkpoint antibodies for systemic delivery directly into the brain. NPs have been used to deliver anti-miR-132 that recover p120RasGAP in the tumor endothelial cells, and have shown reduction in tumor growth in an orthotopic xenograft mouse model of BC [175]. If BBB permeable NPs are not available, regular NPs containing miRNAs can be delivered through the BBB using chemical modifications, partial opening by ultrasound, microwave or electromagnetic field-based thermal translocation of tight junction proteins [176].
Conclusion and future perspective
Both the anatomy and physiology of the brain are very complex; hence, the process of BCBM is enormously complex too. Mechanistic and functional discoveries could expedite the response of BM treatment. Each step of BM is rate-limiting, and miRNA are instrumental in the regulation of every step of metastasis since they are upstream of oncogenes and tumor suppressor genes. All the steps of BMs, starting from the dissociation from primary sites through EMT related genes, survival into the circulation by anoikis resistance genes, brain organotropism, brain niche modulatory genes, and also brain colonization related genes, are all regulated through miRNAs. In the past few years, the focus on BMs has significantly increased as several miRNAs were discovered for initiating steps of metastasis. However, limited research has been done to address questions like: how does miRNA play a role in metastasizing cancer cells to the brain? How can miRNAs breach the BBB? How do cancer cells communicate with an entirely new environment of the brain niche via miRNA? How do astrocytes overcome the defense mechanisms and facilitate the survival of BC cells by altering the miRNA profile? How do miRNAs modulate brain metabolism in favor of cancer cell survival? There are so many unanswered questions in the context of miRNA and BCBM. Therefore, intense research is needed to tackle these problems, to discover better treatment options, to improve BCBM treatment efficacy.
Acknowledgments
This manuscript is supported by financial assistance from the National Institute of Health/NCI Grant R01CA218545. We thank Jeffrey Patterson for editing the manuscript.
Abbreviations
- TNBC
Triple-negative breast cancer
- HER2
Human epidermal growth factor receptor 2
- BCBM
Breast cancer brain metastasis
- ER
Estrogen receptor
- PR
Progesterone receptor
- TGF-β
Transforming growth factor-beta
- TAFs
Tumor-associated fibroblasts
- PAI-1
Plasminogen activator inhibitor-1
- PTHrP
Parathyroid hormone-related protein
- IL-8
Interleukin-8
- IL-6
Interleukin-6
- VEGF
Vascular endothelial growth factor receptor
- NK cells
Natural killer cells
- PDGF
Platelet-derived growth factor
- PMPs
Platelet-derived microparticles
- DAP-12
DNAX-activation protein 12
- CTCs
Circulating tumor cells
- PCDH7
Protocadherin-7
- COX2
Cyclooxygenase-2
- EGFR
Epidermal growth factor receptor
- HBEGF
Heparin binding egf like growth factor
- ST6GALNAC5
ST6N-Acetylgalactosaminide alpha-2,6-sialyltransferase
- BBB
Blood-brain barrier
- ZO-1
Zona occludens 1
- PDPK1
3- Phosphoinositide-dependent protein kinase-1
- PUMA
p53 Upregulated modulator of apoptosis
- TJPs
Tight junction proteins
- RAs
Reactive astrocytes
- PTEN
Phosphatase and tensin homolog
- KISS1
KiSS-1 metastasis suppressor
- XIST
X Inactive specific transcript
- FBP2
Fructose-1,6-bisphosphatases
- GLUT-1
Glucose transporter 1
- PKM2
Pyruvate kinase isozymes
- IGFBP4
Insulin-like growth factor binding protein 4
- CDH1
Cadherin-1
Authors’ contributions
RKK and JAS drafted the manuscript. RKK, JAS, and MWN participated in the design of the review. RKK, JAS, SM, SKB, and MWN critically revised the manuscript. All authors read and approved the final manuscript.
Funding
This manuscript is supported by financial assistance from the National Institute of Health/NCI Grant R01CA218545 to MWN.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Every author is aware of, has agreed to the content of this paper.
Competing interests
SKB is one of the co-founders of Sanguine Diagnostics and Therapeutics, Inc. Other authors declare no competing interests. The authors declared that the submitted manuscript is original, are not under consideration by another journal, and have not been published previously in any form.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Lin X, DeAngelis LM. Treatment of Brain Metastases. J Clin Oncol. 2015;33:3475–3484. doi: 10.1200/JCO.2015.60.9503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witzel I, Oliveira-Ferrer L, Pantel K, Muller V, Wikman H. Breast cancer brain metastases: biology and new clinical perspectives. Breast Cancer Res. 2016;18:8. doi: 10.1186/s13058-015-0665-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28:3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 5.Venur VA, Ahluwalia MS. Targeted Therapy in Brain Metastases: Ready for Primetime? Am Soc Clin Oncol Educ Book. 2016;35:e123–e130. doi: 10.14694/EDBK_100006. [DOI] [PubMed] [Google Scholar]
- 6.Huang Z, Sun B, Wu S, Meng X, Cong Y, Shen G, Song S. A nomogram for predicting survival in patients with breast cancer brain metastasis. Oncol Lett. 2018;15:7090–7096. doi: 10.3892/ol.2018.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin J, Gao Y, Zhang J, Wang L, Wang B, Cao J, Shao Z, Wang Z. Incidence, pattern and prognosis of brain metastases in patients with metastatic triple negative breast cancer. BMC Cancer. 2018;18:446. doi: 10.1186/s12885-018-4371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters S, Bexelius C, Munk V, Leighl N. The impact of brain metastasis on quality of life, resource utilization and survival in patients with non-small-cell lung cancer. Cancer Treat Rev. 2016;45:139–162. doi: 10.1016/j.ctrv.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Graesslin O, Abdulkarim BS, Coutant C, Huguet F, Gabos Z, Hsu L, Marpeau O, Uzan S, Pusztai L, Strom EA, Hortobagyi GN, Rouzier R, Ibrahim NK. Nomogram to predict subsequent brain metastasis in patients with metastatic breast cancer. J Clin Oncol. 2010;28:2032–2037. doi: 10.1200/JCO.2009.24.6314. [DOI] [PubMed] [Google Scholar]
- 10.Sleeman JP. The metastatic niche and stromal progression. Cancer Metastasis Rev. 2012;31:429–440. doi: 10.1007/s10555-012-9373-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WE, Goldbrunner R, Herms J, Winkler F. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med. 2010;16:116–122. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 12.McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Heitz F, Harter P, Lueck HJ, Fissler-Eckhoff A, Lorenz-Salehi F, Scheil-Bertram S, Traut A, du Bois A. Triple-negative and HER2-overexpressing breast cancers exhibit an elevated risk and an earlier occurrence of cerebral metastases. Eur J Cancer. 2009;45:2792–2798. doi: 10.1016/j.ejca.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 14.Gril B, Paranjape AN, Woditschka S, Hua E, Dolan EL, Hanson J, Wu X, Kloc W, Izycka-Swieszewska E, Duchnowska R, Peksa R, Biernat W, Jassem J, Nayyar N, Brastianos PK, Hall OM, Peer CJ, Figg WD, Pauly GT, Robinson C, Difilippantonio S, Bialecki E, Metellus P, Schneider JP, Steeg PS. Reactive astrocytic S1P3 signaling modulates the blood-tumor barrier in brain metastases. Nat Commun. 2018;9:2705. doi: 10.1038/s41467-018-05030-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockman PR, Mittapalli RK, Taskar KS, Rudraraju V, Gril B, Bohn KA, Adkins CE, Roberts A, Thorsheim HR, Gaasch JA, Huang S, Palmieri D, Steeg PS, Smith QR. Heterogeneous blood-tumor barrier permeability determines drug efficacy in experimental brain metastases of breast cancer. Clin Cancer Res. 2010;16:5664–5678. doi: 10.1158/1078-0432.CCR-10-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y, Ma H, Gao C, Lv Y, Chen X, Xu R, Sun M, Liu X, Lu X, Pei X, Li P. Tumor-promoting properties of miR-8084 in breast cancer through enhancing proliferation, suppressing apoptosis and inducing epithelial-mesenchymal transition. J Transl Med. 2018;16:38. doi: 10.1186/s12967-018-1419-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JW, Guan W, Han S, Hong DK, Kim LS, Kim H. MicroRNA-708-3p mediates metastasis and chemoresistance through inhibition of epithelial-to-mesenchymal transition in breast cancer. Cancer Sci. 2018;109:1404–1413. doi: 10.1111/cas.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mansoori B, Mohammadi A, Ghasabi M, Shirjang S, Dehghan R, Montazeri V, Holmskov U, Kazemi T, Duijf P, Gjerstorff M, Baradaran B. miR-142-3p as tumor suppressor miRNA in the regulation of tumorigenicity, invasion and migration of human breast cancer by targeting Bach-1 expression. J Cell Physiol. 2019;234:9816–9825. doi: 10.1002/jcp.27670. [DOI] [PubMed] [Google Scholar]
- 19.Harquail Jason, LeBlanc Nicolas, Ouellette Rodney J, Robichaud Gilles A. miRNAs 484 and 210 regulate Pax-5 expression and function in breast cancer cells. Carcinogenesis. 2019;40(8):1010–1020. doi: 10.1093/carcin/bgy191. [DOI] [PubMed] [Google Scholar]
- 20.Augoff K, Das M, Bialkowska K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 is a broad regulator of beta1-integrin expression and function in cancer cells. Mol Cancer Res. 2011;9:1500–1508. doi: 10.1158/1541-7786.MCR-11-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurmeister S, Baumann M, Balwierz A, Keklikoglou I, Ward A, Uhlmann S, Zhang JD, Wiemann S, Sahin O. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol. 2012;32:633–651. doi: 10.1128/MCB.06212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sossey-Alaoui K, Bialkowska K, Plow EF. The miR200 family of microRNAs regulates WAVE3-dependent cancer cell invasion. J Biol Chem. 2009;284:33019–33029. doi: 10.1074/jbc.M109.034553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sossey-Alaoui K, Pluskota E, Bialkowska K, Szpak D, Parker Y, Morrison CD, Lindner DJ, Schiemann WP, Plow EF. Kindlin-2 Regulates the Growth of Breast Cancer Tumors by Activating CSF-1-Mediated Macrophage Infiltration. Cancer Res. 2017;77:5129–5141. doi: 10.1158/0008-5472.CAN-16-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flores-Perez A, Marchat LA, Rodriguez-Cuevas S, Bautista-Pina V, Hidalgo-Miranda A, Ocampo EA, Martinez MS, Palma-Flores C, Fonseca-Sanchez MA, Astudillo-de la Vega H, Ruiz-Garcia E, Gonzalez-Barrios JA, Perez-Plasencia C, Streber ML, Lopez-Camarillo C. Dual targeting of ANGPT1 and TGFBR2 genes by miR-204 controls angiogenesis in breast cancer. Sci Rep. 2016;6:34504. doi: 10.1038/srep34504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keklikoglou I, Koerner C, Schmidt C, Zhang JD, Heckmann D, Shavinskaya A, Allgayer H, Guckel B, Fehm T, Schneeweiss A, Sahin O, Wiemann S, Tschulena U. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-kappaB and TGF-beta signaling pathways. Oncogene. 2012;31:4150–4163. doi: 10.1038/onc.2011.571. [DOI] [PubMed] [Google Scholar]
- 26.Pakravan K, Babashah S, Sadeghizadeh M, Mowla SJ, Mossahebi-Mohammadi M, Ataei F, Dana N, Javan M. MicroRNA-100 shuttled by mesenchymal stem cell-derived exosomes suppresses in vitro angiogenesis through modulating the mTOR/HIF-1alpha/VEGF signaling axis in breast cancer cells. Cell Oncol (Dordr) 2017;40:457–470. doi: 10.1007/s13402-017-0335-7. [DOI] [PubMed] [Google Scholar]
- 27.Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, Wu S, Han HD, Shah MY, Rodriguez-Aguayo C, Bottsford-Miller J, Liu Y, Kim SB, Unruh A, Gonzalez-Villasana V, Huang L, Zand B, Moreno-Smith M, Mangala LS, Taylor M, Dalton HJ, Sehgal V, Wen Y, Kang Y, Baggerly KA, Lee JS, Ram PT, Ravoori MK, Kundra V, Zhang X, Ali-Fehmi R, Gonzalez-Angulo AM, Massion PP, Calin GA, Lopez-Berestein G, Zhang W, Sood AK. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu Y, Liu H, Lv X, Liu Y, Wang X, Zhang M, Zhang X, Li Y, Lou Q, Li S, Li H. MicroRNA-16-5p overexpression suppresses proliferation and invasion as well as triggers apoptosis by targeting VEGFA expression in breast carcinoma. Oncotarget. 2017;8:72400–72410. doi: 10.18632/oncotarget.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, Yen Y, Wang Y, Marcusson EG, Chu P, Wu J, Wu X, Li AX, Li Z, Gao H, Ren X, Boldin MP, Lin PC, Wang SE. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, Bai Y, Shen Y, Yuan W, Jing Q, Qin Y. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351:157–164. doi: 10.1007/s11010-011-0723-7. [DOI] [PubMed] [Google Scholar]
- 31.Debeb Bisrat G., Lacerda Lara, Anfossi Simone, Diagaradjane Parmeswaran, Chu Khoi, Bambhroliya Arvind, Huo Lei, Wei Caimiao, Larson Richard A., Wolfe Adam R., Xu Wei, Smith Daniel L., Li Li, Ivan Cristina, Allen Pamela K., Wu Wenhui, Calin George A., Krishnamurthy Savitri, Zhang Xiang H., Buchholz Thomas A., Ueno Naoto T., Reuben James M., Woodward Wendy A. miR-141-Mediated Regulation of Brain Metastasis From Breast Cancer. Journal of the National Cancer Institute. 2016;108(8):djw026. doi: 10.1093/jnci/djw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donatelli SS, Zhou JM, Gilvary DL, Eksioglu EA, Chen X, Cress WD, Haura EB, Schabath MB, Coppola D, Wei S, Djeu JY. TGF-beta-inducible microRNA-183 silences tumor-associated natural killer cells. Proc Natl Acad Sci U S A. 2014;111:4203–4208. doi: 10.1073/pnas.1319269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazar S, Goldfinger LE. Platelet Microparticles and miRNA Transfer in Cancer Progression: Many Targets, Modes of Action, and Effects Across Cancer Stages. Front Cardiovasc Med. 2018;5:13. doi: 10.3389/fcvm.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doberstein K, Bretz NP, Schirmer U, Fiegl H, Blaheta R, Breunig C, Muller-Holzner E, Reimer D, Zeimet AG, Altevogt P. miR-21-3p is a positive regulator of L1CAM in several human carcinomas. Cancer Lett. 2014;354:455–466. doi: 10.1016/j.canlet.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 35.Bai Y, Zhang Y, Hua J, Yang X, Zhang X, Duan M, Zhu X, Huang W, Chao J, Zhou R, Hu G, Yao H. Silencing microRNA-143 protects the integrity of the blood-brain barrier: implications for methamphetamine abuse. Sci Rep. 2016;6:35642. doi: 10.1038/srep35642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong L, Yang J, Han Y, Lu Q, Cao J, Syed L. High expression of miR-210 predicts poor survival in patients with breast cancer: a meta-analysis. Gene. 2012;507:135–138. doi: 10.1016/j.gene.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 37.Ma Qingyi, Dasgupta Chiranjib, Li Yong, Huang Lei, Zhang Lubo. MicroRNA-210 Suppresses Junction Proteins and Disrupts Blood-Brain Barrier Integrity in Neonatal Rat Hypoxic-Ischemic Brain Injury. International Journal of Molecular Sciences. 2017;18(7):1356. doi: 10.3390/ijms18071356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reijerkerk A, Lopez-Ramirez MA, van Het Hof B, Drexhage JA, Kamphuis WW, Kooij G, Vos JB, van der Pouw Kraan TC, van Zonneveld AJ, Horrevoets AJ, Prat A, Romero IA, de Vries HE. MicroRNAs regulate human brain endothelial cell-barrier function in inflammation: implications for multiple sclerosis. J Neurosci. 2013;33:6857–6863. doi: 10.1523/JNEUROSCI.3965-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridgway LD, Wetzel MD, Ngo JA, Erdreich-Epstein A, Marchetti D. Heparanase-induced GEF-H1 signaling regulates the cytoskeletal dynamics of brain metastatic breast cancer cells. Mol Cancer Res. 2012;10:689–702. doi: 10.1158/1541-7786.MCR-11-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Sullivan PS, Goodman JC, Gunaratne PH, Marchetti D. MicroRNA-1258 suppresses breast cancer brain metastasis by targeting heparanase. Cancer Res. 2011;71:645–654. doi: 10.1158/0008-5472.CAN-10-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo YY, Feng J, Sanders S, Jin G, Singh R, Vidi PA, Tyagi A, Chan MD, Ruiz J, Debinski W, Pasche BC, Lo HW, Metheny-Barlow LJ, D'Agostino RB, Jr, Watabe K. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer Res. 2018;78:4316–4330. doi: 10.1158/0008-5472.CAN-18-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, Li P, Li M, Wang X, Zhang C, Wang H, Ellis K, Cheerathodi M, McCarty JH, Palmieri D, Saunus J, Lakhani S, Huang S, Sahin AA, Aldape KD, Steeg PS, Yu D. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100–104. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 44.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paterson EL, Kolesnikoff N, Gregory PA, Bert AG, Khew-Goodall Y, Goodall GJ. The microRNA-200 family regulates epithelial to mesenchymal transition. ScientificWorldJournal. 2008;8:901–904. doi: 10.1100/tsw.2008.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grosswendt S, Filipchyk A, Manzano M, Klironomos F, Schilling M, Herzog M, Gottwein E, Rajewsky N. Unambiguous identification of miRNA:target site interactions by different types of ligation reactions. Mol Cell. 2014;54:1042–1054. doi: 10.1016/j.molcel.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park JH, Shin C. MicroRNA-directed cleavage of targets: mechanism and experimental approaches. BMB Rep. 2014;47:417–423. doi: 10.5483/BMBRep.2014.47.8.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yerukala Sathipati S, Ho SY. Identifying a miRNA signature for predicting the stage of breast cancer. Sci Rep. 2018;8:16138. doi: 10.1038/s41598-018-34604-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macha MA, Seshacharyulu P, Krishn SR, Pai P, Rachagani S, Jain M, Batra SK. MicroRNAs (miRNAs) as biomarker(s) for prognosis and diagnosis of gastrointestinal (GI) cancers. Curr Pharm Des. 2014;20:5287–5297. doi: 10.2174/1381612820666140128213117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hashimoto Y, Akiyama Y, Yuasa Y. Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS One. 2013;8:e62589. doi: 10.1371/journal.pone.0062589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cantini Laura, Bertoli Gloria, Cava Claudia, Dubois Thierry, Zinovyev Andrei, Caselle Michele, Castiglioni Isabella, Barillot Emmanuel, Martignetti Loredana. Identification of microRNA clusters cooperatively acting on epithelial to mesenchymal transition in triple negative breast cancer. Nucleic Acids Research. 2019;47(5):2205–2215. doi: 10.1093/nar/gkz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong P, Chen L, Yu M, Tao J, Liu J, Wang Y, Pan H, Zhou W, Wang S. miR-3178 inhibits cell proliferation and metastasis by targeting Notch1 in triple-negative breast cancer. Cell Death Dis. 2018;9:1059. doi: 10.1038/s41419-018-1091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lv ZD, Yang DX, Liu XP, Jin LY, Wang XG, Yang ZC, Liu D, Zhao JJ, Kong B, Li FN, Wang HB. MiR-212-5p Suppresses the Epithelial-Mesenchymal Transition in Triple-Negative Breast Cancer by Targeting Prrx2. Cell Physiol Biochem. 2017;44:1785–1795. doi: 10.1159/000485785. [DOI] [PubMed] [Google Scholar]
- 54.Duhachek-Muggy S, Zolkiewska A. ADAM12-L is a direct target of the miR-29 and miR-200 families in breast cancer. BMC Cancer. 2015;15:93. doi: 10.1186/s12885-015-1108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sayed D, He M, Hong C, Gao S, Rane S, Yang Z, Abdellatif M. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J Biol Chem. 2010;285:20281–20290. doi: 10.1074/jbc.M110.109207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schickel R, Park SM, Murmann AE, Peter ME. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol Cell. 2010;38:908–915. doi: 10.1016/j.molcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu Y, Jiang BQ, Wu Y, Li ZD, Zhuang ZG. Hsa-miR-206 inhibits the migration and invasion of breast cancer by targeting Cx43. Zhonghua Yi Xue Za Zhi. 2013;93:2890–2894. [PubMed] [Google Scholar]
- 58.Aakula A, Kohonen P, Leivonen SK, Makela R, Hintsanen P, Mpindi JP, Martens-Uzunova E, Aittokallio T, Jenster G, Perala M, Kallioniemi O, Ostling P. Systematic Identification of MicroRNAs That Impact on Proliferation of Prostate Cancer Cells and Display Changed Expression in Tumor Tissue. Eur Urol. 2016;69:1120–1128. doi: 10.1016/j.eururo.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 59.Li D, Yang P, Xiong Q, Song X, Yang X, Liu L, Yuan W, Rui YC. MicroRNA-125a/b-5p inhibits endothelin-1 expression in vascular endothelial cells. J Hypertens. 2010;28:1646–1654. doi: 10.1097/HJH.0b013e32833a4922. [DOI] [PubMed] [Google Scholar]
- 60.Ostadrahimi S, Fayaz S, Parvizhamidi M, Abedi-Valugerdi M, Hassan M, Kadivar M, Teimoori-Toolabi L, Asgari M, Shahrokh H, Abolhasani M, Mahdian R, Fard-Esfahani P. Downregulation of miR-1266-5P, miR-185-5P and miR-30c-2 in prostatic cancer tissue and cell lines. Oncol Lett. 2018;15:8157–8164. doi: 10.3892/ol.2018.8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeh TC, Huang TT, Yeh TS, Chen YR, Hsu KW, Yin PH, Lee HC, Tseng LM. miR-151-3p Targets TWIST1 to Repress Migration of Human Breast Cancer Cells. PLoS One. 2016;11:e0168171. doi: 10.1371/journal.pone.0168171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suarez Y, Wang C, Manes TD, Pober JS. Cutting edge: TNF-induced microRNAs regulate TNF-induced expression of E-selectin and intercellular adhesion molecule-1 on human endothelial cells: feedback control of inflammation. J Immunol. 2010;184:21–25. doi: 10.4049/jimmunol.0902369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu XY, Chen D, Gu XY, Ding J, Zhao YJ, Zhao Q, Yao M, Chen Z, He XH, Cong WM. Predicting Value of ALCAM as a Target Gene of microRNA-483-5p in Patients with Early Recurrence in Hepatocellular Carcinoma. Front Pharmacol. 2017;8:973. doi: 10.3389/fphar.2017.00973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei LQ, Liang HT, Qin DC, Jin HF, Zhao Y, She MC. MiR-212 exerts suppressive effect on SKOV3 ovarian cancer cells through targeting HBEGF. Tumour Biol. 2014;35:12427–12434. doi: 10.1007/s13277-014-2560-2. [DOI] [PubMed] [Google Scholar]
- 66.Majumder M, Dunn L, Liu L, Hasan A, Vincent K, Brackstone M, Hess D, Lala PK. COX-2 induces oncogenic micro RNA miR655 in human breast cancer. Sci Rep. 2018;8:327. doi: 10.1038/s41598-017-18612-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kurcon T, Liu Z, Paradkar AV, Vaiana CA, Koppolu S, Agrawal P, Mahal LK. miRNA proxy approach reveals hidden functions of glycosylation. Proc Natl Acad Sci U S A. 2015;112:7327–7332. doi: 10.1073/pnas.1502076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tominaga N, Kosaka N, Ono M, Katsuda T, Yoshioka Y, Tamura K, Lotvall J, Nakagama H, Ochiya T. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat Commun. 2015;6:6716. doi: 10.1038/ncomms7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, Holland EC. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo P, Lan J, Ge J, Nie Q, Guo L, Qiu Y, Mao Q. MiR-26a enhances the radiosensitivity of glioblastoma multiforme cells through targeting of ataxia-telangiectasia mutated. Exp Cell Res. 2014;320:200–208. doi: 10.1016/j.yexcr.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 72.Kaverina N, Borovjagin AV, Kadagidze Z, Baryshnikov A, Baryshnikova M, Malin D, Ghosh D, Shah N, Welch DR, Gabikian P, Karseladze A, Cobbs C, Ulasov IV. Astrocytes promote progression of breast cancer metastases to the brain via a KISS1-mediated autophagy. Autophagy. 2017;13:1905–1923. doi: 10.1080/15548627.2017.1360466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orihuela R, McPherson CA, Harry GJ. Microglial M1/M2 polarization and metabolic states. Br J Pharmacol. 2016;173:649–665. doi: 10.1111/bph.13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, Chow A, O'Connor ST, Li S, Chin AR, Somlo G, Palomares M, Li Z, Tremblay JR, Tsuyada A, Sun G, Reid MA, Wu X, Swiderski P, Ren X, Shi Y, Kong M, Zhong W, Chen Y, Wang SE. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17:183–194. doi: 10.1038/ncb3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim S, Lee E, Jung J, Lee JW, Kim HJ, Kim J, Yoo HJ, Lee HJ, Chae SY, Jeon SM, Son BH, Gong G, Sharan SK, Chang S. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene. 2018;37:2982–2991. doi: 10.1038/s41388-018-0124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaudhuri AD, Kabaria S, Choi DC, Mouradian MM, Junn E. MicroRNA-7 Promotes Glycolysis to Protect against 1-Methyl-4-phenylpyridinium-induced Cell Death. J Biol Chem. 2015;290:12425–12434. doi: 10.1074/jbc.M114.625962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 78.Wang Y, Zhou BP. Epithelial-mesenchymal Transition---A Hallmark of Breast Cancer Metastasis. Cancer Hallm. 2013;1:38–49. doi: 10.1166/ch.2013.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 81.Imani S, Hosseinifard H, Cheng J, Wei C, Fu J. Prognostic Value of EMT-inducing Transcription Factors (EMT-TFs) in Metastatic Breast Cancer: A Systematic Review and Meta-analysis. Sci Rep. 2016;6:28587. doi: 10.1038/srep28587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kveiborg M, Albrechtsen R, Couchman JR, Wewer UM. Cellular roles of ADAM12 in health and disease. Int J Biochem Cell Biol. 2008;40:1685–1702. doi: 10.1016/j.biocel.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 83.Eckert MA, Santiago-Medina M, Lwin TM, Kim J, Courtneidge SA, Yang J. ADAM12 induction by Twist1 promotes tumor invasion and metastasis via regulation of invadopodia and focal adhesions. J Cell Sci. 2017;130:2036–2048. doi: 10.1242/jcs.198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Valastyan S, Weinberg RA. Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci. 2011;124:999–1006. doi: 10.1242/jcs.081513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen X, Zhao M, Huang J, Li Y, Wang S, Harrington CA, Qian DZ, Sun XX, Dai MS. microRNA-130a suppresses breast cancer cell migration and invasion by targeting FOSL1 and upregulating ZO-1. J Cell Biochem. 2018;119:4945–4956. doi: 10.1002/jcb.26739. [DOI] [PubMed] [Google Scholar]
- 86.Dua RS, Gui GP, Isacke CM. Endothelial adhesion molecules in breast cancer invasion into the vascular and lymphatic systems. Eur J Surg Oncol. 2005;31:824–832. doi: 10.1016/j.ejso.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 87.Muller WA. Mechanisms of transendothelial migration of leukocytes. Circ Res. 2009;105:223–230. doi: 10.1161/CIRCRESAHA.109.200717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Deryugina EI, Kiosses WB. Intratumoral Cancer Cell Intravasation Can Occur Independent of Invasion into the Adjacent Stroma. Cell Rep. 2017;19:601–616. doi: 10.1016/j.celrep.2017.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ungefroren H, Sebens S, Seidl D, Lehnert H, Hass R. Interaction of tumor cells with the microenvironment. Cell Commun Signal. 2011;9:18. doi: 10.1186/1478-811X-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zonneville J, Safina A, Truskinovsky AM, Arteaga CL, Bakin AV. TGF-beta signaling promotes tumor vasculature by enhancing the pericyte-endothelium association. BMC Cancer. 2018;18:670. doi: 10.1186/s12885-018-4587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strilic B, Offermanns S. Intravascular Survival and Extravasation of Tumor Cells. Cancer Cell. 2017;32:282–293. doi: 10.1016/j.ccell.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 92.Ponert Jan, Gockel Lukas, Henze Svenja, Schlesinger Martin. Unfractionated and Low Molecular Weight Heparin Reduce Platelet Induced Epithelial-Mesenchymal Transition in Pancreatic and Prostate Cancer Cells. Molecules. 2018;23(10):2690. doi: 10.3390/molecules23102690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jung K, Gupta N, Wang P, Lewis JT, Gopal K, Wu F, Ye X, Alshareef A, Abdulkarim BS, Douglas DN, Kneteman NM, Lai R. Triple negative breast cancers comprise a highly tumorigenic cell subpopulation detectable by its high responsiveness to a Sox2 regulatory region 2 (SRR2) reporter. Oncotarget. 2015;6:10366–10373. doi: 10.18632/oncotarget.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang CY, Yaswen P, Goga A, Werb Z. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526:131–135. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ren D, Zhu X, Kong R, Zhao Z, Sheng J, Wang J, Xu X, Liu J, Cui K, Zhang XH, Zhao H, Wong STC. Targeting Brain-Adaptive Cancer Stem Cells Prohibits Brain Metastatic Colonization of Triple-Negative Breast Cancer. Cancer Res. 2018;78:2052–2064. doi: 10.1158/0008-5472.CAN-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gersuk GM, Westermark B, Mohabeer AJ, Challita PM, Pattamakom S, Pattengale PK. Inhibition of human natural killer cell activity by platelet-derived growth factor (PDGF). III. Membrane binding studies and differential biological effect of recombinant PDGF isoforms. Scand J Immunol. 1991;33:521–532. doi: 10.1111/j.1365-3083.1991.tb02522.x. [DOI] [PubMed] [Google Scholar]
- 97.Ponert JM, Schwarz S, Haschemi R, Muller J, Potzsch B, Bendas G, Schlesinger M. The mechanisms how heparin affects the tumor cell induced VEGF and chemokine release from platelets to attenuate the early metastatic niche formation. PLoS One. 2018;13:e0191303. doi: 10.1371/journal.pone.0191303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Di Raimo T, De Santis E, Coppola L, Rosario D, Andrea M, Angelini F. Circulating tumor cells and the metastatic process: the complexity of malignancy. Journal of Cancer Metastasis and Treatment. 2018;4:54. doi: 10.20517/2394-4722.2018.50. [DOI] [Google Scholar]
- 99.Bendas G, Borsig L. Cancer cell adhesion and metastasis: selectins, integrins, and the inhibitory potential of heparins. Int J Cell Biol. 2012;2012:676731. doi: 10.1155/2012/676731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reymond N, d'Agua BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer. 2013;13:858–870. doi: 10.1038/nrc3628. [DOI] [PubMed] [Google Scholar]
- 101.Wasilewski D, Priego N, Fustero-Torre C, Valiente M. Reactive Astrocytes in Brain Metastasis. Front Oncol. 2017;7:298. doi: 10.3389/fonc.2017.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XH, Lee DJ, Chaft JE, Kris MG, Huse JT, Brogi E, Massague J. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. 2014;156:1002–1016. doi: 10.1016/j.cell.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamanaka S, Olaru AV, An F, Luvsanjav D, Jin Z, Agarwal R, Tomuleasa C, Popescu I, Alexandrescu S, Dima S, Chivu-Economescu M, Montgomery EA, Torbenson M, Meltzer SJ, Selaru FM. MicroRNA-21 inhibits Serpini1, a gene with novel tumour suppressive effects in gastric cancer. Dig Liver Dis. 2012;44:589–596. doi: 10.1016/j.dld.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, Jacob L, Patwa R, Shah H, Xu K, Cross JR, Massague J. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016;533:493–498. doi: 10.1038/nature18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim SW, Choi HJ, Lee HJ, He J, Wu Q, Langley RR, Fidler IJ, Kim SJ. Role of the endothelin axis in astrocyte- and endothelial cell-mediated chemoprotection of cancer cells. Neuro Oncol. 2014;16:1585–1598. doi: 10.1093/neuonc/nou128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim SJ, Kim JS, Park ES, Lee JS, Lin Q, Langley RR, Maya M, He J, Kim SW, Weihua Z, Balasubramanian K, Fan D, Mills GB, Hung MC, Fidler IJ. Astrocytes upregulate survival genes in tumor cells and induce protection from chemotherapy. Neoplasia. 2011;13:286–298. doi: 10.1593/neo.11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Soto MS, Serres S, Anthony DC, Sibson NR. Functional role of endothelial adhesion molecules in the early stages of brain metastasis. Neuro Oncol. 2014;16:540–551. doi: 10.1093/neuonc/not222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Deng H, Song Z, Xu H, Deng X, Zhang Q, Chen H, Wang Y, Qin Y, Li Y. MicroRNA-1185 Promotes Arterial Stiffness though Modulating VCAM-1 and E-Selectin Expression. Cell Physiol Biochem. 2017;41:2183–2193. doi: 10.1159/000475576. [DOI] [PubMed] [Google Scholar]
- 109.Leong HS, Robertson AE, Stoletov K, Leith SJ, Chin CA, Chien AE, Hague MN, Ablack A, Carmine-Simmen K, McPherson VA, Postenka CO, Turley EA, Courtneidge SA, Chambers AF, Lewis JD. Invadopodia are required for cancer cell extravasation and are a therapeutic target for metastasis. Cell Rep. 2014;8:1558–1570. doi: 10.1016/j.celrep.2014.07.050. [DOI] [PubMed] [Google Scholar]
- 110.Williams Karla C., Cepeda Mario A., Javed Sumreen, Searle Karlee, Parkins Katie M., Makela Ashley V., Hamilton Amanda M., Soukhtehzari Sepideh, Kim Yohan, Tuck Alan B., Ronald John A., Foster Paula J., Chambers Ann F., Leong Hon S. Invadopodia are chemosensing protrusions that guide cancer cell extravasation to promote brain tropism in metastasis. Oncogene. 2019;38(19):3598–3615. doi: 10.1038/s41388-018-0667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu K, Fukuda K, Xing F, Zhang Y, Sharma S, Liu Y, Chan MD, Zhou X, Qasem SA, Pochampally R, Mo YY, Watabe K. Roles of the cyclooxygenase 2 matrix metalloproteinase 1 pathway in brain metastasis of breast cancer. J Biol Chem. 2015;290:9842–9854. doi: 10.1074/jbc.M114.602185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Majumder M, Landman E, Liu L, Hess D, Lala PK. COX-2 Elevates Oncogenic miR-526b in Breast Cancer by EP4 Activation. Mol Cancer Res. 2015;13:1022–1033. doi: 10.1158/1541-7786.MCR-14-0543. [DOI] [PubMed] [Google Scholar]
- 113.Drolez Aurore, Vandenhaute Elodie, Delannoy Clément, Dewald Justine, Gosselet Fabien, Cecchelli Romeo, Julien Sylvain, Dehouck Marie-Pierre, Delannoy Philippe, Mysiorek Caroline. ST6GALNAC5 Expression Decreases the Interactions between Breast Cancer Cells and the Human Blood-Brain Barrier. International Journal of Molecular Sciences. 2016;17(8):1309. doi: 10.3390/ijms17081309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wilhelm I, Molnar J, Fazakas C, Hasko J, Krizbai IA. Role of the blood-brain barrier in the formation of brain metastases. Int J Mol Sci. 2013;14:1383–1411. doi: 10.3390/ijms14011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wrobel JK, Toborek M. Blood-brain Barrier Remodeling during Brain Metastasis Formation. Mol Med. 2016;22:32–40. doi: 10.2119/molmed.2015.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen W, Hoffmann AD, Liu H, Liu X. Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precis Oncol. 2018;2:4. doi: 10.1038/s41698-018-0047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stamatovic SM, Johnson AM, Keep RF, Andjelkovic AV. Junctional proteins of the blood-brain barrier: New insights into function and dysfunction. Tissue Barriers. 2016;4:e1154641. doi: 10.1080/21688370.2016.1154641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brooks TA, Hawkins BT, Huber JD, Egleton RD, Davis TP. Chronic inflammatory pain leads to increased blood-brain barrier permeability and tight junction protein alterations. Am J Physiol Heart Circ Physiol. 2005;289:H738–H743. doi: 10.1152/ajpheart.01288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hawkins BT, Davis TP. The blood-brain barrier/neurovascular unit in health and disease. Pharmacol Rev. 2005;57:173–185. doi: 10.1124/pr.57.2.4. [DOI] [PubMed] [Google Scholar]
- 120.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xing F, Sharma S, Liu Y, Mo YY, Wu K, Zhang YY, Pochampally R, Martinez LA, Lo HW, Watabe K. miR-509 suppresses brain metastasis of breast cancer cells by modulating RhoC and TNF-alpha. Oncogene. 2015;34:4890–4900. doi: 10.1038/onc.2014.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Camacho L, Guerrero P, Marchetti D. MicroRNA and protein profiling of brain metastasis competent cell-derived exosomes. PLoS One. 2013;8:e73790. doi: 10.1371/journal.pone.0073790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ferraro GB, Kodack DP, Askoxylakis V, Jain RK. Closing the gap: astrocytes and brain metastasis. Cell Res. 2016;26:973–974. doi: 10.1038/cr.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Frediani JN, Fabbri M. Essential role of miRNAs in orchestrating the biology of the tumor microenvironment. Mol Cancer. 2016;15:42. doi: 10.1186/s12943-016-0525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McNally LR, Welch DR, Beck BH, Stafford LJ, Long JW, Sellers JC, Huang ZQ, Grizzle WE, Stockard CR, Nash KT, Buchsbaum DJ. KISS1 over-expression suppresses metastasis of pancreatic adenocarcinoma in a xenograft mouse model. Clin Exp Metastasis. 2010;27:591–600. doi: 10.1007/s10585-010-9349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Muir AI, Chamberlain L, Elshourbagy NA, Michalovich D, Moore DJ, Calamari A, Szekeres PG, Sarau HM, Chambers JK, Murdock P, Steplewski K, Shabon U, Miller JE, Middleton SE, Darker JG, Larminie CG, Wilson S, Bergsma DJ, Emson P, Faull R, Philpott KL, Harrison DC. AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J Biol Chem. 2001;276:28969–28975. doi: 10.1074/jbc.M102743200. [DOI] [PubMed] [Google Scholar]
- 127.Navenot JM, Wang Z, Chopin M, Fujii N, Peiper SC. Kisspeptin-10-induced signaling of GPR54 negatively regulates chemotactic responses mediated by CXCR4: a potential mechanism for the metastasis suppressor activity of kisspeptins. Cancer Res. 2005;65:10450–10456. doi: 10.1158/0008-5472.CAN-05-1757. [DOI] [PubMed] [Google Scholar]
- 128.Wang XH, Wang TL. MicroRNAs of microglia: Wrestling with central nervous system disease. Neural Regen Res. 2018;13:2067–2072. doi: 10.4103/1673-5374.241444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 1990;39:151–170. doi: 10.1016/0306-4522(90)90229-W. [DOI] [PubMed] [Google Scholar]
- 130.Greter M, Lelios I, Croxford AL. Microglia Versus Myeloid Cell Nomenclature during Brain Inflammation. Front Immunol. 2015;6:249. doi: 10.3389/fimmu.2015.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Leitinger N, Schulman IG. Phenotypic polarization of macrophages in atherosclerosis. Arterioscler Thromb Vasc Biol. 2013;33:1120–1126. doi: 10.1161/ATVBAHA.112.300173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Semin Cancer Biol. 2011;21:139–146. doi: 10.1016/j.semcancer.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 134.Martinez-Outschoorn UE, Lisanti MP, Sotgia F. Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Semin Cancer Biol. 2014;25:47–60. doi: 10.1016/j.semcancer.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 135.Chen J, Lee HJ, Wu X, Huo L, Kim SJ, Xu L, Wang Y, He J, Bollu LR, Gao G, Su F, Briggs J, Liu X, Melman T, Asara JM, Fidler IJ, Cantley LC, Locasale JW, Weihua Z. Gain of glucose-independent growth upon metastasis of breast cancer cells to the brain. Cancer Res. 2015;75:554–565. doi: 10.1158/0008-5472.CAN-14-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Adekola K, Rosen ST, Shanmugam M. Glucose transporters in cancer metabolism. Curr Opin Oncol. 2012;24:650–654. doi: 10.1097/CCO.0b013e328356da72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Choi DC, Chae YJ, Kabaria S, Chaudhuri AD, Jain MR, Li H, Mouradian MM, Junn E. MicroRNA-7 protects against 1-methyl-4-phenylpyridinium-induced cell death by targeting RelA. J Neurosci. 2014;34:12725–12737. doi: 10.1523/JNEUROSCI.0985-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chui MH. Insights into cancer metastasis from a clinicopathologic perspective: Epithelial-Mesenchymal Transition is not a necessary step. Int J Cancer. 2013;132:1487–1495. doi: 10.1002/ijc.27745. [DOI] [PubMed] [Google Scholar]
- 139.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chao Y, Wu Q, Acquafondata M, Dhir R, Wells A. Partial mesenchymal to epithelial reverting transition in breast and prostate cancer metastases. Cancer Microenviron. 2012;5:19–28. doi: 10.1007/s12307-011-0085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Drago-Garcia D, Espinal-Enriquez J, Hernandez-Lemus E. Network analysis of EMT and MET micro-RNA regulation in breast cancer. Sci Rep. 2017;7:13534. doi: 10.1038/s41598-017-13903-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, Wei Y, Hu G, Garcia BA, Ragoussis J, Amadori D, Harris AL, Kang Y. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Madhavan D, Cuk K, Burwinkel B, Yang R. Cancer diagnosis and prognosis decoded by blood-based circulating microRNA signatures. Front Genet. 2013;4:116. doi: 10.3389/fgene.2013.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, Smolsky M, Kim RY, Saria MG, Pastorino S, Kesari S, Krichevsky AM. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14:689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Le MT, Hamar P, Guo C, Basar E, Perdigao-Henriques R, Balaj L, Lieberman J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124:5109–5128. doi: 10.1172/JCI75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nagaishi M, Nakata S, Ono Y, Hirata K, Tanaka Y, Suzuki K, Yokoo H, Hyodo A. Tumoral and stromal expression of Slug, ZEB1, and ZEB2 in brain metastasis. J Clin Neurosci. 2017;46:124–128. doi: 10.1016/j.jocn.2017.08.050. [DOI] [PubMed] [Google Scholar]
- 147.Hu M, Xiong S, Chen Q, Zhu S, Zhou X. Novel role of microRNA-126 in digestive system cancers: From bench to bedside. Oncol Lett. 2019;17:31–41. doi: 10.3892/ol.2018.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xiang LY, Ou HH, Liu XC, Chen ZJ, Li XH, Huang Y, Yang DH. Loss of tumor suppressor miR-126 contributes to the development of hepatitis B virus-related hepatocellular carcinoma metastasis through the upregulation of ADAM9. Tumour Biol. 2017;39:1010428317709128. doi: 10.1177/1010428317709128. [DOI] [PubMed] [Google Scholar]
- 149.Hua Y, Liang C, Miao C, Wang S, Su S, Shao P, Liu B, Bao M, Zhu J, Xu A, Zhang J, Li J, Wang Z. MicroRNA-126 inhibits proliferation and metastasis in prostate cancer via regulation of ADAM9. Oncol Lett. 2018;15:9051–9060. doi: 10.3892/ol.2018.8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xing F, Watabe K. miRNAs as biomarkers for brain metastasis of breast cancer. Biomark Med. 2013;7:387–390. doi: 10.2217/bmm.13.43. [DOI] [PubMed] [Google Scholar]
- 151.Wang H, Yu X, Fan Y, Jiang Y. Multiple treatment modalities for brain metastasis in patients with EGFR-mutant non-small-cell lung cancer. Onco Targets Ther. 2018;11:2149–2155. doi: 10.2147/OTT.S156570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Broekman F, Giovannetti E, Peters GJ. Tyrosine kinase inhibitors: Multi-targeted or single-targeted? World J Clin Oncol. 2011;2:80–93. doi: 10.5306/wjco.v2.i2.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schmid P, Chui SY, Emens LA. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. Reply. N Engl J Med. 2019;380:987–988. doi: 10.1056/NEJMc1900150. [DOI] [PubMed] [Google Scholar]
- 154.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shalaby T, Grotzer MA. Tumor-Associated CSF MicroRNAs for the Prediction and Evaluation of CNS Malignancies. Int J Mol Sci. 2015;16:29103–29119. doi: 10.3390/ijms161226150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Drusco A, Bottoni A, Lagana A, Acunzo M, Fassan M, Cascione L, Antenucci A, Kumchala P, Vicentini C, Gardiman MP, Alder H, Carosi MA, Ammirati M, Gherardi S, Luscri M, Carapella C, Zanesi N, Croce CM. A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS malignancies. Oncotarget. 2015;6:20829–20839. doi: 10.18632/oncotarget.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Noyan S, Gurdal H, Gur Dedeoglu B. Involvement of miR-770-5p in trastuzumab response in HER2 positive breast cancer cells. PLoS One. 2019;14:e0215894. doi: 10.1371/journal.pone.0215894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li Y, Liang Y, Sang Y, Song X, Zhang H, Liu Y, Jiang L, Yang Q. MiR-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of STMN1. Cell Death Dis. 2018;9:14. doi: 10.1038/s41419-017-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhao K, Wang L, Li T, Zhu M, Zhang C, Chen L, Zhao P, Zhou H, Yu S, Yang X. The role of miR-451 in the switching between proliferation and migration in malignant glioma cells: AMPK signaling, mTOR modulation and Rac1 activation required. Int J Oncol. 2017;50:1989–1999. doi: 10.3892/ijo.2017.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 161.Deng YW, Hao WJ, Li YW, Li YX, Zhao BC, Lu D. Hsa-miRNA-143-3p Reverses Multidrug Resistance of Triple-Negative Breast Cancer by Inhibiting the Expression of Its Target Protein Cytokine-Induced Apoptosis Inhibitor 1 In Vivo. J Breast Cancer. 2018;21:251–258. doi: 10.4048/jbc.2018.21.e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tormo E, Ballester S, Adam-Artigues A, Burgues O, Alonso E, Bermejo B, Menendez S, Zazo S, Madoz-Gurpide J, Rovira A, Albanell J, Rojo F, Lluch A, Eroles P. The miRNA-449 family mediates doxorubicin resistance in triple-negative breast cancer by regulating cell cycle factors. Sci Rep. 2019;9:5316. doi: 10.1038/s41598-019-41472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Okuda H, Xing F, Pandey PR, Sharma S, Watabe M, Pai SK, Mo YY, Iiizumi-Gairani M, Hirota S, Liu Y, Wu K, Pochampally R, Watabe K. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013;73:1434–1444. doi: 10.1158/0008-5472.CAN-12-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sun X, Liu J, Xu C, Tang SC, Ren H. The insights of Let-7 miRNAs in oncogenesis and stem cell potency. J Cell Mol Med. 2016;20:1779–1788. doi: 10.1111/jcmm.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hayward SL, Wilson CL, Kidambi S. Hyaluronic acid-conjugated liposome nanoparticles for targeted delivery to CD44 overexpressing glioblastoma cells. Oncotarget. 2016;7:34158–34171. doi: 10.18632/oncotarget.8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dehshahri A, Sadeghpour H. Surface decorations of poly(amidoamine) dendrimer by various pendant moieties for improved delivery of nucleic acid materials. Colloids Surf B Biointerfaces. 2015;132:85–102. doi: 10.1016/j.colsurfb.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 167.Saraiva C, Praca C, Ferreira R, Santos T, Ferreira L, Bernardino L. Nanoparticle-mediated brain drug delivery: Overcoming blood-brain barrier to treat neurodegenerative diseases. J Control Release. 2016;235:34–47. doi: 10.1016/j.jconrel.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 168.Lo YL, Chou HL, Liao ZX, Huang SJ, Ke JH, Liu YS, Chiu CC, Wang LF. Chondroitin sulfate-polyethylenimine copolymer-coated superparamagnetic iron oxide nanoparticles as an efficient magneto-gene carrier for microRNA-encoding plasmid DNA delivery. Nanoscale. 2015;7:8554–8565. doi: 10.1039/C5NR01404B. [DOI] [PubMed] [Google Scholar]
- 169.Carnero E, Sutherland JD, Fortes P. Adenovirus and miRNAs. Biochim Biophys Acta. 2011;1809:660–667. doi: 10.1016/j.bbagrm.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wen MM. Getting miRNA Therapeutics into the Target Cells for Neurodegenerative Diseases: A Mini-Review. Front Mol Neurosci. 2016;9:129. doi: 10.3389/fnmol.2016.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bergen JM, Park IK, Horner PJ, Pun SH. Nonviral approaches for neuronal delivery of nucleic acids. Pharm Res. 2008;25:983–998. doi: 10.1007/s11095-007-9439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Huang Y, Gao X, Chen J. Leukocyte-derived biomimetic nanoparticulate drug delivery systems for cancer therapy. Acta Pharm Sin B. 2018;8:4–13. doi: 10.1016/j.apsb.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mitchell MJ, Castellanos CA, King MR. Surfactant functionalization induces robust, differential adhesion of tumor cells and blood cells to charged nanotube-coated biomaterials under flow. Biomaterials. 2015;56:179–186. doi: 10.1016/j.biomaterials.2015.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Galstyan A, Markman JL, Shatalova ES, Chiechi A, Korman AJ, Patil R, Klymyshyn D, Tourtellotte WG, Israel LL, Braubach O, Ljubimov VA, Mashouf LA, Ramesh A, Grodzinski ZB, Penichet ML, Black KL, Holler E, Sun T, Ding H, Ljubimov AV, Ljubimova JY. Blood-brain barrier permeable nano immunoconjugates induce local immune responses for glioma therapy. Nat Commun. 2019;10:3850. doi: 10.1038/s41467-019-11719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, King PD, Weis SM, Cheresh DA. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kung Y, Lan C, Hsiao MY, Sun MK, Hsu YH, Huang AP, Liao WH, Liu HL, Inserra C, Chen WS. Focused shockwave induced blood-brain barrier opening and transfection. Sci Rep. 2018;8:2218. doi: 10.1038/s41598-018-20672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.