Abstract
Background
Exploration of the bioactive components of bovine milk has gained global interest due to their potential applications in human nutrition and health promotion. Despite advances in proteomics profiling, limited studies have been carried out to fully characterize the bovine milk proteome. This study explored the milk proteome of Jersey and Kashmiri cattle at day 90 of lactation using high-resolution mass spectrometry based quantitative proteomics nano-scale LC-MS/Q-TOF technique. Data are available via ProteomeXchange with identifier PXD017412.
Results
Proteins from whey were fractionated by precipitation into high and low abundant proteins. A total of 81 high-abundant and 99 low-abundant proteins were significantly differentially expressed between Kashmiri and Jersey cattle, clearly differentiating the two breeds at the proteome level. Among the top differentiating proteins, the Kashmiri cattle milk proteome was characterised by increased concentrations of immune-related proteins (apelin, acid glycoprotein, CD14 antigen), neonatal developmental protein (probetacellulin), xenobiotic metabolising enzyme (flavin monooxygenase 3 (FMO3), GLYCAM1 and HSP90AA1 (chaperone) while the Jersey milk proteome presented higher concentrations of enzyme modulators (SERPINA1, RAC1, serine peptidase inhibitor) and hydrolases (LTF, LPL, CYM, PNLIPRP2). Pathway analysis in Kashmiri cattle revealed enrichment of key pathways involved in the regulation of mammary gland development like Wnt signalling pathway, EGF receptor signalling pathway and FGF signalling pathway while a pathway (T-cell activation pathway) associated with immune system regulation was significantly enriched in Jersey cattle. Most importantly, the high-abundant FMO3 enzyme with an observed 17-fold higher expression in Kashmiri cattle milk seems to be a characteristic feature of the breed. The presence of this (FMO3) bioactive peptide/enzyme in Kashmiri cattle could be economically advantageous for milk products from Kashmiri cattle.
Conclusion
In conclusion, this is the first study to provide insights not only into the milk proteome differences between Kashmiri and Jersey cattle but also provides potential directions for application of specific milk proteins from Kashmiri cattle in special milk preparations like infant formula.
Keywords: Jersey, Kashmiri, Milk proteome, FMO3 enzyme
Background
Bovine milk is a valued natural product which delivers a matrix of essential nutrients including growth and immune factors to offspring and a key raw material for human food preparations [1, 2]. Some studies have characterized the bovine milk proteome, its bioactive profile, and the extent of cross reactivity of bovine bioactive milk peptides on various biological functions [3–7]. Milk proteins are generally categorized into three major groups: caseins, whey proteins and milk fat globule membrane proteins [4, 8]. Most of the polypeptides in milk are an essential source of amino acids to neonates [9] and many resist proteolysis [10, 11]. Milk peptides also facilitate absorption of other nutrients in the gastro-intestinal tract, provide humoral immune responses and support intestinal development [12]. Besides, digestion or fermentation of milk proteins also produces a number of bioactive peptides, which contribute as well to the various functional properties of milk [13, 14]. The major proteins in milk are far outnumbered by numerous other minor proteins which play important roles in a wide range of physiological activities including antioxidant activity, post-natal development of new-borns, maturation of the immune system, establishment of symbiotic microflora, and protection against various pathogens [15, 16].
Several studies have characterised the milk proteome in different species and breeds using different quantitative proteomic techniques [7, 16–20]. The differences in the milk proteome profile have been attributed to genetic, management and disease factors [7, 21]). Although the diverse composition and biological functions of bovine milk has been reported extensively [22–24], the comparative abundance of milk proteins in Indian cattle breeds have not been investigated till date. Kashmiri and Jersey cattle are two important milk animals which contribute significantly to the total milk production in the Indian northern state of Kashmir. The Kashmiri cattle is an indigenous breed kept mainly for milk production in the hilly regions of Kashmir. Kashmiri cattle are small, hardy and adapted to the hilly regions of Kashmir. Whereas, Jersey is a well-established dairy breed imported to augment the milk production ability of Kashmiri cattle through cross breeding. We hypothesize that the proteome profile of Kashmiri cattle milk may have special properties or differ from that of the well-established Jersey dairy breed due to its different genetic background and milk producing ability. Therefore, the aim of this study was to study the protein profiles of Kashmiri and Jersey cattle milk which could reveal important protein factors underlying the physiological differences and differences in milk traits between the two breeds.
Results
Proteome profile of bovine milk
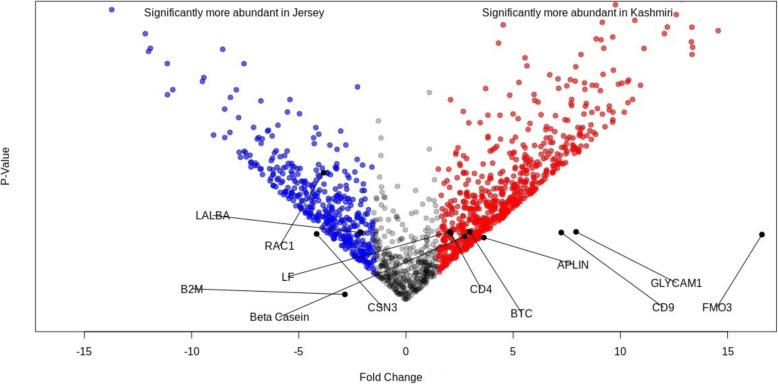
Proteins from whey were fractionated by precipitation into high and low abundant proteins. A total of 180 proteins were differentially expressed (DE) (FDR < 0.1) between Kashmiri and Jersey cattle. Specifically, 91 and 89 proteins were significantly upregulated (FDR < 0.1) in Kashmiri and Jersey cattle, respectively (Additional file 2: Table S2a and S2b, Additional file 3). The most upregulated high abundant proteins (fold change (FC) > 2) were CSN2, CD4 and LF, and low abundant proteins were FMO3, GLYCAM1, APLN and BTC in Kashmiri cattle (Table 1, Fig. 1). Whereas, LALBA, ZNF496, CSN3 and LGB were the most upregulated high abundant proteins and RAC1, B2M and SAR1B were the most upregulated minor milk proteins in Jersey cattle (Table 1).
Table 1.
Significantly upregulated high abundant and low abundant milk proteins in Kashmiri and Jersey cattle
| Accession No. | Protein | Gene ID | FC | P-value | FDR | |
|---|---|---|---|---|---|---|
| Kashmiri Cattle | Significantly upregulated abundant milk proteins | |||||
| J9UHS4 | Beta-casein | CSN2 | 2.74 | 0.044 | 0.055 | |
| Q8HY42 | CD4 antigen | CD4 | 2.09 | 0.039 | 0.043 | |
| E6YCQ7 | Lactoferrin | LF | 2.04 | 0.037 | 0.047 | |
| Significantly upregulated less abundant milk proteins | ||||||
| Q8HYK4 | Flavin-containing monooxygenase 3 | FMO3 | 16.6 | 0.041 | 0.050 | |
| P80195 | Glycosylation-dependent cell adhesion molecule 1 | GLYCAM1 | 7.93 | 0.037 | 0.047 | |
| P30932 | CD9 antigen | CD9 | 7.24 | 0.038 | 0.048 | |
| Q9TUI9 | Apelin | APLN | 3.63 | 0.046 | 0.050 | |
| Q9TTC5 | Probetacellulin | BTC | 2.97 | 0.037 | 0.042 | |
| Q9TRC0 | Enterotoxin-binding glycoprotein PP16K | N/A | 2.91 | 0.038 | 0.048 | |
| Q3SZR3 | Alpha-1-acid glycoprotein | ORM1 | 2.66 | 0.046 | 0.050 | |
| C4PU73 | Serin peptidase inhibitor, clade A | LOC286871 | 2.53 | 0.039 | 0.046 | |
| Q9TS52 | Adipocyte differentiation-related protein | N/A | 2.53 | 0.042 | 0.055 | |
| P46201 | Uterine milk protein | N/A | 2.41 | 0.043 | 0.049 | |
| Q5GN72 | Alpha-1-acid glycoprotein | AGP | 2.07 | 0.037 | 0.040 | |
| Jersey cattle | Significantly upregulated abundant milk proteins | |||||
| P02754 | Beta-lactoglobulin | LGB | 7.24 | 0.037 | 0.051 | |
| A0A140T8A9 | Kappa-casein | CSN3 | 4.17 | 0.04 | 0.046 | |
| F6I8C5 | Zinc finger protein 496 | ZNF496 | 2.33 | 0.037 | 0.061 | |
| G9G9X6 | Alpha-lactalbumin | LALBA | 2.11 | 0.038 | 0.041 | |
| Significantly upregulated less abundant milk proteins | ||||||
| G8FZ88 | ATP synthase subunit A | N/A | 4.09 | 0.037 | 0.047 | |
| P62998 | Ras-related C3 botulinum toxin substrate | RAC1 | 3.85 | 0.044 | 0.067 | |
| P01888 | Beta-2-microglobulin | B2M | 2.85 | 0.039 | 0.041 | |
| Q3T0T7 | GTP-binding protein SAR1b | SAR1B | 2.2 | 0.037 | 0.046 | |
| Q9XSC9 | Transcobalamin-2 | TCN2 | 2.18 | 0.044 | 0.051 | |
| Q95114 | Lactadherin | MFGE8 | 2.11 | 0.039 | 0.049 | |
Fig. 1.
Volcano plot of differentially expressed proteins between Kashmiri and Jersey cattle. Red points indicate more abundant proteins in Kashmiri cattle; blue points indicate more abundant proteins in Jersey cattle
Enriched gene ontology terms of significantly upregulated proteins in Kashmiri and Jersey cattle
Gene ontology (GO) enrichment of significantly upregulated proteins in Kashmiri and Jersey cattle found a total of 4 enriched GO terms in Kashmiri and 4 in Jersey cattle (Table 2). Only extracellular region (GO:0005576) reached significance after FDR correction in both breeds (Table 2).
Table 2.
Gene ontology terms enriched for significantly upregulated proteins in Kashmiri and Jersey cattle
| Functions | Description | GO term | No. of proteins | Protein IDs | Gene IDs | P-value | FDR |
|---|---|---|---|---|---|---|---|
| Kashmiri Cattle | |||||||
| Molecular | Catalytic activity | GO:0003824 | 18 | P19120, P12763, P30122, P11017, Q8MK44, P80209, P62871, Q8HXQ5, Q4GZT4, Q0IIG8, A5PK46, P00794, P80929, Q0VCZ8, Q2UVX4, P80025 | HSPA8, AHSG, CEL, GNB2, DGAT1, CTSD, GNB1, ABCC1, ABCG2, RAB18, PNLIPRP2, CYM, ANG2, ACSL1, C3, LPO | 0.0002 | 0.44 |
| Antioxidant activity | GO:0016209 | 1 | P80025 | LPO | 0.0843 | 0.79 | |
| Cellular | Membrane | GO:0016020 | 4 | P19120, P30122, Q8MK44, P80209, Q8HXQ5, Q4GZT4, Q0IIG8, P00794, P02702, P30932, P18892 | HSPA8, CEL, DGAT1, CTSD, ABCC1, ABCG2, RAB18,CYM,FOLR1,CD9,BTN1A1 | 0.0198 | 0.181 |
| Extracellular region | GO:0005576 | 10 | P46201, P02666, P30122, C4PU73, Q9TTE1, P21214, P80025 | Uterine milk protein, CSN2, CEL, Serin peptidase inhibitor,SERPINA3–1,TGFB2,LPO | 0.00111 | 0.0354 | |
| Jersey Cattle | |||||||
| Molecular | Reproduction | GO:0000003 | 2 | A0A140T8A9, P11151, P02668, A5PK46 | CSN3, LPL, CSN3, | 0.005 | 0.422 |
| Catalytic activity | GO:0003824 | 22 |
Q8HYJ9, Q5E9R3, P11151, Q5E9B1, Q8MK44, P62998, Q2TBH2, Q8HXQ5, Q148J4, F1MN60, |
FMO3, EHD1, LPL, LDHB, DGAT1, RAC1, RRAS, ABCC1, RAB2A, ATP2B2, ANG1, ABCG2, PNLIPRP2, CYM, XDH, LGB, LPO | 0.066 | 0.83 | |
| Antioxidant activity | GO:0016209 | 1 | P80025 | LPO | 0.087 | 0.52 | |
| Cellular | Extracellular region | GO:0005576 | 11 |
P17697, A0A140T8A9, P34955, P46201, P02663, |
CLU, CSN3, SERPINA1, Uterine milk protein, CSN1S2, AZGP1, SERPINC1, SERPINF2, Serin peptidase inhibitor, CSN1S1, CSN3, TGFB2, LPO | 0 | 0 |
Protein categories identified through GO annotation
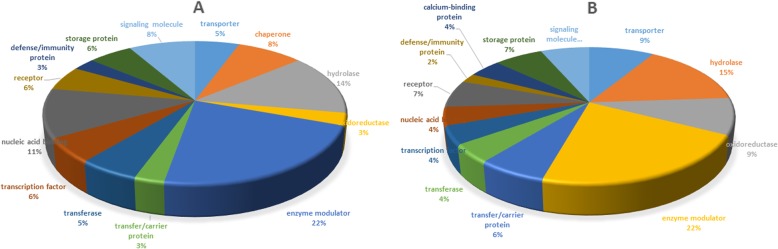
The identified differentially upregulated proteins in Kashmiri and Jersey cattle were categorized according to their GO annotation (Additional file 2: Table S103). Most of the significantly upregulated proteins in both cattle breeds were enzyme modulators (SERPINA3, BTN1A1, SERPINC1, SERPINF2, Serin peptidase inhibitor, RAC1, RRAS, BTN1A1 and uterine milk protein) and hydrolases (GNB2, CTSD, GNB1, PNLIPRP2, CYM) (Fig. 1 a and b). However, proteins belonging to the chaperone classes (HSP90AA1, YWHAB, YWHAZ) were significantly upregulated in Kashmiri cattle only (Fig. 2a and b).
Fig. 2.
Classification of differentially expressed proteins in Kashmiri and Jersey cattle by gene ontology annotation (a) Protein classes (upregulated proteins only) in Kashmiri cattle and (b) Jersey cattle
Enriched pathways by significantly upregulated proteins in Kashmiri and Jersey cattle
Significantly upregulated proteins in Kashmiri and Jersey cattle were enriched to 12 and 4 pathways at uncorrected P < 0.05, respectively (Table 3). When FDR correction was applied, 10 and one proteins remained significant (FDR < 0.1) in Kashmiri and Jersey cattle, respectively (Table 3). Of all the pathways, only EGF receptor signalling pathway was enriched at uncorrected P < 0.05 by significantly upregulated proteins in both breeds.
Table 3.
Enriched pathways by upregulated proteins in Kashmiri and Jersey cattle
| Pathway | Proteins | p-value | FDR | Proteins | Genes |
|---|---|---|---|---|---|
| Kashmiri cattle | |||||
| Beta3 adrenergic receptor signaling pathway (P04379) | 2 | 0.07265 | 0.1719 | P11017, P62871 | GNB2, GNB1 |
| Beta2 adrenergic receptor signaling pathway (P04378) | 2 | 0.09576 | 0.8552 | P11017, P62871 | GNB2, GNB1 |
| Metabotropic glutamate receptor group III pathway (P00039) | 2 | 0.0124 | 0.0723 | P11017, P62871 | GNB2, GNB1 |
| Beta1 adrenergic receptor signaling pathway (P04377) | 2 | 0.00599 | 0.0543 | P11017, P62871 | GNB2, GNB1 |
| 5HT4 type receptor mediated signaling pathway (P04376) | 2 | 0.07387 | 0.1987 | P11017, P62871 | GNB2, GNB1 |
| 5HT2 type receptor mediated signaling pathway (P04374) | 2 | 0.0141 | 0.0792 | P11017, P62871 | GNB2, GNB1 |
| 5HT1 type receptor mediated signaling pathway (P04373) | 2 | 0.09671 |
0.1897 0.049 |
P11017, P62871 | GNB2, GNB1 |
| Integrin signalling pathway (P00034) | 2 | 0.0823 | 0.353 | P63258, E1BBG2 | ACTG1, MICALL1 |
| Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway (P00026) | 2 | 0.0658 | 0.298 | P11017, P62871 | GNB2, GNB1 |
| Wnt signaling pathway (P00057) | 3 | 0.0388 | 0.197 | P63258, P11017, P62871 | GNB2, GNB1, ACTG1 |
| Thyrotropin-releasing hormone receptor signaling pathway (P04394) | 2 | 0.082 | 0.759 | P11017, P62871 | GNB2, GNB1 |
| FGF signaling pathway (P00021) | 2 | 0.0417 | 0.2 | P63103, P68250 | YWHAZ,YWHAB |
| EGF receptor signaling pathway (P00018) | 4 | 0 | 0.041 | P63103, Q95115, P68250, Q9TTC5 | YWHAZ, STAT5A, YWHAB, BTC |
| PI3 kinase pathway (P00048) | 3 | 0.002 | 0.0685 | P63103, P11017, P62871 | GNB2, GNB1, YWHAZ |
| Opioid prodynorphin pathway (P05916) | 2 | 0.00406 | 0.0602 | P11017, P62871 | GNB2, GNB1 |
| Histamine H1 receptor mediated signaling pathway (P04385) | 2 | 0.00647 | 0.0502 | P11017, P62871 | GNB2, GNB1 |
| Enkephalin release (P05913) | 2 | 0.00387 | 0.0701 | P11017, P62871 | GNB2, GNB1 |
| Angiotensin II-stimulated signaling through G proteins and beta-arrestin (P05911) | 2 | 0.00488 | 0.0497 | P11017, P62871 | GNB2, GNB1 |
| CCKR signaling map (P06959) | 2 | 0.0816 | 0.36 | P62871, P68250 | GNB1,YWHAB |
| Metabotropic glutamate receptor group II pathway (P00040) | 2 | 0.00647 | 0.0527 | P11017, P62871 | GNB2, GNB1 |
| Jersey Cattle | |||||
| Integrin signalling pathway (P00034) | 2 | 0.0879 | 0.796 | P62998, Q2TBH2 | RAC1, RRAS |
| EGF receptor signaling pathway (P00018) | 3 | 0.00663 | 0.36 | P62998, Q2TBH2, Q95115 | RAC1, RRAS, STAT5A |
| PDGF signaling pathway (P00047) | 2 | 0.0567 | 0.66 | Q148J4, Q95115 | RAB2A, STAT5A |
| Blood coagulation (P00011) | 3 | 0.000428 | 0.0698 | P34955, P41361, P28800 | SERPINA1, SERPINC1, SERPINF2 |
| CCKR signaling map (P06959) | 2 | 0.0872 | 0.836 | P17697, P62998 | CLU, RAC1 |
| T cell activation (P00053) | 2 | 0.0318 | 0.648 | P01888, P62998 | B2M, RAC1 |
| TGF-beta signaling pathway (P00052) | 2 | 0.0298 | 0.694 | Q2TBH2, P21214 | RRAS, TGFB2 |
Discussion
The present study was designed to characterize and compare the milk proteome of Kashmiri and Jersey cattle. Over the past few decades, interest to reveal the dynamics of milk proteome has grown and there have been remarkable developments in the techniques used for fractionation and identification of proteins [25–27]. In the present study, a combination of fractionation and mass spectrometry techniques were used to comprehensively characterize the milk proteome profiles of Kashmiri and Jersey cattle breeds.
A total of 180 proteins were found to be differentially expressed between Kashmiri and Jersey cattle. Interestingly, 90 and 89 of the differentially expressed proteins were significantly upregulated in Kashmiri and Jersey cattle, respectively. Enzyme modulators were the major class of up-regulated proteins in both Kashmiri (20.51%) and Jersey cattle (14.28%). Hydrolases represented 12.82 and 14.28% of upregulated proteins in Kashmiri and Jersey cattle, respectively. Interestingly, chaperone class of proteins was only observed in milk of Kashmiri cattle. Chaperones help in the folding of newly synthesized proteins and prevent their premature (mis) folding at least until a domain capable of forming a stable structure is synthesized. As expected and in agreement with earlier studies ([26, 27]), the casein and whey fraction proteins were highly expressed in both breeds. However, a different set of high abundant milk proteins were significantly upregulated in each of the breeds. For example, the abundantly expressed proteins beta-casein, lactoferrin and CD4 were significantly upregulated in Kashmiri while beta-lacto globulin, kappa-casein and alpha-lactalbumin were significantly upregulated in Jersey (Table 1). Interestingly, the low abundant proteins FMO3, GLYCAM1, CD9, APLN, BTC, enterotoxin-binding glycoprotein PP16K, ORM1, serin peptidase inhibitor clade A, adipocyte differentiation-related protein and uterine milk protein were significantly upregulated in Kashmiri while ATP synthase subunit A, RAC1, B2M, SAR1B, TCN2 and MFGE8 were upregulated in Jersey. These results indicate a clear distinction as well as wide differences in the proteome profiles between the breeds which could be explained by high selection pressure for milk production traits in Jersey.
The differences in the expression of high abundant proteins between the breeds might confer differential benefits to their milks. For example, different levels of phosphorylation of beta-casein has been reported to affect the availability of calcium and protein micelle stability of milk [28], which might have important consequences on the nutrition and technological properties of milk and milk products. Additionally, other key bioactive proteins identified in this study that are well known to exert beneficial effects on human nutrition and health include lactoferrin, GLYCAM1, betacellulin, apelin, LALBA and serine peptidase inhibitor, etc. Iron sequestering properties of lactoferrin (LF), along with blockade of microbial carbohydrate metabolism and destabilisation of the bacterial cell wall [29, 30], has been shown to produce bactericidal and bacteriostatic effects in a wide range of microorganisms, including gram positive and gram negative bacteria, aerobes, anaerobes, yeasts and parasites [31–33]. Similarly, GLYCAM1 with a 7.93-fold expression in Kashmiri cattle is known to act as an antimicrobial peptide with ability to protect the intestinal mucosal tract of neonates largely due to its lubricating properties [34, 35]. In addition to these, apelin peptides might be involved in maturation of the gastrointestinal tract [36, 37]. Betacellulin (BTC), a key epidermal growth factor (EGF) [38] might regulate the development and maturation of the neonatal gut and immune system [39]. EGFs are major growth promoting factors in human milk [40] but the biological significance of BTC in bovine milk is currently unclear and needs further investigation. However, one plausible explanation for the presence of BTC in bovine milk might be to stimulate the proliferation of the gastrointestinal epithelia in new-borns, as has been proposed for milk-borne EGF and TGF-α (Transforming growth factor alpha) in other species [41]. With respect to Jersey breed, peptides resulting from partial digestion of high abundant proteins such as LALBA, CSN2 and CSN3 in the small intestine may influence gut functions including immune stimulation, mineral and trace element absorption and host defence against infection [42]. Alpha-lactalbumin enhances infant gastrointestinal function [43], motility and antimicrobial activity [44]. CSN3 is readily hydrolysed in calf’s stomach, allowing the formation of a coagulum that can be readily digested [45] and also provides heat stability to milk by stabilising the casein micelle [45]. Moreover, CSN3 prevents infection by disrupting the attachment of pathogens to mucosal cells [46]. CSN3 digestion results in the formation of a glycomacropeptide which in turn enhances mineral absorption [47]. Bovine beta 2-microglobulin (B2M) is an antibacterial protein present in milk fat globules. B2M possesses potent antibacterial activities against Gram positive pathogenic bacteria [48]. Bovine milk is an abundant source of bioavailable B12 vitamin wherein when complexed with transcobalamin, a major vitamin B12 binding protein in cows’ milk [49], stimulates vitamin B12 absorption through intestinal epithelial cells [50]. Lactadherin is secreted by mammary epithelial cells and stored in milk fat globules [51]. Lactadherin, as one of the immune components in bovine milk has been found to prevent rota viral infection in infants by removing the sialic acid from the viral coat [52, 53].
It is worthwhile to note that the low abundant protein, flavin-containing monooxygenase 3 (FMO3) had 16.6 fold expression rate in Kashmiri as compared to Jersey. This is the first report wherein FMO3 has been found to be highly expressed in Kashmiri cattle. Increased presence of FMO3 might be important due to its ability to oxidise trimethylamine (TMA), a compound with fishy odour, to TMAO (Trimethylamine N-oxide), an odourless oxide. Absence of FMO3 leads to fishy flavour in milk due to increased build-up of TMA, and thus might play an important role in maintaining the quality of milk [54–56]. Moreover, FMO3 belongs to a drug metabolising enzyme class with ability to oxidize xenobiotics, pesticides and other foreign inhabitants in body fluids including milk and serum [57–60] and hence presents an efficient defence mechanism in new-borns. The presence of FMO3 at high concentrations in Kashmiri cattle milk can favour utilization of Kashmiri cattle milk in commercial preparations for the promotion of human health and nutritional status. In fact, bio-mining of such bioactive milk protein constituent and marketing it as ingredients may not only serve as a lucrative business for the Indian dairy industry but also in the development of products for consumers with special needs like allergy and milk tolerance.
The GO analysis of significantly up-regulated proteins revealed only one significantly enriched GO term (extracellular region) after FDR correction in both breeds and limited functional overlap was found between the present proteomic data and our earlier transcriptome data [61] indicating the failure of RNA-based analyses to represent completely protein dynamics [62].
Pathway analysis helps in biological interpretation of proteomic and other high-throughput data in cells or organisms [63]. Most of the pathways (Wnt signaling pathway, EGF receptor signaling pathway, FGF signaling pathway, PI3 kinase pathway) significantly enriched by the significantly upregulated proteins in Kashmiri cattle are involved in mammary gland development. Wnt signaling pathway regulates mammary development [64] during various stages of mammary morphogenesis [65]. The proteins enriched in the Wnt signalling pathway were GNB1(G protein subunit beta 1), GNB2 (G protein subunit bBeta 2) and ACTG1(actin gamma 1). ACTG1 plays a critical role in branching and alveolar development of the mammary gland through cytoskeletal remodelling [66]. FGF signalling pathway controls mammary epithelial cell branching and morphogenesis [67] and activates PI3 kinase pathway through phosphorylation [68]. Epidermal growth factor family plays essential roles in regulating cell proliferation, survival and differentiation of mammary epithelial cells through STAT5A, a key non-tyrosine kinase protein indirectly regulated by JAK2/ELF5, insulin growth factor, estrogen, and progesterone signalling pathways [69]. In Jersey cattle, two significantly (p < 0.05) enriched pathways, blood coagulation/coagulation cascades and T cell activation pathways are associated with immune system regulation [70]. SERPINA1, SERPINC1, SERPINF2 are important proteins in blood coagulation pathway whereas, B2M and RAC1 play critical roles in T cell activation pathway. These proteins play fundamental roles in innate immunity in addition to enhancing adaptive immune responses [71]. Altogether, a wide range of proteins were detected in this study including proteins involved in immune response, host defense and milk quality as well as qualitative and quantitative differences in their milk proteome.
Conclusion
A total of 91 and 89 proteins were significantly upregulated in Kashmiri and Jersey cattle, respectively. A different set of high- abundant and low-abundant proteins were significantly upregulated in Kashmiri and Jersey cattle, clearly differentiating the two breeds at the proteome level. Immune-related proteins (CD4, LF and GLYCAM 1) and drug metabolising enzyme (FMO3) were abundantly expressed in Kashmiri cattle milk. The presence of FMO3 at high concentrations in Kashmiri cattle milk could favour its utilization in commercial preparations for human health promotion and consequently serve as a boost for increased business opportunities for the Indian dairy industry.
Methods
Experimental animals and sampling
The ethical clearance was approved by the Institutional Animal Ethics Committee (IAEC) of Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir. A total of three healthy Kashmiri and three Jersey cows in their 3rd lactation from the university dairy farm (Mountain Livestock Research Institute, Share-Kashmir University of Agricultural Sciences and Technology of Kashmir, India) were selected for the study. The animals were kept under similar feeding and management conditions to minimise environmental variation. Fresh milk samples (200 mL) were aseptically collected from all the four quarters (50 mL per quarter) at day 90 in milk (D90), mixed thoroughly, placed on ice and immediately transported to the laboratory for further analysis.
Protein preparation
Milk samples were processed differently for high and low abundance protein analysis. For high-abundance protein analysis, 50 mL of milk was immediately placed on ice after collection followed by centrifugation at 4000×g for 10 min at 4 °C within 2 h of collection. The fat layer was removed and skimmed fraction was stored at − 20 °C. Whereas, for low abundance protein analysis, 0.24 mL (100X) mammalian protease inhibitor cocktail (Sigma, Milwaukee, WI, USA) was added to 50 mL of milk followed by centrifugation at 4000×g for 15 min at 4 °C. The cream layer was removed and the skimmed or whey portion was depleted of casein using a previously described method [72]. Briefly, 60 mM CaCl2 was added to skimmed sample and the pH was adjusted to 4.3 using 30% acetic acid (Fisher Scientific, Fair Lawn, NJ, USA). Samples were then centrifuged at 189,000×g at 4 °C for 70 min and the supernatant was collected and stored at − 80 °C.
Enrichment of low abundance proteins
Low abundance minor proteins were enriched using the ProteoMiner Kit (BioRad Laboratories, Hercules, CA, USA) as per manufacturer’s protocol. Whey samples were placed in individual ProteoMiner columns, mixed thoroughly by shaking (gently) followed by incubation at room temperature for 2 h. Subsequently, samples were washed thoroughly using HPLC grade water to remove excess proteins by centrifugation at 7000 g for 5 min. Low abundance proteins were eluted off the beads by addition of 20 μl 4 x Laemmli sample buffer (8% SDS, 40% glycerol, 250 mM Tris, pH 6.8, 400 mM DTT with trace amount of bromophenol blue).
In-solution digestion of proteins and nano-scale LC/MS analysis on QTOF
The pellets after acetone precipitation (high abundant proteins) or TCA (Trichloroacetic acid)-acetone precipitation (low abundant proteins) were dissolved in 50 mM ammonium bicarbonate (dilution 1:3) and 0.1% SDS. 100 μg of the extracted protein was subjected to in solution trypsin digestion with carbamidomethylation at cysteine (fixed) and oxidation at methionine (variable). The dissolved pellet was treated with 10 μl of 100 mM DTT (Dithiothreitol) followed by incubation on a thermo mixer (Eppendorf ThermoMixer® C,) at 95 °C for 1 h. The sample was treated with 18 μl of 250 mM IDA (Iodoacetamide) and then incubated in the dark for 45 min at room temperature. To stop the IDA reaction, 40 μl DTT was added at room temperature and incubated for 10 min. To this solution, 50 mM ammonium bicarbonate and 0.1% SDS was added to make up the volume to 300 μl. For Enzymatic cleavage of the protein, trypsin in the ratio 50:1 (w/v) was added to sample and incubated on the thermo mixer at 37 °C overnight. To stop the trypsin activity, the peptides were then extracted in 0.1% formic acid followed by incubation at 37 °C for 45 min. The extracted mixture was then centrifuged at 13000 g for 10 min and the supernatant was placed in a separate Eppendorf tube. This supernatant was subjected to speed vac at 45 °C. The resulting peptides were then dissolved in 20 μl of 0.1% formic acid and 10 μL of this solution was used on C18 UPLC column for separation of peptides. The mass spectrometer was operated in positive ion mode, and MS spectra were acquired over a range of 375–1500 m/z. For MS and MS/MS scans, the resolution of the orbitrap fusion was set at 120,000 and 50,000 at 200 m/z, respectively. Data-dependent acquisition mode was set as top speed, and ions were fragmented (10 fragment files collected after every full scan) through higher energy collisional dissociation, and cycle time was 3 s with peptide mass tolerance and fragment mass tolerance of 50 ppm and 100 ppm, respectively. The automatic gain control target values for master scan modes and MS/MS were set to 4e5 and 1e5, respectively. Dynamic exclusion duration was 40 s.
Protein identification and differential expression analysis
The individual peptides MSMS spectra were searched against the Swiss-Prot databases using the Mascot Distiller Search engine (v. 2.6.0) for protein identification and expression analysis was performed with PLGS software (Protein Lynx Global Server, Waters, India) by Sandor’s Lifesciences, Hyderabad, India. The results were filtered based on the peptide Benjaminin and Hochberg corrected p-value < 0.1 (FDR < 0.1) or uncorrected p-value < 0.05. Both unique and razor peptides were selected for protein quantification, protein ratios were calculated as the median of only unique or razor peptides of the protein. All peptide ratios were normalized based on the median ratio. The protein species quantification results were statistically analysed by student’s t-test, and the p-value was corrected by the method of Benjamin and Hochberg FDR analysis. An FDR < 0.1 was considered significant due to the low number of samples analysed.
Gene ontology and pathway analysis
Gene ontology (GO) and pathway enrichment analysis of differentially expressed proteins was accomplished with Gene Ontology Consortium data base (http://www.geneontology.org) (Falcon and Gentleman, 2007). GO terms and KEGG pathways (http://www.genome.jp/kegg/) with FDR < 0.1were considered significantly enriched.
Supplementary information
Additional file 1 : Table S1. Significantly upregulated proteins in Kashmiri cattle (high and low abundant proteins) Table S2. Significantly upregulated proteins in Jersey cattle (high and low abundant proteins). Table S3. Sample chromatograms of individual samples. Table S4-S6. Peptide information of significantly upregulated proteins in Jersey cattle. Table S7-S9. Peptide information of significantly upregulated proteins in Kashmiri cattle.
Additional file 2 : Table S10. Classification of significantly upregulated proteins in Kashmiri and Jersey cattle.
Additional file 3. Total proteome obtained from different samples.
Acknowledgements
The authors would like to thank the staff of the University Dairy Farm of Mountain Livestock Research Institute-Manasbal, Srinagar-Kashmir for care and management of the experimental animals. We also acknowledge the DBT-supported bioinformatics Infrastructuralfacility for data analysis.
Data submission
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [73] partner repository with the dataset identifier PXD017412
Abbreviations
- AGP
α-1-acid glycoprotein
- APLN
Apelin
- B2M
Beta 2-microglobulin
- BTC
Betacellulin
- CSN2
Beta-casein
- CSN3
Kappa-casein
- CYM
Chymosin
- EGF
Epidermal growth factor
- EGR1
Early growth response protein 1
- EHD
EH domain-containing protein 1
- FDR
False discovery rate
- FGF
Fibroblast growth factor
- FMO3
Flavin mono-oxygenase3
- GALNT1
Polypeptide N-Acetylgalactosaminyltransferase
- GLYCAM1
Glycosylation-dependent cell adhesion molecule 1
- GO
Gene ontology
- HSP90AA1
Heat shock protein90AA1
- LALBA
Alpha-lactalbumin
- LC-MS/Q-TOF
Liquid chromatography-mass spectrometry/quantitative time of flight
- LF
Lactoferrin
- LGB
Beta-lactoglobulin
- LPL
Lipoprotein lipase
- LTF
Lactotransferrin
- MEC
Mammary epithelial cell
- PNLIPRP2
Pancreatic lipase related protein 2
- RAC1
Ras-related C3 botulinum toxin substrate 1
- SERPINA1
Serine protease inhibitor1
- TGF-α
Transforming growth factor
- TLR2
Toll like receptor 2
- TMAO
Trimethylamine N-oxide
- ZNF496
Zinc finger protein 496
Authors’ contributions
SMA: Design the experiment and managed the project; SAB: carried out the experiment, analysed the data and drafted the manuscript; MAD and PTM: Sample collection and lab work; EMI-A: interpreted data and reviewed the manuscript, RAS, MM, NAG, TA, NS, HFB and NAG: reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Department of Biotechnology, Ministry of Science and Technology, Government of India under Biotechnology Research and Development scheme. The funding body has no role in the study design and data collection, analysis, interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets generated and analysed during the current study are available as Additional files.
Ethics approval
The ethical clearance was approved by the Institutional Animal Ethics Committee (IAEC) of Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shakil A. Bhat, Email: shakilvet@gmail.com
Syed M. Ahmad, Email: mudasirbio@gmail.com
Eveline M. Ibeagha-Awemu, Email: Eveline.Ibeagha-Awemu@canada.ca
Mohammad Mobashir, Email: m.mobashir@cdslifesciences.com.
Mashooq A. Dar, Email: darmashooq1@gmail.com
Peerzada T. Mumtaz, Email: drtajamulmumtaz@gmail.com
Riaz A. Shah, Email: drriazshah@gmail.com
Tanveer A. Dar, Email: tanveerali@kashmiruniversity.ac.in
Nadeem Shabir, Email: drnadurose@gmail.com.
Hina F. Bhat, Email: bhat.hina@rediffmail.com
Nazir A. Ganai, Email: drnazirahmad@gmail.com
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-6574-4.
References
- 1.Reinhardt TA, Lippolis JD. Bovine milk fat globule membrane proteome. J Dairy Res. 2006;73(4):406–416. doi: 10.1017/S0022029906001889. [DOI] [PubMed] [Google Scholar]
- 2.Séverin S, Wenshui X. Milk biologically active components as nutraceuticals. Crit Rev Food Sci Nutr. 2005;45(7–8):645–656. doi: 10.1080/10408690490911756. [DOI] [PubMed] [Google Scholar]
- 3.Dai W, Chen Q, Wang Q, White RR, Liu J, Liu H. Complementary transcriptomic and proteomic analyses reveal regulatory mechanisms of milk protein production in dairy cows consuming different forages. Sci Rep. 2017;7:44234. doi: 10.1038/srep44234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lönnerdal B. Infant formula and infant nutrition: bioactive proteins of human milk and implications for composition of infant formulas. Am J Clin Nutr. 2014;99(3):712S–717S. doi: 10.3945/ajcn.113.071993. [DOI] [PubMed] [Google Scholar]
- 5.Liao Y, Jiang R, Lönnerdal B. Biochemical and molecular impacts of lactoferrin on small intestinal growth and development during early life. Biochem Cell Biol. 2012;90(3):476–484. doi: 10.1139/o11-075. [DOI] [PubMed] [Google Scholar]
- 6.Reznikov EA, Comstock SS, Yi C, Contractor N, Donovan SM. Dietary Bovine Lactoferrin Increases Intestinal Cell Proliferation in Neonatal Piglets, 2. J Nutr. 2014;144(9):1401–1408. doi: 10.3945/jn.114.196568. [DOI] [PubMed] [Google Scholar]
- 7.Ibeagha-Awemu EM, Ibeagha AE, Messier S, Zhao X. Proteomics, genomics, and pathway analyses of Escherichia coli and Staphylococcus aureus infected milk whey reveal molecular pathways and networks involved in mastitis. J Proteome Res. 2010;9(9):4604–4619. doi: 10.1021/pr100336e. [DOI] [PubMed] [Google Scholar]
- 8.Wada Y, Lönnerdal B. Bioactive peptides derived from human milk proteins—mechanisms of action. J Nutr Biochem. 2014;25(5):503–514. doi: 10.1016/j.jnutbio.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Casado B, Affolter M, Kussmann M. OMICS-rooted studies of milk proteins, oligosaccharides and lipids. J Proteomics. 2009;73(2):196–208. doi: 10.1016/j.jprot.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 10.German JB, Dillard CJ, Ward RE. Bioactive components in milk. Curr Opin Clin Nutr Metab Care. 2002;5(6):653–658. doi: 10.1097/00075197-200211000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Lönnerdal B. Nutritional and physiologic significance of human milk proteins. Am J Clin Nutr. 2003;77(6):1537S–1543S. doi: 10.1093/ajcn/77.6.1537S. [DOI] [PubMed] [Google Scholar]
- 12.Mohanty DP, Mohapatra S, Misra S, Sahu PS. Milk derived bioactive peptides and their impact on human health–a review. Saudi J Biol Sci. 2016;23(5):577–583. doi: 10.1016/j.sjbs.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meisel H. Biochemical properties of peptides encrypted in bovine milk proteins. Curr Med Chem. 2005;12(16):1905–1919. doi: 10.2174/0929867054546618. [DOI] [PubMed] [Google Scholar]
- 14.Ibeagha-Awemu EM, Liu J, Zhao X. Bioactive components in yogurt products. In: Park YW, editor. Bioactive components in Milk and dairy products, 2009. 2009. pp. 235–250. [Google Scholar]
- 15.Ward RE, German JB. Understanding milk’s bioactive components: a goal for the genomics toolbox. J Nutr. 2004;134(4):962S–967S. doi: 10.1093/jn/134.4.962S. [DOI] [PubMed] [Google Scholar]
- 16.Tacoma R, Fields J, Ebenstein DB, Lam YW, Greenwood SL. Characterization of the bovine milk proteome in early-lactation Holstein and Jersey breeds of dairy cows. J Proteomics. 2016;130:200–210. doi: 10.1016/j.jprot.2015.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litwińczuk Z, Król J, Brodziak A, Barłowska J. Changes of protein content and its fractions in bovine milk from different breeds subject to somatic cell count. J Dairy Sci. 2011;94(2):684–691. doi: 10.3168/jds.2010-3217. [DOI] [PubMed] [Google Scholar]
- 18.D'auria E, Agostoni C, Giovannini M, Riva E, Zetterström R, Fortin R, Roncada P. Proteomic evaluation of milk from different mammalian species as a substitute for breast milk. Acta Paediatr. 2005;94(12):1708–1713. doi: 10.1111/j.1651-2227.2005.tb01842.x. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, Bu D, Zhao X, Sun P, Wang J, Zhou L. Proteomic analysis of cow, yak, buffalo, goat and camel milk whey proteins: quantitative differential expression patterns. J Proteome Res. 2013;12(4):1660–1667. doi: 10.1021/pr301001m. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Zhao X, Huang D, Pan X, Qi Y, Yang Y, Cheng G. Proteomic analysis and cross species comparison of casein fractions from the milk of dairy animals. Sci Rep. 2017;7:43020. doi: 10.1038/srep43020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oltenacu PA, Algers B. Selection for increased production and the welfare of dairy cows: are new breeding goals needed? Ambio. 2005;34(4):311–315. doi: 10.1579/0044-7447-34.4.311. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, van Dijk AD, Hettinga K. An interactomics overview of the human and bovine milk proteome over lactation. Proteome Sci. 2016;15(1):1. doi: 10.1186/s12953-016-0110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mol P, Kannegundla U, Dey G, Gopalakrishnan L, Dammalli M, Kumar M, TSK P. Bovine Milk Comparative Proteome Analysis from Early, Mid, and Late Lactation in the Cattle Breed, Malnad Gidda (Bos indicus) Omics. 2018;22(3):223–235. doi: 10.1089/omi.2017.0162. [DOI] [PubMed] [Google Scholar]
- 24.Affolter M, Grass L, Vanrobaeys F, Casado B, Kussmann M. Qualitative and quantitative profiling of the bovine milk fat globule membrane proteome. J Proteome. 2010;73(6):1079–1088. doi: 10.1016/j.jprot.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 25.Murakami K, Lagarde M, Yuki Y. Identification of minor proteins of human colostrum and mature milk by two-dimensional electrophoresis. Electrophoresis. 1998;19(14):2521–2527. doi: 10.1002/elps.1150191427. [DOI] [PubMed] [Google Scholar]
- 26.Yamada M, Murakami K, Wallingford JC, Yuki Y. Identification of low-abundance proteins of bovine colostral and mature milk using two-dimensional electrophoresis followed by microsequencing and mass spectrometry. Electrophoresis. 2002;23(7–8):1153–1160. doi: 10.1002/1522-2683(200204)23:7/8<1153::AID-ELPS1153>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 27.Palmer DJ, Kelly VC, Smit AM, Kuy S, Knight CG, Cooper GJ. Human colostrum: identification of minor proteins in the aqueous phase by proteomics. Proteomics. 2006;6(7):2208–2216. doi: 10.1002/pmic.200500558. [DOI] [PubMed] [Google Scholar]
- 28.Amigo L, Recio I, Ramos M. Genetic polymorphism of ovine milk proteins: its influence on technological properties of milk—a review. Int Dairy J. 2000;10(3):135–149. doi: 10.1016/S0958-6946(00)00034-0. [DOI] [Google Scholar]
- 29.Masson PL, Heremans JF. Lactoferrin in milk from different species. Comp Biochem Physiol. 1971;1:119–129. doi: 10.1016/0305-0491(71)90258-6. [DOI] [PubMed] [Google Scholar]
- 30.Hennart PF, Brasseur DJ, Delogne-Desnoeck JB, Dramaix MM, Robyn CE. Lysozyme, lactoferrin, and secretory immunoglobulin A content in breast milk: influence of duration of lactation, nutrition status, prolactin status, and parity of mother. Am J Clin Nutr. 1991;53(1):32–39. doi: 10.1093/ajcn/53.1.32. [DOI] [PubMed] [Google Scholar]
- 31.Weinberg ED. Iron withholding: a defense against infection and neoplasia. Physiol Rev. 1984;64(1):65–102. doi: 10.1152/physrev.1984.64.1.65. [DOI] [PubMed] [Google Scholar]
- 32.Arnold RR, Russell JE, Champion WJ, Brewer M, Gauthier JJ. Bactericidal activity of human lactoferrin: differentiation from the stasis of iron deprivation. Infect Immun. 1982;35(3):792–799. doi: 10.1128/IAI.35.3.792-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellison RT, III, LaForce FM, Giehl TJ, Boose DS, Dunn BE. Lactoferrin and transferrin damage of the gram-negative outer membrane is modulated by Ca2+ and Mg2+ Microbiology. 1990;136(7):1437–1446. doi: 10.1099/00221287-136-7-1437. [DOI] [PubMed] [Google Scholar]
- 34.Carraway KL, Hull SR. Cell surface mucin-type glycoproteins and mucin-like domains. Glycobiology. 1991;1(2):131–138. doi: 10.1093/glycob/1.2.131. [DOI] [PubMed] [Google Scholar]
- 35.Larocca D, Peterson JA, Walkup G, Urrea R, Ceriani RL. Cloning and sequencing of a complementary DNA encoding a Mr 70,000 human breast epithelial mucin-associated antigen. Cancer Res. 1990;50(18):5925–5930. [PubMed] [Google Scholar]
- 36.Wang G, Kundu R, Han S, Qi X, Englander EW, Quertermous T, Greeley GH., Jr Ontogeny of apelin and its receptor in the rodent gastrointestinal tract. Regul Pept. 2009;158(1–3):32–39. doi: 10.1016/j.regpep.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aydin S. The presence of the peptides apelin, ghrelin and nesfatin-1 in the human breast milk, and the lowering of their levels in patients with gestational diabetes mellitus. Peptides. 2010;31(12):2236–2240. doi: 10.1016/j.peptides.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Dunbar AJ, Priebe IK, Belford DA, Goddard C. Identification of betacellulin as a major peptide growth factor in milk: purification, characterization and molecular cloning of bovine betacellulin. Biochem J. 1999;344(Pt 3):713. doi: 10.1042/bj3440713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grosvenor CE, Picciano MF, Baumrucker CR. Hormones and growth factors in milk. Endocrine Rev. 1993;14(6):710–728. doi: 10.1210/edrv-14-6-710. [DOI] [PubMed] [Google Scholar]
- 40.Carpenter G. Epidermal growth factor is a major growth-promoting agent in human milk. Science. 1980;210(4466):198–199. doi: 10.1126/science.6968093. [DOI] [PubMed] [Google Scholar]
- 41.Ceciliani F, Pocacqua V, Provasi E, Comunian C, Bertolini A, Bronzo V, Sartorelli P. Identification of the bovine $\alpha $1-acid glycoprotein in colostrum and milk. Vet Res. 2005;36(5–6):735–746. doi: 10.1051/vetres:2005029. [DOI] [PubMed] [Google Scholar]
- 42.Lönnerdal B. Human milk: bioactive proteins/peptides and functional properties. In Protein in Neonatal and Infant Nutrition: Recent Updates,2016, (Vol. 86, pp. 97–107). Karger Publishers.
- 43.Berger M, Gray JA, Roth BL. The expanded biology of serotonin. Ann Rev Med. 2009;60:355–366. doi: 10.1146/annurev.med.60.042307.110802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bounous G, Kongshavn PA. Influence of dietary proteins on the immune system of mice. J Nutr. 1982;112(9):1747–1755. doi: 10.1093/jn/112.9.1747. [DOI] [PubMed] [Google Scholar]
- 45.Creamer LK, Plowman JE, Liddell MJ, Smith MH, Hill JP. Micelle stability: κ-casein structure and function. J Dairy Sci. 1998;81(11):3004–3012. doi: 10.3168/jds.S0022-0302(98)75864-3. [DOI] [PubMed] [Google Scholar]
- 46.Strömqvist M, Falk P, Bergström S, Hansson L, Lönnerdal B, Normark S, Hernell O. Human milk kappa-casein and inhibition of Helicobacter pylori adhesion to human gastric mucosa. J Pediatr Gastroenterol Nutr. 1995;21(3):288–296. doi: 10.1097/00005176-199510000-00006. [DOI] [PubMed] [Google Scholar]
- 47.Kelleher SL, Chatterton D, Nielsen K, Lönnerdal B. Glycomacropeptide and α-lactalbumin supplementation of infant formula affects growth and nutritional status in infant rhesus monkeys. Am J Clin Nutr. 2003;77(5):1261–1268. doi: 10.1093/ajcn/77.5.1261. [DOI] [PubMed] [Google Scholar]
- 48.Pringnitz DJ, Butler JE. Quantitation of bovine β2-microglobulin: occurrence in body fluids, on milk fat globules and origin in milk. Mol Immunol. 1985;22(7):779–786. doi: 10.1016/0161-5890(85)90143-9. [DOI] [PubMed] [Google Scholar]
- 49.Fedosov SN, Petersen TE, Nexø E. Transcobalamin from cow milk: isolation and physico-chemical properties. Biochim Biophys Acta (BBA)-Protein Struct Mol Enzymol. 1996;1292(1):113–119. doi: 10.1016/0167-4838(95)00173-5. [DOI] [PubMed] [Google Scholar]
- 50.Hine B, Boggs I, Green R, Miller JW, Hovey RC, Humphrey R, Wheeler TT. Transcobalamin derived from bovine milk stimulates apical uptake of vitamin B12 into human intestinal epithelial cells. J Cell Biochem. 2014;115(11):1948–1954. doi: 10.1002/jcb.24866. [DOI] [PubMed] [Google Scholar]
- 51.Hvarregaard J, Andersen MH, Berglund L, Rasmussen JT, Petersen TE. Characterization of glycoprotein PAS-6/7 from membranes of bovine milk fat globules. FEBS J. 1996;240(3):628–636. doi: 10.1111/j.1432-1033.1996.0628h.x. [DOI] [PubMed] [Google Scholar]
- 52.Yolken RH, Peterson JA, Vonderfecht SL, Fouts ET, Midthun K, Newburg DS. Human milk mucin inhibits rotavirus replication and prevents experimental gastroenteritis. J Clin Invest. 1992;90(5):1984–1991. doi: 10.1172/JCI116078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Becker-Dreps S, Choi WS, Stamper L, Vilchez S, Velasquez DE, Moon SS, Permar SR. Innate immune factors in Mothers’ breast Milk and their lack of association with rotavirus vaccine immunogenicity in Nicaraguan infants. J Pediatr Infect Dis Soc. 2017;6(1):87–90. doi: 10.1093/jpids/piv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lundén A, Gustafsson V, Imhof M, Gauch R, Bosset JOJ. Dairy Res. 2002;69(3):383–390. doi: 10.1017/S002202990200568X. [DOI] [PubMed] [Google Scholar]
- 55.Peter H, Maria K. Studies on the mechanism of hepatic microsomal N-oxide formation. The role of cytochrome P-450 and mixed-function amine oxidase in the N-oxidation of NN-dimethylaniline. Biochem J. 1977;164(3):487. doi: 10.1042/bj1640487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spellacy E, Watts RWE, Goolamali SK. Trimethylaminuria. J Inherit Metab Dis. 1979;2:85–88. doi: 10.1007/BF01805663. [DOI] [PubMed] [Google Scholar]
- 57.Eswaramoorthy S, Bonanno JB, Burley SK, Swaminathan S. Mechanism of action of a flavin-containing monooxygenase. Proc Natl Acad Sci. 2006;103(26):9832–9837. doi: 10.1073/pnas.0602398103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cashman JR, Zhang J. Human flavin-containing monooxygenases. Annu Rev Pharmacol Toxicol. 2006;46:65–100. doi: 10.1146/annurev.pharmtox.46.120604.141043. [DOI] [PubMed] [Google Scholar]
- 59.Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther. 2005;106(3):357–387. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen GP, Ziegler DM. Liver microsome and flavin-containing monooxygenase catalyzed oxidation of organic selenium compounds. Arch Biochem Biophys. 1994;312(2):566–572. doi: 10.1006/abbi.1994.1346. [DOI] [PubMed] [Google Scholar]
- 61.Bhat Shakil Ahmad, Ahmad Syed Mudasir, Ibeagha-Awemu Eveline M., Bhat Basharat A., Dar Mashooq Ahmad, Mumtaz Peerzada Tajamul, Shah Riaz A., Ganai Nazir A. Comparative transcriptome analysis of mammary epithelial cells at different stages of lactation reveals wide differences in gene expression and pathways regulating milk synthesis between Jersey and Kashmiri cattle. PLOS ONE. 2019;14(2):e0211773. doi: 10.1371/journal.pone.0211773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feder ME, Walser JC. The biological limitations of transcriptomics in elucidating stress and stress responses. J Evol Biol. 2005;18(4):901–910. doi: 10.1111/j.1420-9101.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- 63.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Incassati A, Chandramouli A, Eelkema R, Cowin P. Key signaling nodes in mammary gland development and cancer: β-catenin. Breast Cancer Res. 2010;12(6):213. doi: 10.1186/bcr2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu QC, Verheyen EM, Zeng YA. Mammary development and breast cancer: a Wnt perspective. Cancers. 2016;8(7):65. doi: 10.3390/cancers8070065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brisken C, Heineman A, Chavarria T, Elenbaas B, Tan J, Dey SK, et al. Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev. 2000;14(6):650–654. [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X, Martinez D, Koledova Z, Qiao G, Streuli CH, Lu P. FGF ligands of the postnatal mammary stroma regulate distinct aspects of epithelial morphogenesis. Development. 2014;141(17):3352–3362. doi: 10.1242/dev.106732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10(2):116. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 69.Furth PA, Nakles RE, Millman S, Diaz-Cruz ES, Cabrera MC. Signal transducer and activator of transcription 5 as a key signaling pathway in normal mammary gland developmental biology and breast cancer. Breast Cancer Res. 2011;13(5):220. doi: 10.1186/bcr2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oikonomopoulou K, Ricklin D, Ward PA, Lambris JD. Interactions between coagulation and complement—their role in inflammation. Semin Immunopathol. 2012;341:51–165. doi: 10.1007/s00281-011-0280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carroll MC. Complement and humoral immunity. Vaccine. 2008;26:I28–I33. doi: 10.1016/j.vaccine.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kunz C, Lönnerdal B. Human-milk proteins: analysis of casein and casein subunits by anion-exchange chromatography, gel electrophoresis, and specific staining methods. Am J Clin Nutr. 1990;51(1):37–46. doi: 10.1093/ajcn/51.1.37. [DOI] [PubMed] [Google Scholar]
- 73.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 : Table S1. Significantly upregulated proteins in Kashmiri cattle (high and low abundant proteins) Table S2. Significantly upregulated proteins in Jersey cattle (high and low abundant proteins). Table S3. Sample chromatograms of individual samples. Table S4-S6. Peptide information of significantly upregulated proteins in Jersey cattle. Table S7-S9. Peptide information of significantly upregulated proteins in Kashmiri cattle.
Additional file 2 : Table S10. Classification of significantly upregulated proteins in Kashmiri and Jersey cattle.
Additional file 3. Total proteome obtained from different samples.
Data Availability Statement
The datasets generated and analysed during the current study are available as Additional files.