Abstract
Introduction
Recently, the incidence of melanoma has been rising and there is a lack of effective targeted therapies. The regulatory mechanisms of microRNA-1246 (miR-1246) have been found in many cancers, except melanoma. This study focused on the regulatory mechanism of miR-1246 in melanoma development.
Methods
The expression of miR-1246 was assessed using quantitative real-time polymerase chain reaction (RT-qPCR). Cell viability and metastasis were detected by Transwell and MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assays. The protein expression of epithelial mesenchymal transition (EMT) makers was assessed by Western blot analysis. The target gene of miR-1246 was detected using luciferase reporter assay.
Results
MiR-1246 expression was increased in melanoma tissues and cells. In addition, upregulation of miR-1246 promoted cell viability and metastasis in melanoma. Forkhead box protein A2 (FOXA2) was confirmed to be a direct target of miR-1246. And FOXA2 expression was decreased in melanoma and was suppressed by miR-1246. Importantly, upregulation of FOXA2 restored the carcinogenesis of miR-1246 in melanoma.
Conclusion
MiR-1246 promoted cell viability and metastasis in melanoma by inhibiting FOXA2 expression.
Keywords: miR-1246, melanoma, cell viability, cell metastasis, FOXA2
Introduction
Malignant melanoma is a tumor of melanocytes derived from the skin and other organs. Malignant melanoma is more common in adults, and cases of congenital massive pigmented nevus are more common in children.1 Although the incidence of melanoma is low, its malignancy and mortality are high. In addition, melanoma is prone to distant metastases.2 Thus, surgical resection of melanoma that has not yet metastasized should be performed at the early stage. Patients with extensively metastatic melanoma often use chemotherapy and radiation therapy.3 The stage of melanoma is closely related to the prognosis. The 5-year survival rates for stage I, II, III and IV were 94%, 44%, 38%, and 4.6%, respectively. In addition, the thickness of the primary tumor is significantly correlated with the prognosis. The 5-year survival rates of ≤1 mm and >4 mm were 92% and 43%.4 Therefore, early diagnosis and treatment are very important. Biochemical therapy and molecular targeted therapy have broad prospects in the treatment of melanoma.
Recently, microRNA (miRNA) has become a major breakthrough in understanding the regulation of gene expression, and its role in malignant tumors has been extensively explored. MiRNAs mainly degrade or inhibit the expression of target genes by binding to their 3ʹ-untranslated region (3ʹ-UTR).5 In melanoma, miR-29a was found to inhibit the growth, migration and invasion of melanoma cells by directly targeting BMI1.6 It was7 found that miR-155 promoted the proliferation and invasion of uveal melanoma cells by regulating NDFIP1 expression. Especially, it has been reported that miR-1246 was upregulated in metastatic cutaneous melanoma.8 However, the function of miR-1246 has not been illuminated in melanoma. In addition, previous studies have shown that miR-1246 was upregulated in hepatocellular carcinoma, ovarian carcinoma and esophageal squamous cell carcinoma.9–11 However, downregulation of miR-1246 was identified in cervical cancer and prostate cancer.12,13 Functionally, miR-1246 promoted cell proliferation, invasion and drug resistance in breast cancer by targeting CCNG2.14 In contrast, miR-1246 inhibited cell invasion and epithelial mesenchymal transition (EMT) in lung cancer by targeting CXCR4.15 These findings indicate that the role of miR-1246 depends on the type of human cancers. In this study, we mainly investigated the role of miR-1246 in melanoma.
As a member of the forkhead box transcription factor A family, forkhead box protein A2 (FOXA2) has the ability to regulate metabolism and homeostasis.16 Currently, many studies have shown that FOXA2 is involved in the regulation of human cancers. Zhu et al17 found that FOXA2 inhibited the occurrence of gastric cancer in vitro and in vivo. In addition, FOXA2 inhibited tumor metastasis by blocking EMT in human lung cancer.18 The anti-tumor effect of FOXA2 was also detected in pancreatic cancer.19 Epigenetic regulation of FOXA2 has been reported to be involved in the pathogenesis of melanoma.20 However, the relationship between FOXA2 and miR-1246 has not been reported. Therefore, we investigated their interaction in this study. Meanwhile, the function of miR-1246 was also explored in melanoma cells.
Patients and Methods
Clinical Tissues
The tissues used in this experiment were collected from 43 melanoma patients in Weihai Central Hospital Affiliated to Qingdao University. All melanoma patients who participated in the experiment underwent surgery only. Participants provided written informed consents before the study began, and the experiment was approved by the Human Ethics Committee of Weihai Central Hospital Affiliated to Qingdao University. This study is performed in accordance with the Declaration of Helsinki.
Cell Culture and Transfection
Human epidermal melanocytes (HEM, ScienCell Research Laboratories, San Diego, CA, USA) were seeded in melanocyte culture medium. A375 and A2058 human melanoma cell lines (American Type Culture Collection, ATCC, Manassas, VA, USA)) were cultured in dulbecco’s modified eagle medium (DMEM) medium (Invitrogen; Carlsbad, CA, USA) with 10% fetal bovine serum (FBS) (Gibco, Rockville, MD, USA) at 37°C in an atmosphere with 5% CO2. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to transfect miR-1246 mimics, miR-1246 inhibitors, FOXA2 siRNA or FOXA2 plasmid (Generay Biotech, Shanghai, China) to A375 cells, respectively.
RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA isolation was performed using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The cDNA solution was obtained using Prime-Script miRNA cDNA Synthesis kit (TaKaRa, Tokyo, Japan). RT-qPCR was performed using SYBR-Green Premix Ex Taq II (TaKaRa, Tokyo, Japan) on ABI 7500 thermocycler (Applied Biosystems, Foster City, CA, USA). U6 or GAPDH was used as the control for miR-1246 or FOXA2, respectively. The expressions of miR-1246 and FOXA2 were quantified by the 2−∆∆cq method. The primers used in our work were as follows: miR-1246, forward primer: 5ʹ-TGA AGT AGG ACT GGG CAG AGA-3ʹ, reverse primer: 5ʹ-TGT TTG CAA TAG CCC TTT GAG-3ʹ; U6, forward primer: 5ʹ-CTC GCT TCG GCA GCA CA-3ʹ, reverse primer: 5ʹ-AAC GCT TCA CGA ATT TGC GT-3ʹ; FOXA2 forward primer: 5ʹ-ATG CAC TCG GCT TCC AGT AT-3ʹ, reverse primer: 5ʹ-GTT GCT CAC GGA GGA GTA GC-3ʹ; glyceraldheyde 3-phosphate dehydrogenase (GAPDH) forward, 5ʹ-ACA TCG CTC AGA CAC CAT G-3ʹ, reverse, 5ʹ-TGT AGT TGA GGT CAA TGA AGG G-3ʹ.
MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyl Tetrazolium Bromide) Assay
Firstly, transfected A375 cells (4×103 cells/well) were prepared in a 96-well plate. A375 cells were then incubated in fresh medium for 24, 48, 72 or 96 h, respectively. After that, 10 μL of MTT solution (Sigma–Aldrich, St. Louis, MO, USA) was added there, and the cells were further cultured for 4 h. Next, the MTT solution was aspirated and the Formazan solution was added to fully dissolve the crystals. The absorbance of each well was measured with a microscope (Olympus Corp., Tokyo, Japan) at 490 nm.
Transwell Assay
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was coated in the upper chambers for cell invasion. Next, 2 × 104 transfected A375 cells were placed in the upper chamber. A chemoattractant medium containing 10% FBS was added to the lower chamber. The cells were incubated for 12 h at 37°C. The moving cells were fixed and stained. The number of invading cells was observed under a microscope. Matrigel is not required for cell migration assay, and other procedures are the same as cell invasion assay.
Western Blot Analysis
First, we used radioimmunoprecipitation assay (RIPA) buffer (Beyotime, Shanghai, China) to lyse protein samples. Protein samples were separated in 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) protein loading buffer. The protein was then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Next, primary antibodies (Bax, Bcl-2, FOXA2 and GAPDH) were added and the membrane was incubated at 4°C overnight. After washing, corresponding secondary antibodies were added and incubated the membrane for 2 h at room temperature. Finally, protein bands were detected using electrochemiluminescence (ECL) reagent (Millipore, Billerica, MA, USA).
Dual-Luciferase Reporter Assay
The pcDNA3.1 plasmid vectors (Promega, Madison, WI, USA) containing the 3ʹ-UTR of wild-type or mutant FOXA2 was prepared. Next, the above luciferase vectors and miR-1246 mimics were co-transfected into A375 cells. After 48 h, the dual-luciferase reporter assay system (Promega, Madison, WI, USA) was used to assess luciferase activity.
Statistical Analysis
Experimental data were analyzed using Statistical Product and Service Solutions (SPSS) 18.0 (SPSS Inc., Chicago, IL, USA) or Graphpad Prism 6 (La Jolla, CA, USA). Data are shown as mean ± SD (standard deviation). Comparisons between multiple groups were performed using one-way ANOVA test followed by Post Hoc Test (Least Significant Difference). Significant differences were defined as P<0.05.
Results
MiR-1246 Expression Was Increased in Melanoma
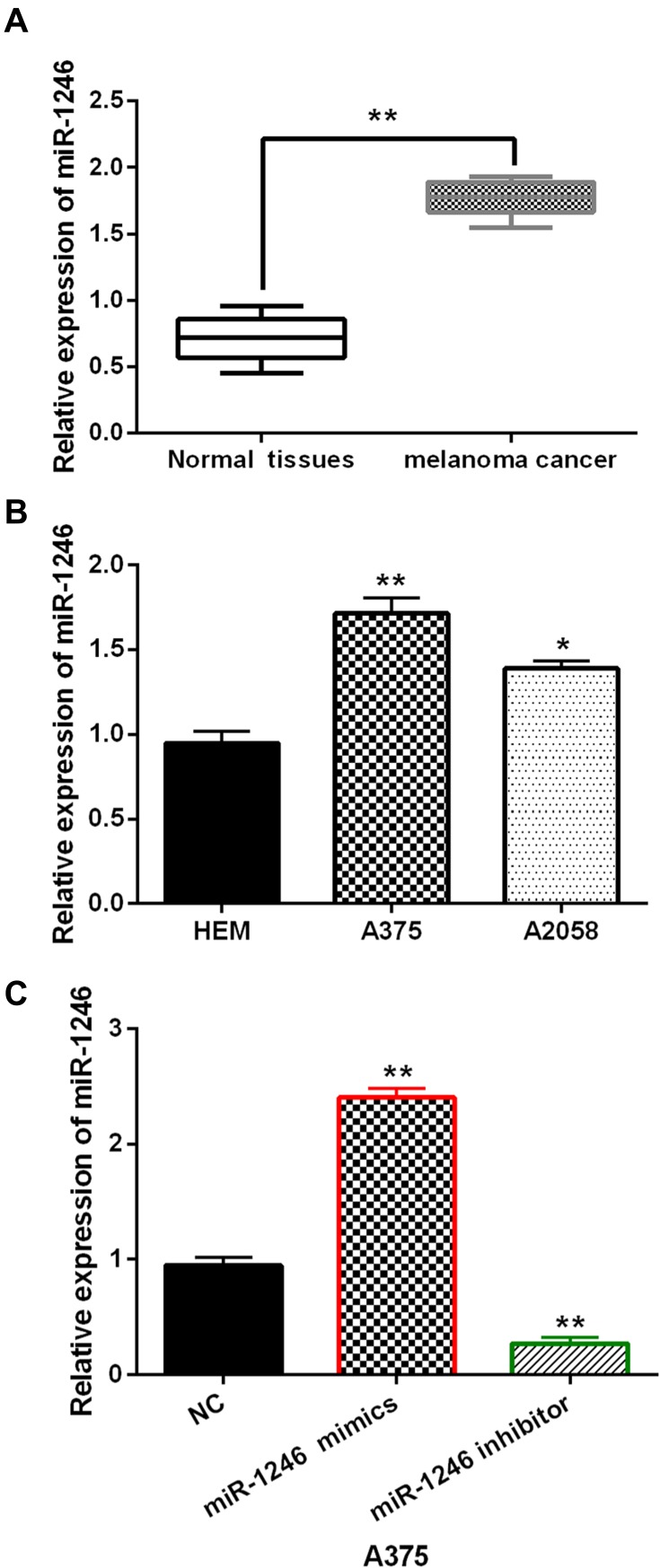
In melanoma tissues and cell lines, the abnormal expression of miR-1246 was observed. We found that miR-1246 expression was significantly increased in melanoma tissues compared to normal tissues (P<0.01, Figure 1A). Similarly, compared to HEM cells, upregulation of miR-1246 was detected in A375 and A2058 cell lines (P<0.01 or 0.05, Figure 1B). The findings suggest that miR-1246 may be involved in the pathogenesis of melanoma. Because the difference in miR-1246 expression was more significant in A375 cells than A2058cells, A375 cells were selected to investigate the function of miR-1246 in melanoma. Next, miR-1246 mimics or inhibitor was transfected into A375 cells. RT-qPCR showed that miR-1246 mimics can promote miR-1246 expression, but miR-1246 inhibitors can reduce its expression in A375 cells (P<0.01, Figure 1C).
Figure 1.
The expression of miR-1246 was increased in melanoma. (A, B) The mRNA miR-1246 expression in melanoma tissues and cell lines. (C) The miR-1246 expression regulated by its mimics or inhibitor in A375 cells. *P<0.05, **P<0.01.
MiR-1246 Promoted Cell Viability and Metastasis in Melanoma
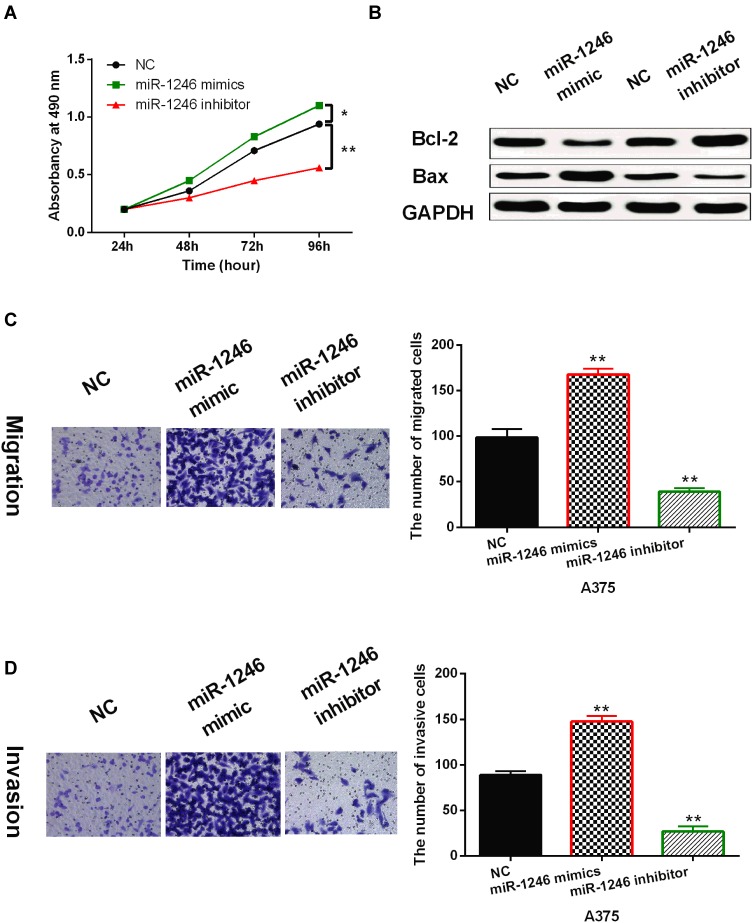
Next, the effects of miR-1246 on cell viability and metastasis were investigated in A375 cells using MTT and Transwell assays. First, it was found that miR-1246 overexpression promoted cell proliferation, while miR-1246 knockdown inhibited cell proliferation in A375 cells (P<0.01, Figure 2A). Then, we investigated how miR-1246 regulates apoptosis-associated proteins (Bcl-2/Bax) in A375 cells. We found that upregulation of miR-1246 suppressed Bax expression and promoted the expression of survival gene Bcl-2 (P<0.01, Figure 2B). In contrast, knockdown of miR-1246 promoted Bax expression and suppressed Bcl-2 expression level (P<0.01, Figure 2B). In addition, Transwell assay showed that miR-1246 mimics promoted cell migration. And knockdown of miR-1246 suppressed cell migration in A375 cells (P<0.01, Figure 2C). Similarly, cell invasion also showed the same results (P<0.01, Figure 2D). Collectively, miR-1246 promoted cell viability and metastasis in melanoma.
Figure 2.
MiR-1246 promoted cell viability and metastasis in melanoma cells. (A) The cell proliferation regulated by miR-1246 mimics or inhibitor in A375 cells. (B) miR-1246 regulated the expressions of Bcl-2/Bax in A375 cells. (C, D) Cell migration and invasion in A375 cells with miR-1246 mimics or inhibitor *P<0.05, **P<0.01.
MiR-1246 Directly Targeted FOXA2
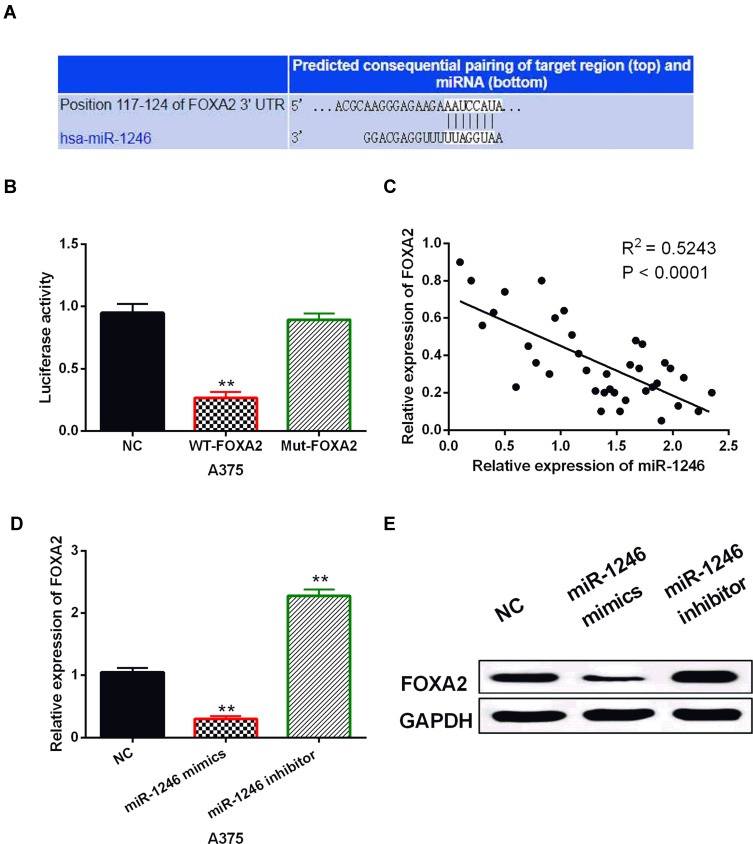
To further illustrate how miR-1246 promotes the progression of melanoma, we explored the targets of miR-1246 in the TargetScan (http://www.targetscan.org/) database. It predicts that miR-1246 has a binding site with the 3ʹ-UTR of FOXA2 (Figure 3A). Luciferase reporter assay showed that miR-1246 overexpression reduced the luciferase activity of Wt-FOXA2, but had no effect on the luciferase activity of Mut-FOXA2 (P<0.01, Figure 3B). In melanoma tissues, a negative correlation was detected between miR-1246 and FOXA2 (P<0.0001, R2=0.5243; Figure 3C). In addition, we found that FOXA2 expression was reduced by miR-1246 mimics and promoted by miR-1246 inhibitor (P<0.01, Figure 3D and E), consistent with the above results. These findings suggested that miR-1246 directly targeted FOXA2 and reverse-regulated FOXA2 expression in melanoma.
Figure 3.
MiR-1246 directly targeted FOXA2. (A) The binding sites between FOXA2 with miR-1246. (B) Luciferase reporter assay. (C) MiR-1246 was negatively correlated with FOXA2 in melanoma tissues. (D, E) FOXA2 expression regulated by miR-1246 mimics or inhibitor in A375 cells **P<0.01.
FOXA2 Was Downregulated in Melanoma
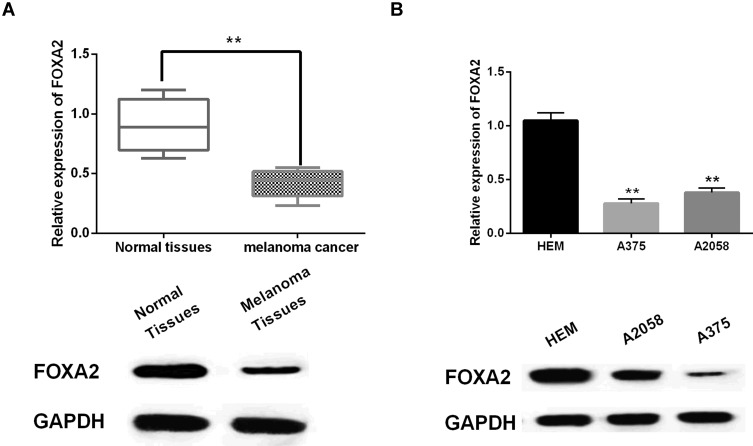
Next, the abnormal expression of FOXA2 was detected in melanoma tissues and cell lines. We found that FOXA2 was downregulated in melanoma tissues compared to normal tissues (P<0.01, Figure 4A). In addition, FOXA2 expression was lower in A375 and A2058 cell lines than in HEM cells (P<0.01, Figure 4B). The results indicate that FOXA2 may be involved in the occurrence of melanoma.
Figure 4.
FOXA2 was downregulated in melanoma. (A, B) The mRNA and protein expression of FOXA2 in melanoma tissues and cell lines **P<0.01.
Upregulation of FOXA2 Restored the Carcinogenesis of miR-1246 in Melanoma
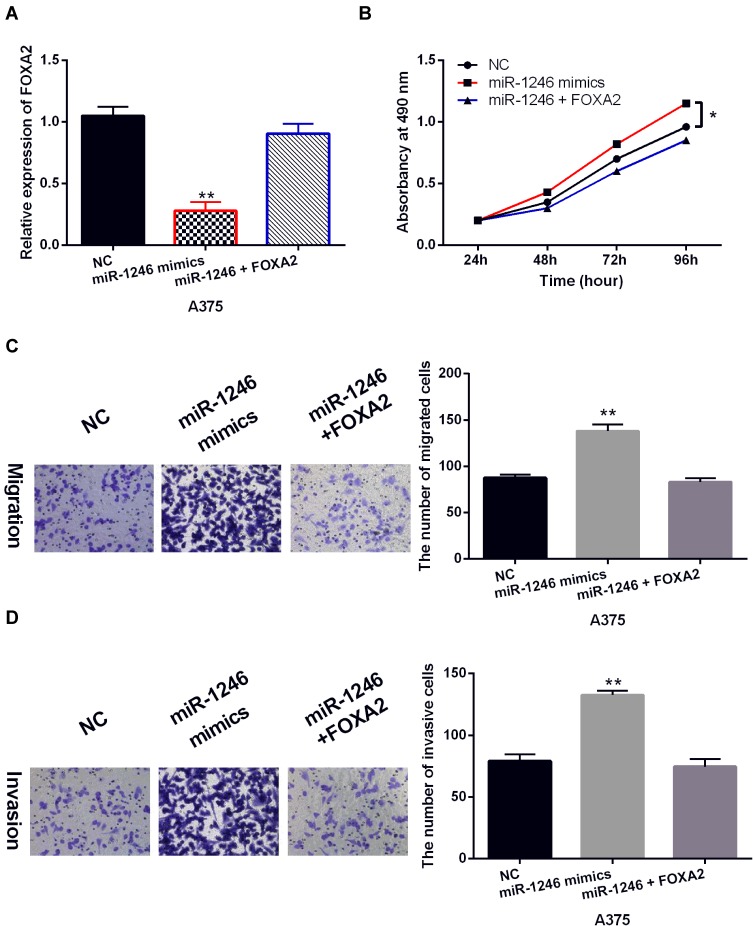
To explore the role of FOXA2 in melanoma, the FOXA2 vector was transfected into A375 cells with miR-1246 mimics. The decreased expression of FOXA2 in A375 cells with miR-1246 mimics was restored by the FOXA2 vector (Figure 5A). Functionally, the FOXA2 vector attenuated the increased ability of cell proliferation induced by miR-1246 mimics (Figure 5B). Similarly, the reversal effect of FOXA2 on cell migration and invasion was also detected in A375 cells with miR-1246 mimics (Figure 5C and D). Briefly, upregulation of FOXA2 restored the carcinogenesis of miR-1246 in melanoma.
Figure 5.
Upregulation of FOXA2 restored the carcinogenesis of miR-1246 in melanoma. (A) The FOXA2 expression in A375 cells with FOXA2 vector and miR-1246 mimics. (B–D) The cell proliferation, migration and invasion were detected in A375 cells with FOXA2 vector and miR-1246 mimics. * P<0.05, **P<0.01.
Discussion
Recently, the important regulatory mechanisms of miR-1246 have been found in human cancers. For example, miR-1246 promoted the proliferation, invasion, and migration of cervical cancer cells by inhibiting THBS2 expression.21 Similarly, miR-1246 has also been reported to induce cell motility and invasion in oral squamous cell carcinoma by regulating DENND2D.22 In addition, miR-1246 was found to promote the progression of non-small cell lung cancer.23 Most studies have shown that miR-1246 plays a role in human cancers by mediating the expressions of target genes such as CCNG2 and CPEB4.24,25 In melanoma, increased expression of miR-1246 was found. The upregulation of miR-1246 was related to the resistance of BRAF inhibitors.26 However, it is unclear how miR-1246 regulates melanoma tumorigenesis. Therefore, we designed this study to further elucidate the regulatory mechanism of miR-1246 in melanoma.
In our study, the carcinogenic effect of miR-1246 was also identified in melanoma. First, we found the increased expression of miR-1246 in melanoma. Overexpression of miR-1246 promoted cell viability and metastasis in melanoma. These results are consistent with the results of previous studies. In addition, miR-1246 suppressed Bax expression and promoted Bcl-2 expression in melanoma cells, which has not been reported in other cancers. Moreover, we also found that FOXA2 is a direct target of miR-1246 and upregulated in melanoma. Meanwhile, FOXA2 expression was inversely correlated with miR-1246 in melanoma. More importantly, upregulation of FOXA2 impaired the carcinogenic effect of miR-1246 in melanoma. These findings indicated that miR-1246 promoted the progression of melanoma by targeting FOXA2.
As a target gene, FOXA2 has been regulated by various miRNAs, such as miR-1291 and miR-590.27,28 Consistent with our results, downregulation of FOXA2 has been reported to serve as a tumor suppressor in lung cancer, breast cancer and pancreatic cancer.18,29,30 In addition, miR-141-3p has been reported to foster cell growth, invasion, and tumorigenesis of cervical cancer by targeting FOXA2.31 MiR-92a was found to promote the proliferation and invasion of hepatocellular carcinoma cells by regulating FOXA2.32 These findings support our conclusion that miR-1246 promotes cell viability and metastasis in melanoma by targeting FOXA2. Nevertheless, miR-1246 must have multiple targets in melanoma cells, and it must also have other downstream targets. Therefore, other potential downstream targets of miR-1246 in melanoma cells deserve further study. In addition, the pigmentation degree of the tested human melanocytes cell line has not been detected. It is important to analyze the correlation between miR-1246 and pigmentation degree. In the future, we will conduct further research to solve the above problems.
Conclusion
In summary, upregulation of miR-1246 was observed in melanoma. In addition, upregulation of miR-1246 promoted cell viability and metastasis in melanoma by downregulating FOXA2. Based on these results, we suspect that miR-1246 may have potential as a diagnostic indicator of melanoma.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Carvajal RD, Spencer SA, Lydiatt W. Mucosal melanoma: a clinically and biologically unique disease entity. J Natl Compr Canc Netw. 2012;10(3):345–356. doi: 10.6004/jnccn.2012.0034 [DOI] [PubMed] [Google Scholar]
- 2.Lopez F, Rodrigo JP, Cardesa A, et al. Update on primary head and neck mucosal melanoma. Head Neck. 2016;38(1):147–155. doi: 10.1002/hed.23872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pessina F, Navarria P, Tomatis S, et al. Outcome evaluation of patients with limited brain metastasis from malignant melanoma, treated with surgery, radiation therapy, and targeted therapy. World Neurosurg. 2017;105:184–190. doi: 10.1016/j.wneu.2017.05.131 [DOI] [PubMed] [Google Scholar]
- 4.Nichols EE, Richmond A, Daniels AB. Tumor characteristics, genetics, management, and the risk of metastasis in uveal melanoma. Semin Ophthalmol. 2016;31(4):304–309. doi: 10.3109/08820538.2016.1154175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;96 Suppl:R40–R44. [PubMed] [Google Scholar]
- 6.Xiong Y, Liu L, Qiu Y, Liu L. MicroRNA-29a inhibits growth, migration and invasion of melanoma A375 cells in vitro by directly targeting BMI1. Cell Physiol Biochem. 2018;50(1):385–397. doi: 10.1159/000494015 [DOI] [PubMed] [Google Scholar]
- 7.Peng J, Liu H, Liu C. MiR-155 promotes uveal melanoma cell proliferation and invasion by regulating NDFIP1 expression. Technol Cancer Res Treat. 2017;16(6):1160–1167. doi: 10.1177/1533034617737923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armand-Labit V, Meyer N, Casanova A, et al. Identification of a circulating microRNA profile as a biomarker of metastatic cutaneous melanoma. Acta Derm Venereol. 2016;96(1):29–34. doi: 10.2340/00015555-2156 [DOI] [PubMed] [Google Scholar]
- 9.Moshiri F, Salvi A, Gramantieri L, et al. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget. 2018;9(20):15350–15364. doi: 10.18632/oncotarget.24601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todeschini P, Salviato E, Paracchini L, et al. Circulating miRNA landscape identifies miR-1246 as promising diagnostic biomarker in high-grade serous ovarian carcinoma: a validation across two independent cohorts. Cancer Lett. 2017;388:320–327. doi: 10.1016/j.canlet.2016.12.017 [DOI] [PubMed] [Google Scholar]
- 11.Takeshita N, Hoshino I, Mori M, et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108(3):644–652. doi: 10.1038/bjc.2013.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, Xie YJ, Xu Q, Chen JX, Shan NC, Zhang Y. Down-regulation of miR-1246 in cervical cancer tissues and its clinical significance. Gynecol Oncol. 2015;138(3):683–688. doi: 10.1016/j.ygyno.2015.06.015 [DOI] [PubMed] [Google Scholar]
- 13.Bhagirath D, Yang TL, Bucay N, et al. microRNA-1246 is an exosomal biomarker for aggressive prostate cancer. Cancer Res. 2018;78(7):1833–1844. doi: 10.1158/0008-5472.CAN-17-2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li XJ, Ren ZJ, Tang JH, Yu Q. Exosomal microRNA miR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell Physiol Biochem. 2017;44(5):1741–1748. doi: 10.1159/000485780 [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Cao L, Zhang Y, Lian H, Sun Z, Cui Y. MicroRNA-1246 inhibits cell invasion and epithelial mesenchymal transition process by targeting CXCR4 in lung cancer cells. Cancer Biomark. 2018;21(2):251–260. doi: 10.3233/CBM-170317 [DOI] [PubMed] [Google Scholar]
- 16.Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63(19–20):2317–2328. doi: 10.1007/s00018-006-6095-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu CP, Wang J, Shi B, et al. The transcription factor FOXA2 suppresses gastric tumorigenesis in vitro and in vivo. Dig Dis Sci. 2015;60(1):109–117. doi: 10.1007/s10620-014-3290-4 [DOI] [PubMed] [Google Scholar]
- 18.Tang Y, Shu G, Yuan X, Jing N, Song J. FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers. Cell Res. 2011;21(2):316–326. doi: 10.1038/cr.2010.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vorvis C, Hatziapostolou M, Mahurkar-Joshi S, et al. Transcriptomic and CRISPR/Cas9 technologies reveal FOXA2 as a tumor suppressor gene in pancreatic cancer. Am J Physiol Gastrointest Liver Physiol. 2016;310(11):G1124–G1137. doi: 10.1152/ajpgi.00035.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu KS, Jo JY, Kim SJ, et al. Epigenetic regulation of the transcription factor Foxa2 directs differential elafin expression in melanocytes and melanoma cells. Biochem Biophys Res Commun. 2011;408(1):160–166. doi: 10.1016/j.bbrc.2011.04.001 [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Yao D, Zhao S, et al. MiR-1246 promotes SiHa cervical cancer cell proliferation, invasion, and migration through suppression of its target gene thrombospondin 2. Arch Gynecol Obstet. 2014;290(4):725–732. doi: 10.1007/s00404-014-3260-2 [DOI] [PubMed] [Google Scholar]
- 22.Sakha S, Muramatsu T, Ueda K, Inazawa J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci Rep. 2016;6:38750. doi: 10.1038/srep38750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang WC, Chin TM, Yang H, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016;7:11702. doi: 10.1038/ncomms11702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin SS, Peng CY, Liao YW, Chou MY, Hsieh PL, Yu CC. miR-1246 targets CCNG2 to enhance cancer stemness and chemoresistance in oral carcinomas. Cancers (Basel). 2018;10:8. doi: 10.3390/cancers10080272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang W, Li H, Luo R. The microRNA-1246 promotes metastasis in non-small cell lung cancer by targeting cytoplasmic polyadenylation element-binding protein 4. Diagn Pathol. 2015;10:127. doi: 10.1186/s13000-015-0366-1 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Kim JH, Ahn JH, Lee M. Upregulation of microRNA-1246 is associated with BRAF inhibitor resistance in melanoma cells with mutant BRAF. Cancer Res Treat. 2017;49(4):947–959. doi: 10.4143/crt.2016.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu MJ, Pan YZ, Qiu JX, Kim EJ, Yu AM. MicroRNA-1291 targets the FOXA2-AGR2 pathway to suppress pancreatic cancer cell proliferation and tumorigenesis. Oncotarget. 2016;7(29):45547–45561. doi: 10.18632/oncotarget.9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salem M, O’Brien JA, Bernaudo S, et al. miR-590-3p promotes ovarian cancer growth and metastasis via a novel FOXA2-versican pathway. Cancer Res. 2018;78(15):4175–4190. doi: 10.1158/0008-5472.CAN-17-3014 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Yang C, Gao W, et al. FOXA2 attenuates the epithelial to mesenchymal transition by regulating the transcription of E-cadherin and ZEB2 in human breast cancer. Cancer Lett. 2015;361(2):240–250. doi: 10.1016/j.canlet.2015.03.008 [DOI] [PubMed] [Google Scholar]
- 30.Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70(5):2115–2125. doi: 10.1158/0008-5472.CAN-09-2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li JH, Zhang Z, Du MZ, et al. microRNA-141-3p fosters the growth, invasion, and tumorigenesis of cervical cancer cells by targeting FOXA2. Arch Biochem Biophys. 2018;657:23–30. doi: 10.1016/j.abb.2018.09.008 [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Wu J, Xie C. miR-92a promotes hepatocellular carcinoma cells proliferation and invasion by FOXA2 targeting. Iran J Basic Med Sci. 2017;20(7):783–790. doi: 10.22038/IJBMS.2017.9010 [DOI] [PMC free article] [PubMed] [Google Scholar]