Abstract
Introduction:
DNA methylation may be one of the biological mechanisms underlying the health benefits of physical activity (PA). Our objective was to determine the association between PA and genome-wide DNA methylation at CpG level.
Methods:
We designed a two-stage epigenome wide association study. In the discovery stage, we used 619 individuals from the REGICOR cohort. Next, we validated the CpGs suggestively associated with PA (p-value <10−5) in two independent populations (n=1,735 and 190, respectively). PA was assessed with validated questionnaires and classified as light (LPA), moderate (MPA), vigorous (VPA), moderate-vigorous (MVPA) and total PA (TPA). We examined linear and non-linear associations and meta-analyzed the results in the three populations. The linear associations were meta-analyzed with a fixed-effects model and the p-values of the non-linear associations with the Stouffer and Fisher methods. We established a p-value threshold that fulfilled Bonferroni criteria over the number of CpGs analyzed (0.05/421,940=1.185·10−7).
Results:
In the meta-analyses, two CpG sites had a statistically significant non-linear association with MVPA. cg24155427 (p-value=1.19·10−9), located in an intergenic region in chromosome 1, has been previously associated with smoking, lupus and aging. cg09565397 (p-value=1.59·10−7), located within DGAT1 in chromosome 8, encodes an enzyme involved in triacylglycerol synthesis and has been associated with body mass index.
Conclusion:
This population-based study identified two new, differentially methylated CpG sites with a non-linear dose-response relationship to MVPA. These associations must be additionally validated and may be considered for further research on the biological mechanisms underlying health benefits of PA.
Keywords: DNA methylation, physical activity, epigenome-wide association study, meta-analysis
INTRODUCTION
Physical activity (PA) is well known to influence health during the lifespan, preventing diseases and improving health outcomes.(1–3) Specifically, PA practice helps to improve lipid profiles, control blood pressure, regulate carbohydrate metabolism and modulate homeostasis and inflammation.(4) On the other hand, insufficient PA is responsible for 9% of premature mortality worldwide, and also for 6% of worldwide disease burden of coronary heart disease (CHD), 7% of type 2 diabetes, 10% of breast cancer, and 10% of colon cancer.(1) Moreover, physical inactivity is one of the fundamental causes of the present obesity epidemic.(5) Thus, for the adult population, current guidelines recommend 150 minutes of moderate-intensity aerobic PA or 75 minutes of vigorous-intensity aerobic PA, or an equivalent combination of both, as a minimum weekly goal.(6)
Despite existing knowledge of the health benefits of PA practice, the biological mechanisms underlying this association are not fully understood.(3) Unraveling these mechanisms would contribute to our knowledge of the pathogenic processes related to the most significant non-communicable diseases and help to identify the molecular links between PA and health.(7) One of the most promising areas that could explain part of the mechanisms involved is epigenetic variability.(8) Epigenetics, a biological mechanism that regulates gene expression, comprises the mitotically heritable changes that do not modify the DNA sequence.(9) Epigenetic variability is not only heritable, but is also related to environmental and lifestyle factors and has been proposed as a potential biomarker of the so-called “exposome”.(10)
The most widely studied epigenetic mechanism, DNA methylation, is one of the putative and dynamic mechanisms underlying complex diseases related to PA.(11) DNA methylation consists of the addition of a methyl group mostly to the cytosines followed by guanines (CpG sites). This dynamic mechanism has been shown to be related to PA in experimental and observational designs.(12) Some studies report that PA practice induces the hypermethylation of some genes related to inflammation and fatty acid metabolism, such as ASC (13) and FASN (14), and the hypomethylation of genes regulating glucose transport and glycolytic rate, such as GLUT4 and GSKA (14). Moreover, these changes seem to be tissue-specific, and different effects have been observed in skeletal muscle and in adipose tissue after a 6-month program.(15,16) Conversely, other studies did not find any association between PA and DNA methylation.(17–20) Recently, van Roekel et al assessed the association between DNA methylation and PA in a prospective population-based epigenome-wide association study (EWAS).(21) They reported one CpG annotated to SAA2 that is related to total PA (including household chores) (p-value=6·10−9) and weaker evidence (p-value <10−5) for 14 additional methylation sites related to total PA and 7 CpG sites associated with leisure-time PA.
Apart from the lack of consistency on the approaches used in those preliminary insights on the link between DNA methylation and PA, the reported studies only considered the scenario of linear associations. However, the relationship between PA and clinical phenotypes, such as CHD risk, was found to be linear only at low levels of PA, fading at higher levels of PA. Thus, non-linear associations involving PA should be analyzed. In this study, we hypothesized that PA could modify DNA methylation patterns, and designed a study that aimed to assess whether PA was associated with DNA methylation either in a linear or non-linear manner, using a representative population sample.
METHODS
Study design and populations
We designed a two-stage (discovery and validation) EWAS involving three independent populations (see Figure, Supplemental Digital Content 1, flowchart of the study).
Discovery stage:
A subsample of 648 individuals was randomly selected from those attending the 2009–2013 follow-up visit of the REGICOR (REgistre GIroní del COR) cohort. This REGICOR population has been described in previous studies by our group.(22)
Validation stage:
Two independent populations were used to validate the CpGs found to be suggestively significant (p<10−5) in the discovery stage. One of the populations included the 2,568 individuals from the Framingham Offspring Study with a blood cell DNA methylation assessment (examination 8; data available from the Genotypes and Phenotypes database, http://dbgap.ncbi.nlm.nih.gov; project number #9047). This population has been described in previous EWAS reports from our group.(23)
The third population included 208 individuals from a case-control study, matched by age and sex, of the REGICOR cohort designed to identify methylation patterns associated with myocardial infarction. Cases were individuals with the event of interest and controls were individuals attending the REGICOR 2009–2013 follow-up visit and not included in the discovery sample. The present analysis included only the control individuals. As in the discovery sample, all participants were of European descent and lived in the geographical area monitored by the REGICOR study.
The study was approved by the local ethics committee (2012/4729/I) and meets the principles expressed in the Declaration of Helsinki and the relevant Spanish legislation. All participants provided informed written consent prior to study inclusion.
Physical activity assessment
Questionnaires for the recording of physical activity:
The amount and intensity of PA practice in the REGICOR cohort was assessed by the Minnesota leisure-time PA questionnaire,(24) validated for the Spanish population.(25)(26) Briefly, from a list of 64 activities, participants marked those they had practiced during the year prior to the visit and a trained interviewer collected information related to the frequency of practice and the duration of each session.
In the Framingham Offspring Study, PA practice was assessed by a standardized validated questionnaire.(27) This questionnaire considers 16 types of leisure-time PA and records the frequency of practice and the number of hours/minutes per day in a 2-week period.
Physical activity variables:
In both populations, we assigned an intensity to each recorded PA, ranging from 2 to 14 metabolic equivalents (METs).(28) Estimated energy expenditure in PA, quantified as METs·min/week,(29) was categorized as light-intensity (<4 METs), moderate-intensity (4–5.9 METs) and vigorous-intensity (≥6 METs), or LPA, MPA, VPA, respectively. These definitions were based on the cutoffs proposed in the original validation of the Minnesota Leisure Time PA questionnaire.(16) Examples of PA defined as LPA, MPA and VPA are included in Supplementary Table 1 (see Table, Supplemental Digital Content 2, types of PA in each category). We also estimated the combination of MPA and VPA (MVPA) and the combination of all three PA intensities, or total PA (TPA). Individuals showing a TPA, MVPA and VPA >15,000 METs·min/week were considered as outliers and excluded. We also excluded any participant with TPA=0 METs·min/week from the main analysis, but included them in a sensitivity analysis.
Assessment of DNA methylation status
Both DNA extraction and methylation assessment methods have been fully described in previous reports.(22) Briefly, DNA was extracted from whole peripheral blood in the REGICOR cohort and from buffy coat in the Framingham Offspring Study. DNA methylation was assessed genome-wide with commercial arrays based on bisulfite conversion of unmethylated cytosines. The Infinium HumanMethylation450 BeadChip (Illumina, CA, USA) was used in the cross-sectional REGICOR study and in the Framingham Offspring Study, according to the standard protocol. This array analyzes over 485,000 CpGs per sample.(30) Conversely, in the case-control study in the REGICOR population we used the Infinium MethylationEPIC BeadChip (Illumina, CA, USA). This array analyzes over 850,000 CpGs per sample, including 439,562 from the Infinium HumanMethylation450 BeadChip.(31)
Analysis and quality control of the raw data has been previously described for the Infinium HumanMethylation450 BeadChip.(22) The DNA from participants of the case-control study was analyzed in 13 batches of the Infinium MethylationEPIC BeadChip in the Genomics and Epigenomics Service of the Bellvitge Institute for Biomedical Research (Barcelona, Spain). Quality control of samples and CpGs in the methylation analysis was similar to that used in the other populations.(22,23) A detailed quality control pipeline for the Illumina MethylationEPIC BeadChip data is available in the Supplemental text appendix (see Text, Supplemental Digital Content 3, quality control for the samples and the CpGs).
Methylation status at each CpG site was reported by M-values, which are more robust statistically than β values.(32) They were calculated according to equation 1:
| Equation 1 |
where:
Mi = intensity of methylated probes,
Ui = intensity of unmethylated probes, and
α = 1; constant offset.
M-values close to 0 mean the CpG site is half-methylated. Positive M-values indicate the presence of more methylated than unmethylated cytosines, while negative M-values denote the opposite ratio.
To remove potential sources of technical variation not related to the underlying biology, we standardized the M-values by batch, as previously reported,(22) using Equation 2:
| Equation 2 |
where:
Z = standardized M-value,
X = M-value for a specific individual,
= mean of M-value for a specific batch, and
n = sample size.
Finally, to avoid the influence of extreme values, we excluded those CpGs with an M-value >4 standard deviations from the mean.
We obtained genomic information of the CpGs using the manifest and annotation provided by Illumina and contained in the corresponding R packages available through the Bioconductor repository (IlluminaHumanMethylation450kanno.ilmn12.hg19 and IlluminaHumanMethylationEPICanno.ilm10b2.hg19).
Covariates assessment
The REGICOR study specifically trained a group of nurses to collect blood samples and sociodemographic (age and sex) and lifestyle (smoking habits) information, and to measure anthropometric variables (height, weight) using validated questionnaires and methods. Measurements and data from the Framingham Offspring Study were obtained from examination 8 through the Genotypes and Phenotypes database (http://dbgap.ncbi.nlm.nih.gov; project number #9047). The procedures used to collect data and blood samples from both populations have been previously described.(23)
We included smoking habit as a covariate in our analysis because of its strong relation to changes in DNA methylation patterns. We categorized participants as current smokers (≥1 cigarette/day or gave up smoking <1 year before the visit), former smokers for 1–5 years prior to the visit, former smokers for >5 years prior to the visit, and never smokers.(22) We excluded from analysis those individuals with no information available regarding their smoking habits.
As DNA methylation patterns also vary according to cell type, we inferred the peripheral blood cell counts using DNA methylations signatures with the FlowSorted.Blood.450k R package available through the Bioconductor repository.(33) Finally, we estimated two surrogate variables for unknown sources of potential confounding using the sva R package.(34)
Statistical analysis
We considered DNA methylation as the outcome variable. Generalized additive models were selected to characterize a non-linear dose-response relationship between PA and DNA methylation using smooth functions of the independent variable of interest (PA). We defined three models according to PA intensities: LPA, MPA and VPA (model 1); LPA and MVPA (model 2); and TPA (model 3). All the models were adjusted for age, sex, smoking status, estimated cell counts, and two surrogate variables. Model 2 was considered as the main analysis. In the secondary analysis, we also included BMI as a potential confounder or mediator variable.
In the validation stage, we analyzed the CpG sites related to any of the PA in a linear or a non-linear trend with an arbitrary p-value <10−5 in the REGICOR discovery cohort as previously described.(22,35) We used a Bonferroni-corrected significance threshold for the number of CpG sites taken forward for replication. Finally, we meta-analyzed the results observed in the discovery and validation cohorts. In this meta-analysis, the p-value threshold fulfilled Bonferroni criteria over the number of CpG sites analyzed in the discovery stage that were also in the Illumina MethylationEPIC Beadchip after quality control was performed (0.05/421,940=1.185·10−7).
Linear associations
We performed a fixed-effects meta-analysis of the effect sizes observed in the discovery stage and in the two validation samples to determine linear associations.
Non-linear associations
As effect sizes were not available, we performed a meta-analysis of the p-values using the metap R package. Following Loughin’s recommendations based on structure and evidence against the null hypothesis,(36) we applied both the Fisher method, which combines p values by the summation of logs (37) and the Stouffer method of summation of z values (38). We considered as validated those CpG sites that were significant according to one of the methods and showing consistent results (statistically significant or close to the Bonferroni p-value threshold) when applying the other one.
RESULTS
Quality control of DNA methylation and physical activity data
The quality control of the DNA methylation data resulted in 428,013 CpGs available to perform the discovery EWAS, 483,656 CpGs available for the validation in the Framingham Offspring Study, and 811,610 CpGs available for the validation in REGICOR. Since different DNA methylation arrays were used, 421,940 CpGs were available for the meta-analysis of the three populations.
Supplementary figure 2 shows the flowchart describing the sample size of the three populations in each step prior to the association analysis (see Figure, Supplemental Digital Content 4, flowchart of the data quality control). After the quality control of the DNA methylation data, we included 646 of 648 individuals from the REGICOR discovery sample; 2,542 of 2,568 individuals from the Framingham Offspring Study; and 195 of the 208 individuals from the REGICOR validation sample.
Due to missing data, our analysis excluded one participant (unknown smoking habits) from the REGICOR discovery sample (n=645), 804 individuals (7 missing smoking data, 797 missing PA data, and 5 missing both variables) in the Framingham population (n=1,738), and 2 participants (missing PA data) from the REGICOR validation sample (n=193). Moreover, we excluded participants with TPA=0 (no PA practice) and outliers, considered as a potential reporting error. Consequently, we performed the analysis with 619 participants in the REGICOR discovery sample, 1,735 in the Framingham population, and 190 in the REGICOR validation sample. Sample sizes in the secondary and the sensitivity analyses are included in the Supplemental text appendix (see Text, Supplemental Digital Content 3, results of the quality control of data).
The main sociodemographic and clinical characteristics of the three populations used in the main analyses are shown in Table 1.
Table 1.
Descriptive characteristics of the participants in the discovery stage (cross-sectional study of the REGICOR cohort) and the validation stage (controls from the case-control study of the REGICOR cohort and participants from the Framingham Offspring Study) of the main analysis.
| Variables | Discovery population | Validation populations | ||||
|---|---|---|---|---|---|---|
| REGICOR - cross-sectional | REGICOR - case-control | Framingham Offspring Cohort | ||||
| N=619 | NAs | N=190 | NAs | N=1,735 | NAs | |
| Age | 63.1 (11.7) | 0 | 63.4 (6.90) | 0 | 66.3 (9.00) | 0 |
| Sex, female, n (%) | 309 (49.9) | 0 | 98 (51.6) | 0 | 950 (54.8) | 0 |
| Smokers, n (%) | 101 (16.3) | 0 | 16 (8.42) | 0 | 181 (10.4) | 0 |
| BMI, kg/m2*‡ | 26.9 (4.00) | 2 | 28.8 (5.05) | 0 | 28.4 (5.42) | 3 |
| Obesity, n (%) | 123 (19.9) | 2 | 68 (35.8) | 0 | 573 (33.1) | 3 |
| Total cholesterol, mg/dl* | 208 (36.8) | 0 | 197 (29.1) | 0 | 186 (37.4) | 1 |
| LDL cholesterol, mg/dl*‡ | 135 (32.4) | 3 | 127 (26.2) | 1 | 105 (31.6) | 2 |
| HDL cholesterol, mg/dl*‡ | 53.0 (12.3) | 0 | 50.0 (10.5) | 0 | 57.4 (18.2) | 2 |
| Triglycerides, mg/dl† | 89.0 [66.0;121] | 0 | 90.5 [67.2;122] | 0 | 102 [74.0;144] | 1 |
| Cholesterol treatment, n (%) | 149 (24.1) | 2 | 545 (28.6) | 1 | 767 (44.3) | 4 |
| SBP, mmHg*‡ | 131 (18.5) | 0 | 135 (18.3) | 0 | 126 (17.3) | 1 |
| DBP, mmHg*‡ | 76.0 (9.84) | 0 | 78.5 (9.86) | 0 | 71.8 (10.2) | 3 |
| HTN, n (%)‡ | 288 (46.7) | 2 | 113 (59.5) | 0 | 1,021 (59.0) | 5 |
| HTN treatment, n (%)‡ | 188 (30.5) | 3 | 70 (36.8) | 0 | 877 (50.7) | 4 |
| Glucose, mg/dl† | 93.0 [86.5;102] | 0 | 93.2 [87.0;105] | 0 | 102 [95.0;112] | 2 |
| Diabetes, n (%) | 60 (9.74) | 3 | 33 (17.4) | 0 | 274 (16.7) | 94 |
| Diabetes treatment, n (%) | 42 (6.82) | 3 | 20 (10.5) | 0 | 90 (5.47) | 89 |
| Light PA, METs/week†‡ | 448 [53.5;985] | 0 | 336 [0.00;839] | 0 | 105 [84.0;147] | 0 |
| Moderate PA, METs/week†‡ | 105 [0.00;839] | 0 | 630 [0.00;2,098] | 0 | 968 [367;1,889] | 0 |
| Vigorous PA, METs/week†‡ | 420 [49.0;1,274] | 0 | 444 [97.9;1,192] | 0 | 22.5 [0.00;420] | 0 |
| Moderate vigorous PA, METs/week†‡ | 1,114 [240;2,199] | 0 | 1,691 [546;3,035] | 0 | 1,250 [484;2,445] | 0 |
| Total PA, METs/week†‡ | 1,771 [912;3,211] | 0 | 2,199 [1,163;3,907] | 0 | 1,357 [583;2,561] | 0 |
Mean (Standard Deviation).
Median (Interquartile Range).
BMI, Body Mass Index; Obesity, defined as BMI≥30 kg/m2; LDL, Low-Density Lipoprotein; HDL, High-Density Lipoprotein; SBP, Systolic Blood Pressure; DBP, Diastolic Blood Pressure; HTN, Hypertension, defined as previous treatment or SBP≥140 mmHg or DBP≥90 mmHg (or self-reported in the REGICOR case-control study); Diabetes, defined as previous treatment or glycaemia≥126mg/dl (or self-reported in the REGICOR case-control study); PA, physical activity, categorized as light, moderate and vigorous intensity physical activities, as a combination of moderate and vigorous intensity physical activities, and as the total sum of the three intensities of physical activity, all reported in metabolic equivalents of tasks (METs) per week.
Discovery Stage of the EWAS
Defining an arbitrary p-value threshold of 10–5 as previously reported,(23,35) in the main analysis we identified 100 differentially methylated CpGs associated with PA in model 1 (see Table, Supplemental Digital Content 5, associations between methylation at CpGs and PA variables): 4 and 29 differentially methylated CpGs in linear and non-linear association with LPA, respectively; 7 and 29 differentially methylated CpGs in linear and non-linear association with MPA, respectively; and 16 and 17 differentially methylated CpGs in linear and non-linear association with VPA, respectively. Supplementary figure 3 shows the Manhattan and QQ plots (see Figure, Supplemental Digital Content 6, results of the associations between methylation at CpGs and PA variables in the discovery stage of model 1).
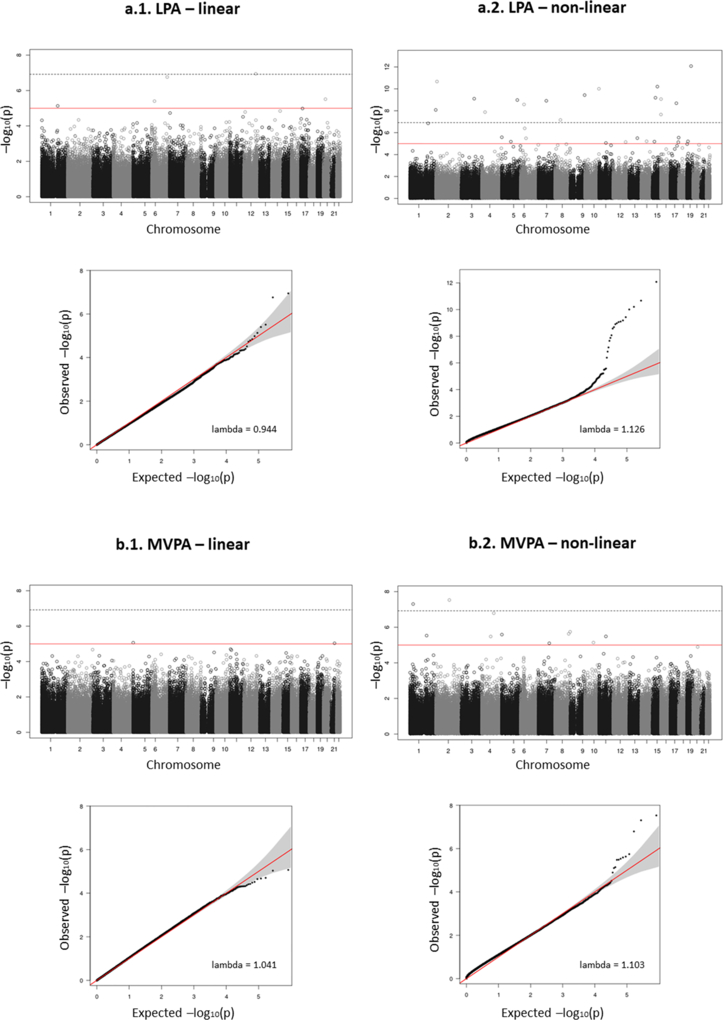
In model 2, we found 47 differentially methylated CpGs associated with PA (see Table, Supplemental Digital Content 7, associations between methylation at CpGs and PA variables): 5 and 29 differentially methylated CpGs in linear and non-linear association with LPA, respectively; and 2 and 11 differentially methylated CpGs in linear and non-linear association with MVPA, respectively. Figure 1 shows the Manhattan and QQ plots (lambdas between 0.944 and 1.126).
Figure 1. Manhattan plots and QQ plots of the associations between DNA methylation and PA variables in the discovery stage of model 2.
Plots are given for: (a) light-intensity PA (LPA), and (b) moderate-vigorous-intensity PA (MVPA). For each PA, the linear (a.1, b.1) and non-linear (a.2, b.2) p-values from the main analysis are represented.
Finally, in model 3, we discovered 11 differentially methylated CpGs related to PA (see Table, Supplemental Digital Content 8, associations between methylation at CpGs and PA variables): 4 and 7 differentially methylated CpGs in linear and non-linear association with TPA. Supplementary figure 4 shows the Manhattan and QQ plots (see Figure, Supplemental Digital Content 9, results of the associations between methylation at CpGs and PA variables in the discovery stage of model 3).
In total, we discovered 118 unique differentially methylated CpGs associated with PA (33 and 87 in linear and non-linear association, respectively). These CpGs were located in 81 different genes and 36 intergenic regions.
Validation Stage of the EWAS and Meta-Analysis
We separately assessed the association between PA and the initially identified CpG sites in the Framingham population and the REGICOR validation sample, and thereafter meta-analyzed the results from all three populations. We could not validate cg20134658 in any population as this CpG was discarded in the quality control.
Linear associations
From 33 CpGs linearly associated with PA in the discovery sample, we could assess 29 and 33 CpGs in the REGICOR validation sample and in Framingham, respectively. Considering a Bonferroni-corrected p-value threshold (p-value<1.72·10–3 and <1.52·10–3, respectively), we validated cg09565397 (DGAT1) as associated with MVPA and TPA in the REGICOR validation sample (p-value= 1.11·10–4 and 3.87·10–4, respectively). However, we did not validate any CpG associated with any PA in the Framingham sample.
In the fixed-effects meta-analysis of the three studies, none of the CpG sites initially identified as showing a linear relationship to PA were validated (see Tables, Supplemental Digital Content 10–12, results of the meta-analyses of models 1–3, respectively).
Non-linear associations
From 87 CpG in non-linear association with PA in the discovery, we could assess 83 and 86 CpGs in the REGICOR validation sample and in Framingham, respectively. Considering a Bonferroni-corrected p-value threshold (p-value<6.02·10–4 and p-value<5.81·10–4, respectively), we validated cg24155427 as associated with MVPA in the REGICOR validation sample (p-value=2.49·10–4). However, we did not validate any CpG associated with any PA in the Framingham sample.
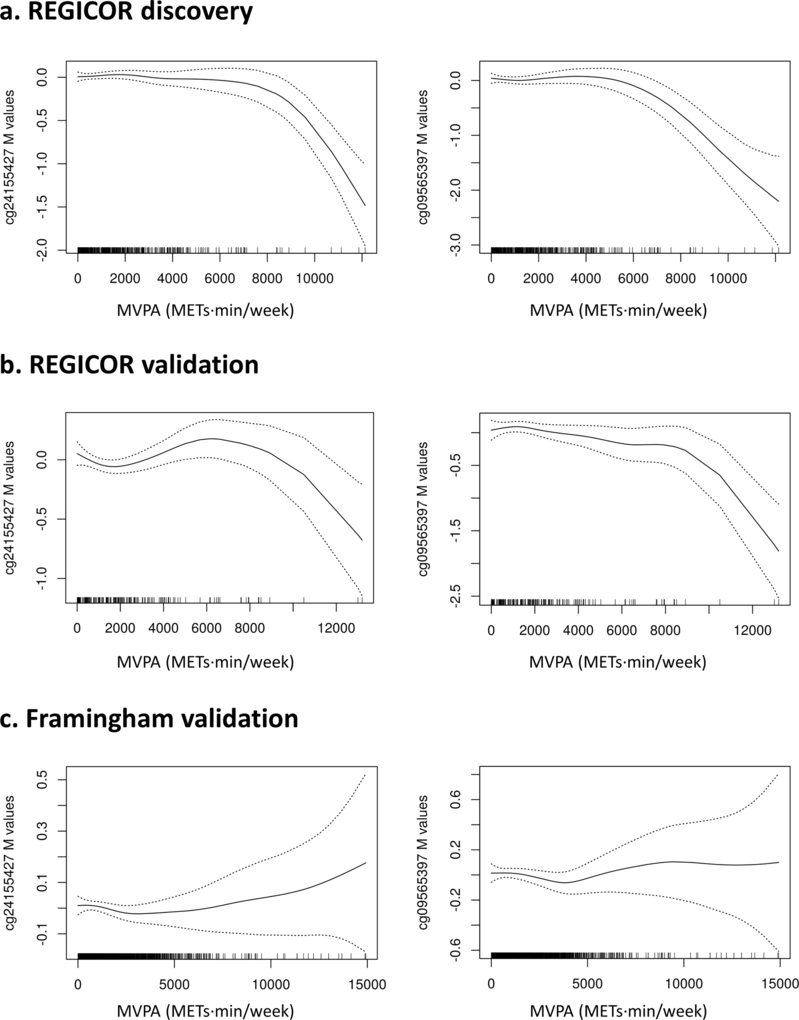
In the meta-analysis of the p-values of the three studies, we validated two differentially methylated CpGs in non-linear association with MVPA: cg24155427 and cg09565397 (Table 2; see Table, Supplemental Digital Content 13, results of the meta-analyses of models 1–3). These CpGs are located in two loci: an intergenic region in chromosome 1 and DGAT1 in chromosome 8. Figure 2 shows the plots of the association between PA and DNA methylation of these CpGs in each population.
Table 2.
Differentially methylated CpGs non-linearly associated with physical activity (PA). P-values of the significant associations with moderate-vigorous-intensity PA (MVPA) are given for the discovery, validation and meta-analysis stage of the main analysis. P-values of the meta-analysis with the Fisher and Stouffer methods are provided. Significant associations in the discovery population are those with a p-value <10−5, while significant associations in the meta-analysis are those with a p-value <1.185·10−7.
| Genomic information | Discovery | Validation | Meta-analysis | |||||
|---|---|---|---|---|---|---|---|---|
| CpG | Chr* | Position (bp) | Gene | P-value | P-valueREG | P-valueFOS | P-valueFisher | P-valueStouffer |
| cg24155427 | 1 | 31,242,051 | NA | 4.97·10−08 | 2.49·10−04 | 2.55·10−01 | 1.19·10−09 | 2.29·10−08 |
| cg09565397 | 8 | 145,543,014 | DGAT1 | 1.81·10−06 | 1.23·10−03 | 2.91·10−01 | 1.59·10−07 | 1.06·10−06 |
CpG, CpG ID; Chr, chromosome location; Position (bp), basepair genomic position; NA, non-annotated gene; REG, REGICOR; FOS, Framingham Offspring Cohort.
Figure 2. Smooth splines and standard errors bands for the association between PA and methylation levels (M-values) for cg24155427 and cg09565397 in each population (a, discovery; b, REGICOR validation; c, Framingham).
Smooth splines and standard errors bands are given for the significant associations in the meta-analysis of the p-values from the main analysis; i.e. MVPA in model 2. Generalized additive models were adjusted for sex, age, smoking status, estimated cell counts and two surrogate variables.
Secondary and sensitivity analyses
The results of the secondary analysis including BMI in the multivariate models and those of the sensitivity analyses including participants with TPA=0, were similar to the main results. Supplementary tables 5b–7b show the results for the linear associations in the secondary analysis (see Table, Supplemental Digital Content 10–12, results of the meta-analyses of models 1–3, respectively). Supplementary tables 8b and 8c show the results for the non-linear associations in the secondary and sensitivity analyses, respectively (see Table, Supplemental Digital Content 13, results of the meta-analyses of models 1–3).
DISCUSSION
In this two-stage EWAS, we found two CpG sites showing differential methylation in association with PA, specifically MVPA. The analysis of the dose-response pattern suggests a non-linear relationship between PA and methylation levels of the reported CpG.
The non-linear dose-response relationship between DNA methylation and PA is consistent with previous studies focused on clinical phenotypes such as CHD risk, in which low levels of PA were associated with CHD risk reduction, but at higher levels of PA, risk reduction tended to reach a plateau.(39) However, in our results only high levels of MVPA were associated with DNA methylation patterns. On one hand, these results highlight the importance of analyzing both the linear and non-linear association between PA and health phenotypes. On the other hand, given the unexpected non-linear pattern, we cannot discard the possibility of false positive findings; this should be validated in other studies.
The two validated CpG sites
A non-linear association was observed between MVPA and the two validated CpG sites, neither of which had previously been associated with either PA or metabolic-related traits, to our knowledge. Interestingly, these CpG sites were associated with PA in the REGICOR discovery and validation populations but not in the Framingham sample. This difference could be related to the type of questionnaires each study used to assess PA practice or to other differences between these two populations. Although we cannot disregard the possibility of false positive results, the consistency of the association patterns observed in the two REGICOR samples supports the legitimacy of these findings.
One of the validated CpG sites, cg24155427, is located in an intergenic region of chromosome 1. It was found to be differentially methylated in association with inflammation-related states: smoking,(40) systemic lupus erythematosus(41) and aging in neutrophils of HIV patients(42).
The second validated CpG site, cg09565397, was associated with MVPA in a non-linear trend. The p-value of the meta-analysis was close to the Bonferroni-corrected p-value threshold and the association was highly significant in the REGICOR population used for validation. To our knowledge, this CpG site was not found as differentially methylated in association to any trait. It is located within the coding region of DGAT1, which encodes an enzyme involved in triacylglycerol synthesis and associated with body height.(43) Regular aerobic PA practice is well known to decrease plasma triglyceride levels,(44) and our study provides new insights on the potential role of DNA methylation as a mediator between PA and triglycerides metabolism. Moreover, DGAT1 levels are regulated by the AMP-activated protein kinase (AMPK) cascade.(45) Preclinical models have suggested that this cascade mediates some of the protective mechanisms of PA.(46) This result should be replicated in other studies.
Replication of previously reported EWAS results
We checked for any overlap of methylation sites recently reported to be related to PA by van Roekel et al. We used our discovery sample to validate the 7 CpGs reportedly associated with leisure-time PA. Among them, we found cg11031064 to be in linear association with MVPA (p-value=1.19 10−3). However, this CpG was not related to leisure-time PA in either the FOS or REGICOR validation samples, or in the meta-analysis of the three populations (data not shown). We were unable to replicate the association of cg10266336, associated with TPA (including household chores) by van Roekel et al (p-value=6.0·10−9), as this CpG had been discarded in the quality control. The discrepancies between the two studies could be due to differences in study design, PA assessment and classification, and statistical approach.
Strengths
The main strengths of this study are the use of population-based samples and validated instruments to assess PA, as well as the two-stage EWAS strategy with a discovery population and two independent validation populations. We used standardized protocols to assess the variables included in the analyses and to remove non-biological sources of variation in the case of DNA methylation. Moreover, we used M-values as the DNA methylation measurements, which better identify differentially methylated CpG sites, compared to Beta values.(32) Our analysis was further adjusted for confounding variables and non-measured confounding (surrogate) variables. We also considered BMI as a covariate and performed a sensitivity analysis including individuals having reported TPA=0. Lastly, this is the largest well-characterized human study in which a systematic study of methylation patterns related to PA has been conducted in general population.
Limitations
Some limitations of this study should be noted. First, some CpGs analyzed in the discovery and the Framingham population could not be examined in the REGICOR validation population due to the use of a different methylation array. Second, the measurement of PA was based on questionnaires; although validated, questionnaires provide less precise and objective information, compared to accelerometers.(47) Also, we only considered leisure-time PA and did not investigate sedentary behavior. Third, our results are based on populations of European origin and cannot be extrapolated to other groups. Fourth, due to the cross-sectional study design, we cannot infer causality of the association between DNA methylation and PA. Fifth, we observed a slight inflation in the results in the discovery population; however, in the associations with MVPA the lambda was 1.103 and the inflation appeared not substantial based on visual assessment of the QQ plot. Finally, the study has the intrinsic EWAS limitations, such as the type of sample used or the population size.
CONCLUSION
This study identified two loci showing a non-linear dose-response relationship between differential methylation and reported MVPA intensity. These findings must be further validated and both the effect of these loci on gene expression and the causality of these associations must be determined. These differentially methylated loci also could be studied in association with disease incidence.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elaine M. Lilly, PhD, for her critical reading and revision of the English text.
SOURCE OF FUNDING
This project was funded by the Carlos III Health Institute–European Regional Development Fund (FIS PI12/00232; FIS PI15/00051, CIBERCV, CIBERESP, CIBERONC), and the Government of Catalonia through the Agency for Management of University and Research Grants (2014SGR240). S. Sayols-Baixeras was funded by the Instituto de Salud Carlos III-Fondos FEDER (IFI14/00007). A. Fernández-Sanlés was funded by the Spanish Ministry of Economy and Competitiveness (BES-2014–069718).
The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195 and HHSN268201500001I). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. Additional funding for SABRe was provided by Division of Intramural Research, NHLBI, and Center for Population Studies, NHLBI.
Footnotes
CONFLICT OF INTEREST
No conflict of interest to declare. The results of the present study do not constitute endorsement by the American College of Sports Medicine. The authors declare that the results of the study are presented clearly and honestly, and without fabrication, falsification, or inappropriate data manipulation.
List of Supplemental Digital Content
Supplemental Digital Content 1.pdf
Supplemental Digital Content 2.xlsx
Supplemental Digital Content 3.pdf
Supplemental Digital Content 4.pdf
Supplemental Digital Content 5.xlsx
Supplemental Digital Content 6.pdf
Supplemental Digital Content 7.xlsx
Supplemental Digital Content 8.xlsx
Supplemental Digital Content 9.pdf
Supplemental Digital Content 10.xlsx
Supplemental Digital Content 11.xlsx
Supplemental Digital Content 12.xlsx
Supplemental Digital Content 13.xlsx
REFERENCES
- 1.Lee I-M, Shiroma EJ, Lobelo F, et al. Effect of physical inactivity on major non-communicable diseases worldwide: an analysis of burden of disease and life expectancy. Lancet. 2012;380(9838):219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elosua R, Redondo A, Segura A, et al. Dose–response association of physical activity with acute myocardial infarction: Do amount and intensity matter? Prev Med. 2013;57(5):567–72. [DOI] [PubMed] [Google Scholar]
- 3.Neufer PD, Bamman MM, Muoio DM, et al. Understanding the Cellular and Molecular Mechanisms of Physical Activity-Induced Health Benefits. Cell Metab. 2015;22(1):4–11. [DOI] [PubMed] [Google Scholar]
- 4.Fiuza-Luces C, Santos-Lozano A, Joyner M, et al. Exercise benefits in cardiovascular disease: beyond attenuation of traditional risk factors. Nat Rev Cardiol. 2018;15(12):731–43. [DOI] [PubMed] [Google Scholar]
- 5.Bray GA. Diabetes and Obesity--Time Bombs to Be Defused. Diabetes Care. 2015;38(11):1997–9. [DOI] [PubMed] [Google Scholar]
- 6.Kahlmeier S, Wijnhoven TMA, Alpiger P, Schweizer C, Breda J, Martin BW. National physical activity recommendations: systematic overview and analysis of the situation in European countries. BMC Public Health. 2015;15(1):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawley JA, Hargreaves M, Joyner MJ, Zierath JR. Integrative Biology of Exercise. Cell. 2014;159(4):738–49. [DOI] [PubMed] [Google Scholar]
- 8.Skipper M, Eccleston A, Gray N, et al. Presenting the Epigenome Roadmap. Nature. 2015;518(7539):313–313. [DOI] [PubMed] [Google Scholar]
- 9.Russo VEA, Martienssen RA, Riggs AD. Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press; 1996. 692 p. [Google Scholar]
- 10.Petronis A Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465(7299):721–7. [DOI] [PubMed] [Google Scholar]
- 11.Jin Z, Liu Y. DNA methylation in human diseases. Genes Dis. 2018;5(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grazioli E, Dimauro I, Mercatelli N, et al. Physical activity in the prevention of human diseases: role of epigenetic modifications. BMC Genomics. 2017;18(Suppl 8):802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakajima K, Takeoka M, Mori M, et al. Exercise Effects on Methylation of ASC Gene. Int J Sports Med. 2010;31(9):671–5. [DOI] [PubMed] [Google Scholar]
- 14.Rowlands DS, Page RA, Sukala WR, et al. Multi-omic integrated networks connect DNA methylation and miRNA with skeletal muscle plasticity to chronic exercise in Type 2 diabetic obesity. Physiol Genomics. 2014;46(20):747–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nitert MD, Dayeh T, Volkov P, et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61(12):3322–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rönn T, Volkov P, Davegårdh C, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9(6):e1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang FF, Santella RM, Wolff M, Kappil MA, Markowitz SB, Morabia A. White blood cell global methylation and IL-6 promoter methylation in association with diet and lifestyle risk factors in a cancer-free population. Epigenetics. 2012;7(6):606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peplonska B, Bukowska A, Wieczorek E, Przybek M, Zienolddiny S, Reszka E. Rotating night work, lifestyle factors, obesity and promoter methylation in BRCA1 and BRCA2 genes among nurses and midwives. PLoS One. 2017;12(6):e0178792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quach A, Levine ME, Tanaka T, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging. 2017;9(2):419–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gale CR, Marioni RE, Čukić I, et al. The epigenetic clock and objectively measured sedentary and walking behavior in older adults: the Lothian Birth Cohort 1936. Clin Epigenetics. 2018;10(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Roekel EH, Dugué P-A, Jung C-H, et al. Physical Activity, Television Viewing Time, and DNA Methylation in Peripheral Blood. Med Sci Sport Exerc. 2018;51(3):1. [DOI] [PubMed] [Google Scholar]
- 22.Sayols-Baixeras S, Lluís-Ganella C, Subirana I, et al. Identification of a new locus and validation of previously reported loci showing differential methylation associated with smoking. The REGICOR study. Epigenetics 2015;10(12):1156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayols-Baixeras S, Subirana I, Lluis-Ganella C, et al. Identification and validation of seven new loci showing differential DNA methylation related to serum lipid profile: an epigenome-wide approach. The REGICOR study. Hum Mol Genet. 2016;25(20):ddw285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor HL, Jacobs DR, Schucker B, Knudsen J, Leon AS, Debacker G. A questionnaire for the assessment of leisure time physical activities. J Chronic Dis. 1978;31(12):741–55. [DOI] [PubMed] [Google Scholar]
- 25.Elosua R, Marrugat J, Molina L, Pons S, Pujol E. Validation of the Minnesota Leisure Time Physical Activity Questionnaire in Spanish men. The MARATHOM Investigators. Am J Epidemiol. 1994;139(12):1197–209. [DOI] [PubMed] [Google Scholar]
- 26.Elosua R, Garcia M, Aguilar A, Molina L, Covas MI, Marrugat J. Validation of the Minnesota Leisure Time Physical Activity Questionnaire In Spanish Women. Investigators of the MARATDON Group. Med Sci Sports Exerc. 2000;32(8):1431–7. [DOI] [PubMed] [Google Scholar]
- 27.Kannel WB, Sorlie P. Some Health Benefits of Physical Activity. Arch Intern Med. 1979;139(8):857. [PubMed] [Google Scholar]
- 28.Ainsworth BE, Haskell WL, Herrmann SD, et al. 2011 Compendium of Physical Activities. Med Sci Sport Exerc. 2011;43(8):1575–81. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs DR, Ainsworth BE, Hartman TJ, Leon AS. A simultaneous evaluation of 10 commonly used physical activity questionnaires. Med Sci Sports Exerc. 1993;25(1):81–91. [DOI] [PubMed] [Google Scholar]
- 30.Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. [DOI] [PubMed] [Google Scholar]
- 31.Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8(3):389–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du P, Zhang X, Huang C-C, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics; 2010;11(1):587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Story Jovanova O, Nedeljkovic I, Spieler D, Walker RM, Liu C, Luciano M, et al. DNA Methylation Signatures of Depressive Symptoms in Middle-aged and Elderly Persons: Meta-analysis of Multiethnic Epigenome-wide Studies. JAMA psychiatry. 2018;75(9):949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loughin TM. A systematic comparison of methods for combining p-values from independent tests. Comput Stat Data Anal. 2004;47(3):467–85. [Google Scholar]
- 37.Fisher RA. Statistical Methods for Research Workers. Oliver & Boyd; 1925. 336 p. [Google Scholar]
- 38.Stouffer A, Suchman EA, DeVinney LC, Star SA, Williams RMJ. The American soldier, vol 1: Adjustment during army life. Princeton University Press; 1949. 600 p. [Google Scholar]
- 39.Sattelmair J, Pertman J, Ding EL, Kohl HW, Haskell W, Lee I-M. Dose Response Between Physical Activity and Risk of Coronary Heart Disease. Circulation. 2011;124(7):789–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joehanes R, Just AC, Marioni RE, et al. Epigenetic Signatures of Cigarette Smoking. Circ Cardiovasc Genet. 2016;9(5):436–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imgenberg-Kreuz J, Carlsson Almlöf J, Leonard D, et al. DNA methylation mapping identifies gene regulatory effects in patients with systemic lupus erythematosus. Ann Rheum Dis. 2018;77(5):736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross AM, Jaeger PA, Kreisberg JF, et al. Methylome-wide Analysis of Chronic HIV Infection Reveals Five-Year Increase in Biological Age and Epigenetic Targeting of HLA. Mol Cell. 2016;62(2):157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhatt-Wessel B, Jordan TW, Miller JH, Peng L. Role of DGAT enzymes in triacylglycerol metabolism. Arch Biochem Biophys. 2018;655:1–11. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Xu D. Effects of aerobic exercise on lipids and lipoproteins. Lipids Health Dis. BioMed Central; 2017;16(1):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee T-W, Bai K-J, Lee T-I, Chao T-F, Kao Y-H, Chen Y-J. PPARs modulate cardiac metabolism and mitochondrial function in diabetes. J Biomed Sci. 2017;24(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2017;19(2):121–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skender S, Ose J, Chang-Claude J, et al. Accelerometry and physical activity questionnaires - a systematic review. BMC Public Health. 2016;16:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.