Abstract
Purpose
Preterm birth (PTB) is a major cause of neonatal mortality. The vaginal microbiome is associated with PTB, but results vary across racial/ethnic populations. Some evidence suggests gestational age affects this association. We investigated these associations in a novel population, conducting a post hoc analysis assessing if associations differed between women swabbed at different gestational ages.
Methods
We compared vaginal microbiomes from women with PTB (N=25) to a random sample of women with term births (N=100) among participants in the Pregnancy Outcomes, Maternal and Infant Study (PrOMIS), conducted in Lima, Peru. Using DADA2 we identified taxa from 16S DNA sequencing and used Dirichlet multinomial mixture models to group into community state types (CSTs).
Results
If gestational age at sampling was not considered, no CST, (diverse, Lactobacillus-dominated or L. iners-dominated) was associated with PTB. Among women sampled before 12 weeks’ gestation, women with Lactobacillus-dominated CST’s were less likely to have a PTB than those with a diverse CST. Among those swabbed between 12 to 16 weeks’ gestation the reverse was true.
Conclusions
Our study supports previous literature suggesting that what constitutes a healthy vaginal microbiome varies by race/ethnicity. Longitudinal studies are necessary to disentangle affects of vaginal microbiome differences over gestation.
Keywords: Premature Birth, Microbiota, Host-microbial interactions, Molecular epidemiology
Introduction
Preterm birth (PTB) is a major cause of neonatal mortality worldwide (1–3). Infants born preterm are more likely to suffer from respiratory distress syndrome, necrotizing enterocolitis, and intraventricular hemorrhage and developmental delays (2,4,5). Twenty-five to thirty percent of preterm births are attributed to intrauterine infection and subsequent immune response (4,6,7). The most common pathway to intrauterine infection is ascent from the vagina and cervix (4); bacterial vaginosis (BV) is associated with 1.5- to 3-fold increases risk for PTB (4). However, interventions targeting BV have been unsuccessful (6,8).
Next Generation Sequencing technologies make it possible to characterize complex microbial communities associated with PTB (9–12). However, findings from studies deploying these novel methods have been inconsistent, due in part to differences in study design, populations studied, and bioinformatics and statistical analytical techniques (2,13–15). Nonetheless, available evidence suggests that the vaginal microbiome among pregnant women is characterized by increased community stability and decreased community diversity when compared with their non-pregnant counterparts (2,16). For example, Aagaard et al. reported that the Shannon diversity (a measure of species diversity in a community) of the vaginal microbiome decreased with increasing gestational age among pregnant women (16). This observation was corroborated by Stout et al. using a cohort mostly of African American women. However, following a comparison of a predominantly Caucasian cohort to a predominantly African American cohort Callahan et al. concluded that PTB microbiota associations are population dependent (13). Specifically, L. crispatus and all Lactobacillus species occurred less frequently in the American-American cohort, although in both cohorts there was apparent exclusion of Gardnerella vaginalis by L. crispatus.
The patterns of microbiota changes during pregnancy also may be predictive of PTB. Stout et al. showed that women with subsequent PTB had a significantly greater decrease in vaginal microbiome diversity, richness and evenness between the first and second trimester compared to those with term deliveries - by the third trimester, there were no differences in the vaginal microbiome by term status (17). Fettweis et al. found that among women with PTB, taxa associated with BV tended to decrease in abundance throughout pregnancy, while L. crispatus increased. Fettweis et al. also noted that race affected changes over time in the vaginal microbiome during pregnancy, with Caucasian women exhibiting more stable microbiomes, although G. vaginalis increased among Caucasian women with PTB (18). On balance, available evidence suggests that gestational age-specific changes in the vaginal microbiome may be predictive of term status, but these findings vary by race/ethnicity. Whether these alterations are risk factors or risk markers of PTB remains uncertain.
Given the available evidence, we conducted a pilot case-control study, nested within a well-characterized prospective cohort of pregnant Peruvian women, to describe the association of maternal early-pregnancy vaginal microbiome signatures, determined using 16S rRNA gene sequence-based methods, with PTB. Our study adds to the growing literature suggesting modifications of the association between changes in the vaginal microbiota during pregnancy and risk of PTB by race/ethnicity.
Materials and Methods
Study population
The sample for the present study was drawn from participants enrolled in the Pregnancy Outcomes, Maternal and Infant Study (PrOMIS) Cohort who provided vaginal swabs (N=785) between October 2013 and May 2014. Details of the PrOMIS cohort have been described previously (19). Briefly, the study population consists of women attending prenatal care clinics at the Instituto Nacional Materno Perinatal (INMP) in Lima, Peru. Women who initiated prenatal care prior to 16 weeks’ gestation were eligible; if they were younger than 18 years of age, did not speak and read Spanish, or had completed more than 16 weeks’ gestation they were excluded. Enrolled consenting participants were interviewed by trained research personnel using a structured questionnaire to elicit information regarding maternal socio-demographic, lifestyle characteristics, and medical and reproductive histories. All participants provided written informed consent. The institutional review boards of the INMP, Lima, Peru and the Harvard School of Public Health Office of Human Research Administration, Boston, MA approved all procedures used in this study.
Of the 785 selected participants, 35 women spontaneously delivered or had premature rupture of membranes and delivered at less than 37 weeks of gestation. After excluding women with multiple pregnancies, stillbirth, medical complications of pregnancy including pre-eclampsia, a total of 25 PTB cases (gestational age at delivery < 37 completed weeks) remained for microbiome analyses. A total of 529 women had term delivery, single live birth and no medical complications (i.e., pre-eclampsia and no placental abruption). Randomly sampled 100 women who delivered at term (≥37 weeks of gestation; mean ± SD = 39.0 ± 1.0) were selected as term controls (ratio of 4:1). The 100 participants selected for this analysis did not differ when compared to all participants in the PrOMIS cohort.
Sample collection
Maternal vaginal swabs were self-collected using (i) dry Dacron sterile polyester (Starplex Scientific Starswab II Collection and Transport Systems) and (ii) ESwab (Copan Liquid Amies Elution Swab Collection and Transport System) swabs at 9 weeks of gestation, on average. The ESwab swabs were placed in the ESwab tube containing 1ml of modified Liquid Amies solution. The swabs were immediately stored on ice and transported to the laboratory for storage at −80°C prior to processing. The second swab was used to create a vaginal smear on the glass slide for BV analysis.
Definition of preterm birth
We defined PTB according to the American College of Obstetricians and Gynecologists (ACOG) guidelines (20). Gestational age was determined using the last menstrual period and confirmed by ultrasound examination, conducted prior to 20 weeks of gestation. Using information collected from maternal medical records, we categorized PTB cases according to the three pathophysiological groups previously described (i.e., spontaneous PTB, preterm premature rupture of membranes, and medically induced PTB) (21,22). Women who delivered prior to 37 completed weeks of gestation as a result of medical intervention were not eligible for this study.
Sequencing
The 16S rRNA gene V4 variable region PCR primers 515/806 with barcode on the forward primer were used in a 30 cycle PCR using the HotStarTaq Plus Master Mix Kit (Qiagen, USA) under the following conditions: 94°C for 3 minutes, followed by 28 cycles of 94°C for 30 seconds, 53°C for 40 seconds and 72°C for 1 minute, after which a final elongation step at 72°C for 5 minutes was performed. After amplification, PCR products were checked in 2% agarose gel to determine the success of amplification and the relative intensity of bands. Multiple samples were pooled together (e.g., 100 samples) in equal proportions based on their molecular weight and DNA concentrations. Pooled samples were purified using calibrated Ampure XP beads. Then the pooled and purified PCR products were used to prepare DNA libraries by following Illumina TruSeq DNA library preparation protocol. Sequencing was performed at MR DNA (www.mrdnalab.com, Shallowater, TX, USA) on a MiSeq following the manufacturer’s guidelines. Reagent controls using certified DNAfree water were run through library preparation and PCR and did not generate libraries. For quality control, samples submitted for sequencing included a random sample of technical replicates. All library preparation and sequencing were done at MR DNA.
Bioinformatics
Reads were demultiplexed using idemp and then trimmed of adapters, quality-trimmed, and quality-filtered using DADA2. Of 8,811,003 total reads, 8,320,983 passed quality filtering. Error rates were learned on a subset of the samples and used to infer sequence variants separately for each run, then paired ends were merged. Runs were then merged, chimeras were removed, and taxonomy was assigned using the Silva v132 database (23) (Supplemental Methods). Technical replicates were visually examined for differences (Supplemental Figure 2). For a single sample sequenced in triplicate, one replicate did not match the other two replicates and was excluded from further analysis. All other replicates were similar to each other and counts from replicates were summed. We removed any amplicon sequence variants (ASVs) that were not bacterial and collapsed ASVs assigned to the same species together. We conducted diversity analyses on rarefied and unrarefied samples. For the community state type (CST) and ALDEX2 analyses, we filtered phyla occurring in <1 sample on average and ASVs present at < 00.5% relative abundance in every sample.
Statistical analysis
We created Dirichlet multinomial mixture models using R v3.3.2 and the DirichletMultinomial v1.6.0 package (24) to assign all samples to CSTs. We determined the number of CSTs by comparing the Laplace approximation of the negative log models and identifying the point at which an increase in Dirichlet components resulted in minor reductions in model fit. We compared the results of the community state typing to results from a complete linkage hierarchical clustering method using Euclidian distances on the matrix of microbe relative abundances.
We used logistic models to test the association between PTB and CST, controlling for parity and Mestizo ethnicity, as these factors may influence vaginal microbiota and risk of preterm delivery. To test if individual taxa were differentially abundant between term and preterm samples, we used R v3.3.2 and the ALDEX2 package (25). Logistic models and ALDEX2 analyses were performed on the entire sample as well as separately among those sampled before 12-weeks’ gestation (N=55) and those sampled at 12- to 16-weeks’ gestation (N=69). As sensitivity analyses, we 1) included a parameter modeling dispersion for these models and 2) excluded non-Mestizo individuals due to a lack of non-Mestizo cases sampled at 12- to 16-weeks’ gestation (Supplemental Tables 5–8). We calculated measures of multiplicative and additive effect modification (Supplemental Tables 9–11) (26,27). Additionally, we examined key dominating Lactobacillus sp. for patterns of exclusion with Gardnerella sp., as suggested by Callahan et al. (13), stratified by gestational age at sampling.
Results
Study population
At an alpha=0.05, cases and controls were not statistically significant for any patient demographic or microbial features in the unstratified analysis (Table 1). Only 24.0% of cases were educated beyond high school, while 47.0% of controls were (p=0.075). Additionally, 8.00% of cases were underweight in early pregnancy, while no controls were (p=0.066).
Table One:
Prevalence of selected socio-demographic, medical and reproductive health variables by preterm birth status in a sample of Peruvian women
| Term N=100 N(%) or Mean (sd) | Preterm N=25 N(%) or Mean (sd) | p-value | |
|---|---|---|---|
| Community state type: | 0.811 | ||
| Diverse | 50 (50.0%) | 12 (48.0%) | |
| Lactobacillus ASV2 dominated | 26 (26.0%) | 8 (32.0%) | |
| Lactobacillus iners dominated | 24 (24.0%) | 5 (20.0%) | |
| Gestational age at swab: | 0.791 | ||
| < 12 weeks | 54 (54.5%) | 15 (60.0%) | |
| > or = 12 to 16 weeks | 45 (45.5%) | 10 (40.0%) | |
| Shannon Diversity | 1.35 (0.73) | 1.17 (0.72) | 0.283 |
| Maternal age | 27.9 (6.15) | 30.1 (7.37) | 0.165 |
| Maternal age (categories): | 0.808 | ||
| 18 to 19 | 6 (6.00%) | 1 (4.00%) | |
| 20 to 29 | 57 (57.0%) | 12 (48.0%) | |
| 30 to 34 | 19 (19.0%) | 6 (24.0%) | |
| 35 and older | 18 (18.0%) | 6 (24.0%) | |
| Education: | 0.075 | ||
| >12th grade | 47 (47.0%) | 6 (24.0%) | |
| 7th to 12th grade | 48 (48.0%) | 18 (72.0%) | |
| < or = 6th grade | 5 (5.00%) | 1 (4.00%) | |
| Mestizo: | 0.432 | ||
| No | 26 (26.0%) | 4 (16.0%) | |
| Yes | 74 (74.0%) | 21 (84.0%) | |
| Married: | 0.349 | ||
| No | 23 (23.0%) | 3 (12.0%) | |
| Yes | 77 (77.0%) | 22 (88.0%) | |
| Employment: | 0.964 | ||
| No | 41 (41.0%) | 11 (44.0%) | |
| Yes | 59 (59.0%) | 14 (56.0%) | |
| Trouble paying for basics: | 0.754 | ||
| No | 50 (50.0%) | 11 (44.0%) | |
| Yes | 50 (50.0%) | 14 (56.0%) | |
| Planned pregnancy: | 0.349 | ||
| No | 50 (51.0%) | 16 (64.0%) | |
| Yes | 48 (49.0%) | 9 (36.0%) | |
| Early pregnancy BMI: | 0.066 | ||
| <18.5 | 0 (0.00%) | 2 (8.00%) | |
| 18.5–24.9 | 47 (47.0%) | 11 (44.0%) | |
| 25–29.9 | 35 (35.0%) | 6 (24.0%) | |
| > or = 30 | 17 (17.0%) | 5 (20.0%) | |
| Missing | 1 (1.00%) | 1 (4.00%) | |
| Nulliparous: | 1.000 | ||
| Parous | 46 (46.5%) | 12 (48.0%) | |
| Nulliparous | 53 (53.5%) | 13 (52.0%) | |
| Bacterial vaginosis (Hay-Ison criteria): | 0.669 | ||
| I | 15 (15.0%) | 6 (24.0%) | |
| Missing | 3 (3.00%) | 0 (0.00%) | |
| N | 56 (56.0%) | 14 (56.0%) | |
| VB | 26 (26.0%) | 5 (20.0%) |
The second CST’s dominating organism - labeled ASV2 - was identified as either L. acidophilusor L. crispatus by a BLAST search.
Mothers sampled at or after 12- to 16-weeks’ gestation were slightly younger than mothers sampled before 12-weeks’ gestation, with respective mean ages of 26.9 vs 29.6 (p = 0.019). No other differences were statistically significant (Supplemental Table 1). Cases sampled at or after 12-weeks’ gestation were less likely to be educated beyond high school than controls (10.0% vs 53.3%, p= 0.018). The same trend existed in those sampled before 12-weeks’ gestation, but the difference was not statistically significant (p=0.562). All cases among non-Mestizo women had vaginal samples from before 12-weeks’ gestation (Supplemental Table 2).
Microbial diversity
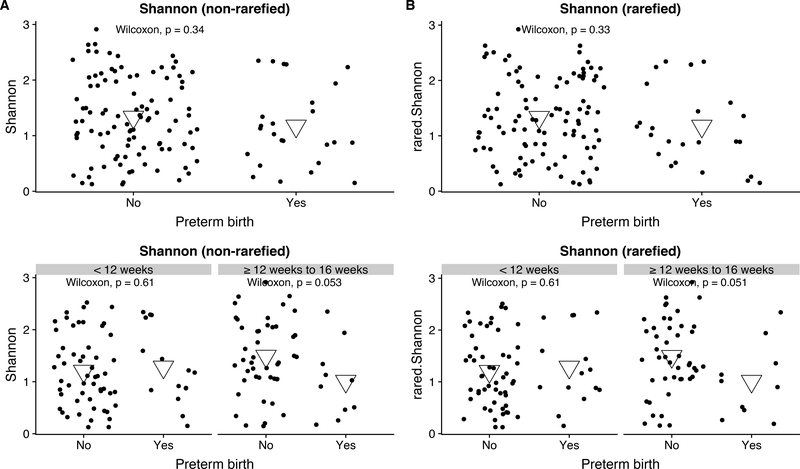
When considered altogether, there were no differences in alpha diversity measures by case status. After stratifying by trimester of microbiome assessment, cases had lower alpha diversity than controls among those sampled at or after 12-weeks’ gestation (respective mean Shannon 1.01 vs 1.49, p=0.053) – regardless of whether samples were rarefied (Figure 1). The same relationship held when limiting the sample to Mestizo women (Supplemental Figure 3). Microbial communities did not cluster together by PTB status or gestational age at sampling on an NMDS plot of Bray-Curtis distances (Supplemental Figure 4).
Figure 1: Alpha diversity of vaginal communities in pregnant women who delivered <37 weeks (n=25) or at term (n=100) both unstratified and stratified by gestational age at vaginal swab (<12 weeks 15 preterm birth, 54 term births; ≥12 weeks 10 preterm births, 45 term births).
Violin plots showing the Shannon diversity of vaginal swabs by term status both when unstratified and stratified by gestational age at gestational swab. Violins show distribution of Shannon diversity when microbial samples have A) not been rarefied to even depth and B) have been rarefied to even depth. Means shown as large, inverted triangles.
Community state types
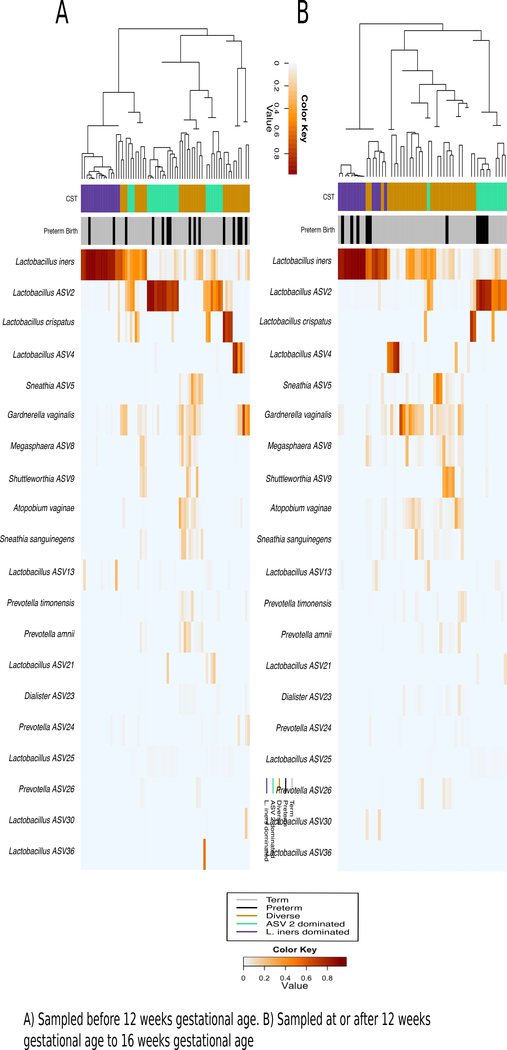
Three CSTs emerged from Dirichilet multinomial modeling. These included 1) a more diverse CST, 2) a CST dominated by a Lactobacillus species (hereafter referred to as Lactobacillus ASV2) and 3) a CST dominated by L. iners (Figure 2). Lactobacillus ASV2 was identified as either L. acidophilus or L. crispatus by a BLAST search.
Figure 2: Dendogram clustering method compares well to CST clustering; samples cluster by CST and preterm status in swabs from at or after 12 weeks to 16 weeks’ gestational age but only by CST in swabs taken before 12 weeks’ gestational age.
Heatmap showing correlations between bacterial oligotypes and individual samples, with a dendogram derived from a Euclidian matrix clustering technique on the left-hand side. Dendogram clustering is in accordance with Dirichlet multinomial derived community state types (colored bar under the dendrogram) in both trimesters of sampling, however, swabs only cluster by preterm status (shown in black on bar directly on the heatmap) in the second trimester swabs.
Microbial characteristics by subject and swab characteristics
In the unstratified sample, only classification using the Hay-Ison criteria was associated with CST (p<0.001), with more individuals in the diverse CST classified as intermediate or BV (Supplemental Table 3). The same association with Hay-Ison criteria existed in both strata of gestational age at swab. Among those sampled at or after 12-weeks gestation, more unplanned pregnancies existed in the L. iners-dominated CST than in other CSTs (p=0.01) (Supplemental Table 4).
Association with PTB
Overall, no CST was associated with PTB in crude or adjusted logistic models (Table 2). When stratified by timing of microbiome assessment, there was a marginal association (p<0.10) with the Lactobacillus-ASV2-dominated CST in the fully adjusted (OR 4.65, 95% CI: (0.81, 29.96)) model (Table 3). An effect modification analysis suggests the presence of effect-measure modification on the multiplicative (Ratio of ORs, Lactobacillus ASV2 dominated vs diverse CST (95% CI): 10.41 (1.21, 101.18); p=0.04, L. iners dominated vs diverse CST (95% CI): 9.47 (0.86, 129.05); p=0.07) but not additive scale (Supplemental Tables 9–11).
Table 2:
Results of logistic regression for preterm birth status (Odds ratios and 95% CI) in a sample of Peruvian women
| Model 1a OR (95% CI)d 25 cases / 100 controls | Model 2b OR (95% CI)d 25 cases / 99 controls | Model 3c OR (95% CI)d 25 cases / 99 controls | |
|---|---|---|---|
| Lactobacillus ASV2e dominated (reference Diverse community) | 1.28 (0.45, 3.5) | 1.25 (0.43, 3.49) | 1.27 (0.44, 3.56) |
| L. iners dominated (reference Diverse community) | 0.87 (0.25, 2.64) | 0.85 (0.25, 2.59) | 0.87 (0.25, 2.68) |
Unadjusted
Adjusted for parity status (one individual missing parity status)
Adjusted for parity and Mestizo status (one individual missing parity status)
Profile likelihood-based confidence intervals reported
The second CST’s dominating organism - labeled ASV2 - was identified as either L. acidophilus or L. crispatus by a BLAST search.
Table 3:
Results of logistic regression for preterm birth status stratified by gestational age at vaginal swab (Odds ratios and 95% CI) in a sample of Peruvian women
| Sampled < 12 weeks’ gestation | Sampled >12 weeks to 16 weeks’ gestation | |||||
|---|---|---|---|---|---|---|
| Model 1a OR (95% CI)d 15 cases / 54 controls | Model 2b OR (95% CI)d 15 cases / 53 controls | Model 3c OR (95% CI)d 15 cases / 53 controls | Model 1a OR (95% CI)d 10 cases / 45 controls | Model 2b OR (95% CI)d 10 cases / 45 controls | Model 3c OR (95% CI)d 10 cases / 45 controls | |
| Lactobacillus ASV2 dominatede (reference Diverse community) | 0.49 (0.12, 1.78) | 0.49 (0.12, 1.82) | 0.49 (0.11, 1.81) | 5.33T (0.97, 32.91) | 5.06T (0.9, 31.91) | 4.65T (0.81, 29.96) |
| L. iners dominated (reference Diverse community) | 0.33 (0.05, 1.54) | 0.31 (0.04, 1.45) | 0.31 (0.04, 1.45) | 2.80 (0.45, 17.45) | 2.71 (0.43, 17.02) | 3.48 (0.52, 23.92) |
p<0.10
Unadjusted
Adjusted for parity status (one individual missing parity status; one individual missing gestational age at swab)
Adjusted for parity and Mestizo status (one individual missing parity status; one individual missing gestational age at swab)
Profile likelihood-based confidence intervals reported
The second CST’s dominating organism - labeled ASV2 - was identified as either L. acidophilus or L. crispatus by a BLAST search.
Alternative clustering technique
In dendrograms showing clustering and accompanying heatmaps showing composition, CSTs and the hierarchical clustering technique clustered samples similarly (Figure 2). Samples collected during 12- to 16-weeks’ gestation – but not those collected before 12-weeks gestation – clustered by CST and preterm status.
Joint distribution of key taxa
We examined the joint distribution of Lactobacillus species with Gardnerella species. Consistent with the analysis by Callahan et al., we observed that Gardnerella species displayed exclusionary patterning with L. crispatus and a co-occurring pattern with L. iners (13). In our sample, Lactobacillus ASV2 appeared to have an exclusionary pattern with Gardnerella (Supplemental Figure 7).
Individual taxa
No taxa associated with PTB in unstratified or stratified analysis after Benjamini-Hochberg correction for multiple testing. Prior to correction for multiple testing, two ASVs were associated (p<0.05) in the first trimester (Supplemental Figures 8–9).
Discussion
We conducted a 16S rRNA gene taxonomic screen characterizing the vaginal microbiome during the first and early second trimester of gestation among 25 PTB cases and 100 term controls sampled from a well characterized cohort of pregnant Peruvian women. We identified three community states: one dominated by L. iners, one by another Lactobacillus species and one more diverse. Vaginal microbial communities did not associate with PTB overall. The direction of associations between Lactobacillus-dominated communities and PTB differed between women sampled at different gestational ages. The finding that vaginal microbiomes do not associate with PTB is consistent with some results of studies conducted among non-Caucasian women (2,13). Thus, our results support the hypothesis that Lactobacillus-dominated community types are not universal indicators of a healthy pregnant vaginal microbiome in all populations.
The types of vaginal microbial communities in our sample are similar to those reported elsewhere, including communities dominated by Lactobacillus species and more diverse communities. The Peruvian women in our sample have vaginal microbiota more similar to African American and Hispanic US women than to Caucasian or Asian women (28,29).
In the unstratified analysis, we found no association between vaginal communities and PTB. This contradicts previous literature suggesting a protective association between Lactobacillus-dominated vaginal communities and PTB (13–15). However, studies among non-Caucasian women have reported either weaker or null associations of Lactobacillus-dominated communities with PTB (2,13,30). In a primarily African American cohort, Romero et al. reported no associations of microbial CSTs with PTB (2). Callahan et al. found no statistically significant differences in relative abundances of Lactobacillus or Gardnerella by term status in a cohort of predominately African American women although they did report statistically significant difference of L. crispatus (13). Among a mixed cohort of both African American and Caucasian women, Fettweis et al. reported term women had higher abundances of L. crispatus than preterm women, but when stratified by race, these differences were greater among Caucasian women than among African American women (18). Vaginal microbiota and PTB rates differ by race and geographic locale; the association between the vaginal microbiota and PTB also appears to differ, and race/locale should be considered in evaluations of this association.
In our study, Lactobacillus-dominated communities had an inverse association with PTB among women swabbed before 12 weeks’ but a direct association among those swabbed at or after 12 weeks’ gestation. We observed effect modification on the multiplicative scale. These findings fit with observations in longitudinal studies – for example, Stout et al. found greater decreases in vaginal microbiome diversity over pregnancy in women with PTB than term birth (17) and Fettweis et al. found increases in L. crispatus abundance over the course of pregnancy among African American women who delivered preterm (18). However, our analysis was post-hoc, taking advantage of variation in timing of vaginal swab collection and we lacked repeated measures of the vaginal microbiome over pregnancy. Therefore, we cannot disentangle causal from noncausal mechanisms of the observed effect heterogeneity. It is possible that the women sampled before 12 weeks’ are not exchangeable with those sampled at or after 12 weeks’ gestation. Although we controlled for important confounding factors, we cannot exclude the possibility of residual confounding by unmeasured variables such as administration of vaginal progesterone during pregnancy or placement of cervical cerclage.
Our study has other limitations that should also be considered. Our negative controls had very low biomass and were not sequenced. Thus, we cannot assess low-level contaminants. However, low-level contamination is unlikely to have severely affected our CSTs, which were characterized by typical, high-abundance vaginal organisms. Additionally, we could not confidently assign a species level identification to the Lactobacillus sequence dominating our second community. Confidence intervals in the stratified analysis were large, due in part to the relatively small sample size – sample size may have impacted our power to detect differences. Future studies exploring the association between serial measures of vaginal microbiome across pregnancy and risk of PTB are warranted.
Our study has several strengths. First, our study was conducted in a well-characterized Peruvian cohort, and is one of only a handful of such studies in Latin America; the results highlight similarities and differences in pregnant vaginal microbiomes in different countries. Second, the determination of vaginal microbiomes collected during the first and early-second trimester of gestation served to clarify the temporal relationship between changing microbiome community characteristics and subsequent risk of PTB. Third, our use of high-throughput 16S rRNA gene sequence-based methods for microbial profiling allowed for simultaneously investigating the entire microbial community.
In conclusion, in this cohort of Peruvian women, a Lactobacillus-dominated CST did not appear protective for PTB. We found no association between microbial communities and PTB but did observe potential effect heterogeneity between vaginal communities and PTB by gestational age at microbiome assessment. Our findings may indicate the importance of considering race/ethnicity, geographic locale, and timing of vaginal sampling in studies of PTB and vaginal microbiomes.
Supplementary Material
Acknowledgements and Funding
This research was supported by an award from the Eunice Kennedy Shriver Institute of Child Health and Human Development (R01-HD-059835).
Paper Presentation Information: An earlier version of the work was presented at the 2018 American Epidemiology Society Meeting, Baltimore, Maryland.
List of Abbreviations
- PTB
Preterm birth
- BV
Bacterial vaginosis
- 16S rRNA
16 subunit ribosomal ribonucleic acid
- PrOMIS
Pregnancy Outcomes, Maternal and Infant Study
- INMP
Instituto Nacional Materno Perinatal
- CST
Community state type
- ASV(s)
Amplicon Sequence Variant(s)
Footnotes
Disclosure Statement: The authors have no conflicts of interest to disclose
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller A-B, Narwal R, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012. June;379(9832):2162–72. [DOI] [PubMed] [Google Scholar]
- 2.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Bieda J, et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome [Internet]. 2014. May;2(1):18 Available from: 10.1186/2049-2618-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frey HA, Klebanoff MA. The epidemiology, etiology, and costs of preterm birth. Semin Fetal Neonatal Med. 2016. April;21(2):68–73. [DOI] [PubMed] [Google Scholar]
- 4.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008. January;371(9606):75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.for Disease Control C, Prevention. CDC. Preterm Birth [Internet]. 2016. Available from: Available [Google Scholar]
- 6.Lamont RF, Nhan-Chang C-L, Sobel JD, Workowski K, Conde-Agudelo A, Romero R. Treatment of abnormal vaginal flora in early pregnancy with clindamycin for the prevention of spontaneous preterm birth: a systematic review and metaanalysis. Am J Obstet Gynecol [Internet]. 2011;205(3):177–90. Available from: 10.1016/j.ajog.2011.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polettini J, Cobo T, Kacerovsky M, Vinturache AE, Laudanski P, Peelen MJCS, et al. Biomarkers of spontaneous preterm birth: a systematic review of studies using multiplex analysis. J Perinat Med. 2017. January;45(1):71–84. [DOI] [PubMed] [Google Scholar]
- 8.Brocklehurst P, Gordon A, Heatley E, Milan SJ. Antibiotics for treating bacterial vaginosis in pregnancy. Cochrane Database Syst Rev. 2013. January;(1):CD000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foxman B, Wen A, Srinivasan U, Goldberg D, Marrs CF, Owen J, et al. Mycoplasma, bacterial vaginosis-associated bacteria BVAB3, race, and risk of preterm birth in a high-risk cohort. Am J Obs Gynecol. 2014. March;210(3):226.e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mysorekar IU, Cao B. Microbiome in parturition and preterm birth. Semin Reprod Med. 2014. January;32(1):50–5. [DOI] [PubMed] [Google Scholar]
- 11.Petricevic L, Domig KJ, Nierscher FJ, Sandhofer MJ, Fidesser M, Krondorfer I, et al. Characterisation of the vaginal Lactobacillus microbiota associated with preterm delivery. Sci Rep [Internet]. 2014;4:5136 Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4038809/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox C, Eichelberger K. Maternal microbiome and pregnancy outcomes. Fertil Steril. 2015. December;104(6) :135 8–63. [DOI] [PubMed] [Google Scholar]
- 13.Callahan BJ, DiGiulio DB, Goltsman DSA, Sun CL, Costello EK, Jeganathan P, et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc Natl Acad Sci [Internet]. 2017;114(37):9966–71. Available from: http://www.pnas.org/content/114/37/9966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A. 2015. September; 112(3 5):11060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kindinger LM, Bennett PR, Lee YS, Marchesi JR, Smith A, Cacciatore S, et al. The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome [Internet]. 2017. January;5(1):6 Available from: 10.1186/s40168-016-0223-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aagaard K, Riehle K, Ma J, Segata N, Mistretta T-A, Coarfa C, et al. A Metagenomic Approach to Characterization of the Vaginal Microbiome Signature in Pregnancy. PLoS One [Internet]. 2012;7(6):e36466−−. Available from: 10.1371/journal.pone.0036466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stout MJ, Zhou Y, Wylie KM, Tarr PI, Macones GA, Tuuli MG. Early pregnancy vaginal microbiome trends and preterm birth. Am J Obs Gynecol. 2017. September;217(3):356.e1–356.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, et al. The vaginal microbiome and preterm birth. Nat Med [Internet]. 2019;25(6):1012–21. Available from: 10.1038/s41591-019-0450-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrios YV, Sanchez SE, Nicolaidis C, Garcia PJ, Gelaye B, Zhong Q, et al. Childhood Abuse and Early Menarche among Peruvian Women. J Adolesc Health [Internet]. 2015;56(2):197–202. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4306809/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The American College of Obstetricians and Gynecologists. Management of Preterm Labor: Practice Bulletin Number 171. 2016. [Google Scholar]
- 21.Savitz DA, Blackmore CA, Thorp JM. Epidemiologic characteristics of preterm delivery: Etiologic heterogeneity. Am J Obstet Gynecol [Internet]. 1991;164(2):467–71. Available from: 10.1016/S0002-9378(11)80001-3 [DOI] [PubMed] [Google Scholar]
- 22.Williams M, Mittendorf R, G. Stubblefield P, Lieberman E, Schoenbaum S, R. Monson R Cigarettes, Coffee, and Preterm Premature Rupture of the Membranes. Vol. 135, American journal of epidemiology. 1992. 895–903 p. [DOI] [PubMed] [Google Scholar]
- 23.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies JGF. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. 2013. p. D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan M DirichletMultinomial: Dirichlet-Multinomial Mixture Model Machine Learning for Microbiome Data. 2017. [Google Scholar]
- 25.Fernandes AD, Macklaim JM, Linn TG, Reid G, Gloor GB. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS One. 2013;8(7):e67019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathur MB, VanderWeele TJ. R Function for Additive Interaction Measures. Epidemiology [Internet]. 2018. January;29(1):e5–6. Available from: https://www.ncbi.nlm.nih.gov/pubmed/28901974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.VanderWeele T Explanation in Causal Inference : Methods for Mediation and Interaction [Internet]. New York, UNITED STATES: Oxford University Press, Incorporated; 2015. Available from: http://ebookcentral.proquest.com/lib/umichigan/detail.action?docID=1920742 [Google Scholar]
- 28.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci [Internet]. 2011;108(Supplement 1):4680–7. Available from: http://www.pnas.org/content/108/Supplement_1/4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P, et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 2007. June;1(2):121–33. [DOI] [PubMed] [Google Scholar]
- 30.Hyman RW, Fukushima M, Jiang H, Fung E, Rand L, Johnson B, et al. Diversity of the vaginal microbiome correlates with preterm birth. Reprod Sci. 2014. January;21(1):32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.