Abstract
This Viewpoint addresses the question of whether, after years of declining use, the time is ripe for renewed investigations into the aminoglycoside class of antibiotics for development of novel therapeutic agents for the treatment of drug-resistant bacterial infections, particularly of the Gram-negative type. The reasons underlying the decline in use of the aminoglycosides are briefly considered and found to be outweighed by the ever-increasing clinical need for improved antibacterials with which to combat modern day multidrug resistant pathogens. The potential of the aminoglycosides builds on their well-established PK/PD, mechanisms of action, toxicity, and resistance, and the extensive existing SAR databases which permit rational, informed drug design. When coupled with the power of modern synthetic organic chemistry and improved funding scenarios, these multiple attributes open the door for the development of structurally novel, potent and less toxic aminoglycosides to address the pressing societal problem of MDR infectious disease.
Keywords: Multidrug resistant infections, aminoglycoside modifying enzymes, ribosomal methyltransferases, NDM-1, ototoxicity, orphan disease
1. The problem
Aminoglycoside antibiotics (AGAs) are listed by the WHO as critically important antimicrobials for human therapy. Their high efficacy, broad-spectrum antibacterial activity in combination with unmatched rapid bactericidal potency, lack of drug-related allergy, little protein binding and minimal drug metabolism, absence of interaction with other pharmaceutical agents and with the host’s intestinal microbiome, are features that combine to make aminoglycosides a potent and powerful choice for treatment of serious infections by Gram-negative pathogens. Profound clinical experience with this antibiotic class, accumulated since their introduction in the 1950’s, and correspondingly predictable absorption, distribution, metabolism, and excretion (ADME) mitigate the risk in new AGA development
The development of potent antibacterial compound classes in the 1970’s and 1980’s (3rd generation cephalosporins, carbapenems, and fluoroquinolones) coupled with concerns about toxicity and the need for IV administration, shifted the interest away from the AGAs, and reduced their application in the clinic. However, the global emergence of antibiotic drug resistance has nullified our arsenal of potent broad-spectrum antibacterials leading to an ever-increasing global public health crisis. Multidrug-resistant Gram-negative pathogens are of particular concern as they have evolved resistance to all major antibiotic classes including carbapenems, 3rd and 4th generation cephalosporins, fluoroquinolones, and aminoglycosides. Widespread antimicrobial resistance now limits the therapeutic options for these pathogens to only a very few antibiotics. Last resort drugs such as tigecycline and colistin are however compromised by significant adverse side effects and plagued by limited efficacy, and are increasingly challenged by emerging resistance. Infectious diseases, caused by multidrug-resistant (MDR) and extremely drug-resistant (XDR) pathogens, carbapenemase-producing Enterobacteriaceae (CPE) and carbapenemase-producing Acinetobacter baumannii (CPA), are a major cause of morbidity and mortality worldwide. We face the evolution of pathogens for which no effective antimicrobial therapy is possible. The description of the NDM-1 strain in 2010 was probably the single most discomforting observation in this respect.1 Corresponding strains have mainly been observed in Asia, e.g. India, China, Pakistan, the Middle East, and the South Americas. For the time being North America and Europe have been little hit by NDM-1 strains, but a recent outbreak of NDM-1 in Tuscany, Italy, involving 7 hospitals and 350 patients2 testifies to the rapid and global spread of multidrug-resistant Gram-negative pathogens, not obeying artificial state or natural continental borders.
2. Aminoglycoside antibiotics
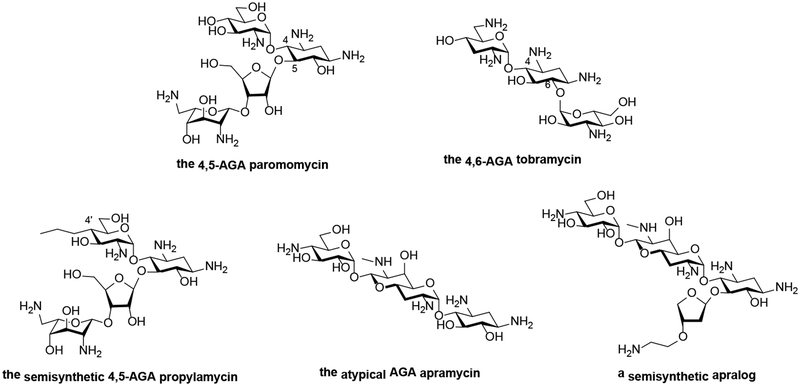
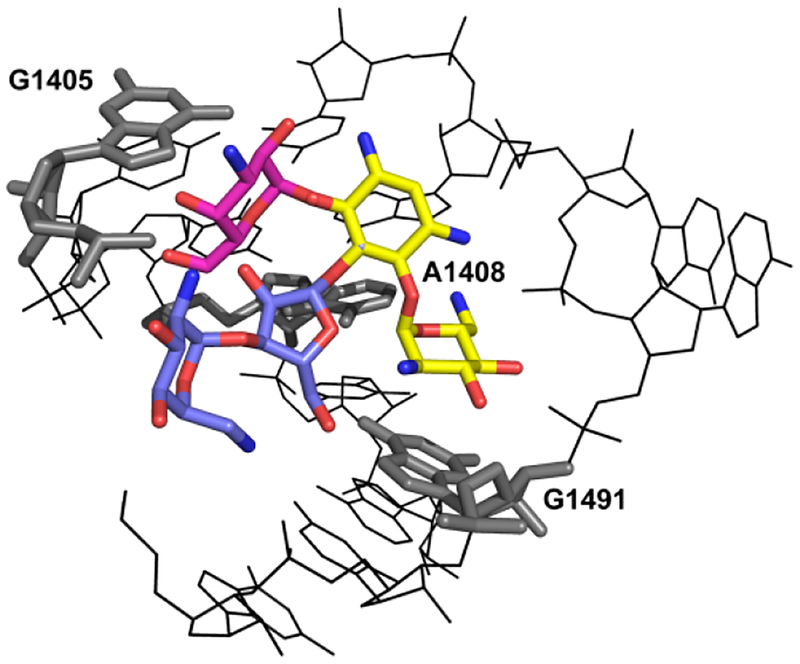
Classical AGAs are exemplified by the 2-deoxystreptamines which share a common neamine core, to which additional sugars are attached at positions 5- or 6-, resulting in compounds categorized as 4,5- or 4,6-aminoglycosides (Fig. 1A). AGAs inhibit the essential process of protein synthesis by targeting the bacterial ribosome, one of the most effective targets in drug history. AGAs bind to helix 44 of 16S rRNA, which is part of the decoding A-site of the small ribosomal subunit, resulting in mRNA misreading and translocation inhibition (Fig. 1B).3, 4 All 2-deoxystreptamines in clinical use for treatment of systemic bacterial infections are 4,6-aminoglycosides.
Figure 1.
A Chemical structures of 4,5-, and 4,6-aminoglycosides, of propylamycin, apramycin, and an apralog.
B View of the three-dimensional structure of the A-site loop within rRNA helix 44, the drug binding pocket, complexed with 4,5- and 4,5-disubstituted AGAs: the common neamine core is denoted in yellow; ring III of the 4,6-compounds (kanamycin) is denoted in red; rings III and IV of the 4,5-compounds (paromomycin) are denoted in blue. Indicated are the polymorphic residues (1408, 1491) and G1405, the target for methylation by RMTases.
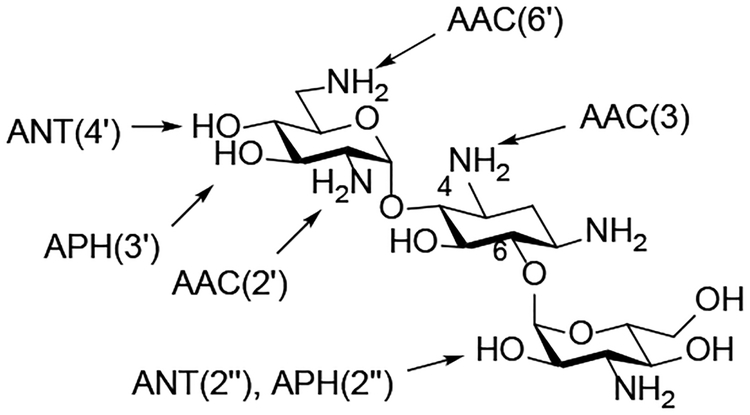
C AMEs Acting on the 4,6-AGA Kanamycin B
3. Aminoglycoside toxicity
Selectivity is of particular concern for antibiotics targeting protein synthesis as the ribosome is present in all three domains of life, and aminoglycosides are no exception. Recent evidence converges on mitochondrial function as a key element in aminoglycoside toxicity, in particular ototoxicity.5 Thus, the picture emerges that aminoglycoside toxicity is mainly mechanism-of-action-related with limitations in ribosomal target selectivity the main denominator here. Fortunately, AGA pharmacokinetics are notably different for the human host versus the bacterial pathogen. Bactericidal activity is peak concentration-dependent while aminoglycoside transport into eukaryotic cells is limited and readily saturated, with host toxicity being mainly trough level-dependent. Consequently, AGA toxicity can be minimized and bactericidal activity enhanced by administration of a single daily dose, combining high peak concentrations with low trough levels, for a period of not exceeding 10–14 days.
4. Aminoglycoside resistance
Clinically relevant AGAs are increasingly challenged by emerging resistance. Two general mechanisms exist which confer resistance to aminoglycoside antibiotics: aminoglycoside-modifying enzymes (AMEs) and target-modifying enzymes (ribosomal methyltransferases, RMTases).
AMEs modify specific hydroxy or amino substituents on the compound’s scaffold and are comprised of three different families: aminoglycoside acetyltransferases (AACs), aminoglycoside phosphotransferases (APHs), and aminoglycoside nucleotidyl transferases (ANTs). AME nomenclature consists of a three letter code to identify the activity (AAC, APH, ANT), followed by a number in parentheses that specifies the site of modification (e.g. 3, 6’, 3’), and a roman number that describes – if present – the isoform (e.g. I, IIa), see Fig. 1C for a scheme of AMEs.6
RMTases modify the drug binding pocket by methylation, in particular at N7 of G1405. While AMEs have been known for more than 50 years, RMTases were first described in a clinical pathogen some 15 years ago.7 A number of enzymes, all carrying out the same modification of G1405 have since been reported in a variety of Gram-negative species, including ArmA, RmtA, RmtB, RmtC, RmtD, RmtE, RmtF, RmtG and RmtH. Two features of RMTases are particularly relevant: without exception they affect all 4,6-AGAs, and they are frequently associated with carbapenemases and CTX-M-type ESBLs.8
5. Possibilities for drug development
With many hundreds of semi-synthetic AGAs described in the open and patent literature over more than half a century, there is an extensive recently reviewed knowledge base of chemical methods for their manipulation.9 This knowledge base is a useful guide to modern day medicinal chemists seeking to develop next generation compounds, but it is mostly limited to the regioselective functionalization of amino and hydroxyl groups through the exploitation of protecting group chemistry.10 However, it is the combination of this know-how with the power of modern synthetic organic chemistry that enables the development of novel structures and compositions of matter such as will be required to generate commercially viable next generation compounds.
The many hundreds of described AGAs also provide much data with which to construct structure activity relationships, whether directed at activity, toxicity, or resistance.11 This SAR data is all the more powerful when informed by up-to-date understanding of AGA interactions with the drug binding pockets on the target ribosomes derived from state of the art X-ray crystallographic and site-directed mutagenesis studies.
Optimization of target selectivity, which is predictive of AGA toxicity, is guided by cell-free translation assays using wild-type bacterial ribosomes, mutant recombinant bacterial ribosomes carrying single point mutations characteristic of eukaryotic residues in small subunit rRNA,12 and genetically engineered chimeric hybrid ribosomes, which incorporate the complete eukaryotic rRNA h44 of either mitochondrial or cytosolic ribosomes.13
Coupled with in-vitro models to rapidly assess target selectivity, and broad MIC testing of activity in resistant and wild-type bacterial strains, the substantial existing SAR and chemical know-how permit the informed design, synthesis, and evaluation of next generation AGAs with relatively high levels of confidence.
5.1. 4,5-aminoglycosides
The 4,5-aminoglycosides are an excellent choice of parental AGAs for further development as – in contrast to 4,6-aminoglycosides – they are not affected by RMTases. Recent developments have resulted in the discovery of novel modifications to the 4,5-AGA framework that reduce ototoxicity in ex-vivo and in-vivo models, while at the same time suppressing the action of common AMEs, and without compromising levels of antibacterial activity.14–16
Most pertinently, the recent discovery of propylamycin,17 the 4’-deoxy-4’-C-propyl derivative of paromomycin (Fig 1A) with increased antibacterial activity and reduced ototoxicity, clearly indicates that, in spite of the large numbers of AGAs prepared in past discovery programs there remains considerable scope for improvement. This is especially the case when the power of modern synthetic chemistry is applied to make novel modifications, as with propylamycin.
5.2. Plazomicin
Plazomicin – 1-N-(4-amino-2S-hydroxybutyroyl)-6’-N-(2-hydroxyethyl)-sisomicin – is a AGA developed by Achaogen and introduced to the market as Zemdri in summer 2018. It is a prime example of exploitation of the chemical knowledge and existing SAR to develop a novel compound with reduced susceptibility to AME resistance determinants by the combination of two well-established modifications into a single compound.18
Unfortunately, Zemdri failed commercially because it was only approved for the treatment of complicated urinary tract infections (cUTI), which at present has only a limited patient population in North America. Approval for treatment of the more widespread blood stream infections (BSI), which offers broader prospects for commercial success, was withheld on the basis of a prematurely terminated clinical trial. Additionally, as a classical 4,6-AGA, Zemdri is not active against RMTase-carrying pathogens including the NDM-1 and related strains of CREs.
5.3. Apramycin
Apramycin is a unique member of the 2-deoxystreptamine class of aminoglycoside antibiotics characterized by a mono-substituted 2-deoxystreptamine ring that carries an unusual bicyclic moiety (Fig. 1A). Apramycin shows broad-range antibacterial activity, including Gram-negative pathogens. Because of its unique structure apramycin evades almost all AMEs found in MDR and XDR Gram-negative bacteria, including the RMTases frequently encountered in carbapenemase-producing clinical isolates, e.g. CPE, CPA.19–21 The only resistance mechanism that confers non-susceptibility to apramycin is the aminoglycoside 3-N-acetyltransferase subtype IV, AAC(3)-IV.21
As for all 2-deoxystreptamine AGAs apramycin binds to the ribosomal decoding A-site, i.e., h44 in small subunit rRNA. However, and in contrast to AGAs currently in clinical practice, chimeric ribosomes with humanized small subunit rRNA h44 revealed that apramycin exhibits exquisite selectivity for the bacterial ribosome over mitochondrial ribosomes. This was reinforced by ex-vivo cochlea explant and in-vivo ototoxicity studies, which testified to a low ototoxic potential of apramycin.22
Despite the widespread agricultural use of apramycin in Asia, clinical isolates from a global collection of roughly 1,100 genetically diverse CPE strains, including more than 200 clinical isolates from Asia, have demonstrated the scarcity of AAC(3)-IV.21 The lack of coexistence of carbapenemase and AAC(3)-IV production has important and far-reaching clinical implications, as a combination of a carbapenem with apramycin as empirical treatment for severe bacterial infections, including sepsis, BSI, hospital-acquired and ventilator-associated pneumonia (HAP, VAP), can be expected to provide broad and efficacious coverage of Gram-negative pathogens, including obnoxious carbapenemase-producing MDR and XDR strains, the most challenging and still unmet clinical need.
On the basis of these characteristics and extensive preclinical profiling, including pharmacokinetics/pharmacodynamics and animal in-vivo efficacy and toxicity data, apramycin received clearance from the European authorities for human evaluation, ultimately entering Phase 1 First-in-Human studies in October 2019. This development of apramycin by the Swiss biotech start-up Juvabis AG in collaboration with the ND4BB European Gram-Negative Antibacterial Engine (ENABLE) Consortium, funded by the Innovative Medicines Initiative (IMI), serves to highlight the importance of public-private partnerships in the early stages of antibacterial drug development. Needless to say apramycin also provides an excellent framework on which to elaborate future generations of AGAs, such as the apralogs (Figure 1A), that retain its almost ideal properties and at the same time do not fall victim to the AAC(3)-IV resistance determinant.23
6. Clinical development
The main problem in drug development is not only activity, but toxicity – the nightmare scenario of having a billion dollar investment wasted due to unforeseen toxicity problems (mainly off-target), which are only recognized in clinical phase 3 or even later (recent examples: phase 3 stop for Murepavadin in 2019, withdrawal of Telithromycin from the market in 2007). Fortunately, for AGAs, in-vivo efficacy can largely be predicted on the basis of in-vitro MIC data, owing to the wealth of PK/PD data available for this compound class. Indeed, AGAs offer unique advantages, unmet by any other antibiotic class. Toxicity is largely mechanism-of-action-related and reflects limitations in target selectivity. Cell-free ribosomal translation assays using chimeric hybrid ribosomes can be used early in the drug development process to select for compounds with a suitable target selectivity profile, followed by subsequent testing of corresponding compounds in established preclinical models of toxicity (in-vitro cochleotoxicity using cochlear explants, in-vivo nephrotoxicity in mice or rats). Aminoglycosides offer great freedom to operate in terms of chemical synthesis, as numerous chemical substitutions can be done without resulting in unforeseen off-target effects. In addition, the pharmacology of this class of compounds is well-known facilitating rapid progress to clinical trials and all-in-all the risk of having to withdrawn an active compound in clinical phase 3 or even later due to unforeseen side-effects is negligible.
7. Business models of antibiotic drug development
Science is one thing, but bringing a drug into the market is another. In the end: isn’t it all about bringing a compound into the market for patient benefit?
Regrettably, not just for AGAs, but for antibiotic drug development as a whole, neither big pharma nor venture capitalists show any interest in developing focused antibacterials targeting multidrug-resistant pathogens. Thus, while there is a great societal need and antibiotic resistance is a frequent topic at G20 summits (see also Davos declaration), the reality is that the main players guiding pharma development remain absent, and this for a simple reason: the profit margin is too low to be of interest. Until this Catch 22 situation is resolved antibiotic drug development will depend mainly upon academic groups working in partnership with public consortia e.g. ENABLE (with its demonstrated success taking apramycin into Phase I), Carbex, Barda, and GARDP, albeit these latter organizations are not readily accessible to academic groups because of their business models.
Different business models are needed. Given the yet relatively low numbers of “non-treatable” multidrug-resistant Gram-negative pathogens in the North Americas and Europe, we suggest that a Target Product Profile (TPP) “life-threatening multidrug-resistant pathogen” should qualify as an orphan disease. This would avoid the problems with the limited reimbursement currently available for antibiotics and would leverage development of antimicrobials interesting for start-ups and investors alike. In addition, incentives, e.g. successful development and FDA approval of an antibacterial compound addressing “multidrug-resistant pathogens” may, in exchange for extended patent life of a blockbuster drug, render antibacterial development more attractive for big pharma.
Conclusion
The ever-increasing societal threat of widespread multidrug-resistant Gram-negative bacterial infections, the inadequacy of current therapeutics, and the numerous advantages of the aminoglycoside antibiotics together ensure that it is time for resurrection of aminoglycosides as antibacterials. Modern synthetic chemistry enables development novel classes of aminoglycosides that display enhanced activity, while avoiding inactivation by multiple resistance determinants and minimizing toxicity. The stagnant funding climate for antibacterial development however remains a hurdle to success and is in urgent need of attention.
Acknowledgements
Work in the authors’ laboratories is supported by the European Commission (ENABLE, JPIAMR), the National Institutes of Health, USA (AI123352), and the Swiss National Science Foundation.
Footnotes
Transparency declaration
The authors are co-founders of and have an equity interest in Juvabis AG, a biotech start-up operating in the area of aminoglycoside antibiotics.
References
- 1.Kumarasamy KK; Toleman MA; Walsh TR; Bagaria J; Butt F; Balakrishnan R; Chaudhary U; Doumith M; Giske CG; Irfan S; Krishnan P; Kumar AV; Maharjan S; Mushtaq S; Noorie T; Paterson DL; Pearson A; Perry C; Pike R; Rao B; Ray U; Sarma JB; Sharma M; Sheridan E; Thirunarayan MA; Turton J; Upadhyay S; Warner M; Welfare W; Livermore DM; Woodford N, Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis 2010, 10, 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ECDC European Centre for Disease Prevention and Control. Regional Outbreak of New Delhi Metallo-beta-lactamase Producing Carbapenen-resistant Enterobacteriaceae, Italy, 2018–2019; ECDC: Stockholm, 2019. [Google Scholar]
- 3.Davies J; Gorini L; Davies BD, Misreading of RNA Codewords Induced by Aminoglycoside Antibiotics. Mol. Pharmacol 1965, 1, 93–106. [PubMed] [Google Scholar]
- 4.Cabañas MJ; Vázquez D; Modolell J, Inhibition of Ribosomal Translocation by Aminoglycoside Antibiotics. Biochem. Biophys. Res. Comm 1978, 83, 991–997. [DOI] [PubMed] [Google Scholar]
- 5.Hobbie SN; Akshay S; Kalapala SK; Bruell C; Shcherbakov D; Böttger EC, Genetic Analysis of Interactions with Eukaryotic rRNA Identify the Mitoribosome as Target in Aminoglycoside Ototoxicity. Proc. Natl. Acad. Sci., USA 2008, 105, 20888–20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vakulenko SB; Mobashery S, Versatility of Aminoglycosides and Prospects for Their Future. Clin. Microbiol. Rev 2003, 16, 430–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galimand M; Courvalin P; Lambert T, Plasmid-Mediated High-Level Resistance to Aminoglycosides in Enterobacteriaceae Due to 16S rRNA Methylation. Antimicrob. Agent. Chemother 2003, 47, 2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doi Y; Wachino JI; Arakawa Y, Aminoglycoside Resistance: The Emergence of Acquired 16S Ribosomal RNA Methyltransferases. Infect. Dis. Clin. North Am 2016, 30, 523–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandrika NT; Garneau-Tsodikova S, Comprehensive review of chemical strategies for the preparation of new aminoglycosides and their biological activities. Chem. Soc. Rev 2018, 47, 1189–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkov-Zrihen Y; Fridman M, Synthesis of Aminoglycosides. In Modern Synthetic Methods in Carbohydrate Chemistry; From Monosaccharides to Complex Glycoconjugates, Werz DB; Vidal S, Eds. Wiley: Weinheim, 2014; pp 161–190. [Google Scholar]
- 11.Bacot-Davis VR; Bassenden AV; Berghuis AM, Drug-target networks in aminoglycoside resistance: hierarchy of priority in structural drug design. Med. Chem. Commun 2016, 7, 103–113. [Google Scholar]
- 12.Hobbie SN; Bruell C; Kalapala S; Akshay S; Schmidt S; Pfister P; Böttger EC, A genetic model to investigate drug–target interactions at the ribosomal decoding site. Biochimie 2006, 88, 1033–1043. [DOI] [PubMed] [Google Scholar]
- 13.Hobbie SN; Kalapala SK; Akshay S; Bruell C; Schmidt S; Dabow S; Vasella A; Sander P; Böttger EC, Engineering the rRNA Decoding Site of Eukaryotic Cytosolic Ribosomes in Bacteria. Nucl. Acids Res 2007, 35, 6086–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Fernandez D; Shcherbakov D; Matt T; Leong NC; Kudyba I; Duscha S; Boukari H; Patak R; Dubbaka SR; Lang K; Meyer M; Akbergenov R; Freihofer P; Vaddi S; Thommes P; Ramakrishnan V; Vasella A; Böttger EC, 4’-O-Substitutions Determine Aminoglycoside Selectivity at the Drug Target Level. Nature Commun 2014, 5, 3112/doi: 10.1038/ncomms4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsushita T; Chen W; Juskeviciene R; Teo Y; Shcherbakov D; Vasella A; Böttger EC; Crich D, Influence of 4′-O-Glycoside Constitution and Configuration on Ribosomal Selectivity of Paromomycin. J. Am. Chem. Soc 2015, 137, 7706–7717. [DOI] [PubMed] [Google Scholar]
- 16.Sati GC; Sarpe VA; Furukawa T; Mondal S; Mantovani M; Hobbie SN; Vasella A; Böttger EC; Crich D, Modification at the 2’-Position of the 4,5-Series of 2-Deoxystreptamine Aminoglycoside Antibiotics to Resist Aminoglycoside Modifying Enzymes and Increase Ribosomal Target Selectivity. ACS Infect. Dis 2019, 5, 1718–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsushita T; Sati GC; Kondasinghe N; Pirrone MG; Kato T; Waduge P; Kumar HS; Cortes Sanchon A; Dobosz-Bartoszek M; Shcherbakov D; Juhas M; Hobbie SN; Schrepfer T; Chow CS; Polikanov YS; Schacht J; Vasella A; Böttger EC; Crich D, Design, Multigram Synthesis, and in Vitro and in Vivo Evaluation of Propylamycin: A Semisynthetic 4,5-Deoxystreptamine Class Aminoglycoside for the Treatment of Drug-Resistant Enterobacteriaceae and Other Gram-Negative Pathogens. J. Am. Chem. Soc 2019, 141, 5051–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aggen JB; Armstrong ES; Goldblum AA; Dozzo P; Linsell MS; Gliedt MJ; Hildebrandt DJ; Feeney LA; Kubo A; Matias RD; Lopez S; Gomez M; Wlasichuk KB; Diokno R; Miller GH; Moser HE, Synthesis and Spectrum of the Neoglycoside ACHN-490. Antimicrob. Agent. Chemother 2010, 54, 4636–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith KP; Kirby JE, Evaluation of apramycin activity against carbapenem-resistant and -susceptible strains of Enterobacteriaceae. Diagn. Microbiol. Infect. Dis 2016, 86, 439–441. [DOI] [PubMed] [Google Scholar]
- 20.Kang AD; Smith KP; Eliopoulos GM; Berg AH; McCoy C; Kirby JE, In vitro Apramycin Activity against multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Diagn. Microbiol. Infect. Dis 2017, 88, 188–191. [DOI] [PubMed] [Google Scholar]
- 21.Juhas M; Widlake E; Teo J; Huseby DL; Tyrrell JM; Polikanov Y; Ercan O; Petersson A; Cao S; Aboklaish AF; Rominski A; Crich D; Böttger EC; Walsh TR; Hughes DE; Hobbie SN, In-vitro Activity of Apramycin Against Multidrug-, Carbapenem-, and Aminoglycoside-Resistant Enterobacteriaceae and Acinetobacter baumannii. J. Antimicrob. Chemother 2019, 74, 944–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matt T; Ng CL; Lang K; Sha S-H; Akbergenov R; Shcherbakov D; Meyer M; Duscha S; Xie J; Dubbaka SR; Perez-Fernandez D; Vasella A; Ramakrishnan V; Schacht J; Böttger EC, Dissociation of Antibacterial Activity and Aminoglycoside Ototoxicity in the 4-Monosubstituted 2-Deoxystreptamine Apramycin. Proc. Natl. Acad. Sci., USA 2012, 109, 10984–10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quirke JCK; Rajasekaran P; Sarpe VA; Sonousi A; Osinnii I; Gysin M; Haldimann K; Fang Q-J; Shcherbakov D; Hobbie SN; Sha S-H; Schacht J; Vasella A; Böttger EC; Crich D, Apralogs: Apramycin 5-O-Glycosides and Ethers with Improved Antibacterial Activity and Ribosomal Selectivity and Reduced Susceptibility to the Aminoacyltranserferase (3)-IV Resistance Determinant. J. Am. Chem. Soc DOI: 10.1021/jacs.9b11601. [DOI] [PMC free article] [PubMed] [Google Scholar]