Abstract
Unique among human viruses, Kaposi’s sarcoma-associated herpesvirus (KSHV) encodes several homologs of cellular interferon regulatory factors (vIRFs). Since KSHV expresses multiple factors that can inhibit IFN signaling and promote virus production, it is still unclear to what extent vIRFs specifically contribute to these processes during KSHV infection. To study the function of vIRFs during viral infection, we engineered 3xFLAG-tagged-vIRF and vIRF-knockout (KO) recombinant KSHV clones, which were utilized to test vIRF expression, as well as their requirement for viral replication, virus production, and inhibition of the type I IFN pathway in different models of lytic KSHV infection. Our data show that all vIRFs can be expressed as lytic viral proteins, yet were dispensable for KSHV production and inhibition of type I IFN. Nevertheless, as vIRFs were able to suppress IFN-stimulated antiviral genes, vIRFs may still promote the viral lytic cycle in the presence of an ongoing antiviral response.
Keywords: KSHV, vIRFs, lymphatic endothelial cells, IFN signaling, viral replication, BAC16, viral immune evasion, interferon-stimulated genes
Introduction
The interferon regulatory factors (IRF) are a family of transcription factors with pleiotropic roles in the maturation and response of the immune system. The cellular IRFs are also involved in the regulation of the DNA damage response, cell cycle, cell proliferation, cell death, and have roles in tumorigenesis (Tamura et al., 2008). Yet, IRFs are probably best known for their ability to control interferon (IFN) and inflammatory responses downstream of pattern recognition receptors, thereby linking pathogen surveillance to immune response (Honda and Taniguchi, 2006). Therefore, not only do IRFs promote normal cell growth, but they also contribute to innate and adaptive immunity (Jefferies, 2019), all of which are essential to the control of viral infections. However, viruses have evolved a number of strategies to inhibit or exploit the functions of different cellular IRFs to promote viral infection (Marsili et al., 2016).
Kaposi’s sarcoma-associated herpesvirus (KSHV) is a large, double-stranded DNA virus of the gammaherpesvirus family. The clinical manifestations of KSHV infection include Kaposi’s sarcoma (Chang et al., 1994), primary effusion lymphoma (PEL) (Cesarman et al., 1995), multicentric Castleman disease (MCD) (Soulier et al., 1995), and KSHV inflammatory cytokine syndrome (Polizzotto et al., 2012; Uldrick et al., 2010). Following primary infection, KSHV establishes lifelong infection in humans, which is supported by a broad arsenal of viral immunomodulatory factors that protect the infected cells from the detection by the host immune system. Most of these viral immunomodulatory factors are expressed during the lytic cycle, presumably due to heightened immune sensing and effector responses (Aresté and Blackbourn, 2009; Lee et al., 2012). A few prominent examples of KSHV immunomodulatory proteins include immediate-early genes RTA and ORF45, both of which robustly inhibit the type I IFN pathway (Yu and Hayward, 2010; Yu et al., 2005; Zhu et al., 2002), ORF64, which represses RIG-I signaling (Inn et al., 2011), ORF52, which represses cGAS/STING signaling (Wu et al., 2015), ORF63, which decreases caspase 1 activation and subsequent IL-1β production (Gregory et al., 2011), and K3/K5, which are involved in the degradation of human leukocyte antigen (HLA) class I molecules as well as intercellular adhesion molecule (ICAM)-1 in a stage-specific manner (Brulois et al., 2014; Tomescu et al., 2003).
KSHV is unique among all human viruses in that it encodes four homologs of the cellular IRFs, named the viral interferon regulatory factors (vIRF), which are clustered into a single genomic locus (Burysek et al., 1999; Koch and Schulz, 2017; Lubyova and Pitha, 2000; Moore et al., 1996; Russo et al., 1996). All of the vIRFs are expressed during the lytic cycle (Cousins and Nicholas, 2014; Cunningham et al., 2003; Jenner et al., 2001b; Lubyova and Pitha, 2000), although vIRF3 can also be expressed in latently infected PEL cells (Rivas et al., 2001). A substantial body of work generated over the last 20 years has demonstrated that each of the vIRFs can suppress IFN signaling, though likely at different steps (Areste et al., 2009; Gao et al., 1997; Hwang et al., 2017; Jacobs et al., 2013; Koch and Schulz, 2017; Lubyova and Pitha, 2000; Ma et al., 2015). In addition to the inhibition of IFN production, vIRFs may also regulate the IFN effector response, which is the second phase of the IFN pathway (Baresova et al., 2013; Mutocheluh et al., 2011a). Once IFN binds to an IFN receptor, it can activate JAK/STAT signaling cascades that lead to the induction of hundreds of interferon-stimulated genes (ISG), many of which are globally repressive to viral replication (Schoggins, 2014). While vIRFs may evade the ISG response by blocking IFN production in the first phase of the pathway, vIRFs can also block the ISG response downstream of IFN receptor engagement (Mutocheluh et al., 2011a). In tandem with blocking IFN production, such a strategy would complement the viral immune evasion abilities of KSHV, especially if certain ISG functions could be commandeered to benefit the virus. In addition to the regulation of IFN signaling pathway, vIRFs are also involved in deregulating normal apoptotic pathways (Nakamura et al., 2001; Seo et al., 2001; Shin et al., 2006). Though vIRF1 seems to be a bone fide viral oncoprotein, the other vIRFs may also act as oncoproteins due to their known connections to several tumor-promoting pathways such as blocking the activity of p53 (Gao et al., 1997; Jacobs and Damania, 2011). Therefore, a more comprehensive understanding of vIRFs would not only provide a clearer picture of herpesvirus immune evasion strategies, but could also provide valuable insight into the mechanisms of vIRF-regulated oncogenesis.
Since KSHV encodes multiple factors that can inhibit IFN signaling, it is still unclear to what extent vIRFs contribute to the repression of type I IFN signaling during KSHV infection (Ma et al., 2015). In addition, vIRFs have also been shown to be directly involved in the regulation of viral lytic gene expression, whereby they may facilitate KSHV lytic replication (Park et al., 2007; Xi et al., 2012). However, despite extensive investigation of vIRFs, there are still no comparative studies using genetic analysis to test how all vIRFs affect virus production and IFN signaling during lytic KSHV infection. Therefore, to study the function of vIRFs in the context of viral infection, we engineered 3xFLAG-tagged-vIRF and vIRF-knockout (KO) recombinant KSHV clones and used them to analyze vIRF expression and their requisite for viral replication, virus production, and the inhibition of the type I IFN pathway during lytic KSHV infection.
Materials and Methods
Cell lines and primary cells
293T (ATCC) and iSLK (obtained from Jae Jung at the University of Southern California) cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (P/S). The characteristics of the iSLK cell line has been described previously (Myoung and Ganem, 2011). Primary, adult human dermal lymphatic microvascular endothelial cells (HDLMEC) were purchased from Lonza (CC-2810) and cultured in microvascular endothelial cell growth media containing 5% FBS and growth factors (CC-3202). HDLMECs were used between passages 6 and 9 for experiments.
Chemicals and antibodies
Doxycycline (Dox), sodium butyrate (NaB), and phosphonoacetic acid (PAA) were purchased from Sigma. PAA was used at 100 μM to inhibit KSHV replication. Recombinant human IFNβ was from Peprotech (300–02BC). The following antibodies were used in our study: anti-FLAG (F1804, Sigma), anti-tubulin (GTU-88, Sigma), anti-LANA (13–210-100, Advanced Biotechnologies), anti-ORF45 (sc-53883, Santa Cruz), anti-K8 (sc-57889, Santa Cruz), anti-K8.1 (sc-65446, Santa Cruz), anti-ORF26 (NBP1–47357, Novus Biologicals), anti-vIRF3 (NB200–167, Novus Biologicals), anti-CBP (sc-369, Santa Cruz), anti-IRF3 (sc-33641, Santa Cruz), and anti-pIRF3 (S396) (4947S, Cell Signaling). Anti-RTA and anti-ORF6 antibodies were generously provided by Dr. Yoshihiro Izumiya (University of California, Davis) and Dr. Gary Hayward (Johns Hopkins University), respectively.
Generation of recombinant KSHV BAC16 clones
The vIRF-recombinant KSHV clones were constructed by bacterial artificial chromosome (BAC)-based homologous recombination using KSHV BAC16 in the E. coli strain GS1783, as previously described (Brulois et al., 2012; Tischer et al., 2006). All recombination steps were verified by Sanger sequencing and restriction enzyme digestion of the BAC clones followed by pulsed-field gel electrophoresis analysis. The primers used for BAC recombination are listed in Table 1.
Table 1. Primers used for generating recombinant BAC16 clones.
All primers listed are given in 5’ to 3’ orientation.
| Oligo name | Oligo sequence in 5’ to 3’ orientation | Purpose |
|---|---|---|
| vIRF1_bacF | CACTGGACATTGCGGCGCGAGCTAGTCTGGTTGCGGGACAATGGACTACAAAGACCATGA | BAC16–3xFLAG-vIRF1 |
| vIRF1_bacR | CCCCAGGCGCCCCAAAAGGGTTCGGTCTTTGGCCTGGGTCTCCACCCTTGTCATCGTCATCCTTGTAATCGATGTCATGAAACCAATTAACCAATTCTGATTAG | BAC16–3xFLAG-vIRF1 |
| vIRF1stop_bacR | CCCCAGGCGCCCCAAAAGGGTTCGGTCTATTGGCCTGGGTCTCCACCCTTGTCATCGTCATCCTTGTAATCGATGTCATGAAACCAATTAACCAATTCTGATTAG | BAC16–3xFLAG-vIRF1 knockout |
| vIRF2_bacF | ACGGAAAAGGTGTTTTGTGTCGTGGCTTTTGCCTAAAAAGATGGACTACAAAGACCATGA | BAC16–3xFLAG-vIRF2 |
| vIRF2_bacR | TAATAAAGTCCGTGAGCCATTCCGACTCCGTGTAGCGAGGTCCACCCTTGTCATCGTCATCCTTGTAATCGATGTCATGAAACCAATTAACCAATTCTGATTAG | BAC16–3xFLAG-vIRF2 |
| vIRF2stop_bacR | TAATAAAGTCCGTGAGCCTATTCCGACTCCGTGTAGCGAGGTCCACCCTTGTCATCGTCATCCTTGTAATCGATGTCATGAAACCAATTAACCAATTCTGATTAG | BAC16–3xFLAG-vIRF2 knockout |
| vIRF3–3xF_bacF | CGTCATCAAGCTCAGATTGCGTTCAGTTACATGTGATGACGGTGGAATGGACTACAAAGACCATGA | BAC16-vIRF3–3xFLAG |
| vIRF3–3xF_bacR | CCACAGCCCGTCAAACCACAGGGACCCTGTTGGCTGACTACTACTTGTCATCGTCATCCTTGTAATCGATGTCATGATCTTTAAACCAATTAACCAATTCTGATTAG | BAC16-vIRF3–3xFLAG |
| vIRF3stop_bacF | TGACAGGTCAACATGGCGGGACGCAGGCTTACCTGGATTTGCTAGCGTTTATTGTAGGTGCTTTGGAGGATGACGACGATAAGTAGGG | BAC16-vIRF3 knockout |
| vIRF3stop_bacR | CAAAGGATATTTATCAGAGTCCAAAGCACCTACAATAAACGCTAGCAAATCCAGGTAAGCCTGCGTAACCAATTAACCAATTCTGATTAG | BAC16-vIRF3 knockout |
| vIRF4_bacF | TGCCTCAAAGACGAACGCCGATCGGTTTCTGTGTCGGACCATGGACTACAAAGACCATGA | BAC16–3xFLAG-vIRF4 |
| vIRF4_bacR | TAATCCATAACGTGGCCCATTCTGAGCCACCGGCTTTAGGTCCACCCTTGTCATCGTCATCCTTGTAATCGATGTCATGAAACCAATTAACCAATTCTGATTAG | BAC16–3xFLAG-vIRF4 |
| vIRF4stop_bacR | TAATCCATAACGTGGCCCTATTCTGAGCCACCGGCTTTAGGTCCACCCTTGTCATCGTCATCCTTGTAATCGATGTCATGAAACCAATTAACCAATTCTGATTAG | BAC16–3xFLAG-vIRF4 knockout |
| vIRF2stop_bacF2 | ACGGAAAAGGTGTTTTGTGTCGTGGCTTTTGCCTAAAAAGATGCCTCGCTACACGGAGTCAGGATGACGACGATAAGTAGGG | vIRF2 knockout in BAC16-QKO |
| vIRF2stop_bacR2 | TAAAGTCCGTGAGCCTATTCCGACTCCGTGTAGCGAGGCATCTTTTTAGGCAAAAGCCACGAACCAATTAACCAATTCTGATTAG | vIRF2 knockout in BAC16-QKO |
| vIRF4stop_bacF2 | TGCCTCAAAGACGAACGCCGATCGGTTTCTGTGTCGGACCATGCCTAAAGCCGGTGGCTCAGGATGACGACGATAAGTAGGG | vIRF4 knockout in BAC16-QKO |
| vIRF4stop_bacR2 | TCCATAACGTGGCCCTATTCTGAGCCACCGGCTTTAGGCATGGTCCGACACAGAAACCGATAACCAATTAACCAATTCTGATTAG | vIRF4 knockout in BAC16-QKO |
| vIRFdel_bacF | CTCCCTCCCATAACAATACGGTGTAGGCATTTTGTATTATCTTTTTAGGCAAAAGCCACGAGGATGACGACGATAAGTAGGG | BAC16-ΔvIRF |
| vIRFdel_bacR | ACGGAAAAGGTGTTTTGTGTCGTGGCTTTTGCCTAAAAAGATAATACAAAATGCCTACACAACCAATTAACCAATTCTGATTAG | BAC16-ΔvIRF |
BAC16 DNA transfection and establishment of iSLK-BAC16 cell lines
BAC16 DNA and its derivatives were isolated from 5 ml of bacterial culture and the miniprep DNA was resuspended in 40 μl of RNase/DNase-free water. To make iSLK-BAC16 cell lines, iSLK cells were seeded at 2×105 cells per well in a 6-well plate, and the next day the cells were transfected with BAC16 DNA using FuGENE HD (Promega). The transfection complexes were prepared by combining 155 μl of Opti-MEM with 10 μl of BAC16 miniprep DNA and 10 μl of FuGENE HD. After 10 min of incubation at room temperature, the transfection complexes were added to the iSLK cells. Two days after transfection, the cells were trypsinized and re-seeded in complete DMEM containing 300 μg/ml hygromycin B. One week later the hygromycin B concentration was increased to 500–1000 μg/ml and GFP+/hygromycin B resistant iSLK-BAC16 cell lines were established by approximately one month of hygromycin B selection. The 293T-BAC16 cell lines were generated with the same procedure, except that 300 μg/ml hygromycin B was used for selection and establishing the stable cell lines.
KSHV production, lytic infection, and virus titering
To produce a large batch of KSHV, the iSLK-BAC16 cell lines were grown in 12–24 of 150-mm cell culture dishes while treating with 1 μg/ml Dox and 1 mM NaB for 84 hours. The supernatant was then collected and cleared of cell debris by centrifugation (2000 rpm, 5 min), filtered through a 0.45 μm PES filter, and concentrated about 100–200-fold by ultracentrifugation (27000 rpm, 3 hrs, 10°C).
For primary lytic infection of iSLK cells, the cells were pretreated with 1 μg/ml Dox for 6 hr and then infected with KSHV by spinoculation in the presence of 1 μg/ml Dox and 1 mM NaB. Lytic infection of HDLMECs was performed as described previously (Golas et al., 2019). MOI of 1 was used for KSHV infection. For both primary lytic infection models, after 2 hpi, the media were removed, the cells were washed once with PBS, and the cells were harvested to check initial infection (input vDNA level) or were provided fresh media and cultured until the indicated time points. For IFN pretreatment, 500 IU/ml recombinant IFNβ was added to HDLMECs for 24 hrs prior to KSHV infection. After 2 hours of KSHV infection, the cells were washed with PBS, and fresh media was added without IFNβ, and cultured until the indicated time points.
When the virus production of different cell lines was tested, 3×105 of iSLK-BAC16 cells per well in 6-well culture plates were induced by treating the cells with 1 μg/ml Dox and 1 mM NaB. At 84–96 hours post-induction the supernatants were collected and an equivalent amount of supernatant was used to infect 293T cells by spinoculation (2000 rpm, 45 min, 30°C). The GFP-positive cells were quantified by flow cytometry at 24 hpi.
Flow cytometry and viability assay
For 293T cells, the cells were pooled, washed in FACS buffer (1% FBS with 4 mM EDTA in PBS), fixed in 4% paraformaldehyde, then washed and resuspended in FACS buffer. For HDLMECs, the cells were pooled, washed in FACS buffer, stained with live/dead discrimination dye (ThermoFisher, L10119) according to the manufacturer’s protocol, fixed in 2% paraformaldehyde, then washed and resuspended in FACS buffer. All GFP and live/dead signals were quantified on an LSR-II or an LSR-Fortessa flow cytometer (BD Biosciences). Following the removal of doublet cells, flow cytometry data were analyzed by FlowJo.
Immunofluorescence analysis
Cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized by 0.5% Triton X-100 for 5 minutes, washed twice with washing buffer (PBS with 0.2% Tween-20), and then incubated in blocking buffer (5% FBS, 0.2% fish skin gelatin, and 0.2% Tween-20 in PBS) for 30 minutes. Antibodies were diluted in blocking buffer and used to stain the cells for 1–2 hours at room temperature. Subsequently, the cells were washed three times with washing buffer and then incubated with anti-rabbit Alexa Fluor 568 antibody (Invitrogen) for 1 hour at room temperature. Cells were washed with washing buffer three times and then stained with DAPI to visualize the nuclei. For GFP imaging and the immunofluorescence analyses, a Revolve fluorescence microscope (Echo Laboratories) was used.
Immunoprecipitation
For immunoprecipitation, cells were lysed in ice-cold Co-IP buffer (50 mM Tris-HCl pH 7.5, 1 mM EDTA pH 8.0, 150 mM NaCl, and 0.5% NP40) in the presence of 1 mM PMSF, protease inhibitor cocktail (1X), and phosphatase inhibitor cocktail (Sigma) and then incubated on ice for 30 min. Lysates were pre-cleared by incubating with Protein A Sepharose and rotating overnight. The next day, the pre-cleared lysates were incubated with primary antibody for 3 hours by rotating at 4°C. Then the samples were incubated with Sepharose Protein A/G beads for an additional 2 hours. Subsequently, the beads were pelleted and washed three times in Co-IP buffer, and then resuspended in Laemmli buffer containing beta-mercaptoethanol for immunoblot analysis.
Measuring Type I IFN production
Type I IFN reporter bioassay was performed using HEK-Blue IFN-α/β cells (Invivogen) according to the manufacturer’s protocol. Quantification of the enzymatic reaction of the reporter product was achieved at 640 nm using a SpectraMax-M2 plate reader (Molecular Devices). For direct quantification of IFNβ protein in the supernatant, an IFNβ immunoassay was executed according to the manufacturer’s protocol (Invitrogen, ProcartaPlex Simplex Kit, EPX01A-12088–901). IFNβ immunoassay was quantified on a Luminex 200 instrument. For experiments using Sendai virus (SeV) to induce IFN, the cells were infected with 1 HA/ml of SeV Cantell strain (Charles River Laboratories).
RT-qPCR and qPCR analyses
Total RNA was extracted from cells by using Trizol (Sigma). DNase I-treated RNA (0.5–1 μg) was then reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad). The sequences of gene-specific primers used in qPCR are listed in Table 2. Relative gene expression was calculated by using the 2−ΔCt method where the expression of 18S gene was used for normalization. For the RT-qPCR array, an RT2 Profiler PCR Array composed of 84 target genes was used (Qiagen, Human Type I Interferon Response, PAHS-016Z) following the manufacturer’s recommended protocol. Relative gene expression was first normalized to actin, which was included in the array, then analyzed using the 2−ΔΔCt method by comparing ΔvIRF to WT KSHV-infected cells.
Table 2. RT-qPCR primers for gene expression analysis.
All primers listed are given in 5’ to 3’ orientation.
| Gene target | Primer sequence in 5’ to 3’ orientation |
|---|---|
| For RT-qPCR | |
| vIRF1_F | ATACACAACACCCAATTCCC |
| vIRF1_R | CGCTGGTTTTTGACTACCCAG |
| vIRF2_F | TCATGGCTGGTTCCTGCGTCA |
| vIRF2_R | TCCGTAGTGAGTTCTAACCAC |
| vIRF3_F | AGCCGTACACTGTGTTGATAC |
| vIRF3_R | CACGATTCATAGTGAGAAACA |
| vIRF4_F | CTAGTGTCACTGCGTCGCGTA |
| vIRF4_R | CAGGACATTTGTCAAAGGAGC |
| RTA_F | TTGCCAAGTTTGTACAACTGCT |
| RTA_R | ACCTTGCAAAGACCATTCAGAT |
| ORF25_F | ACAGTTTATGGCACGCATAGTG |
| ORF25_R | GGTTCTCTGAATCTCGTCGTGT |
| ORF36_F | ATTGCCAACGACCTGATGCA |
| ORF36_R | ACTCCAGTCCAGCTGCAGCA |
| ORF45_F | CCATACAGCGACCCTGATGA |
| ORF45_R | CCGATTCTCTGACTCAATACT |
| ORF59_F | AACCGCAGTTCGTCAGGACCACCA |
| ORF59_R | CCTTAGCCACTTAAGTAGGAATG |
| IFN|3_F | CAGCAATTTTCAGTGTCAGAAGC |
| IFNp_R | TCATCCTGTCCTTGAGGCAGT |
| ISG15_F | CTCTGAGCATCCTGGTGAGGAA |
| ISG15_R | AAGGTCAGCCAGAACAGGTCGT |
| 18S_F | TTCGAACGTCTGCCCTATCAA |
| 18S_R | GATGTGGTAGCCGTTTCTCAGG |
| For DNA qPCR | |
| ORF11_F | GGCACCATACAGCTTCTACGA |
| ORF11_R | CGTTTACTACTGCACACTGCA |
| HS1_F | TTCCTATTTGCCAAGGCAGT |
| HS1_R | CTCTTCAGCCATCCCAAGAC |
To measure the viral DNA load of KSHV-infected cells, the cells were lysed in RIPA buffer, sonicated (1 cycle, 30 sec) using a Bioruptor Pico instrument (Diagenode), and the total DNA was isolated by using phenol-chloroform extraction. 10 ng of total DNA was used to measure the viral copy number relative to host DNA. Viral DNA was amplified with KSHV ORF11-specific primers while the host DNA was amplified using HS1-specific primers (Toth et al., 2016) (Table 2). Analysis for PCR graphs was compiled from at least three independent experiments.
Statistical Methods
An unpaired Student’s two-tailed t-test was employed for all experiments. Statistical significance was determined if p<0.05.
Results
Construction of recombinant KSHV clones expressing 3xFLAG-tagged vIRFs
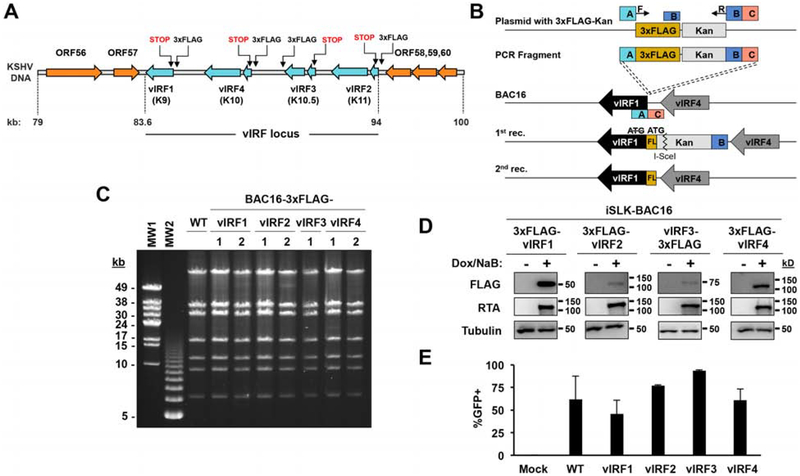
In order to test the contribution of vIRFs of KSHV to lytic viral production and the inhibition of the type I IFN pathway during viral infection, we constructed a panel of recombinant KSHV clones, using homologous recombination, targeting each of the four vIRFs. First, we 3xFLAG-tagged each vIRF in BAC16 KSHV to be able to detect the expression of each vIRF using FLAG antibody during the lytic cycle of viral infection (Fig. 1A and B). While vIRF1, vIRF2, and vIRF4 were 3xFLAG-tagged at their N-terminus, vIRF3 was 3xFLAG-tagged at its C-terminus because we had found that N-terminal epitope tagging seemed to interrupt normal vIRF3 expression from the KSHV genome. To ensure that the 3xFLAG-vIRF BAC16 clones were free from any unwanted genomic rearrangements after the 3xFLAG insertion, we performed pulsed-field gel electrophoresis (PFGE) of SbfI-digested BAC16 DNAs. Figure 1C demonstrates that the enzymatic digestion pattern of the viral DNAs of the 3xFLAG-tagged vIRF BAC16 clones matched that of wild-type (WT) BAC16, verifying that the recombinant viruses were devoid of any confounding genomic rearrangements, which can occur during the homologous recombination procedure. Next, we transfected the 3xFLAG-tagged vIRF BAC16 clones into iSLK cells to establish stable cell lines carrying KSHV in latency. The lytic cycle of KSHV can triggered by treating iSLK-BAC16 cells with Dox, which induces the transgenic expression of RTA, a key viral transcription factor that is required for KSHV lytic replication (Myoung and Ganem, 2011). Figure 1D shows that the expression of each 3xFLAG-tagged vIRF could be detected by FLAG immunoblot during KSHV lytic reactivation. Interestingly, while vIRF3 is known to be expressed as a latent viral protein in KSHV-infected PEL cells (Rivas et al., 2001), we could detect its induction in iSLK-BAC16 cells during lytic reactivation. Finally, we tested whether we could produce infectious KSHV from the 3xFLAG-tagged vIRF BAC16 clones in iSLK cells. For this, we harvested supernatants from lytically reactivated iSLK-BAC16 cell lines after 4 days post-induction and transferred them onto 293T cells. Since BAC16 expresses GFP from a constitutive promoter in infected cells, we used flow cytometry to quantify the number of GFP+ 293T cells 24 hrs after supernatant transfer to assess the relative amount of infectious virus produced. As seen in Figure 1E, virus production of the 3xFLAG-tagged vIRF BAC16 clones was comparable to that of WT KSHV. Taken together, we could successfully generate KSHV clones expressing 3xFLAG-tagged vIRFs and showed that each of them produced infectious viruses in a comparable amount to WT KSHV. Importantly, these 3xFLAG-tagged vIRF KSHV clones will facilitate the study of endogenous vIRFs in KSHV-infected cells, which has been hampered by the lack of availability or variation in quality of vIRF-specific antibodies.
Figure 1. Construction of recombinant KSHV clones expressing 3xFLAG-tagged vIRFs.
(A) Schematic showing the vIRF locus within the KSHV genome and the sites targeted for homologous recombination to engineer 3xFLAG-vIRF and vIRF-KO recombinant KSHV clones. (B) Diagram of the homologous recombination showing the two-step procedure, which was used to make FLAG-tagged vIRF or vIRF knockout clones using BAC16 KSHV. Shown is an example targeting vIRF1 where a 3xFLAG epitope tag was fused to the 5’ end of vIRF1. The ATG start codon of vIRF1 was deleted and instead, ATG of the 3xFLAG epitope tag was utilized. (C) PFGE analysis of SbfI-digested WT and 3xFLAG-vIRF BAC16 DNAs. (D) iSLK cells latently infected by 3xFLAG-vIRF recombinant KSHV were treated with 1 μg/ml Dox and 1 mM NaB to induce lytic reactivation. vIRF protein expression was analyzed from cell lysates by immunoblot at 48 hpi. (E) To assess infectious KSHV production, an equal amount of supernatant from lytically reactivated iSLK-BAC16 vIRF cell lines at 4 dpi was used to infect an equivalent number of 293T cells. BAC16 encodes GFP under the control of the EF1α cellular promoter. The resulting GFP-positive 293T cells at 24 hpi were quantified as readout of virus production using flow cytometry. Error bars represent standard deviation (n=3). Molecular Weight (MW) markers: MW1 for λ DNA-Mono Cut Mix, MW2 for 1 kbp DNA ladder.
Kinetics of vIRF expression during lytic reactivation of KSHV
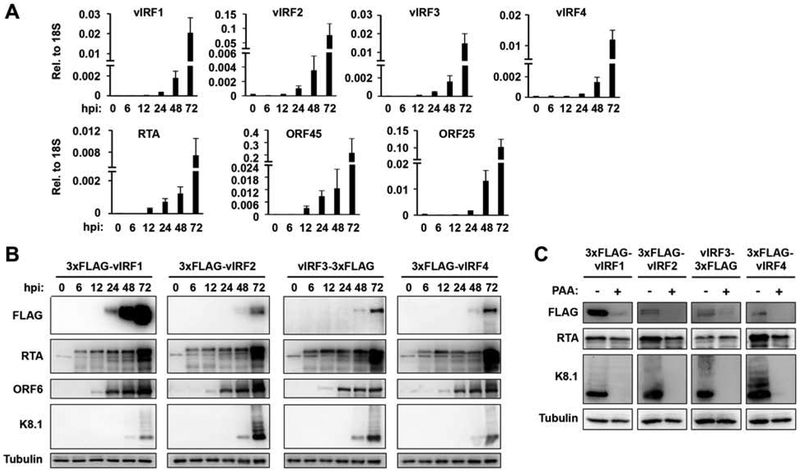
Previous studies have shown that vIRF1, vIRF2, and vIRF4 are expressed as lytic genes during lytic reactivation of KSHV, while vIRF3 is expressed as a latent gene in PEL and MCD samples (Cunningham et al., 2003; Koch and Schulz, 2017; Nakamura et al., 2003). These conclusions have been drawn based on the detection of the vIRF mRNA transcripts or by analyzing vIRF protein expression using different antibodies during the life cycle of KSHV. However, the use of different vIRF-specific detection reagents has resulted in some conflicting data about when vIRFs are expressed during the viral life cycle, and whether or not vIRF3 expression is restricted to KSHV-infected B cells. Therefore, the 3xFLAG-tagged vIRF KSHV clones allowed us to examine, and directly compare for the first time, the endogenous protein expression of the different vIRFs in infected cells by using the same antibody. To this end, we induced lytic reactivation of KSHV in iSLK cells, harboring WT BAC16 or the different 3xFLAG-tagged vIRF BAC16 clones, and measured both the mRNA and protein expression of the vIRFs at 0 hpi (latency), 6, 12, 24, 48, and 72 hpi (Fig. 2). The RT-qPCR analysis of viral mRNA expression in iSLK-BAC16 cells showed that all four vIRFs were greatly upregulated upon lytic reactivation following the gene expression pattern of ORF45 (early) and ORF25 (late) viral genes, (Fig. 2A). Immunoblot analysis of vIRF expression revealed that while vIRF1 could be detected as early as 24 hpi, the other vIRFs could be seen only from 48 hpi (Fig. 2B). The protein expression of vIRF1 correlated closest with that of ORF6 (early), whereas the other vIRFs could be detected only when the late KSHV protein K8.1 was produced. Because the same antibody was used to test vIRF protein levels, we were able to conclude that vIRF1 was produced the earliest and most abundantly during KSHV lytic replication compared to the other vIRFs. Because of the robust protein expression of vIRFs is correlated with the enrichment of K8.1, we tested if vIRFs can be induced as late KSHV genes. To test this, we used PAA to inhibit viral DNA replication during KSHV lytic reactivation, which is known to abrogate late gene expression. We found that while the protein expression of vIRF2 and vIRF4 were abolished by PAA suggesting that they display late gene expression kinetics, the expression of vIRF1 and vIRF3 could still be detected in PAA-treated reactivated iSLK-BAC16 cells. Collectively, these data showed that although the expression pattern of vIRFs is distinct, all vIRFs are lytic factors in iSLK cells. While the gene transcription of vIRFs was induced similarly during the lytic cycle, the level and timing of their protein expression differed in iSLK cells. This indicates that there may be post-transcriptional and/or post-translational regulations that can control the levels of vIRF proteins during KSHV lytic reactivation.
Figure 2. Kinetics of vIRF expression during lytic KSHV reactivation.
To test vIRF expression, 3xFLAG-vIRF BAC16 KSHV was lytically reactivated from latently infected iSLK cell lines using 1 μg/ml Dox and 1 mM NaB and harvested cell lysates at various time points post-induction. (A) vIRF mRNA expression was analyzed by RT-qPCR. Additional viral genes RTA (immediate-early), ORF45 (early), and ORF25 (late) were included as controls. Error bars represent standard deviation (n=3). (B) vIRF protein expression was analyzed by immunoblot using FLAG antibody. Additional viral proteins RTA (IE), ORF6 (early), and K8.1 (late) were included as controls. (C) The iSLK-3xFLAG-vIRF BAC16 cell lines were induced by Dox and NaB for 60 hours in the absence (−) or the presence (+) of 100 uM PAA. The expression of vIRFs was detected by FLAG antibody. The late KSHV protein K8.1 was used as positive control for the PAA treatment.
Construction of vIRF knockout KSHV mutants
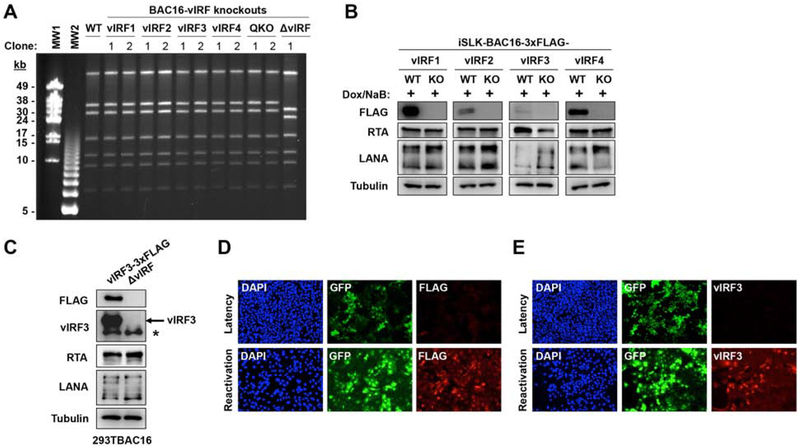
To investigate the effect of vIRFs on KSHV lytic replication and virus production, we generated a series of vIRF-knockout (KO) KSHV mutants for comparative genetic analysis. We made single vIRF-KO mutants by creating a STOP codon and a frame shift mutation in the 5’ end of each 3xFLAG-tagged vIRF gene in BAC16. We also constructed a quadruple vIRF-KO (QKO) BAC16 KSHV mutant by combining the four point mutations used to make the single vIRF-KOs. In parallel to this approach, we also generated a vIRF locus deletion (ΔvIRF) BAC16 KSHV mutant. PFGE analysis of the SbfI-digested DNAs of the vIRF-KO BAC16 KSHV clones showed that they were identical to that of WT BAC16 KSHV, with the exception of the ΔvIRF mutant, which lacks a nearly 10-kb DNA segment due to the deletion of the vIRF locus (Fig. 3A). This assay verified that the vIRF-KO mutants were devoid of any genomic rearrangements, which can sometimes occur inadvertently during the construction of BAC16 mutants by homologous recombination. Importantly, immunoblot analyses confirmed that vIRFs were not expressed in lytically reactivated iSLK cells harboring the vIRF-KO BAC16 mutants (Fig. 3B). Collectively, these data show that we could successfully generate vIRF-knockout BAC16 KSHV clones for functional analyses.
Figure 3. Construction of recombinant KSHV clones lacking vIRFs.
(A) WT and vIRF-KO BAC16 DNAs were digested with SbfI and analyzed by PFGE. (B) iSLK cells latently infected by 3xFLAG-vIRF or derivative vIRF-KO recombinant BAC16 clones were treated with 1 μg/ml Dox and 1 mM NaB to induce lytic reactivation for 48 hpi. The loss of vIRF protein expression was confirmed using FLAG-specific immunoblots. RTA and LANA were included as KSHV viral protein controls. (C) To confirm vIRF3 as a lytic protein, 293T cell lines carrying either BAC16-vIRF3–3xFLAG or BAC16-ΔvIRF were treated with 3 mM NaB to induce lytic reactivation. vIRF3 protein expression was determined using FLAG- and vIRF3-specific immunoblots. The asterisk marks a non-specific band in the vIRF3 immunoblot. (D) Immunofluorescence analysis of the expression of vIRF3 using FLAG antibody in latent and reactivated (60 hpi) iSLK-BAC16-vIRF3–3xFLAG cells. (E) Immunofluorescence analysis of vIRF3 expression using vIRF3 antibody in latent and reactivated (60 hpi) iSLK-BAC16 cells.
It is worthwhile to note that a number of previous studies have showed that vIRF3 is a latent viral protein expressed predominantly in PEL and MCD samples (Baresova et al., 2013; Rivas et al., 2001). However, our data again demonstrated that vIRF3 can also be induced as a lytic viral protein (Fig. 1D, 2, and 3B). The lytic expression of vIRF3 was further confirmed by vIRF3-specific immunoblot in lytically reactivated KSHV-infected 293T cells (Fig. 3C). Immunofluorescence analysis also showed that vIRF3 can be detected only in lytically reactivated iSLK-BAC16 cells (Fig. 3D, E). These results support our conclusion that vIRF3 can be expressed as a lytic viral protein in epithelial cell lines.
vIRFs are not essential for virus production following lytic reactivation
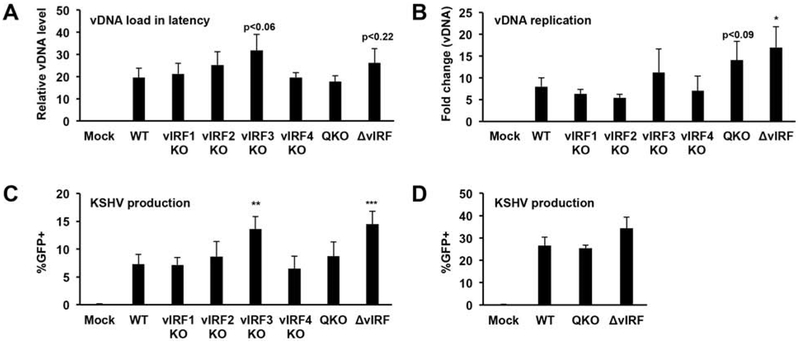
Next, we investigated whether or not vIRFs were required for KSHV lytic replication and virus production following lytic reactivation from latency. For this, we transfected BAC16 DNA into iSLK cells to establish stable iSLK cell lines carrying the different vIRF-KO mutants. We found that the lytic reactivation of these cell lines produced various amount of viruses (data not shown). We note that the establishment of stable iSLK-BAC16 cell lines requires more than one month of hygromycin selection after BAC16 DNA transfection, during which time the cell lines can undergo changes that may influence their responsiveness to lytic reactivation, potentially leading to inaccurate measurement of virus production by the different cell lines. To circumvent this problem, we titered WT and vIRF-KO BAC16 viruses derived from the transfected iSLK cell lines and then used the same amount of virus to establish new iSLK-BAC16 cell lines by infection (Fig. 4). Using viral DNA-specific qPCR we determined that the viral DNA load during latency was comparable across these cell lines (Fig. 4A). While lytic reactivation resulted in similar level of viral DNA replication in most of the vIRF-KO iSLK-BAC16 cell lines relative to the WT control, iSLK-BAC16-ΔvIRF showed slight increase in viral DNA replication (Fig. 4B). In addition, we found that iSLK-BAC16-vIRF3-KO and iSLK-BAC16-ΔvIRF cell lines produced slightly more infectious virions than from iSLK-BAC16-WT (Fig. 4C). These results were in line with our observation that the iSLK-BAC16-ΔvIRF cell line established by viral DNA transfection also produced more viruses relative to iSLK-BAC16 cells (Fig. 4D). Taken together, we found that although vIRFs were not essential to produce virus following lytic reactivation, they may affect the amount of KSHV produced in reactivated iSLK-BAC16 cells.
Figure 4. Analyzing the role of vIRFs for virus production during lytic reactivation.
The iSLK-BAC16 cell lines in panel A-C were made by infection. (A) Viral DNA load was quantified in latently infected cells at 10 dpi by qPCR. (B) Lytic reactivation was induced with 1 μg/ml Dox and 1 mM NaB for 4 days and viral DNA in the cells was quantified by qPCR. (C) KSHV production was assessed by counting the number of GFP+ 293T cells following infection as described in Fig. 1E. (D) Measuring KSHV production from iSLK-BAC16 cell lines created by transfection of viral DNA. Error bars represent standard deviation (n=3). T-tests were performed between WT and the indicated mutants (*p<0.05, **p<0.01, *** p<0.001).
Testing the requirement of vIRFs for KSHV production following de novo lytic infection
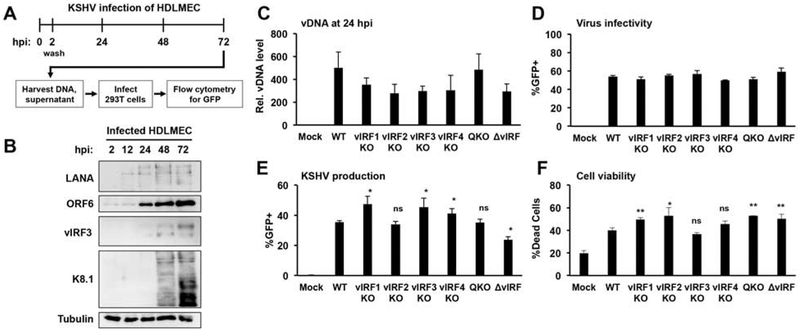
We used primary human dermal lymphatic microvascular endothelial cells (HDLMEC) and an iSLK lytic infection model to test whether vIRFs are required for virus production following lytic de novo infection (Fig. 5 and 6). We have recently demonstrated that HDLMECs are naturally permissive to KSHV lytic replication following de novo infection (Golas et al., 2019). We can detect robust lytic viral protein expression, viral DNA replication, and virus production in HDLMECs in the first 3 days of KSHV de novo infection (Fig. 5A, B, and (Golas et al., 2019)). To test whether vIRFs affect virus production in HDLMECs, we infected the cells with WT KSHV, single vIRF-KO, QKO, and ΔvIRF KSHV mutants for 72 hours (Fig. 5C–F). We found that the efficiency of HDLMEC infection was comparable for the different viruses (Fig. 5C and D) and that they all were able to produce a comparable amount of infectious virions (Fig. 5E). We also analyzed the cell viability of the infected cells at 72 hpi. We found that the viability of vIRF-KO KSHV-infected HDLMECs was slightly reduced in comparison to WT KSHV-infected cells (Fig. 5F). Although we observed minor changes in cell viability, since the different vIRF-KO BAC16 mutants produced a comparable amount of virus relative to WT BAC16 KSHV, these changes in cell death did not seem to affect virus production upon vIRF-KO KSHV infection.
Figure 5. Testing the requirement of vIRFs for KSHV production following de novo lytic infection of HDLMECs.
(A) Schematic for the analysis of de novo lytic infection of HDLMECs. (B) Immunoblot analysis of viral protein expression in WT BAC16-infected cells. (C) Input viral DNA level was determined by qPCR at 24 hpi. (D) Infectivity was measured by flow cytometry analysis of GFP+ cells at 24 hpi. (E) KSHV production from infected HDLMECs at 72 hpi was determined by GFP flow cytometry. (F) Total cell viability of KSHV-infected HDLMECs was analyzed by flow cytometry at 72 hpi using a fixable, dead cell discrimination dye. Error bars represent standard deviation (n=3). ns: non-significant. T-tests were performed between WT and the indicated mutants (*p<0.05, **p<0.01, *** p<0.001).
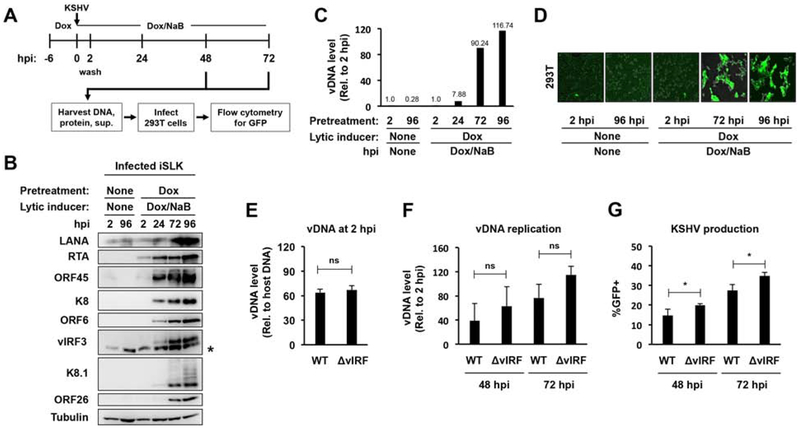
Figure 6. Testing the requirement of vIRFs for KSHV production following lytic infection of iSLK-preRTA cells.
(A) Schematic for the analysis of de novo lytic infection of iSLK-preRTA cells. Note that RTA was pre-expressed in iSLK cells for 6 hours before KSHV infection. (B) Immunoblot analysis of viral protein expression. The asterisk marks non-specific band. (C) Viral DNA replication was measured by qPCR. (D) Representative GFP images of infected 293T cells. (E) Viral DNA level at 2 hpi was measured by qPCR. (F) Viral DNA replication in cells was measured by qPCR, which was calculated relative to 2 hpi. (G) Measuring KSHV production using GFP flow cytometry. Error bars represent standard deviation (n=3). (ns: non-significant, *p<0.05).
To further test the virus production ability of the BAC16-ΔvIRF mutant following de novo lytic infection, we also tested it in an iSLK lytic infection system (iSLK-preRTA). Figure 6A shows the setup of the iSLK-preRTA lytic infection model. Before infecting iSLK cells with KSHV, we pretreated the cells with Dox for 6 hours to pre-express the RTA transgene (the master lytic transactivator of KSHV) while during infection, we added both Dox and the histone deacetylase inhibitor sodium butyrate (NaB) to the cells. In doing so, we created a condition in iSLK cells that, following de novo infection, prevented KSHV from going into latency, and instead propelled the virus into lytic replication. Indeed, while the infection of untreated iSLK cells with KSHV led to viral latency, the infection of the Dox/NaB-treated iSLK cells resulted in robust lytic viral protein expression, viral DNA replication, and the production of infectious virions, which are all hallmarks of successful lytic viral infection (Fig. 6B–D).
We used the iSLK-preRTA lytic infection model to determine if vIRFs are necessary for virus production following lytic infection. For this, we infected Dox-treated iSLK cells with either WT or BAC16-ΔvIRF KSHV, and then determined the viral DNA load in the Dox/NaB-treated, infected cells at 2, 48, and 72 hpi as well as measured the virus production at 48 and 72 hpi (Fig. 6E–G). Importantly, while the viral DNA load was comparable in the WT and ΔvIRF-infected cells at 2 hpi (Fig. 6E), BAC16-ΔvIRF infection tended to show slightly higher lytic viral DNA replication (Fig. 6F) and slightly increased virus production (Fig. 6G). Nevertheless, collectively these results suggest that vIRFs do not have any major effects on KSHV production in the epithelial cell line iSLK and in primary endothelial cells (HDLMECs).
Analyzing the effect of vIRFs on the induction of IFNβ expression during KSHV lytic reactivation
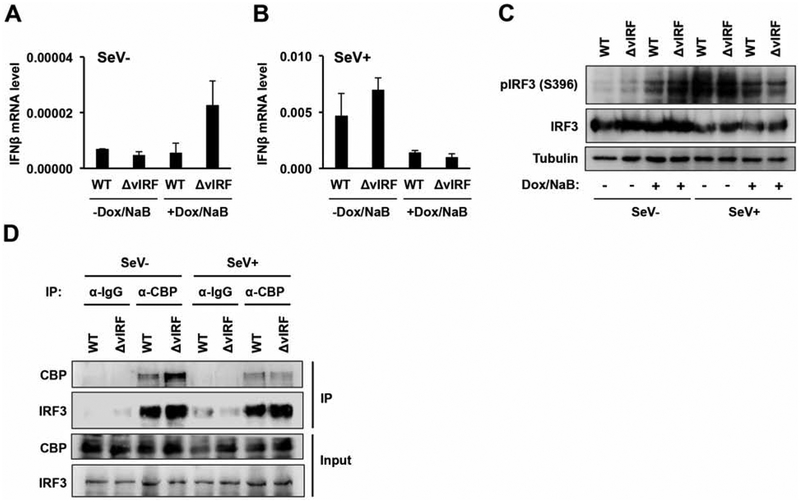
It is still unclear to what degree vIRFs contribute to the suppression of IFNβ expression and the dysregulation of the IFN-induced signaling pathway in KSHV-infected cells because several KSHV proteins, other than vIRFs, can also inhibit the activation of IFN signaling. Therefore, we analyzed how the lack of vIRFs affected IFNβ expression in KSHV-infected cells as well as IRF3 phosphorylation and the interaction of IRF3 with the histone acetyltransferase CBP, which are hallmark features of efficient activation of the IFNβ pathway (Fig. 7) (Jefferies, 2019). We found that while we could detect comparable IFNβ expression in latent iSLK-BAC16 and iSLK-BAC16-ΔvIRF cells, there was a 4-fold increase in IFNβ expression in reactivated iSLK-BAC16-ΔvIRF in comparison to iSLK-BAC16 cells; however, the overall level of IFNβ in both latent and reactivation was very low (Fig. 7A). We also tested the effect of vIRFs on Sendai virus (SeV)-induced IFNβ expression. For this, we infected iSLK-BAC16 and iSLK-BAC16-ΔvIRF cells with SeV for 8 hours after 40 hours of lytic KSHV reactivation (Fig. 7B). SeV infection robustly increased IFNβ expression by nearly 700-fold in both latent iSLK-BAC16 and iSLK-BAC16-ΔvIRF cells; however, IFNβ expression was similarly suppressed in both cell lines during lytic reactivation. In the absence of SeV infection, we observed increased phosphorylation of IRF3 at serine 396 (pIRF3 S396) in reactivated iSLK-BAC16-ΔvIRF cells compared to the reactivated iSLK-BAC16 cells, which correlated with the 4-fold increase in IFNβ gene expression in the absence of vIRFs (Fig. 7B and C). In contrast, while SeV infection induced robust IRF3 phosphorylation in both latent iSLK-BAC16 and iSLK-BAC16-ΔvIRF cells, interestingly; IRF3 phosphorylation was reduced in both cell lines upon lytic reactivation, which was in agreement with the reduced IFNβ expression. Taken together, these results indicate that although vIRFs may be involved in reducing viral lytic reactivation-induced phosphorylation of IRF3 and IFNβ expression, they seem to be dispensable for reducing SeV-induced IFN expression during lytic KSHV reactivation.
Figure 7. vIRFs do not disrupt the CBP-IRF3 interaction and are not needed for efficient silencing of type I IFN expression during lytic reactivation.
iSLK-BAC16 and iSLK-BAC16-ΔvIRF cell lines were lytically reactivated using 1 μg/ml Dox and 1 mM NaB for 40 hpi and then challenged with SeV infection. (A) IFNβ gene expression in the absence of SeV. (B) IFNβ gene expression after SeV (1 HA/ml) infection for 8 hours. Error bars represent standard deviation (n=3). (C) Analyzing the expression of phosphorylated IRF3 (pIRF3 S396) in cell lysates. (D) CBP and control IgG immunoprecipitations testing for CBP and IRF3.
Next, we also tested whether vIRFs have any effects on the formation of CBP/IRF3 protein complex, which is important for activation of the IFNβ promoter. Previous studies have shown that vIRF1 overexpression in KSHV-free cells can inhibit the binding of histone acetyltransferases such as CBP to IRF3 (Li et al., 2000; Lin et al., 2001), which was suggested to be one of the mechanisms involved in vIRF1-mediated repression of IFNβ expression. Strikingly, we did not see any global changes in CBP/IRF3 interaction between iSLK-BAC16-ΔvIRF and iSLK-BAC16 cells during lytic reactivation, regardless of whether or not the cells were infected with SeV to enhance the robustness of IFNβ induction (Fig. 7D). These data suggest that while vIRFs may contribute to the repression of IFNβ expression, there are still other viral factors besides vIRFs that can still efficiently block IFNβ expression in KSHV-infected cells during viral lytic reactivation.
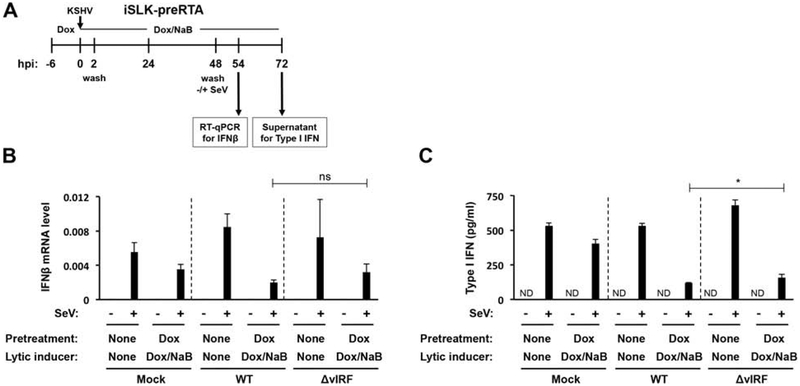
Impact of vIRFs on type I IFN production during KSHV lytic infection
To determine if vIRFs are necessary for the inhibition of IFN production during primary lytic infection, we used both the iSLK-preRTA and HDLMEC lytic infection models (Fig. 8 and 9). Figure 8A shows the setup of the iSLK-preRTA experiment. After 6 hours of RTA pre-expression, the cells were infected with WT or ΔvIRF BAC16 for 48 hours, which was followed by SeV infection for 6 or 24 hours. We chose 48 hours post-KSHV infection to add SeV to the cells because, at this time point, all vIRFs would be expressed, thereby allowing us to assess their effect on the inhibition of IFN production in KSHV-infected cells. Incredibly, we found that the robust expression and secretion of SeV-induced IFN was comparably reduced by both WT and ΔvIRF BAC16 lytic infection (Fig. 8B and C).
Figure 8. vIRFs are dispensable for efficient silencing of type I IFN production during de novo lytic infection of iSLK-preRTA cells.
(A) Setup of the experiment. iSLK-preRTA cells were infected with WT or ΔvIRF KSHV for 48 hours followed by SeV (1 HA/ml) infection for 6 hours. (B) The early interferon response was analyzed by quantifying IFNβ mRNA 6 hours after SeV infection. (C) The late interferon response was analyzed by assessing the amount of IFNα/β secreted into the supernatant by type I IFN reporter bioassay 24 hours after SeV infection. The concentration of type I IFN was determined using an IFNβ standard curve. Error bars represent standard deviation (n=3). T-test was performed between SeV-treated WT and ΔvIRF samples (ND: not detected, ns: non-significant, *p<0.05).
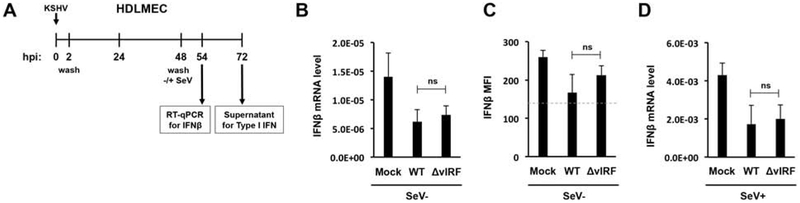
Figure 9. vIRFs are not needed for efficient suppression of type I IFN production during primary lytic infection of HDLMECs.
(A) Schematic of the experiment. Primary HDLMECs were infected by WT or ΔvIRF KSHV for 48 hours followed by SeV infection for 6 hours. (B) IFNβ gene expression in the absence of SeV infection was analyzed by RT-qPCR at 54 hpi. (C) IFNβ in the supernatant was measured by bead-based immunoassay. (D) IFNβ gene expression in the presence of SeV infection was measured by RT-qPCR at 54 hpi. Dashed line indicates the limit of assay detection. Error bars represent standard deviation (n=3). ns: non-significant.
Since RTA overexpression and/or NaB treatment alone also reduced IFN production slightly in iSLK-preRTA cells (Fig. 8B and C), we used HDLMECs to determine the effect of the lack of vIRFs on basal and SeV-induced IFN production during natural lytic KSHV infection (Fig. 9). First, HDLMECs were infected with WT or ΔvIRF BAC16 KSHV for 48 hrs and were subsequently treated with SeV for 6 or 24 hours (Fig. 9A). The results showed that BAC16-ΔvIRF could reduce both the basal level of IFNβ (Fig. 9B and C) and the SeV-induced IFNβ expression (Fig. 9D) similarly to WT KSHV infection. These findings are in line with the results of the iSLK-preRTA experiments (Fig. 8). Based on these data we concluded that while vIRFs may contribute to the inhibition of IFN production in KSHV-infected cells, they are not essential to it. As there are several viral factors other than vIRFs that can also inhibit the IFN signaling pathway in KSHV-infected cells, our data reinforce the idea that KSHV has functional redundancy built into its genome to ensure vigorous repression of the type I IFN pathway. Presumably, this redundancy would benefit KSHV if some of the viral anti-IFN factors were not expressed or became inactivated under certain conditions.
Reduced lytic gene expression and increased ISG expression in ΔvIRF KSHV-infected endothelial cells treated with IFNβ
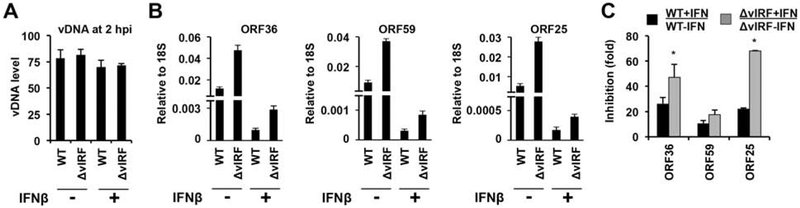
Type I IFN can induce hundreds of ISGs in cells triggering an antiviral state, which can block the replication of viruses and provide protection to host cells against viral infections (Jefferies, 2019). However, viral factors may overcome the IFN-induced antiviral effect to promote viral infection. Thus, we tested whether or not vIRFs can play any role in KSHV infection of lymphatic endothelial cells treated by IFNβ (Fig. 10). To this end, HDLMECs were pretreated with IFNβ for 24 hours, which was followed by infection with WT or BAC16-ΔvIRF KSHV for 24 hours. Figure 10A shows that KSHV infection was comparable between the IFNβ-treated and untreated cells. Analysis of the expression of viral genes (ORF36, ORF59, and ORF25) revealed that while lytic gene expression was increased in BAC16-ΔvIRF-infected cells compared to WT BAC16-infected cells, the IFNβ treatment significantly reduced viral gene expression in both cases (Fig. 10B). When we compared the fold reduction of viral gene expression in WT and ΔvIRF BAC16-infected cells upon IFNβ treatment, we observed that lytic gene expression was reduced to a greater extent by IFNβ in BAC16-ΔvIRF-infected cells (Fig. 10C).
Figure 10. IFNβ-driven reduction in lytic viral gene expression is more pronounced in BAC16-ΔvIRF-infected cells.
Primary HDLMECs were pretreated with IFNβ (500 IU/ml) for 24 hours or left untreated and then were infected by WT or BAC16-ΔvIRF KSHV. (A) KSHV vDNA level was analyzed by qPCR at 2 hpi. (B) RT-qPCR analysis of KSHV gene expression in WT and ΔvIRF KSHV-infected cells at 24 hpi in the absence or presence of IFNβ pretreatment. Error bars represent standard deviation (n=3). T-test was performed between WT and ΔvIRF samples (*p<0.05). (C) Fold inhibition of viral lytic gene expression in the presence of IFNβ relative to samples devoid of IFNβ treatment.
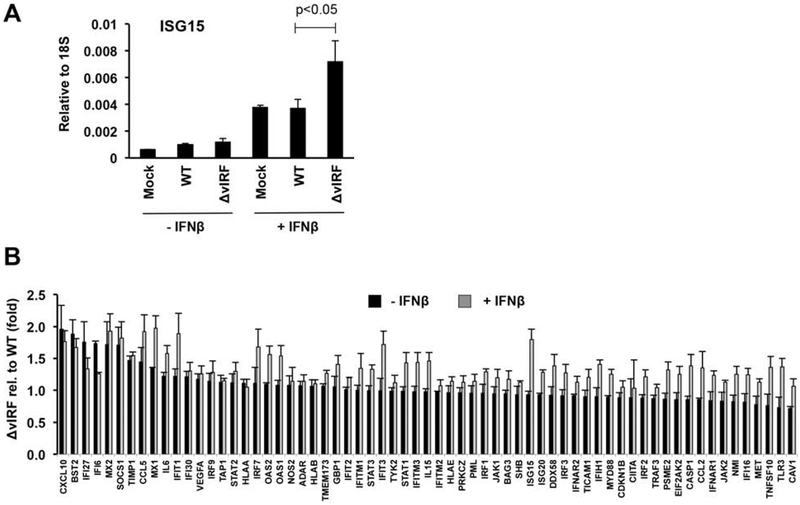
We found that the increased sensitivity of BAC16-ΔvIRF to IFNβ can be due to the elevated expression of ISGs such as ISG15, which has been shown to be inhibitory for lytic KSHV replication (Jacobs et al., 2015) (Fig. 11A). We also tested the expression changes of 84 human type I IFN response genes upon WT and BAC16-ΔvIRF infection both in the absence and presence of IFNβ (Fig. 11B). We calculated the differential host gene expression between WT and BAC16-ΔvIRF infection in the absence and presence of IFNβ and found that many other ISGs besides ISG15 were also increased in BAC16-ΔvIRF-infected cells compared to WT BAC16-infected cells in the presence of IFNβ (Fig. 11B). These results suggest that the lack of vIRFs can result in increased ISG expression, which in turn leads to the reduced lytic viral gene expression. Thus, vIRFs may play a role in facilitating lytic infection of cells that are in an antiviral state triggered by type I IFN.
Figure 11. Increased ISG expression in BAC16-ΔvIRF KSHV-infected endothelial cells in the presence of IFNβ.
Samples described in Fig 10 were used for analyzing the expression of ISGs. (A) RT-qPCR measurement of ISG15 gene expression. (B)Type I IFN pathway-related gene expressions were determined by RT-qPCR array. Error bars represent standard deviation (n=3).
Discussion
In this study, we presented for the first time a comparative analysis of the KSHV vIRFs regarding their protein expression, their requirement for viral lytic replication and virus production, and their necessity for inhibition of the host type I IFN pathway in different models of lytic KSHV infection. Using 3xFLAG-tagged vIRF recombinant KSHV clones, we directly compared the protein expression of vIRFs, which revealed that vIRF1 was expressed first and the most abundantly among vIRFs during lytic KSHV reactivation. In addition, we demonstrated that vIRF3, which is typically considered as a latent factor in PEL cells, can also be expressed as a true lytic factor in line with recent observations (Xiang et al., 2018). Using vIRF-KO KSHV mutants we found, surprisingly, that vIRFs were dispensable for KSHV production and inhibition of type I IFN expression. However, we showed that vIRFs can play a role in suppressing IFN-stimulated antiviral genes during KSHV infection, which may contribute to sustaining the viral lytic cycle in the presence of IFN.
Our finding, that vIRF3 is expressed during the lytic cycle of KSHV in epithelial and endothelial cells, proves that all four KSHV vIRFs can be lytic proteins. Initially, vIRF3 was shown to function as a latent factor in PEL cells, which is not detectable in KS biopsies (Rivas et al., 2001). These previous findings have been recently revisited and challenged by new results. Interestingly, when KSHV was lytically reactivated from a KSHV-infected stable B cell line (BJAB), vIRF3 expression was inducible, which result is contrast to studies in PEL cells, which constitutively express vIRF3 as a latent factor (Kati et al., 2013). In addition, a recent study has shown that vIRF3 can be expressed in up to 40% of KS biopsies (Lee et al., 2018) while another study also showed vIRF3 expression during lytic reactivation in iSLK-BAC16 cells (Xiang et al., 2018). By using HDLMECs, we have extended these results by demonstrating that vIRF3 is naturally expressed during the lytic cycle upon primary lytic KSHV infection. Thus, given this growing body of work on vIRF3, it may be reasonable to re-classify vIRF3 from a latent factor to a lytic factor that gained expression during the KSHV latent cycle in PEL cells.
Previous studies have shown that vIRFs may exert disparate effects on the KSHV lytic cycle by modulating the functions of distinct viral and cellular factors that are involved in the regulation of immune response pathways or viral gene expression. vIRF1 has been reported as an enhancer of viral lytic reactivation in endothelial cells by inhibiting viral replication-induced apoptosis (Choi and Nicholas, 2010), in epithelial cells by blocking the cGAS-STING DNA sensing pathway (Ma et al., 2015), and in PEL cells by modulating ISG15 conjugation and IFN response (Jacobs et al., 2015). In addition, vIRF1 was also shown to be able to block MAVS-regulated antiviral signaling and modulate mitophagy, which can also play a role in promoting the lytic cycle of KSHV (Hwang and Choi, 2016; Vo et al., 2019). Also, it was determined that vIRF4 can promote viral lytic replication by downregulating c-Myc expression and cooperating with RTA in the activation of some lytic promoters (Lee et al., 2013; Xi et al., 2012). In contrast, vIRF2 and vIRF3 were found to be repressors of viral lytic replication, which was determined by using vIRF2-KO BAC16 (Koch et al., 2019) and vIRF3-KO BAC16 (Xiang et al., 2018). Thus, it was surprising that we found that loss of vIRFs had no effect on productive KSHV infection in the context of RTA-driven lytic reactivation (Fig. 4), naturally lytic, primary infection of HDLMECs (Fig. 5) or RTA-driven primary lytic infection of iSLK epithelial cells (Fig. 6). The difference between our results and previous studies can be, in part, accounted for how the expression of vIRFs was inhibited in the different studies. We generated a STOP codon/frameshift mutation at the 5’ end of each vIRF, while previous studies have mutated the ATG start codon or used shRNAs to eliminate the expression of vIRFs. Regardless of how the vIRF knockout KSHV mutants were made, each study including ours, showed loss of the expression of full-length vIRFs, yet with different outcomes. We speculate that there may be vIRF isoforms produced by alternative splicing, internal AUG or alternative start codon usage, which can still be expressed even if the full-length vIRF expression was abrogated by point mutations. It is possible that these isoforms may still be functional to promote KSHV replication and/or suppress type I IFN production. In fact, it was reported that a number of unspliced and alternatively spliced RNA transcripts can be produced in the vIRF locus, which can potentially result in distinct vIRF isoforms (Bruce et al., 2017; Jenner et al., 2001a). Koch et al. also reported that vIRF2 may have several isoforms whose translation are initiated from internal start codons, which can explain the different phenotypes of vIRF2 knockouts, which were obtained by STOP codon mutation or gene deletion, in the regulation of lytic KSHV gene expression (Koch et al., 2019). Alternative translational or internal start codon usage is a common strategy of viruses to increase their coding capacity, which has also been reported for KSHV. For example, LANA, one of the latent viral proteins of KSHV, has been shown to use such strategies to generate multiple isoforms, which can function in different subcellular compartments of infected cells (Toptan et al., 2013; Zhang et al., 2016). If each vIRF has multiple isoforms as shown for vIRF2, the only way of abrogating the expression of both the full length and the isoforms of vIRFs completely is by deleting the genes or using shRNAs. We chose to make point mutation in the 5’ end of vIRF genes to avoid any effect on the expression of neighboring genes. However, we did make a vIRF locus deletion, which produced a comparable amount of virus relative to WT KSHV. We speculate that the positive and negative effects of vIRFs on KSHV replication and virus production may counteract one another, resulting in a phenotype similar to WT. We note, however, that our data are unique in that we have defined, for the first time, the contribution of the entire vIRF locus to virus production during natural lytic infection in HDLMECs (Golas et al., 2019). Our findings indicate that none of the full-length vIRFs are required for virus production under the conditions used in our study, which was confirmed by using ΔvIRF and QKO KSHV, as well as by single vIRF-KO KSHV mutants.
Since vIRFs may help KSHV to evade IFN-mediated immunity (Baresova et al., 2013; Jacobs and Damania, 2011; Lee et al., 2009), we also investigated whether vIRFs are necessary to reduce IFN expression during lytic KSHV infection. We found that while WT KSHV efficiently suppressed type I IFN expression at both the mRNA and protein levels, BAC16-ΔvIRF could still suppress type I IFN expression to a comparable extent (Fig. 7–9). These data indicate that vIRFs are redundant with other KSHV factors for the evasion of type I IFN production during lytic viral infection, at least in the cell lines we used in our study. Considering the evolutionary benefit of mitigating IFN production, it is not surprising that KSHV encodes several factors for this function. For example, LANA, ORF36, ORF45, ORF50 (RTA), ORF52, ORF64, and K8 are a few of the KSHV factors that have been demonstrated to suppress IFN expression (Hwang et al., 2009; Inn et al., 2011; Lefort et al., 2007; Wu et al., 2015; Yu and Hayward, 2010; Yu et al., 2005; Zhang et al., 2016; Zhu et al., 2002). However, we cannot rule out a cell-type specific benefit to KSHV for vIRFs such as in B cells or monocytes (Hwang and Choi, 2016; Jacobs et al., 2013; Lee et al., 2015). Unfortunately, primary infection of these cell types is not only notoriously inefficient, but would also lead to latency, making them unsuitable for testing the role of vIRFs as lytic factors during lytic primary infection (Bechtel et al., 2003). However, vIRFs might be advantageous to KSHV in vivo. Though no other human virus encodes homologs of the cellular IRFs, other gammaherpesviruses related to KSHV in non-human primates do encode vIRFs such as rhesus macaque rhadinovirus (RRV) and retroperitoneal fibromatosis herpesvirus of pig-tailed macaques (RFHV) (Koch and Schulz, 2017). Rhesus macaques infected by ΔvIRF RRV had lower viral loads in whole blood samples compared to WT RRV during the acute viral replication period, with ΔvIRF RRV even falling below the limit of detection in 6 out of 8 rhesus macaques for the entire study compared to none of the WT RRV-infected rhesus macaques at peak viral replication (Robinson et al., 2012). Robinson et al. also detected type I IFN more frequently in ΔvIRF RRV-infected rhesus macaque plasma. Therefore, the contributions of vIRFs to KSHV in vivo may be similar to those of RRV.
Importantly, we have found that KSHV vIRFs may contribute to the control of the IFN-stimulated effector response in the context of KSHV infection (Fig. 10 and 11). This conclusion extends previous observations that have been made for vIRF1 (Gao et al., 1997; Li et al., 1998; Zimring et al., 1998) and vIRF2 (Burýsek and Pitha, 2001; Fuld et al., 2006; Mutocheluh et al., 2011b) and would seem to fit with work that found that vIRF1 could extend B cell proliferation under conditions of IFN treatment (Flowers et al., 1998). Additionally, an elegant study by Springgay et al. demonstrated that RRV vIRFs are able to regulate the IFN effector response by impairing promyelocytic leukemia (PML)-associated responses and select ISG expressions (Springgay et al., 2019). It will be interesting to see if KSHV vIRFs can also utilize this, or related mechanisms in the context of viral infection as some KSHV vIRFs have already been identified to co-localize with PML nuclear bodies (Hossain et al., 2018).
In addition to disrupting IFN signaling, multiple vIRFs may also be needed to regulate different immune response pathways under various conditions, which can indirectly affect IFN production. For instance, vIRF1 was shown to bind to the vIL-6 promoter resulting in vIL-6 expression, which can activate interleukin (IL)-6 signaling (Park et al., 2007). This could allow vIRF1 (or other vIRFs) to drive inflammatory and/or pro-proliferative pathways that promote KSHV survival, yet may also affect viral-associated tumorigenesis. Interestingly, the secretion of vIL-6 can also repress the IFN-stimulated effector response (Chatterjee et al., 2002). Yet, a more comprehensive and unbiased characterization of the functions of vIRFs will be required during KSHV infection in different cell types using genomics approaches. Such analysis will lead not only to a more detailed understanding of vIRFs in immunity, but perhaps also to their distinct roles in DNA damage (Shin et al., 2006), apoptosis (Cousins and Nicholas, 2014), lymphangiogenesis (Lee et al., 2018), or yet uncharacterized cellular pathways.
Highlights.
We constructed 3xFLAG-tagged and vIRF-knockout KSHV clones for functional analyses.
vIRF1 is expressed earliest and strongest among vIRFs during the lytic cycle.
vIRFs are dispensable for KSHV production in vitro.
Inhibition of type I IFN expression does not require vIRFs during lytic infection.
vIRFs may play a role in the repression of the induction of ISGs.
Acknowledgement
We would like to sincerely thank Drs. Laurence Morel and Rolf Renne (UF College of Medicine) for their valuable comments to our IFN experiments and Dr. Shannon Wallet (UNC Chapel Hill) for providing us with HEK-Blue IFN-α/β cells. We also thank Dr. Frank Gibson (UF, Oral Biology Department) and the UF Center for Immunology and Transplantation for providing access to their equipment including flow cytometers and data analysis programs. We also thank the members of the Toth lab for their valuable comments and discussions. This study was supported in part by NIH grants R01AI132554 and R03DE028029 as well as UF Research Opportunity Seed Fund. GJG and SJJ were supported by NIH Training Grant T90DE021990.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
There is no conflict of interest.
References
- Aresté C, Blackbourn DJ, 2009. Modulation of the immune system by Kaposi’s sarcoma-associated herpesvirus. Trends Microbiol 17, 119–129. [DOI] [PubMed] [Google Scholar]
- Areste C, Mutocheluh M, Blackbourn DJ, 2009. Identification of caspase-mediated decay of interferon regulatory factor-3, exploited by a Kaposi sarcoma-associated herpesvirus immunoregulatory protein. The Journal of biological chemistry 284, 23272–23285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baresova P, Pitha PM, Lubyova B, 2013. Distinct roles of Kaposi’s sarcoma-associated herpesvirus-encoded viral interferon regulatory factors in inflammatory response and cancer. J Virol 87, 9398–9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtel JT, Liang Y, Hvidding J, Ganem D, 2003. Host range of Kaposi’s sarcoma-associated herpesvirus in cultured cells. Journal of virology 77, 6474–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AG, Barcy S, DiMaio T, Gan E, Garrigues HJ, Lagunoff M, Rose TM, 2017. Quantitative Analysis of the KSHV Transcriptome Following Primary Infection of Blood and Lymphatic Endothelial Cells. Pathogens 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulois K, Toth Z, Wong LY, Feng P, Gao SJ, Ensser A, Jung JU, 2014. Kaposi’s sarcoma-associated herpesvirus K3 and K5 ubiquitin E3 ligases have stage-specific immune evasion roles during lytic replication. J Virol 88, 9335–9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU, 2012. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J Virol 86, 9708–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burýsek L, Pitha PM, 2001. Latently expressed human herpesvirus 8-encoded interferon regulatory factor 2 inhibits double-stranded RNA-activated protein kinase. J Virol 75, 2345–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burysek L, Yeow WS, Pitha PM, 1999. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2). J Hum Virol 2, 19–32. [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM, 1995. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332, 1186–1191. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS, 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 266, 1865–1869. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Osborne J, Bestetti G, Chang Y, Moore PS, 2002. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 298, 1432–1435. [DOI] [PubMed] [Google Scholar]
- Choi YB, Nicholas J, 2010. Bim nuclear translocation and inactivation by viral interferon regulatory factor. PLoS pathogens 6, e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins E, Nicholas J, 2014. Molecular biology of human herpesvirus 8: novel functions and virus-host interactions implicated in viral pathogenesis and replication. Recent Results Cancer Res 193, 227–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C, Barnard S, Blackbourn DJ, Davison AJ, 2003. Transcription mapping of human herpesvirus 8 genes encoding viral interferon regulatory factors. J Gen Virol 84, 1471–1483. [DOI] [PubMed] [Google Scholar]
- Flowers CC, Flowers SP, Nabel GJ, 1998. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor confers resistance to the antiproliferative effect of interferon-alpha. Mol Med 4, 402–412. [PMC free article] [PubMed] [Google Scholar]
- Fuld S, Cunningham C, Klucher K, Davison AJ, Blackbourn DJ, 2006. Inhibition of interferon signaling by the Kaposi’s sarcoma-associated herpesvirus full-length viral interferon regulatory factor 2 protein. J Virol 80, 3092–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SJ, Boshoff C, Jayachandra S, Weiss RA, Chang Y, Moore PS, 1997. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene 15, 1979–1985. [DOI] [PubMed] [Google Scholar]
- Golas G, Alonso JD, Toth Z, 2019. Characterization of de novo lytic infection of dermal lymphatic microvascular endothelial cells by Kaposi’s sarcoma-associated herpesvirus. Virology 536, 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SM, Davis BK, West JA, Taxman DJ, Matsuzawa S, Reed JC, Ting JP, Damania B, 2011. Discovery of a viral NLR homolog that inhibits the inflammasome. Science 331, 330–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Taniguchi T, 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 6, 644–658. [DOI] [PubMed] [Google Scholar]
- Hossain MG, Ohsaki E, Honda T, Ueda K, 2018. Importance of Promyelocytic Leukema Protein (PML) for Kaposi’s Sarcoma-Associated Herpesvirus Lytic Replication. Front Microbiol 9, 2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang KY, Choi YB, 2016. Modulation of Mitochondrial Antiviral Signaling by Human Herpesvirus 8 Interferon Regulatory Factor 1. Journal of virology 90, 506–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Kim KS, Flano E, Wu TT, Tong LM, Park AN, Song MJ, Sanchez DJ, O’Connell RM, Cheng G, Sun R, 2009. Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell host & microbe 5, 166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SW, Kim D, Jung JU, Lee HR, 2017. KSHV-encoded viral interferon regulatory factor 4 (vIRF4) interacts with IRF7 and inhibits interferon alpha production. Biochem Biophys Res Commun 486, 700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inn KS, Lee SH, Rathbun JY, Wong LY, Toth Z, Machida K, Ou JH, Jung JU, 2011. Inhibition of RIG-I-mediated signaling by Kaposi’s sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J Virol 85, 10899–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SR, Damania B, 2011. The viral interferon regulatory factors of KSHV: immunosuppressors or oncogenes? Front Immunol 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SR, Gregory SM, West JA, Wollish AC, Bennett CL, Blackbourn DJ, Heise MT, Damania B, 2013. The viral interferon regulatory factors of Kaposi’s sarcoma-associated herpesvirus differ in their inhibition of interferon activation mediated by toll-like receptor 3. J Virol 87, 798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SR, Stopford CM, West JA, Bennett CL, Giffin L, Damania B, 2015. Kaposi’s Sarcoma-Associated Herpesvirus Viral Interferon Regulatory Factor 1 Interacts with a Member of the Interferon-Stimulated Gene 15 Pathway. J Virol 89, 11572–11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies CA, 2019. Regulating IRFs in IFN Driven Disease. Front Immunol 10, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner RG, Alba MM, Boshoff C, Kellam P, 2001a. Kaposi’s sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. Journal of virology 75, 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner RG, Albà MM, Boshoff C, Kellam P, 2001b. Kaposi’s sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J Virol 75, 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kati S, Tsao EH, Gunther T, Weidner-Glunde M, Rothamel T, Grundhoff A, Kellam P, Schulz TF, 2013. Activation of the B cell antigen receptor triggers reactivation of latent Kaposi’s sarcoma-associated herpesvirus in B cells. Journal of virology 87, 8004–8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S, Damas M, Freise A, Hage E, Dhingra A, Ruckert J, Gallo A, Kremmer E, Tegge W, Bronstrup M, Brune W, Schulz TF, 2019. Kaposi’s sarcoma-associated herpesvirus vIRF2 protein utilizes an IFN-dependent pathway to regulate viral early gene expression. PLoS pathogens 15, e1007743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S, Schulz TF, 2017. Rhadinoviral interferon regulatory factor homologues. Biol Chem 398, 857–870. [DOI] [PubMed] [Google Scholar]
- Lee HR, Amatya R, Jung JU, 2015. Multi-step regulation of innate immune signaling by Kaposi’s sarcoma-associated herpesvirus. Virus research 209, 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HR, Brulois K, Wong L, Jung JU, 2012. Modulation of Immune System by Kaposi’s Sarcoma-Associated Herpesvirus: Lessons from Viral Evasion Strategies. Front Microbiol 3, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HR, Doganay S, Chung B, Toth Z, Brulois K, Lee S, Kanketayeva Z, Feng P, Ha T, Jung JU, 2013. KSHV vIRF4 targets cellular IRF4 and Myc gene expressions to facilitate lytic replication. Journal of virology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HR, Kim MH, Lee JS, Liang C, Jung JU, 2009. Viral interferon regulatory factors. J Interferon Cytokine Res 29, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HR, Li F, Choi UY, Yu HR, Aldrovandi GM, Feng P, Gao SJ, Hong YK, Jung JU, 2018. Deregulation of HDAC5 by Viral Interferon Regulatory Factor 3 Plays an Essential Role in Kaposi’s Sarcoma-Associated Herpesvirus-Induced Lymphangiogenesis. MBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort S, Soucy-Faulkner A, Grandvaux N, Flamand L, 2007. Binding of Kaposi’s sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J Virol 81, 10950–10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Damania B, Alvarez X, Ogryzko V, Ozato K, Jung JU, 2000. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol Cell Biol 20, 8254–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Lee H, Guo J, Neipel F, Fleckenstein B, Ozato K, Jung JU, 1998. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor. J Virol 72, 5433–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Genin P, Mamane Y, Sgarbanti M, Battistini A, Harrington WJ, Barber GN, Hiscott J, 2001. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20, 800–811. [DOI] [PubMed] [Google Scholar]
- Lubyova B, Pitha PM, 2000. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J Virol 74, 8194–8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B, 2015. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proceedings of the National Academy of Sciences of the United States of America 112, E4306–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsili G, Perrotti E, Remoli AL, Acchioni C, Sgarbanti M, Battistini A, 2016. IFN Regulatory Factors and Antiviral Innate Immunity: How Viruses Can Get Better. J Interferon Cytokine Res 36, 414–432. [DOI] [PubMed] [Google Scholar]
- Moore PS, Boshoff C, Weiss RA, Chang Y, 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274, 1739–1744. [DOI] [PubMed] [Google Scholar]
- Mutocheluh M, Hindle L, Areste C, Chanas SA, Butler LM, Lowry K, Shah K, Evans DJ, Blackbourn DJ, 2011a. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor-2 inhibits type 1 interferon signalling by targeting interferon-stimulated gene factor-3. The Journal of general virology 92, 2394–2398. [DOI] [PubMed] [Google Scholar]
- Mutocheluh M, Hindle L, Aresté C, Chanas SA, Butler LM, Lowry K, Shah K, Evans DJ, Blackbourn DJ, 2011b. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor-2 inhibits type 1 interferon signalling by targeting interferon-stimulated gene factor-3. J Gen Virol 92, 2394–2398. [DOI] [PubMed] [Google Scholar]
- Myoung J, Ganem D, 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J Virol Methods 174, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Li M, Zarycki J, Jung JU, 2001. Inhibition of p53 tumor suppressor by viral interferon regulatory factor. J Virol 75, 7572–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU, 2003. Global changes in Kaposi’s sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. Journal of virology 77, 4205–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee MS, Yoo SM, Jeong KW, Lee D, Choe J, Seo T, 2007. Identification of the DNA sequence interacting with Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor 1. J Virol 81, 12680–12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizzotto MN, Uldrick TS, Hu D, Yarchoan R, 2012. Clinical Manifestations of Kaposi Sarcoma Herpesvirus Lytic Activation: Multicentric Castleman Disease (KSHV-MCD) and the KSHV Inflammatory Cytokine Syndrome. Front Microbiol 3, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas C, Thlick AE, Parravicini C, Moore PS, Chang Y, 2001. Kaposi’s sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. Journal of virology 75, 429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BA, O’Connor MA, Li H, Engelmann F, Poland B, Grant R, DeFilippis V, Estep RD, Axthelm MK, Messaoudi I, Wong SW, 2012. Viral interferon regulatory factors are critical for delay of the host immune response against rhesus macaque rhadinovirus infection. Journal of virology 86, 2769–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS, 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci U S A 93, 14862–14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, 2014. Interferon-stimulated genes: roles in viral pathogenesis. Curr Opin Virol 6, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo T, Park J, Lee D, Hwang SG, Choe J, 2001. Viral interferon regulatory factor 1 of Kaposi’s sarcoma-associated herpesvirus binds to p53 and represses p53-dependent transcription and apoptosis. Journal of virology 75, 6193–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YC, Nakamura H, Liang X, Feng P, Chang H, Kowalik TF, Jung JU, 2006. Inhibition of the ATM/p53 signal transduction pathway by Kaposi’s sarcoma-associated herpesvirus interferon regulatory factor 1. Journal of virology 80, 2257–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L, 1995. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 86, 1276–1280. [PubMed] [Google Scholar]
- Springgay LK, Fitzpatrick K, Park B, Estep RD, Wong SW, 2019. Rhesus Macaque Rhadinovirus Encodes a Viral Interferon Regulatory Factor To Disrupt Promyelocytic Leukemia Nuclear Bodies and Antagonize Type I Interferon Signaling. Journal of virology 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T, 2008. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 26, 535–584. [DOI] [PubMed] [Google Scholar]
- Tischer BK, von Einem J, Kaufer B, Osterrieder N, 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40, 191–197. [DOI] [PubMed] [Google Scholar]
- Tomescu C, Law WK, Kedes DH, 2003. Surface downregulation of major histocompatibility complex class I, PE-CAM, and ICAM-1 following de novo infection of endothelial cells with Kaposi’s sarcoma-associated herpesvirus. J Virol 77, 9669–9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toptan T, Fonseca L, Kwun HJ, Chang Y, Moore PS, 2013. Complex alternative cytoplasmic protein isoforms of the Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 generated through noncanonical translation initiation. Journal of virology 87, 2744–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth Z, Papp B, Brulois K, Choi YJ, Gao SJ, Jung JU, 2016. LANA-Mediated Recruitment of Host Polycomb Repressive Complexes onto the KSHV Genome during De Novo Infection. PLoS Pathog 12, e1005878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldrick TS, Wang V, O’Mahony D, Aleman K, Wyvill KM, Marshall V, Steinberg SM, Pittaluga S, Maric I, Whitby D, Tosato G, Little RF, Yarchoan R, 2010. An interleukin-6-related systemic inflammatory syndrome in patients co-infected with Kaposi sarcoma-associated herpesvirus and HIV but without Multicentric Castleman disease. Clin Infect Dis 51, 350–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo MT, Smith BJ, Nicholas J, Choi YB, 2019. Activation of NIX-mediated mitophagy by an interferon regulatory factor homologue of human herpesvirus. Nat Commun 10, 3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, Konrad A, Neipel F, Sturzl M, Whitby D, Li H, Zhu F, 2015. Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein. Cell Host Microbe 18, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi X, Persson LM, O’Brien MW, Mohr I, Wilson AC, 2012. Cooperation between viral interferon regulatory factor 4 and RTA to activate a subset of Kaposi’s sarcoma-associated herpesvirus lytic promoters. Journal of virology 86, 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q, Ju H, Li Q, Mei SC, Chen D, Choi YB, Nicholas J, 2018. Human Herpesvirus 8 Interferon Regulatory Factors 1 and 3 Mediate Replication and Latency Activities via Interactions with USP7 Deubiquitinase. Journal of virology 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Hayward GS, 2010. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 33, 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang SE, Hayward GS, 2005. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22, 59–70. [DOI] [PubMed] [Google Scholar]
- Zhang G, Chan B, Samarina N, Abere B, Weidner-Glunde M, Buch A, Pich A, Brinkmann MM, Schulz TF, 2016. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proceedings of the National Academy of Sciences of the United States of America 113, E1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FX, King SM, Smith EJ, Levy DE, Yuan Y, 2002. A Kaposi’s sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc Natl Acad Sci U S A 99, 5573–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimring JC, Goodbourn S, Offermann MK, 1998. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol 72, 701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]