Abstract
Jumbo squid (Dosidicus gigas) is one of the largest cephalopods, and represents an important economic fishery in several regions of the Pacific Ocean, from southern California in the United States to southern Chile. Large and considerable discards of this species, such as skin, have been reported to constitute an important source of potential by-products. In this paper, a shotgun proteomics approach was applied for the first time to the characterization of the jumbo squid (Dosidicus gigas) skin proteome. A total of 1004 different peptides belonging to 219 different proteins were identified. The final proteome compilation was investigated by integrated in-silico studies, including gene ontology (GO) term enrichment, pathways, and networks studies. Potential new valuable bioactive peptides such as antimicrobial, bioactive collagen peptides, antihypertensive and antitumoral peptides were predicted to be present in the jumbo squid skin proteome. The integration of the global proteomics results and the bioinformatics analysis of the jumbo squid skin proteome show a comprehensive knowledge of this fishery discard and provide potential bioactive peptides of this marine by-product.
Keywords: Dosidicus gigas, squid, skin, by-product, shotgun proteomics, mass spectrometry, protein-based bioinformatics, bioactive peptides
1. Introduction
Marine by-products are the body parts of marine species that are removed before they reach the final consumer in order to improve their preservation, reduce the shipping weight, and increase the quality of the main product [1,2]. These organic materials are the main concern for current fishery management policies and legislation because they represent a significant source of valuable compounds such as proteins, minerals and lipids. In fact, from 2019 new regulations of fishery landing in the European Commission (EU) (European Commission Regulation (EU) No 1380/2013) oblige to keep and not discard all the species that are caught that are subjected to quota as well as underutilized commercial species [3]. For this reason, valorization solutions of marine discards biomasses have to be implemented. These new potential bioactive compounds could be used for human nutrition, as well as for their functional properties for nutraceutical, pharmaceutical, and cosmeceuticals industries [4,5,6,7].
Jumbo squid (Dosidicus gigas), also known as Humboldt squid, is one of the largest cephalopods and lives in the waters of the Humboldt Current in the eastern Pacific Ocean. It represents an important economic fishery resource in a wide number of countries such as Chile, Peru, Japan, and Mexico [8]. Nevertheless, only the jumbo squid mantle is marketed. During its processing, large amounts (up to 60% of whole weight) of squid off-products, such as skin, heads, fins, tentacles, and guts are generated and discarded [9].
By-products of the jumbo squid have recently attracted great attention due to the discovery of the presence of several relevant bioactive compounds. These include valuable and profitable bio-ingredients such as chitin, chitosan, collagen, gelatin, and pigments [10,11,12,13,14].
Particularly, the skin constitutes a significant sub-product in the jumbo squid fishery industry. Skin is actually a biological cooperative tissue formed by four different tissue types (epithelial, connective, muscle, and nerve tissues). Peptides derived from a tryptic hydrolysate of jumbo squid skin exhibited strong inhibition of lipid peroxidation that was much higher than the natural antioxidant α-tocopherol [15]. Skin molecules as xanthommatin also showed in vitro antioxidant effects [16]. Additionally, cytotoxic, antimicrobial, anti-biofilm, angiotensin converting enzyme (ACE)-inhibitory peptides, and anti-tumoral properties have been demonstrated for skin ink and the hydrolyzed skin of different squid species [14,17,18]. Recently, the inclusion on ice of a jumbo squid skin extract led to a remarkable microbial inhibition and a significant shelf life extension during fish chilled storage [19,20]. However, the global characterization of proteins and peptides from jumbo skin proteome has not been investigated to date.
Proteomics, as the discipline for the large-scale analysis of proteins of a particular biological system, has greatly contributed to the assessment of quality, safety, and bioactivity of seafood products [21,22,23,24]. In a shotgun proteomics approach, a mixture of proteins is digested with a protease (i.e., trypsin), and the resulting mixture of peptides is then analyzed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) [25]. Using database searching programs, like SEQUEST [26] or Mascot [27], fragmentation spectra obtained are assigned to putative peptide sequences and the assignments are then validated with programs like PeptideProphet [28] or Percolator [29]. The identification of these peptides allows for the identification of proteins present in the complex mixture.
Additionally, potential bioactive proteins and peptides can be characterized by protein-based bioinformatics tools. Such software includes programs to simulate in-silico proteolysis and to predict the physicochemical properties of the released peptides (i.e., antihypertensive, antimicrobial, immunomodulatory). Several bioactive peptide databases are available online such as APD3 [30], BioPep [31], BioPD [32], BioPepDB [33], CAMP [34], PPIP [35], starPepDB [36] and StraPep [37].
Therefore, the present work focuses for the first time on the global characterization of the jumbo squid (Dosidicus gigas) skin proteome using a shotgun proteomic approach. Meanwhile, a combination of different protein-based bioinformatics programs is carried out to determine potential bioactive peptides of this marine discard.
2. Results and Discussion
2.1. Jumbo Squid (Dosidicus gigas) Skin Proteome
A shotgun proteomics analysis for the jumbo squid (Dosidicus gigas) skin proteome is presented in this work, to our knowledge, for the first time. This repository was created merging a total of 6559 identified spectra (PSMs) from 1004 different peptides belonging to 219 different non-redundant annotated proteins from the different sample replicates (n = 4) (Supplementary Tables S1–S3). Table 1 summarizes the list of the non-redundant annotated proteins of the jumbo squid skin proteome (n = 219). This discovery stage was based on the LC-MS/MS analysis and SEQUEST-HT search of the tryptic digestions for the global protein extracts from the skin of each jumbo squid specimens studied (A–D replicates).
Table 1.
Jumbo squid (Dosidicus gigas) skin proteome (FDR < 1%). See Supplementary Tables S1–S3 for complete information.
| N | Accession | Description | Gene | Uni. Pep. | PSM | Cov. (%) |
|---|---|---|---|---|---|---|
| 1 | A0A1Y1DCG9 | Paramyosin OS = Dosidicus gigas | DgPm | 17 | 46 | 22 |
| 2 | A0A2Z5EQ31 | Symplectin/biotinidase-like protein OS = Dosidicus gigas | sympp | 1 | 2 | 3 |
| 3 | A0A0P0UX03 | Hemocyanin subunit 1 OS = Todarodes pacificus | Tphcy | 116 | 3007 | 38 |
| 4 | A0A077B1P8 | Hemocyanin subunit 2 OS = Euprymna scolopes | HCY2 | 10 | 1608 | 24 |
| 5 | A0A077B6R8 | Hemocyanin subunit 1 OS = Euprymna scolopes | HCY1 | 13 | 1437 | 19 |
| 6 | T2F8L5 | Hemocyanin OS = Sepiella maindroni | HCY1 | 8 | 1544 | 18 |
| 7 | W6CNR9 | Hemocyanin subunit 3 OS = Sepia officinalis | HCY3 | 10 | 1035 | 13 |
| 8 | A0A1Q2SJF4 | Hemocyanin-like protein OS = Uroteuthis edulis | hc | 8 | 746 | 14 |
| 9 | F1ADJ4 | Myosin heavy chain OS = Todarodes pacificus | MYH | 16 | 456 | 15 |
| 10 | I0JGT9 | Actin I OS = Sepia officinalis | ACTI | 11 | 202 | 53 |
| 11 | G4V4Y8 | Myosin heavy chain isoform C OS = Doryteuthis pealeii | MYH | 3 | 411 | 12 |
| 12 | A4D0I0 | Hemocyanin subunit 1 OS = Todarodes pacificus | Tphcy | 6 | 174 | 50 |
| 13 | A0A0P0UX01 | Hemocyanin subunit2 OS = Todarodes pacificus | Tphcy | 4 | 171 | 51 |
| 14 | A0A0L8G4B4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22000685mg | 27 | 53 | 13 |
| 15 | V6A729 | Myosin heavy chain isoform A OS = Octopus bimaculoides | MYH | 2 | 348 | 8 |
| 16 | Q2V0V2 | Tropomyosin OS = Todarodes pacificus | tp-tm | 27 | 127 | 46 |
| 17 | A0A0L8GFI1 | Spectrin beta chain OS = Octopus bimaculoides | OCBIM_22034275mg | 24 | 72 | 12 |
| 18 | I7H9I6 | Haemocyanin OS = Nautilus pompilius | hc | 1 | 532 | 5 |
| 19 | A0A075IT96 | Heat shock protein 70 OS = Sepiella maindroni | HSP70 | 3 | 59 | 23 |
| 20 | A0A0L8HMH4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22011261mg | 12 | 35 | 3 |
| 21 | E7CLR5 | Hemocyanin (Fragment) OS = Spirula spirula | HCY1 | 1 | 315 | 12 |
| 22 | A0A0L8IA52 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22026555mg | 1 | 49 | 18 |
| 23 | A0A0L8GPG8 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22030693mg | 11 | 59 | 17 |
| 24 | A0A0L8FFZ3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22022789mg | 2 | 394 | 30 |
| 25 | A0A0L8H027 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22024964mg | 8 | 48 | 5 |
| 26 | A0A0L8G0V9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22003270mg | 6 | 32 | 16 |
| 27 | Q06270 | Intermediate filament protein OS = Nototodarus sloanii | OCBIM_22025455mg | 9 | 38 | 18 |
| 28 | Q76EJ2 | Cathepsin D OS = Todarodes pacificus | tpaD | 9 | 49 | 22 |
| 29 | P08052 | Myosin regulatory light chain LC-2, mantle muscle OS = Todarodes pacificus | MYL | 8 | 23 | 50 |
| 30 | A0A0L8HC80 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22017953mg | 8 | 16 | 5 |
| 31 | A0A0L8G3E9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22001601mg | 1 | 31 | 11 |
| 32 | P30842 | Omega-crystallin OS = Nototodarus sloanii | N/A | 5 | 22 | 9 |
| 33 | Q68LN1 | Filamin OS = Euprymna scolopes | OCBIM_22031719mg | 4 | 20 | 34 |
| 34 | A0A0L8FU30 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22007941mg | 1 | 12 | 33 |
| 35 | A0A0L8I9I4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22028792mg | 1 | 18 | 22 |
| 36 | A0A0L8FNC4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22013362mg | 5 | 12 | 4 |
| 37 | Q6E216 | Tropomysin-like protein OS = Todarodes pacificus | ATRP | 5 | 9 | 26 |
| 38 | A0A0L8HDP4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22016840mg | 3 | 27 | 5 |
| 39 | A0A0L8FVD0 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22007411mg | 4 | 15 | 27 |
| 40 | A0A0L8GWE3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22026600mg | 3 | 13 | 12 |
| 41 | A0A0L8HKK9 | Fructose-bisphosphate aldolase OS = Octopus bimaculoides | OCBIM_22013272mg | 3 | 21 | 7 |
| 42 | A0A0L8FP56 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22013360mg | 1 | 16 | 3 |
| 43 | G1CW44 | Triosephosphate isomerase OS = Enteroctopus dofleini | OCBIM_22037419mg | 1 | 27 | 11 |
| 44 | G1CW45 | Triosephosphate isomerase OS = Euprymna scolopes | OCBIM_22037419mg | 1 | 8 | 19 |
| 45 | A0A0L8GN79 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22030767mg | 2 | 11 | 9 |
| 46 | A0A0L8FZT7 | Protein disulfide-isomerase OS = Octopus bimaculoides | OCBIM_22003356mg | 3 | 17 | 8 |
| 47 | A0A0L8H0K3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22024969mg | 3 | 8 | 7 |
| 48 | A0A0L8GNQ0 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22030666mg | 2 | 5 | 10 |
| 49 | A0A0L8IA72 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025549mg | 4 | 8 | 7 |
| 50 | A0A0L8IAK7 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025100mg | 5 | 9 | 1 |
| 51 | A0A0L8HDG9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22017348mg | 3 | 4 | 20 |
| 52 | Q86DP6 | Malate dehydrogenase (Fragment) OS = Sepia officinalis | Mdh | 3 | 7 | 11 |
| 53 | P05945 | Myosin catalytic light chain LC-1, mantle muscle OS = Todarodes pacificus | MYL | 2 | 6 | 19 |
| 54 | A0A0L8GQL2 | Tubulin beta chain OS = Octopus bimaculoides | OCBIM_22029847mg | 3 | 8 | 8 |
| 55 | A0A0L8HMP5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22011994mg | 3 | 10 | 16 |
| 56 | A0A0L8IAD9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025091mg | 3 | 3 | 6 |
| 57 | A0A0L8FJA0 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22017780mg | 2 | 6 | 3 |
| 58 | A0A0L8G425 | Adenosylhomocysteinase OS = Octopus bimaculoides | OCBIM_22000532mg | 3 | 6 | 7 |
| 59 | A0A0L8FXP2 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22004658mg | 3 | 3 | 5 |
| 60 | A0A0L8I198 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22039192mg | 3 | 8 | 19 |
| 61 | A0A0L8I871 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22028797mg | 2 | 7 | 18 |
| 62 | A0A2S1FRU3 | Elongation factor 1-alpha OS = Callistoctopus minor | EEF1A1 | 4 | 6 | 7 |
| 63 | A0A0L8FFD9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22023810mg | 2 | 3 | 2 |
| 64 | A0A0L8I874 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22028979mg | 2 | 6 | 16 |
| 65 | A0A0L8FK19 | Tubulin alpha chain OS = Octopus bimaculoides | OCBIM_22016917mg | 2 | 5 | 3 |
| 66 | A0A0K0WTY3 | Arginine kinase OS = Sepia pharaonis | AK | 4 | 7 | 7 |
| 67 | A0A0L8GXA0 | Glucosamine-6-phosphate isomerase OS = Octopus bimaculoides | OCBIM_22026276mg | 1 | 3 | 9 |
| 68 | F8V2T7 | Sodium/potassium-transporting ATPase subunit alpha OS = Bathypolypus arcticus | OCBIM_22028074mg | 2 | 4 | 2 |
| 69 | A0A0L8H4W4 | Proteasome subunit alpha type OS = Octopus bimaculoides | OCBIM_22022293mg | 2 | 3 | 10 |
| 70 | A0A0L8GSZ5 | Histone H4 OS = Octopus bimaculoides | OCBIM_22029078mg | 2 | 5 | 10 |
| 71 | A0A0L8GDJ1 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22035502mg | 2 | 4 | 6 |
| 72 | A0A159BRC2 | ColAa OS = Sepia pharaonis | N/A | 2 | 6 | 1 |
| 73 | A0A0L8FIB5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22020215mg | 1 | 2 | 5 |
| 74 | A0A0L8G4U5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22000359mg | 2 | 4 | 6 |
| 75 | Q9NL93 | G protein a subunit o class OS = Octopus vulgaris | OvGao | 2 | 5 | 6 |
| 76 | A0A0L8IG11 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22004528mg | 2 | 8 | 11 |
| 77 | A0A0L8GG89 | Proteasome subunit alpha OS = Octopus bimaculoides | OCBIM_22033871mg | 2 | 3 | 9 |
| 78 | A0A0L8H716 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22020867mg | 2 | 12 | 11 |
| 79 | A0A0S1U346 | Triosephosphate isomerase OS = Amphioctopus fangsiao | OCBIM_22037419mg | 1 | 3 | 18 |
| 80 | A0A0L8H4E7 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22022663mg | 2 | 4 | 6 |
| 81 | A0A0L8I919 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22027793mg | 1 | 7 | 5 |
| 82 | A0A0L8HN83 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22010679mg | 1 | 1 | 3 |
| 83 | A0A0L8ICB5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22019476mg | 2 | 4 | 4 |
| 84 | A0A0L8FMD3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22014986mg | 1 | 2 | 4 |
| 85 | A0A0L8H0E1 | Sorting nexin OS = Octopus bimaculoides | OCBIM_22024936mg | 1 | 5 | 3 |
| 86 | A0A0L8IA39 | Tubulin alpha chain OS = Octopus bimaculoides | OCBIM_22026381mg | 1 | 2 | 3 |
| 87 | A0A0L8IG73 | Malic enzyme OS = Octopus bimaculoides | OCBIM_22004207mg | 1 | 1 | 3 |
| 88 | A0A0L8H635 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22021483mg | 1 | 2 | 8 |
| 89 | A0A0L8GYT6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22026168mg | 1 | 3 | 10 |
| 90 | A0A0L8GFD5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22034343mg | 1 | 2 | 3 |
| 91 | A0A0L8HKN4 | Ornithine aminotransferase OS = Octopus bimaculoides | OCBIM_22012517mg | 1 | 4 | 3 |
| 92 | A0A0L8G0I6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22003454mg | 2 | 2 | 4 |
| 93 | A0A0L8HE61 | AP complex subunit beta OS = Octopus bimaculoides | OCBIM_22016805mg | 1 | 1 | 1 |
| 94 | A0A0L8HMS6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22011048mg | 1 | 3 | 3 |
| 95 | A0A0L8FWD6 | Calcium-transporting ATPase OS = Octopus bimaculoides | OCBIM_22006279mg | 2 | 6 | 2 |
| 96 | A0A0L8GP54 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22030838mg | 1 | 2 | 7 |
| 97 | A0A0L8G9P1 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22037676mg | 1 | 4 | 9 |
| 98 | A0A0L8HTA6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22007620mg | 1 | 4 | 6 |
| 99 | A0A0L8IAN9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025097mg | 1 | 1 | 8 |
| 100 | A0A0L8HCU8 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22018310mg | 1 | 3 | 2 |
| 101 | A0A0A7NZU2 | Putative chitotriosidase OS = Euprymna scolopes | Chia | 1 | 1 | 4 |
| 102 | A0A0L8G3Z0 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22000581mg | 1 | 3 | 4 |
| 103 | A0A0L8I836 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22028993mg | 1 | 3 | 3 |
| 104 | A0A0L8IDP3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22014847mg | 1 | 1 | 4 |
| 105 | A0A0L8FZ08 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22004461mg | 1 | 1 | 1 |
| 106 | A0A0L8GZM9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025211mg | 1 | 4 | 2 |
| 107 | A0A193PD55 | Chitinase OS = Todarodes pacificus | TpChi | 1 | 2 | 2 |
| 108 | Q8IS80 | 60S acidic ribosomal protein OS = Euprymna scolopes | OCBIM_22035130mg | 1 | 3 | 19 |
| 109 | A0A0L8FQ90 | Serine/threonine-protein phosphatase OS = Octopus bimaculoides | OCBIM_22011907mg | 1 | 1 | 4 |
| 110 | A0A0L8FIY8 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22018177mg | 1 | 3 | 13 |
| 111 | A0A0L8I107 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22039276mg | 1 | 2 | 4 |
| 112 | A0A0L8G4M6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22000216mg | 1 | 2 | 0 |
| 113 | A0A0L8GLC5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22031874mg | 1 | 3 | 8 |
| 114 | A0A0L8HDX1 | Superoxide dismutase OS = Octopus bimaculoides | OCBIM_22016770mg | 1 | 2 | 6 |
| 115 | A0A0L8HU31 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22005978mg | 1 | 2 | 3 |
| 116 | Q8SWQ7 | Non-muscle myosin II heavy chain OS = Doryteuthis pealeii | MYH | 1 | 1 | 1 |
| 117 | B8Q2 × 2 | G alpha q subunit OS = Euprymna scolopes | COI | 1 | 1 | 5 |
| 118 | A0A0L8G1S2 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22001882mg | 1 | 1 | 3 |
| 119 | A0A0L8HAV5 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22019117mg | 1 | 1 | 7 |
| 120 | A0A0L8IDX1 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22013485mg | 1 | 4 | 4 |
| 121 | A0A0L8GRX5 | Histone H2B OS = Octopus bimaculoides | OCBIM_22029075mg | 1 | 1 | 6 |
| 122 | A0A0L8FS75 | Proteasome subunit alpha type OS = Octopus bimaculoides | OCBIM_22010113mg | 1 | 2 | 4 |
| 123 | A0A0L8FRK2 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22010655mg | 1 | 2 | 6 |
| 124 | A0A0L8GZX1 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025682mg | 1 | 5 | 1 |
| 125 | A0A0L8G456 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22000796mg | 1 | 1 | 6 |
| 126 | A0A0L8FF63 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22024380mg | 1 | 1 | 10 |
| 127 | A0A0L8H8U9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22020735mg | 1 | 1 | 5 |
| 128 | A0A0L8I5N4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22033390mg | 1 | 2 | 3 |
| 129 | A0A0L8I398 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22037157mg | 1 | 1 | 11 |
| 130 | A0A0L8GP93 | Nicotinamide-nucleotide adenylyltransferase OS = Octopus bimaculoides | OCBIM_22030204mg | 1 | 1 | 6 |
| 131 | A0A0L8IIH3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025740mg | 1 | 3 | 0 |
| 132 | A0A0L8GZD4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025455mg | 1 | 1 | 1 |
| 133 | A0A0L8HQW9 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22008430mg | 1 | 4 | 2 |
| 134 | A0A0L8G2Z7 | Small ubiquitin-related modifier OS = Octopus bimaculoides | OCBIM_22001102mg | 1 | 1 | 11 |
| 135 | A0A0L8G8L3 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22038063mg | 1 | 2 | 2 |
| 136 | O46345 | S-syntaxin OS = Doryteuthis pealeii | STX1 | 1 | 1 | 3 |
| 137 | A0A0L8GDD2 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22036000mg | 1 | 1 | 2 |
| 138 | C4N147 | Sodium/calcium exchanger regulatory protein 1 OS = Doryteuthis pealeii | SLC8A1 | 1 | 4 | 7 |
| 139 | A0A0L8FJE4 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22017696mg | 1 | 2 | 2 |
| 140 | A0A0L8I067 | Kinesin-like protein OS = Octopus bimaculoides | OCBIM_22000619mg | 1 | 1 | 1 |
| 141 | A0A0L8FYB6 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22005155mg | 1 | 1 | 1 |
| 142 | A0A0L8GUV0 | Serine/threonine-protein phosphatase OS = Octopus bimaculoides | OCBIM_22027338mg | 1 | 1 | 2 |
| 143 | A0A0L8GJ12 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22032700mg | 1 | 2 | 1 |
| 144 | A0A0L8GLG2 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22032112mg | 1 | 1 | 1 |
| 145 | A0A0L8GY97 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22026356mg | 1 | 2 | 2 |
| 146 | Q27Q56 | Hemocyanin subunit 2 OS = Sepia officinalis | HCY2 | 1 | 961 | 7 |
| 147 | A0A161HPY5 | Actin OS = Crassostrea brasiliana | ACTI | 3 | 96 | 38 |
| 148 | D2YZ90 | Beta actin OS = Idiosepius paradoxus | ACTI | 2 | 95 | 37 |
| 149 | K1QFR9 | Spectrin beta chain OS = Crassostrea gigas | CGI_10013845 | 1 | 34 | 4 |
| 150 | C1KC83 | Heat shock cognate protein 70 OS = Haliotis diversicolor | HSP70 | 1 | 37 | 16 |
| 151 | A0A2C9K1T4 | Uncharacterized protein OS = Biomphalaria glabrata | 106078167 | 1 | 68 | 13 |
| 152 | A0A0B7B7H2 | Uncharacterized protein OS = Arion vulgaris | ORF162822 | 1 | 40 | 11 |
| 153 | A0A2T7NLR4 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_17913 | 1 | 18 | 5 |
| 154 | K1RH58 | Alpha-actinin, sarcomeric OS = Crassostrea gigas | CGI_10003110 | 1 | 43 | 10 |
| 155 | A0A2P1H676 | Heat shock protein 70 OS = Diplodon chilensis | HSP70 | 1 | 36 | 12 |
| 156 | K1PMY9 | Calmodulin OS = Crassostrea gigas | CGI_10006482 | 1 | 22 | 13 |
| 157 | A0A2T7NGU8 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_18553 | 5 | 12 | 29 |
| 158 | Q564J1 | Haemocyanin OS = Aplysia californica | hc | 2 | 927 | 2 |
| 159 | A0A2T7NV41 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_15545 | 4 | 18 | 25 |
| 160 | E7DS67 | Actin (Fragment) OS = Gonospira metablata | ACTI | 1 | 38 | 18 |
| 161 | K1RBG6 | Actin-1/3 OS = Crassostrea gigas | CGI_10017112 | 1 | 39 | 8 |
| 162 | P02595 | Calmodulin OS = Patinopecten sp. | CAM | 1 | 16 | 30 |
| 163 | V6A758 | Myosin heavy chain isoform C OS = Sepia officinalis | MYH | 1 | 17 | 16 |
| 164 | A0A0B7BLG3 | Uncharacterized protein OS = Arion vulgaris | ORF192624 | 3 | 23 | 2 |
| 165 | K1PPW8 | Coatomer subunit beta OS = Crassostrea gigas | CGI_10006442 | 2 | 8 | 7 |
| 166 | A0A210R0F2 | Fructose-bisphosphate aldolase OS = Mizuhopecten yessoensis | KP79_PYT16607 | 2 | 8 | 6 |
| 167 | A0A2T7PZW7 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_01565 | 1 | 6 | 1 |
| 168 | A0A0B7B4N1 | Uncharacterized protein OS = Arion vulgaris | ORF158201 | 1 | 10 | 4 |
| 169 | A0A210QY92 | Coatomer subunit beta’ OS = Mizuhopecten yessoensis | KP79_PYT21841 | 1 | 5 | 5 |
| 170 | V3ZPS1 | Uncharacterized protein OS = Lottia gigantea | LOTGIDRAFT_222012 | 2 | 9 | 12 |
| 171 | E3VWM3 | Fructose-bisphosphate aldolase OS = Meretrix meretrix | FBA | 1 | 20 | 4 |
| 172 | A0A2T7PSV4 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_03483 | 2 | 10 | 11 |
| 173 | A0A0B7AZA8 | Uncharacterized protein OS = Arion vulgaris | ORF148015 | 2 | 10 | 19 |
| 174 | K7WKX6 | Fructose-bisphosphate aldolase OS = Haliotis rufescens | FBA | 1 | 3 | 9 |
| 175 | A0A2T7NF32 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_20261 | 1 | 5 | 4 |
| 176 | A0A2T7NMW4 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_18325 | 2 | 4 | 5 |
| 177 | K1QZU8 | Calcium-transporting ATPase OS = Crassostrea gigas | CGI_10023684 | 1 | 2 | 1 |
| 178 | A0A0L8IAE8 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22025089mg | 1 | 2 | 8 |
| 179 | A0A2C9KC89 | Uncharacterized protein OS = Biomphalaria glabrata | 106056965 | 2 | 5 | 3 |
| 180 | A0A210R746 | Ras-related protein Rab-6A OS = Mizuhopecten yessoensis | KP79_PYT20147 | 1 | 9 | 11 |
| 181 | A0A0B6Z4Q3 | Uncharacterized protein OS = Arion vulgaris | ORF48472 | 2 | 12 | 8 |
| 182 | A0A2T7PZP4 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_01513 | 1 | 4 | 3 |
| 183 | A0A2C9JIZ4 | Uncharacterized protein OS = Biomphalaria glabrata | 106056849 | 1 | 9 | 13 |
| 184 | K1PTH4 | ADP-ribosylation factor OS = Crassostrea gigas | CGI_10020174 | 1 | 2 | 1 |
| 185 | Q6PTL0 | Triosephosphate isomerase OS = Nucula proxima | OCBIM_22037419mg | 1 | 5 | 6 |
| 186 | A0A2C9JZR8 | Uncharacterized protein OS = Biomphalaria glabrata | 106074442 | 1 | 2 | 2 |
| 187 | A0A2C9JIA9 | Uncharacterized protein OS = Biomphalaria glabrata | 106056539 | 1 | 6 | 4 |
| 188 | A0A385NHM7 | Glutathione S-transferase OS = Tegillarca granosa | GST | 1 | 8 | 5 |
| 189 | A0A210QUP5 | Malic enzyme OS = Mizuhopecten yessoensis | KP79_PYT06884 | 1 | 1 | 3 |
| 190 | V3YXF9 | Adenosylhomocysteinase OS = Lottia gigantea | LOTGIDRAFT_184532 | 1 | 2 | 3 |
| 191 | A0A210QGP4 | Chitotriosidase-1 OS = Mizuhopecten yessoensis | KP79_PYT06201 | 1 | 1 | 3 |
| 192 | A0A210QHE1 | Adenosylhomocysteinase OS = Mizuhopecten yessoensis | KP79_PYT14445 | 1 | 4 | 3 |
| 193 | A0A210PIA6 | Ornithine aminotransferase OS = Mizuhopecten yessoensis | KP79_PYT16913 | 1 | 3 | 3 |
| 194 | K1QQB6 | 40S ribosomal protein S14 OS = Crassostrea gigas | CGI_10011151 | 1 | 4 | 9 |
| 195 | A0A2C9KEN8 | Tubulin alpha chain OS = Biomphalaria glabrata | 106069694 | 1 | 2 | 3 |
| 196 | A0A2T7PWT6 | Serine/threonine-protein phosph OS = Pomacea canaliculata | C0Q70_00460 | 1 | 1 | 3 |
| 197 | A0A0B7AJW7 | Fructose-bisphosphate aldolase OS = Arion vulgaris | ORF124546 | 1 | 8 | 4 |
| 198 | A0A2C9L7N6 | Uncharacterized protein OS = Biomphalaria glabrata | 106080319 | 1 | 49 | 4 |
| 199 | A0A210QTZ1 | Peptidyl-prolyl cis-trans OS = Mizuhopecten yessoensis | KP79_PYT00632 | 1 | 2 | 6 |
| 200 | A0A2I7M8C2 | Go protein alpha subunit OS = Argopecten irradians | N/A | 1 | 4 | 3 |
| 201 | K1R2G8 | Titin OS = Crassostrea gigas | CGI_10016808 | 1 | 2 | 0 |
| 202 | K1QVD7 | Neuronal acetylcholine receptor subunit non-alpha-2 OS = Crassostrea gigas | CGI_10016138 | 1 | 2 | 1 |
| 203 | K1Q7G5 | Ficolin-2 OS = Crassostrea gigas | CGI_10026202 | 1 | 2 | 3 |
| 204 | A0A2C9K9W9 | Uncharacterized protein OS = Biomphalaria glabrata | 106068683 | 1 | 1 | 1 |
| 205 | A0A0B6ZP87 | Uncharacterized protein OS = Arion vulgaris | ORF71130 | 1 | 3 | 4 |
| 206 | V4AP92 | Elongation factor 1-alpha OS = Lottia gigantea | LOTGIDRAFT_239271 | 1 | 2 | 2 |
| 207 | A0A2T7PU69 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_03920 | 1 | 4 | 4 |
| 208 | V3ZN51 | Staphylococcal nuclease domain-cont. OS = Lottia gigantea | LOTGIDRAFT_235720 | 1 | 3 | 1 |
| 209 | A0A2T7PSF5 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_03333 | 1 | 2 | 0 |
| 210 | K1PQD4 | Phosphoglucomutase-1 OS = Crassostrea gigas | CGI_10011818 | 1 | 1 | 2 |
| 211 | A0A0B7BF17 | Uncharacterized protein OS = Arion vulgaris | ORF179770 | 1 | 3 | 2 |
| 212 | A0A2T7Q0W0 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_01928 | 1 | 1 | 3 |
| 213 | A0A0L8I692 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22034637mg | 1 | 4 | 19 |
| 214 | K1PQ79 | Copine-3 OS = Crassostrea gigas | CGI_10011897 | 1 | 3 | 1 |
| 215 | K1PWB9 | EH domain-containing protein 1 OS = Crassostrea gigas | CGI_10005813 | 1 | 1 | 4 |
| 216 | A0A2T7Q016 | Uncharacterized protein OS = Pomacea canaliculata | C0Q70_01636 | 1 | 1 | 2 |
| 217 | V4AKV4 | Calcium-transporting ATPase OS = Lottia gigantea | LOTGIDRAFT_208914 | 1 | 3 | 1 |
| 218 | A0A2T7NL99 | Proteasome subunit beta OS = Pomacea canaliculata | C0Q70_17739 | 1 | 2 | 4 |
| 219 | A0A0L8HWW8 | Uncharacterized protein OS = Octopus bimaculoides | OCBIM_22003772mg | 1 | 2 | 2 |
N (Identification Number); FDR (False Discovery Rate); Uni. Pep. (Unique Peptides); PSMs (Peptide Spectrum Matches); Cov. (Protein Coverage).
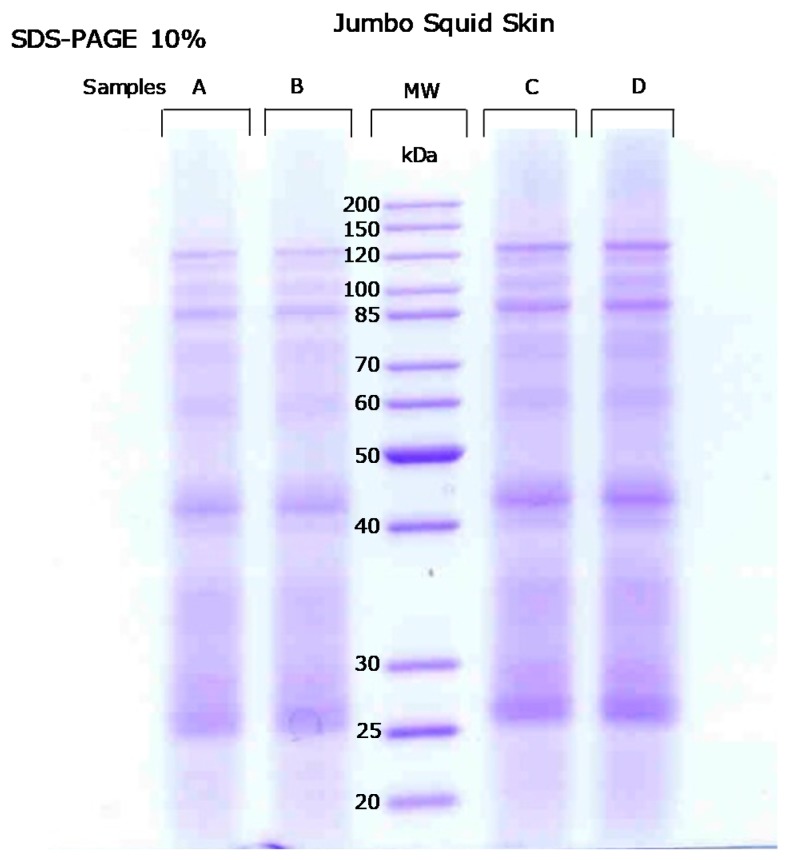
Additionally, to visualize and corroborate the intact protein extraction of the jumbo squid skin fraction, complete protein extracts of the four replicates (A–D) were separated by SDS-PAGE 10% (Figure 1). This gel illustrates that all replicate extracts show the same protein weight distribution.
Figure 1.
SDS-PAGE 10% profiles of the extracted proteins of jumbo squid skin samples (A–D replicates). MW denotes molecular weight.
To our knowledge, this is the most comprehensive dataset of peptides and proteins for jumbo squid (D. gigas) skin identified to date. This valuable protein repository will add new and significant information to the universal public protein databases and could be very useful for new investigations of this marine by-product. Raw data and analyses outputs are publicly available in MassIVE data repository (https://massive.ucsd.edu/) (Reference: MSV000084702).
We need to take into account the difficulties and limitations of working with un-sequenced organisms as in the case of D. gigas. Thus, due to the fact that in the universal UniprotKB protein database only 40 different proteins for D. gigas are registered (Cytochrome c oxidase subunit 1, subunit 3; Cytochrome b; NADH-ubiquinone oxidoreductase chain 2, chain 4, chain 5; Cytochrome c oxidase subunit 2; ATP synthase subunit a; Histone H3; Chitin binding beak protein 1, 2, 3, 4; NADH dehydrogenase subunit 4L, subunit 2; ATP synthetase subunit 8; Paramyosin; Histidine rich beak protein 1, protein 2, protein 3; Suckerin-1, -2, -3, -4, -5, -6, -7, -8, -9, -10, -12, -13, -14, -15, -16, -17, -18, -20, -21; Symplectin/biotinidase-like protein), we decided to perform the protein identification using Proteome Discoverer 2.2 using a global database according to phylogenetic similarity for the class “Cephalopoda”. This class presents 40,780 entries, these including the 40 different proteins for D. gigas in order to increase the number of protein identifications. In Table 1, assignments for D. gigas protein are indicated in the first lines (Paramyosin and Symplectin/biotinidase-like protein). Many of the protein assignments are uncharacterized proteins (n = 109 proteins; n = 1393 PSMs) that may change with future Cephalopoda and D. gigas specific databases updates.
Thus, the final global dataset of the jumbo squid skin proteome was subsequently investigated by protein-based bioinformatics, like gene ontologies, pathways, network analyses and by prediction of potential bioactive peptides to gather more functional insights.
2.2. Functional Analysis: Gene Ontologies and Pathways Analysis
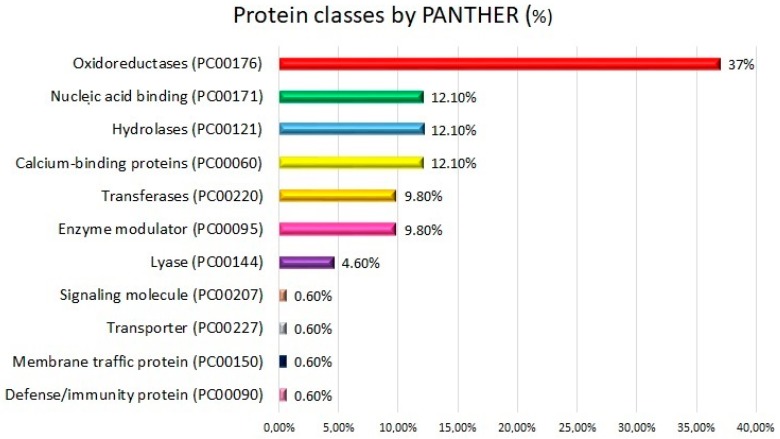
PANTHER analysis revealed the presence of 11 different protein classes in the jumbo squid skin proteome (Figure 2). The most prominent classes were oxidoreductases (37.0%), nucleic acid binding proteins (12.1%), hydrolases (12.1%), calcium-binding proteins (12.1%), transferases (9.8%), and enzyme modulator (9.8%). Thus, in the jumbo squid skin, oxidoreductases are mainly involved in the energetic metabolism, antioxidant defense and cephalopod coloration [38]. Another significant protein class is that of calcium-binding proteins, which are involved in muscle relaxation and nervous transmission in the marine skin species [39,40].
Figure 2.
Protein classes of the jumbo squid skin proteome identified by shotgun proteomics and categorized by PANTHER (http://pantherdb.org/).
KEGG pathway analysis was carried out by comparing the input data with the background of the Octopus bimaculoides genome by DAVID version 6.8 program (https://david.ncifcrf.gov/home.jsp); this cephalopod species is the most phylogenetically closest included in DAVID software. KEGG showed that most of the identified proteins were involved in metabolic pathways (cysteine and methionine metabolism), endocytosis/phagosome, RNA transport, protein methylation, and calcium homeostasis (Table 2).
Table 2.
KEGG pathway analysis of the jumbo squid skin proteome by DAVID.
| KEGG Pathway | p-Value |
|---|---|
| Metabolic pathways (cysteine and methionine metabolism) | 4.53 × 10−4 |
| Endocytosis/phagosome | 1.05 × 10−2 |
| RNA transport | 2.24 × 10−2 |
| Protein methylation | 3.46 × 10−2 |
| Calcium homeostasis | 1.00 × 10−1 |
The study of functional domains by InterPro performed by DAVID software revealed that the top protein motifs corresponded to small GTP-binding protein domains, heat shock protein 70, small GTPase superfamily, proteasome, P-loop containing nucleoside triphosphate hydrolase and EF-hand-like domains (Table 3). These EF-hand domains corresponded to calcium-binding domains in concordance with the calcium homeostasis pathway discovered for the calcium-binding proteins, which correspond to 12.1% of the total jumbo squid skin proteome.
Table 3.
Functional InterPro motifs by DAVID.
| InterPro Motifs | p-Value |
|---|---|
| Small GTP-binding protein domain | 3.1 × 10−4 |
| Heat shock protein 70, conserved site | 8.5 × 10−4 |
| Small GTPase superfamily | 8.6 × 10−4 |
| Proteasome, alpha-subunit, N-terminal domain | 1.3 × 10−3 |
| P-loop containing nucleoside triphosphate hydrolase | 8.3 × 10−3 |
| EF-hand-like domain | 2.9 × 10−2 |
| Ubiquitin | 3.4 × 10−2 |
2.3. Network Analysis
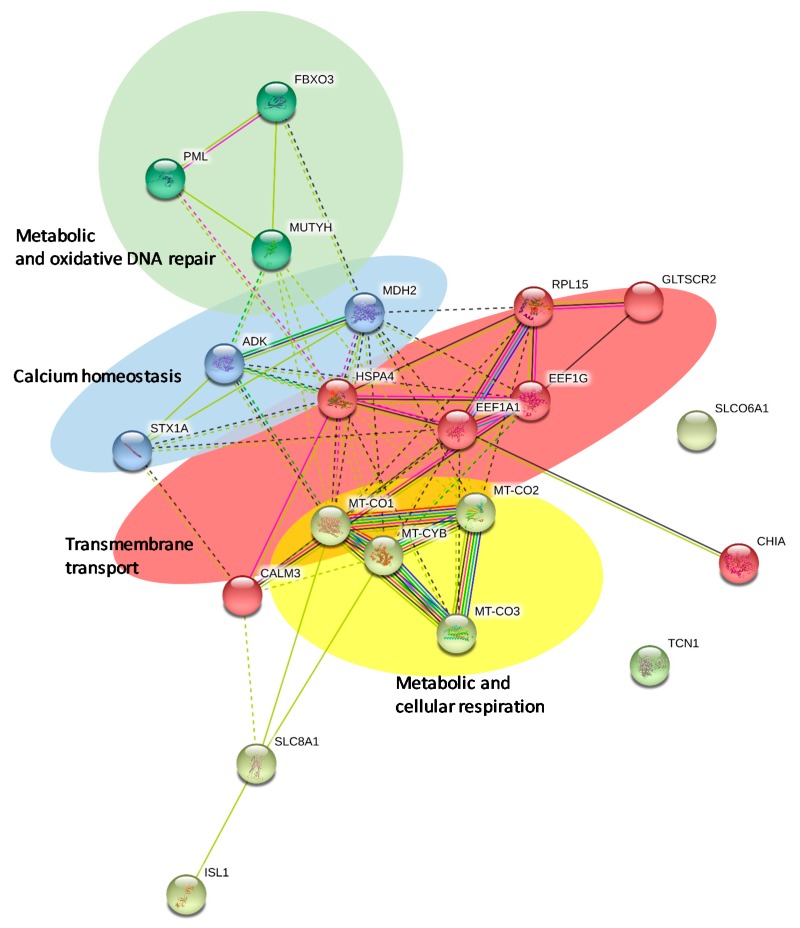
Network analysis was created merging all the proteins identified for the jumbo squid skin proteome using the STRING software (v.11.0) (https://string-db.org/). A specific organism was not selected (organism Auto-detect) because the genome of D. gigas is not available in the STRING software. According to MCL inflation clustering (MCL = 3), 21 nodes (proteins) and 61 edges (interactions) were obtained (Figure 3).
Figure 3.
Protein network for the jumbo squid skin proteome using the STRING (v.11.0) software. Physical direct interactions are represented with continuous lines and functional interactions with interrupted lines.
Physical direct interactions are represented with continuous lines and functional interactions with interrupted lines. The topological analysis of this network demonstrated mainly four different sub-networks. Two of them are relevant sub-networks implicated in metabolic and oxidative cellular respiration (Figure 3 in green and yellow).
Other relevant sub-network is composed of three nodes and is referred as calcium homeostasis (Figure 3 in blue). The results of this sub-network are in concordance with one of the top protein classes categorized previously by PANTHER and DAVID (Figure 2 and Table 2).
Other relevant sub-network is referred as transmembrane transport proteins (Figure 3, in red), as was obtained previously by PANTHER (Figure 2).
Finally, this network represents to date the first most comprehensive interactomic map for the jumbo squid skin proteome.
2.4. Putative Bioactive Peptides
Bioactive peptides are inactive when they are part of parent protein, but become active when released due to the action of enzymes. Thus, bioactive peptides encrypted in the parent jumbo squid skin proteome (n = 219) were predicted using different in-silico software. Thus, protein hydrolysates with pepsin and trypsin were performed in-silico using the MS-Digest program. No missed cleavages and a minimum of six residues per peptide were selected as parameters. Thus, the predicted peptides after every enzymatic digestion (pepsin and trypsin) are presented in Supplementary Table S4.
The first enzymatic digestion using pepsin released a total of 5077 different peptides (6–39 amino acid residues). This enzyme cleaves the proteins at Phe, Tyr, Trp, and Leu residues in positions P1 and P1’ [41]. Compared with the most used and conventional BIOPEP database, no bioactive peptides were identified probably because none squid bioactive peptide is included in the database. However, by using PeptideRanker (http://distilldeep.ucd.ie/PeptideRanker/), the complete list of potential bioactive peptides was ranked using the N-to-1 neural network probability [42], which predicts the peptides that may be more bioactive (Supplementary Table S4). Among them, 18 peptides with a PeptideRanker score higher than 0.9 (7–30 amino acid residues) were selected as potential bioactive peptides (Table 4). The majority of the results corresponded to collagen ColAa proteins, hemocyanin subunit proteins and different uncharacterized proteins.
Table 4.
Selected potential bioactive peptides of the jumbo squid skin proteome predicted by in-silico digestions with pepsin.
| Proteins | Peptides | PeptideRanker Score | Anti-Microbial Peptide (AMP) | Discriminant Score for AMP |
|---|---|---|---|---|
| ADP-ribosylation factor OS = Crassostrea gigas | SPSPKQMVSCPVCGL | 0.915222 | Non-AMP | 0.043 |
| Collagen ColAa OS = Sepia pharaonis | PGDPGPVGRTGPMGL | 0.934847 | Non-AMP | 0.003 |
| Collagen ColAa OS = Sepia pharaonis | RGPPGPPGL | 0.912657 | Non-AMP | 0.030 |
| Heat shock protein 70 OS = Sepiella maindroni | GGMPGGMPGGMPGGMPNF | 0.92432 | AMP | 0.504 |
| Hemocyanin OS = Sepiella maindroni | KKPMMPF | 0.932566 | AMP | 0.978 |
| Hemocyanin OS = Sepiella maindroni | PNQPMRPF | 0.920777 | AMP | 0.983 |
| Hemocyanin subunit 1 OS = Todarodes pacificus | NDPMRPF | 0.923312 | AMP | 0.795 |
| Hemocyanin subunit 2 OS = Sepia officinalis | SDPMRPF | 0.938433 | AMP | 0.879 |
| Uncharacterized protein OS = Octopus bimaculoides | CPCMGRF | 0.985441 | AMP | 0.622 |
| Uncharacterized protein OS = Octopus bimaculoides | GGPPGMPPF | 0.973279 | Non-AMP | 0.208 |
| Uncharacterized protein OS = Octopus bimaculoides | GRCVMCNCNKHSSTCDPQTGKCVNCQHNTL | 0.969319 | Non-AMP | 0.238 |
| Uncharacterized protein OS = Octopus bimaculoides | GSCVPCNCNGF | 0.952459 | AMP | 0.745 |
| Uncharacterized protein OS = Octopus bimaculoides | QPPQCCPSKGGSF | 0.943546 | AMP | 0.687 |
| Uncharacterized protein OS = Octopus bimaculoides | GSWGNGNRW | 0.915802 | Non-AMP | 0.403 |
| Uncharacterized protein OS = Octopus bimaculoides | PPPSKRF | 0.911736 | AMP | 0.983 |
| Uncharacterized protein OS = Biomphalaria glabrata | PPPPQPVGGGGGNRW | 0.955862 | Non-AMP | 0.092 |
| Uncharacterized protein OS = Biomphalaria glabrata | SRSPPRPF | 0.904351 | AMP | 0.993 |
| Uncharacterized protein OS = Pomacea canaliculata | HDGDGPRPCCF | 0.93215 | Non-AMP | 0.031 |
Regarding tryptic digestion, this enzyme predicted the release of a total of 8042 different peptides (6–45 amino acid residues) (Supplementary Table S4). This enzyme preferentially cleaves the proteins at Lys and Arg residues in position P1 except for the case in which Pro is found in position P1’ [41]. Using a PeptideRanker score higher than 0.9, a total of 73 tryptic peptides (7–30 amino acid residues) were selected as potential bioactive peptides (Table 5). The majority of such peptides corresponded to calcium-transporting ATPase, collagen ColAa proteins, hemocyanin proteins, myosin heavy chain, titin and different uncharacterized proteins.
Table 5.
Selected potential bioactive peptides of the jumbo squid skin proteome predicted by in-silico digestions with trypsin.
| Proteins | Peptides | PeptideRanker Score | Anti-Microbial Peptide (AMP) | Discriminant Score for AMP |
|---|---|---|---|---|
| ADP-ribosylation factor OS = Crassostrea gigas | CPICYDFMHTAMILPECSHTFCSFCIR | 0.902646 | Non-AMP | 0.160 |
| Calcium-transporting ATPase OS = Octopus bimaculoides | FSDDYPGFF | 0.970864 | Non-AMP | 0.006 |
| Calcium-transporting ATPase OS = Crassostrea gigas | FLQFQLTVNCVAVMVAFFGACIINDSPLK | 0.979848 | Non-AMP | 0.281 |
| Calcium-transporting ATPase OS=Lottia gigantea | FADAPFMK | 0.93747 | Non-AMP | 0.014 |
| Calmodulin OS = Crassostrea gigas | GAFFVFDR | 0.915228 | Non-AMP | 0.003 |
| Chitinase OS = Todarodes pacificus | MLAVSLLFLLAIGGVSSAGHR | 0.976725 | AMP | 0.746 |
| Chitotriosidase OS = Euprymna scolopes | MASTFATVFGVLSLCFLGLHLTNGEYK | 0.984749 | Non-AMP | 0.106 |
| Coatomer subunit beta’ OS = Mizuhopecten yessoensis | YCLCLFR | 0.924855 | AMP | 0.579 |
| Collagen ColAa OS = Sepia pharaonis | GPPGIPGLPGPK | 0.93716 | AMP | 0.504 |
| Collagen ColAa OS = Sepia pharaonis | GPPGPPGLK | 0.913133 | Non-AMP | 0.119 |
| Collagen ColAa OS = Sepia pharaonis | AGPPGFPGTPGPK | 0.907398 | AMP | 0.682 |
| Ficolin-2 OS = Crassostrea gigas | DQDNDMYVSDNCGILFPSGWWHR | 0.901865 | Non-AMP | 0.008 |
| Fructose-bisphosphate aldolase OS = Mizuhopecten yessoensis | KPWALTFSFGR | 0.93422 | Non-AMP | 0.123 |
| Hemocyanin OS = Aplysia californica | MVGYLGQALMALLLLALSNAALVR | 0.993669 | Non-AMP | 0.380 |
| Hemocyanin OS = Aplysia californica | FEPNPFFSGK | 0.924588 | Non-AMP | 0.093 |
| Hemocyanin OS = Aplysia californica | VACCLHGMPVFPHWHR | 0.903581 | Non-AMP | 0.106 |
| Hemocyanin OS = Nautilus pompilius | MATHWHSLLLFSLQLLVFTYATSDPTNIR | 0.97599 | Non-AMP | 0.008 |
| Hemocyanin OS = Sepiella maindroni | GSPIGVPYWDWTKPMK | 0.917605 | Non-AMP | 0.027 |
| Hemocyanin-like protein OS = Uroteuthis edulis | TNFFFLALIATVWLGNAETETETSK | 0.90323 | Non-AMP | 0.062 |
| Hemocyanin subunit 1 OS = Euprymna scolopes | VFVGFLLHGFGSSAYATFDICNDAGECR | 0.96087 | Non-AMP | 0.233 |
| Hemocyanin subunit 1 OS = Euprymna scolopes | LNHLPLLCLAVILTLWMSGSNTVNGNLVR | 0.926117 | Non-AMP | 0.287 |
| Hemocyanin subunit 1 OS = Euprymna scolopes | VFAGFLFMGIK | 0.904542 | AMP | 0.865 |
| Hemocyanin subunit 2 OS = Euprymna scolopes | VFAGFWFHGIK | 0.943 | AMP | 0.506 |
| Hemocyanin subunit 2 OS = Sepia officinalis | VFGGFWLHGIK | 0.907156 | AMP | 0.739 |
| Hemocyanin subunit 3 OS = Sepia officinalis | TSFLFLAFVATSWFVYAVTASK | 0.905214 | Non-AMP | 0.136 |
| Malate dehydrogenase OS = Sepia officinalis | DLFNTNASIVANLADACAQYCPK | 0.965037 | Non-AMP | 0.251 |
| Myosin heavy chain isoform A OS = Octopus bimaculoides | YQSGFIYTYSGLFCVAINPYR | 0.956725 | Non-AMP | 0.024 |
| Myosin heavy chain OS = Todarodes pacificus | NWEWWR | 0.951523 | Non-AMP | 0.478 |
| Myosin II heavy chain OS = Doryteuthis pealeii | NWQWWR | 0.973264 | AMP | 0.959 |
| Myosin II heavy chain OS = Doryteuthis pealeii | YYSGLIYTYSGLFCVVVNPYK | 0.939159 | Non-AMP | 0.032 |
| Neuronal acetylcholine receptor subunit non-alpha-2 OS = Crassostrea gigas | LLIDLCLSVLVTTLAIVSLYFYDMSDSR | 0.904075 | Non-AMP | 0.015 |
| Peptidyl-prolyl cis-trans isomerase OS = Mizuhopecten yessoensis | MAGAGIGCVLLFLLPALLSAGK | 0.996478 | Non-AMP | 0.159 |
| Phosphoglucomutase-1 OS = Crassostrea gigas | DGLWAVLAWLSVLANQNCSVEECIK | 0.991266 | AMP | 0.904 |
| Protein disulfide-isomerase OS = Octopus bimaculoides | NVFIEFYAPWCGHCK | 0.907443 | Non-AMP | 0.053 |
| S-syntaxin OS = Doryteuthis pealeii | IAILVCLVILVLVIVSTVGGVFGG | 0.965343 | Non-AMP | 0.000 |
| Titin OS = Crassostrea gigas | DGSWQNLVTVLGCLKPQFVNLQR | 0.974127 | AMP | 0.724 |
| Titin OS = Crassostrea gigas | GYPPPIISWYR | 0.917986 | Non-AMP | 0.074 |
| Tubulin alpha chain OS = Octopus bimaculoides | FVDWCPTGFK | 0.923256 | Non-AMP | 0.010 |
| Uncharacterized protein OS = Arion vulgaris | APDFIFYAPR | 0.921198 | Non-AMP | 0.009 |
| Uncharacterized protein OS = Octopus bimaculoides | FLQFQLTVNVVAVLVAFFGACTINVSI | 0.978717 | AMP | 0.916 |
| Uncharacterized protein OS = Octopus bimaculoides | YYTFFVTIFLFATTLCSTIPKPK | 0.984914 | Non-AMP | 0.012 |
| Uncharacterized protein OS = Octopus bimaculoides | LFPAFGFGAR | 0.94902 | AMP | 0.505 |
| Uncharacterized protein OS = Octopus bimaculoides | ATMLGAQGNIFFASLSCCCLILSCS | 0.999233 | AMP | 0.879 |
| Uncharacterized protein OS = Octopus bimaculoides | SGPFYIFSGGMPR | 0.939205 | Non-AMP | 0.089 |
| Uncharacterized protein OS = Octopus bimaculoides | EFSMMFR | 0.931708 | Non-AMP | 0.001 |
| Uncharacterized protein OS = Octopus bimaculoides | YGSCVPCNCNGFSNDCDPVTGECIDCQR | 0.980617 | Non-AMP | 0.243 |
| Uncharacterized protein OS = Octopus bimaculoides | HNPEGCISCFCMGVTEFCTSTSR | 0.964134 | Non-AMP | 0.083 |
| Uncharacterized protein OS = Octopus bimaculoides | APMVELCECPQGYTGVSCQECSPGYSR | 0.963828 | Non-AMP | 0.012 |
| Uncharacterized protein OS = Octopus bimaculoides | GCGCSAGQFECQNGLCINENK | 0.930153 | AMP | 0.982 |
| Uncharacterized protein OS = Octopus bimaculoides | EECMSCFCFK | 0.918951 | AMP | 0.982 |
| Uncharacterized protein OS = Octopus bimaculoides | NSEYGFACFCPQGFAGYQCDTVGER | 0.906197 | AMP | 0.576 |
| Uncharacterized protein OS = Octopus bimaculoides | MIIYILSLAGVALGVYFLSCVR | 0.995663 | Non-AMP | 0.008 |
| Uncharacterized protein OS = Octopus bimaculoides | MILTIFACLMALDIELNTSNSIQEE | 0.968187 | Non-AMP | 0.026 |
| Uncharacterized protein OS = Octopus bimaculoides | AIGALVDACGPGLCPDWADWAPK | 0.948884 | AMP | 0.774 |
| Uncharacterized protein OS = Octopus bimaculoides | QGDWTCPNPACGNNNFGWR | 0.9572 | Non-AMP | 0.286 |
| Uncharacterized protein OS = Octopus bimaculoides | GGFGGGGGGGGGMGGDR | 0.928063 | Non-AMP | 0.065 |
| Uncharacterized protein OS = Octopus bimaculoides | GFFEDDYDEYGGGYGGGMGFGGLNR | 0.944869 | Non-AMP | 0.143 |
| Uncharacterized protein OS = Octopus bimaculoides | LDDGDACLLDMGTEYCCYASDITCSYPVNGK | 0.968621 | Non-AMP | 0.056 |
| Uncharacterized protein OS = Octopus bimaculoides | MAFYTILNVVTIVLLIIVGQCR | 0.998628 | Non-AMP | 0.031 |
| Uncharacterized protein OS = Octopus bimaculoides | GGSFGFNFR | 0.969779 | Non-AMP | 0.355 |
| Uncharacterized protein OS = Octopus bimaculoides | NSTDVCNCSIYVGLFPCNECTK | 0.994975 | Non-AMP | 0.462 |
| Uncharacterized protein OS = Octopus bimaculoides | PPSPPIYFR | 0.946483 | Non-AMP | 0.226 |
| Uncharacterized protein OS = Octopus bimaculoides | CFLCATGTGTSIEVLALVTIGWCLLHATGTR | 0.96344 | AMP | 0.768 |
| Uncharacterized protein OS = Octopus bimaculoides | FDFFYK | 0.96245 | Non-AMP | 0.032 |
| Uncharacterized protein OS = Octopus bimaculoides | FSPIPFLFCTISGTCNFATR | 0.95134 | AMP | 0.505 |
| Uncharacterized protein OS = Octopus bimaculoides | FWELTECCPHQCLEWLSNLVTR | 0.933791 | Non-AMP | 0.106 |
| Uncharacterized protein OS = Octopus bimaculoides | DAFCSSPNFNSWLK | 0.922125 | Non-AMP | 0.058 |
| Uncharacterized protein OS = Octopus bimaculoides | NGYEEDDALIGLLNLCTAILK | 0.917521 | Non-AMP | 0.479 |
| Uncharacterized protein OS = Octopus bimaculoides | DYFWLVCR | 0.911557 | Non-AMP | 0.001 |
| Uncharacterized protein OS = Biomphalaria glabrata | QGELGDCWLLAAVASLTCNPK | 0.919385 | AMP | 0.783 |
| Uncharacterized protein OS = Biomphalaria glabrata | SPPRPFEWK | 0.905581 | Non-AMP | 0.006 |
| Uncharacterized protein OS = Pomacea canaliculata | SVFNIPPNCFSEMM | 0.908085 | Non-AMP | 0.003 |
| Uncharacterized protein OS = Pomacea canaliculata | SCLMGHGSLFGAGAGSLHLQAIAALK | 0.919795 | Non-AMP | 0.315 |
It is known that the employment of collagenous residues obtained from jumbo squid skin after hydrolysis with pepsin exhibit a good gelatin gel-forming ability including the absence of color, opacity and high-puncture deformation [43]. The collagen alpha chains proteins determined in this study were characterized as belonging to type-I. Additionally, jumbo squid skin collagen was explored to enhance the anti-damage and anti-osteoporosis activity in osteoblast cells [44,45]. Thus, potential pepsin (PGDPGPVGRTGPMGL, RGPPGPPGL) and tryptic (GPPGIPGLPGPK, GPPGPPGLK, AGPPGFPGTPGPK) bioactive collagen peptides determined in this study may be used to stimulate the regeneration of joint cartilages in patients with chronic joint symptoms (Table 4 and Table 5). GELITA® and CH-Alpha® are examples of commercial products containing collagen hydrolysates.
Hemocyanins are the oxygen transporters of cephalopods and mollusks. These proteins play important immune-related roles as antimicrobial, antiviral, agglutinative and antitumor proliferation of cancer cells [46]. In fact, hemocyanin of marine mollusks (Megathura crenulata and Concholepas concholepas) has showed significant antitumor effects of breast, pancreas and prostate cancer cells [47,48]. Although, no previous studies are available related to the use of jumbo squid hemocyanin from a bioactive and immunotherapeutic point of view, it can be considered that the potential pepsin (KKPMMPF, PNQPMRPF, NDPMRPF, SDPMRPF) and tryptic (MVGYLGQALMALLLLALSNAALVR, FEPNPFFSGK, VACCLHGMPVFPHWHR, MATHWHSLLLFSLQLLVFTYATSDPTNIR, GSPIGVPYWDWTKPMK, TNFFFLALIATVWLGNAETETETSK, VFVGFLLHGFGSSAYATFDICNDAGECR, LNHLPLLCLAVILTLWMSGSNTVNGNLVR, VFAGFLFMGIK, VFAGFWFHGIK, VFGGFWLHGIK, TSFLFLAFVATSWFVYAVTASK) bioactive hemocyanin peptides determined in this study may be used in the future as an antitumor therapy for cancer cells (Table 4 and Table 5).
Calcium-transporting ATPase protein is an important regulator of the Ca2+ concentration in the cells and extracellular space. It is necessary for the cell signaling and for the nerve transmission of the squid axons [49]. Potential tryptic (FSDDYPGFF, FLQFQLTVNCVAVMVAFFGACIINDSPLK, FADAPFMK) bioactive calcium-transporting ATPase peptides determined in this study may be used in a future to investigate the in vitro axon stimulation (Table 5).
Myosin heavy chain is one of the major components of the muscle that participates in the muscle contraction as well as in a wide variety of non-muscular cells movements. Previous studies identified different ACE-inhibitory peptides from alcalase hydrolysis of a protein concentrate recovered from a cuttlefish (Sepia officinalis) industrial manufacturing effluent [17]. In fact, several potential bioactive peptides had a proline residue in one of the last positions of C-terminal which promotes enzyme binding (YQSGFIYTYSGLFCVAINPYR, YYSGLIYTYSGLFCVVVNPYK) [50] (Table 5). However, these results need to be further investigated because this is neither sufficient nor essential to confer bioactivity.
Titin (also known as connectin) is a giant protein that works as a molecular spring for the passive elasticity of tissues. The degradation of this protein is one of the major reasons for quality changes in fresh raw squid tissues [51]. Potential tryptic (DGSWQNLVTVLGCLKPQFVNLQR, GYPPPIISWYR) bioactive titin peptides determined in this study may be used as potential biomarkers of quality changes or processing time in squid products (Table 5).
The antimicrobial activity of jumbo squid skin crude pigments extracts has been recently demonstrated [52]. In the present work, antimicrobial peptides (AMPs) were identified using the CAMP (Collection of Anti-Microbial Peptides) database (http://www.bicnirrh.res.in/antimicrobial/) and applying the DAC score (Discriminate Analysis Classifier score) [34]. Table 4 and Table 5 show the potential anti-microbial bioactive peptides. A total of 16 pepsin peptides and 20 tryptic peptides with anti-microbial peptides were predicted. Among them, seven anti-microbial peptides (four pepsin and three tryptic) were encrypted in the hemocyanin parent protein (KKPMMPF, PNQPMRPF, NDPMRPF, SDPMRPF, VFAGFLFMGIK, VFAGFWFHGIK, VFGGFWLHGIK), two anti-microbial tryptic peptides in the collagen parent protein (GPPGIPGLPGPK, AGPPGFPGTPGPK), one anti-microbial tryptic peptide in the myosin heavy chain protein (NWQWWR) and one anti-microbial tryptic peptide in the titin protein (DGSWQNLVTVLGCLKPQFVNLQR).
All these potential bioactive peptides need to be validated by further bioactivity assays using synthetic versions of the peptides. Nevertheless, compared with the classical approaches, the bioinformatics methods are faster and lower-cost alternatives that predict and reduce the number of potential targets to be investigated.
3. Materials and Methods
3.1. Chemicals and Reagents
Bicinchoninic acid (BCA), dithiothreitol (DTT), sodium dodecyl sulphate (SDS), Tris-HCl, and the protease inhibitor phenylmethylsulphonyl fluoride (PMSF) were purchased from Sigma (St. Louis, MO, USA). Ammonium persulphate (APS), bromophenol blue and N,N,N′,N′-tetramethylethylenediamine (TEMED) were purchased from GE Healthcare Science (Uppsala, Sweden). Acrylamide and bis N,N′-methylene-bis-acrylamide were obtained from Bio-rad (Hercules, CA, USA). Glycerol was obtained from Merck (Darmstadt, Germany). Sequencing grade porcine trypsin was purchased from Promega (Madison, WI, USA). All other chemicals were reagent/analytical grade and water was purified using a Milli-Q system (Millipore, Billerica, MA, USA).
3.2. Jumbo Squids
Jumbo squid (D. gigas) specimens were harvested off the coast of Kino Bay, Mexico. Specimens were degutted and major beheaded on site, and the skins bagged and placed in alternate layers of ice-squid-ice in a portable cooler, and transported to the laboratory. Time between capture and arrival at the laboratory did not exceed 12 h.
3.3. Skin Protein Samples
A total of 0.25 g of lyophilized jumbo squid skin were homogenized in 4 mL of lysis buffer (10 mM Tris-HCl buffer pH 7.2, 5 mM of PMSF) on ice for 6 cycles of 5 s pulses in a sonicator device (Werke, Germany). Samples were centrifuged at 40,000× g for 20 min at 4 °C in a J221-M centrifuge (Beckman, Palo Alto, CA, USA). The supernatant proteins were recovered and stored at −80 °C until used. Protein concentration in the protein extracts was determined by the bicinchoninic acid (BCA) method (Sigma Chemical Co., St. Louis, MI, USA).
3.4. SDS-Polyacrylamide Gel Electrophoresis
Squid skin proteins were separated on 10% (v/v) polyacrylamide gels (acrylamide/N,N′-ethylene-bis-acrylamide, 200:1) with a stacking gel of 4% polyacrylamide. A total of 25 µg of proteins in Laemmli buffer were boiled for 5 min at 100 °C and separated per well in a Mini-PROTEAN 3 cell (Bio-Rad, Hercules, CA, USA). The running buffer consisted of an aqueous solution, composed by 1.44% (w/v) glycine, 0.67% Tris-base, and 0.1% SDS. Running conditions were 80 V for the first 20 min and then 120 V until the end of the electrophoresis. PageRuler unstained protein ladder was also used as molecular weight (MW) indicator (Thermo Fisher Scientific, San Jose, CA, USA).
Gels were stained overnight with Coomassie dye PhastGel Blue R-350 (GE Healthcare, Uppsala, Sweden). Scanned Coomassie-stained gels were analysed by means of the 1-d gel electrophoresis analysis software LabImage 1D (Kapelan Bio-Imaging Solutions, Halle, Germany).
3.5. In-Solution Protein Digestion with Trypsin
A total of 100 μg of jumbo squid skin protein extract were denatured in 8 M urea and then reduced with 5 mM TCEP (Pierce, Thermo Fisher Scientific) for 30 min at 37 °C. After alkylation with 50 mM iodoacetamide (Pierce, Thermo Fisher Scientific) in 25 mM ammonium bicarbonate pH 8.25 for 60 min at room temperature in dark, samples being diluted 4-fold with 25 mM ammonium bicarbonate pH 8.25 to decrease the urea concentration. Proteins were digested with trypsin (Promega) (1:100 protease-to-protein ratio) overnight at 37 °C.
3.6. Shotgun LC-MS/MS Analysis
Peptides were acidified with formic acid, cleaned on a C18 MicroSpinTM column (The Nest Group, South-borough, MA) and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a Proxeon EASY-nLC II liquid chromatography system (Thermo Fisher Scientific, San Jose, CA, USA) coupled to a LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Peptide separation (1 µg) was done on a RP column (EASY-Spray column, 50 cm × 75 µm ID, PepMap C18, 2 µm particles, 100 Å pore size, Thermo Fisher Scientific) with a 10-mm pre-column (Accucore XL C18, Thermo Fisher Scientific) using 0.1% formic acid (mobile phase A) and 98% acetonitrile (98% ACN) with 0.1% formic acid (mobile phase B). A 120 min linear gradient from 5 to 35% B, at a flow rate of 300 nL min−1, was used. A spray voltage of 1.95 kV and a capillary temperature of 230 °C were used for ionization. The peptides were analyzed in positive mode (1 µscan; 400–1600 amu), followed by 10 data-dependent collision-induced dissociation (CID) MS/MS scans (1 µscans), using a normalized collision energy of 35% and an isolation width of 3 amu. Dynamic exclusion for 30 s after the second fragmentation event was applied and unassigned charged ions were excluded from the analysis.
A total of four replicates (n = 4) were analyzed independently.
3.7. Processing of the Mass Spectrometry Data
All the MS/MS spectra were analyzed using SEQUEST-HT (Proteome Discoverer 2.2 package, Thermo Fisher Scientific) against the Cephalopoda UniProt/TrEMBL database (release 2018_11; 40,780 entries). The following restrictions were used: tryptic cleavage with up to 2 missed cleavage sites and tolerances of 0.8 Da for parent ions and 0.6 Da for MS/MS fragment ions. Carbamidomethylation of Cys (C*) was considered as a fixed modification. The permissible variable modifications were: methionine oxidation (Mox) and acetylation of the N-terminus of the protein (N-Acyl). The results were subjected to statistical analysis with the Percolator algorithm to keep a false discovery rate (FDR) below 1%.
3.8. Functional Gene Ontologies and Pathways Analysis
The final list of non-redundant protein IDs was submitted to PANTHER program (http://www.pantherdb.org/), for the classification based on two main types of annotations: protein class and biological process. A statistical significance of representation for the analysis was also provided.
KEGG pathway analysis was performed by comparing the input data with the background of the Octopus bimaculoides genome by DAVID version 6.8 (https://david.ncifcrf.gov/home.jsp). Functional domains by InterPro Motifs were also obtained using DAVID version 6.8 software.
3.9. Network Analysis
Network analysis was performed submitting the protein dataset to the STRING (Search Tool for the Retrieval of Interacting Genes) software (v.11.0) (http://stringdb.org/) [53]. This is a large database of known and predicted protein interactions. Proteins were represented with nodes and the interactions with continuous lines to represent direct interactions (physical), while indirect ones (functional) were presented by interrupted lines. To minimize false positives as well as false negatives, all interactions tagged as “low-confidence” (<0.4) in STRING software have been eliminated from the analysis. Cluster networks were created using the MCL inflation algorithm which is included in the STRING website and a value of 3 was selected for all the analyses.
3.10. Bioactive Peptides Prediction
Bioactive peptides encrypted in the parent jumbo squid skin proteome were predicted combining different in-silico protein hydrolysates using pepsin and trypsin enzymes. For that, all the proteolytic digestions were performed in-silico using the MS-Digest software, which is included in ProteinProspector v.5.24.0 website (http://prospector.ucsf.edu/prospector/mshome.htm).
To evaluate the results, all the potential peptides were ranked using the PeptideRanker software (http://bioware.ucd.ie/~testing/biowareweb/) using the N-to-1 neural network probability to predict which peptides can be more bioactive [42]. In addition, all the potential peptides were compared with previous databases that included known bioactive peptides, such as BIOPEP (http://www.uwm.edu.pl/biochemia/index.php/pl/biopep/) and CAMP (http://www.bicnirrh.res.in/antimicrobial/).
4. Conclusions
In this study, a shotgun proteomics strategy was applied for the first time for the characterization of the jumbo squid skin proteome. A total of 1004 different peptides belonging to 219 different proteins were identified. The final proteome compilation was investigated using different in-silico studies, including GO term enrichment, pathways and networks studies. The most prominent protein classes were oxidoreductases, calcium-binding proteins, hydrolases, nucleic acid binding, enzyme modulation, transferases involved in metabolic pathways (cysteine and methionine metabolism), endocytosis/phagosome, RNA transport, protein methylation, and calcium homeostasis. The first most comprehensive interactomic network map for the jumbo squid skin proteome was built up containing 21 nodes and 61 interactions. Most of the jumbo squid skin proteins were grouped under pathways and networks referring to metabolic and oxidative metabolism, calcium homeostasis, transmembrane transport and metabolic and cellular respiration. Moreover, potential valuable bioactive peptides were predicted after different in-silico digestions with pepsin and trysin. Antimicrobial, bioactive collagen peptides, antihypertensive, and antitumor properties were predicted to be present in the jumbo squid skin proteome. The integration of the global proteomics results and the bioinformatics analysis of the jumbo squid skin proteome show a comprehensive knowledge of this fishery discard and provide potential bioactive peptides of this marine by-product.
Acknowledgments
We are grateful to Lorena Barros (IIM-CSIC, Vigo, Spain) for her excellent technical assistance during the experiments.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/18/1/31/s1, Table S1: Peptide Spectrum Matches (PSMs), Table S2: Peptide Groups, Table S3: Proteins, Table S4: Potential bioactive peptides predicted after pepsin or trypsin digestion.
Author Contributions
M.C. and J.M.E.-B. performed experiments and analyzed data. M.C. wrote the manuscript. J.M.E.-B. and S.P.A. conceptualized, designed the research, revised and corrected the paper. All authors agreed with the final submitted version. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ramón Areces Foundation (XVII National Grant), GAIN-Xunta de Galicia Project (IN607D 2017/01) and by CONACyT-Mexico under grant 2174. M.C. is supported by the Ramón y Cajal Contract (Ministry of Science, Innovation and Universities of Spain).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Rustad T., Storrø I., Slizyte R. Possibilities for the utilization of marine by-products. Int. J. Food Sci. Technol. 2011;46:2001–2014. doi: 10.1111/j.1365-2621.2011.02736.x. [DOI] [Google Scholar]
- 2.Blanco M., Vázquez J.A., Pérez-Martín R.I., Sotelo C.G. Hydrolysates of fish skin collagen: An opportunity for valorizing fish industry byproducts. Mar. Drugs. 2017;15:131. doi: 10.3390/md15050131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Commission . Regulation (EU) No 1380/2013 of the European Parliament and the Council of 11 December 2013 on the Common Fisheries Policy, Amending Council Regulations (EC) No 1954/2003 and (EC) No 1224/2009 and Repealing Council Regulations (EC) No 2371/2002 and (EC) No 639/2004 and Council Decision 2004/585/EC. European Commission; Brussels, Belgium: 2013. [Google Scholar]
- 4.Carrera M., Cañas B., Gallardo J.M. The sarcoplasmic fish proteome: Pathways, metabolic networks and potential bioactive peptides for nutritional inferences. J. Proteomics. 2013;78:211–220. doi: 10.1016/j.jprot.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Venkatesan J., Anil S., Kim S.K., Shim M.S. Marine fish proteins and peptides for cosmeceuticals: A review. Mar. Drugs. 2017;15:143. doi: 10.3390/md15050143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez A., Blanco M., Correa B., Pérez-Martín R.I., Sotelo C.G. Effect of fish collagen hydrolysates on type I collagen mRNA levels of human dermal fibroblast culture. Mar. Drugs. 2018;16:144. doi: 10.3390/md16050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vázquez J.A., Meduíña A., Durán A.I., Nogueira M., Fernández-Compás A., Pérez-Martín R.I., Rodríguez-Amado I. Production of valuable compounds and bioactive metabolites from by-products of fish discards using chemical processing, enzymatic hydrolysis, and bacterial fermentation. Mar. Drugs. 2019;17:139. doi: 10.3390/md17030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Food and Agriculture Organisation of the United Nations Global Production Statistics-Fisheries and Aquaculture. [(accessed on 27 May 2017)]; Available online: http://www.fao.org/fishery/statistics/global-aqua.
- 9.Ezquerra-Brauer J.M., Aubourg S. Recent trends for the employment of jumbo squid (Dosidicus gigas) by products as a source of bioactive compounds with nutritional, functional and preservative applications: A review. Int. J. Food Sci. Technol. 2019;54:987–998. doi: 10.1111/ijfs.14067. [DOI] [Google Scholar]
- 10.Mäthger L.M., Denton E.J., Marshall N.J., Hanlon R.T. Mechanisms and behavioral functions of structural coloration in cephalopods. J. R. Soc. Interface. 2009;6:S149–S163. doi: 10.1098/rsif.2008.0366.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deravi L.F., Magyar A.P., Sheehy S.P., Bell G.R., Mäthger L.M., Senft S.L., Wardill T.J., Lane W.S., Kuzirian A.M., Hanlon R.T., et al. The structure-function relationships of a natural nanoscale photonic device in cuttlefish chromatophores. J. R. Soc. Interface. 2014;11:20130942. doi: 10.1098/rsif.2013.0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aubourg S.P., Torres-Arreola W., Trigo M., Ezquerra-Brauer J.M. Partial characterization of jumbo squid skin pigment extract and its antioxidant potential in a marine oil system. Eur. J. Lipid Sci. Technol. 2016;118:1293–1304. doi: 10.1002/ejlt.201500356. [DOI] [Google Scholar]
- 13.Mosquera M., Giménez B., Montero P., Gómez-Guillén M.C. Incorporation of liposomes containing squid tunic ACE-inhibitory peptides into fish gelatin. J. Sci. Food Agric. 2016;96:769–776. doi: 10.1002/jsfa.7145. [DOI] [PubMed] [Google Scholar]
- 14.Shahidi S., Jamili S., Ghavam Mostafavi P., Rezaie S., Khorramizadeh M. Assessment of the inhibitory effects of ficin-hydrolyzed gelatin derived from squid (Uroteuthis duvauceli) on breast cancer cell lines and animal model. Iran. J. Allergy Asthma Immunol. 2018;17:436–452. doi: 10.18502/ijaai.v17i5.302. [DOI] [PubMed] [Google Scholar]
- 15.Mendis E., Rajapakse N., Byun H.G., Kim S.K. Investigation of jumbo squid (Dosidicus gigas) skin gelatin peptides for their in vitro antioxidant effects. Life Sci. 2005;77:2166–2178. doi: 10.1016/j.lfs.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 16.Chan-Higuera J.E., Santacruz-Ortega H.D.C., Carbonell-Barrachina A.A., Burgos-Hernández A., Robles-Sánchez R.M., Cruz-Ramírez S.G., Ezquerra-Brauer J.M. Xanthommatin is behind the antioxidant activity of the skin of Dosidicus gigas. Molecules. 2019;24:3420. doi: 10.3390/molecules24193420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amado I.R., Vázquez J.A., Gónzález P., Esteban-Fernández D., Carrera M., Piñeiro C. Identification of the major ACE-inhibitory peptides produced by enzymatic hydrolysis of a protein concentrate from cuttlefish wastewater. Mar. Drugs. 2014;12:1390–1405. doi: 10.3390/md12031390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar P., Kannan M., ArunPrasanna V., Vaseeharan B., Vijavakumar S. Proteomic analysis of crude squid ink isolated from Sepia esculenta for their antimicrobial, antibiofilm and cytotoxic properties. Microb. Pathog. 2018;116:345–350. doi: 10.1016/j.micpath.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 19.Ezquerra-Brauer J.M., Miranda J.M., Cepeda A., Barros-Velázquez J., Aubourg S.P. Effect of jumbo squid (Dosidicus gigas) skin extract on the microbial activity in chilled mackerel (Scomber scombrus) LWT-Food Sci. Technol. 2016;72:134–140. doi: 10.1016/j.lwt.2016.04.024. [DOI] [Google Scholar]
- 20.Ezquerra-Brauer J.M., Miranda J.M., Chan-Higuera J.E., Barros-Velázquez J., Aubourg S.P. New icing media for quality enhancement of chilled hake (Merluccius merlucius) using a jumbo squid (Dosidicus gigas) skin extract. J. Sci. Agric. 2017;97:3412–3419. doi: 10.1002/jsfa.8192. [DOI] [PubMed] [Google Scholar]
- 21.Carrera M., Cañas B., Gallardo J.M. Proteomics for the assessment of quality and safety of fishery products. Food Res. Int. 2013;54:972–979. doi: 10.1016/j.foodres.2012.10.027. [DOI] [Google Scholar]
- 22.Stryiński R., Mateos J., Pascual S., González A.F., Gallardo J.M., Łopieńska-Biernat E., Medina I., Carrera M. Proteome profiling of L3 and L4 Anisakis simplex development stages by TMT-based quantitative proteomics. J. Proteomics. 2019;201:1–11. doi: 10.1016/j.jprot.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Gallardo J.M., Carrera M., Ortea I. Proteomics in food science. In: Cifuentes A., editor. Foodomics: Advanced Mass Spectrometry in Modern Food Science and Nutrition. John Wiley & Sons Inc.; Hoboken, NJ, USA: 2013. pp. 125–165. [Google Scholar]
- 24.Carrera M., Cañas B., Gallardo J.M. Advanced proteomics and systems biology applied to study food allergy. Curr. Opin. Food Sci. 2018;22:9–16. doi: 10.1016/j.cofs.2017.12.001. [DOI] [Google Scholar]
- 25.Carrera M., González-Fernández A., Magadán S., Mateos J., Pedrós L., Medina I., Gallardo J.M. Molecular characterization of B-cell epitopes for the major fish allergen, parvalbumin, by shotgun proteomics, protein-based bioinformatics and IgE-reactive approaches. J. Proteomics. 2019;200:123–133. doi: 10.1016/j.jprot.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Eng J.K., McCormack A.L., Yates J.R., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 27.Perkins D.N., Pappin D.J.C., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 28.Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 29.Kall L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 30.Wang G., Li X., Wang Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;4:D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwaniak A., Dziuba J., Niklewicz M. The BIOPEP database—A tool for the in silico method of classification of food proteins as the source of peptides with antihypertensive activity. Acta Aliment. Hung. 2005;34:417–425. doi: 10.1556/AAlim.34.2005.4.9. [DOI] [Google Scholar]
- 32.Shi L., Zhang Q., Rui W., Lu M., Jing X., Shang T., Tang J. BioPD: A web-based information center for bioactive peptides. Regul. Pept. 2004;120:1–3. doi: 10.1016/j.regpep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Li Q., Zhang C., Chen H., Xue J., Guo X., Liang M., Chen M. BioPepDB: An integrated data platform for food-derived bioactive peptides. Int. J. Food Sci. Nutr. 2018;69:963–968. doi: 10.1080/09637486.2018.1446916. [DOI] [PubMed] [Google Scholar]
- 34.Thomas S., Karnik S., Barai R.S., Jayaraman V.K., Idicula-Thomas S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010;38:D774–D780. doi: 10.1093/nar/gkp1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rong M., Zhou B., Zhou R., Liao Q., Zeng Y., Xu S., Liu Z. PPIP: Automated software for identification of bioactive endogenous peptides. J. Proteome Res. 2019;18:721–727. doi: 10.1021/acs.jproteome.8b00718. [DOI] [PubMed] [Google Scholar]
- 36.Aguilera-Mendoza L., Marrero-Ponce Y., Beltran J.A., Tellez Ibarra R., Guillen-Ramirez H.A., Brizuela C.A. Graph-based data integration from bioactive peptide databases of pharmaceutical interest: Towards an organized collection enabling visual network analysis. Bioinformatics. 2019;35:4739–4747. doi: 10.1093/bioinformatics/btz260. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Yin T., Xiao X., He D., Xue Z., Jiang X., Wang Y. StraPep: A structure database of bioactive peptides. Database (Oxford) 2018 doi: 10.1093/database/bay038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinoshita Y., Yoshioka T., Kato S., Konno K. Color development of squid skin as affected by oxygen concentrations. J. Food Sci. 2009;74:S142–S146. doi: 10.1111/j.1750-3841.2009.01105.x. [DOI] [PubMed] [Google Scholar]
- 39.Celio M.R., Heizmann C.W. Calcium-binding protein parvalbumin is associated with fast contracting muscle fibres. Nature. 1982;297:504–506. doi: 10.1038/297504a0. [DOI] [PubMed] [Google Scholar]
- 40.Nelson T.J., Cavallaro S., Yi C.L., McPhie D., Schreurs B.G., Gusey P.A., Favit A., Zohar O., Kim J., Beushausen S. Calexcitin: A signaling protein that binds calcium and GTP, inhibits potassium channels, and enhances membrane excitability. Proc. Natl. Acad. Sci. USA. 1996;93:13808–13813. doi: 10.1073/pnas.93.24.13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keil B. Specificity of proteolysis. 1st ed. Springer-Verlag; Berlin/Heidelberg, Germany: 1992. [Google Scholar]
- 42.Mooney C., Haslam N.J., Pollastri G., Shields D.C. Towards the improved discovery and design of functional peptides: Common features of diverse classes permit generalized prediction of bioactivity. PLoS ONE. 2012;7:e45012. doi: 10.1371/journal.pone.0045012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giménez B., Gómez-Estaca J., Alemán A., Gómez-Guillén M.C., Montero P. Physico-chemical and film forming properties of giant squid (Dosidicus gigas) gelatin. Food Hydrocoll. 2009;23:585–592. doi: 10.1016/j.foodhyd.2008.07.003. [DOI] [Google Scholar]
- 44.Cai J., Li Y., Zhang Y., Tong Q., Wang F., Su X. Protective effects of collagen extracted from Dosidicus gigas skin on MC3T3-E1 cell induced by H2O2. J. Chin. Inst. Food Sci. Technol. 2015;15:6–12. [Google Scholar]
- 45.Cai J., Li Y., Quan J., Lin J., Zhang Y., Wang F., Su X. Effect of collagen peptide extracted from Dosidicus gigas skin on proliferation, differentiation and calcification of MC3T3-E1 cell induced by Cd. J. Chin. Inst. Food Sci. Technol. 2015;15:18–24. [Google Scholar]
- 46.Liu S., Aweya J.J., Zheng L., Wang F., Zheng Z., Zhong M., Lun J., Zhang Y. A Litopenaeus vannamei hemocyanin-derived antimicrobial peptide (peptide B11) attenuates cancer cells’ proliferation. Molecules. 2018;23:3202. doi: 10.3390/molecules23123202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atala A. This month in investigative urology. J. Urol. 2006;176:2335–2336. doi: 10.1016/j.juro.2006.09.002. [DOI] [Google Scholar]
- 48.McFadden D.W., Riggs D.R., Jackson B.J., Vona-Davis L. Keyhole limpet hemocyanin, a novel immune stimulant with promising anticancer activity in Barrett’s esophageal adenocarcinoma. Am. J. Surg. 2003;186:552–555. doi: 10.1016/j.amjsurg.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 49.Dipolo R. Ca pump driven by ATP in squid axons. Nature. 1978;274:390–392. doi: 10.1038/274390a0. [DOI] [PubMed] [Google Scholar]
- 50.Cheung H.S., Wang F.L., Ondetti M., Sabo E., Cushman D. Binding of peptide substrates and inhibitors of angiotensin-converting enzyme: Importance of the COOH-terminal dipeptide sequences. J. Biol. Chem. 1980;255:401–407. [PubMed] [Google Scholar]
- 51.Kasamatsu C., Kimura S., Kagawa M., Hatae K. Identification of high molecular weight proteins in squid muscle by western blotting analysis and postmortem rheological changes. Biosci. Biotechnol. Biochem. 2004;68:1119–1124. doi: 10.1271/bbb.68.1119. [DOI] [PubMed] [Google Scholar]
- 52.Chan-Higuera J.E., Carbonell-Barrachina A.A., Cárdenas-López J.L., Kačániová M., Burgos-Hernández A., Ezquerra-Brauer J.M. Jumbo squid (Dosidicus gigas) skin pigments: Chemical analysis and evaluation of antimicrobial and antimutagenic potential. J. Microbiol. Biotech. Food Sci. 2019;9:349–353. doi: 10.15414/jmbfs.2019.9.2.349-353. [DOI] [Google Scholar]
- 53.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.