Abstract
Loss of function in tumor suppressor genes is commonly associated with the onset/progression of cancer and treatment resistance. The p53 tumor suppressor gene, a master regulator of diverse cellular pathways, is frequently altered in various cancers, for example, in ~36% of hepatocellular carcinomas (HCCs) and ~68% of non–small cell lung cancers (NSCLCs). Current methods for restoration of p53 expression, including small molecules and DNA therapies, have yielded progressive success, but each has formidable drawbacks. Here, a redox-responsive nanoparticle (NP) platform is engineered for effective delivery of p53-encoding synthetic messenger RNA (mRNA). We demonstrate that the synthetic p53-mRNA NPs markedly delay the growth of p53-null HCC and NSCLC cells by inducing cell cycle arrest and apoptosis. We also reveal that p53 restoration markedly improves the sensitivity of these tumor cells to everolimus, a mammalian target of rapamycin (mTOR) inhibitor that failed to show clinical benefits in advanced HCC and NSCLC. Moreover, cotargeting of tumor-suppressing p53 and tumorigenic mTOR signaling pathways results in marked antitumor effects in vitro and in multiple animal models of HCC and NSCLC. Our findings indicate that restoration of tumor suppressors by the synthetic mRNA NP delivery strategy could be combined together with other therapies for potent combinatorial cancer treatment.
INTRODUCTION
The mammalian target of rapamycin (mTOR) is a serine/threonine kinase that regulates major cell functions such as growth and proliferation in physiological and pathological conditions (1). Dysregulation of the mTOR signaling pathway has been reported for a wide range of cancers including liver and lung cancers (2–4). Everolimus (RAD001) is an effective mTOR inhibitor that has been clinically approved for several types of cancers, such as advanced kidney cancer and pancreatic neuroendocrine tumor. However, everolimus failed to improve survival in patients with other advanced cancers, such as hepatocellular carcinoma (HCC) or non–small cell lung cancer (NSCLC) (5–8). Previous studies have proposed several mechanisms underlying the variable response or resistance to everolimus in different tumor cells (9, 10), including the activation of prosurvival autophagy (11–13) and the dysregulation of apoptotic pathways [for example, up-regulation of antiapoptotic protein B cell lymphoma 2 (BCL-2)] (14). Combining everolimus with autophagy or BCL-2 inhibitors improved antitumor efficacy, but these inhibitors could also induce undesired toxicities by interfering with physiological processes in normal cells (15–17).
In parallel to the gain of protumorigenic functions such as the mTOR signaling pathway, cancer is also frequently associated with the inactivation of tumor suppressors. p53 is one of the most widely altered tumor suppressor genes in numerous cancers. For example, the loss of p53 function has been widely detected in ~36% of HCC and ~68% of NSCLC, according to The Cancer Genome Atlas (TCGA) database in the cBio Cancer Genomics Portal (http://cbioportal.org) (18). p53 regulates many important cellular pathways. As a transcription factor, p53 can activate its downstream genes in response to oncogenic signals (19), such as pro-apoptotic proteins BAX (BCL-2–associated X protein) and PUMA (p52 up-regulated modulator of apoptosis) (20). p53 also acts as a cell cycle checkpoint guard to induce cell cycle arrest (21) and participates in DNA replication and repair to protect genomic integrity (22). In addition, cytoplasmic (but not nuclear) p53 inhibits the activation of protective autophagy that may contribute to the tolerance to chemotherapies (23, 24). Therefore, the restoration of p53 expression could potentially not only inhibit tumor growth by inducing cell apoptosis and cell cycle arrest but also sensitize p53-deficient cancers to the mTOR inhibitor everolimus.
Two different strategies have been widely explored for p53 reactivation: (i) the use of small molecules to disrupt the p53-MDM2 (mouse double minute 2 homolog) interaction and release p53 or to restore wild-type function to mutant p53 by covalent modification of its core domain (25–28) and (ii) the restoration of a functional copy via viral or nonviral DNA transfection (29–31). Although these attempts have exhibited some successes, each has formidable limitations. For instance, small molecular compounds are likely ineffective when the tumor suppressor gene has been deleted, and p53-DNA–based gene therapies have the potential risk of genomic integration and mutagenesis (32, 33). Here, we demonstrate the feasibility of using mRNA to reconstitute p53 expression in p53-deficient HCC and NSCLC with redox-responsive lipid-polymer hybrid nanoparticles (NPs) engineered for effective delivery of synthetic mRNA (fig. S1A). Because mRNA functions in the cytoplasm, this strategy avoids the requirement of nuclear localization and the risk of insertional mutagenesis associated with DNA (34, 35). Our results demonstrate that treatment of p53-null Hep3B HCC and H1299 NSCLC cells with the p53-mRNA hybrid NPs inhibited tumor cell growth by inducing cell apoptosis and G1-phase cell cycle arrest. The p53-mRNA NPs also sensitized these tumor cells to everolimus, presumably via p53 restoration–mediated regulation of the autophagy pathway (fig. S1B), resulting in synergistic antitumor efficacy in vitro and in vivo.
RESULTS
Engineering and characterization of synthetic mRNA NPs
We used in vitro transcription to synthesize enhanced green fluorescent protein (EGFP) mRNA and p53-mRNA (fig. S1A). The 5′ terminal of mRNA was designed with an untranslated region to enhance the translational initiation of the mRNA (fig. S2). Anti-reverse cap analog (ARCA) capping of 3′-O-Me-m7G(5′)ppp(5′)G (fig. S3) and enzymatic polyadenylation were further used to modify the mRNA to increase its stability and translation efficiency. To reduce mRNA immunostimulation, 5-methylcytidine-5′-triphosphate and pseudouridine-5′-triphosphate were used to replace regular CTP and UTP (36, 37). We next used a robust self-assembly approach (38–40) to engineer lipid-polymer hybrid NPs for effective loading of the chemically modified mRNA, by using a cationic lipid-like molecule G0-C14, a hydrophobic redox-responsive cysteine-based poly(disulfide amide) (PDSA), and two lipid-poly(ethylene glycol) (lipid-PEG) compounds (fig. S4). The cationic G0-C14 was used for mRNA complexation and to facilitate its cytosolic transport (40), and the PDSA was chosen to form a stable NP core under normal physiological conditions while providing a rapid triggered release of payloads in tumor cells with high intracellular concentration of glutathione (GSH) (41–43). Both 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)] (DMPE-PEG) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)] (DSPE-PEG) were coated onto the surface of the hybrid NPs to simultaneously achieve a relatively long circulation time and high tumor cell uptake through a de-PEGylation effect (39). As shown in fig. S5A, mRNA could be effectively condensed with G0-C14 at a weight ratio (G0-C14/mRNA) of 10% (w/w) or above, with no effect of the dimethylformamide solvent used for NP formulation on the integrity of mRNA. We prepared the redox-responsive hybrid NPs at the G0-C14/mRNA weight ratio of 15, and the engineered mRNA NPs showed an average size of ~125 nm and were stable under physiological conditions (fig. S5B). As characterized by transmission electron microscopy (TEM) (Fig. 1A), the solid PDSA polymer core contributed to the formation of a rigid and stable nanostructure in (pH 7.4) phosphate-buffered saline (PBS) while efficiently responding to dithiothreitol (DTT; a reductive agent) by rapid disassembly of the NPs for release of mRNA (fig. S5C). The redox-triggered sufficient release of payloads could potentially contribute to more effective therapeutic activities (41–47). The evaluation and selection of mRNA NP formulations are provided in figs. S6 to S8 and table S1.
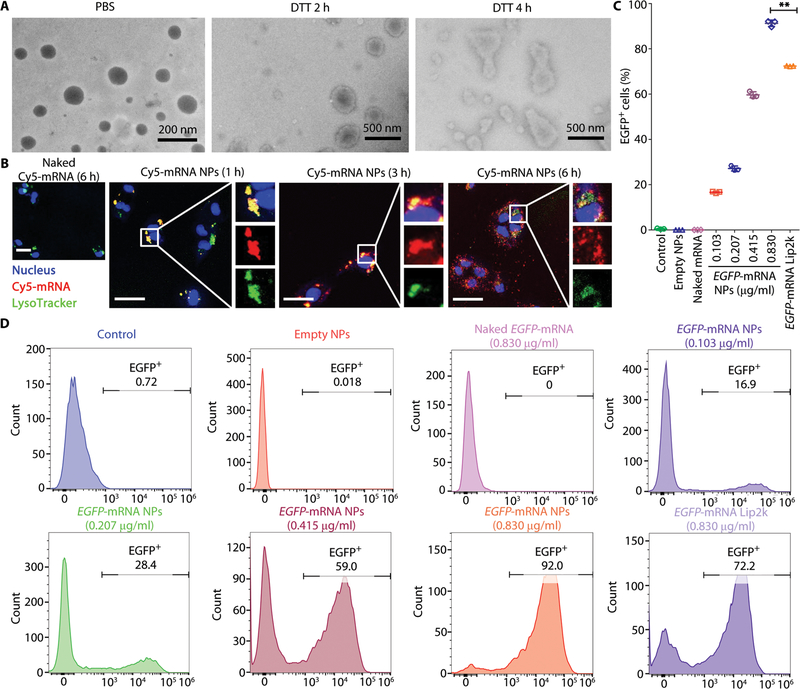
Fig. 1. In vitro transfection efficiency of the redox-responsive mRNA NPs in p53-null Hep3B cells.
(A) Transmission electron microscopy (TEM) images of the hybrid mRNA NPs before incubation (in PBS) or after incubation in 10 mM DTT for 2 or 4 hours at 37°C. (B) Confocal laser scanning microscopy (CLSM) images of p53-null Hep3B cells after incubation with naked Cy5-labeled mRNA (red) for 6 hours and with engineered Cy5-labeled mRNA NPs for 1, 3, or 6 hours. Endosomes were stained by LysoTracker Green (green), and nuclei were stained by 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bars, 50 μm. (C) In vitro transfection efficiency (percentage of EGFP-positive cells) was determined by flow cytometry. Data shown as means ± SEM (n = 3), and statistical significance was determined using two-tailed t test (**P < 0.01). (D) Histogram analysis of the in vitro transfection efficiency by FlowJo software.
We next examined the cytosolic delivery of mRNA using the engineered NPs in vitro. As shown in Fig. 1B and fig. S9, the NPs could effectively transport Cy5-labeled mRNA into the cytoplasm in a time-dependent manner. Most of the internalized mRNA NPs first colocalized with LysoTracker Green at 1 hour. After 3 hours of incubation, some of Cy5-labeled mRNA entered the cytoplasm, and at 6 hours after incubation, a large amount of them escaped from endosomes and diffused into the cytoplasm. In comparison, naked mRNA could not readily enter the cells after 6 hours of incubation. The efficient cytosolic delivery of mRNA with the hybrid NPs could be observed in both p53-null HCC (Hep3B) and NSCLC (H1299) cells.
To further check the transfection efficacy in vitro, EGFP-mRNA was chosen as a model mRNA. The high transfection efficiency of the EGFP-mRNA NPs can be directly visualized by confocal laser scanning microscopy (CLSM), with considerable green fluorescence detected in both NP-transfected and commercial transfection agent Lipofectamine 2000 (Lip2k)–transfected cells (fig. S10). To quantitatively analyze mRNA transfection, EGFP expression in Hep3B and H1299 cells was measured by flow cytometry (Fig. 1, C and D, and fig. S11). The EGFP expression showed a dose-dependent increase (EGFP-mRNA concentration from 0.103 to 0.830 μg/ml). Moreover, the percentage of EGFP-positive cells was significantly higher for the NP-transfected cells than for Lip2k-transfected cells at the concentration of 0.830 μg/ml (P < 0.01), indicating a better transfection efficacy with the NP-mediated strategy in both Hep3B and H1299 cells. When using N-ethylmaleimide (Nem) to quench intracellular GSH, we noticed a marked decrease of EGFP expression by the mRNA NPs (fig. S12), indicating that the redox-triggered mRNA release within the tumor cells may lead to better bioactivity. Moreover, no obvious in vitro cytotoxicity was observed in Hep3B and H1299 cells with all the tested concentrations of EGFP-mRNA NPs via AlamarBlue assay (fig. S13). These results suggested the potential of the engineered hybrid NPs for synthetic mRNA delivery to restore tumor suppressor p53 in p53-null tumor cells.
Hybrid mRNA NP–mediated p53 restoration in p53-null HCC and NSCLC cells
To examine the mRNA NP strategy for restoration of tumor suppressor p53 in p53-null Hep3B and H1299 cells, immunofluorescence (IF) staining and Western blot (WB) were performed to check the p53 protein expression in both cell lines after treatment with p53-mRNA NPs. The IF results showed that p53 proteins were mainly expressed in the cytoplasm of both cell lines (Fig. 2A and fig. S14). WB results also demonstrated that the expression of p53 protein was obviously increased in both cells after NP treatment (fig. S15). Next, we tested whether the p53-mRNA NPs could restore the suppressing function of p53 in p53-null tumor cells. After incubation with different doses of p53-mRNA NPs, strong cytotoxicity was observed in a dose-dependent manner in Hep3B (Fig. 2B) and H1299 (fig. S16A) cells. Colony formation was also markedly inhibited in both cells treated with p53-mRNA NPs versus empty NPs, further demonstrating p53 restoration–mediated antitumor activities (Fig. 2C and fig. S16B). Meanwhile, apoptosis was measured using the annexin V (AnnV) and propidium iodide (PI) costaining method, followed by flow cytometry analysis. As can be seen in Fig. 2 (D and E) and fig. S17, cell apoptosis greatly increased after treatment with p53-mRNA NPs at the concentrations of 0.415 and 0.830 μg/ml in Hep3B and H1299 cells, whereas empty NPs and naked mRNA did not induce apoptosis.
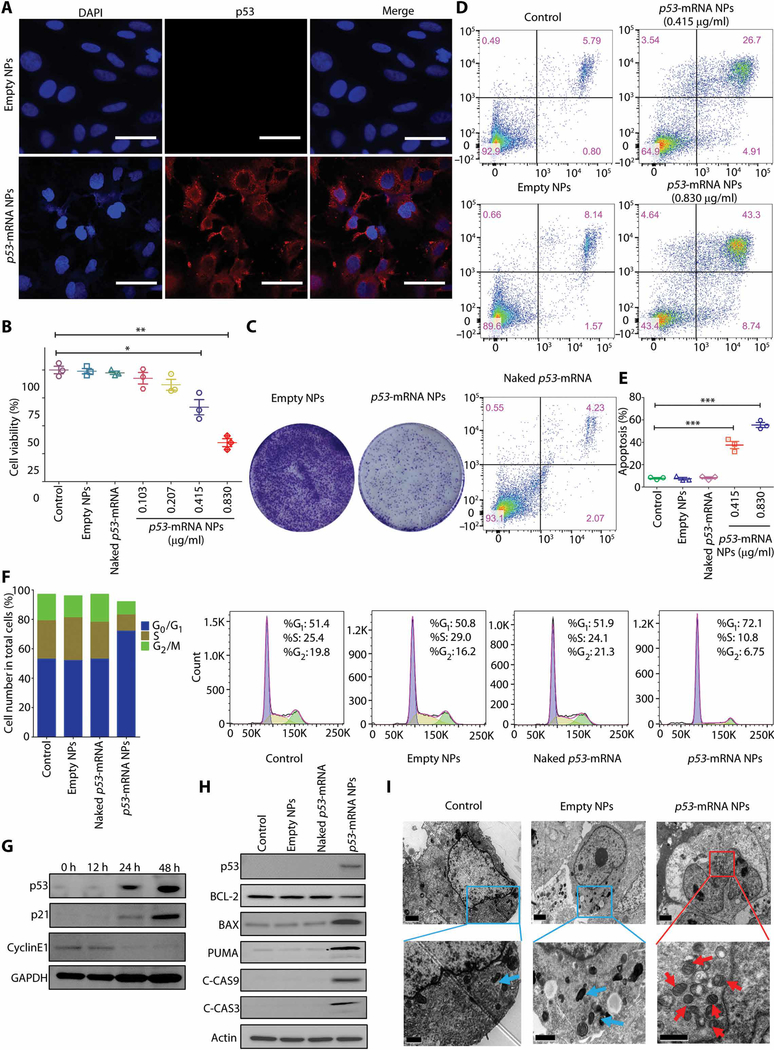
Fig. 2. Restoration of p53 functions in p53-null Hep3B cells by the mRNA NPs and in vitro mechanisms for p53 restoration–mediated antitumor effect.
(A) Immunofluorescence (IF) staining of p53 in the p53-null Hep3B cells treated by empty NP or p53-mRNA NPs. Scale bars, 50 μm. (B) Viability of the p53-null Hep3B liver cancer cells after treatment with PBS, empty NPs, naked p53-mRNA (0.830 μg/ml), or p53-mRNA NPs (mRNA concentrations: 0.103, 0.207, 0.415, or 0.830 μg/ml) by alarmBlue assay. Statistical significance was determined using two-tailed t test (*P < 0.05, **P < 0.01). (C) Colony formation assays of Hep3B cells after treatment with empty NPs versus p53-mRNA NPs in six-well plates. (D) Apoptosis of Hep3B cells as determined by flow cytometry after treatment with empty NPs, naked p53-mRNA, or p53-mRNA NPs. (E) Histogram analysis of the cell apoptosis (%) by FlowJo software. Data shown as means ± SEM (n = 3), and statistical significance was determined using two-tailed t test (***P < 0.001). (F) Cell cycle distributions of Hep3B cells after treatment with PBS, empty NPs, naked p53-mRNA, or p53-mRNA NPs (mRNA concentration: 0.830 μg/ml). (G) Western blot (WB) analysis of cell cycle–related protein expression (p21 and CyclinE1) after treatment with p53-mRNA NPs (mRNA concentration: 0.830 μg/ml). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the loading control. (H) WB analysis of the mitochondrial apoptotic signaling pathway in p53-null Hep3B cells treated with PBS, empty NPs, naked p53-mRNA, or p53-mRNA NPs (mRNA concentration: 0.830 μg/ml). BCL-2, BAX, PUMA, C-CAS9, and C-CAS3 proteins were detected. Actin was used as the loading control. (I) TEM images of the mitochondrial morphology in Hep3B cells from control, empty NPs, and p53-mRNA NP groups (mRNA concentration: 0.830 μg/ml; blue arrow, normal mitochondria; red arrow, swelling mitochondria). Scale bars, 2 μm for the top images and 1 μm for the enlarged images (bottom).
In addition, the cell cycle phase distribution was studied upon treatment with p53-mRNA NPs in Hep3B and H1299 cells. Figure 2F shows that Hep3B cells treated with p53-mRNA NPs had a larger G1 population (72.1%) compared with ~50% in the control, empty NPs, or naked mRNA groups. Concomitant decreases were observed in S and G2 phases after p53-mRNA NP treatment compared with the control, empty NPs, or naked mRNA groups. Similar results were observed in H1299 cells (fig. S18), suggesting that p53 restoration could effectively induce G1-phase cell cycle arrest to inhibit cell proliferation. We also investigated the signaling pathways involved in cell cycle regulation by evaluating the cell cycle–related proteins in Hep3B cells (Fig. 2G). The restoration of p53 functions by mRNA NPs resulted in the up-regulation of p21 and the down-regulation of CyclinE1 from 12 to 48 hours, and it blocked the cell cycle at the G1 phase.
To further assess the in vitro antitumor mechanisms of the p53-mRNA NPs in p53-null Hep3B and H1299 cells, we performed WB studies to verify the effects of p53 on the apoptosis pathway. As shown in Fig. 2H and fig. S19, p53-mRNA NPs efficiently activated PUMA to initiate the cleaved caspase-9 (C-CAS9)– and C-CAS3–induced apoptosis pathway. We further confirmed this pathway through TEM analysis of mitochondrial morphology change, which is usually a common phenomenon for this apoptosis pathway (48, 49). Consistent with the WB results, we observed increased numbers of swollen mitochondria (red arrows) in the cytoplasm of Hep3B and H1299 cells after treatment with p53-mRNA NPs (Fig. 2I and fig. S20), as compared to the control and empty NP groups. These results indicated that p53 restoration by our mRNA NPs causes mitochondrial depolarization and swelling, further confirming the initiation of cellular apoptosis. Moreover, we designed and tested a mutant p53-R175H-mRNA (table S2) as another control mRNA. As shown in fig. S21, treatment with p53-R175H-mRNA NPs induced the expression of mutant p53 in both Hep3B and H1299 cells. However, neither p21 nor C-CAS3 was detected after NP treatment. The expression of the mutant p53 also did not cause cytotoxicity.
p53 restoration sensitizes p53-null HCC and NSCLC cells to mTOR inhibitor everolimus
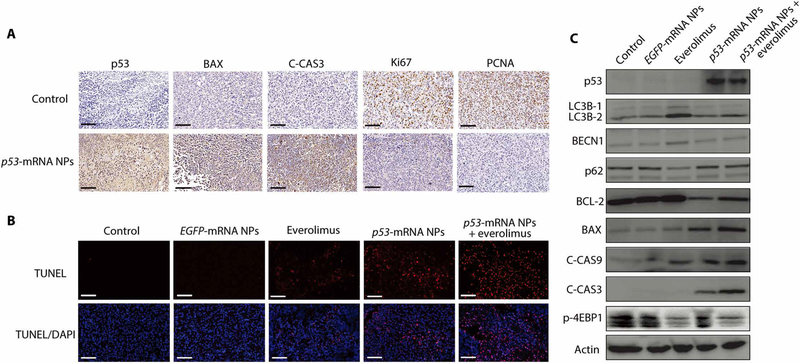
To examine the effects of p53 restoration on everolimus activity, we first measured the cytotoxicity of this mTOR inhibitor in p53-null Hep3B and H1299 cells and explored its effect on the mTOR pathway. Figure 3A and fig. S22A indicate relative insensitivity of Hep3B and H1299 to everolimus, with more than 50% of cells still alive at 64 nM. Although the mTOR pathway targets (p-mTOR and p-p70S6K) were substantially blocked by increasing everolimus concentrations (Fig. 3B and fig. S22B), there was no notable decrease in cell viability. We then examined the effect of everolimus on the autophagy pathway. According to the method previously reported (50), the extent of autophagy can be measured by the ratio of LC3B-2/actin on WB. With the increase of everolimus concentration, we observed up-regulation of LC3B-2 and higher LC3B-2/actin ratios by WB (Fig. 3C). The increased number of autophagosomes by TEM and increased fluorescence intensity of GFP-LC3B by CLSM were also consistent with the activation of autophagy by everolimus in Hep3B and H1299 cells (Fig. 3, D and E, and fig. S23).
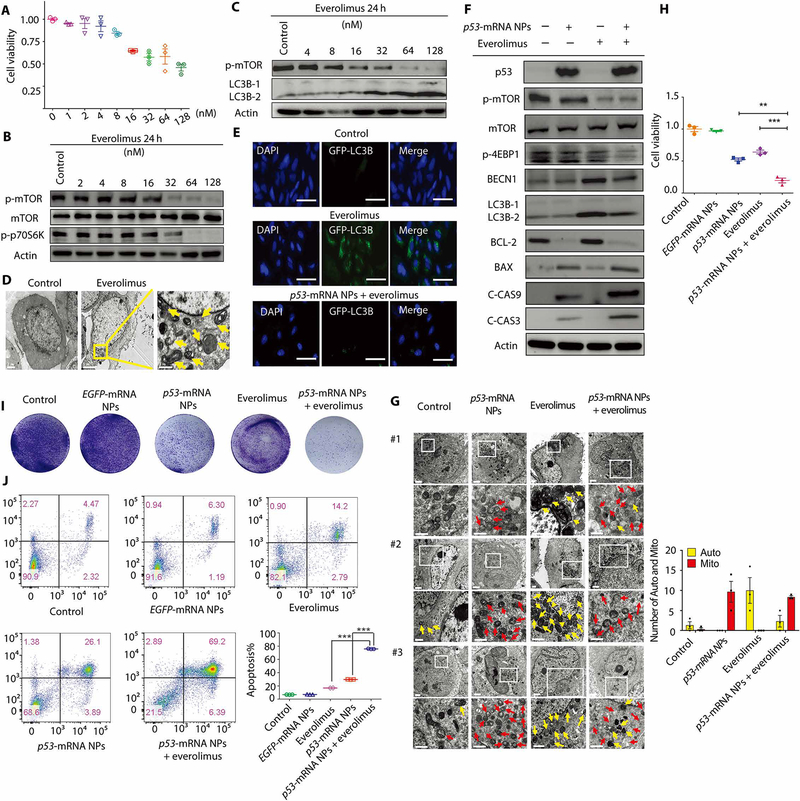
Fig. 3. Mechanisms of the p53-mRNA NP–mediated sensitization to everolimus in p53-null Hep3B cells.
(A) Viability of Hep3B cells after treatment with everolimus, as measured by AlamarBlue assay. Data shown as means ± SEM (n = 3). (B) WB analysis of total mTOR, p-mTOR, and p-p70S6K after treatment with everolimus at different concentrations. Actin was measured as the loading control. (C) WB analysis of p-mTOR, LC3B-1, and LC3B-2. Actin was measured as the loading control. (D) TEM images of Hep3B cells before and after 24 hours of treatment with everolimus (32 nM). Autophagosomes were labeled by yellow arrows. Scale bars (left to right), 2, 5, and 1 μm. (E) CLSM images of GFP-LC3–transfected Hep3B cells from different treatment groups. Scale bars, 50 μm. (F) WB analysis of p53, p-mTOR, total m-TOR, p-4EBP1, BECN1, LC3B-1, LC3B-2, BCL-2, BAX, C-CAS9, and C-CAS3 in Hep3B cells after different treatments. Actin was used as the loading control. (G) Left: TEM images of Hep3B cells in control, p53-mRNA NPs, everolimus, and p53-mRNA NPs + everolimus groups (mRNA concentration: 0.415 μg/ml; everolimus concentration: 32 nM). Scale bars, 2 μm for the raw images and 1 μm for the enlarged images. Yellow arrows, autophagosomes; red arrows, mitochondria. Right: Statistical analysis of the numbers of autophagosomes (Auto; yellow) and swollen mitochondria (Mito; red) after different treatments. (H) Viability of Hep3B cells in different groups (control, EGFP-mRNA NPs, p53-mRNA NPs, everolimus, or p53-mRNA NPs + everolimus) as measured by AlamarBlue assay (mRNA concentration: 0.415 μg/ml; everolimus concentration: 32 nM). Data shown as means ± SEM (n = 3), and statistical significance was determined using two-tailed t test (**P < 0.01, ***P < 0.001). (I) Colony formation of Hep3B cells in different treatment groups in six-well plates. (J) Flow cytometry analysis of the cell apoptosis (AnnV+PI− and AnnV+PI+). The percentage of apoptotic Hep3B cells was shown in the histogram. Statistical significance was determined using two-tailed t test (***P < 0.001).
Next, we examined whether the p53-mRNA NPs could inhibit the autophagy induced by everolimus. Both the CLSM and WB results in Fig. 3 (E and F) demonstrated that treatment with p53-mRNA NPs markedly reduced autophagy activation in p53-null Hep3B cells. We also observed the reduced number of autophagosomes (yellow arrows) in the “p53-mRNA NPs + everolimus” group as compared to the everolimus alone group by TEM (Fig. 3G). Moreover, we tested whether, in the presence of everolimus, the p53-mRNA NPs could still restore the apoptotic pathway in Hep3B cells, similar to those shown in Fig. 2. As can be seen in Fig. 3 (F and G), the up-regulated expression of C-CAS3/9 and increased number of swollen mitochondria (red arrows) suggested the successful activation of the apoptotic pathway after treatment with p53-mRNA NPs. Similar results could also be observed in p53-null H1299 cells (figs. S23C to S25).
Motivated by the results showing inhibition of the autophagy pathway and activation of the apoptotic pathway, we next determined whether the p53-mRNA NPs could sensitize Hep3B and H1299 cells to everolimus. As measured by AlamarBlue assay (Fig. 3H and fig. S26A), everolimus showed a moderate therapeutic effect (with ~70% viability in Hep3B cells and more than 80% viability in H1299 cells), whereas cotreatment with everolimus and p53-mRNA NPs showed strong in vitro antitumor effects in both cell lines (with ~19% viability in Hep3B cells and ~14% viability in H1299 cells). The EGFP-mRNA NPs were used as control NPs and did not show cytotoxicity. We also calculated the combination index (CI) using a reported method (51, 52) to assess whether there was a synergistic effect of the combination treatment. The CI value of p53-mRNA NPs + everolimus treatment was 1.71 in Hep3B cells and 1.74 in H1299 cells, indicating the presence of a synergistic effect (CI > 1) in both cell lines. The colony formation assay also showed a marked reduction in live cells after cotreatment with p53-mRNA NPs and everolimus (Fig. 3I and fig. S26B). Consistent with the above, flow cytometry analysis of apoptosis demonstrated that everolimus induced moderate apoptotic cell death, whereas cotreatment with everolimus and p53-mRNA NPs effectively augmented apoptosis (Fig. 3J and fig. S27). To investigate the synergistic effect, we tested whether the inhibition of BCL-2 may also contribute to the improvement in everolimus sensitivity, as previously reported with small cell lung cancer H-510 cells (14). Two strategies (small molecular inhibitor venetoclax and small interfering RNA) were used to target BCL-2 and combine with everolimus. Both approaches showed moderate combinatorial antitumor effect from BCL-2 inhibition together with high-dose everolimus (figs. S28 and S29), indicating that BCL-2 inhibition may not contribute to the improved everolimus sensitivity in p53-null Hep3B or H1299 cells. These results suggest that the synthetic mRNA NP–mediated p53 restoration can sensitize p53-null HCC and NSCLC cells to everolimus, presumably by inhibiting the activation of prosurvival autophagy.
Furthermore, we explored the possible mechanisms of how p53 restoration inhibits the protective autophagy. As shown in the quantitative real-time polymerase chain reaction (qRT-PCR) results (fig. S30 and table S3), the intervention of NPs effectively increased the expression of p53-mRNA compared to the groups without NP treatment in both cell lines. The increased p53-mRNA expression was also accompanied by clear inhibition of ULK1, ATG7, BECN1, and ATG12 mRNA expression (fig. S31) but showed no obvious effects on the mRNA expression of DRAM1, ISG20L1, and SESN1 (fig. S32). These results indicate that the autophagy-related genes ULK1, ATG7, BECN1, and ATG12 may be involved in the p53-mRNA NP–mediated inhibition of autophagy activation. We also examined two p53 target genes, TIGAR (TP53-induced glycolysis and apoptosis regulator) and AMPKα. TIGAR is a p53-regulated gene that can be rapidly activated in response to cellular stress (53). TIGAR can inhibit autophagy in a transcription-independent manner (54, 55). Consistent with previous studies (54–56), both our PCR and WB results (figs. S33 and S34) demonstrated that the expression of cytoplasmic p53 via p53-mRNA NPs activated the expression of TIGAR. The WB data also indicated the suppression of the 5′ adenosine monophosphate-activated protein kinase (AMPK) signaling pathway (23, 57), which can induce transcription-independent inhibition of autophagy (58). On the basis of these results, we proposed a possible mechanism (fig. S35) of how p53 tumor suppressor inhibits the protective autophagy and thus improves the sensitivity of p53-null tumor cells to everolimus.
p53 restoration sensitizes p53-null HCC and NSCLC xenograft models to everolimus
The lipid-PEG layer plays a critical role in controlling the cell uptake, pharmacokinetics (PK), and tumor accumulation of the hybrid lipid-polymer NPs (38, 39). We prepared the hybrid mRNA NPs with three different DSPE-PEG/DMPE-PEG ratios (NP25, NP50, and NP75 shown in table S1). We evaluated PK of the three Cy5-labeled mRNA NPs delivered by intravenous injection into healthy BALB/c mice. Naked Cy5-mRNA was used as a control. Figure 4A shows that naked mRNA was cleared within a few minutes, whereas the hybrid NPs effectively extended the circulation half-life (t1/2) of mRNA (NP25, t1/2 < 30 min; NP50, t1/2 ≈ 30 min; NP75, t1/2 ≈ 1 hour). In addition, ~40% of NP75 were still circulating in blood at 2 hours after administration. We then examined the biodistribution (BioD) and tumor accumulation of these NPs. Athymic nude mice carrying Hep3B xenograft were treated with naked Cy5-mRNA, Cy5-mRNA NP25, Cy5-mRNA NP50, or Cy5-mRNA NP75 by intravenous injection. As revealed in Fig. 4B and fig. S36, the fluorescent signal of naked Cy5-mRNA was barely detectable in the tumor at 24 hours after injection. Among the three different NPs, NP75 exhibited the highest tumor accumulation, which may be attributable to its long circulation, and was thus used for all the following in vivo studies. A comparable NP accumulation was also observed in H1299 xenograft tumors (fig. S37), which may be due to the abundant blood vessels in these two tumor models (fig. S38).
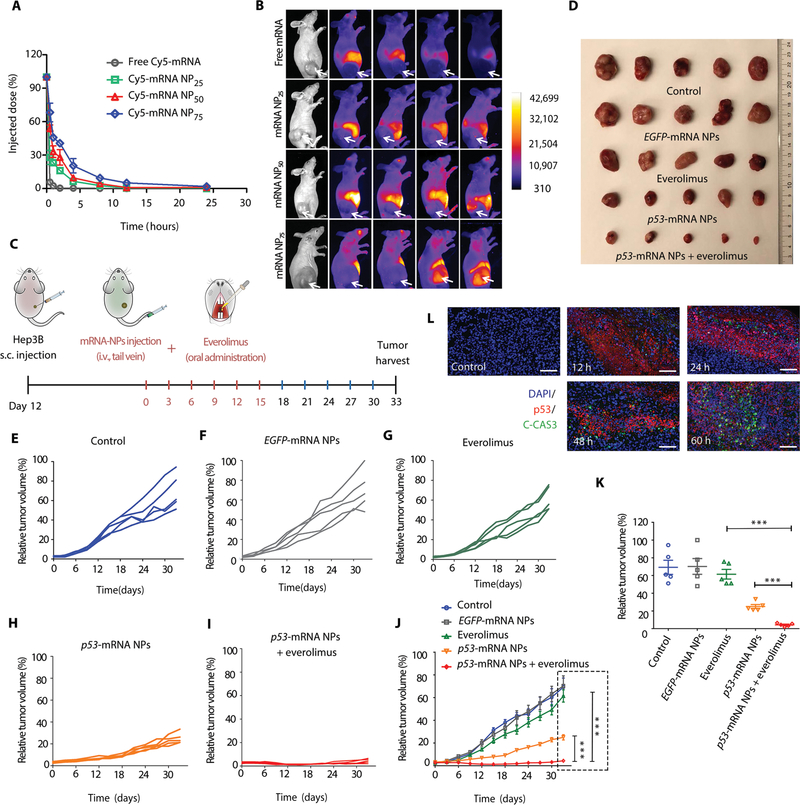
Fig. 4. Antitumor effects of p53-mRNA NPs are synergistic with everolimus in p53-null HCC xenograft model.
(A) Blood circulation profiles of naked Cy5-labeled mRNA and Cy5-labeled mRNA NPs (at an mRNA dose of 750 μg/kg of animal weight). NP25, NP50, and NP75 represent three different ratios of DSPE-PEG/DMPE-PEG (25:75, 50:50, and 75:25) hybrid in the lipid-PEG layer of hybrid NPs. Data shown as means ± SEM (n = 3). (B) Time-lapse near-infrared fluorescence imaging of nude mice bearing p53-null HCC xenograft tumors after intravenous injection of free Cy5-mRNA, Cy5-mRNA NP25, Cy5-mRNA NP50, or Cy5-mRNA NP75. The tumors were annotated with white arrows. (C) Scheme of tumor inoculation [subcutaneous (s.c.)] and treatment schedule in Hep3B tumor–bearing athymic nude mice. Twelve days after tumor inoculation, mice were treated with PBS [intravenous (i.v.)], EGFP-mRNA NPs (intravenous), p53-mRNA NPs (intravenous), everolimus (oral), or p53-mRNA NPs (intravenous) + everolimus (oral) every 3 days for six rounds (mRNA dose, 750 μg/kg; everolimus dose, 5 mg/kg). Tumors from different groups were harvested 18 days after the last treatment. (D) Photos of excised tumors from mice bearing Hep3B xenografts in different treatment groups on day 33 (n = 5). (E to I) Individual tumor growth kinetics in (E) control, (F) EGFP-mRNA NPs, (G) everolimus, (H) p53-mRNA NPs, and (I) p53-mRNA NPs + everolimus group (n = 5). (J) Average tumor growth kinetics for all treatment groups. Data shown as means ± SEM (n = 5), and significance was determined using two-tailed t test (***P < 0.001). (K) Average tumor volumes at experimental endpoint (day 33) in all groups. Data shown as means ± SEM (n = 5), and statistical significance was determined using two-tailed t test (***P < 0.001). (L) IF images of p53 (red) and C-CAS3 (green) costained Hep3B tumor sections at 12, 24, 48, and 60 hours after intravenous injection of p53-mRNA NPs. PBS (60 hours after intravenous injection) was used as control group. Scale bars, 100 μm.
To validate the therapeutic efficacy of the p53-mRNA NPs and their ability to sensitize tumors to everolimus, we performed in vivo studies in immunocompromised athymic nude mice bearing p53-null Hep3B xenografts (Fig. 4C). The p53-mRNA NPs were systemically injected via tail vein every 3 days for six treatments. Meanwhile, everolimus was administered orally right after each intravenous injection of NPs. PBS and EGFP-mRNA NPs were used as controls. Hep3B tumor–bearing mice treated with PBS and EGFP-mRNA NPs showed similarly rapid tumor growth, whereas everolimus alone showed moderate antitumor activity (Fig. 4, D to K, and fig. S39A). The p53-mRNA NPs demonstrated a potent effect on suppressing the growth of Hep3B tumors. Cotreatment with everolimus and p53-mRNA NPs greatly enhanced the therapeutic efficacy compared to the treatment with everolimus alone or p53-mRNA NPs at the end point of this study. The CI value was 5.08, indicating a potent synergistic effect of everolimus in combination with p53-mRNA NPs in vivo. No obvious change in body weight was observed in any groups (fig. S39B). In addition, the combination treatment was highly effective in vivo in p53-null H1299 xenograft tumors (fig. S40). The CI value was 2.87 for the combination of everolimus with p53-mRNA NPs. The cotreatment even resulted in regression of the H1299 tumors. Moreover, the p53 restoration strategy also worked in the immunocompetent mouse tumor model of p53-null RIL-175, as evidenced by the inhibition of tumor growth after treatment with murine p53-mRNA NPs (figs. S41 and S42).
To better understand the in vivo mechanisms underlying this antitumor effect, we tested p53 expression in p53-null Hep3B tumor sections obtained at different time points (12, 24, 48, and 60 hours) after three injections of p53-mRNA NPs by IF analysis (PBS treatment was used as control). Figure 4L shows p53 protein expression in tumor sections at all these time points, and the signals were still clear at 60 hours after treatment. We also detected up-regulated signals of C-CAS3, indicating the apoptosis pathway activated by these p53-mRNA NPs. PBS control group did not show any signal of p53 or C-CAS3. Furthermore, we performed immunohistochemistry (IHC) analysis and confirmed the high expression of p53 in p53-null Hep3B tumor sections (Fig. 5A), along with the high expression of C-CAS3 after treatment with p53-mRNA NPs. These results indicated the activation of the apoptotic pathway, consistent with the in vitro results. We also observed that the restored p53 proteins were mainly located in the cytoplasm of Hep3B and H1299 cells in vivo (figs. S43 and S44). Tumor cell proliferation was assessed by Ki67 (proliferation marker) and PCNA (proliferating cell nuclear antigen) expression, both of which were decreased after treatment with p53-mRNA NPs. In addition, TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) assay in tumor sections (Fig. 5B) confirmed that p53-mRNA NP treatment activated the apoptosis pathway. Furthermore, p53 restoration–mediated sensitization to everolimus was examined in vivo. Proteins from Hep3B tumors in different treatment groups were extracted and analyzed by WB. As shown in Fig. 5C, everolimus induced autophagy, as indicated by the expression of LC3B-2 relative to actin (50), as well as the increase in Beclin 1 (BECN1), whereas the cotreatment with p53-mRNA NPs reduced autophagy activation to levels comparable to the control groups. Apoptosis (C-CAS9 and C-CAS3) was enhanced in the p53-mRNA NPs + everolimus group. We also analyzed the mTOR and autophagic pathways in a p53-null NSCLC xenograft model via IHC studies (fig. S45). The expression of major proteins (p53, TIGAR, LC3B, Ki67, and C-CAS3) involved in the pathways discussed above was verified in the H1299 tumor sections. Treatment with p53-mRNA NPs resulted in the expressions of p53 and TIGAR and inhibited the LC3B (autophagy marker) expression induced by everolimus. The down-regulation of Ki67 and up-regulation of C-CAS3 indicated activation of the apoptosis pathway.
Fig. 5. In vivo mechanisms underlying the p53-mRNA NP–mediated sensitization of p53-null HCC xenograft model to everolimus.
(A) Immunohistochemistry (IHC) images from tumor sections of Hep3B tumor–bearing xenograft mice before and after treatment with p53-mRNA NPs (mRNA dose, 750 μg/kg). The protein expressions of p53, apoptotic markers (BAX and C-CAS3), and proliferation markers (Ki67 and PCNA) were evaluated by IHC staining (blue: nucleus; brown: p53, BAX, C-CAS3, Ki67, or PCNA). Scale bars, 100 μm. (B) CLSM images of fixed tumor tissues with the TUNEL staining (blue: nucleus; red: apoptosis) from PBS, EGFP-mRNA NPs, p53-mRNA NPs, everolimus, and p53-mRNA NPs + everolimus groups. Scale bars, 100 μm. (C) WB analysis of p53, LC3B-1, LC3B-2, BECN1, p62, BCL-2, BAX, C-CAS9, C-CAS3, and p-4EBP1 in the Hep3B xenograft tumors after different treatments. Actin was used as the loading control.
In vivo therapeutic efficacy in p53-null orthotopic HCC model and disseminated NSCLC model
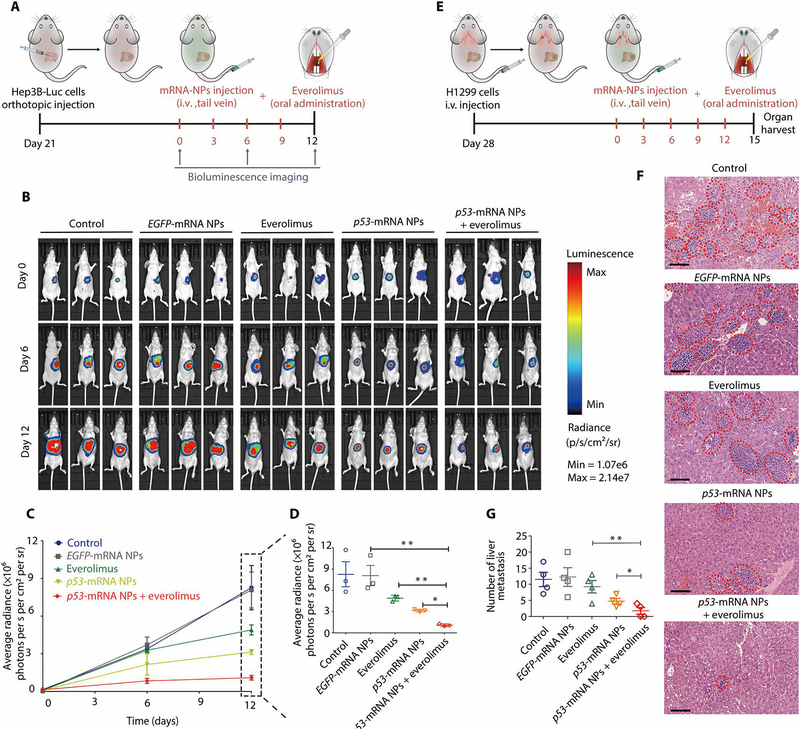
To further evaluate the therapeutic efficacy of p53-mRNA NPs in combination with everolimus, we established a p53-null orthotopic model of HCC by injecting luciferase-expressing Hep3B (Hep3B-Luc) cells into the left lobe of the livers of immunodeficient nude mice. Tumor growth was monitored by detecting the average radiance of the tumor sites through bioluminescence imaging. Twenty-one days later, mice were randomly divided into different groups and treated with PBS, EGFP-mRNA NPs, everolimus, p53-mRNA NPs, or p53-mRNA NPs + everolimus every 3 days (Fig. 6A). Everolimus was orally administered, whereas PBS and all NPs were given by intravenous injection. Bioluminescence imaging was performed on days 0, 6, and 12. As shown in Fig. 6B, everolimus somewhat inhibited the growth of orthotopic tumors, as compared to the PBS and EGFP-mRNA NP groups. p53-mRNA NPs effectively reduced the orthotopic tumor burden, and cotreatment with p53-mRNA NPs and everolimus showed the strongest therapeutic effect in the orthotopic model (Fig. 6C).
Fig. 6. Therapeutic efficacy in the p53-null orthotopic HCC tumors and the liver metastases of p53-null NSCLC.
(A) Scheme of tumor inoculation and different treatments in luciferase-expressing Hep3B (Hep3B-Luc) orthotopic tumor–bearing nude mice. Twenty-one days after tumor inoculation, mice were treated with PBS (intravenous), EGFP-mRNA NPs (intravenous), p53-mRNA NPs (intravenous), everolimus (oral), or p53-mRNA NPs (intravenous) + everolimus (oral) every 3 days for four rounds (mRNA dose, 750 μg/kg; everolimus dose, 5 mg/kg). (B) Bioluminescence images of the Hep3B-Luc orthotopic tumor–bearing nude mice at days 0, 6, and 12. (C) Average radiance [×106 photons per second (s) per cm2 per steradian (sr)] of tumor burden determined by bioluminescence imaging at different time points. (D) Average radiance of tumor burden at the endpoint (day 12). Data shown as means ± SEM (n = 3), and statistical significance was determined using two-tailed t test (*P < 0.05, **P < 0.01). (E) Scheme of tumor inoculation and different treatments in p53-null H1299 metastatic tumor–bearing nude mice. Twenty-eight days after tumor inoculation, mice were treated with PBS (intravenous), EGFP-mRNA NPs (intravenous), p53-mRNA NPs (intravenous), everolimus (oral), or p53-mRNA NPs (intravenous) + everolimus (oral) every 3 days for five rounds (mRNA dose, 750 μg/kg; everolimus dose, 5 mg/kg). Organs from different groups were harvested three days after the final treatment. (F) Histological examination of liver tissues from each group by H&E staining. The metastatic lesions (red dotted ovals) were identified as cell clusters with darkly stained nuclei. Scale bars, 100 μm. (G) The number of metastatic nodules in the liver from each group. One liver was randomly selected from each group with a blind method, and the liver section from each group was divided into four regions for counting of the metastasis nodules. Data shown as means ± SEM (n = 4 regions), and statistical significance was determined using two-tailed t test (*P < 0.05, **P < 0.01).
We also used an experimental liver metastasis model to evaluate this combination strategy by intravenous injection of the H1299 NSCLC cells into immunodeficient mice via the tail vein. Four weeks later, all the mice were randomly assigned to different groups and treated with PBS, EGFP-mRNA NPs, everolimus, p53-mRNA NPs, or p53-mRNA NPs + everolimus every 3 days (Fig. 6D). After five rounds of treatment, all mice were euthanized, and their livers were collected to detect metastases (Fig. 6, E to G). Numerous metastatic nodules were detected in the livers from the PBS and EGFP-mRNA NP groups, and everolimus showed moderate effects. In comparison, p53-mRNA NPs effectively reduced the number of metastatic nodules, whereas cotreatment with p53-mRNA NPs and everolimus showed the most profound therapeutic effect.
In vivo safety of p53-mRNA NPs and their combination with everolimus
To evaluate the in vivo safety of p53-mRNA NPs and their combination with everolimus, various organs (heart, kidneys, liver, lungs, and spleen) were harvested at the end point (day 33) of the Hep3B xenograft study, followed by section and hematoxylin and eosin (H&E) staining (fig. S46A). No obvious histological differences were detected in the sections of organs from all the treatment groups, indicating no notable toxicity. We also performed serum biochemistry analysis and whole blood panel tests. A series of parameters were tested (fig. S46B), including alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN), red blood cells (RBCs), white blood cells (WBCs), hemoglobin (Hb), mean corpuscular Hb concentration (MCHC), mean corpuscular Hb (MCH), hematocrit (HCT), and lymphocyte count (LY). These parameters did not show significant differences between the groups treated with PBS, p53-mRNA NPs, and p53-mRNA NPs + everolimus. Moreover, we further performed IHC analysis for the expressions of p53 and C-CAS3 in major organs (heart, liver, spleen, lungs, and kidneys) and tumors. As can be seen in fig. S47, p53 was mainly expressed in the tumor and liver, which is consistent with our BioD results (with the NP delivery platform, mRNA had higher accumulation in the tumor and liver). The restoration of p53 in p53-null HCC tumors resulted in effective expression of C-CAS3, consistent with in vitro studies. In addition, no obvious expression of C-CAS3 was observed in normal tissues including the liver, which is consistent with H&E staining results. Moreover, blood serum concentrations of immunotoxicity markers such as interferon-γ (IFN-γ), tumor necrosis–α (TNF-α), interleukin-12 (IL-12), and IL-6 were in the normal range at 24 hours after treatment with either empty NPs or p53-mRNA NPs (fig. S48). These results indicated that no observable innate immune responses were caused by the mRNA NPs at the tested time point.
DISCUSSION
The p53 gene is a critical tumor suppressor gene involved in the majority of cancers (59, 60). The clinical data from TCGA show that both HCC and NSCLC patients with high expression of p53 have much longer overall survival and/or progression-free survival than those with low p53 expression (61, 62). With its diverse functions (such as regulation of cell cycle checkpoints, apoptosis, senescence, and DNA repair), p53 restoration has long been considered an attractive anticancer strategy (63–65). Various methods have been developed to reactivate p53 functions, which can be summarized in the two categories of small molecular compounds (25–27) and DNA therapeutics (29, 30). Small molecular inhibitors, such as RITA (reactivation of p53 and induction of tumor cell apoptosis), Nutlin, and MI-319, have shown high binding potency and selectivity for MDM2 in the treatment of HCC and other cancers (66–68). Other small molecules like CP-31398 have also been developed to target mutant p53 and reactivate its normal functions (69, 70). Encouraging clinical outcomes are being continually generated with compounds such as RG7112, MI-773, and APR-246 in different cancers. For example, the phase 1 trial of RG7112 (an MDM2 antagonist) has demonstrated clinical responses in hematologic malignancies (71). MI-773 (SAR405838; an HDM2 antagonist) was shown to be safe with preliminary antitumor activity in locally advanced or metastatic solid tumors (72). In addition, combination treatment with APR-246 and azacitidine resulted in responses in all patients with TP53-mutant myelodysplastic syndromes and acute myeloid leukemia in a phase 1b/2 study (73). Despite these efforts and the progress in clinical trials (32), this method is likely to be ineffective when the suppressor gene has been deleted. For DNA therapeutics, several candidates using adenoviral vectors are in clinical trials, with Gendicine approved in China in 2003 (74). Advexin, another Adp53 vector, however, failed in the phase 3 trials (75). Considering the low transduction rate of p53 gene via Adp53 (76), some tumor-specific, replication-competent CRAdp53 vectors (AdDelta24-p53, SG600-p53, ONYX 015, OBP-702, and H101) have been developed to induce higher p53 expression and antitumor effect. SGT-53, a cationic liposome encapsulating p53 plasmid, is also in clinical trials for solid tumors (31). Although Gendicine and H101 have been approved for head and neck cancers in China (76), they are not widely used, presumably due to the limitations of intratumoral injection. Furthermore, gene therapy for systemic cancer treatment still has several potential risks, including (i) host immune responses and preexisting antiviral immunity resulting in the neutralization of efficacy, modification of PK and pharmacodynamics, and allergic responses; and (ii) potential genotoxicity owing to integration in the host genome (33).
The use of synthetic mRNA has recently attracted considerable attention owing to its distinctive features. For example, it does not require nuclear entry for transfection activity and has a negligible chance of integrating into the host genome, thus avoiding potentially detrimental genotoxicity (34, 35). Chemical modification of mRNA molecules has also enhanced their stability and decreased activation of innate immune responses (37). Whereas the use of mRNA to restore tumor suppressors seems straightforward and highly promising, effective systemic delivery of mRNA to tumors remains a major challenge. Nanotechnology has shown promise to improve cytosolic delivery of various RNA therapeutics into tumor cells (77, 78), and different NP systems have been developed for systemic mRNA delivery (79–81), particularly to the liver for genetic and infectious diseases (82–88). However, little efforts have been reported on systemic delivery of mRNA for restoration of tumor suppressors.
We had developed a lipid-polymer hybrid mRNA NP platform composed of poly(lactic-co-glycolic acid) (PLGA) and successfully applied it for in vivo restoration of tumor suppressor phosphatase and tensin homolog deleted on chromosome 10 (PTEN) in prostate cancer (40). Considering the fact that the concentration of reductive agent GSH in tumor cells could be about 100- to 1000-fold higher than that in the extracellular fluids (89), redox-responsive NP platforms have emerged for effective intracellular delivery (41–47), which may be particularly beneficial for biomacromolecules that need to be released into the cytoplasm for therapeutic effects. In this work, we incorporated redox-responsive polymer PDSA in our hybrid NP platform, which showed a fast mRNA release in the presence of reductive agent DTT and resulted in excellent mRNA transfection. In addition, the reduced EGFP protein expression after the quenching of intracellular GSH by Nem also suggested that redox-responsive NPs might be more potent for mRNA delivery than nonresponsive NPs. In addition to the polymer core, the surface lipid-PEG layer also plays an important role in controlling the performance (cellular uptake and PK) of the hybrid NPs for delivery of RNA therapeutics by serum albumin–mediated de-PEGylation (38, 39). For instance, DSPE-PEG contributes to a long circulation life and high tumor circulation due to its slow dissociation from NPs, whereas DMPE-PEG contributes to a high cellular uptake and excellent in vitro performance of the hybrid NPs due to its quick de-PEGylation kinetics. Here, we further blended the two lipid-PEG molecules by changing the DSPE-PEG/DMPE-PEG ratio for different in vitro or in vivo applications. To maximize the tumor accumulation, the lipid-PEG layer of NPs needs to be relatively stable (with a slow de-PEGylation kinetic profile) to enable a relatively long circulation time. Therefore, a high ratio of DSPE-PEG (75%, w/w) to the total lipid-PEGs on the surface layer was designed for systemic delivery of mRNA. Compared with the PLGA-based NPs coated with a layer of single lipid-PEG (40), the PDSA-based NPs coated with a layer of hybrid lipid-PEGs are more adjustable for on-demand applications.
Previous studies (11–13) have shown that activation of autophagy by mTOR inhibitors including everolimus may be an undesired effect because it acts as a resistance mechanism that limits drug efficacy. The incorporation of autophagy inhibitors could prevent resistance to mTOR inhibitors and enhance their therapeutic efficacy. For example, a dual mTORC1 and mTORC2 inhibitor, OSI-027, was reported to induce protective autophagy, whereas disruption of this pathway with chloroquine (autophagy inhibitor) contributed to apoptotic cell death (90). Both selective knockdown of autophagy genes (ATG3, ATG5, and ATG7) and pretreatment with hydroxychloroquine (autophagy inhibitor) also contributed to activating the mitochondrial apoptotic pathway and improving everolimus activity, sensitizing mantle cell lymphoma to everolimus (10). p53 plays a dual role in control of autophagy: (i) nuclear p53 can induce autophagy through transcriptional effects, whereas (ii) cytoplasmic p53 can act as a master repressor of autophagy (57, 91). In this work, we observed that the p53 proteins restored by mRNA NPs are mainly located in the cytoplasm of both Hep3B and H1299 cells in vitro and in vivo. In addition, we observed that everolimus-induced autophagy activation was effectively inhibited by mRNA NP–based restoration of p53, further demonstrating the expression of p53 proteins mainly in the cytoplasm.
In summary, we demonstrate that p53 restoration by synthetic mRNA NPs can inhibit autophagy, thus providing a strategy for sensitizing p53-null tumor cells to everolimus, and simultaneously activate apoptosis and cell cycle arrest. The redox-responsive p53-mRNA NPs enhanced the therapeutic responses to everolimus in p53-null HCC and NSCLC in vitro and in vivo. A synergistic antitumor effect was also observed in multiple animal models of both HCC and NSCLC with the combinatorial treatment, which might be explained by (i) the mild therapeutic effect of everolimus, (ii) cytoplasmic p53-mediated inhibition of autophagy and sensitization to the mTOR inhibitor, and (iii) the simultaneous activation of apoptosis by p53 restoration. The synthetic mRNA NP–based p53 restoration strategy might therefore revive this U.S. Food and Drug Administration–approved mTOR inhibitor for clinical translation in p53-deficient HCC and NSCLC patients. Note that this study is limited to HCC and NSCLC cell lines with p53 deficiency. More efforts are needed to answer whether this combination strategy may work for other p53-deficient cancers or for cancers with different p53 mutations. In addition, the current formulation method might not be amenable for large-scale synthesis of mRNA NPs. For further preclinical development and evaluation of translational potential of this mRNA NP system, the scalability of NP formulation and the systematic optimization of NP physicochemical properties should be considered. Compared to the bulk synthesis approach used herein, microfluidic-based strategies have exhibited the capability for more controllable, homogeneous, and reproducible formulation of NPs, as well as the feasibility of mass production (92–94). It is thus expected that the use of microfluidic platforms might facilitate the identification of optimal mRNA NP formulations suitable for preclinical pharmacology and toxicology evaluation in large animals. To maximize the therapeutic effect of this strategy, the heterogeneity and complexity of tumors should also be considered. Careful identification of what cancer type(s) may benefit most from such combination treatment is still needed, as well as the selection of tumors with high enhanced permeability and retention effect to receive mRNA NPs (77, 95). We expect that this mRNA NP approach could be applied to many other tumor suppressors and rationally combined with other therapeutic modalities for effective combinatorial cancer treatment.
MATERIALS AND METHODS
Study design
This study aimed to explore an mRNA-based strategy for restoring tumor suppressor p53 in p53-null HCC and NSCLC cells and to evaluate whether p53 reactivation would sensitize these tumor cells to mTOR inhibition for more effective combination treatment. We addressed this objective by (i) developing a redox-responsive p53-mRNA NP platform that showed the feasibility of p53 restoration in p53-deficient Hep3B and H1299 cells, (ii) demonstrating antitumor effects of the p53-mRNA NPs that can induce cell apoptosis and G1-phase cell cycle arrest, and (iii) revealing that p53 reactivation can sensitize tumor cells to mTOR inhibitor everolimus. The therapeutic efficacy and safety of the combination of p53-mRNA NPs with everolimus were thoroughly evaluated in vivo.
Four animal models, including xenograft models of p53-null HCC and NSCLC, orthotopic model of p53-null HCC, and disseminated model of p53-null NSCLC, were used to evaluate antitumor effects of this combinatorial strategy. The animals were randomly assigned to the study groups. The experimentalists were not blinded during the study.
Animals
All the in vivo studies were conducted following the animal protocols approved by the Institutional Animal Care and Use Committees on animal care (Brigham and Women’s Hospital and Hangzhou Normal University). The animal studies were performed under strict regulations and pathogen-free conditions in the animal facilities of Brigham and Women’s Hospital or Hangzhou Normal University. Female athymic nude mice (4 to 6 weeks old), wild-type BALB/c mice (6 weeks old), and female C57BL/6 mice (4 weeks old) were purchased from Charles River Laboratories or Zhejiang Medical Academy Animal Center. Mice were raised for at least 1 week before the start of the experiments to acclimatize them to the environment and food of the animal facilities.
PK and BioD studies
For the in vivo PK study, healthy BALB/c mice (6 weeks old, n = 3 per group) were intravenously injected with naked Cy5-mRNA, Cy5-mRNA NP25, Cy5-mRNA NP50, or Cy5-mRNA NP75 via tail vein. At predetermined time intervals (0, 0.5, 1, 2, 4, 8, 12, and 24 hours), retro-orbital vein blood was obtained in a heparin-coated capillary tube. The wound was gently pressed for 1 min to stop the bleeding. Fluorescence intensity of Cy5-mRNA was measured by a microplate reader. PK was assessed by measuring the percentage of Cy5-mRNA in blood at these time points after getting rid of the background and normalization to the initial time point (0 hours). For the BioD study, p53-null Hep3B xenograft–bearing athymic nude mice were intravenously injected with naked Cy5-mRNA, Cy5-mRNA NP25, Cy5-mRNA NP50, or Cy5-mRNA NP75 (at an mRNA dose of 750 μg/kg of animal weight) via tail vein (n = 3 per group). After 24 hours, all the mice were euthanized, and the dissected organs and tumors were visualized using a Syngene PXi imaging system (Synoptics Ltd.).
In vivo therapeutic efficacy in p53-null
HCC xenograft tumor model
To establish the HCC xenograft tumor model, ~1 × 107 p53-null Hep3B liver cancer cells in 100 μl of PBS mixed with 100 μl of Matrigel (BD Biosciences) were implanted subcutaneously on the right flank (near the liver) of female athymic nude mice. Mice were monitored for tumor growth every other day according to the animal protocol. When the tumor volume reached ~100 mm3, the mice were randomly divided into five groups (n = 5), which received treatment with PBS, EGFP-mRNA NPs, everolimus, p53-mRNA NPs, or p53-mRNA NPs + everolimus. The mRNA NPs used for the in vivo therapeutic studies had 75% (w/w) of DSPE-PEG in the lipid-PEG layer. The human p53-mRNA sequence is shown in table S2. The EGFP-mRNA NPs or p53-mRNA NPs were injected via tail vein at an mRNA dose of 750 μg/kg, whereas the everolimus was orally administered at 5 mg/kg every 3 days for six rounds of treatment. The day that first treatment was performed was designated as day 0. Tumor size was measured using a caliper every 3 days from day 0 to day 33, and the average tumor volume (mm3) was calculated as: 4π/3 × (tumor length/2) × (tumor width/2)2. Relative tumor volume (%) was calculated and presented according to a reported method (96). The largest tumor volume from the mouse at the end of this study was defined as 100%. The body weights of all the mice were also recorded over this period.
In vivo therapeutic efficacy in p53-null
NSCLC xenograft tumor model
To establish the xenograft tumor mouse model, ~5 × 106 H1299 lung cancer cells in 100 μl of PBS mixed with 100 μl of Matrigel (BD Biosciences) were implanted subcutaneously on the left fore (near the lung) of female athymic nude mice. Mice were monitored for tumor growth every other day according to the animal protocol. When the tumor volume reached ~100 mm3, the mice were randomly divided into five groups (n = 5), which received treatment with PBS, EGFP-mRNA NPs, everolimus, p53-mRNA NPs, or p53-mRNA NPs together with everolimus. The engineered mRNA NPs used for the in vivo therapeutic studies have 75% (w/w) of DSPE-PEG in the lipid-PEG layer. The EGFP-mRNA NPs or p53-mRNA NPs were injected via tail vein at an mRNA dose of 750 μg/kg, whereas the everolimus was orally administered at 5 mg/kg every 3 days for six treatments. The day that the first treatment was performed was designated as day 0. Tumor size was measured using a caliper every 3 days from day 0 to day 18, and the average tumor volume (mm3) was calculated as: 4π/3 × (tumor length/2) × (tumor width/2)2. Relative tumor volume (%) was calculated and presented according to a reported method (96). The largest tumor volume from the mouse at the end of this study was defined as 100%.
In vivo therapeutic efficacy of murine p53-mRNA NPs in immunocompetent mice
To establish the immunocompetent mouse tumor model, ~1 × 106 of p53-null RIL-175 mouse HCC cells in 100 μl of PBS mixed with 100 μl of Matrigel (BD Biosciences) were implanted subcutaneously on the right flank (near the liver) of female C57BL/6 mice. Mice were monitored for tumor growth every other day according to the animal protocol. When the tumor volume reached ~100 mm3, the mice were randomly divided into three groups (n = 5), which received treatment with PBS, EGFP-mRNA NPs, or murine p53-mRNA NPs. The mRNA NPs used for the in vivo therapeutic studies had 75% (w/w) of DSPE-PEG in the lipid-PEG layer. The mouse p53-mRNA sequence is shown in table S2. The EGFP-mRNA NPs or murine p53-mRNA NPs were intravenously injected via tail vein at an mRNA dose of 750 μg/kg, every 3 days for six rounds of treatment. The day that first treatment was performed was designated as day 0. Tumor size was measured using a caliper every 3 days from day 0 to day 18, and the average tumor volume (mm3) was calculated as: 4π/3 × (tumor length/2) × (tumor width/2)2. Relative tumor volume (%) was calculated and presented according to a reported method (96).
In vivo mechanisms underlying the p53-mRNA NP–mediated sensitization to everolimus
To verify the in vivo mechanisms underlying this p53-mRNA NP–mediated strategy, mice bearing p53-null Hep3B liver xenografts were treated with p53-mRNA NPs via tail vein injection at an mRNA dose of 750 μg/kg every 3 days for three rounds of treatment. The mice were euthanized at 12, 24, 48, or 60 hours after the last injection of p53-mRNA NPs, and the tumors were harvested for sections. Mice bearing p53-null Hep3B liver xenografts and intravenously injected with PBS were used as controls and euthanized at 60 hours after the last injection. The expression of p53 and C-CAS3 was monitored via IF detection. Moreover, tumor sections from both the PBS group and p53-mRNA NP group (60 hours after the last injection) were analyzed by IHC. The expression of p53, tumor cell apoptosis markers (BAX and C-CAS3), and proliferation markers (Ki67 and PCNA) was further assessed. In addition, tumors obtained from all the groups (control, EGFP-mRNA NPs, everolimus, p53-mRNA NPs, or p53-mRNA NPs + everolimus) in the above-mentioned therapeutic study using the p53-null Hep3B liver xenograft model were further sectioned for a TUNEL apoptosis assay and lysed for WB studies to detect the expression of p53, LC3B-2, BECN1, p62, p-4EBP1, C-CAS9, and C-CAS3.
In vivo therapeutic efficacy in p53-null orthotopic HCC model
To establish the orthotopic HCC model, Hep3B-Luc cells were used. Six-week-old female athymic nude mice were obtained from Zhejiang Medical Academy Animal Center. Animal studies were conducted following the protocol approved by the Institutional Animal Ethics Committee of Hangzhou Normal University. First, anterior abdominal exposure was made, and a cotton swab with iodine volts was used to sterilize this area. A 1-cm-long midline incision was made along the anterior abdominal wall below the xiphoid after anesthesia by isoflurane, and ~5 × 106 p53-null Hep3B-Luc cells in 50 μl of PBS were injected into the left lobe of the livers of the athymic nude mice (30 in total). The injection depth was not deeper than 2 mm. The inner and outer layers of the abdominal cavity were sutured one by one after tumor cell inoculation. Three weeks later, 15 mice (incidence rate of orthotopic HCC model, 50%) were randomly assigned to five groups (n = 3 per group), which received treatment with PBS, EGFP-mRNA NPs, everolimus, p53-mRNA NPs, or p53-mRNA NPs together with everolimus. The EGFP-mRNA NPs or p53-mRNA NPs were injected via tail vein at an mRNA dose of 750 μg/kg, whereas everolimus was orally administered at 5 mg/kg every 3 days for four rounds of treatment. The first treatment was performed at day 0. On day 12, all the mice were euthanized. Mice were monitored for tumor growth by bioluminescent in vivo imaging every 6 days (days 0, 6, and 12). To do this, these mice were injected intraperitoneally with D-luciferin substrate (150 mg/kg; PerkinElmer, catalog no. 122799) and imaged by an IVIS Lumina S5 (PerkinElmer) imaging system.
In vivo therapeutic efficacy in p53-null disseminated NSCLC model
To establish the experimental disseminated metastatic model, ~1 × 106 p53-null H1299 cells in 100 μl of PBS were injected via tail vein into female athymic nude mice. Four weeks after the intravenous injection of tumor cells, mice were randomly divided into five groups (n = 5), which received treatment with PBS, EGFP-mRNA NPs, everolimus, p53-mRNA NPs, or p53-mRNA NPs together with everolimus. The EGFP-mRNA NPs or p53-mRNA NPs were injected via tail vein at an mRNA dose of 750 μg/kg, whereas everolimus was orally administered at 5 mg/kg every 3 days for five rounds of treatment. The first treatment was performed at day 0. On day 15, all the mice were euthanized, and one liver was randomly selected from each group for H&E staining. The liver section from each group was divided into four regions for calculation of the metastasis numbers (fig. S49).
Immune response detection by the enzyme-linked immunosorbent assay
Female BALB/c mice (6 weeks old, n = 3 per group) were intravenously injected with PBS, empty NPs, or p53-mRNA NPs (750 μg mRNA/kg). Serum samples were collected after 24 hours of treatment. Representative cytokines (TNF-α, IFN-γ, IL-6, and IL-12) were detected by enzyme-linked immunosorbent assay (PBL Biomedical Laboratories and BD Biosciences) according to the manufacturers’ instructions.
In vivo toxicity evaluation
To evaluate in vivo toxicity, major organs were harvested at the end point of different tumor models (p53-null Hep3B liver xenograft tumor model and liver metastases of p53-null H1299 lung tumor model), followed by section and H&E staining to evaluate the histological differences. In addition, blood was drawn retro-orbitally, and serum was isolated from p53-null Hep3B liver xenograft tumor model at the end of the efficacy experiment. Various parameters including ALT, AST, BUN, RBCs, WBCs, Hb, MCHC, MCH, HCT, and LY were tested to assess for toxicity.
Statistical analysis
Statistical analysis was carried out by GraphPad Prism 7 software to perform two-tailed t test or one-way analysis of variance (ANOVA). All studies were performed at least in triplicate unless otherwise stated. Error bars indicate SEM. A value of P < 0.05 is considered statistically significant, where all statistically significant values shown in the figures are indicated as: *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Fig. S1. Study summary.
Fig. S2. The structure schematic of synthetic mRNA.
Fig. S3. The chemical structure of 3′-O-Me-m7G(5′)ppp(5′)G ARCA cap.
Fig. S4. Chemicals for NP synthesis.
Fig. S5. Characterization of the engineered hybrid mRNA NPs.
Fig. S6. Size of EGFP-mRNA NPs and Luc-mRNA NPs with various formulations.
Fig. S7. Encapsulation efficiency of EGFP-mRNA NPs and Luc-mRNA NPs with various formulations.
Fig. S8. Normalized luminescence intensity of Hep3B cells after treatment with various Luc-mRNA NP formulations at the mRNA dose of 0.830 μg/ml.
Fig. S9. Endosomal escape of mRNA NPs.
Fig. S10. Transfection efficacy verified by CLSM imaging.
Fig. S11. Transfection efficacy verified by flow cytometry.
Fig. S12. Transfection efficacy after quenching intracellular GSH.
Fig. S13. In vitro toxicity of the synthetic EGFP-mRNA NPs.
Fig. S14. IF staining of p53 in p53-null H1299 cells.
Fig. S15. WB analysis of p53 protein expression.
Fig. S16. In vitro therapeutic efficacy of the synthetic p53-mRNA NPs in p53-null H1299 cells.
Fig. S17. Apoptosis of p53-null H1299 cells as determined by flow cytometry after different treatments.
Fig. S18. G1-phase cell cycle arrest induced by p53-mRNA NPs.
Fig. S19. WB analysis of apoptotic signaling pathway in p53-null H1299 cells after different treatments.
Fig. S20. TEM images of mitochondrial morphology in p53-null H1299 cells after different treatments.
Fig. S21. In vitro toxicity of the mutant p53-R175H-mRNA NPs.
Fig. S22. Cytotoxicity of everolimus in p53-null H1299 cells.
Fig. S23. Effect of everolimus on autophagy activation in p53-null H1299 cells.
Fig. S24. WB analysis of autophagy and apoptotic signaling pathways in p53-null H1299 cells.
Fig. S25. Analysis of the autophagosomes and swollen mitochondria in p53-null H1299 cells after different treatments.
Fig. S26. In vitro therapeutic efficacy of the combination of p53-mRNA NPs with everolimus in p53-null H1299 cells.
Fig. S27. In vitro apoptosis of p53-null H1299 cells after different treatments.
Fig. S28. In vitro toxicity of the combination of everolimus with venetoclax.
Fig. S29. In vitro toxicity of the combination of everolimus with siBcl-2.
Fig. S30. The relative mRNA expression of p53.
Fig. S31. The relative mRNA expression of ULK1, ATG7, BECN1, and ATG12.
Fig. S32. The relative mRNA expression of DRAM1, ISG20L1, and SESN1.
Fig. S33. The relative mRNA expression of TIGAR.
Fig. S34. WB analysis of AMPK and TIGAR pathways.
Fig. S35. Schematic representation of the possible mechanism by which p53 tumor suppressor inhibits protective autophagy and sensitizes tumor cells to everolimus.
Fig. S36. BioD of different mRNA NPs in HCC xenograft tumor model.
Fig. S37. BioD of different mRNA NPs in NSCLC xenograft tumor model.
Fig. S38. Blood vessel staining in tumor sections.
Fig. S39. Efficacy and safety of different treatments in HCC xenograft model.
Fig. S40. Antitumor effects of p53-mRNA NPs are synergistic with everolimus in NSCLC xenograft model.
Fig. S41. Murine p53 restoration in p53-null murine liver cancer RIL-175 cells.
Fig. S42. Therapeutic efficacy of murine p53-mRNA NPs in immunocompetent mice bearing p53-null RIL-175 tumors.
Fig. S43. Expression of p53 protein in HCC xenograft model after treatment with p53-mRNA NPs.
Fig. S44. Expression of p53 protein in NSCLC xenograft model after treatment with p53-mRNA NPs.
Fig. S45. IHC images from tumor sections of H1299 tumor–bearing mice before and after treatment with p53-mRNA NPs.
Fig. S46. In vivo toxicity of the p53-mRNA NP–mediated strategy for everolimus rescue assessed by histopathological and hematological analysis.
Fig. S47. IHC images from major organs and tumor sections of the HCC xenograft model.
Fig. S48. Evaluation of immune responses after the treatment with mRNA NPs.
Fig. S49. Scans of the liver metastases from different treatment groups in Fig. 6.
Table S1. Compositions of different NP formulations.
Table S2. Different p53-mRNA sequences used in this study.
Table S3. Primer sequences for qRT-PCR.
Acknowledgments
We thank Dana-Farber/Harvard Cancer Center (P30 CA06516) for the use of the Rodent Histopathology Core for H&E staining service and Harvard Medical School Neurobiology Imaging Facility (P30 NS072030) for imaging and analysis service.
Funding: This work was, in part, supported by the U.S. NIH grant R01 CA200900 (to J.S.), Key Project of Zhejiang province Ministry of Science and Technology no. 2015C03055 (to T.X.), and Key Project of Hangzhou Ministry of Science and Technology no. 20162013A07 (to T.X.). J.S. is a recipient of the Lung Cancer Discovery Award from the American Lung Association. W.T. is a recipient of the U.S. METAvivor Early Career Investigator Award (no. 2018A020560). N.Y.K. acknowledges the support of NEU CaNCURE program (NCI R25CA174650). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Competing interests: N.K., W.T., O.C.F., and J.S. are inventors on a U.S. patent application (62778215) entitled “Devices and methods for treatment of tumor suppressor-deficient cancers” and filed by Brigham and Women’s Hospital related to the technology disclosed herein. O.C.F. declares financial interests in Selecta Biosciences, Tarveda Therapeutics, and Seer. D.G.D. received honoraria from Bristol-Myers Squibb; consulting fees from Bayer, Tilos, and twoXAR; and research funding from Merrimack, Leap Therapeutics, Bayer, Bristol-Myers Squibb, and Exelixis. No reagents or funding from these companies were used in the study reported here. All other authors declare that they have no competing interests.
Footnotes
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. All reasonable requests for collaboration involving materials used in the research will be fulfilled provided that a written agreement is executed in advance between Brigham and Women’s Hospital and the requester.
SUPPLEMENTARY MATERIALS
stm.sciencemag.org/cgi/content/full/11/523/eaaw1565/DC1
Materials and Methods
View/request a protocol for this paper from Bio-protocol.
REFERENCES AND NOTES
- 1.Laplante M, Sabatini DM, mTOR signaling in growth control and disease. Cell 149, 274–293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dancey J, mTOR signaling and drug development in cancer. Nat. Rev. Clin. Oncol 7, 209–219 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Yoshizawa A, Fukuoka J, Shimizu S, Shilo K, Franks TJ, Hewitt SM, Fujii T, Cordon-Cardo C, Jen J, Travis WD, Overexpression of phospho-eIF4E is associated with survival through AKT pathway in non–small cell lung cancer. Clin. Cancer Res 16, 240–248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, Tovar V, Roayaie S, Minguez B, Sole M, Battiston C, van Laarhoven S, Fiel MI, Di Feo A, Hoshida Y, Yea S, Toffanin S, Ramos A, Martignetti JA, Mazzaferro V, Bruix J, Waxman S, Schwartz M, Meyerson M, Friedman SL, Llovet JM, Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology 135, 1972–1983.e11 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu AX, Kudo M, Assenat E, Cattan S, Kang Y-K, Lim HY, Poon RTP, Blanc J-F, Vogel A, Chen C-L, Dorval E, Peck-Radosavljevic M, Santoro A, Daniele B, Furuse J, Jappe A, Perraud K, Anak O, Sellami DB, Chen L-T, Effect of everolimus on survival in advanced hepatocellular carcinoma after failure of sorafenib: The EVOLVE-1 randomized clinical trial. JAMA 312, 57–67 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Besse B, Leighl N, Bennouna J, Papadimitrakopoulou VA, Blais N, Traynor AM, Soria J-C, Gogov S, Miller N, Jehl V, Johnson BE, Phase II study of everolimus–erlotinib in previously treated patients with advanced non-small-cell lung cancer. Ann. Oncol 25, 409–415 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Tarhini A, Kotsakis A, Gooding W, Shuai Y, Petro D, Friedland D, Belani CP, Dacic S, Argiris A, Phase II study of everolimus (RAD001) in previously treated small cell lung cancer. Clin. Cancer Res 16, 5900–5907 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Ohtsu A, Ajani JA, Bai Y-X, Bang Y-J, Chung H-C, Pan H-M, Sahmoud T, Shen L, Yeh K-H, Chin K, Muro K, Kim YH, Ferry D, Tebbutt NC, Al-Batran S-E, Smith H, Costantini C, Rizvi S, Lebwohl D, Van Cutsem E, Everolimus for previously treated advanced gastric cancer: Results of the randomized, double-blind, phase III GRANITE-1 study. J. Clin. Oncol 31, 3935–3943 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Reilly T, McSheehy PMJ, Biomarker development for the clinical activity of the mTOR inhibitor everolimus (RAD001): Processes, limitations, and further proposals. Transl. Oncol 3, 65–79 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carew JS, Kelly KR, Nawrocki ST, Mechanisms of mTOR inhibitor resistance in cancer therapy. Target. Oncol 6, 17–27 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Rosich L, Xargay-Torrent S, López-Guerra M, Campo E, Colomer D, Roué G, Counteracting autophagy overcomes resistance to everolimus in mantle cell lymphoma. Clin. Cancer Res 18, 5278–5289 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Jiang Q, Zhu P, Chen Y, Xie X, Du Z, Jiang L, Tang W, NPRL2 enhances autophagy and the resistance to Everolimus in castration-resistant prostate cancer. Prostate 79, 44–53 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Lee SC, Kim KH, Kim OH, Lee SK, Hong HE, Choi BJ, Jeong W, Kim SJ, Everolimus plus Ku0063794 regimen promotes anticancer effects against hepatocellular carcinoma cells through the paradoxical inhibition of autophagy. Cancer Res. Treat 50, 1023–1038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marinov M, Ziogas A, Pardo OE, Tan LT, Dhillon T, Mauri FA, Lane HA, Lemoine NR, Zangemeister-Wittke U, Seckl MJ, Arcaro A, AKT/mTOR pathway activation and BCL-2 family proteins modulate the sensitivity of human small cell lung cancer cells to RAD001. Clin. Cancer Res 15, 1277–1287 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Lessene G, Czabotar PE, Colman PM, BCL-2 family antagonists for cancer therapy. Nat. Rev. Drug Discov 7, 989–1000 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Wagner KU, Claudio E, Rucker III EB, Riedlinger G, Broussard C, Schwartzberg PL, Siebenlist U, Hennighausen L, Conditional deletion of the Bcl-x gene from erythroid cells results in hemolytic anemia and profound splenomegaly. Development 127, 4949–4958 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Kimura T, Takabatake Y, Takahashi A, Isaka Y, Chloroquine in Cancer Therapy: A Double-Edged Sword of Autophagy. Cancer Res. 73, 3–7 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW, Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan KD, Galbraith MD, Andrysik Z, Espinosa JM, Mechanisms of transcriptional regulation by p53. Cell Death Differ. 25, 133–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W, Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho CC, Siu WY, Lau A, Chan WM, Arooz T, Poon RYC, Stalled replication induces p53 accumulation through distinct mechanisms from DNA damage checkpoint pathways. Cancer Res. 66, 2233–2241 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, Nannmark U, Samara C, Pinton P, Vicencio JM, Carnuccio R, Moll UM, Madeo F, Paterlini-Brechot P, Rizzuto R, Szabadkai G, Pierron G, Blomgren K, Tavernarakis N, Codogno P, Cecconi F, Kroemer G, Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol 10, 676–687 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q, Wang X, He C, Pan H, Autophagy and chemotherapy resistance: A promising therapeutic target for cancer treatment. Cell Death Dis. 4, e838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bykov VJN, Issaeva N, Zache N, Shilov A, Hultcrantz M, Bergman J, Selivanova G, Wiman KG, Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J. Biol. Chem 292, 19607 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller PA, Vousden KH, Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 25, 304–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bykov VJN, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G, Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med 8, 282–288 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA, In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004). [DOI] [PubMed] [Google Scholar]
- 29.van Beusechem VW, van den Doel PB, Grill J, Pinedo HM, Gerritsen WR, Conditionally replicative adenovirus expressing p53 exhibits enhanced oncolytic potency. Cancer Res. 62, 6165–6175 (2002). [PubMed] [Google Scholar]
- 30.Guan YS, Liu Y, Zhou XP, Li X, He Q, Sun L, p53 gene (Gendicine) and embolisation overcame recurrent hepatocellular carcinoma. Gut 54, 1318–1319 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Pirollo KF, Nemunaitis J, Leung PK, Nunan R, Adams J, Chang EH, Safety and efficacy in advanced solid tumors of a targeted nanocomplex carrying the p53 gene used in combination with docetaxel: A phase 1b study. Mol. Ther 24, 1697–1706 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheok CF, Verma CS, Baselga J, Lane DP, Translating p53 into the clinic. Nat. Rev. Clin. Oncol 8, 25–37 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Kapoor R, Klueter T, Wilson JM, Challenges in the gene therapy commercial ecosystem. Nat. Biotechnol 35, 813–815 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Van Tendeloo VF, Ponsaerts P, Berneman ZN, mRNA-based gene transfer as a tool for gene and cell therapy. Curr. Opin. Mol. Ther 9, 423–431 (2007). [PubMed] [Google Scholar]
- 35.Ulmer JB, Mason PW, Geall A, Mandl CW, RNA-based vaccines. Vaccine 30, 4414–4418 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Azimi SM, Sheridan SD, Ghannad-Rezaie M, Eimon PM, Yanik MF, Combinatorial programming of human neuronal progenitors using magnetically-guided stoichiometric mRNA delivery. eLife 7, e31922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kormann MS, Hasenpusch G, Aneja MK, Nica G, Flemmer AW, Herber-Jonat S, Huppmann M, Mays LE, Illenyi M, Schams A, Griese M, Bittmann I, Handgretinger R, Hartl D, Rosenecker J, Rudolph C, Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol 29, 154–157 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Zhu X, Xu Y, Solis LM, Tao W, Wang L, Behrens C, Xu X, Zhao L, Liu D, Wu J, Zhang N, Wistuba II, Farokhzad OC, Zetter BR, Shi J, Long-circulating siRNA nanoparticles for validating Prohibitin1-targeted non-small cell lung cancer treatment. Proc. Natl. Acad. Sci. U.S.A 112, 7779–7784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu X, Tao W, Liu D, Wu J, Guo Z, Ji X, Bharwani Z, Zhao L, Zhao X, Farokhzad OC, Shi J, Surface De-PEGylation controls nanoparticle-mediated siRNA delivery in vitroand in vivo. Theranostics 7, 1990–2002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Islam MA, Xu Y, Tao W, Ubellacker JM, Lim M, Aum D, Lee GY, Zhou K, Zope H, Yu M, Cao W, Oswald JT, Dinarvand M, Mahmoudi M, Langer R, Kantoff PW, Farokhzad OC, Zetter BR, Shi J, Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat. Biomed. Eng 2, 850–864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, Zhao L, Xu X, Bertrand N, Choi WII, Yameen B, Shi J, Shah V, Mulvale M, MacLean JL, Farokhzad OC, Hydrophobic cysteine poly(disulfide)-based redox-hypersensitive nanoparticle platform for cancer theranostics. Angew. Chem. Int. Ed 54, 9218–9223 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Ling X, Chen X, Riddell IA, Tao W, Wang J, Hollett G, Lippard SJ, Farokhzad OC, Shi J, Wu J, Glutathione-scavenging poly(disulfide amide) nanoparticles for the effective delivery of pt(IV) prodrugs and reversal of cisplatin resistance. Nano Lett. 18, 4618–4625 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu X, Wu J, Liu S, Saw PE, Tao W, Li Y, Krygsman L, Yegnasubramanian S, Marzo AMD, Shi J, Bieberich CJ, Farokhzad OC, redox-responsive nanoparticle-mediated systemic RNAi for effective cancer therapy. Small 14, e1802565 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Y, Aimetti AA, Langer R, Gu Z, Bioresponsive materials. Nat. Rev. Mater 2, 16075 (2016). [Google Scholar]
- 45.Zhao M, Biswas A, Hu B, Joo KI, Wang P, Gu Z, Tang Y, Redox-responsive nanocapsules for intracellular protein delivery. Biomaterials 32, 5223–5230 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Meng F, Hennink WE, Zhong Z, Reduction-sensitive polymers and bioconjugates for biomedical applications. Biomaterials 30, 2180–2198 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Cheng R, Feng F, Meng F, Deng C, Feijen J, Zhong Z, Glutathione-responsive nano-vehicles as a promising platform for targeted intracellular drug and gene delivery. J. Control. Release 152, 2–12 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Mancini M, Anderson BO, Caldwell E, Sedghinasab M, Paty PB, Hockenbery DM, Mitochondrial proliferation and paradoxical membrane depolarization during terminal differentiation and apoptosis in a human colon carcinoma cell line. J. Cell Biol 138, 449–469 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desagher S, Martinou J-C, Mitochondria as the central control point of apoptosis. Trends Cell Biol. 10, 369–377 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Chauhan S, Ahmed Z, Bradfute SB, Arko-Mensah J, Mandell MA, Won Choi S, Kimura T, Blanchet F, Waller A, Mudd MH, Jiang S, Sklar L, Timmins GS, Maphis N, Bhaskar K, Piguet V, Deretic V, Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. Nat. Commun 6, 8620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yokoyama Y, Dhanabal M, Griffioen AW, Sukhatme VP, Ramakrishnan S, Synergy between angiostatin and endostatin: Inhibition of ovarian cancer growth. Cancer Res. 60, 2190–2196 (2000). [PubMed] [Google Scholar]
- 52.Tao W, Ji X, Zhu X, Li L, Wang J, Zhang Y, Saw PE, Li W, Kong N, Islam MA, Gan T, Zeng X, Zhang H, Mahmoudi M, Tearney GJ, Farokhzad OC, Two-dimensional antimonene-based photonic nanomedicine for cancer theranostics. Adv. Mater 30, 1802061 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bensaad K, Tsuruta A, Selak MA, Vidal MNC, Nakano K, Bartrons R, Gottlieb E, Vousden KH, TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Xie J-M, Li B, Yu HP, Gao QG, Li W, Wu HR, Qin ZH, TIGAR has a dual role in cancer cell survival through regulating apoptosis and autophagy. Cancer Res. 74, 5127–5138 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Bensaad K, Cheung EC, Vousden KH, Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 28, 3015–3026 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sui X, Jin L, Huang X, Geng S, He C, Hu X, p53 signaling and autophagy in cancer: A revolutionary strategy could be developed for cancer treatment. Autophagy 7, 565–571 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Green DR, Kroemer G, Cytoplasmic functions of the tumour suppressor p53. Nature 458, 1127–1130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meley D, Bauvy C, Houben-Weerts JHPM, Dubbelhuis PF, Helmond MTJ, Codogno P, Meijer AJ, AMP-activated protein kinase and the regulation of autophagic proteolysis. J. Biol. Chem 281, 34870–34879 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J, Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet 47, 505–511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogelstein B, Lane D, Levine AJ, Surfing the p53 network. Nature 408, 307–310 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Cancer Genome Atlas Research, Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169, 1327–1341.e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cancer N Genome Atlas Research, Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T, Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Mandinova A, Lee SW, The p53 pathway as a target in cancer therapeutics: Obstacles and promise. Sci. Transl. Med 3, 64rv61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harajly M, Zalzali H, Nawaz Z, Ghayad SE, Ghamloush F, Basma H, Zainedin S, Rabeh W, Jabbour M, Tawil A, Badro DA, Evan GI, Saab R, p53 restoration in induction and maintenance of senescence: Differential effects in premalignant and malignant tumor cells. Mol. Cell. Biol 36, 438–451 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang S, Zhou L, Hong B, van den Heuvel APJ, Prabhu VV, Warfel NA, Kline CLB, Dicker DT, Kopelovich L, el-Deiry WS, Small-molecule NSC59984 restores p53 pathway signaling and antitumor effects against colorectal cancer via p73 activation and degradation of mutant p53. Cancer Res. 75, 3842–3852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panka DJ, Liu Q, Geissler AK, Mier JW, Effects of HDM2 antagonism on sunitinib resistance, p53 activation, SDF-1 induction, and tumor infiltration by CD11b+/Gr-1+ myeloid derived suppressor cells. Mol. Cancer 12, 17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Turnbull AP, Ioannidis S, Krajewski WW, Pinto-Fernandez A, Heride C, Martin ACL, Tonkin LM, Townsend EC, Buker SM, Lancia DR, Caravella JA, Toms AV, Charlton TM, Lahdenranta J, Wilker E, Follows BC, Evans NJ, Stead L, Alli C, Zarayskiy VV, Talbot AC, Buckmelter AJ, Wang M, McKinnon CL, Saab F, McGouran JF, Century H, Gersch M, Pittman MS, Marshall CG, Raynham TM, Simcox M, Stewart LMD, McLoughlin SB, Escobedo JA, Bair KW, Dinsmore CJ, Hammonds TR, Kim S, Urbé S, Clague MJ, Kessler BM, Komander D, Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature 550, 481–486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bykov VJN, Selivanova G, Wiman KG, Small molecules that reactivate mutant p53. Eur. J. Cancer 39, 1828–1834 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Foster BA, Coffey HA, Morin MJ, Rastinejad F, Pharmacological rescue of mutant p53 conformation and function. Science 286, 2507–2510 (1999). [DOI] [PubMed] [Google Scholar]
- 71.Andreeff M, Kelly KR, Yee K, Assouline S, Strair R, Popplewell L, Bowen D, Martinelli G, Drummond MW, Vyas P, Kirschbaum M, Iyer SP, Ruvolo V, Gonzalez GMN, Huang X, Chen G, Graves B, Blotner S, Bridge P, Jukofsky L, Middleton S, Reckner M, Rueger R, Zhi J, Nichols G, Kojima K, Results of the phase I trial of RG7112, a small-molecule MDM2 antagonist in Leukemia. Clin. Cancer Res 22, 868–876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Weger VA, de Jonge M, Langenberg MHG, Schellens JHM, Lolkema M, Varga A, Demers B, Thomas K, Hsu K, Tuffal G, Goodstal S, Macé S, Deutsch E, A phase I study of the HDM2 antagonist SAR405838 combined with the MEK inhibitor pimasertib in patients with advanced solid tumours. Brit. J. Cancer 120, 286–293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sallman DA, DeZern A, Sweet K, Steensma DP, Cluzeau T, Sekeres M, Garcia-Manero G, Roboz G, McLemore A, McGraw K, Puskas J, Zhang L, Bhagat C, Graber A, Al Ali NH, Padron E, Tell R, Lancet JE, Fenaux P, List AF, Komrokji RS, Phase Ib/II combination study of APR-246 and azacitidine (AZA) in patients with TP53 mutant myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML). Cancer Res. 78, CT068 (2018). [Google Scholar]
- 74.Pearson S, Jia H, Kandachi K, China approves first gene therapy. Nat. Biotechnol 22, 3–4 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Osborne R, Ark floats gene therapy’s boat, for now. Nat. Biotechnol 26, 1057–1059 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Chen GX, Zhang S, He XH, Liu SY, Ma C, Zou XP, Clinical utility of recombinant adenoviral human p53 gene therapy: Current perspectives. Onco. Targets Ther 7, 1901–1909 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi J, Kantoff PW, Wooster R, Farokhzad OC, Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 17, 20–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG, Non-viral vectors for gene-based therapy. Nat. Rev. Genet 15, 541–555 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Xiong Q, Lee GY, Ding J, Li W, Shi J, Biomedical applications of mRNA nanomedicine. Nano Res. 11, 5281–5309 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Islam MA, Reesor EK, Xu Y, Zope HR, Zetter BR, Shi J, Biomaterials for mRNA delivery. Biomater. Sci 3, 1519–1533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hajj KA, Whitehead KA, Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater 2, 17056 (2017). [Google Scholar]
- 82.Prieve MG, Harvie P, Monahan SD, Roy D, Li AG, Blevins TL, Paschal AE, Waldheim M, Bell EC, Galperin A, Ella-Menye JR, Houston ME, Targeted mRNA therapy for ornithine transcarbamylase deficiency. Mol. Ther 26, 801–813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schrom E, Huber M, Aneja M, Dohmen C, Emrich D, Geiger J, Hasenpusch G, Herrmann-Janson A, Kretzschmann V, Mykhailyk O, Pasewald T, Oak P, Hilgendorff A, Wohlleber D, Hoymann HG, Schaudien D, Plank C, Rudolph C, Kubisch-Dohmen R, Translation of angiotensin-converting enzyme 2 upon liver- and lung-targeted delivery of optimized chemically modified mRNA. Mol. Ther. Nucleic Acids 7, 350–365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramaswamy S, Tonnu N, Tachikawa K, Limphong P, Vega JB, Karmali PP, Chivukula P, Verma IM, Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. U.S.A 114, E1941–E1950 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.An D, Schneller JL, Frassetto A, Liang S, Zhu X, Park JS, Theisen M, Hong SJ, Zhou J, Rajendran R, Levy B, Howell R, Besin G, Presnyak V, Sabnis S, Murphy-Benenato KE, Kumarasinghe ES, Salerno T, Mihai C, Lukacs CM, Chandler RJ, Guey LT, Venditti CP, Martini PGV, Systemic Messenger RNA Therapy as a Treatment for Methylmalonic Acidemia. Cell Rep. 21, 3548–3558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, Zhu H, Siegwart DJ, Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int 56, 1059–1063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang C, Mei M, Li B, Zhu X, Zu W, Tian Y, Wang Q, Guo Y, Dong Y, Tan X, A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 27, 440–443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li B, Luo X, Deng B, Wang J, McComb DW, Shi Y, Gaensler KML, Tan X, Dunn AL, Kerlin BA, Dong Y, An orthogonal array optimization of lipid-like nanoparticles for mRNA Delivery in vivo. Nano Lett. 15, 8099–8107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Castellani P, Balza E, Rubartelli A, Inflammation, DAMPs, tumor development, and progression: A vicious circle orchestrated by redox signaling. Antioxid. Redox Signal 20, 1086–1097 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Carayol N, Vakana E, Sassano A, Kaur S, Goussetis DJ, Glaser H, Druker BJ, Donato NJ, Altman JK, Barr S, Platanias LC, Critical roles for mTORC2- and rapamycin-insensitive mTORC1-complexes in growth and survival of BCR-ABL-expressing leukemic cells. Proc. Natl. Acad. Sci. U.S.A 107, 12469–12474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tasdemir E, Maiuri MC, Morselli E, Criollo A, D’Amelio M, Djavaheri-Mergny M, Cecconi F, Tavernarakis N, Kroemer G, A dual role of p53 in the control of autophagy. Autophagy 4, 810–814 (2008). [DOI] [PubMed] [Google Scholar]
- 92.Valencia PM, Farokhzad OC, Karnik R, Langer R, Microfluidic technologies for accelerating the clinical translation of nanoparticles. Nat. Nanotechnol 7, 623–629 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lim J-M, Swami A, Gilson LM, Chopra S, Choi S, Wu J, Langer R, Karnik R, Farokhzad OC, Ultra-high throughput synthesis of nanoparticles with homogeneous size distribution using a coaxial turbulent jet mixer. ACS Nano 8, 6056–6065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim Y, Lee Chung B, Ma M, Mulder WJM, Fayad ZA, Farokhzad OC, Langer R, Mass production and size control of lipid-polymer hybrid nanoparticles through controlled microvortices. Nano Lett. 12, 3587–3591 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miller MA, Gadde S, Pfirschke C, Engblom C, Sprachman MM, Kohler RH, Yang KS, Laughney AM, Wojtkiewicz G, Kamaly N, Bhonagiri S, Pittet MJ, Farokhzad OC, Weissleder R, Predicting therapeutic nanomedicine efficacy using a companion magnetic resonance imaging nanoparticle. Sci. Transl. Med 7, 314ra183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ventura A, Vassall A, Robinson E, Filler R, Hanlon D, Meeth K, Ezaldein H, Girardi M, Sobolev O, Bosenberg MW, Edelson RL, Extracorporeal photochemotherapy drives monocyte-to-dendritic cell maturation to induce anticancer immunity. Cancer Res. 78, 4045–4058 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Study summary.
Fig. S2. The structure schematic of synthetic mRNA.
Fig. S3. The chemical structure of 3′-O-Me-m7G(5′)ppp(5′)G ARCA cap.
Fig. S4. Chemicals for NP synthesis.
Fig. S5. Characterization of the engineered hybrid mRNA NPs.
Fig. S6. Size of EGFP-mRNA NPs and Luc-mRNA NPs with various formulations.
Fig. S7. Encapsulation efficiency of EGFP-mRNA NPs and Luc-mRNA NPs with various formulations.
Fig. S8. Normalized luminescence intensity of Hep3B cells after treatment with various Luc-mRNA NP formulations at the mRNA dose of 0.830 μg/ml.
Fig. S9. Endosomal escape of mRNA NPs.
Fig. S10. Transfection efficacy verified by CLSM imaging.
Fig. S11. Transfection efficacy verified by flow cytometry.
Fig. S12. Transfection efficacy after quenching intracellular GSH.
Fig. S13. In vitro toxicity of the synthetic EGFP-mRNA NPs.
Fig. S14. IF staining of p53 in p53-null H1299 cells.
Fig. S15. WB analysis of p53 protein expression.
Fig. S16. In vitro therapeutic efficacy of the synthetic p53-mRNA NPs in p53-null H1299 cells.
Fig. S17. Apoptosis of p53-null H1299 cells as determined by flow cytometry after different treatments.
Fig. S18. G1-phase cell cycle arrest induced by p53-mRNA NPs.
Fig. S19. WB analysis of apoptotic signaling pathway in p53-null H1299 cells after different treatments.
Fig. S20. TEM images of mitochondrial morphology in p53-null H1299 cells after different treatments.
Fig. S21. In vitro toxicity of the mutant p53-R175H-mRNA NPs.
Fig. S22. Cytotoxicity of everolimus in p53-null H1299 cells.
Fig. S23. Effect of everolimus on autophagy activation in p53-null H1299 cells.
Fig. S24. WB analysis of autophagy and apoptotic signaling pathways in p53-null H1299 cells.
Fig. S25. Analysis of the autophagosomes and swollen mitochondria in p53-null H1299 cells after different treatments.
Fig. S26. In vitro therapeutic efficacy of the combination of p53-mRNA NPs with everolimus in p53-null H1299 cells.
Fig. S27. In vitro apoptosis of p53-null H1299 cells after different treatments.
Fig. S28. In vitro toxicity of the combination of everolimus with venetoclax.
Fig. S29. In vitro toxicity of the combination of everolimus with siBcl-2.
Fig. S30. The relative mRNA expression of p53.
Fig. S31. The relative mRNA expression of ULK1, ATG7, BECN1, and ATG12.
Fig. S32. The relative mRNA expression of DRAM1, ISG20L1, and SESN1.
Fig. S33. The relative mRNA expression of TIGAR.
Fig. S34. WB analysis of AMPK and TIGAR pathways.
Fig. S35. Schematic representation of the possible mechanism by which p53 tumor suppressor inhibits protective autophagy and sensitizes tumor cells to everolimus.
Fig. S36. BioD of different mRNA NPs in HCC xenograft tumor model.
Fig. S37. BioD of different mRNA NPs in NSCLC xenograft tumor model.
Fig. S38. Blood vessel staining in tumor sections.
Fig. S39. Efficacy and safety of different treatments in HCC xenograft model.
Fig. S40. Antitumor effects of p53-mRNA NPs are synergistic with everolimus in NSCLC xenograft model.
Fig. S41. Murine p53 restoration in p53-null murine liver cancer RIL-175 cells.
Fig. S42. Therapeutic efficacy of murine p53-mRNA NPs in immunocompetent mice bearing p53-null RIL-175 tumors.
Fig. S43. Expression of p53 protein in HCC xenograft model after treatment with p53-mRNA NPs.
Fig. S44. Expression of p53 protein in NSCLC xenograft model after treatment with p53-mRNA NPs.
Fig. S45. IHC images from tumor sections of H1299 tumor–bearing mice before and after treatment with p53-mRNA NPs.
Fig. S46. In vivo toxicity of the p53-mRNA NP–mediated strategy for everolimus rescue assessed by histopathological and hematological analysis.
Fig. S47. IHC images from major organs and tumor sections of the HCC xenograft model.
Fig. S48. Evaluation of immune responses after the treatment with mRNA NPs.
Fig. S49. Scans of the liver metastases from different treatment groups in Fig. 6.
Table S1. Compositions of different NP formulations.
Table S2. Different p53-mRNA sequences used in this study.
Table S3. Primer sequences for qRT-PCR.