SUMMARY
How ubiquitous transcription factors (TFs) coordinate temporal inputs from broadly expressed epigenetic factors to control cell fate remains poorly understood. Here, we uncover a molecular relationship between p53, an abundant embryonic TF, and WDR5, an essential member of the MLL chromatin modifying complex, that regulates mouse embryonic stem cell fate. Wild-type Wdr5 or transient Wdr5 knockout promotes a distinct pattern of global chromatin accessibility and spurs neuroectodermal differentiation through an RbBP5-dependent process in which WDR5 binds to, and activates transcription of, neural genes. Wdr5 rescue after its prolonged inhibition targets WDR5 to mesoderm lineage-specifying genes, stimulating differentiation toward mesoderm fates in a p53-dependent fashion. Finally, we identify a direct interaction between WDR5 and p53 that enables their co-recruitment to, and regulation of, genes known to control cell proliferation and fate. Our results unmask p53-dependent mechanisms that temporally integrate epigenetic WDR5 inputs to drive neuroectoderm and mesoderm differentiation from pluripotent cells.
In Brief
How ubiquitous chromatin-associated proteins and transcription factors (TFs) regulate cell fate determination is poorly understood. Li et al. show that regulation of the broadly expressed TF p53 by the chromatin-associated protein WDR5 is required for neuroectoderm versus mesoderm lineage determination in mouse embryonic stem cells (ESCs).
Graphical Abstract

INTRODUCTION
How ubiquitous epigenetic factors and transcription factors cooperate in a time-dependent manner to direct cell fate remains largely unexplored. In embryonic stem cells (ESCs), WDR5 is highly expressed and decreases during differentiation but remains present in somatic cells (Ang et al., 2011). Broad WDR5 expression is thought to be related to its “epigenetic house-keeping function”: methylation of lysine 4 on histone H3 (H3K4me) via the KMT2 (MLL) histone methyltransferase family. This histone mark is linked to transcription and, thus, WDR5 contributes to regulation of gene expression (Dou et al., 2006; Rao and Dou, 2015).
WDR5 interacts with OCT4, CTCF, or lncRNA to facilitate induced pluripotent stem cell reprogramming and maintains ESC identity (Ang et al., 2011; Yang et al., 2014). Wdr5 haploin-sufficiency in fetal mesoderm triggers somitogenesis defects and WDR5 overexpression accelerates Wnt-mediated osteoblast differentiation (Gori et al., 2006; Vilhais-Neto et al., 2017). WDR5 point mutations are linked to congenital heart defects (Zaidi et al., 2013; Zhu et al., 2017) and disrupted speech development (Eising et al., 2019) in humans. Still, the role of WDR5 in cell fate determination immediately following exit from pluripotency remains enigmatic.
Like WDR5, p53 is enriched in ESCs and decreases during differentiation (Lin et al., 2005). p53 is ubiquitous in the mouse embryo up to day 11 (Lin et al., 2005; Schmid et al., 1991; Rogel et al., 1985). Its abundance and enhanced stability in ESCs suggest roles that may contrast from its function in somatic cells, in which p53 is degraded rapidly by MDM2, and regulates the DNA damage response (Haupt et al., 1997; Sabapathy et al., 1997; Giaccia and Kastan, 1998). Moreover, p53 hyperactivation occurs in several developmental syndromes (e.g., CHARGE and others) that feature neuroectoderm (NE) defects (Bowen and Attardi, 2019).
Here, we identify a regulatory role for WDR5 on the activity of p53 during key ESC cell fate transitions. Molecularly, we found that WDR5 regulates p53 stability and directly interacts with p53 during ESC specification. Intact Wdr5 activity or brief Wdr5 inhibition leads to a distinct chromatin landscape in which WDR5 directly targets NE genes and favors transcription of NE lineage-specifying genes and differentiation of NE organoids. In contrast, prolonged Wdr5 inhibition causes dysregulated ribosomal protein (RP) gene expression and enhanced p53 stability, which leads to p53 activation. Increased p53 activity promotes mesoderm specification, as well as a global chromatin accessibility landscape that is permissive for mesoderm differentiation. Wdr5 rescue redirects WDR5 to mesoderm lineage-identity genes, which promotes differentiation toward contractile cardiogenic and hematopoietic mesoderm fates in a p53-dependent manner. This WDR5-p53 cell fate pathway offers a previously unrecognized example of how a broadly expressed epigenetic factor and embryonically abundant protein coordinate ESC lineage specification and differentiation in a time-dependent manner.
RESULTS
The WDR5-RbBP5 Interaction Surface Controls Rx+ NE Differentiation
WDR5 is essential for mouse ESC (mESC) self-renewal and viability (Ang et al., 2011). To determine whether WDR5 temporally regulates NE differentiation from ESCs, we used CRISPR-Cas9 and piggyBac transposon-based strategies to create “rescue” complementation ESC lines in which endogenous mWdr5 is replaced by a Tet-On, doxycycline (Dox)-inducible HA-tagged, human hWDR5, allowing temporal control of WDR5 expression. hWDR5 protein is 100% identical to mWDR5 (Li et al., 2019). This approach allows mWdr5 editing without altering hWDR5. We used Rx-GFP ESC lines, which contain GFP reporter knocked in to the endogenous NE-specific Rax/Rx promoter; these lines efficiently generate Rx+ NE organoids via serum-free embryoid body (EB)-like aggregates with quick reaggregation (SFEBq) culture (Assawachananont et al., 2014; Eiraku et al., 2011). We derived four independently targeted, hWDR5Dox;mWdr5KO (WDR5Dox;Wdr5KO) Rx-GFP mESC lines (Figures S1A–S1F). Interestingly, while Wdr5 transcripts decreased during NE differentiation, Wdr5 mRNA was expressed in both Rx+ and Rx− EBs (Figure S1A). hWDR5 expression did not alter Rx+ NE induction in wild-type (WT) ESCs (Figures S1B and S1C). In Wdr5KO EBs, Dox removal decreased hWDR5 (Figure S1G) and global histone marks H3K4Me2/3, but not H3K4Me1 (Figure S1H), consistent with the histone-modifying function of WDR5 (Wysocka et al., 2005).
Next, we assessed the role of WDR5 in NE differentiation. Impaired cell proliferation and Rx+ NE differentiation of Wdr5KO EBs was rescued by Dox-induced WDR5 in a dose-dependent manner (Figures 1A and 1B). To test whether interaction of known WDR5 co-factors (Wysocka et al., 2005), MLL1 and RbBP5, with WDR5 is essential for NE differentiation, we expressed FLAG-tagged WT or mutant WDR5 into WDR5Dox;Wdr5KO ESCs (Figure S11). Surprisingly, Wdr5KO EBs expressing WDR5-MLL1 mutants (WDR5F133Y or WDR5I305V) proliferated and differentiated to Rx+ NE normally (Figures 1C and 1D). Wdr5KO EBs constitutively expressing WDR5-RbBP5-binding mutants WDR5N225A, WDR5L240K, WDR5V268E, or WDR5Q289E showed reduced NE differentiation (Figures 1C and 1D) and Rx mRNA (Figure 1E). Defective NE differentiation of WDR5-RbBP5-binding mutants, except for WDR5Q289E, could not be fully rescued with WT WDR5 induced by Dox at 12 h following NE differentiation (T12h; Figures 1C–1F). We next derived dexamethasone (Dex) and Dox double-inducible Wdr5KO ESCs (Figures S2A and S2B), in order to express WDR5-RbBP5 interaction mutants at similar levels. Dex treatment induced translocation of cytoplasmic WT WDR5 to the nucleus in Wdr5KO ESCs, which maintained ESC pluripotency and self-renewal (data not shown). Both Dex-induced nuclear WT-WDR5 and Dox-inducible WT-WDR5 supported NE proliferation in Wdr5KO EBs in monolayer and suspension-based NE differentiation culture conditions (Figures S2C and S2D). However, Dox-induced expression of WDR5-RbBP5-binding mutants, expressed at similar levels (Figure S2B) in Wdr5KO EBs, showed defective NE differentiation (Figures 1G, S2C, and S2D). Together, we find specific interaction surfaces of WDR5 and RbBP5 are required for Rx+ NE differentiation.
Figure 1. WDR5 Induces Rx+ Neuroectoderm Differentiation via Its RbBP5 Interaction Surface.
(A and B) WDR5 regulates NE proliferation and differentiation in a dose-dependent manner. WT and WDR5Dox;Wdr5KO ESCs were maintained in the Dox-containing ESC media. ESCs were resuspended in SFEBq differentiation (EB day 0) in different concentrations of Dox. Cell proliferation at EB day 4 (A) and Rx-GFP positive NE at EB day 5 (B) were determined.
(C and D) Proliferation and NE differentiation in WDR5Dox; Wdr5KO ESCs stably transfected with FLAG-tagged WT or different WDR5 mutants. Upon differentiation, Dox was removed (no Dox) or added back 12 h later (2.0 ug/ml, T12h). Cell proliferation was determined at EB day 4 and relative cell proliferation was normalized to the respective group with Dox as 1.0 (D). Percentage of Rx-GFP+ NE cells at EB day 5 was determined by flow cytometry (E).
(E) Effects of WDR5-RbBP5 interaction mutants with or without Dox-induced WT WDR5 rescue on NE differentiation were determined by Rax mRNA qRT-PCR (EB day 6).
(F) WT, but not WDR5-RbBP5 interaction mutant WDR5Q289E, retained capacity to induce Rax-GFP+ NE in Wdr5KO EBs. Representative day 6 EBs were recorded under microscope using bright field or fluorescence channel. Scale bars: 50 μm.
(G) Expression of Dox-inducible WDR5-RbBP5 interaction mutants was not able to induce Rx-GFP+ NE differentiation. WDR5Dex; Wdr5KO ESCs (Δ26), maintained in dexamethasone (Dex) containing ES media, which enabled Dex-inducible WDR5 expression to maintain ESC self-renewal, were stably transfected with Dox-inducible forms of various HA-tagged WDR5 mutants. Upon differentiation, Dex was removed and Dox was added 12 h later (2.0 ug/ml, T12h). Percentage of Rx-GFP+ NE cells at EB day 7 was determined by flow cytometry.
EV: empty vector backbone control. Data in (A)–(F), (G), (H), and (I) represent mean ± SD (n ≥ 3). **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.001.
See also Figures S1 and S2.
Timing of WDR5 Expression in ESCs Mediates an NE versus Mesoderm Fate Decision
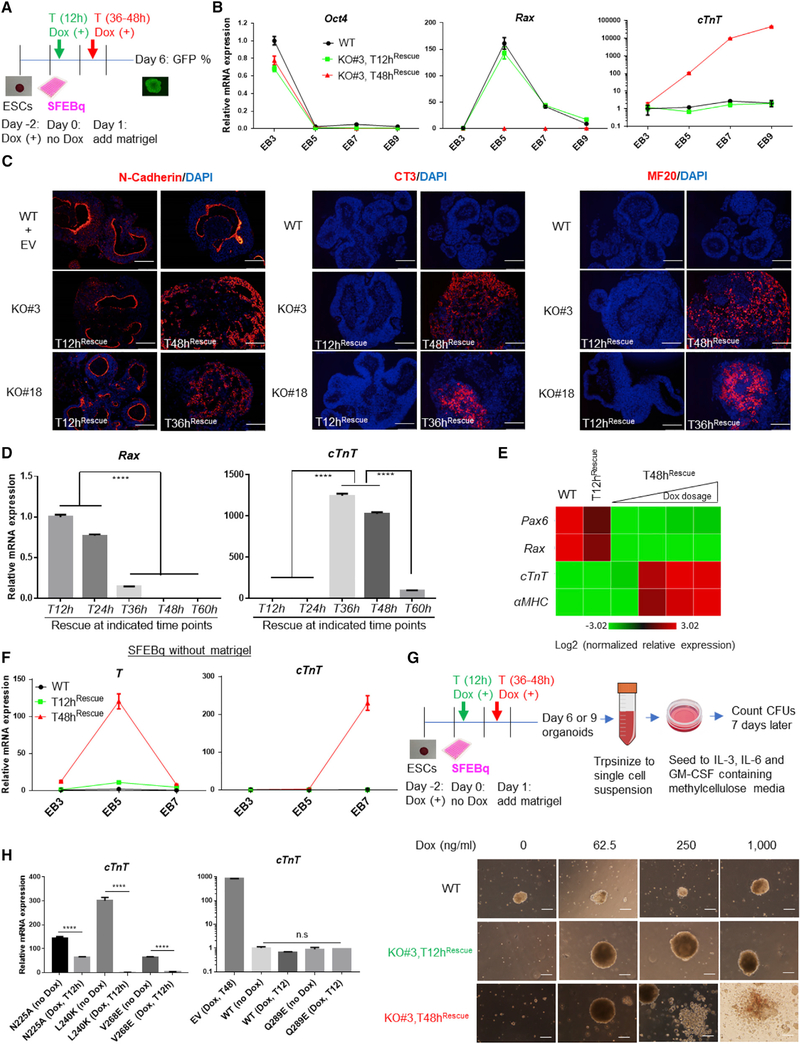
To determine whether WDR5 exerts time-dependent effects on lineage-specific ESC differentiation, we rescued Wdr5KO EBs with Dox-inducible WDR5 at consecutive 12-h intervals (Figure 2A). Induction of WDR5 from T12–24h after differentiation (hereafter termed “early void,” “early rescue,” or brief inhibition [of WDR5]) maintained EB proliferation and stimulated NE differentiation (Figures S3A and S3B). However, WDR5 induction at T36–48h after differentiation (hereafter named as “extended void,” “late rescue,” or prolonged inhibition) maintained Wdr5KO EB growth but impaired NE formation. Unlike WT or early-void groups, virtually all extended-void EBs spontaneously contracted by days 8–9, suggesting cardiogenic mesoderm (MES) differentiation. Time-course qRT-PCR analysis showed induced cardiomyocyte-lineage transcripts in three independent WDR5Dox;Wdr5KO ESC lines in extended-void conditions such as Ncad (Honda et al., 2006), cTnT, αMHC, βMHC, and Myocd mRNA (Figures 2B and S3C–S3E). Cardiogenic-lineage-specific proteins such as intracellular N-cadherin, cTnT (CT3), and myosin heavy chain (MF20) were also present (Figure 2C). In contrast, WT and early-void groups showed induction of NE-specific Rx mRNA (Figures 2B and S3D), Moreover, N-cadherin exhibited NE-specific apical localization in WT or Wdr5KO EBs with Dox-inducible WDR5 in early-void groups (Figure 2C) (Eiraku and Sasai, 2011). Time-course western blotting analyses confirmed expression of exogenous WDR5 protein in Wdr5 knockout (KO) EBs with Dox-inducible early and extended voids (Figure S3F). Cardiogenic MES differentiation was both WDR5 time (Figure 2D) and dose dependent (Figure 2E), indicating that MES induction is specific to WDR5 and not simply a “default” pathway for ESC differentiation. Further, generation of cardiogenic MES was genetically driven by Wdr5 and independent of external cues specific to SFEBq differentiation such as retinoic acid receptor antagonism, or specific lots of matrigel or KO serum replacement (Figure 2F; data not shown). Induction of the MES master transcription factor (TF) T/Brachyury in Wdr5KO EBs with extended voids (Figure 2F) prompted us to examine whether other MES lineages may form in these conditions. When Wdr5KO EBs with extended voids were seeded into methylcellulose-based differentiation medium at day 6 or 9 for secondary hematopoietic differentiation (Figure 2G, upper panel), colony-forming units of granulocyte-macrophage progenitor cells (CFU-GM, CFU-G, and CFU-M) arose, which were not observed in Wdr5KO EBs with early voids (Figure 2G, lower panel; Figures S3G and S3H). These data demonstrate that an extended void in Wdr5KO EBs stimulates ESCs toward cardiogenic and hematopoietic MES fates. This time-dependent ESC fate transition to MES occurs in SFEBq conditions, which does not contain the exogenous cytokines or fetal calf serum usually required for ESC-derived MES differentiation (Wataya et al., 2008; Eiraku et al., 2008, 2011). These findings demonstrate that the length of time of Wdr5 inhibition, followed by subsequent WDR5 rescue, determines whether ESCs differentiate toward NE versus MES-specific lineages. Since continuous and early voids (T12h) stimulate NE differentiation, and an extended void (T36–48h) promotes cardiogenic or hematopoietic MES formation, we further conclude that EB day 2 (T48h) defines a crucial period of lineage specification during which differentiating ESCs make an NE versus MES fate decision.
Figure 2. Prolonged Wdr5 Inhibition Triggers Mesoderm Differentiation.
(A) A schematic outline of SFEBq differentiation in WT, and WDR5Dox;Wdr5KO ESCs with early or extended void/rescue of Dox-inducible WDR5.
(B) qRT-PCR mRNA analysis of pluripotent (Oct4), NE (Rax/Rx), and cardiac mesoderm (MES) markers (cTnT) during time-course SFEBq differentiation of WT, early (T12hRescue) and extended void (T36h or 48hRescue)/rescue of WDR5 in Wdr5KO EBs.
(C) Immunofluorescence staining on day 9 EB sections with neuroepithelial marker (N-cadherin), cardiac specific troponin T (CT3), and sarcomere myosin (MF20) in WT, early and extended void/rescue of WDR5 in mWdr5KO EBs. DAPI was counterstained for nuclei. Scale bars: 100 μm.
(D) Critical time points for WDR5-mediated Rax/Rx+ NE and cardiogenic MES fate choice in Wdr5KO EBs were determined by Rax and cTnT mRNA expression (EB day 6).
(E) Dose-dependent effects of WDR5 in Wdr5KO EBs during WDR5 extended void on cardiogenic MES induction (cTnTand αMHC) and NE inhibition (Pax6 and Rax). Samples were collected on EB day 6.
(F) Cardiogenic MES induction by WDR5 extended void is not restricted to specific NE differentiation conditions. WT and Wdr5KO EBs with WDR5 early or extended void were differentiated using SFEBq methods without Matrigel and Rx-inducing NE differentiation agent AGN. Induction of MES markers T (Brachyury) and cTnT was determined by qRT-PCR.
(G) MES induction by prolonged deletion of Wdr5 leads to hematopoietic differentiation. Upper panel: a schematic description of secondary hematopoietic cell colony-forming unit (CFU) assay using primary differentiating SFEBq organoids as test cells. Lower panel: the representative pictures of resultant secondary EBs and all CFU were presented at day 9 after secondary differentiation. Dox dose for primary differentiation: 2.0 ug/ml. Scale bars: 100 μm.
(H) Effects of WDR5-RbBP5 interaction mutants with or without Dox-inducible WT WDR5 void on cardiogenic MES-specific transcription were determined by cTnT qRT-PCR (EB day 6).
Data in (B), (D), (F), and (I) represent mean ± SD (n ≥ 3). ****p ≤ 0.001.
See also Figure S3.
The WDR5-RbBP5 Interaction Surface Regulates Mesoderm-Related Transcription
We next asked whether the WDR5-RbBP5 interaction surface regulates cardiogenic MES induction. Wdr5KO EBs expressing constitutive WDR5N225A, L240K or V268E mutants showed ~50û300-fold induction of cTnT mRNA (Figure 2H, left panel). This pro-cardiogenic MES induction effect was inhibited by concomitant rescue of WT WDR5 at T12h (Figure 2H, left panel). However, WDR5Q289E had similar capacity as WT WDR5 to repress cTnT mRNA upregulation in Wdr5KO EBs (Figure 2H, right panel). Thus, while the WDR5-RbBP5 interaction promotes NE fates (Figure 1C), it only partially restrains transcription of cardiogenic MES-related genes.
WDR5 Regulates Overlapping and Distinct Direct Target Genes in Divergent Cell Lineages
To identify molecular targets for WDR5 in Rx+ NE and cardiogenic MES lineages, we performed RNA sequencing (RNA-seq) from EB day 6 RNA isolated from four groups: WT, Wdr5KO, Wdr5KO with early (T12hRescue), or extended (T48hRescue) void. The majority of differentially expressed transcripts arose, and overlapped (75%–80%; Figure 3A), in WT versus T48hRescue (2,758), and T12hRescue versus T48Rescue (2,937) groups (Figure S4A), compared to those of WT versus T12hRescue groups (593; Figure S4A). These data indicate that transcriptomes of WT and T12hRescue groups were comparable and harbored RNA profiles distinct from T48hRescue and Wdr5KO groups (Figure S4B). Gene Ontology (GO) themes in the T12hRescue group included NE development, while those of the T48hRescue group emphasized MES specification (Figure 3B). RNA-seq analysis indicated that trophectoderm markers (Cdx2 and Gata3) and endoderm markers (Gata4 and Gata6) were upregulated in T48hRescue group (data not shown). Time-course qRT-PCR analysis showed that Cdx2 and Gata3 mRNA were transiently upregulated in T36–48hRescue groups during differentiation, to a greater degree than in the WT and T12hRescue groups (Figure 3C; data not shown), consistent with a previous report (Ang et al., 2011). However, the lack of concomitant endoderm-specific Foxa2 upregulation suggested that Gata4 upregulation suggests formation of lateral mesoderm rather than endoderm in T36–48hRescue groups (Figure 3C) (Rojas et al., 2005).
Figure 3. Early and Extended Void of WDR5 in Wdr5KO EBs Activates Overlapping and Distinct Lineage-Determination Target Genes.
(A) Venn diagram of differentially expressed genes among groups for WT, Wdr5KO with WDR5 early (T12hRescue), or extended void (T48hRescue) EBs (day 6).
(B) GO and heatmap clustering analysis of differentially expressed genes in day 6 Wdr5KO EBs with early (T12hRescue) versus extended (T48hRescue) void of WDR5.
(C) Validation of RNA-seq data by qRT-PCR analysis of markers for trophectoderm (Cdx2 and Gata3) and endoderm (Gata4 and Foxa2) in WT or Wdr5KO EBs with early or extended WDR5 void during time-course SFEBq differentiation.
(D) Metaprofiles (upper panels) and heatmaps (lower panels) of HA-WDR5 ChIP-seq signals demonstrating overlapping and distinct WDR5 bound peaks in day 6 Wdr5KO EBs with T12hRescue and T48hRescue settings using all peaks existing in the T12hRescue group (left panel) or T48hRescue group (right panel) as reference, respectively.
(E) Heatmap demonstrating WDR5 direct target genes in day 6 Wdr5KO EBs with T12hRescue and T48hRescue groups.
See also Figure S4.
To define direct target genes of WDR5 in NE and MES lineages, we performed HA-WDR5 chromatin immunoprecipitation sequencing (ChIP-seq) in Wdr5KO EBs with T12hRescue and T48hRescue at EB day 6. We found that in both groups, WDR5 bound to intergenic, intron, and promoter regions (Figure S4C). Using peak centers of HA-WDR5-bound targets in the T12hRescue or T48hRescue group (Figure 3D) as references, we found that WDR5 bound both overlapping and distinct targets. In both NE organoids and MES-enriched EBs, we identified distinct de novo and known WDR5 target DNA motifs, including consensus sites shared with c-MYC, consistent with a previous report (Figure S4D) (Thomas et al., 2015). Integration of ChIP-seq with RNA-seq datasets revealed WDR5-bound targets that were associated with >2-fold differential gene expression (Figure S4E). Such WDR5 direct target genes in the T12hRescue group (123 genes: 76 and 47 genes down- and upregulated, respectively) included NE-related genes such as Sox2, Nestin, Sfrp2, and others (Figures 3E and S4E). In the T48hRescue group (45 genes: 25 and 20 genes down- and upregulated, respectively), these included MES-associated genes such as Pabpc4l, Nr3c1, Egln3, and others (Figures 3E and S4E). 22 direct WDR5 target genes (10 and 12 genes down- and upregulated, respectively) overlapped in both the NE and MES lineages, including Efcab12, Esrrb, Brn2, and 8 RP genes such as Rpl5 and Rpl37 (Figure S4E). These data show that WDR5 targets both overlapping and distinct genes in NE organoids and MES-enriched EBs.
WDR5 Regulates Target Gene Expression with Dynamic Chromatin Accessibility during ESC Lineage Specification
WDR5 is a core member of the MLL chromatin-modifying complex and is essential for H3K4me3 formation (Figure S1H) (Dou et al., 2006; Rao and Dou, 2015). Having established that WDR5 regulates differentiation of Rx+ NE and cardiogenic MES lineages at EB day 6, we sought to understand chromatin-related events that precede the formation of these distinct lineages, in WT EBs and immediately before T48hRescue in Wdr5KO EBs, during the lineage specification period (EB day 2). RNA-seq analyses at EB day 2 revealed that ~60% of differentially expressed genes were downregulated in Wdr5KO EBs (Figure 4A). We used CUT&RUN (cleavage under targets and release using nuclease sequencing) to define the genomic targets of WDR5 and H3K4me3 in WT and Wdr5KO EBs at day 2 (Figure 4B) (Meers et al., 2019). H3K4me3 at WT WDR5-bound chromatin targets were reduced in Wdr5KO EBs (Figure 4C). Genome-wide integration of WDR5-bound peaks from CUT&RUN with transcripts from RNA-seq data elucidated direct WDR5 target genes during EB day 2 lineage specification (Figure 4D). In Wdr5KO EBs, we detected 33 WDR5 direct target genes. Of 33 genes, 20 were downregulated including Rpl29 and E2f3, which are genes essential for NE proliferation or differentiation while 13 genes were upregulated including Nodal, which is required for MES specification. Interestingly, differentially expressed transcripts of WDR5 direct target genes in day 2 WT and Wdr5KO EBs overlapped with those of day 6 WT and Wdr5KO EBs (Figure S5A).
Figure 4. Prolonged Inhibition of Wdr5 Leads to Dysregulated Target Gene Expression during mESC Lineage Specification.
(A) Volcano plots of down- and upregulated genes upon prolonged Wdr5 deletion (EB day 2) determined by RNA-seq. ≥ 2-fold higher or lower differentially expressed genes are colored red and blue, respectively. KO-A and KO-B represents EBs differentiating from two independent Wdr5KO ESC clones.
(B) Heatmaps showing WDR5 and H3K4Me3 DNA binding peaks determined by CUT&RUN sequencing. WDR5-bound peaks in WT EBs that decreased due to Wdr5 deletion in day 2 EBs (Wdr5KO;p53KO DKO cells; see below) were called for H3K4Me3 peaks in WT and Wdr5KO EBs at same time point (day 2).
(C) Metaprofiles comparing mean RPKM of H3K4Me3 peaks centered on WDR5-bound loci (WT) in day 2 WT and Wdr5KO EBs.
(D) Heatmap displaying differential mRNA expression of WDR5 target genes upon prolonged Wdr5 deletion (EB day 2).
(E) Track views of ATAC-seq peak dynamics for a representative NE gene (Rax) and a cardiac MES gene (cTnT) in WT, Wdr5KO EBs with T12h and T48h WDR5 void during time-course differentiation. The y axes were set as the same scale by group autoscale in Integrative Genomics Viewer.
(F) Metaprofiles showing correlation of ATAC-seq peaks with H3K4Me3 CUT&RUN sequencing peaks for day 2 WT and Wdr5KO EBs. Genome-wide H3K4Me3 peaks lost by Wdr5 prolonged deletion were furtherly called for ATAC-seq change.
(G) GO analysis of closing and opening peaks by ATAC-seq analysis in Wdr5KO EBs at day 2.
See also Figure S5.
As Wdr5 deletion led to both down- and upregulated direct target genes, we posited that WDR5-mediated canonical (transcriptional activation) and non-canonical H3K4me3 (transcriptional repression) chromatin mechanisms contribute to WDR5-mediated target gene regulation (Zhang et al., 2016). To this end, we performed ATAC-seq to determine whether WDR5 regulates “open and closed” chromatin accessibility regions at NE and MES lineage-identity genes. Genome-wide analyses revealed that genes with loss of H3K4me3 mark correlated, to some extent, with reduced chromatin accessibility in day 2 Wdr5KO EBs (Figure 4F). We observed increased (“open”) chromatin accessibility at the NE-specific gene Rx transcription start site (TSS) region as early as EB day 4 in WT and T12hRescue groups, indicating proper NE specification (Figure 4E). In contrast, chromatin accessibility at the Rx TSS region remained inaccessible (“closed”) during the day 2–6 differentiation period in Wdr5KO EBs with the T48hRescue group. For the MES-specific gene cTnT/Tnnt2, opening peaks were observed at its TSS in Wdr5KO EBs with T48hRescue until EB day 6 differentiation, but not in WT and T12hRescue groups (Figure 4E).
We next sought to gain insights into WDR5-dependent, chromatin accessibility during lineage specification (EB day 2). Decreased chromatin accessibility in Wdr5KO EBs at day 2 was associated with NE-pathways (Figures 4G and S5B) and motif analysis of those closing peaks correlated with the NE master regulator SOX1 (Figure S5C). Increased chromatin accessibility in Wdr5KO EBs at day 2 was associated with Hippo, Wnt, and p53-signaling pathways (Figures 4G and S5B) and motif analysis of those open peaks correlated with transcription factor NF-YB and YY1 (Figure S5C). Indeed, activation of p53 and Wnt signaling pathways, and the transcription factor YY1, have been previously reported to promote MES differentiation (Wang et al., 2017; Gregoire et al., 2017). Hyperactivation of the Hippo pathway and overexpression of YAP impairs neural differentiation in mESCs (Lian et al., 2010), which is consistent with defective NE differentiation in Wdr5KO EBs. Collectively, these data show that prolonged Wdr5 inactivation triggers alterations in chromatin accessibility and target gene expression that are permissive for MES differentiation and repressive for NE induction.
p53 Signaling Contributes to the NE versus Cardiogenic MES Cell Fate Decision
p53 signaling emerged as a common theme in GO analyses of opening peaks in ATAC-seq (Figure 4G) and of RNA-seq in Wdr5KO EBs at days 2 and 6, respectively (Figures 4G and S5D). Indeed, the p53 direct target gene Cdkn1a mRNA in Wdr5KO EBs was upregulated at days 2 and 3 (Figure 6C). To test whether p53 activation regulates Wdr5-mediated NE and MES fate choice, we edited p53 via CRISPR-Cas9 to derive WDR5Dox;Wdr5KO Rx-GFP ESC lines (Figures S6A and S6B). p53 and Wdr5 double-knockout (DKO) EBs showed reduced expression of p53 direct target genes Cdkn1a and Bax (Figures 6C and S6C). While p53 deletion rescued cell proliferation defects in Wdr5KO EBs at EB day 4 (Figure 5A), neither Rx+ NE nor cardiogenic MES arose from DKO EBs (Figure 5B; data not shown), suggesting that p53 harbors proliferation-independent effects on cell fate.
Figure 6. WDR5 Regulates p53-Dependent Gene Regulation during ESC Specification.
(A) Metaprofiles for ATAC-seq chromatin accessibility in closing (left panel) and opening (right panel) peaks in day 2 Wdr5KO EBs partially rescued by p53 deletion.
(B) Boxplots for downregulated (left panel) and upregulated (right panel) genes from RNA-seq show that transcriptional defects in day 2 Wdr5KO EBs (versus WT) are partially rescued by p53 deletion. For downregulated genes, WT versus KO-A or KO-B: p = 2.2e–16. KO-A versus KO-A derived DKO-A: p = 0.009. KO-B versus KO-B derived DKO-B: p = 2.2e–16. For upregulated genes, WT versus KO-A or KO-B: p = 2.2e–16. KO-A versus KO-A derived DKO-A: p = 2.2e–16. KO-B versus KO-B derived DKO-B: p = 2.2e–16.
(C) qRT-PCR validation of partially rescued genes E2f3 and Cdkn1a in day 2 or day 3 Wdr5KO EBs by p53 deletion. Data represent mean ± SD (n ≥ 3). **p ≤ 0.01 and ***p ≤ 0.001.
(D) Metaprofiles displaying H3K4Me3 peaks centered at WDR5-bound loci (WT) were reduced in day 2 Wdr5KO EBs, but were partially reversed (rescued) by p53 deletion (DKO EBs).
Figure 5. p53 Deletion Represses Cardiogenic MES Induction Linked to Extended Wdr5 Inhibition and Partially Rescues NE Differentiation.
(A) Cell proliferation in day 4 Wdr5KO and DKO EBs with or without WDR5 void.
(B) NE differentiation in WT, Wdr5KO, and DKO EBs without WDR5 void (no Dox), or with early (Dox, T12h) or extended (Dox, T44h) WDR5 void, was determined by flow cytometry for Rx(+)-GFP cells at day 6.
(C) Rx+ NE differentiation by early WDR5 void in DKO EBs is WDR5 dose-dependent. Samples were analyzed at EB day 6.
(D) Representative images (EB day 7) of Rx+ NE and cardiogenic MES fate choice in WT, Wdr5KO, and DKO EBs with early or extended WDR5 void. Scale bars: 50 μm.
(E) qRT-PCR analysis of NE (Rax/Rx and Pax6) and MES (cTnT) specific markers in WT, Wdr5KO, and DKO EBs with early or extended WDR5 void. Samples were collected on EB day 10.
(F) Metaprofiles and heatmaps of ATAC-seq closing and opening peaks in WT, Wdr5KO, and DKO EBs with early or extended WDR5 void. Samples were analyzed at EB day 6.
Data in (A)–(C) and (E) represent mean ± SD (n ≥ 3). ****p ≤ 0.001.
See also Figure S6.
Next, we added Dox (WDR5 rescue) to DKO EBs at T12h or T36–48h, which allowed us to determine whether p53 regulates WDR5-driven NE versus MES fate choice. Similar to Wdr5KO EBs, early void (Dox, T12h) in DKO EBs stimulated Rx+ NE differentiation (Figures 5B and 5D; Videos S1, S3, and S5). Rx+ NE induction was WDR5 dose dependent (Figure 5C). Notably, unlike Wdr5KO EBs with an extended void, which induces cardiogenic MES differentiation (Figure 5E; Video S4), DKO EBs with extended voids (Dox, T36–48h) did not form cardiogenic MES (Figure 5E; Video S6). Instead, Rx+ NE differentiation occurred (Figures 5B, 5D, 5E, and S6D). Corroborating these findings, EB day 6 ATAC-seq analyses revealed that NE-specific, open, unique peaks (chromatin accessible regions) in WT EBs that had closed (reduced chromatin accessibility) in Wdr5KO EBs with T48hRescue were restored to an open state in DKO EBs (Figure 5F). Similarly, unique, opening peaks associated with MES specification in Wdr5KO EBs with T48hRescue had closed in DKO EBs (Figure 5F). Thus, p53 controls the cardiogenic MES versus Rx+ NE fate choice in Wdr5KO EBs.
WDR5 Regulates p53-Dependent Chromatin Accessibility, Histone Methylation, and Transcription during ESC Lineage Specification
Next, we wondered whether WDR5 and p53 regulate common target genes during ESC lineage specification. We compared EB day 2 ATAC-seq chromatin accessibility profiles of control (Wdr5KO with T12hRescue), Wdr5KO, and DKO EBs. p53 deletion reversed closed peaks observed in day 2 Wdr5KO EBs (increased chromatin accessibility at regions of previously decreased accessibility) when compared to control (Figure 6A). Compared to the T12hRescue group, opening peaks in Wdr5KO EBs modestly reverted (“closed”) upon p53 deletion (Figure 6A).
To determine whether the above changes in chromatin accessibility lead to transcriptional changes, RNA-seq was performed on day 2 WT, Wdr5KO, or DKO EBs from two independent ESC lines. Among 736 downregulated genes in Wdr5KO EBs, 63% (464) of the genes were rescued by p53 deletion in DKO EBs (Figures S6E, 6B, and 6C). Among these genes, Bcl2l14 is a direct p53 target gene and downregulation or mutation of genes including Fras1, Thy1, Meis1, and Vax2OS has been reported in NE defects including eye development, which is consistent with phenotypes observed later in day 6 Wdr5KO EBs (Figure 1B). Among 440 upregulated genes in Wdr5KO EBs, 60% (262) of the genes were rescued by p53 deletion in DKO EBs (Figures 6B and S6E). Among these genes, Cdkn1a is a known p53 target gene (Figure 6C) and Wnt3 is a p53 direct target that in ESCs promotes MES differentiation (Wang et al., 2017). Genes essential for MES specification such as Mesp2, Lefty1, Nodal, or Pitx2 were upregulated in Wdr5KO EBs but were repressed by p53 deletion in DKO EBs. Interestingly, reduced H3K4me3 at WT WDR5-bound peaks in Wdr5KO EBs was partially rescued by p53 deletion (Figure 6D). Thus, WDR5 regulates p53-dependent chromatin accessibility, H3K4me3, and transcription during ESC specification.
WDR5 Regulates p53 Protein Stability and Directly Interacts with p53 during ESC Specification
Next, we sought to explore the mechanism(s) by which WDR5 regulates p53. We found that Wdr5 extended void did not alter p53 transcripts (Figure 7A) (Sun et al., 2015; Xie et al., 2017). We noticed that ChIP-seq peaks representing WDR5 bound to RP genes were overrepresented (Figures S4E and S7A; data not shown). Downregulation of a subset of RP genes was confirmed by RNA-seq (Figure S7B). It is known that reduced RP gene transcription triggers defects in ribosome biogenesis, which upregulates p53 activity via MDM2 inhibition (RP-MDM2-p53 pathway) (Liu et al., 2016). Wdr5 deletion (EB day 2) and an extended void (EB day 6) led to downregulation of WDR5-bound RP genes including Rpl5, Rpl24, and Rpl29 (Figures 7B, 7C, and S7A). These data suggest that p53 activation due to Wdr5 deletion can be, in part, due to perturbation of WDR5-bound, RP target genes.
Figure 7. WDR5 Controls Posttranscriptional Regulation of p53 and Directly Interacts with p53 during ESC Lineage Specification.
(A) qRT-PCR analysis of p53 mRNA in WT and Wdr5KO EBs at days 2 and 3.
(B) Track views of WDR5-bound peaks at TSS regions of Rpl5 and Rpl29 gene in day 2 WT and DKO EBs determined by CUT&RUN sequencing. The y axes were set as the same scale by group autoscale in Integrative Genomics Viewer.
(C) qRT-PCR validation of mRNA expression of ribosomal protein (RP) genes in day 2 WT and Wdr5KO EBs.
(D) Western blot showing p53 levels in day 2 WT and Wdr5KO EBs treated with cycloheximide (CHX, 50 μg/ml) for different time points. Right panel: quantification of p53 levels relative to loading control β-actin. Relative p53 level at time 0 of each group was set as 1.0.
(E) Physical interaction of endogenous WDR5 and p53 determined by immunoprecipitation (IP). EB day 2 WT and DKO cell lysates were precipitated with p53 antibody. IP elutions were probed with WDR5 and p53 antibodies, respectively. β-actin served as input loading control.
(F) Heatmaps showing p53-bound peaks determined by CUT&RUN sequencing on day 2 WT and DKO EBs.
(G) Track views of WDR5 and p53 co-bound peaks at the TSS region of Rap2b gene in day 2 WT and DKO EBs, as determined by CUT&RUN. The y axes were set as the same scale by group autoscale in Integrative Genomics Viewer.
(H) qRT-PCR showing upregulation of Rap2b mRNA in day 2 Wdr5KO EBs and partial rescue (downregulation) of Rap2b transcription due to p53 deletion. Data in (A), (C), and (H) represent mean ± SD (n ≥ 3). **p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.001.
See also Figure S7.
While p53 transcripts during lineage specification of WT and Wdr5KO EBs were comparable (Figure 7A), Wdr5KO EBs harbored elevated p53 protein levels (Figure 7D). We treated WT and Wdr5KO EBs with the protein translation inhibitor cycloheximide (CHX), which showed that p53 stability was enhanced in Wdr5KO EBs (Figure 7D). How does WDR5 regulate p53 stability? Post-translational modifications (PTMs) such as serine (S), phosphorylation (p), and lysine (K) acetylation (Ac) stabilize p53 (Hafner et al., 2019). Indeed, we found that Wdr5 deletion in Wdr5KO EBs increased p53 PTMs at p-p53S15, p-p53S392, Ac-p53K305, and Ac-p53K379; WDR5 rescue reduced these PTMs (Figure S7C). Moreover, using immunofluorescence, we observed that prolonged deletion of Wdr5 led to increased nuclear p53 (Figures S7D and S7E). Together, Wdr5 deletion triggers PTMs that stabilize p53 and increase nuclear p53 activity, as evidenced by enhanced p53 stability and increased transcription of p53 direct target genes (Figure 6C).
Finally, by immunoprecipitation, we found that endogenous WDR5 and p53 physically interact during ESC specification (Figure 7E). We performed p53 CUT&RUN at EB day 2 and integrated the data with that of WDR5 CUT&RUN at EB day 2 (Figure 7F). We found co-recruitment of WDR5 and p53 to Rap2b, Irf2bp2, and Sox2 (Figure 7G). qRT-PCR confirmed that these direct, joint target genes displayed differential mRNA expression in WT and Wdr5KO EBs, and that p53 deletion partially rescued defective transcription of these genes in Wdr5KO EBs (Figures 7H and S7F). Notably, Rap2b and Irf2bp2 are known direct targets of p53; these genes regulate cell proliferation (Zhang et al., 2013; Koeppel et al., 2009). p53 CUT&RUN of WT EBs at day 2 revealed that p53-bound motifs at target loci are shared with motifs bound by pro-MES TF NKX2.5 (data not shown). Thus, WDR5 directly interacts with p53 and regulates p53 activity via direct and indirect PTMs during ESC specification.
DISCUSSION
Transcription factors expressed at a particular time and location, and at proper magnitude during development, have traditionally been the focus of studies of cell fate determination. In contrast, our study, using an ESC-to-organoid platform, unmasks time-dependent roles for the ubiquitous epigenetic protein and TF, WDR5 and p53, respectively, to control ESC fate choice.
Several WDR5-driven mechanisms regulate ESC differentiation. First, specific interaction sites that mediate WDR5-RbBP5 binding are required for NE differentiation. Second, WDR5 regulates p53 stability. WDR5 targets and transcriptionally activates a subset of RP genes. Wdr5 deletion leads to reduced transcription of these RP genes, triggering p53 activation via RP-MDM2-p53 pathway and to PTMs that increase p53 stability. Indeed, while this work was under review, a report independently confirmed our findings, which showed that RP genes are direct WDR5 targets and that WDR5 inhibition triggers p53 upregulation via the RP-MDM2-p53 pathway in leukemia cells (Aho et al., 2019). Third, during lineage specification, Wdr5 deletion derepresses the WDR5 direct target gene and MES TF Nodal; induced p53 activation from Wdr5 loss promotes MES lineage specification. p53 targets motifs on loci shared with another MES TF NKX2.5 (data not shown). Thus, WDR5 suppresses cardiogenic MES and promotes NE-specific chromatin accessibility, transcription, and cell fate in a p53-dependent manner. Indeed, impaired H3K4me3 deposition, chromatin accessibility, transcription, and NE specification in Wdr5KO EBs are partially rescued when p53 is concurrently deleted. Finally, WDR5 directly interacts with p53 and both are recruited to target genes related to NE fate (Sox2) and cell proliferation (Rap2b and Irf2bp2).
Given that RbBP5, MYC, or KANSL2 bind to the same WDR5 interaction surface in mutually exclusive manner, a central role for this binding surface on WDR5 on the regulation of NE fate is supported from RbBP5 and MYC loss-of-function assays from our study and others (Thomas et al., 2015). RbBP5 depletion in ESCs leads to NE differentiation defects and deletion of c/n-Myc inhibits NE fates from mESCs (Jiang et al., 2011; Smith et al., 2010). Since we observed enrichment of MYC-associated DNA motifs at WDR5 direct targets in NE-lineage cells at EB day 6, the interplay of WDR5, RbBP5, and MYC on chromatin to regulate NE lineage-identity genes warrants further study.
p53 family members have been shown to regulate NE versus MES cell fate choice during development. In Xenopus, p53 promotes MES via an interaction with XFDL156, which represses ectoderm formation (Sasai et al., 2008). In mESCs, p53/p63/p73 contribute to MES differentiation by integration of WNT-TCF and Nodal-SMAD signaling (Wang et al., 2017). Indeed, we show that during prolonged Wdr5 inhibition, p53 deletion partially rescues Rx+ NE fate and inhibits cardiogenic MES differentiation. In the context of WDR5-dependent regulation, p63 and p73 do not appear to share redundant functions with p53, as p53 deletion alone is sufficient to stimulate NE and repress MES. Interestingly, p53-associated human syndromes and analogous mouse models feature p53 hyperactivation and NE defects (Bowen and Attardi, 2019; Bowen et al., 2019; Van Nostrand et al., 2014). Importantly, many of the p53-dependent, embryonic NE defects can be partially rescued by p53 deletion, similar to our work (Bowen et al., 2019; Van Nostrand et al., 2014). Still, whether our ESC-based study on the WDR5-p53 cell fate pathway relates to altered developmental events in vivo remains unknown.
Several pieces of evidence suggest that an instructive mechanism contributes to the WDR5-p53 cell fate pathway during lineage specification. First, Wdr5 deletion during a narrow time window (36–48 h after ESC exit from pluripotency) stimulates cardiogenic MES induction, highlighting that a short, 12-h “pulse” of Wdr5 inhibition initiates MES differentiation. Second, cell proliferation rates did not significantly differ in WT WDR5, Wdr5KO, or DKO EBs that undergo WDR5 rescue at 36–48 h, and which differentiate to Rx+ NE, cardiogenic MES, or Rx+ NE, respectively. Third, despite rescue of impaired cell proliferation in Wdr5KO EBs expressing mutant WDR5L240K, N225A, V268E by WT WDR5, the cardiogenic MES fate is maintained. Fourth, prolonged Wdr5 inhibition rapidly triggers upregulation of a subset of genes permissive for MES induction. Notably, Wnt3 is a key p53 direct target gene that promotes MES fate specification from ESCs (Wang et al., 2017). Further, we found that WDR5 directly binds the MES master regulator Nodal, and that Nodal is rapidly derepressed within 36–48 h of Wdr5 loss during ESC specification. These findings further support an instructive role for WDR5-mediated cardiogenic MES induction (Kitajima et al., 2000).
Here, we define a physical interaction between WDR5 and p53 and find that they jointly bind and regulate common target genes related to cell proliferation (Rap2b and Irf2bp2) and NE fate (Sox2). Whether co-recruitment of WDR5 and p53 “draws” these directly interacting proteins away from other known canonical or non-canonical partners with which WDR5 and p53 interact individually, allowing for transcription of NE genes and suppression of non-NE genes, is a concept that ought to be explored in future studies. Further, it will be important to determine the function of WDR5-p53 protein-protein interaction in p53-associated human syndromes that feature congenital NE defects (Bowen and Attardi, 2019; Van Nostrand et al., 2014; Bowen et al., 2019). Finally, our work informs future translational studies that investigate potential off-target/teratogenic effects of WDR5 inhibitors used for WT and mutant p53-associated cancers, a field that has attracted a $1 billion investment by industry (AP, 2019).
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
LEAD CONTACT AND MATERIALS AVAILABILITY
Please direct any requests for further information or regents generated in current manuscript to the lead contact, Dr. Rajesh C. Rao, MD (rajeshr@umich.edu). All plasmids and ESC cell lines generated are shared for research and educational purposes under Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
Rx:GFP K/I EB5 mouse ESCs, a subline of mouse embryonic stem cell line, EB5 (129/Ola), in which GFP gene is knocked-in under Rax gene promoter, was ordered from RIKEN Cell Bank (Ibaraki, Japan) (Wataya et al., 2008). Rx:GFP ESCs were maintained in leukemia inhibitory factor (LIF, ESG1107, Millipore) containing ES media [GMEM supplemented with 2.5 glutamine (GIBCO), 10% knockout serum replacement, KSR (GIBCO), 1% ES qualified fetal calf serum (FCS), 1 mM sodium pyruvate (Sigma), 0.1 mM non-essential amino acid (Sigma) and 0.1 mM of 2-mercaptoethnal (Sigma)].
METHOD DETAILS
ESC Maintenance and Induced Differentiation
Rx:GFP ESCs were routinely maintained at 37°C, 5% CO2 in ESC media in the presence of 2,000 IU/ml LIF. Induced differentiation to Rax (+) neuroectoderm via 3D (serum-free culture of embryoid-body-like aggregate with quick reaggregation, SFEBq) organoid methods was following a protocol as described previously. Briefly, 5,000 cells were re-suspended in 100 μl of GMEM based differentiation media supplemented with 5% KSR, 1 mM sodium pyruvate, 0.1 mM non-essential amino acid and 0.1 mM of 2-mercaptoethnal and 0.1 μM of retinoic acid receptor antagonist AGN 193109 Sodium Salt (AGN, Santa Cruz) and seeded on each well of 96U ultralow attachment plate (ThermoFisher Scientific) on day 0. On day 1, 50 μl of 2% Matrigel solution (v/v, Corning) was added to embryonic body (EB) suspension. NE differentiation was sustained in 96U plate for 6 to 9 days and differentiation efficiency was monitored by GFP via fluorescent microscopy (OLYPUS IX73) and flow cytometry (BD LSR II). For differentiation of ESCs using SFEBq method without matrigel and AGN, 5,000 cells were re-suspended in 150 μl of GMEM based differentiation media with 10% KSR as described elsewhere (Kamiya et al., 2011). For secondary hematopoietic cell differentiation, day 6 or 9 SFEBq organoids were re-suspended and trypsinized to single cell suspension. 1×105 cells were seeded to methylcellulose based semisolid media M3234 (Stem Cell Technologies) in the presence of IL-3 (10ng/ml), SCF (10ng/ml) and GM-CSF (10ng/ml). After 9 days of differentiation, resultant colony forming units (CFU) were counted under microscope (OLYPUS IX73). For NE differentiation under adherent monolayer condition, we followed a protocol as described elsewhere (Ying et al., 2003).
Plasmids and ESC Transfection
Plasmids including pPBCAG-rtTM2-IN, transposase and Doxcycline (Dox) inducible PiggyBac pPBhCMV1cHApA were kindly provided by Dr. Hitoshi Niwa (Kumamoto University, Japan). WT or mutant forms of WDR5 (F133Y, N225A, L240K, V268E, Q289E and I305V) with Flag or HA tags were subcloned into to Dox inducible pPBhCMV1cHApA plasmid, all-in-one Dox inducible Piggybac TREG/Tet3G (subcloning from Addgene #97421 plasmid by removing dCas9 and APEX2 cassettes), or non-inducible PiggyBac plasmid (Addgene, #48754). For Dexamethasone inducible WT-WDR5 plasmid, glucocorticoid receptor ligand binding domain (GRBD) was amplified using PCR from pPyCAG-cGR-IP plasmid (catalog #RDB10440, RIKEN) and Flag-WDR5 and GRBD inframe infusion DNAs were subcloned to non-inducible Piggybac plasmid. For mESC transfection, 20 μg endotoxins free plasmid DNA was electroporated to 5×106 ESCs using mouse ES cell nucleofector® kit (Lonza). Antibiotics resistance ESC colonies were selected for 5 days and pooled or single cell derived clones were picked for further characterization. To generate CRISPR-Cas9 mediated Wdr5 knockout (KO) plasmid, Wdr5 guide RNA sequence (TGTGAAGTTCAGCCCCAATG) was sub-cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 plasmid (Addgene, #62988). To generate Wdr5 KO ESC clones harboring with Dox inducible exogenous human WT WDR5 rescue platform (WDR5Dox;Wdr5KO) or Dex inducible exogenous human WT WDR5 rescue platform (WDR5Dex; Wdr5KO), Dox or Dex inducible WDR5 pooled ESC population were subjected to 2nd round of transfection with Wdr5 KO plasmid and selected with puromycin (2 μg/ml) in the presence of Dox (2 μg/ml) or Dex (20 μM). Single cell derived ES clones were picked up, expanded, and maintained in presence of Dox or Dex. Sanger sequencing was used to determine DNA editing at expected sites with homozygous Indel mutations with frameshift will be selected and confirmed with western blotting to use as Wdr5 KO ESC clones. To generate CRISPR-Cas9 mediated p53 KO plasmids, two independent p53 guide RNA sequences AAAATGTCTCCTGGCTCAGA or ATAAGCCTGAAAATGTCTCC were sub-cloned into pSpCas9(BB)-2A-Hygro (PX459) V2.0 plasmid. Two independent WDR5Dox;Wdr5KO ESC lines were transfected with p53 KO plasmids and selected with 200 ug/ml hygromycin in the absence of Dox (2 ug/ml). Single cell clones were picked up and Wdr5 plus p53 double knockout (DKO) ESCs were confirmed with DNA Sanger sequencing for homozygous frameshift mutation and western blotting.
RNA Isolation, Reverse Transcription, and Quantitative Real-Time PCR (RT-qPCR)
Trizol reagent (ThermoFisher Scientific) and RNeasy mini kit (QIAGEN) were utilized for RNA isolation from ESCs or EBs. 1.0 μg total RNA was reversely transcribed to cDNA using high-capacity RNA-to-cDNA kit (ThermoFisher Scientific). RT-qPCR was performed using iQ SYBR green supermix (Bio-Rad). Gapdh, β-actin was used as internal control for normalization. Primer sequences for real-time PCR were available upon request. Data were automatically analyzed using Bio-Rad CFX manager software using ΔΔCt method.
Whole-Cell Lysate Preparation, Histone Extraction, Western Blotting, and Immunoprecipitation
ESCs or EBs were lysed with RIPA buffer (Pierce) in the presence of EDTA free protease inhibitor cocktail (Sigma). Histone extraction on EBs was performed using a histone extraction kit (Abcam) as described previously (Khan et al., 2015). Protein concentration was determined using Pierce BCA protein assay kit (ThermoFisher Scientific). 1.0 μg of histone extracts, or 10 μg of whole cell lysate was resolved on 4%–20% precast gel (Bio-Rad) was transferred to 0.45 μm PVDF membrane (Millipore). For immunoprecipitation, day 2 WT and Wdr5KO EBs were harvested and lysed in RIPA buffer with EDTA free protease inhibitor cocktail (Sigma). 1 mg sonicated cell lysates were subjected to immunoprecipitation using WDR5 antibody (Bethyl). The following primary antibodies were used for probing: anti-HA (1:10,000, Abcam, ab9110), anti-WDR5 (1:5000, R&D), anti-WDR5 (1:5,000, Bethyl), anti-Flag (1:2,000, Sigma), anti-H3 (1:10,000, Abcam), anti-H3K4Me1 (1:5,000, Millipore), anti-H3K4Me2 (1:10,000, Millipore), anti-H3K4Me3 (1:10,000, Abcam), anti-P53 (1:5,000, Cell Signaling), anti-Tubulin (1:10,000, Cell signaling), anti-β-Actin (1:10,000, Cell signaling), anti-Phospho-p53 (Ser15) Antibody (1:5,000, Cell Signaling), anti-Phospho-p53 (Ser392) Antibody (1:5,000, Abcam), anti-Acetyl-p53 (K305) Antibody (1:5,000, Abcam), anti-Actyl-p53 (K379) Antibody (1:5,000, Cell Signaling).
Immunohistochemistry, Cytospin, and Immunofluorescence Staining
Day 9 EBs were harvested and fixed with 4% paraformaldehyde at 4°C overnight. EBs were sent to ULAM (Unit for laboratory animal medicine) at University of Michigan for paraffin processing, embedding, and sectioning. Antigen unmasking on slides was using citric acid methods as prescribed previously (Gage and Camper, 1997). Immunostaining was performed by combining of VectaStain ABC-HRP kit (Vector laboratories) and TSA Kit #24, with HRP-Streptavidin and Alexa Fluor 568 Tyramide (ThermoFisher Scientific). The primary antibodies were used for staining: anti-N-cadherin (mouse/monoclonal/1:2000, BD), anti- cardiac troponin T (CT3 clone, mouse/monoclonal/1:100, Developmental Studies Hybridoma Bank) and anti-myosin heavy chain antibody (MF20 clone, mouse/monoclonal/1:100, Developmental Studies Hybridoma Bank). For cytospin, Day 2 WT and Wdr5KO EBs were harvested and trypsinized to single cell suspension. Cells were attached to EZ cytofunnels (ThermoFisher) slides using Cytospin 3 (Shandon). Cells were fixed with 100% methanol and immunostaining was performed using p53 antibody (NCL-L-P53-CM5P, Leica biosystems) at 1:100 dilution. Counter nuclear staining was performed with DAPI (Molecular Probes). Control sections were incubated without primary antibodies. 4 representative pictures from different field were recorded under fluorescence microscope (Olympus DP73).
Flow Cytometry
2 to 4 EBs (organoids) were combined and dissociated to single cell suspension by 0.25% trypsin-EDTA (Invitrogen). DMEM media with 10% FBS (Sigma, v/v) was used to inactivate trypsin and cells were subjected to spin down at 300 g for 5 min. Cell pellet was washing once using D-PBS without calcium and magnesium and re-suspended in 200 μl of D-PBS in V-bottom plate. Cells were immediately transferred to LSR-II flowcytometer (BD) for data recording and analysis. The dead cells in the cell population were gated out and percentage of GFP (+) cells were calculated on live cells.
ATAC-Seq
Time-course ATAC-Seq on SFEBq organoids with duplicates at day 2, 4 and 6 were performed using Nextera DNA sample prep kit (Illumina) by following a protocol described elsewhere. 5×104 cells were washed with cold D-PBS and resuspended in 50 μl of cold lysis buffer (CHP-118-C, Boston Bioproducts) and pelleted. Transposition reaction by Tn5 transposase was incubated in TD buffer at 37°C for 30 min. After purification with MinElute Kit (QIAGEN), transposed DNA fragments were amplified through PCR using bar-coded Nextera PCR primers. Extra number of PCR cycles were added by qPCR as described and amplified DNA products were subjected to another round of DNA purification using MinElute Kit. Libraries were sent to University of Michigan DNA Sequencing Core for quality control test based on Bioanalyzer and sequenced using HiSeq 2500 or 4000 with pair-end reads.
RNA-Seq
RNA isolation for RNA-seq was performed by combination of Trizol reagent (ThermoFisher Scientific) and RNeasy mini kit (QIAGEN). Day 2 [WT, Wdr5 KO groups (KO#7 or KO#18 without Dox for 48 h), Wdr5 and p53 KO groups (DKO#7–11 or DKO#18–17 without Dox for 48 h)] or Day 6 EB RNA samples [WT group, KO group (Wdr5 KO #3 without Dox), Wdr5 KO #3 with early (Dox added at 12 h after differentiation, T12h group) and late (Dox added at 48 h after differentiation, T48h group) WDR5 rescue] were sent to University of Michigan DNA Sequencing Core for RNA quality analysis, library construction, and sequencing using Illumina HiSeq 4000 Libraries platform. Duplicate samples for each group were subjected to two round of independent library preparation and sequencing to avoid sample and batch effect of RNA-seq.
ChIP-Seq
Chromatin Immunoprecipitation (ChIP) assay were performed using a slightly modified version of ChIP Assay Kit (Millipore) based protocol as described previously. Day 6 EBs [Wdr5KO #3 with early (Dox added at 12 h after differentiation, T12h group, total cell number 3×107) and late (Dox added at 48 h after differentiation, T48h group, total cell number 4×107) hWDR5 rescue were fixed with Disuccinimidyl glutarate crosslinker (COVAChem) at final concentration of 2 μM at room temperate for 30 min. Cells were further subjected to crosslink with 1% paraformaldehyde (Sigma) at room temperature at 10 min and the reaction was quenched using glycine at final concentration of 0.125M. The cells pellets were further sonicated to 300–500 bp in SDS lysis buffer by using a Biorupter 300 (Diagenode). 15 μg ChIP grade anti-HA antibody (ab9110, Abcam) was used for ChIP assay. The ChIP-DNA were sent to University of Michigan DNA Sequencing Core for quality control, library preparation, and sequencing using Illumina HiSeq 4000 Libraries platform.
CUT&RUN
Day 2 WT, Wdr5KO and DKO EBs were harvested and trypsinized to single cell suspension. 2×106 cells were prepared for Cut & Run experiment per antibody as previously described (Skene and Henikoff, 2017; Skene et al., 2018) and the following antibodies were used: WDR5 antibody (Bethyl), P53 antibody (NCL-L-P53-CM5P, Leica biosystems) and H3K4me3 antibody (Abcam). Protein A–micrococcal nuclease (pA-MN) fusion protein were generously provided by Dr. Steven Henikoff.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Analysis
All experiments were independently repeated with different mWdr5 KO ESC clones at least for 2 to 4 times with similar results, and data from one representative experiment are presented unless otherwise stated. Two-tailed unpaired Student’s t test was applied using GraphPad Prism (version 7.00) to determine whether the observed differences were statistically significant. Changes were considered statistically significant when p value less than 0.05.
Bioinformatics Analysis
Reads Mapping and Coverage
All RNA-seq data were mapped to the mm10 genome for mouse, and hg38 genome for human, using Tophat2 (2.1.1) (Kim et al., 2013), which was shown to be accurate alignment of transcriptomes in the presence of insertions, deletions, and gene fusions. Then, duplicated reads for pair-end data were removed, but not single-end data by SAMtools (v1.5) (Li et al., 2009). All ChIP-seq, ATAC-seq, and CUT&RUN sequencing data were mapped to the mm10 genome for mouse by using Bowtie2 (v2–2.2.4) with parameters “-q–phred33–very-sensitive -p 10” (Langmead and Salzberg, 2012), which is ultrafast and memory-efficient tool for aligning sequencing reads to long reference sequences. Then, we removed any duplicated reads for both pair-end and single-end data using SAMtools (v1.5) (Li et al., 2009). For all sequencing datasets, the bigwig files for visualization in Integrative Genomics Viewer (IGV) (Thorvaldsdóttir et al., 2013) were generated from BAM files by using “bamCoverage” from deepTools (Ramírez et al., 2014) with parameters “–ignoreDuplicates–normalizeUsingRPKM–skipNonCoveredRegions–binSize 50.” The bigwig files for IP/input ratio were generated from BAM files by using deepTools2 (v2.5.0) (Ramírez et al., 2016) with command “bamCompare -b1 ChIP-bam -b2 Input-bam-ignoreDuplicates–minMappingQuality 30–normalizeUsing RPKM –binSize 20–operation ratio–scaleFactorsMethod None -p 20.” Whereas, for ATAC-seq and CUT&RUN datasets, the bigwig files were generated from BAM files by using deepTools2 (v2.5.0) (Ramírez et al., 2016) with command “bamCoverage–outFileFormat bigwig–ignoreForNormalization chrM chrX chrY–skip-NAs-bam bowtie2.rmdup.bam-outFileName output.bigwig–normalizeUsing RPKM–minMappingQuality 30–binSize 20–smooth-Length 60–ignoreDuplicates–scaleFactor 1-numberOfProcessors 2.”BAM files of mapping results were merged for the same sample using SAMtools and converted to BED format by using BEDTools (Quinlan and Hall, 2010). Peaks of regulatory regions were called for each sample by using MACS (v 1.4.2) (Zhang et al., 2008) from bed files of ChIP-seq with parameters “-w -S -p 0.00001 -g mm.” The input signal was used as the control to call peaks for the ChIP-seq dataset. The heatmap plot of signals centered on peaks was implicated by deepTools2. The heatmap plot of signals centered on peaks was implicated by deepTools2 subcommand plotHeatmap.
Peak Calling and Annotation
BAM files of mapping results were merged for the same sample using SAMtools and converted to BED format by using BEDTools (Quinlan and Hall, 2010). Peaks of regulatory regions were called for each sample by using MACS (v 1.4.2) (Zhang et al., 2008) from datasets of ChIP-seq with parameters “-w -S -p 0.00001.” The input signal was used as the control to call peaks for the ChIP-seq dataset. The heatmap plot of signals centered on peaks and gene promoters was implicated by deepTools2 (v2.5.0) (Ramírez et al., 2016). Peak annotation was performed by using HOMER (v4.9.1) (Heinz et al., 2010) with default parameters. Motif analysis on peak regions was performed with HOMER function findMotifsGenome.pl with parameters “-size 50 -mask.”
DATA AND CODE AVAILABILITY
The datasets generated during this study are available at Gene Expression Omnibus (GEO) repository with accession numbers GEO: GSE116153, GSE116154, GSE116155, and GSE140899.
The graphical abstract was created with Biorender.com.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-WDR5 | Bethyl | Cat# A302-429A, RRID:AB_1944302 |
| Anti-WDR5 | Bethyl | Cat# A302-430A, RRID:AB_1944300 |
| Anti-WDR5 | R & D Systems | Cat# AF5810, RRID:AB_2215565 |
| Anti-HA | Abcam | Cat# ab9110, RRID:AB_307019 |
| anti-FLAG | Sigma | Cat# F3165, RRID:AB_259529 |
| anti-H3 | Abcam | Cat# ab1791, RRID:AB_302613 |
| anti-H3K4Me1 | Millipore | Cat# 07-436, RRID:AB_310614 |
| anti-H3K4Me2 | Millipore | Cat# 07-030, RRID:AB_11213050 |
| anti-H3K4Me3 | Abcam | Cat# ab8580, RRID:AB_306649 |
| anti-p53 | Cell Signaling Technology | Cat# 32532, RRID:AB_2757821 |
| anti-p53 | Leica Biosystems | Cat# NCL-L-P53-CM5P |
| anti-α/β-Tubulin | Cell Signaling Technology | Cat# 2148, RRID:AB_2288042 |
| Anti-β-Actin | Abcam | Cat# ab49900, RRID:AB_867494 |
| anti-p53 (acetyl K305) | Abcam | Cat# ab109396, RRID:AB_10861725 |
| anti-p53 (phospho S392) | Abcam | Cat# ab33889, RRID:AB_776988 |
| anti-Phospho-p53 (Ser15) | Cell Signaling Technology | Cat# 12571, RRID:AB_2714036 |
| anti-N-cadherin | BD Biosciences | Cat# 610920, RRID:AB_2077527 |
| anti-myosin, sarcomere (MHC) antibody | DSHB | Cat# MF 20, RRID:AB_2147781 |
| anti-troponin T (cardiac/slow) antibody | DSHB | Cat# ct3, RRID:AB_528495 |
| Anti-p53, acetyl (Lys379) | Cell Signaling Technology | Cat# 2570, RRID:AB_823591 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technology | Cat# 7076, RRID:AB_330924 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat# 7074, RRID:AB_2099233 |
| Donkey Anti-Goat IgG HRP Affinity Purified PAb antibody | R & D Systems | Cat# HAF109, RRID:AB_357236 |
| Peroxidase IgG Fraction Monoclonal Mouse Anti-Rabbit IgG, light chain specific | Jackson ImmunoResearch Labs | Cat# 211-032-171, RRID:AB_2339149 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| AGN 193109 Sodium Salt | Santa Cruz Biotechnology | Cat# sc-210768 |
| Dexamethasone | Sigma | Cat# D4902 |
| Cycloheximide solution | Millipore | Cat# C4859 |
| KnockOut Serum Replacement | GIBCO | Cat# 10828028 |
| Matrigel® Growth Factor Reduced (GFR) Basement Membrane Matrix | CORNING | Cat# 354230 |
| Glasgow’s MEM (GMEM) | GIBCO | Cat# 11710-035 |
| Sodium Pyruvate (100 mM) | GIBCO | Cat# 11360070 |
| MEM Non-Essential Amino Acids Solution (100X) | GIBCO | Cat# 11140050 |
| TRIzol | Thermo Fischer | Cat# 15596026 |
| iQ SYBR® Green Supermix | Bio-Rad | Cat# 1708880 |
| TaqMan Gene Expression Master Mix | Thermo Fischer | Cat# 4369016 |
| RIPA Lysis and Extraction Buffer | Thermo Fischer | Cat# 89900 |
| Cold lysis buffer | Boston Bioproducts | Cat# CHP-118-C |
| Disuccinimidyl glutarate crosslinker | COVAChem | Cat# 13301 |
| Dynabeads Protein G for Immunoprecipitation | Thermo Fisher | Cat# 10003D |
| VECTASHIELD® Hardset Antifade Mounting Medium with DAPI | Vector Laboratories | Cat# H-1500 |
| Protein A-micrococcal nuclease | Dr. Steve Henikoff | N/A |
| Critical Commercial Assays | ||
| Nunclon Sphera Microplates | Thermo Fisher | Cat# 174925 |
| In-Fusion® HD Cloning Plus | Takara | Cat# 638909 |
| Mouse ES cell nucleofector® kit | Lonza | Cat# VPH-1001 |
| ZymoPURE II Plasmid Midiprep Kit | Zymo | Cat# D4200 |
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| High-capacity RNA-to-cDNA kit | Thermo Fisher | Cat# 4387406 |
| Histone extraction kit | Abcam | Cat# ab113476 |
| Pierce BCA Protein Assay Kit | Thermo Fisher | Cat# 23225 |
| Invitrogen Molecular Probes TSA Kit 24 | Thermo Fisher | Cat# T20934 |
| Mouse on Mouse (M.O.M.®) Elite® Peroxidase Kit | Vector Laboratories | Cat# PK-2200 |
| Nextera DNA sample prep kit | Illumina | Cat# 15028212 |
| MinElute PCR Purification Kit | QIAGEN | Cat# 28004 |
| Chromatin Immunoprecipitation (ChIP) Assay Kit | Millipore | Cat# 17-295 |
| Deposited Data | ||
| ATAC-seq Data | This Study | GSE116153 |
| RNA-seq Data | This Study | GSE116155 |
| ChIP-seq Data | This Study | GSE116154 |
| CUT&RUN Data | This Study | GSE140899 |
| Experimental Models: Cell Lines | ||
| Rx-GFP K/I EB5 (sex: Male [XY]) | RIKEN | Cat# AES0145 |
| Oligonucleotides | ||
| Wdr5 guide RNA:TGTGAAGTTCAGCCCCAATG | This Study | N/A |
| p53 guide RNA #1:AAAATGTCTCCTGGCTCAGA | This Study | N/A |
| p53 guide RNA #2:ATAAGCCTGAAAATGTCTCC | This Study | N/A |
| Recombinant DNA | ||
| pPBCAG-rtTM2-IN | Dr. Hitoshi Niwa (Kumamoto University, Japan) | N/A |
| Transposase pBase | Dr. Hitoshi Niwa (Kumamoto University, Japan) | N/A |
| pPBCMV1cHApA | This Study | N/A |
| pPBCMV1-WT-WDR5-HA | This Study | N/A |
| pPBCMV1-L240K-WDR5-HA | This Study | N/A |
| pPBCMV1-V268E-WDR5-HA | This Study | N/A |
| pPBCMV1-Q289E-WDR5-HA | This Study | N/A |
| pPBTREG/Tet3G-FLAG-WT-WDR5 | This Study | N/A |
| pPBTREG/Tet3G-FLAG-N225A-WDR5 | This Study | N/A |
| pPBTREG/Tet3G-FLAG-L240K-WDR5 | This Study | N/A |
| pPBTREG/Tet3G-FLAG-V268E-WDR5 | This Study | N/A |
| pPBTREG/Tet3G-FLAG-Q289E-WDR5 | This Study | N/A |
| PiggyBac-FLAG-WT-WDR5-GRBD | This Study | N/A |
| PiggyBac-FLAG-WT-WDR5 | This Study | N/A |
| PiggyBac-FLAG-F133Y-WDR5 | This Study | N/A |
| PiggyBac-FLAG-I305V-WDR5 | This Study | N/A |
| PiggyBac-FLAG-N225A-WDR5 | This Study | N/A |
| PiggyBac-FLAG-L240K-WDR5 | This Study | N/A |
| PiggyBac-FLAG-V268E-WDR5 | This Study | N/A |
| PiggyBac-FLAG-Q289E-WDR5 | This Study | N/A |
| Software and Algorithms | ||
| Image lab Version 5.2.1 | Bio-Rad | https://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z |
| GraphPad Prism Version 7.00 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| SAMtools Version 1.5 | Li et al., 2009 | http://www.htslib.org/ |
| Bowtie2 Version 2–2.2.4 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| deepTools Version 2.5.0 | Ramírez et al., 2014 | https://deeptools.readthedocs.io/en/develop/ |
| BEDTools | Quinlan and Hall, 2008; PMID: 20110278 | https://bedtools.readthedocs.io/en/latest/ |
| MACS Version 1.4.2 | Zhang et al., 2008 | https://github.com/taoliu/MACS/ |
| HOMER Version 4.9.1 | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
Highlights.
WDR5-RbBP5 interaction surface controls ESC-to-neuroectoderm organoid differentiation
Wdr5 inhibition stabilizes p53 and activates p53 signaling
Transient Wdr5 inhibition stimulates ESC differentiation toward mesoderm fates via p53
ACKNOWLEDGMENTS
This research was supported by the National Eye Institute (K08EY026654, to R.C.R), the National Cancer Institute (P30CA046592, to the University of Michigan Comprehensive Cancer Center), and Research to Prevent Blindness (to the University of Michigan Kellogg Eye Center and R.C.R.). R.C.R. received funding from the Beatrice & Reymont Paul Foundation, March Hoops to Beat Blindness, the Taubman Institute, the Leonard G. Miller Endowed Professorship and Ophthalmic Research Fund at the Kellogg Eye Center, and the Grossman, Elaine Sandman, Marek and Maria Spatz (endowed), Greenspon, Dunn, Avers, Boustikakis, Sweiden, and Terauchi research funds.
Footnotes
DECLARATION OF INTERESTS
Portions of this work were used as the basis for provisional U.S. patent (no. 62/618,937) and international patent applications (no. PCT/US19/14192). The authors declare no other competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.12.039.
REFERENCES
- Aho ER, Wang J, Gogliotti RD, Howard GC, Phan J, Acharya P, Macdonald JD, Cheng K, Lorey SL, Lu B, et al. (2019). Displacement of WDR5 from Chromatin by a WIN Site Inhibitor with Picomolar Affinity. Cell Rep. 26, 2916–2928.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. (2011). Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145, 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AP. (2019). MaRS Innovation’s Triphase Accelerator Announces New Partnership with Celgene for First-in-class WDR5 Leukemia Therapy, AP, February 1, 2019. https://apnews.com/ef2dfde807375dd594325c4c5da5db99.
- Assawachananont J, Mandai M, Okamoto S, Yamada C, Eiraku M, Yonemura S, Sasai Y, and Takahashi M (2014). Transplantation of embryonic and induced pluripotent stem cell-derived 3D retinal sheets into retinal degenerative mice. Stem Cell Reports 2, 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen ME, and Attardi LD (2019). The role of p53 in developmental syndromes. J. Mol. Cell Biol 11, 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen ME, Mcclendon J, Long HK, Sorayya A, Van Nostrand JL, Wysocka J, and Attardi LD (2019). The Spatiotemporal Pattern and Intensity of p53 Activation Dictates Phenotypic Diversity in p53-Driven Developmental Syndromes. Dev. Cell 50, 212–228.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, and Roeder RG (2006). Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol 13, 713–719. [DOI] [PubMed] [Google Scholar]
- Eiraku M, and Sasai Y (2011). Mouse embryonic stem cell culture for generation of three-dimensional retinal and cortical tissues. Nat. Protoc 7, 69–79. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, and Sasai Y (2008). Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K,Adachi T, and Sasai Y (2011). Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56. [DOI] [PubMed] [Google Scholar]
- Eising E, Carrion-Castillo A, Vino A, Strand EA, Jakielski KJ, Scerri TS, Hildebrand MS, Webster R, Ma A, Mazoyer B, et al. (2019). A set of regulatory genes co-expressed in embryonic human brain is implicated in disrupted speech development. Mol. Psychiatry 24, 1065–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage PJ, and Camper SA (1997). Pituitary homeobox 2, a novel member of the bicoid-related family of homeobox genes, is a potential regulator of anterior structure formation. Hum. Mol. Genet 6, 457–464. [DOI] [PubMed] [Google Scholar]
- Giaccia AJ, and Kastan MB (1998). The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 12, 2973–2983. [DOI] [PubMed] [Google Scholar]
- Gori F, Friedman LG, and Demay MB (2006). Wdr5, a WD-40 protein, regulates osteoblast differentiation during embryonic bone development. Dev. Biol 295, 498–506. [DOI] [PubMed] [Google Scholar]
- Gregoire S, Li G, Sturzu AC, Schwartz RJ, and Wu SM (2017). YY1 Expression Is Sufficient for the Maintenance of Cardiac Progenitor Cell State. Stem Cells 35, 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner A, Bulyk ML, Jambhekar A, and Lahav G (2019). The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol 20, 199–210. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, and Oren M (1997). Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M, Kurisaki A, Ohnuma K, Okochi H, Hamazaki TS, and Asashima M (2006). N-cadherin is a useful marker for the progenitor of cardiomyocytes differentiated from mouse ES cells in serum-free condition. Bio-chem. Biophys. Res. Commun 351, 877–882. [DOI] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, and Roeder RG (2011). Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell 144, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya D, Banno S, Sasai N, Ohgushi M, Inomata H, Watanabe K, Kawada M,Yakura R, Kiyonari H, Nakao K, et al. (2011). Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature 470, 503–509. [DOI] [PubMed] [Google Scholar]
- Khan M, Walters LL, Li Q, Thomas DG, Miller JM, Zhang Q, Sciallis AP, Liu Y, Dlouhy BJ, Fort PE, et al. (2015). Characterization and pharmacologic targeting of EZH2, a fetal retinal protein and epigenetic regulator, in human retinoblastoma. Lab. Invest 95, 1278–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S, Takagi A, Inoue T, and Saga Y (2000). MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development 127, 3215–3226. [DOI] [PubMed] [Google Scholar]
- Koeppel M, van Heeringen SJ, Smeenk L, Navis AC, Janssen-Megens EM, and Lohrum M (2009). The novel p53 target gene IRF2BP2 participates in cell survival during the p53 stress response. Nucleic Acids Res. 37, 322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Mao F, Zou Z, DenDekker A, Xu J, Hou S, Dou Y, and Rao RC (2019). p53 Coordinates Temporal WDR5 Inputs During Neuroectoderm and Mesoderm Differentiation of Mouse Embryonic Stem Cells. Dev. Cell, Published online January 4, 2019. 10.2139/ssrn.3307529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, et al. (2010). The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 24, 1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, and Xu Y (2005). p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol 7, 165–171. [DOI] [PubMed] [Google Scholar]
- Liu Y, Deisenroth C, and Zhang Y (2016). RP-MDM2-p53 Pathway: Linking Ribosomal Biogenesis and Tumor Surveillance. Trends Cancer 2, 191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers MP, Bryson T, and Henikoff S (2019). A streamlined protocol and analysis pipeline for CUT&RUN chromatin profiling. bioRxiv. 10.1101/569129. [DOI] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, and Manke T (2014). deep-Tools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44 (W1), W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RC, and Dou Y (2015). Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 15, 334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogel A, Popliker M, Webb CG, and Oren M (1985). p53 cellular tumor antigen: analysis of mRNA levels in normal adult tissues, embryos, and tumors. Mol. Cell. Biol 5, 2851–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, and Black BL (2005). Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development 132, 3405–3417. [DOI] [PubMed] [Google Scholar]
- Sabapathy K, Klemm M, Jaenisch R, and Wagner EF (1997). Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J. 16, 6217–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai N, Yakura R, Kamiya D, Nakazawa Y, and Sasai Y (2008). Ectodermal factor restricts mesoderm differentiation by inhibiting p53. Cell 133, 878–890. [DOI] [PubMed] [Google Scholar]
- Schmid P, Lorenz A, Hameister H, and Montenarh M (1991). Expression of p53 during mouse embryogenesis. Development 113, 857–865. [DOI] [PubMed] [Google Scholar]
- Skene PJ, and Henikoff S (2017). An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff JG, and Henikoff S (2018). Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc 13, 1006–1019. [DOI] [PubMed] [Google Scholar]
- Smith KN, Singh AM, and Dalton S (2010). Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell 7, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Bell JL, Carter D, Gherardi S, Poulos RC, Milazzo G, Wong JW, Al-Awar R, Tee AE, Liu PY, et al. (2015). WDR5 Supports an N-Myc Transcriptional Complex That Drives a Protumorigenic Gene Expression Signature in Neuroblastoma. Cancer Res. 75, 5143–5154. [DOI] [PubMed] [Google Scholar]
- Thomas LR, Wang Q, Grieb BC, Phan J, Foshage AM, Sun Q, Olejniczak ET, Clark T, Dey S, Lorey S, et al. (2015). Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC. Mol. Cell 58, 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand JL, Brady CA, Jung H, Fuentes DR, Kozak MM, Johnson TM, Lin CY, Lin CJ, Swiderski DL, Vogel H, et al. (2014). Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature 514, 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhais-Neto GC, Fournier M, Plassat JL, Sardiu ME, Saraf A, Garnier JM, Maruhashi M, Florens L, Washburn MP, and Pourquié O (2017). The WHHERE coactivator complex is required for retinoic acid-dependent regulation of embryonic symmetry. Nat. Commun 8, 728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zou Y, Nowotschin S, Kim SY, Li QV, Soh CL, Su J, Zhang C, Shu W, Xi Q, et al. (2017). The p53 Family Coordinates Wnt and Nodal Inputs in Mesendodermal Differentiation of Embryonic Stem Cells. Cell Stem Cell 20, 70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wataya T, Ando S, Muguruma K, Ikeda H, Watanabe K, Eiraku M, Kawada M, Takahashi J, Hashimoto N, and Sasai Y (2008). Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proc. Natl. Acad. Sci. USA 105, 11796–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, and Allis CD (2005). WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121, 859–872. [DOI] [PubMed] [Google Scholar]
- Xie Q, Li Z, and Chen J (2017). WDR5 positively regulates p53 stability by inhibiting p53 ubiquitination. Biochem. Biophys. Res. Commun 487, 333–338. [DOI] [PubMed] [Google Scholar]
- Yang YW, Flynn RA, Chen Y, Qu K, Wan B, Wang KC, Lei M, and Chang HY (2014). Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife 3, e02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, and Smith A (2003). Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol 21, 183–186. [DOI] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. (2013). De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, et al. (2016). Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557. [DOI] [PubMed] [Google Scholar]
- Zhang X, He Y, Lee KH, Dubois W, Li Z, Wu X, Kovalchuk A, Zhang W, and Huang J (2013). Rap2b, a novel p53 target, regulates p53-mediated pro-survival function. Cell Cycle 12, 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Fu Y, Nettleton M, Richman A, and Han Z (2017). High throughput in vivo functional validation of candidate congenital heart disease genes in Drosophila. eLife 6, e22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available at Gene Expression Omnibus (GEO) repository with accession numbers GEO: GSE116153, GSE116154, GSE116155, and GSE140899.
The graphical abstract was created with Biorender.com.